ABSTRACT
Viral hemorrhagic septicemia virus (VHSV) is a pathogenic fish rhabdovirus found in discrete locales throughout the Northern Hemisphere. VHSV infection of fish cells leads to upregulation of the host's virus detection response, but the virus quickly suppresses interferon (IFN) production and antiviral gene expression. By systematically screening each of the six VHSV structural and nonstructural genes, we identified matrix protein (M) as the virus' most potent antihost protein. Only M of VHSV genotype IV sublineage b (VHSV-IVb) suppressed mitochondrial antiviral signaling protein (MAVS) and type I IFN-induced gene expression in a dose-dependent manner. M also suppressed the constitutively active simian virus 40 (SV40) promoter and globally decreased cellular RNA levels. Chromatin immunoprecipitation (ChIP) studies illustrated that M inhibited RNA polymerase II (RNAP II) recruitment to gene promoters and decreased RNAP II C-terminal domain (CTD) Ser2 phosphorylation during VHSV infection. However, transcription directed by RNAP I to III was suppressed by M. To identify regions of functional importance, M proteins from a variety of VHSV strains were tested in cell-based transcriptional inhibition assays. M of a particular VHSV-Ia strain, F1, was significantly less potent than IVb M at inhibiting SV40/luciferase (Luc) expression yet differed by just 4 amino acids. Mutation of D62 to alanine alone, or in combination with an E181-to-alanine mutation (D62A E181A), dramatically reduced the ability of IVb M to suppress host transcription. Introducing either M D62A or D62A E181A mutations into VHSV-IVb via reverse genetics resulted in viruses that replicated efficiently but exhibited less cytotoxicity and reduced antitranscriptional activities, implicating M as a primary regulator of cytopathicity and host transcriptional suppression.
IMPORTANCE Viruses must suppress host antiviral responses to replicate and spread between hosts. In these studies, we identified the matrix protein of the deadly fish novirhabdovirus VHSV as a critical mediator of host suppression during infection. Our studies indicated that M alone could block cellular gene expression at very low expression levels. We identified several subtle mutations in M that were less potent at suppressing host transcription. When these mutations were engineered back into recombinant viruses, the resulting viruses replicated well but elicited less toxicity in infected cells and activated host innate immune responses more robustly. These data demonstrated that VHSV M plays an important role in mediating both virus-induced cell toxicity and viral replication. Our data suggest that its roles in these two processes can be separated to design effective attenuated viruses for vaccine candidates.
KEYWORDS: MAVS, matrix protein, VHSV, transcriptional inhibition
INTRODUCTION
Higher eukaryotes have evolved complex innate immune systems that serve as the first line of defense against pathogens like bacteria, fungi, and viruses. Host cells detect conserved pathogen-associated molecular patterns (PAMPs) via germ line-encoded pattern recognition receptors (PRRs) (1), which, once activated, initiate signaling cascades to produce antipathogenic factors, such as type I interferons (IFNs) and other proinflammatory cytokines (2). The retinoic acid-inducible gene 1 (RIG-I)-like helicases (RLHs), including RIG-I, melanoma differentiation-associated factor 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), are cytoplasmic PRRs, expressed in both immune and nonimmune cells, which are essential for detection of intracellular RNA products, primarily of viral origin (3). Upon activation, both RIG-I and MDA5 recruit and activate MAVS (mitochondrial antiviral signaling protein; also called IPS-1/Cardif/VISA) (4), leading to activation of downstream signaling molecules and induction of type I IFNs and other double-stranded RNA (dsRNA)/virally regulated genes (5–7). Secreted IFNs binds to the cognate type I IFN receptor (IFNAR) complex and activate signal transducer and activator of transcription (STAT)-dependent signaling cascades that lead to transcription of IFN-stimulated genes (ISGs) (8). ISG proteins impact a variety of cellular functions, including transcriptional and translational regulation, pro- and antiapoptotic processes, cell signaling, etc., and work together to establish an antiviral state (9). Perturbation of the viral detection or IFN response pathways leads to enhanced sensitivity to most viruses.
The IFN system is highly conserved from mammals down to bony fish (10–12). Teleost IFNs are similar to mammalian type I IFNs based on coding sequences and crystalline structure analysis, although the fish genes differ from their mammalian counterparts by the inclusion of introns (13–15). All of the critical signaling molecules in the viral detection and IFN response pathways, including RLHs, Janus kinase (JAK), and STAT signaling molecules, and a number of traditional ISGs, such as Mx (13, 16–18), have been cloned from multiple fish species. These studies suggest that the innate antiviral pathways and proteins in teleost fish have many regulatory features in common with their mammalian orthologues.
Viral hemorrhagic septicemia virus (VHSV) causes severe disease and mortality among more than 90 marine and freshwater fish species worldwide (19, 20). VHSV is a bullet-shaped, enveloped, nonsegmented, negative-sense, single-stranded RNA virus in the Novirhabdovirus genus of the Rhabdoviridae family (20). Its 11-kb viral genome contains 6 genes encoding (in order, from 3′ to 5′) nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), nonvirion protein (NV), and RNA-dependent RNA polymerase (L) (21, 22). Replication occurs entirely in the cytoplasm by means of a combination of virally encoded and host-derived factors. VHSV isolates are classified in four genotypes (designated I to IV) based on phylogenetic analysis (23). Each group is endemic to specific geographic regions, with freshwater strains included in genotypes I and IV, and each appears to infect regional fish species (24). Genotype I is further divided into five sublineages (Ia to Ie), with the Ia strain being responsible for most outbreaks in European freshwater rainbow trout farms (20, 23, 25, 26). Genotype IV viruses are further divided into three sublineages: IVa, IVb, and IVc (24). In 2005, VHSV-IVb was first isolated from muskellunge (Esox masquinongy) from Lake Ontario and subsequently found in an archived sample from Lake St. Clair, Ontario, dating back to 2003 (isolate MI03GL) (27). VHSV-IVb caused massive die-offs among many freshwater species during the next decade and continues to pose a potential threat to both fish farming and the sport fishing industry in the Great Lakes watershed (28–30). VHSV-IVb has been isolated from at least 31 fish species, including muskellunge, yellow perch, and walleye (28), and has been detected in all five of the Laurentian Great Lakes (31, 32). However, unlike the European Ia sublineage, IVb has low pathogenicity for rainbow trout (33). Despite intensive management and surveillance, VHSV is still detectable among many asymptomatic fish in the Great Lakes region (34).
As with other viruses, VHSV must suppress or evade components of the host antiviral responses in order to propagate. Studies have suggested that NV from the IVb sublineage suppressed apoptosis (35) and that NV from the Ia sublineage suppressed innate immune responses (36, 37). M has been implicated in cellular apoptosis and transcriptional suppression in the related fish novirhabdovirus infectious hematopoietic necrosis virus (IHNV) (38). Several M variants have been found for VHSV-IVb (39), but little else is known about VHSV-IVb antihost processes.
We have undertaken a systematic study of VHSV-IVb proteins to identify which might contribute to antihost activities. VHSV infection of fish cells led to activation of the virus detection system, but the virus quickly suppressed IFN production and antiviral gene expression. By screening each of the six viral structural and nonstructural genes, we have identified M as the most potent antihost protein expressed by VHSV-IVb. Comparative studies with M isolated from other VHSV strains or other fish novirhabdoviruses identified mutations that reduced antitranscriptional function. In particular, point mutations of the aspartic acid residue at position 62 (D62G or D62A) and/or the glutamic acid residue at position 181 (E181A) had combinatorial effects on M antitranscriptional potential (D62A E181A). When reverse engineered into recombinant viruses, the single and double mutations in M led to reduced antihost activities that altered viral cytopathicity and antiviral gene expression. These data suggested that VHSV M is a pivotal component of the VHSV suppression of host antiviral responses and that targeting M for mutation has the potential to undermine this antihost function without destroying the viral replication function of M.
RESULTS
VHSV inhibits IFN antiviral signaling.
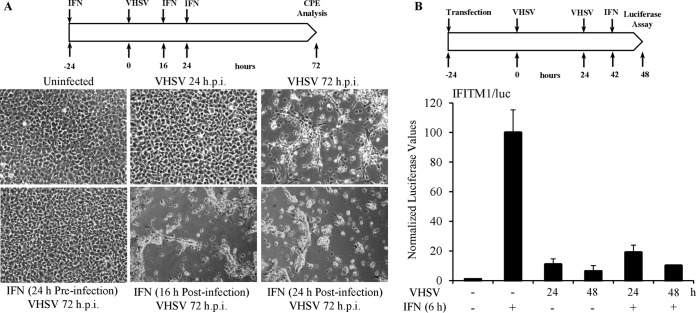
To study the effect of VHSV infection on host innate immune detection and IFN production, epithelioma papulosum cyprinid (EPC) cells were treated with or without IFN (EPC-derived type I IFN) either 24 h prior to or 16 or 24 h postinfection (h.p.i.). VHSV-IVb was used at a multiplicity of infection (MOI) of 1, and cellular cytopathic effects (CPE) were assessed at 72 h.p.i. (Fig. 1A). Infection resulted in significant CPE, which were completely prevented by IFN pretreatment (Fig. 1A). In contrast, IFN added 16 or 24 h.p.i. was ineffective at blocking viral CPE, despite the fact that no cell death was observed at the time of IFN addition (Fig. 1A). These data suggest that upon infection, VHSV produces a factor or factors that are capable of shutting down host IFN-mediated antiviral responses. To further dissect the interplay between viral infection and cellular IFN production, EPC cells were transiently transfected with a luciferase reporter under the control of the IFN-induced transmembrane 1 (IFITM1) promoter. Cells were left uninfected or infected with VHSV for 24 or 48 h and untreated or treated with IFN for the final 6 h prior to the luciferase (Luc) assay (Fig. 1B). As expected, viral infection alone led to a 6- to 10-fold increase in IFN-induced transcription from the IFITM1 promoter compared to that in uninfected cells (Fig. 1B). However, this induction was far less than that observed with IFN treatment alone. More importantly, viral infection suppressed responsiveness to exogenous IFN by 80 to 90% compared to IFN treatment alone (Fig. 1B). These data provide evidence that VHSV is capable of inhibiting IFN production in host cells and/or blocking the cells' response to IFN.
FIG 1.
