Abstract
Merkel cell polyomavirus (MCPyV) causes the highly aggressive and relatively rare skin cancer known as Merkel cell carcinoma (MCC). MCPyV also causes a lifelong yet relatively innocuous infection and is one of 14 distinct human polyomaviruses species. Although polyomaviruses typically do not cause illness in healthy individuals, several can cause catastrophic diseases in immunocompromised hosts. MCPyV is the only polyomavirus clearly associated with human cancer. How MCPyV causes MCC and what oncogenic events must transpire to enable this virus to cause MCC is the focus of this essay.
This article is part of the themed issue ‘Human oncogenic viruses’.
Keywords: Merkel cell polyomavirus, Merkel cell carcinoma, T-antigen
1. Introduction
(a). Common infection and a rare cancer
Perhaps the most striking feature about Merkel cell polyomavirus (MCPyV) is how frequently it causes a lifelong though relatively innocuous infection yet how rarely it causes the highly aggressive skin cancer known as Merkel cell carcinoma (MCC). When MCPyV was first identified in MCC tumour specimens in 2008, it was only the fifth human polyomavirus to be identified at that time [1]. Its discovery quickly led to the realization that although MCPyV was likely to be causal in MCC, it was a typical polyomavirus, infecting most people at an early age. What has come into sharper focus is that although some of the now 14 human polyomaviruses can cause exceptionally catastrophic diseases, MCPyV is the only one clearly associated with cancer. Indeed, MCPyV has been classified by the World Health Organization-International Agency for Research on Cancer as probably carcinogenic to humans (Group 2A) [2]. How MCPyV causes MCC and what oncogenic events must transpire to enable this virus to cause MCC is the focus of this essay.
A pathogenic cause for MCC was first suspected when it was reported that the incidence of MCC was greater than 10-fold in HIV-1 AIDS patients compared with the general population [3]. In addition, the risk for developing MCC is increased in patients with medically induced immunosuppression for autoimmune conditions such as rheumatoid arthritis and solid organ transplantation [4–6]. Given the increased risk by immunosuppression for developing MCC, Huichen Feng and Masahiro Shuda in the laboratory of Yuan Chang and Patrick Moore began a search for a pathogenic cause for MCC. They performed whole transcriptome sequencing of several MCC tumours and searched for pathogens by first subtracting all human genes from their analysis. In the remaining sequences, novel transcripts distantly related to polyomaviruses were detected in a MCC tumour. Complete sequencing of the viral genome led to the determination that it corresponded to a new human polyomavirus [1]. They determined that MCPyV DNA was clonally integrated into the genome of MCC tumour cells when they observed an identical Southern blot integration pattern for a primary tumour and a metastatic lymph node from the same patient. They detected MCPyV by PCR and Southern blotting in eight of 10 tested MCC tumours, indicating that most but not all MCC contained MCPyV. These results supported the model that MCPyV contributed to MCC in a manner similar to human papillomavirus (HPV) in cervical cancer [7].
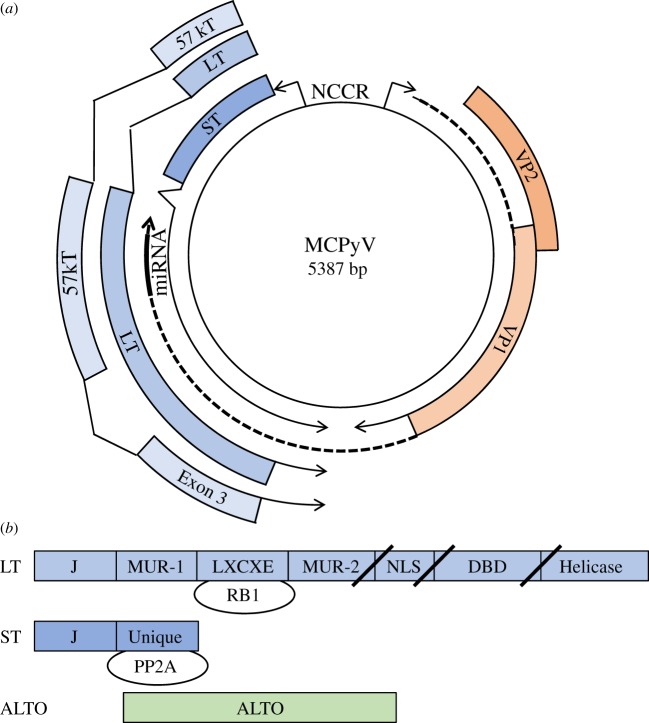
MCPyV is a typical mammalian polyomavirus with a small (5386 bp) circular double-stranded DNA genome (figure 1a) [8]. There are two transcriptional units with an early region that yields four spliced mRNAs encoding four proteins including large T-antigen (LT), 57 kT, small T-antigen (ST) and ALTO (alternate frame of the large T open reading frame) and a late region encoding two viral coat proteins, VP1 and VP2, and a microRNA that targets the T-antigen transcripts [9–12]. Although the late region of MCPyV does not encode VP3, a third viral coat protein that uses an internal translation start site within VP2, most other polyomaviruses encode a VP3 [13]. The non-coding control region (NCCR) contains distinct promoters for the early and late genes, an enhancer and the viral origin of replication [14]. Based on its similarity to other polyomaviruses, it is likely that MCPyV LT forms two hexamers that bind in head-to-head fashion to the origin and serves to melt, twist and unwind the viral DNA and recruit cellular DNA polymerases to enable viral replication [15–18].
Figure 1.
(a) Circular map of MCPyV includes early region genes for LT, ST and 57 kT and late region for VP1, VP2 and miRNA. The non-coding control region (NCCR) contains a bidirectional promoter and the viral origin of replication. Exon 3 of 57 kT is depicted and is in the same reading frame as LT. ALTO is not depicted. (b) Linear maps of LT, ST and ALTO. LT and ST share an N-terminal DNAJ or J domain. LT also contains the LXCXE or RB-binding motif, MCPyV-unique region (MUR) -1 and -2, nuclear localization signal (NLS), DNA-binding domain (DBD) and helicase domain. In MCC, mutations in LT result in truncations after the LXCXE, NLS or DBD and depicted by slashes. ST contains a unique region not shared with LT that binds to protein phosphatase 2A (PP2A). ALTO is expressed in an alternative reading frame from LT.
(b). Polyomavirus-associated diseases
MCPyV is one of 14 distinct human polyomaviruses species [8,19]. Primary infection with MCPyV does not cause any discernable signs or symptoms [20]. Polyomaviruses typically do not cause illness in healthy individuals although several viruses are associated with disease in immunocompromised hosts (table 1).
Table 1.