VHSV-IVb infection blocks IFN responsiveness. (A) EPC cells were left untreated or treated with IFN (10 U/ml) and infected with VHSV-IVb (MOI = 0.1) at the different time points indicated. Cytopathic effects (CPE) were assessed at 72 h.p.i. (B) EPC cells were transfected with a luciferase reporter driven by the IFITM1 promoter (IFITM1/luc) for 24 h, followed by infection with VHSV-IVb for 24 or 48 h, with or without EPC IFN treatment. Luciferase values were quantified 6 h later and normalized to that of the uninfected, IFN-treated control.
VHSV-IVb M inhibits host gene expression.
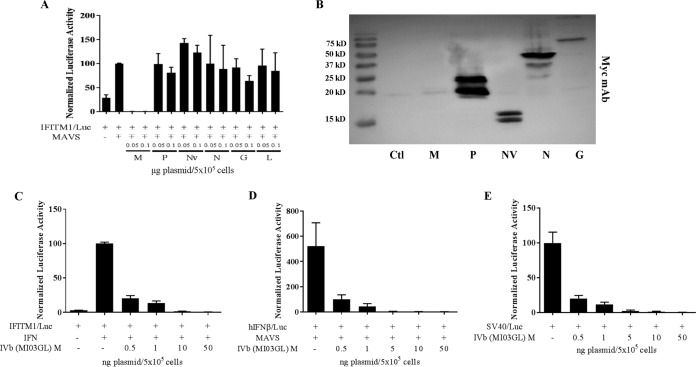
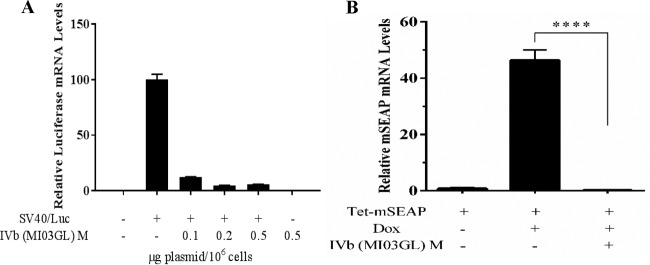
To identify viral proteins involved in the inhibitory effect of VHSV on host IFN responsiveness, each of the six VHSV structural and nonstructural genes was cloned into the pcDNA3.1 expression plasmid and then cotransfected along with IFITM1/luc and EPC-derived MAVS into EPC cells. Ectopic MAVS expression was sufficient to induce endogenous IFN expression and release, which fed back on the cells to activate the IFITM1 promoter. Although several of the viral genes had subtle effects on IFITM1 induction, only the pcDNA3.1 M plasmid (pCD-M) potently decreased IFITM1 promoter activation (Fig. 2A). Conversely, NV was able to upregulate luciferase activity from the IFITM1 promoter. Although each of the Myc-tagged VHSV genes was clearly expressed in the EPC cells, VHSV M was consistently reduced in expression due to the feedback inhibition of the plasmid promoter (Fig. 2B). To determine whether the impact of M was downstream of IFN expression, cells were transiently transfected with pCD-M and IFITM1/luc and then treated with IFN for 24 h. VHSV-IVb M potently inhibited luciferase expression with a 50% inhibitory concentration (IC50) of less than 0.5 ng of plasmid/5 × 105 cells (Fig. 2C). For analysis of pathways upstream of IFN, EPC cells were cotransfected with MAVS, pCD-M, and an IFN promoter-luciferase reporter plasmid (IFN/luc) for 24 h. M again potently blocked induction of luciferase activity in a dose-dependent manner (Fig. 2D), with 80% inhibition observed at concentrations of just 0.5 ng of pCD-M/5 × 105 cells. The potent inhibition of both IFN and ISG transcriptional induction suggested that M either selectively targeted components of both pathways or was a general inhibitor of gene transcription or translation. To test this, pCD-M was cotransfected with an unrelated, constitutively active simian virus 40 (SV40) promoter-luciferase reporter (SV40/luc). VHSV M again potently inhibited SV40 promoter activity in a dose-dependent manner (Fig. 2E), suggesting that M inhibits a general step in cellular protein expression, either transcription or translation. To clarify whether M functioned as a transcriptional or translational inhibitor, cells again were cotransfected with pCD-M and SV40/luc plasmids and luciferase mRNA was quantified via reverse transcription-quantitative PCR (RT-qPCR) (Fig. 3A). M coexpression induced a dose-dependent decrease in luciferase mRNA expression, strongly indicating an impact on cellular RNA transcription or half-life. Since the impact of M on luciferase mRNA was not as potent as that observed when measuring luciferase activity (compare Fig. 3A to Fig. 2E), we reasoned either that M exhibited secondary effects on mRNA translation or that luciferase mRNA expression began in cells before the cotransfected M could elicit an inhibitory effect. Thus, to measure the impact of M on transcriptional initiation instead of preexisting steady mRNA levels, we cotransfected pCD-M with a regulatable plasmid bearing a mouse SEAP (secreted embryonic alkaline phosphatase) reporter gene under the transcriptional control of a tetracycline-responsive element (Tet). After 24 h, transfected cells were left unstimulated or were treated with doxycycline for 24 h prior to RNA extraction and RT-qPCR analysis of SEAP mRNA levels. Under these conditions, SEAP mRNA induction was completely inhibited by M coexpression (Fig. 3B). Taken together, these data suggest that the primary antihost effect of VHSV-IVb M is to inhibit host cellular transcription during viral infection.
FIG 2.
VHSV-IVb M inhibits host promoter activation. (A) EPC cells were cotransfected with 0.4 μg of the IFITM1/luc construct, EPC-derived MAVS (0.3 μg), and plasmids encoding each of the VHSV genes (0.05 or 0.1 μg), followed by luciferase assay 48 h later. Luciferase values were normalized to those with IFITM1/luc plus MAVS. Plasmid concentrations in all samples were equalized with an empty vector. (B) EPC cells (1 × 106) were transfected with 2 μg of expression plasmids for the indicated VHSV proteins expressed in frame with a C-terminal Myc epitope tag. After 48 h, cell lysates were separated by SDS-PAGE and immunoblotted for protein expression with a Myc MAb. Note that VHSV M is less highly expressed because of its ability to inhibit its own expression from the RNAP II-directed CMV promoter. (C) EPC cells were cotransfected with IFITM1/luc (0.4 μg) and various concentrations of a VHSV-IVb M expression plasmid (0.5 to 50 ng) for 24 h and then treated or not treated with EPC IFN for 24 h, followed by luciferase assay. Luciferase values were normalized to that for IFITM1/luc plus IFN. (D) EPC cells were cotransfected with human IFN (hIFN)/luc (0.4 μg), MAVS (0.3 μg), and various concentrations of a VHSV-IVb M expression plasmid (pCD-M; 0.5 to 50 ng) for 24 h, followed by luciferase assay. Luciferase values were normalized to that for IFN/luc plus MAVS. (E) EPC cells were cotransfected with a SV40/luc construct (0.4 μg) and various concentrations of pCD-M (0.5 to 50 ng) for 24 h, followed by luciferase assay. Luciferase values were normalized to that for SV40/luc alone. Ctl, control.
FIG 3.
VHSV-IVb M inhibits host mRNA expression. (A) EPC cells (1 × 106) were cotransfected with or without an SV40/luc construct (0.8 μg) and various concentrations of a VHSV-IVb M expression plasmid (0.1 to 0.5 μg) for 24 h, followed by RNA isolation. Luciferase mRNA levels were measured by RT-qPCR. Data were normalized to β-actin mRNA levels. (B) Tet-mSEAP was cotransfected or not cotransfected with IVb M (0.1 μg) into EPC cells for 24 h and then treated with doxycycline (Dox) for 24 h, followed by RNA isolation and RT-qPCR using the mSEAP primer. ****, P < 0.001.
VHSV M blocks nascent cellular RNA synthesis.
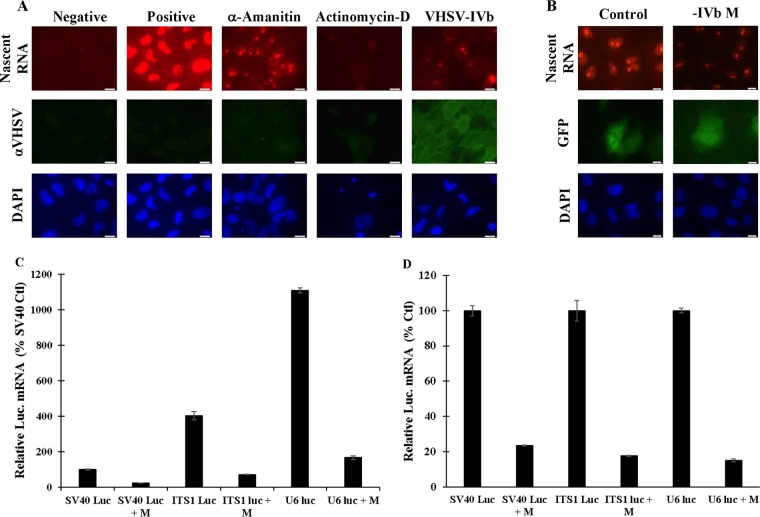
Previous studies focused on the antihost role of vesicular stomatitis virus (VSV) M and demonstrated direct inhibition of host transcription (40). To determine whether VHSV M behaved similarly, we utilized an analog of uracil, 5-ethynyl uridine (5-EU), to study active (nascent) cellular RNA synthesis. 5-EU contains an alkyne that can react with an azide-modified fluorophore to give fluorescent signal (41, 42). EPC cells were left untreated, treated with α-amanitin (1 μg/ml) or actinomycin D (1 μg/ml), or infected with VHSV-IVb (MOI, 1) for 24 h and then pulsed with 5-EU for 2 h, labeled with the Click-iT reagent, and imaged. Untreated, labeled cells showed nuclear RNA staining, with strong puncta representing rRNA synthesis. α-Amanitin, which inhibits predominantly RNAP II-mediated transcription at the dose used (43), showed strongly suppressed nuclear labeling, albeit with residual rRNA nucleolar staining (Fig. 4A). In contrast, actinomycin D, which inhibits RNAP I, II, and III and thus served as a control for complete transcriptional inhibition, suppressed all RNA synthesis in treated cells. 5-EU staining in VHSV-infected cells mimicked the pattern observed with α-amanitin treatment, although residual nucleolar staining in VHSV-infected cells was demonstrably less than in α-amanitin-treated cells (Fig. 4A). These data suggested that VHSV-IVb potently inhibited the activities of all three RNA polymerases to various degrees. To assess a direct role for M in the observed virus-dependent inhibition of host transcription, we cotransfected pCD-M with a cytomegalovirus (CMV)-regulated green fluorescent protein (GFP) plasmid (CMV/GFP, used as a marker of transfection) into EPC cells for 24 h and then labeled cells with 5-EU. M-transfected cells exhibited decreased nascent cellular RNA staining compared to GFP-only-transfected control cells (Fig. 4B). The nascent cellular RNA reduction mirrored that observed upon viral infection, implicating M as a contributing viral protein.