Human polyomaviruses and associated diseases.
| no. | name | alternate names | year | disease | GenBank/NCBI |
|---|---|---|---|---|---|
| 1 | BKPyV | BK, BKV | 1971 [21] | polyomavirus-associated nephropathy haemorrhagic cystitis interstitial cystitis |
AB211374.1 |
| 2 | JCPyV | JC, JCV | 1971 [22] | progressive multifocal leukoencephalopathy granule cell neuropathy |
NC_001699.1 |
| 3 | KIPyV | KI, KIV | 2007 [23] | EF127908.1 | |
| 4 | WUPyV | WU, WUV | 2007 [24] | EF444554.1 | |
| 5 | MCPyV | MCV, Merkel cell polyomavirus | 2008 [1] | Merkel cell carcinoma | NC_010277.2 |
| 6 | HPyV6 | 2010 [25] | pruritic and dyskeratotic dermatoses | NC_014406.1 | |
| 7 | HPyV7 | 2010 [25] | pruritic and dyskeratotic dermatoses | NC_014407.1 | |
| 8 | TSPyV | TSV | 2010 [26] | trichodysplasia spinulosa | NC_014361.1 |
| 9 | HPyV9 | 2011 [27] | NC_015150.1 | ||
| 10 | HPyV10 | MWPyV, Malawi polyomavirus | 2012 [28] | JX262162.1 | |
| 11 | StLPyV | St Louis polyomavirus | 2012 [29] | JX463184.1 | |
| 12 | HPyV12 | 2013 [30] | NC_020890.1 | ||
| 13 | NJPyV | HPyV13, New Jersey polyomavirus | 2014 [31] | NC_024118.1 | |
| 14 | LIPyV | LI, Lyon IARC polyomavirus | 2017 [19] | NC_034253.1 |
BK polyomavirus (BKPyV) virus is the cause of polyomavirus-associated nephropathy (PVAN) in renal transplant patients undergoing immune suppressive therapy to prevent rejection of their allograft kidney transplant [21]. PVAN may result from infection of the transplant recipient with a different strain of BKPyV that accompanied the transplanted kidney [32]. BKPyV can also cause haemorrhagic cystitis in haematopoietic stem cell transplant patients [33]. In addition, BKPyV has been associated with interstitial cystitis with bladder ulcerations and bladder pain syndrome [34,35]. Large-scale sequencing of 131 urothelial bladder cancers identified integrated copies of BKPyV in one tumour while another had integrated copies of HPV16 [36]. Whether BKPyV or HPV16 contributed to the oncogenesis of either tumour was not further explored.
JC polyomavirus (JCPyV) causes progressive multifocal leukoencephalopathy (PML) [22]. PML is characterized by lytic infection of oligodendrocytes and astrocytes with JCPyV that causes a variety of neurological symptoms including ataxia, paresis, dementia and blindness [37]. The incidence of PML increased during the AIDS epidemic but now is frequently associated with immune suppressive therapy for multiple sclerosis [37,38]. JCPyV can also infect neural cells and cause a distinct illness called granule cell neuropathy [39].
Several polyomaviruses have been identified on the skin. MCPyV, HPyV6, HPyV7 and TSPyV DNA can be isolated from skin and eyebrow hair [25,40]. In severely immunocompromised patients, HPyV6 and HPyV7 can cause a pruritic and dyskeratotic dermatoses characterized by brown plaque skin lesions and hyperproliferation of keratinocytes [41,42]. TSPyV causes a severe form of hair folliculitis known as Trichodysplasia spinulosa in solid organ transplant recipients [26]. It has been suggested that trichodysplasia spinulosa may reflect a primary infection with TSPyV because it occurs in relatively young individuals [43].
WUPyV and KIPyV have been isolated from respiratory secretions particularly in children and infants with severe pulmonary symptoms [23,24,44]. Although it is not clear if WUPyV and KIPyV cause pneumonia, WUPyV has been detected in respiratory epithelial cells from a transplanted lung in a patient with Job syndrome [26,45]. HPyV10 and STLPyV have been isolated from stool and may contribute to infectious forms of diarrhoea [28,29,46]. NJPyV was originally isolated from a pancreas transplant patient with severe immunosuppression, retinal blindness and vasculitic myopathy [31].
It is not known what cells normally support MCPyV replication because MCPyV LT expression has not yet been detected by immunohistochemistry (IHC) in any normal human tissue. If healthy skin supports MCPyV replication, then cells within hair follicles such as seen with TSPyV in the trichodysplasia spinulosa syndrome or in keratinocytes with HPyV6 and HPyV7 in dyskeratotic dermatoses. Alternatively, a recent report demonstrated that cultures of primary human skin fibroblasts could support MCPyV replication [47]. It should be noted that papillomavirus infection is dependent on breaks in the intact epithelium permitting access of the papillomavirus to the basement membrane. A similar mechanism has not been described for MCPyV or any other polyomavirus. Instead, polyomaviruses bind specific cell-surface glycans to target cells for infection [48,49].
(c). Lifelong infections with polyomaviruses
Evidence for persistent infection by a specific polyomavirus is reflected in serum antibodies against the corresponding polyomavirus coat protein VP1. The polyomavirus virion is comprised of 72 pentamers of VP1 together with VP2 on the inner surface of each VP1 pentamer [50]. When expressed in bacteria or yeast, VP1 will spontaneously form pentamers or virus-like particles that generates a useful capture antigen to detect antibodies in serum specific for each human polyomavirus [51,52].
Based on the VP1 serology assay, it has been inferred that the initial exposure to MCPyV likely occurs in early childhood because the seroprevalence is lower in children and higher in adults. An intriguing study from Cameroon examined serology against the MCPyV VP1 pentamer in 196 children from birth to 5 years of age (YOA) [53]. Significant titres against MCPyV were detected in newborns but these titres gradually decreased to undetectable levels by 16 months of age. Maternal-derived antibodies likely account for the seropositivity in newborns that gradually declined during the first year of life. These antibodies were likely to be effective in preventing primary infection. By 18 months of age when the maternal antibodies were no longer present, children were susceptible to de novo infection and could mount an antibody response of their own. Beginning at 18 months of age, an increasing fraction of children became positive until approximately 80% tested positive by 5 YOA [53]. In a separate cohort from the same study, the strongest correlation of seropositivity was observed between siblings of similar ages suggesting that siblings likely were exposed to MCPyV at the same time and by each other. Similar results were reported from a population study in Australia that investigated the serology of several cutaneous polyomaviruses including MCPyV, HPyV6, HPyV7 and TSPyV as well as BKPyV. Children below six months displayed seropositivity rates for all viruses studied comparable to that found in adults with rates decreasing after six months of age then starting to increase by 2–3 YOA and continuing to increase with age [54].
Several additional studies support the increasing risk with age for exposure and persistent infection by MCPyV and other polyomaviruses. Seroprevalence of 10 human polyomaviruses were assessed from a population-based skin cancer case–control study conducted in New Hampshire, USA [55]. The overall seropositivity for MCPyV in this study was 70.4%. Of note, all participants were seropositive for at least one polyomavirus and the overall study population had evidence for infection with a mean of 7.3 different polyomaviruses. A study of five polyomaviruses conducted in Italy with participants aged 1–100 YOA found that the seroprevalence for MCPyV rapidly increased with age, from 41.7% in children age 1–4 YOA to 87.6% in 15–19 YOA and remained relatively frequent in adulthood (79.0–96.2%) [56]. Seroprevalence studies of MCPyV performed in China (61% overall) and the Czech Republic (63%) yielded similar results with an increasing trend with age [57,58].
Antibodies to MCPyV LT and ST are usually not present in healthy individuals but can be detected in patients with virus-associated MCC. Antibodies to the common region of MCPyV ST and LT were present in half of patients with MCC and in less than 1% of healthy individuals [59]. Importantly, antibody titres to LT greatly decrease upon definitive treatment of the MCC and can be used as a biomarker to follow disease status [59]. Of note, MCC patients often have higher titres of antibodies to VP1 than normal healthy individuals [60].
(d). How rare is Merkel cell carcinoma?
In part, due to its rarity, it has been challenging to accurately measure the incidence of MCC. The tumour was first described in 1972 by Cyril Toker as a trabecular carcinoma of the skin exhibiting carcinoid features [61]. Later, using an electron microscope, Toker reported the presence of membrane-bound granules containing dense cores within the tumour cells, a feature indistinguishable from tumour cells of neural crest origin. He also noted that MCC cells had an appearance similar to normal Merkel cells, a skin cell that also contained neurosecretory granules [62]. The tumour name was eventually changed from trabecular carcinoma to MCC to reflect the similarity in appearance of tumour cells to normal Merkel cells [63,64]. Later, IHC staining was recognized to be useful in distinguishing MCC from other more common neuroendocrine tumours such as small cell lung carcinoma. In particular, IHC staining for cytokeratin 20 (CK20, KRT20) could distinguish MCC from other skin tumours and could also detect normal Merkel cells in the basal layer of the skin epidermis [65].
In addition to the availability of widely accepted IHC markers, the reporting of MCC increased in 1986 when the ninth edition of the International Classification of Diseases and Related Health Problems (ICD-9) first recognized MCC and distinguished it from the other more common skin cancers such as basal cell carcinoma and squamous cell carcinoma. The updated ICD-10-CM, used to indicate the diagnosis for reimbursement purposes, included specific codes for locations and laterality of MCC [66].
To obtain a measure of the incidence of MCC, several studies used data collected for the surveillance, epidemiology and end results (SEER) program. Using data collected between 1986 and 2001, a total of 1124 cases of MCC were identified yielding a rate of 0.15 cases per 100 000 person-years (PY) in 1986 and 0.44 cases per 100 000 PY in 2001, a threefold overall increase [67]. The incidence of MCC in the SEER database was again analysed in 2015 with a total of 5211 cases between 1986 and 2011. In this analysis, the incidence in 1986 was 0.22 per 100 000 PY and increased more than threefold to 0.79 per 100 000 PY in 2011 [68]. Epidemiologic studies covering similar time periods from other countries also found an increase in MCC incidence. A study of the Netherlands Cancer Registry found a doubling of the incidence of MCC between 1993 and 2007 [69]. An analysis of the Danish Cancer Registry revealed that the incidence increased 5.4-fold between 1986 and 2003 [70]. Whether the incidence of MCC is increasing due to wider recognition by physicians, improved diagnosis and reporting to cancer registries or to a change in risk factors is not known. Now, 45 years after its original description, the availability of CK20 IHC diagnostic markers together with codified categorization for reporting have contributed to a more accurate assessment of the incidence of MCC. In addition, the discovery of MCPyV in MCC and good response rates to checkpoint blockade immunotherapy has brought still wider recognition of this disease and perhaps increased reporting [71,72].
The low incidence of MCC contrasts markedly with the other skin cancers. Although the incidence of non-melanotic skin cancers (NMSCs) is not typically counted in cancer registries, the Girona (Spain) Cancer registry has collected data on all non-melanoma skin cancers since 1994. In this database, the incidence rate of basal cell carcinoma was 113 per 100 000 PY, squamous cell carcinoma was 38, melanoma 8.76 and MCC was 0.28 per 100 000 PY [73].
A variety of immunosuppressive conditions appear to increase the risk for developing MCC including co-morbidity with other cancers [74–77]. For example, an analysis of the SEER database from 1992 to 2007 for patients with chronic lymphocytic leukaemia (CLL) and non-Hodgkin lymphoma (NHL) found an incidence ratio of 8.2 for MCC in CLL patients and 3.2 in NHL patients [78]. A large study of the serology of patients with CLL found lower titres of MCPyV VP1 at early stages of disease [79]. These patients were likely to be infected with MCPyV but perhaps were unable to mount an effective antibody response to ongoing infection.
Medical-induced immunosuppression for a variety of conditions also appears to increase the risk for developing MCC. For example, a recent study of skin cancer in adult solid organ transplant patients found an increased risk for developing squamous cell carcinoma, melanoma and MCC. They observed two cases of MCC in 10 649 adult transplant recipients over the 2-year study period for an incidence rate of 2 MCC per 100 000 PY, 25-fold higher than expected [80]. MCC has been reported in patients with a variety of autoimmune conditions including rheumatoid arthritis, psoriatic arthritis and Wegener's granulomatosis [4,81–83]. While these patients have been treated with a variety of immunosuppressive therapies including cyclophosphamide, azathioprine, cyclosporine and etanercept, it is not clear if the autoimmune disease or the therapy contributes to the increased risk for developing MCC.
(e). How does a common virus cause a rare cancer?
The original report from the Moore and Chang lab outlined several important features about virus-positive MCC. Approximately, 80% of all MCC contain clonally integrated copies of the virus [1]. Viral integration involved mutations that result in the truncation of LT with deletion of its DNA binding and helicase domains as well as a growth suppressing domain in the carboxyl-terminus encoded by Exon 3 of 57 kT loss but preservation of the LXCXE motif that binds and inactivates RB [84–86]. The virus-positive MCC tumours typically express ST and the truncated LT and not the late genes VP1 and VP2. Importantly, most virus-positive MCC maintain expression of ST and truncated LT, supporting their role in the initiation and maintenance of MCC.
What percentage of MCC tumours contain integrated MCPyV is not clear although 80% is a reasonable estimate based on many different reports. The original study that identified MCPyV in MCC used Southern blotting with 32P-phosphate labelled viral probe to confirm MCPyV DNA integration into the tumour genome [1]. Very few, if any, studies since then have used radiolabelled Southern blotting to detect integrated viral DNA in tumour DNA. Another approach used in the original study was PCR amplification of viral DNA from tumour DNA. However, PCR amplification of viral DNA is not always reliable for several reasons. The viral DNA has undergone multiple mutations and re-arrangements during integration that at a minimum result in truncation of LT and can reduce primer recognition [84]. In addition, the integrated viral DNA may have undergone amplification that could introduce additional mutations to the T-antigen genes [87]. There may even be some strain differences in MCPyV common to different parts of the world that could impede detection of integrated viral by PCR [88]. Another challenge to PCR detection arises because most studies use DNA isolated from formalin-fixed paraffin-embedded tumour sections that can result in degradation of DNA.
Several sets of PCR primer have been developed to improve detection of MCPyV in MCC. However, there is often not good agreement between primer sets making it difficult to conclude whether MCPyV is present and, if so, how many integrated copies are present [1,89,90]. Based on the clonality of the virus-positive tumours, it is expected that there should be at least one copy of virus in each tumour cell. However, tumour preparations are not typically pure thereby reducing the apparent copy number. The TPO (thyroid peroxidase) gene has been used to normalize host cell DNA content and determine viral copy number because its location on chromosome 2q25 is not typically involved in copy number alterations in MCC [91]. Alternatively, primers to RNaseP have been used to normalize viral copy number to the host genome [92]. Because the sensitivity of PCR is so high and the specificity of primer sets might not be perfect, it is possible to estimate very low copy number in tumour samples. A recent study established a threshold of one DNA viral copy per 100 cells (0.01) as a cut-off to consider an MCC positive for MCPyV [90].
An alternative approach to determine if an MCC tumour contains MCPyV is to perform IHC with antibodies specific for LT. For LT, monoclonal antibodies CM2B4 and Ab3 have been reported [84,89]. A recent study showed that CM2B4 has slightly lower sensitivity compared with Ab3 (0.882 versus 0.983) but much higher specificity (0.943 versus 0.453) [90]. This study called a MCC tumour as virus-positive when at least one antibody (CM2B4 or Ab3) stained tumour cells positive and the quantitative PCR value was at least 0.01 virus copy per cell or when both antibodies were positive and the PCR determined copy number was less than 0.01. Antibodies to ST have also been used to detect the presence of MCPyV in MCC. Indeed, ST has been detected in tumours that were negative for LT [93].
In addition to the presence or absence of viral DNA and protein expression, it has become clearer that the tumour genomes differ significantly between virus-positive and virus-negative MCC. Recent sequencing studies have revealed striking differences between these two types of MCC. Virus-positive MCC typically contains few somatic mutations and copy number alterations. By contrast, virus-negative MCC shows a very high frequency of DNA mutations associated with UV damage typically seen in other sun-exposure associated skin cancers such as melanoma, basal cell carcinoma and squamous cell carcinoma [87,94–98]. The contrasting mutational profile between virus-positive and virus-negative MCC may provide clues into the oncogenic events necessary to generate the tumour.