FIG 4.
VHSV M blocks nascent cellular RNA synthesis. (A) EPC cells were left untreated, treated with 2 μg/ml α-amanitin or 1 μg/ml actinomycin D, or infected with VHSV-IVb (MOI = 1) for 24 h and then cultured in 100 μM 5-EU for 2 h. 5-EU was visualized with Alexa Fluor 594 via a click chemistry reaction. VHSV-transfected cells were visualized using a polyclonal anti-VHSV antibody, followed by goat anti-rabbit FITC secondary staining. (B) EPC cells were cotransfected with 0.4 μg GFP and 0.1 μg IVb M for 24 h and then labeled with 100 μM 5-EU for 2 h. 5-EU was visualized with Alexa Fluor 594 via a click chemistry reaction. (C) EPC cells were transfected with an SV40/luc, ITS1/luc, or U6/luc reporter construct with 0.25 μg of pCD-M per 1 × 106 cells for 24 h, followed by total RNA extraction and reverse transcription to cDNA. Luciferase mRNA levels were quantified by RT-qPCR and normalized to EPC β-actin mRNA levels. (D) The data in panel C are expressed as a percentage of the control values for each construct, demonstrating that relative levels of inhibition were similar for all.
To more precisely define which transcriptional responses might be inhibited by M, we tested VHSV M inhibitory potency on RNAP I, II, and III promoters in cell-based luciferase assays. The SV40/luc reporter was used again as an indicator of RNAP II-dependent transcription. A human U6/luc reporter was employed to monitor RNAP III-dependent transcription, and a rainbow trout rRNA intergenic sequence region (ITS-1/luc) reporter was used to assess RNAP I-dependent transcription. Luciferase mRNA levels were measured by RT-qPCR since RNAP I and III do not promote efficient protein production. VHSV-IVb M inhibited the activities of all three promoters, but the RNAP II-regulated promoter appeared to be slightly more sensitive to the inhibitory effects of M than were the RNAP I and RNAP III promoters (Fig. 4C and D). Previous studies had implicated VSV M in suppression of all three RNA polymerases, with different efficacies (44), and our studies suggest that VHSV-IVb M is similarly effective in blocking RNAP I- to III-dependent transcription.
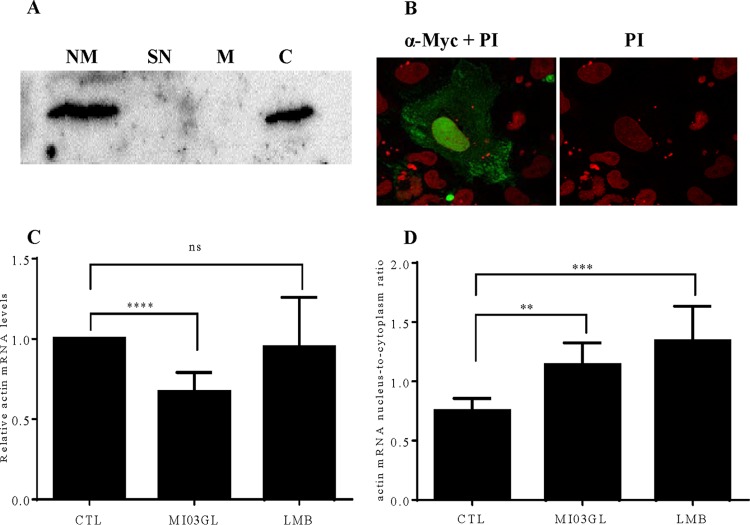
Subcellular localization studies using the Myc/His-tagged M revealed both nuclear and cytoplasmic localization, consistent with a proposed role in both viral packaging and host transcriptional suppression (Fig. 5A and B). Previous studies on VSV M had indicated a role for M in blocking the nuclear export of mRNAs (45–48), in addition to its direct inhibition of cellular transcription. To determine whether VHSV infection alters the subcellular distribution of cellular mRNAs, EPC cells (5 × 106) either were left uninfected or were infected with the VHSV-IVb strain (MOI, 1) for 24 h. Cells were separately treated with leptomycin B (LMB) for 3 h as a control for nuclear export inhibition. After treatment or infection, cell pellets were spiked with 1 × 105 human embryonic kidney 293 (HEK 293) cells to serve as an internal control. The cell mixtures then were separated into nuclear and cytoplasmic fractions, and RNA was isolated from each. RT-qPCR was then performed to assess the subcellular distribution of fish β-actin mRNA in the uninfected, infected, or LMB-treated cells. To control for variability in fractionation fidelity or RNA extraction efficiency, human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was quantified in parallel and used to normalize the β-actin mRNA values. Since the human cells had not been infected or treated with LMB, the human GAPDH mRNA distributions in the spiked HEK 293 cells were expected to be identical for all samples, and the human primers were designed and validated not to cross-react with EPC GAPDH cDNA (data not shown). VHSV infection resulted in a decrease in overall β-actin mRNA levels but had only a moderate impact on the ratio of nuclear to cytoplasmic β-actin mRNA compared to that in uninfected cells (Fig. 5C and D). In contrast, LMB altered dramatically the proportion of mRNA in the nucleus relative to that in the cytoplasm (Fig. 5C and D). From these data, we cannot rule out a minor role for M in altering mRNA subcellular localization, but its primary antihost effect appears to be the inhibition of nascent transcription.
FIG 5.
VHSV IVb M localizes to the nucleus and cytoplasm and alters host mRNA dynamics. (A) Cells transiently transfected with pCD-M were fractionated into nuclear membrane (lane NM), soluble nuclear (lane SN), mitochondrial (lane M), and cytoplasmic (lane C) fractions using differential centrifugation. Lysates were run on a 15% SDS-PAGE gel and transferred to PVDF. IVb M was detected with a 1:1,000-diluted Myc antibody (Cell Signaling). (B) EPC cells were transfected with pCD-M for 24 h. Fixed/permeabilized cells were incubated with anti-Myc primary antibody and then goat anti-rabbit secondary antibody conjugated with FITC. Cells were counterstained with propidium iodide (PI) and viewed using a confocal microscope (magnification, ×100). (C) EPC cells were left uninfected or were infected with VHSV-IVb virus (MOI of 1) for 24 h or treated with leptomycin B (LMB) for 3 h. After infection/treatment, the EPC cell pellets were spiked with 1 × 105 HEK 293 cells before fractionation, which was followed by RNA isolation and RT-qPCR. (D) Total β-actin mRNA levels and ratios of nuclear fish actin mRNA to cytoplasmic fish actin mRNA were calculated. β-Actin values were normalized to human GAPDH values to control for differences in isolation efficiency under conditions in which nuclear/cytoplasmic levels of the human transcript were not altered by treatment. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
VHSV-IVb M disrupted RNAP II activity and recruitment.
To gain further insight into the mechanism by which M inhibits host transcription, we used chromatin immunoprecipitation (ChIP) to assess the impact of VHSV-IVb M on the recruitment of RNAP II to a core promoter sequence. EPC cells were cotransfected with Tet-mSEAP (as described for Fig. 3) and pCD-M for 24 h and then left untreated or treated with doxycycline for 4 h, after which ChIP analyses were performed using an RNAP II antibody and primers representing TATA-proximal primers. RNAP II was constitutively bound to the promoter, but binding was augmented upon doxycycline treatment (Fig. 6A). With pCD-M cotransfection, both uninduced and induced RNAP II binding was significantly decreased (Fig. 6A).
FIG 6.
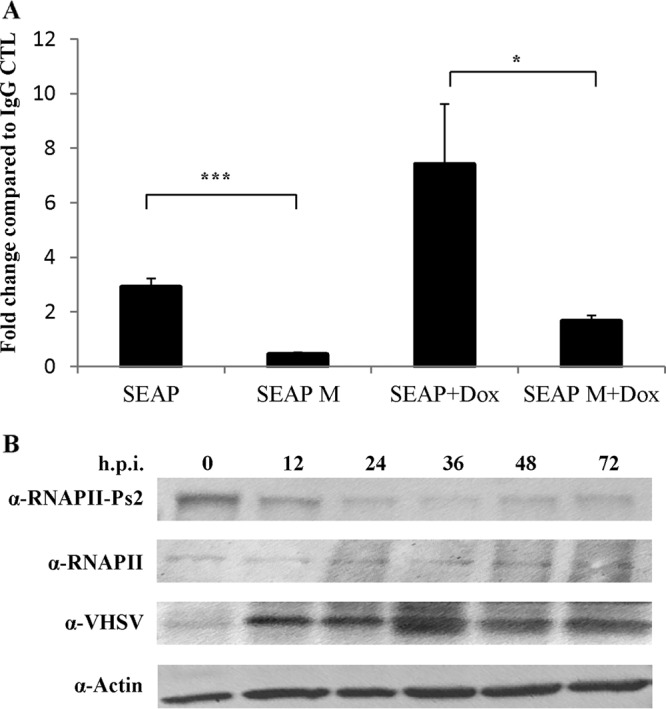
VHSV-IVb M blocks RNAP II recruitment and activation. (A) EPC cells were transfected with a Tet-regulated SEAP plasmid and pCD-M for 24 h and were then left untreated or were induced with 2 μg/ml doxycycline for 4 h. After cross-linking, chromatin was immunoprecipitated with antibodies to IgG and RNAP II and analyzed by RT-qPCR using primers specific for the minimal CMV promoter. RNAP II recruitment was normalized to that in the IgG control. *, P < 0.05; **, P < 0.01; ***, P < 0.005. (B) EPC cells were infected with VHSV-IVb virus (MOI = 5) for 0 to 72 h. Cell lysates were separated by PAGE, and immunoblots were probed with antibodies recognizing the RNA polymerase II CTD repeat YSPTSPS (α-RNAPII-Ps2), total RNAP II, β-actin, and VHSV structural proteins, followed by HRP secondary antibodies and chemiluminescent detection. The bottom panel shows just the M band from the VHSV immunoblot.
The C-terminal domain (CTD) of RPB1, the largest RNAP II subunit, consists of 25 to 52 tandem copies of a conserved YSPTSPS heptapeptide repeat (49). During transcript elongation, the RNAP II CTD is phosphorylated, predominantly at the Ser2 and Ser5 residues, and these modifications indicate different phases of transcription and RNAP II activity (50–52). Early in the transition from preinitiation to elongation, the CTD is phosphorylated primarily on Ser5 residues. During elongation, phosphorylation occurs mainly on Ser2 residues to generate elongation-proficient RNAP II, and by the 3′ end of the gene, CTD phosphorylation is dominated by Ser2 residues. To address which phase of transcription was impacted, we infected EPC cells with VHSV for 0 to 72 h and assessed total and Ser2-phosphorylated RNAP II levels by immunoblotting. Ser2 phosphorylation decreased during the course of a VHSV infection, and interestingly, total RNAP II exhibited a mobility shift from a slower-migrating form to a more rapidly migrating band over the course of the infection (Fig. 6B). Together, these data suggest that VHSV-IVb M inhibits host cellular transcription to suppress host immune responses by disrupting RNAP II recruitment and subsequent phosphorylation, thereby preventing transcriptional initiation and elongation.
Comparative studies on fish rhabdoviral M proteins.
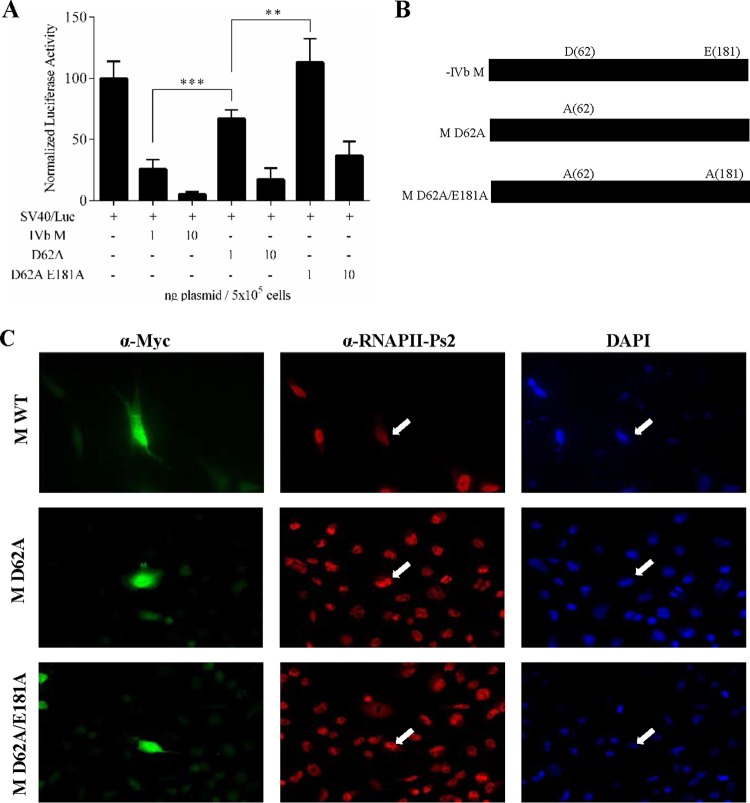
Many of the observed VHSV-IVb M effects on EPC cells mirrored those observed previously with VSV M in mammalian cells (44). To extend our studies beyond IVb M, we assessed the effects of M proteins from various VHSV sublineages, as well as M proteins from the related fish rhabdoviruses infectious hematopoietic necrosis virus (IHNV), spring viremia of carp virus (SVCV), and snakehead rhabdovirus (SHRV), in cell-based luciferase inhibition assays. Each M gene was cloned into pcDNA3.1 and cotransfected with SV40/luc into EPC cells for 24 h, after which time luciferase assays were performed. Like VHSV-IVb M, all rhabdoviral M proteins inhibited luciferase expression to various degrees, although M proteins from SVCV and SHRV exhibited less activity than M proteins in VHSV and IHNV (Fig. 7A) and, in the case of SHRV, required higher plasmid concentrations to exhibit inhibition in these cells (data not shown). Subsequently, Western blotting was used to confirm the expression of rhabdoviral M proteins (Fig. 7B). Although all VHSV strain M proteins had activities similar to that of IVb M, one important exception was a variant M from a VHSV-Ia strain (F1), which exhibited significantly less inhibition than M from the IVb strain when nascent RNA synthesis was monitored (Fig. 7C). This Ia M variant differed from IVb M at only four amino acid positions (T9I, D62G, E181A, and V198A). In order to determine which of these changes impacted antitranscriptional activity, we mutated these same residues in various combinations within the VHSV-IVb M background. The two residues that impacted activity the most were at positions 62 and 181. Reverse mutations (G62D, G62D, and A181E) within the Ia (F1) backbone rescued the antitranscriptional efficacy of M (data not shown), whereas IVb M proteins that were mutated singly at position 62 or doubly at positions 62 and 181 (D62A, D62A, and E181A) each experienced about a 90% reduction in efficacy (Fig. 8A and B). The effect of these mutations on global host transcription was also assessed by measuring RNAP II phosphorylation. For these studies, EPC cells were transfected with either wild-type (WT) or mutant M Myc-His-encoding plasmids for 48 h, and RNAP II CTD serine-2 phosphorylation was assessed using an RNAP II phosphoserine 2-specific antibody (anti-RNAP II-Ps2) in immunofluorescence experiments. Cells expressing either of the VHSV M constructs were visualized using an anti-Myc monoclonal antibody (MAb) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody. These studies showed that cells expressing the mutant M proteins exhibited more-prominent RNAP II-Ps2 staining than the RNAP II-Ps2 staining seen in WT-M-expressing cells (Fig. 8C). These data suggested that the D62A or D62A E181A M mutants were not as effective as WT M at inhibiting RNAP II phosphorylation.
FIG 7.
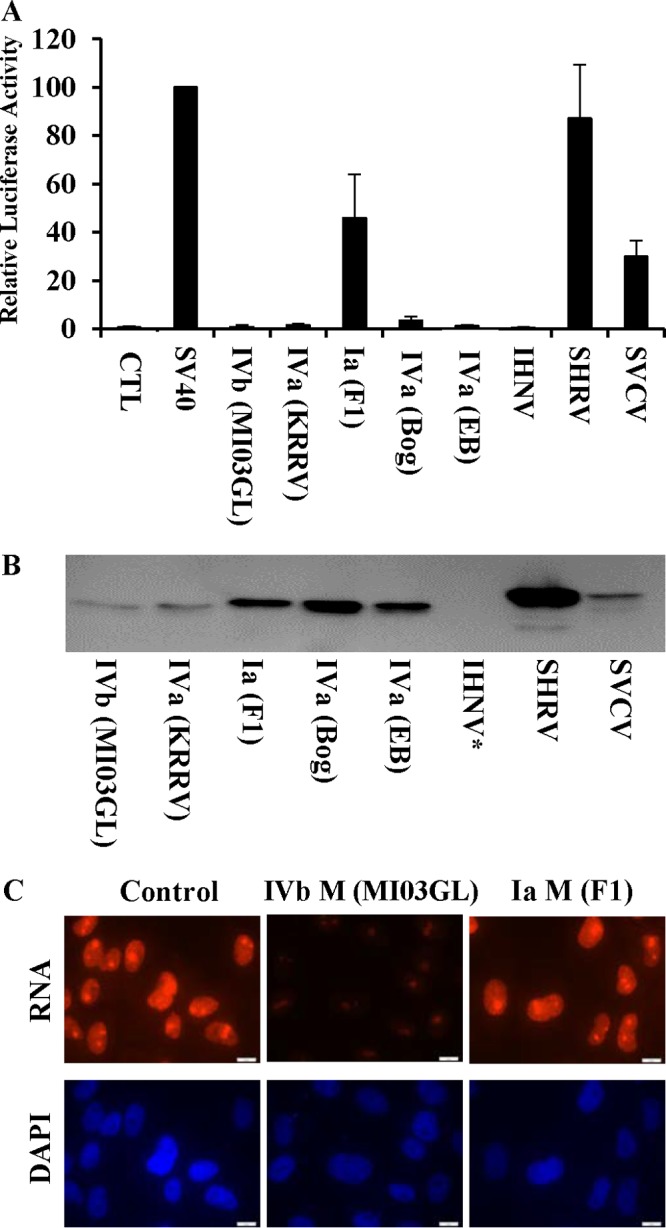
Comparison of fish rhabdoviral M proteins. (A) M proteins from various VHSV strains and substrains (IVb, MI03GL; KRRV, isolate of a IVa strain from Japan; F1, isolate of a Ia strain from Egtved, Denmark; Bog, NA-5 isolate from the Bogachiel River, WA; EB, EB 7 isolate from Elliot Bay, WA) and from IHNV, SVCV, and SHRV were cotransfected with SV40/luc into EPC cells for 24 h, after which time luciferase assays were performed. (B) Immunoblot analysis of transfected viral M proteins (1.0 μg per 1 × 106 cells) in EPC lysates, detected using anti-myc monoclonal antibody followed by HRP goat anti-mouse antibodies and visualization using chemiluminescence. *, IHNV M was so effective at shutting down transcription (including its own) that subsequent studies with higher protein amounts were required to detect its expression (data not shown). (C) EPC cells were left untreated or transfected with 0.1 μg of VHSV-IVb M or VHSV-Ia (F1) M expression plasmids for 24 h and then labeled with 100 μM 5-EU for 2 h. 5-EU was visualized with Alexa Fluor 594 via a Click-iT chemistry reaction.
FIG 8.
Impact of amino acid changes in VHSV-IVb M on transcriptional inhibitory function. (A) EPC cells were cotransfected with wild-type IVb M or M mutants (D62A or D62A E181A M) along with SV40/luc for 24 h, followed by luciferase assay. **, P < 0.01; ***, P < 0.005. (B) Schematic of the M mutants tested in panel A. (C) Immunofluorescence microscopy of cells transfected with WT or mutant M expression plasmids. Cells expressing M were identified by staining them with an anti-Myc (α-Myc) MAb (green), and the level of active RNAP II in M-expressing cells (white arrow) was qualitatively assessed by counterstaining the cells with anti-RNAP II-Ps2 polyclonal antibody (red). Phosphorylation of serine-2 of the RNAP II CTD is a hallmark of an active, elongating RNAP II. All samples were counterstained with DAPI. Images are representative of multiple Myc-positive clones for each transfection.