An additional feature of MCC is that it often presents in advanced age. A variety of institutional based and national cancer registry studies have reported the age at diagnosis for MCC at 69 YOA and higher. A report analysing 6908 MCC cases in the National Cancer Data Base (NCDB), a national tumour registry for the USA, found the median age at diagnosis was 76 years (range: 20–90 years) [99]. In a separate study of 282 cases of MCC, where the presence of virus was established by IHC with both Ab3 and CM2B4 as well as PCR detection, the median age at diagnosis was 71 years for both the virus-negative and virus-positive groups [90]. While lifelong exposure to UV radiation may be required to introduce the numerous mutations found in virus-negative tumour DNA, it is less clear why virus-positive MCC also typically occurs in the elderly.
An important feature of virus-positive MCC is that the tumour maintains expression of LT and ST antigen. The original report describing the discovery of MCPyV in MCC demonstrated that LT mRNA was expressed in tumour cells [1]. In all cases reported to date, the LT is truncated preserving the N-terminal J domain and LXCXE motif but losing the DNA binding and helicase domains. In addition, the C-terminal 100 residues of LT also contains a growth inhibitory domain that is not expressed in MCC [86]. Some of the tumour-derived mutant LTs retained the nuclear localizations signal (NLS) while others lost it [100,101]. When full-length MCPyV LT was expressed in MCC cell lines that contain the truncated LT, a specific DNA-damage response was observed. The full-length LT likely bound to integrated copies of the MCPyV origin of replication and induced in situ replication of the integrated viral DNA leading to the DNA-damage response [84,85]. MCPyV LT may interact with BRD4 to promote viral replication [102].
Most virus-positive MCC contain an intact retinoblastoma suppressor gene (RB1) while virus-negative MCC almost invariably contain mutations that disrupt RB function [96]. This can be explained by expression of the MCPyV LT that contains the LXCXE motif, capable of binding to RB protein and inactivating its function [103]. RB is capable of restricting cell-cycle progression by binding to and repressing the E2F family of transcription factors. E2F serves to transactivate expression of genes required for entry into S phase and for DNA replication [104]. When RB1 is mutated or when MCPyV LT is present, RB is incapable of repressing E2F-dependent gene expression and cells fail to stop in G1/S.
A recent publication provides strong genetic evidence that the target of the truncated LT is RB1 [105]. LoKe cells are a virus-positive MCC cell line with deletion of RB1 gene and no expression of RB protein. The LoKe cell line was derived from a metastasis of an MCC that recurred 3 years after the original primary tumour. Of note, the primary tumour, the recurrent metastasis as well as the LoKe cell line all contained a homozygous focal deletion of the RB1 gene [105]. Furthermore, LoKe cells continue to proliferate when LT is knocked down by shRNA. By contrast, knockdown of LT in other virus-positive MCC cell lines with wild-type RB1 causes a growth arrest that could be rescued by co-knockdown of RB1. This result strongly supports the model that LT serves to inactivate the RB tumour suppression function and that in the absence of RB1, there is no genetic requirement for LT (figure 1b). LT also binds to VPS39 (Vam6) but this interaction is unlikely to contribute to cellular transformation and tumorigenesis [106]. LT has two unique domains (MUR-1 and MUR-2) that surround the LXCXE motif and have minimal contributions to transformation [107].
Virus-positive MCC tumours also express MCPyV ST. Mammalian polyomaviruses all contain ST with a J domain shared with LT and a unique region that contains two zinc fingers. Although the exact molecular functions of MCPyV ST are not well understood, it appears to make important contributions to MCC. MCPyV ST binds to the protein phosphatase PP2A comprising the scaffold Aα (PPP2R1A) and Aβ (PPP2R1B) and catalytic subunits (PPP2CA, PPP2CB) [93,108]. Most polyomavirus ST can bind to PP2A although the exact subunits may differ. For example, SV40 ST can only bind to PP2A-Aα while mouse polyomavirus ST can bind to both Aα and Aβ [109]. In general, it is thought that ST displaces the PP2A B subunit from the holo-enzyme and reduces PP2A activity directed to specific substrates. To date, no phosphatase substrates perturbed by MCPyV ST have been identified. The ST unique region contains two zinc fingers that may be used to bind iron. When it was produced in bacteria, MCPyV ST coordinated iron sulfur [2Fe-2S] and [4Fe-4S] [110]. Whether MCPyV ST binds Fe or Zn in mammalian cells is not clear.
MCPyV ST has an additional domain distinct from PP2A binding known as the LT stabilizing domain or LSD. This unique region of MCPyV ST is not well conserved in ST from other polyomaviruses. The LSD motif in ST appears to increase the levels of MCPyV LT and contributes to increased viral replication at least in part by increasing LT levels [111]. Evidence for a role of MCPyV ST in MCPyV replication includes its ability to translocate to viral DNA replication centres within the nucleus in the presence of the viral origin and LT [110]. The LT stabilizing activity may reflect ST's ability to perturb the function of FBXW7, a component of the cullin RING ligase family of ubiquitin ligases. It should be noted that SV40 LT can bind to FBXW7 and inhibit its ubiquitin ligase activity resulting in increased levels of Cyclin E and Myc [112]. The ST LSD domain also binds to CDC20 and CDH1, substrate recognition components of the anaphase promoting complex [113].
MCPyV ST can increase levels of 4EBP1 phosphorylation that in turn promotes increased protein translation [93]. Significantly, the PP2A-binding activity of MCPyV ST was not required to increase levels of phospho-4EBP1. MCPyV ST binding to CDC20 may contribute to increased 4EBP1 phosphorylation [113]. Expression of ST can promote significant changes in gene expression including induction of pro-glycolytic genes and can induce aerobic glycolysis when expressed in fibroblasts [114]. Whether the ability of ST to induce a Warburg effect in cells is linked to the LSD motif, PP2A binding or 4EBP1 phosphorylation is not known.
MCPyV ST has oncogenic activity that can be distinguished from that contributed by MCPyV LT. For example, ST alone can transform Rat-1 fibroblasts [93]. ST can cooperate with truncated LT to transform human fibroblasts [86]. ST can cause tumours when expressed in mice as a sole transgene [115,116]. Importantly, these examples of ST-induced cellular transformation and transgenic tumorigenesis did not require PP2A binding because mutations in ST that disrupted binding to PP2A were fully functional. Combined expression of MCPyV ST with truncated LT in mice keratinocytes led to hyperplasia, hyperkeratosis and acanthosis of the skin as well as papillomas [117].
While genomic sequencing has revealed that virus-negative MCC has evidence of a high degree of UV damage, this does not exclude a role for UV exposure in the development of virus-positive MCC. The relative lack of UV damaged DNA in virus-positive MCC indicates that the aetiologies are clearly different, suggesting that the precursor to virus-negative MCC was a recipient of lifelong intense UV exposure while the virus-positive MCC were not exposed to the same degree or for as long. However, UV exposure could affect the immune response to virus-negative and virus-positive NCC aetiology. The effect of UV radiation in the pathogenesis of MCC has been suggested to be more likely a result of immune modulation than direct effects on DNA itself [118]. It was reported that the early promoter of MCPyV responds to UV exposure and that levels of ST mRNA increased in UV exposed skin from a healthy human volunteer [119]. Of note, UV exposure was found to increase the risk of wart and squamous cell skin carcinoma development in otherwise healthy mice infected with mouse papillomavirus [120].
In addition to expression of MCPyV ST and truncated LT, virus-positive MCC tumours contain additional mutations that activate the PI3 K pathway (HRAS, KRAS, PIK3CA, PTEN and TSC1). Some of these mutations are also seen in virus-negative tumours. By contrast, most virus-negative MCC contain many more mutations in tumour suppressors and oncogenes including RB1, TP53, NOTCH1-4, chromatin modifying enzymes such as ASXL1, MLL2-KMT2D, and MLL3-KMT2C, ARID1A, ARID1B KMT2A, KMT2C, KMT2D, SMARCA4, KAT6A/B) and DNA-damage pathways (ATM, MSH2, BRCA1, BRCA2, BCOR) [95,97,98]. These observations indicate that the MCPyV viral oncogenes contribute the major oncogenic component to virus-positive MCC and that a limited number of additional somatic mutations are required to cooperate with the virus to generate this cancer.
2. Conclusion
As indicated by serology testing for antibodies specific to the VP1 coat protein of MCPyV, primary infection occurs as early as the first year or two of life and persists for an entire lifetime. Infection with MCPyV apparently is permissive with several additional polyomaviruses as there does not appear to be much cross-reactivity between antibodies. Infection with MCPyV does not appear to cause any symptoms either at the time of initial infection or at later times. While it is likely that MCPyV can replicate on the skin, it is not clear if keratinocytes, hair follicles, dermal fibroblasts or perhaps B lymphocytes or other immune cells that reside in the skin support viral replication. Although the initial infection may occur in childhood, virus-positive MCC typically does not occur until 70 YOA. In an apparent rare event, the MCPyV genome becomes integrated within precursors of MCC. The integration patterns are quite specific, including maintenance of expression for the early region genes ST and the N-terminus of LT retaining the LXCXE or RB-binding motif and not the viral DNA binding, helicase or exon 3 domains. Given the increased risk for MCC in a variety of immunocompromised conditions, it is likely the overall titres of MCPyV or perhaps its distribution in tissues or cell types contributes to the increased risk for MCC. In addition to specific integration pattern, virus-positive MCC are often accompanied by specific mutations in tumour oncogenes and tumour suppressor genes. The combination of nearly ubiquitous presence of a benign and innocuous virus with an exceedingly rare integration pattern and perhaps rarity of precursor cell may account for the relatively rarity of MCC. Recently, two immune checkpoint blockade inhibitors targeting PD-1 (PDCD1) and PD-L1 (CD274) have shown good therapeutic activity in MCC [71,72]. Given the availability of new therapeutic agents against this highly aggressive cancer, it is likely that the number of reported MCC cases will continue to increase.
Acknowledgement
Reety Arora provided critical reading of this manuscript.
Data accessibility
This article has no additional data.
Competing interests
A research project in the DeCaprio laboratory outside the submitted work is supported by Constellation Pharmaceuticals.
Funding
This work was supported in part by U.S. Public Health Service grants R01CA63113, R01CA173023 and P01CA050661 and the DFCI Helen Pappas Merkel Cell Research Fund and the Claudia Adams Barr Program in Cancer Research to J.A.D.
References
- 1.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319, 1096–1100. ( 10.1126/science.1152586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvard V, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Straif K. 2012. Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol. 13, 339–340. ( 10.1016/S1470-2045(12)70125-0) [DOI] [PubMed] [Google Scholar]
- 3.Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. 2002. Merkel cell carcinoma and HIV infection. Lancet 359, 497–498. ( 10.1016/S0140-6736(02)07668-7) [DOI] [PubMed] [Google Scholar]
- 4.Gooptu C, Woollons A, Ross J, Price M, Wojnarowska F, Morris PJ, Wall S, Bunker CB. 1997. Merkel cell carcinoma arising after therapeutic immunosuppression. Br. J. Dermatol. 137, 637–641. ( 10.1111/j.1365-2133.1997.tb03802.x) [DOI] [PubMed] [Google Scholar]
- 5.Lanoy E, Engels EA. 2010. Skin cancers associated with autoimmune conditions among elderly adults. Br. J. Cancer 103, 112–114. ( 10.1038/sj.bjc.6605733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke CA, et al. 2015. Risk of Merkel cell carcinoma after solid organ transplantation. J. Natl Cancer Inst. 107, dju382 ( 10.1093/jnci/dju382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durst M, Gissmann L, Ikenberg H, zur Hausen H. 1983. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl Acad. Sci. USA 80, 3812–3815. ( 10.1073/pnas.80.12.3812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck CB, et al. 2016. The ancient evolutionary history of polyomaviruses. PLoS Pathog. 12, e1005574 ( 10.1371/journal.ppat.1005574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo GJ, Chen CJ, Sullivan CS. 2009. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 383, 183–187. ( 10.1016/j.virol.2008.11.001) [DOI] [PubMed] [Google Scholar]
- 10.Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. 2011. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J. Clin. Virol. 52, 272–275. ( 10.1016/j.jcv.2011.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter JJ, Daugherty MD, Qi X, Bheda-Malge A, Wipf GC, Robinson K, Roman A, Malik HS, Galloway DA. 2013. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc. Natl Acad. Sci. USA 110, 12 744–12 749. ( 10.1073/pnas.1303526110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theiss JM, Gunther T, Alawi M, Neumann F, Tessmer U, Fischer N, Grundhoff A. 2015. A comprehensive analysis of replicating Merkel cell polyomavirus genomes delineates the viral transcription program and suggests a role for mcv-miR-M1 in episomal persistence. PLoS Pathog. 11, e1004974 ( 10.1371/journal.ppat.1004974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schowalter RM, Buck CB.. 2013. The Merkel cell polyomavirus minor capsid protein. PLoS Pathog. 9, e1003558 ( 10.1371/journal.ppat.1003558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. 2009. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J. Virol. 83, 12 118–12 128. ( 10.1128/JVI.01336-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. 2011. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS ONE 6, e22468 ( 10.1371/journal.pone.0022468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. 2011. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J. Mol. Biol. 409, 529–542. ( 10.1016/j.jmb.2011.03.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yardimci H, Wang X, Loveland AB, Tappin I, Rudner DZ, Hurwitz J, van Oijen AM, Walter JC. 2012. Bypass of a protein barrier by a replicative DNA helicase. Nature 492, 205–209. ( 10.1038/nature11730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gai D, Wang D, Li SX, Chen XS. 2016. The structure of SV40 large T hexameric helicase in complex with AT-rich origin DNA. eLife 5, e18129 ( 10.7554/eLife.18129) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Gheit T, et al. 2017. Isolation and characterization of a novel putative human polyomavirus. Virology 506, 45–54. ( 10.1016/j.virol.2017.03.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolstov YL, Knauer A, Chen JG, Kensler TW, Kingsley LA, Moore PS, Chang Y. 2011. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg. Infect. Dis. 17, 1371–1380. ( 10.3201/eid1708.110079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner SD, Field AM, Coleman DV, Hulme B. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1, 1253–1257. ( 10.1016/S0140-6736(71)91776-4) [DOI] [PubMed] [Google Scholar]