Recombinant VHSVs harboring M mutations.
Recombinant VHSVs harboring the D62A and D62A E181A mutations were generated using a reverse-genetics system (53). Recombinant viruses containing the mutant M genes were viable and were compared to recombinant WT (rWT) VHSV in a series of cell-based studies. To assess viral replication, EPC cells were infected for 0, 18, 36, 54, 72, or 96 h at an MOI of 0.1 or 1.0 with either D62A, D62A E181A, or rWT VHSV. Media and cells were harvested at each time point. A viral-yield assay was used to compare the abilities of mutant viruses to replicate to the ability of the rWT virus to replicate by determining their titers in the medium from each time point in 1:10 serial dilutions on Bluegill Fry (BF-2) cells. At an MOI of 0.1, replication of the rWT virus peaked at 54 h, while that of M D62A and M D62A E181A viruses peaked at 96 h and 72 h, respectively (Fig. 9A). M D62A virus replication was similar to that of the rWT virus at 72 h and then exceeded rWT titers at 96 h (Fig. 9A). M D62A E181A virus yielded titers that were several orders of magnitude less than that of the rWT or M D62A virus (Fig. 9A). However, at an MOI of 1, all three viruses reached similar titers after 96 h (Fig. 9B), suggesting that other factors affecting viral replication, such as viral cytotoxicity and restriction by the innate immune response, were impacted by these mutations.
FIG 9.
Replication of wild-type versus M D62A and M D62A E181A mutant viruses. Viral titers from media harvested from cells infected at the indicated time points at an MOI of 0.1 (A) or 1.0 (B) are shown. Error bars reflect SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 for mutant virus results in a comparison with the rWT virus results at the same time point.
Transcriptional inhibition by rWT VHSV and mutants.
The abilities of the rWT M- and mutant-M-containing viruses to suppress host transcription were assessed first by using the constitutively active SV40/luc reporter. EPC cells were transfected with SV40/luc for 6 h and then infected for 24 h with each virus (MOI = 0.1 or 1), after which luciferase activity was quantified. The rWT virus was 2- to 2.5-fold-more effective at inhibiting luciferase activity than the M D62A and M D62A E181A viruses (Fig. 10A), mirroring the impact of the transfected M proteins (Fig. 8C). To examine the effects of mutant-M and rWT viruses on class II-specific gene expression, EPC cells were infected with each of the three viruses for 24 h at an MOI of 1. Cells were subjected to analysis by immunofluorescence microscopy using an RNAP II phospho-serine-2-specific antibody as a surrogate for class II gene transcriptional activity, with values quantified by a mean grey-scale intensity within the nuclei (Fig. 10B and C). Anti-VHSV staining, carried out in parallel cultures (due to secondary-antibody conflict), indicated that virtually all cells were infected. Only the rWT virus significantly inhibited RNAP II phospho-serine-2 staining, suggesting that the antitranscriptional effects of M D62A and M D62A E181A viruses were markedly reduced compared to that of the rWT VHSV.
FIG 10.
Inhibition of host responses by wild-type versus M D62A and M D62A E181A mutant viruses. (A) EPC cells were transfected with SV40/luc for 6 h and then infected with the indicated viruses at an MOI of 0.1 or 1.0. Luciferase activity was quantified 24 h after infection, and data were normalized by the Bradford assay. Error bars reflect SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 in a comparison with the rWT virus at the same MOI. (B) Cells were left uninfected or were infected with the indicated virus and then subjected to immunostaining with either anti-VHSV antibody or anti-RNAP II phosphoserine 2 antibody, with DAPI counterstaining. Note that column 1 (αVHSV) is from a parallel culture that was treated in the same way as the right two columns and that the DAPI images correspond to the adjacent anti-RNAP II-Ps2 images. (C) Quantification of the mean grey-scale intensity found within the nuclei of virus-infected cells stained with the RNAP II phosphoserine 2 antibody. Error bars reflect SEM. ***, P < 0.001 in a comparison with uninfected controls. (D) The sulforhodamine B (SRB) assay was used to measure the viability of EPC cells infected with rWT or mutant M viruses at 96 h postinfection at the indicated MOIs. Error bars reflect SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 for mutant virus results in a comparison with rWT results at the same MOI. (E) EPC cells were infected with the indicated viruses at an MOI of 0.1, and monolayers were overlaid with methylcellulose (0.75%). At 48 or 96 h.p.i., cells were fixed with 10% formalin, stained with crystal violet, and imaged using phase-contrast microscopy (bar = 100 μm).
Cytopathic effects of rWT VHSV and mutants.
The reduced transcriptional inhibitory activities of the M D62A and M D62A E181A mutant viruses suggested that induction of host cell cytopathicity was likely to be less severe for the mutant viruses than for rWT VHSV. To assess the impact of viral titer on cytopathicity, EPC cells were infected with each of the viruses for 96 h at MOIs ranging from 0.0001 to 10. After that time, cells were fixed in trichloroacetic acid and stained with sulforhodamine B to quantify viable cells. This assay showed that the mutant viruses induced significantly fewer cytopathic effects at an MOI of <1.0 (Fig. 10D). Interestingly, in EPC cells infected with mutant M viruses, plaques that initially appeared similar to those for rWT virus formed, but over the course of the experiment, the mutant virus plaques remained smaller than those generated by the rWT virus and in some cases refilled with cells after initial formation (Fig. 10E).
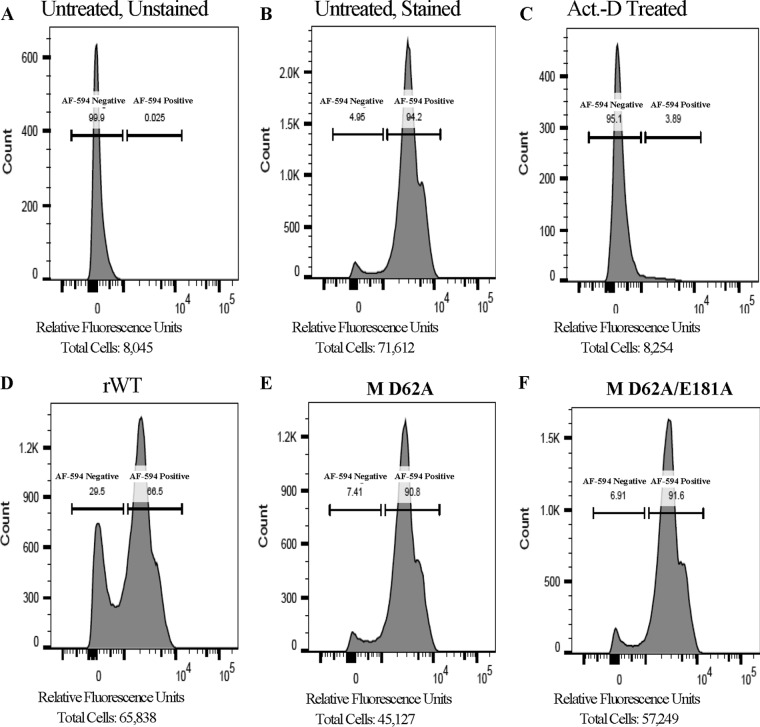
To monitor global cellular RNA synthesis, we again used the Click-iT chemistry reaction with Alexa Fluor 594 (AF-594) and 5-EU and then assessed incorporation with both fluorescence microscopy and flow cytometry. EPC cells were left untreated or were treated with actinomycin D (1 h prior to 5-EU addition) or infected with mutant or WT viruses (24 h). The 5-EU was added for the last hour of treatment/infection, after which labeled cells were subjected to flow cytometry to quantify 5-EU-positive versus -negative cells (Fig. 11). To ensure robust infection, cells on coverslips from each treatment were subjected to immunofluorescence microscopy using an anti-VHSV primary and an FITC-conjugated goat anti-rabbit secondary antibody after the Click-iT reaction was completed (data not shown). Flow cytometric data suggested that the mutant-M-containing viruses were roughly 80% less effective at inhibiting host transcription than rWT VHSV.
FIG 11.
Mutant M viruses elicit reduced transcriptional inhibition. EPC cells were left untreated and unstained (A), untreated and stained (B), treated with actinomycin D (C), or infected with the indicated viruses at an MOI of 1 for 24 h prior to 5-EU labeling of nascent RNA (D to F). 5-EU was conjugated to Alexa Fluor 594 by a Click-iT reaction and flow data were collected. The percentage of cells scored as AF-594 negative and AF-594 positive are shown as an inset within each histogram.
Innate immune activation by rWT and mutant VHSVs.
To determine how differences in transcriptional inhibition impacted antiviral activity release, media collected from control or infected cells were UV irradiated to inactivate live virus and subjected to IFN bioassay (Fig. 12). Both mutant M viruses elicited enhanced antiviral activity (expressed in units of IFN [uIFN] per ml) compared to that of rWT VHSV. The mutant with M D62A induced 1.9- and 22.5-fold-more uIFN/ml than rWT at MOIs of 0.1 and 1, respectively, whereas the M D62A E181A virus induced 2.25- and 49.4-fold-more uIFN/ml than rWT virus at MOIs of 0.1 and 1, respectively (Fig. 12).
FIG 12.
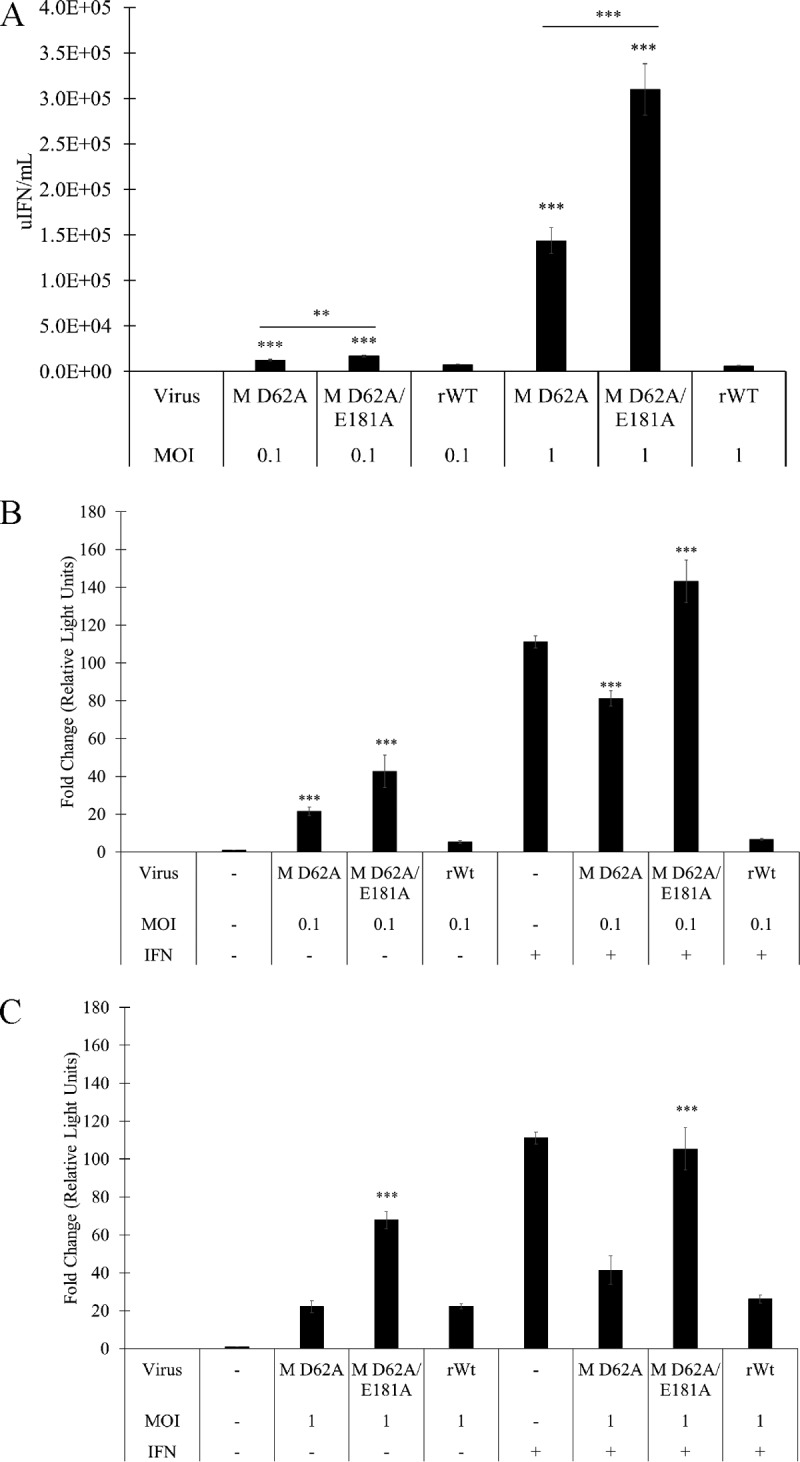
Regulation of host IFN responses by wild-type versus M D62A and M D62A E181A mutant viruses. (A) Media collected at the indicated times postinfection were used in an antiviral assay to assess the antiviral activity released; values are quantified as the number of antiviral units (uIFN) per milliliter as discussed in Materials and Methods. Error bars reflect SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 in a comparison of mutant virus results and rWT results at the same MOI. (B) EPC cells were transfected with an Mx-1/luc reporter and then infected for 24 h with rWT or mutant viruses at MOI of 0.1 and then left untreated (−) or treated (+) with IFN for an additional 18 h. Luciferase activity was quantified and normalized to protein levels in cell lysates by using a Bradford assay. (C) EPC cells were transfected and treated like those in panel B but were infected with rWT or mutant viruses at an MOI of 1 before IFN treatment and luciferase activity quantification. Data are expressed as fold changes in relative light units from the uninfected, untreated control. Error bars reflect SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 in a comparison of mutant virus results and rWT results or in a pairwise comparison, indicated by horizontal bars above the samples of interest.
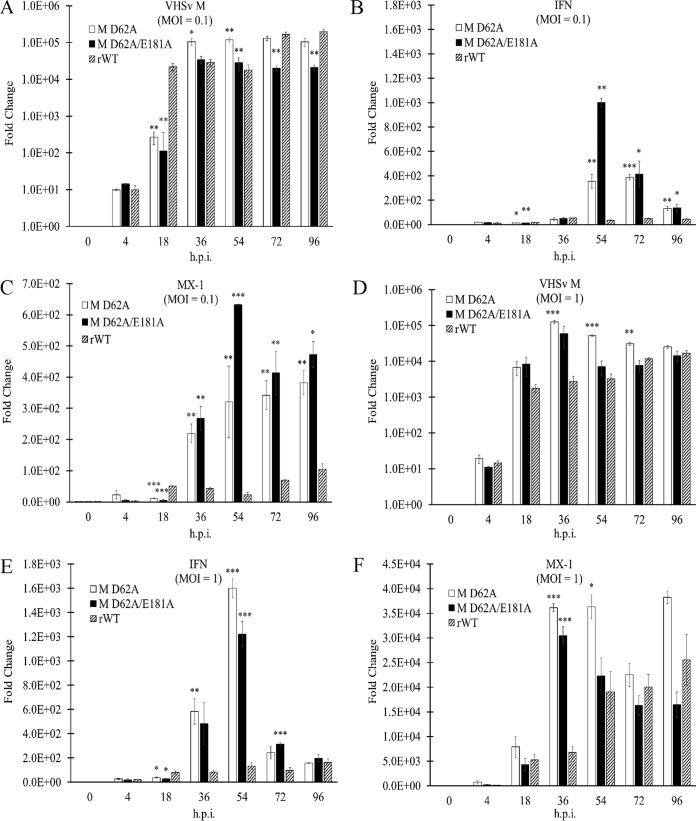
To determine whether the enhanced antiviral activity correlated with altered innate immune gene expression, EPC cells were infected with the M D62A, M D62A E181A, or rWT virus for 96 h at MOIs of 0.1 and 1, with both media and cells collected at 0, 4, 18, 36, 54, 72, and 96 h.p.i. cDNA was made from RNA extracted from the collected cells that had been spiked with 1.0 ng of in vitro-transcribed GFP RNA for normalization, since virus infection alone leads to suppression of host RNA synthesis and impacts reference gene expression. Expression of IFN, IFN-responsive Mx-1, and VHSV RNA was quantified with RT-qPCR (Fig. 13). At both MOIs, the M D62A virus induced more IFN and Mx-1 transcription than the M D62A E181A and rWT viruses. However, when these values were normalized to the amount of viral RNA, the M D62A E181A virus induced 5- and 4.5-fold-more IFN and Mx-1 mRNA, respectively, than the M D62A virus at an MOI of 1. The mutant M viruses also consistently induced 10-fold-more IFN mRNA than rWT virus at MOIs of 0.1 and 1, respectively. These data suggested that the mutant M viruses led to greater expression of IFN throughout the course of infection than did the rWT virus.
FIG 13.
Regulation of innate immune genes by the wild-type versus the M D62A and M D62A E181A mutant viruses. RT-qPCR results from RNA harvested at the indicated times postinfection at an MOI of 0.1 (A to C) or an MOI of 1 (D to F). Synthesized cDNAs were assessed for VHSV RNA (A, D), EPC type I IFN (B), (E), and fish Mx-1 (C, F). Data were normalized to those for a spiked internal control as described in Materials and Methods and are presented as a fold changes in expression from expression in untreated controls. Error bars reflect SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 in a comparison of mutant virus results and rWT results at the same time point.
DISCUSSION
Most viruses have evolved mechanisms to inhibit the expression or functions of host innate immune genes during virus replication (54, 55). Inhibition of host transcription is a common strategy used by RNA viruses that replicate entirely within cytoplasm. Shutting down host transcription not only frees up cellular translational machinery that can be used for the biosynthesis of viral gene products but also inhibits host antiviral responses by preventing the synthesis of antiviral proteins. One clear example of an antihost protein found within the rhabdovirus family is VSV M, which potently inhibits host gene expression, thereby suppressing host antiviral responses, including upregulation of type I IFNs (56, 57). VSV M also may block mRNA export from the nucleus (47, 48, 58). These antihost functions of VSV M are separable from its critical role in viral assembly and budding but are still essential for efficient viral replication (59–61). Salmonid rhabdovirus IHNV M also inhibits host-directed gene expression (38).
Here we report that VHSV-IVb infection suppressed host IFN-mediated antiviral responses, with expression of M alone capable of inhibiting MAVS-mediated IFN expression, as well as IFN-mediated transcriptional responses, in cell-based luciferase assays (Fig. 2). Importantly, viral infection or transfected VHSV M also inhibited transcription from a constitutively active SV40 promoter-driven luciferase construct (Fig. 3), suggesting that VHSV M acts similarly to other rhabdoviral M proteins in shutting down general transcription and/or posttranscriptional events. This possibility was enforced and clarified by the observation that VHSV infection or ectopic expression of M led to decreased nascent RNA transcription by the host. Our results support the hypothesis that VHSV M inhibits host transcription as a means of promoting viral dissemination.
VSV M blocked host transcription directed by all three host RNA polymerases (RNAP I to III [44]). We tested the inhibitory potency of VHSV M on three different luciferase constructs driven by the RNAP I-dependent Atlantic salmon ITS1 promoter, the RNAP II-dependent SV40 promoter, and the human RNAP III-dependent U6 promoter, respectively. VHSV M cotransfection inhibited transcription mediated by all three RNA polymerases (Fig. 4). Interestingly, 5-EU staining of nascent RNA synthesis in VHSV-infected and M-transfected cells resembled the pattern of α-amanitin treatment (Fig. 4). Since α-amanitin targets RNAP II and III, but not RNAP I, this suggests that M may target all three host RNAPs through a common mechanism, which may not be equally efficacious in all instances. Since continued rRNA synthesis benefits the virus, it is possible that residual RNAP I activity is an evolutionary outcome of otherwise-indiscriminate host RNAP suppression.