- 22.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1, 1257–1260. ( 10.1016/S0140-6736(71)91777-6) [DOI] [PubMed] [Google Scholar]
- 23.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J. Virol. 81, 4130–4136. ( 10.1128/JVI.00028-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaynor AM, et al. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3, e64 ( 10.1371/journal.ppat.0030064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7, 509–515. ( 10.1016/j.chom.2010.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 6, e1001024 ( 10.1371/journal.ppat.1001024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. 2011. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 85, 4586–4590. ( 10.1128/JVI.02602-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA.. 2012. Complete genome sequence of a tenth human polyomavirus. J. Virol. 86, 10887 ( 10.1128/JVI.01690-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim ES, et al. 2013. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436, 295–303. ( 10.1016/j.virol.2012.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korup S, et al. 2013. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS ONE 8, e58021 ( 10.1371/journal.pone.0058021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra N, et al. 2014. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J. Infect. Dis. 210, 1595–1599. ( 10.1093/infdis/jiu250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pastrana DV, Ray U, Magaldi TG, Schowalter RM, Cuburu N, Buck CB. 2013. BK polyomavirus genotypes represent distinct serotypes with distinct entry tropism. J. Virol. 87, 10 105–10 113. ( 10.1128/JVI.01189-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arthur RR, Shah KV, Baust SJ, Santos GW, Saral R. 1986. Association of BK viruria with hemorrhagic cystitis in recipients of bone marrow transplants. N. Engl. J. Med. 315, 230–234. ( 10.1056/NEJM198607243150405) [DOI] [PubMed] [Google Scholar]
- 34.Winter BJ, O'Connell HE, Bowden S, Carey M, Eisen DP. 2015. A case control study reveals that polyomaviruria is significantly associated with interstitial cystitis and vesical ulceration. PLoS ONE 10, e0137310 ( 10.1371/journal.pone.0137310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Aa F, Beckley I, de Ridder D. 2014. Polyomavirus BK—a potential new therapeutic target for painful bladder syndrome/interstitial cystitis? Med. Hypotheses 83, 317–320. ( 10.1016/j.mehy.2014.06.004) [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322. ( 10.1038/nature12965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowen LN, Smith B, Reich D, Quezado M, Nath A. 2016. HIV-associated opportunistic CNS infections: pathophysiology, diagnosis and treatment. Nat. Rev. Neurol. 12, 662–674. ( 10.1038/nrneurol.2016.149) [DOI] [PubMed] [Google Scholar]
- 38.Lima MA, Koralnik IJ. 2005. New features of progressive multifocal leukoencephalopathy in the era of highly active antiretroviral therapy and natalizumab. J. Neurovirol. 11(Suppl. 3), 52–57. ( 10.1080/13550280500513325) [DOI] [PubMed] [Google Scholar]
- 39.Koralnik IJ, Wuthrich C, Dang X, Rottnek M, Gurtman A, Simpson D, Morgello S. 2005. JC virus granule cell neuronopathy: a novel clinical syndrome distinct from progressive multifocal leukoencephalopathy. Ann. Neurol. 57, 576–580. ( 10.1002/ana.20431) [DOI] [PubMed] [Google Scholar]
- 40.Hampras SS, et al. 2015. Merkel cell polyomavirus (MCV) T-antigen seroreactivity, MCV DNA in eyebrow hairs, and squamous cell carcinoma. Infect. Agents Cancer 10, 35 ( 10.1186/s13027-015-0030-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho J, et al. 2015. Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J. Infect. Dis. 211, 1560–1565. ( 10.1093/infdis/jiu524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen KD. 2017. Human polyomavirus 6 and 7 are associated with pruritic and dyskeratotic dermatoses. J. Am. Acad. Dermatol. 76, 932–940.e3. ( 10.1016/j.jaad.2016.11.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Meijden E, et al. 2016. Primary polyomavirus infection, not reactivation, as the cause of trichodysplasia spinulosa in immunocompromised patients. J. Infect. Dis. 215, 1080–1084. ( 10.1093/infdis/jiw403) [DOI] [PubMed] [Google Scholar]
- 44.Bergallo M, Costa C, Terlizzi ME, Astegiano S, Curtoni A, Solidoro P, Delsedime L, Cavallo R. 2010. Quantitative detection of the new polyomaviruses KI, WU and Merkel cell virus in transbronchial biopsies from lung transplant recipients. J. Clin. Pathol. 63, 722–725. ( 10.1136/jcp.2010.077966) [DOI] [PubMed] [Google Scholar]
- 45.Siebrasse EA, et al. 2015. WU polyomavirus in respiratory epithelial cells from lung transplant patient with Job syndrome. Emerg. Infect. Dis. 21, 103–106. ( 10.3201/eid2101.140855) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li K, Zhang C, Zhao R, Xue Y, Yang J, Peng J, Jin Q. 2015. The prevalence of STL polyomavirus in stool samples from Chinese children. J. Clin. Virol. 66, 19–23. ( 10.1016/j.jcv.2015.02.017) [DOI] [PubMed] [Google Scholar]
- 47.Liu W, Yang R, Payne AS, Schowalter RM, Spurgeon ME, Lambert PF, Xu X, Buck CB, You J. 2016. Identifying the target cells and mechanisms of Merkel cell polyomavirus infection. Cell Host Microbe. 19, 775–787. ( 10.1016/j.chom.2016.04.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schowalter RM, Pastrana DV, Buck CB. 2011. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 7, e1002161 ( 10.1371/journal.ppat.1002161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neu U, et al. 2012. Structures of Merkel cell polyomavirus VP1 complexes define a sialic acid binding site required for infection. PLoS Pathog. 8, e1002738 ( 10.1371/journal.ppat.1002738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurdiss DL, Morgan EL, Thompson RF, Prescott EL, Panou MM, Macdonald A, Ranson NA.. 2016. New structural insights into the genome and minor capsid proteins of BK polyomavirus using cryo-electron microscopy. Structure 24, 528–536. ( 10.1016/j.str.2016.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chromy LR, Pipas JM, Garcea RL. 2003. Chaperone-mediated in vitro assembly of polyomavirus capsids. Proc. Natl Acad. Sci. USA 100, 10 477–10 482. ( 10.1073/pnas.1832245100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norkiene M, Stonyte J, Ziogiene D, Mazeike E, Sasnauskas K, Gedvilaite A.. 2015. Production of recombinant VP1-derived virus-like particles from novel human polyomaviruses in yeast. BMC Biotechnol. 15, 68 ( 10.1186/s12896-015-0187-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martel-Jantin C, et al. 2013. Merkel cell polyomavirus infection occurs during early childhood and is transmitted between siblings. J. Clin. Virol. 58, 288–291. ( 10.1016/j.jcv.2013.06.004) [DOI] [PubMed] [Google Scholar]