The mechanism of VHSV M transcriptional inhibition remains unclear. Previous studies of VSV M suggested that the TATA-binding protein (TBP) subunit TFIID was a potential target of M. TFIID isolated from VSV-infected cells was inactive in an in vitro transcription assay, but transcription activity could be reconstituted by adding purified recombinant TBP to the experimental system (40). Our ChIP assay results implicated an M-dependent suppression of both basal and doxycycline-induced recruitment of RNAP II to the minimal CMV promoter region of a Tet-mSEAP reporter gene (Fig. 6). Taken together, our data suggest that VHSV M inhibited RNAP II-directed transcription by interrupting RNAP II promoter binding, perhaps by targeting one or more basal transcription factors. However, it remains uncertain whether this inhibition is direct, or indirect via suppression of regulatory pathways involved in RNA polymerase synthesis and subcellular transport. Future studies will need to address this question; however, a possible role for VHSV M in perturbation of nuclear import/export was revealed in our RNA localization studies (Fig. 5). Although not conclusive, the increased ratios of nuclear to cytoplasmic RNA in VHSV-infected cells suggest additional similarities to VSV M function (47, 48) and provide further evidence that the observed effects of M on luciferase inhibition using our various constructs can be attributed to pretranslational effects.
M proteins from a variety of VHSV strains and several closely related fish viruses, including IHNV, SHRV, and SVCV, all inhibited SV40 promoter activity in EPC cells, which is consistent with the suppression of host protein expression being a conserved role of M among rhabdoviruses. Interestingly, a novel M clone isolated from a VHSV F1 strain (Ia substrain) sample was less potent than other M proteins in inhibiting transcription (Fig. 7). When comparing this F1 M sequence with that of WT VHSV-IVb M, just four amino acid differences (T9I, D62G, E181A, and V198A) were present. We tested a range of targeted mutations at these positions and found that G62D conversion in the Ia M clone enhanced function to approximately that of IVb M (data not shown). To further investigate the impact of the M alternatives (using more-traditional alanine substitutions), two mutant M constructs (M D62A and M D62A E181A) were made within the IVb M background. Both exhibited decreased ability to inhibit cellular gene expression compared to that of WT VHSV-IVb M (Fig. 8). The predicted secondary structure of VHSV M is quite similar to that of VSV M, even though amino acid conservation between the two is low (data not shown). The aspartic acid residue at position 62 is conserved across multiple VSV and VHSV strains (62). In VSV M, this aspartic acid (D92) is located between helices α1 and α2 and is surface exposed (62). It may play a critical role in the antihost function of M by serving in a structural capacity or as a site for protein-protein interactions with host factors involved in regulating transcription. Regardless, conservation of this aspartic acid residue across viruses is striking and worthy of additional investigations.
The two M point mutants (M D62A and M D62A E181A) that exhibited significantly reduced antitranscriptional potential in cell-based studies (Fig. 8) were incorporated into an rWT VHSV backbone, using a reverse genetic system. The primary goal was to determine if the M mutants would support M proteins critical functions in viral replication and, if so, whether antihost functions of the resulting viruses were impacted. Both the M D62A and M D62A E181A viruses were viable, which allowed us to investigate the abilities of these mutant viruses to replicate, elicit cytotoxic effects, inhibit host transcription, and/or otherwise modulate host gene expression. The M D62A E181A virus exhibited reduced replicative capabilities at an MOI of 0.1 compared to those of the WT and D62A viruses (Fig. 9). These data suggested that the D62A E181A mutation, but not the D62A mutation alone, elicited a restrictive effect on propagation, perhaps implicating E181 in viral packaging or budding. For VSV M, F208 is structurally homologous to VHSV M E181 and helps coordinate the positioning of a superiorly located α-helix that forms the border of the hydrophobic pocket that VSV M utilizes in multimerization during viral skeleton formation (63). Thus, loss of electron density at this position might allow for a larger degree of freedom in the superior α-helix, leading to a malformed hydrophobic pocket, deficient polymerization, and thus reduced replication. However, the most striking effects of the D62A and D62A E181A mutants were their reduced abilities to suppress transcriptional responses in EPC cells (Fig. 8, 10, 11, and 13). As such, an alternative explanation for the reduced replication of the double mutant might be that enhanced IFN production in infected cells was capable of restricting propagation so effectively that the virus could not spread far beyond the initially infected cells, particularly if replication was even slightly delayed.
Although the D62A virus fared better than the double mutant in viral-yield assays, its inability to suppress gene expression led to decreased cytopathicity and plaque spread at low MOIs (Fig. 10). Since the D62 residue is highly conserved among rhabdoviruses, we predict that it is structurally or functionally important. Molecular modeling predicted that residue D62 of VHSV M is in a random coil located proximal to the globular domain (64). Mutations within this region impacted the localization of VSV M (65), suggesting that the D62A mutation may have affected the ability of M D62A mutants to localize properly, as opposed to causing a more general loss in structural integrity. Other functions have been ascribed to the orthologous region of VSV M, including regulation of membrane association (66) and protein turnover (59), although both of those studies utilized larger deletions or multiple amino acid mutations than the point mutation used here. Outside this specific coil domain, the N terminus of VSV M has been implicated in other aspects of host inhibition, including association with RAE1/NUP98 (45, 48) or suppression of eukaryotic initiation factor 2α (eIF2α) phosphorylation (M51R) (67). Whether VHSV M also engages these conserved proteins and/or whether D62A mutations impact these same cellular functions, directly or indirectly, will have to await future studies.
Overall, our data provide insight into the various and critical roles of VHSV M in viral replication and host suppression. The findings are consistent with many previous studies of VSV M and confirm a conserved role for M in host suppression among many rhabdoviruses. Our results show that, as with VSV M, the antihost functions of VHSV M can be uncoupled from the viral packaging functions and as such lay the groundwork for studies directed toward developing disabled or attenuated recombinant viruses useful in host response and/or immunization studies, as well as more in-depth functional analyses of the mechanisms behind the observed biological effects of VHSV M and its mutants.
MATERIALS AND METHODS
Cell lines and culture conditions.
Epithelioma papulosum cyprinid (EPC) cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD; CRL-2872). The cells were grown in Eagle's minimum essential medium (EMEM) (Fisher) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) and 1% penicillin-streptomycin (Invitrogen) (complete EMEM) at 22°C in a 5%-CO2-enriched environment. Human embryonic kidney 293 (HEK-293) cells were purchased from the ATCC (CRL-1573) and grown in complete Dulbecco's MEM at 37°C in a 5%-CO2-enriched environment. α-Amanitin (Santa Cruz Biotechnology, Inc., CA) and actinomycin D (Sigma-Aldrich, St. Louis, MO) were used at a final concentration of 1 μg/ml, while leptomycin B (LMB) was used at 10 μg/ml.
Plasmids.
Expression vectors for EPC MAVS and EPC IFN were obtained from Michel Brémont (French National Institute for Agricultural Research, Jouy-en-Josas, France), the VHSV L expression plasmid was reported previously (53), the Tet-mSEAP construct was obtained from Fan Dong (University of Toledo, Toledo, OH, USA), and the IFN-luciferase reporter was obtained from John Hiscott (Istituto Pasteur-Fondazione Cenci Bolognetti, Rome, Italy). Other plasmids used included the IFN-induced transmembrane 1 (IFITM1)/luc plasmid (derived from pDW9-27CD2), SV40/β-galactosidase (β-Gal) (Promega), and SV40/luc (modified from SV40/β-Gal). VHSV-IVb M, NV, G, and N coding sequences were PCR amplified with appropriate primers (Table 1). All fragments were cloned into pcDNA 3.1(−)myc/His A (Invitrogen) or p3xFLAG-CMV-14 (Sigma-Aldrich). IHNV, SVCV, and SHRV M cDNAs were PCR cloned from viral stocks using targeted primers (Table 1).
TABLE 1.
Primers for PCR analysis and cloning
| Primera | Sequence (5′→3′) | Restriction site |
|---|---|---|
| mSEAP se | GACCCTGCTCAGGACCCTC | |
| mSEAP as | GATTTGCCATCCTCAGCCTTG | |
| U6 promoter se | TCTCTATCGATAGGTACCTTTCCCATGATTCCTTCATATTTG | KpnI |
| U6 promoter as | CAGTACCGGAATGCCAAGCTTCGTCCTTTCCACAAGATATATAAAG | HindIII |
| CMV se | CGTTTAGTGAACCGTCAGATCG | |
| CMV as | CCGGTGTCTTCTATGGAGGTCA | |
| VHSv IVb M se | ACGAATTCATGGCTCTATTCAAAAGAAAGCGCACCATCCTG | EcoRI |
| VHSv IVb M as | ACGGTACCCCGGGGTCGGACAGAG | KpnI |
| IVb M mid se | ACAAGCTTCAAGATAGCTGAAGC | HindIII |
| IVb M mid as | ACAAGCTTGTGATCAGGGTTTTG | HindIII |
| VHSvNV se | ACGAATTCATGACGACCCAGTCGGCAC | EcoRI |
| VHSvNV as | ACGGTACCTGGGGGAGATTCGGAGCCA | KpnI |
| VHSvN se | CAGAATTCATGGAAGGAGGAATC | EcoRI |
| VHSvN as | GTGGTACCATCAGAGTCCTCG | KpnI |
| VHSvG se | ACGAATTGATGGAATGGAATACTT | EcoRI |
| VHSvG as | GTGGTACCGACCATCTGGCT | KpnI |
| VHSvP se | CAGAATTCATGACTGATATTGAGAT | EcoRI |
| VHSvP as | GTGGTACCCTCTAACTTGTCCA | KpnI |
| F1 M se | ACGAATTCATGGCTCTGTTCAAAAGAAAGCGCATCATCC | EcoRI |
| F1 M H as | ACAAGCTTGGTACCCCGGGGCCG | HindIII |
| F1 M K as | ACGGTACCCCGGGGCCGGGCAGAGGGGG | KpnI |
| D62A se | TCTCTGTGAAGCTCAACATCCT | |
| D62A as | AGGATGTTGAGCTTCACAGAGA | |
| SVCV M se | CAGAATTCATGTCTACTCTAAGAAAG | EcoRI |
| SVCV M as | CAGGTACCATCTCCCATGAACAGGGA | KpnI |
| SHRV M se | CAGAATTCATGGCAGAATCGATCGAG | EcoRI |
| SHRV M as | CAGGTACCCTTTCTTGAGGACTCGTT | KpnI |
| IHNV M se | ACGAATTCATGTCTATTTTCAAGAGAGC | EcoRI |
| IHNV M as | CTTGGTACCTTTTTCCTTCCCCCGCTTTTCGG | KpnI |
| Virus down se | ACGGATCCAAAACGCAGATCAG | |
| Virus down as | AGGGGTGAGTATACAGTGGAGT | |
| VHS clone se | AAGCTAGCACAAAAAACATGGCTCTATTCA | NheI |
| VHS clone as | TTCAGCTGGTTGTGTACACAAA | PvuII |
| EPC IFN se | TGGGTGGAAAATATCCTGAG | |
| EPC IFN as | CTCCTTATGTGATGGCTGGT | |
| Fish MX-1 se | ATTAACCTGGTTGTGGTGCCATGC | |
| Fish MX-1 as | TACCACTGTCCCTTCAGTGCCTTT | |
| Fish β-actin se | AGACATCAGGGTGTCATGGTTGGT | |
| Fish β-actin as | GGGGTGCTCCTCTGGGGCAA | |
| GFP se | ATGGTGAGCAAGGGCGAGGA | |
| GFP as | TAGCGGCTGAAGCACTGCACGCC |
se, sense; as, antisense.