- 54.van der Meijden E, Bialasiewicz S, Rockett RJ, Tozer SJ, Sloots TP, Feltkamp MC.. 2013. Different serologic behavior of MCPyV, TSPyV, HPyV6, HPyV7 and HPyV9 polyomaviruses found on the skin. PLoS ONE 8, e81078 ( 10.1371/journal.pone.0081078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gossai A, et al. 2016. Seroepidemiology of human polyomaviruses in a US population. Am. J. Epidemiol. 183, 61–69. ( 10.1093/aje/kwv155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicol JT, Robinot R, Carpentier A, Carandina G, Mazzoni E, Tognon M, Touzé A, Coursaget P. 2013. Age-specific seroprevalences of Merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and trichodysplasia spinulosa-associated polyomavirus. Clin. Vaccine Immunol. 20, 363–368. ( 10.1128/CVI.00438-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C, et al. 2014. Seroprevalence of Merkel cell polyomavirus in the general rural population of Anyang, China. PLoS ONE 9, e106430 ( 10.1371/journal.pone.0106430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sroller V, et al. 2014. Seroprevalence rates of BKV, JCV, and MCPyV polyomaviruses in the general Czech Republic population. J. Med. Virol. 86, 1560–1568. ( 10.1002/jmv.23841) [DOI] [PubMed] [Google Scholar]
- 59.Paulson KG, et al. 2016. Viral oncoprotein antibodies as a marker for recurrence of Merkel cell carcinoma: a prospective validation study. Cancer 123, 1464–1474. ( 10.1002/cncr.30475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. 2009. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 5, e1000578 ( 10.1371/journal.ppat.1000578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toker C. 1972. Trabecular carcinoma of the skin. Arch. Dermatol. 105, 107–110. ( 10.1001/archderm.1972.01620040075020) [DOI] [PubMed] [Google Scholar]
- 62.Tang CK, Toker C. 1978. Trabecular carcinoma of the skin: an ultrastructural study. Cancer 42, 2311–2321. [DOI] [PubMed] [Google Scholar]
- 63.Toker C. 1982. Trabecular carcinoma of the skin. A question of title. Am. J. Dermatopathol. 4, 497–500. ( 10.1097/00000372-198212000-00003) [DOI] [PubMed] [Google Scholar]
- 64.Rywlin AM. 1982. Malignant Merkel-cell tumor is a more accurate description than trabecular carcinoma. Am. J. Dermatopathol. 4, 513–515. ( 10.1097/00000372-198212000-00007) [DOI] [PubMed] [Google Scholar]
- 65.Agelli M, Clegg LX, Becker JC, Rollison DE. 2010. The etiology and epidemiology of Merkel cell carcinoma. Curr. Probl. Cancer 34, 14–37. ( 10.1016/j.currproblcancer.2010.01.001) [DOI] [PubMed] [Google Scholar]
- 66.Moshiri AS, Nghiem P. 2014. Milestones in the staging, classification, and biology of Merkel cell carcinoma. J. Natl Compr. Cancer Netw. 12, 1255–1262. ( 10.6004/jnccn.2014.0123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodgson NC. 2005. Merkel cell carcinoma: changing incidence trends. J. Surg. Oncol. 89, 1–4. ( 10.1002/jso.20167) [DOI] [PubMed] [Google Scholar]
- 68.Fitzgerald TL, Dennis S, Kachare SD, Vohra NA, Wong JH, Zervos EE. 2015. Dramatic increase in the incidence and mortality from Merkel cell carcinoma in the United States. Am. Surg. 81, 802–806. [DOI] [PubMed] [Google Scholar]
- 69.Reichgelt BA, Visser O. 2011. Epidemiology and survival of Merkel cell carcinoma in the Netherlands. A population-based study of 808 cases in 1993–2007. Eur. J. Cancer 47, 579–585. ( 10.1016/j.ejca.2010.11.002) [DOI] [PubMed] [Google Scholar]
- 70.Lyhne D, Lock-Andersen J, Dahlstrom K, Drzewiecki KT, Balslev E, Muhic A, Krarup-Hansen A. 2011. Rising incidence of Merkel cell carcinoma. J. Plast. Surg. Hand Surg. 45, 274–280. ( 10.3109/2000656X.2011.613233) [DOI] [PubMed] [Google Scholar]
- 71.Nghiem PT, et al. 2016. PD-1 Blockade with pembrolizumab in advanced Merkel-cell carcinoma. N. Engl. J. Med. 374, 2542–2552. ( 10.1056/NEJMoa1603702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaufman HL, et al. 2016. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 17, 1374–1385. ( 10.1016/S1470-2045(16)30364-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tejera-Vaquerizo A, Descalzo-Gallego MA, Otero-Rivas MM, Posada-Garcia C, Rodriguez-Pazos L, Pastushenko I, Marcos-Gragera R, García-Doval I. 2016. Skin cancer incidence and mortality in Spain: a systematic review and meta-analysis. Actas Dermosifiliogr. 107, 318–328. ( 10.1016/j.ad.2015.12.008) [DOI] [PubMed] [Google Scholar]
- 74.Quaglino D, Di Leonardo G, Lalli G, Pasqualoni E, Di Simone S, Vecchio L, Ventura T. 1997. Association between chronic lymphocytic leukaemia and secondary tumours: unusual occurrence of a neuroendocrine (Merkell cell) carcinoma. Eur. Rev. Med. Pharmacol. Sci. 1, 11–16. [PubMed] [Google Scholar]
- 75.Ziprin P, Smith S, Salerno G, Rosin RD. 2000. Two cases of Merkel cell tumour arising in patients with chronic lymphocytic leukaemia. Br. J. Dermatol. 142, 525–528. ( 10.1046/j.1365-2133.2000.03370.x) [DOI] [PubMed] [Google Scholar]
- 76.Vlad R, Woodlock TJ. 2003. Merkel cell carcinoma after chronic lymphocytic leukemia: case report and literature review. Am. J. Clin. Oncol. 26, 531–534. ( 10.1097/01.coc.0000037108.86294.5E) [DOI] [PubMed] [Google Scholar]
- 77.Koljonen V, Kukko H, Pukkala E, Sankila R, Bohling T, Tukiainen E, Sihto H, Joensuu H. 2009. Chronic lymphocytic leukaemia patients have a high risk of Merkel-cell polyomavirus DNA-positive Merkel-cell carcinoma. Br. J. Cancer 101, 1444–1447. ( 10.1038/sj.bjc.6605306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brewer JD, Shanafelt TD, Call TG, Cerhan JR, Roenigk RK, Weaver AL, Otley CC. 2015. Increased incidence of malignant melanoma and other rare cutaneous cancers in the setting of chronic lymphocytic leukemia. Int. J. Dermatol. 54, e287–e293. ( 10.1111/ijd.12564) [DOI] [PubMed] [Google Scholar]
- 79.Robles C, et al. 2015. Seroreactivity against Merkel cell polyomavirus and other polyomaviruses in chronic lymphocytic leukaemia, the MCC-Spain study. J. Gen. Virol. 96, 2286–2292. ( 10.1099/vir.0.000167) [DOI] [PubMed] [Google Scholar]
- 80.Garrett GL, et al. 2017. Incidence of and risk factors for skin cancer in organ transplant recipients in the United States. JAMA Dermatol. 153, 296–303. ( 10.1001/jamadermatol.2016.4920) [DOI] [PubMed] [Google Scholar]
- 81.Cirillo F, Buononato M, Lima G, Cafaro I, Alquati P. 2003. Clinical experience on eight cases of Merkel cell carcinoma. Tumori 89, 146–151. [DOI] [PubMed] [Google Scholar]
- 82.de Giorgi V, Benemei S, Grazzini M, Lotti T, Geppetti P.. 2011. Rapid growth of Merkel cell carcinoma during etanercept treatment of psoriatic arthritis: cause or coincidence? Acta Derm. Venereol. 91, 354–355. ( 10.2340/00015555-1038) [DOI] [PubMed] [Google Scholar]
- 83.Stone JH, et al. 2006. Solid malignancies among patients in the Wegener's granulomatosis etanercept trial. Arthritis Rheum. 54, 1608–1618. ( 10.1002/art.21869) [DOI] [PubMed] [Google Scholar]
- 84.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. 2008. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl Acad. Sci. USA 105, 16 272–16 277. ( 10.1073/pnas.0806526105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li J, Wang X, Diaz J, Tsang SH, Buck CB, You J. 2013. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 87, 9173–9188. ( 10.1128/JVI.01216-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. 2013. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J. Virol. 87, 6118–6126. ( 10.1128/JVI.00385-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Starrett GJ, et al. 2017. Merkel cell polyomavirus exhibits dominant control of the tumor genome and transcriptome in virus-associated Merkel cell carcinoma. MBio 8, e02079-16 ( 10.1128/mBio.02079-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martel-Jantin C, et al. 2014. Molecular epidemiology of Merkel cell polyomavirus: evidence for geographically related variant genotypes. J. Clin. Microbiol. 52, 1687–1690. ( 10.1128/JCM.02348-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rodig SJ, et al. 2012. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J. Clin. Invest. 122, 4645–4653. ( 10.1172/JCI64116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moshiri A, et al. 2017. Polyomavirus-negative Merkel cell carcinoma: a more aggressive subtype based on analysis of 282 cases using multi-modal tumor virus detection. J. Invest. Dermatol. 137, 819–827. ( 10.1016/j.jid.2016.10.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Gele M, Speleman F, Vandesompele J, Van Roy N, Leonard JH. 1998. Characteristic pattern of chromosomal gains and losses in Merkel cell carcinoma detected by comparative genomic hybridization. Cancer Res. 58, 1503–1508. [PubMed] [Google Scholar]
- 92.Guastafierro A, Feng H, Thant M, Kirkwood JM, Chang Y, Moore PS, Shuda M. 2013. Characterization of an early passage Merkel cell polyomavirus-positive Merkel cell carcinoma cell line, MS-1, and its growth in NOD scid gamma mice. J. Virol. Methods 187, 6–14. ( 10.1016/j.jviromet.2012.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. 2011. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Invest. 121, 3623–3634. ( 10.1172/JCI46323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong SQ, et al. 2015. UV-associated mutations underlie the etiology of MCV-negative Merkel cell carcinomas. Cancer Res. 75, 5228–5234. ( 10.1158/0008-5472.CAN-15-1877) [DOI] [PubMed] [Google Scholar]
- 95.Harms PW, et al. 2015. The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 75, 3720–3727. ( 10.1158/0008-5472.CAN-15-0702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harms PW, et al. 2016. Next generation sequencing of cytokeratin 20-negative Merkel cell carcinoma reveals ultraviolet-signature mutations and recurrent TP53 and RB1 inactivation. Mod. Pathol. 29, 240–248. ( 10.1038/modpathol.2015.154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goh G, et al. 2015. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget 7, 3403–3415. ( 10.18632/oncotarget.6494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gonzalez-Vela MD, et al. 2017. Shared oncogenic pathways implicated in both virus-positive and UV-induced Merkel cell carcinomas. J. Invest. Dermatol. 137, 197–206. ( 10.1016/j.jid.2016.08.015) [DOI] [PubMed] [Google Scholar]
- 99.Bhatia S, et al. 2016. Adjuvant radiation therapy and chemotherapy in Merkel cell carcinoma: survival analyses of 6908 cases from the National Cancer Data Base. J. Natl Cancer Inst. 108, djw042 ( 10.1093/jnci/djw042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laude HC, et al. 2010. Distinct Merkel cell polyomavirus molecular features in tumour and non tumour specimens from patients with Merkel cell carcinoma. PLoS Pathog. 6, e1001076 ( 10.1371/journal.ppat.1001076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nakamura T, et al. 2010. Nuclear localization of Merkel cell polyomavirus large T antigen in Merkel cell carcinoma. Virology 398, 273–279. ( 10.1016/j.virol.2009.12.024) [DOI] [PubMed] [Google Scholar]
- 102.Wang X, Li J, Schowalter RM, Jiao J, Buck CB, You J.. 2012. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog. 8, e1003021 ( 10.1371/journal.ppat.1003021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Borchert S, Czech-Sioli M, Neumann F, Schmidt C, Wimmer P, Dobner T, Grundhoff A, Fischer N. 2014. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J. Virol. 88, 3144–3160. ( 10.1128/JVI.02916-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fischer M, Grossmann P, Padi M, DeCaprio JA. 2016. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 44, 6070–6086. ( 10.1093/nar/gkw523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hesbacher S, et al. 2016. RB1 is the crucial target of the Merkel cell polyomavirus large T antigen in Merkel cell carcinoma cells. Oncotarget 7, 32 956–32 968. ( 10.18632/oncotarget.8793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu X, Hein J, Richardson SC, Basse PH, Toptan T, Moore PS, Gjoerup OV, Chang Y. 2011. Merkel cell polyomavirus large T antigen disrupts lysosome clustering by translocating human Vam6p from the cytoplasm to the nucleus. J. Biol. Chem. 286, 170 79–17 090. ( 10.1074/jbc.M110.192856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Houben R, Angermeyer S, Haferkamp S, Aue A, Goebeler M, Schrama D, Hesbacher S. 2015. Characterization of functional domains in the Merkel cell polyoma virus large T antigen. Int. J. Cancer 136, E290-E300. ( 10.1002/ijc.29200) [DOI] [PubMed] [Google Scholar]
- 108.Kwun HJ, Shuda M, Camacho CJ, Gamper AM, Thant M, Chang Y, Moore PS. 2015. Restricted protein phosphatase 2a targeting by Merkel cell polyomavirus small T antigen. J. Virol. 89, 4191–4200. ( 10.1128/JVI.00157-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Andrabi S, Hwang JH, Choe JK, Roberts TM, Schaffhausen BS. 2011. Comparisons between murine polyomavirus and Simian virus 40 show significant differences in small T antigen function. J. Virol. 85, 10 649–10 658. ( 10.1128/JVI.05034-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsang SH, Wang R, Nakamaru-Ogiso E, Knight SA, Buck CB, You J. 2016. The oncogenic small tumor antigen of Merkel cell polyomavirus is an iron-sulfur cluster protein that enhances viral DNA replication. J. Virol. 90, 1544–1556. ( 10.1128/JVI.02121-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. 2013. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe. 14, 125–135. ( 10.1016/j.chom.2013.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Welcker M, Clurman BE. 2005. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J. Biol. Chem. 280, 7654–7658. ( 10.1074/jbc.M413377200) [DOI] [PubMed] [Google Scholar]
- 113.Shuda M, Velasquez C, Cheng E, Cordek DG, Kwun HJ, Chang Y, Moore PS. 2015. CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc. Natl Acad. Sci. USA 112, 5875–5882. ( 10.1073/pnas.1505787112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Berrios C, et al. 2016. Merkel cell polyomavirus small T antigen promotes pro-glycolytic metabolic perturbations required for transformation. PLoS Pathog. 12, e1006020 ( 10.1371/journal.ppat.1006020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shuda M, et al. 2015. Merkel cell polyomavirus small T antigen induces cancer and embryonic Merkel cell proliferation in a transgenic mouse model. PLoS ONE 10, e0142329 ( 10.1371/journal.pone.0142329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Verhaegen ME, et al. 2015. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J. Invest. Dermatol. 135, 1415–1424. ( 10.1038/jid.2014.446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spurgeon ME, Cheng J, Bronson RT, Lambert PF, DeCaprio JA. 2015. Tumorigenic activity of Merkel cell polyomavirus T antigens expressed in the stratified epithelium of mice. Cancer Res. 75, 1068–1079. ( 10.1158/0008-5472.CAN-14-2425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dowlatshahi M, et al. 2013. Tumor-specific T cells in human Merkel cell carcinomas: a possible role for Tregs and T-cell exhaustion in reducing T-cell responses. J. Invest. Dermatol. 133, 1879–1889. ( 10.1038/jid.2013.75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mogha A, Fautrel A, Mouchet N, Guo N, Corre S, Adamski H, Watier E, Misery L, Galibert M-D. 2010. Merkel cell polyomavirus small T antigen mRNA level is increased following in vivo UV-radiation. PLoS ONE 5, e11423 ( 10.1371/journal.pone.0011423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Uberoi A, Yoshida S, Frazer IH, Pitot HC, Lambert PF. 2016. Role of ultraviolet radiation in papillomavirus-induced disease. PLoS Pathog. 12, e1005664 ( 10.1371/journal.ppat.1005664) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.