Luciferase driven by a human U6 promoter reporter construct (pGL3-U6-Luc) was generated by subcloning the human U6 promoter from the pLKO.1 puro vector (Addgene plasmid 10879) into the pGL3 luciferase reporter vector (Promega E1751) upstream of the luciferase gene using Gibson Assembly (NEB E5520). The rainbow trout RNAP I promoter luciferase construct (pGL3-ITS1-Luc) was generated by subcloning rainbow trout rRNA intergenic sequence region 1 (ITS1) into the pGL3 luciferase reporter vector between two HindIII sites.
Recombinant virus production.
Construction of a full-length infectious clone of VHSV has been described earlier (68). This clone was used as a backbone to introduce desired mutations in the M gene, which is flanked by unique NheI and PvuII restriction sites in the full-length clone. To construct plasmid pVHSV-D62A, primers were designed to effect the Asp-to-Ala mutation at amino acid position 62 (D62A) in M and used in PCR along with the flanking NheI forward and PvuII reverse primers to amplify a PCR fragment of 712 bp. The obtained PCR product was subjected to DNA sequencing to confirm the presence of the introduced mutation (D62A). This fragment was doubly digested with NheI and PvuII restriction enzymes and cloned between the NheI-PvuII sites of the full-length VHSV clone. To construct plasmid pVHSV-D62A E181A, another set of primers was used to effect the Glu-to-Ala mutation at position 181 of M. Using the single mutant cDNA fragment as a template, another PCR was carried out with the flanking NheI forward and PvuII reverse primers to obtain a PCR product of 712 bp. DNA of the PCR product was sequenced to confirm the presence of two mutations (D62A E181A). This product was doubly digested with NheI-PvuII and cloned into the full-length VHSV clone, as described above.
To generate recombinant viruses, EPC cells were transfected with plasmids pVHSV-D62A and pVHSV-D62A E181A, along with the support plasmids, using a protocol described previously (69). Briefly, plasmid pVHSV or its derivatives were diluted in 500 μl of Opti-MEM medium. Next, Lipofectamine LTX reagent (Invitrogen) was added according to the manufacturer's instructions and incubated for 30 min at room temperature. The plasmid-Lipofectamine reaction mixture was added to the EPC monolayer in a six-well plate without replacing the growth medium. The transfection mixture was removed after 8 h of incubation at 28°C, and the transfected cells were washed and maintained in Eagle's MEM containing 10% FBS at 14°C for 5 days. The cell monolayer was observed for the development of virus-induced CPE. After 5 days of incubation, the cells were submitted to three cycles of freeze-thawing. The supernatant was clarified at 8,000 × g in a microcentrifuge and used to inoculate fresh cell monolayers in T-25 flasks at 14°C. The supernatant was harvested and clarified for further characterization of the recombinant viruses. To verify that recovered viruses contain the introduced mutations, genomic RNA was extracted from partially purified virus using an RNeasy minikit (Qiagen, Hilden, Germany) and subjected to reverse transcription to obtain cDNA fragments of the VHSV genome, which were sequenced completely to confirm the presence of introduced mutations in the M gene.
VHSV amplification and purification.
The VHSV-IVb MI03GL isolate was kindly provided by James Winton (United States Geological Survey, Seattle, WA). The VHSV F1 strain was kindly provided by Gale Kurath (United States Geological Survey, Seattle, WA). VHSV-IVb isolates and recombinant viruses were amplified for subsequent purification by infecting a confluent monolayer of BF-2 cells in a 15-cm tissue culture dish with a 1:1,000 (vol/vol) dilution of unpurified virus stock in serum-free EMEM. Viral adsorption was allowed to proceed for 1 h before virus-containing medium was replaced with complete EMEM. Virus was cultured until the onset of cytopathicity was observed (72 h). Virus-containing media and attached cells were subjected to a freeze-thaw cycle before the removal of cell debris by low-speed centrifugation (4,000 × g, 30 min). The resulting supernatant was clarified using a 0.22-μm syringe tip filter and then subjected to ultracentrifugation through a 25% (wt/vol) sucrose pad at 25,000 rpm for 3 h at 4°C. The virus-containing pellet was resuspended overnight at 4°C in phosphate-buffered saline (PBS). The titers of virus stocks were determined by 1:10 serial dilution using confluent EPC cells and then divided into 100-μl aliquots and stored at −80°C until use.
Virus yield and IFN bioassays.
To assess viral replication, EPC cells were infected for 0, 4, 18, 36, 54, 72, or 96 h at an MOI of 0.1 or 1.0 with either M D62A, M D62A E181A, or rWT VHSV. Media and cells were harvested at each time point. A viral-yield assay was used to compare the abilities of mutant viruses to replicate to the level of rWT virus by determining the titer of the medium from each time point in 1:10 serial dilutions on BF-2 cells. At 72 h.p.i., plaques were counted and a final viral concentration in numbers of PFU per milliliter was calculated for each time point. Antiviral assays were completed using UV-irradiated medium (70 mJ/cm2) from each time point, which was applied overnight to EPC cells in 1:3 serial dilutions. Cells treated with irradiated medium were subjected to virus challenge using sucrose-purified rWT VHSV for 72 h and then were fixed and stained with crystal violet. Stained wells were dried overnight, and dye was dissolved with 30% (vol/vol) acetic acid for spectrophotometric quantification. Absorbance values of treated wells were normalized to values obtained by untreated and uninfected wells, and 1 unit of IFN was defined as the amount necessary to provide 50% protection from virus CPE.
Transfection.
EPC cell transfections were performed by using PolyJet reagent (SignaGen, Gaithersburg, MD) by following the manufacturer's instructions. Briefly, plasmids were mixed with PolyJet in serum-free EMEM for 20 min and then added to cells in serum-free medium. Media were changed to complete medium after 3 h of incubation. Plasmid concentrations in all transfection experiments were equalized between samples by inclusion of an empty vector.
Click-iT RNA Alexa Fluor 594 imaging and immunofluorescence microscopy.
Click-iT reactions were performed using a Click-iT RNA imaging kit (Invitrogen). For fluorescence microscopy, cells were seeded on poly-l-lysine-coated coverslips in a 6-well culture dish and grown to ∼70% confluence. For flow cytometry, cells were plated identically but without coverslips. Wells were then infected with either rWT, M D62A, or M D62A E181A virus at an MOI of 1.0 for 24 h. At 23 h.p.i., an uninfected well was treated with actinomycin D (1 μg/ml) for 1 h before 5-ethynyl uridine (5-EU) was added to all wells to a final concentration of 1 mM. Cells were allowed to incorporate 5-EU for 1 h before monolayers were washed with PBS and coverslips removed. Cells remaining on the plate were harvested by the addition of Versene (0.02% [wt/vol] EDTA in PBS) and centrifuged at 700 × g for 5 min at 4°C. Cells on coverslips and in Eppendorf tubes were fixed and permeabilized by the addition of fixation buffer (4.0% [wt/vol] paraformaldehyde in Tris-buffered saline with Tween 20, pH 7.5 [TBST]) for 30 min at room temperature. Cells were washed once with 100 mM glycine in PBS to quench paraformaldehyde-induced auto-fluorescence and then once in PBS. Cells were incubated in Click-iT reaction cocktail for 30 min at room temperature in the dark and then washed with Click-iT reaction rinse buffer. For immunofluorescent costaining, cells on coverslips were blocked for 30 min at room temperature (1% bovine serum albumin [BSA] in PBS) and then with primary antibody (in 1% BSA in PBS) for 1 h. Cells were washed in PBS 3 times for 5 min each, and then FITC-conjugated secondary antibody conjugate (in 1% BSA in PBS) was added for 1 h at room temperature. After a PBS wash, the coverslips were mounted to slides with ProLong Gold antifade mountant with DAPI (4′,6-diamidino-2-phenylindole; Life Technologies, Carlsbad, CA) for 24 h and then imaged on an Olympus IX81 inverted fluorescence microscope. For flow cytometric analysis, cells were pelleted and then resuspended in 1% (wt/vol) BSA and incubated at 4°C in the dark until flow cytometry data collection using a BD Scientific LSRFortessa cell analyzer using the allophycocyanin (APC) 650/660-nm excitation/emission filter. Flow data were then analyzed using FlowJo single-cell analysis software v.10 (BD Biosciences, San Jose, CA).
Luciferase assays/β-Gal assays.
Cells were transfected with the appropriate plasmids in 12-well tissue culture plate at a density of ∼70% for 24 or 48 h. After medium removal, cells were washed with PBS twice and then lysed for 15 min on ice in 150 μl of 5× cell culture lysis reagent (diluted to 1× in water) (Promega, Madison, WI). Half of the clarified lysate was used to assess luciferase activity, while the other half was used for β-Gal activity determination using 50 μl of β-Gal buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 2 mM MgSO4, 2.6 μM ortho-nitrophenyl-β-d-galactopyranoside, 3.2 μl β-mercaptoethanol). The mixture was incubated for 1 h at 37°C, and then absorbance was read at 414 nm using a plate reader (SpectraMax; Molecular Devices, Sunnyvale, CA). The luciferase reading was normalized to the β-Gal reading. To obtain a fold induction value, the value of each sample was normalized to the value of the negative control.
Cell fractionation.
Cells transiently transfected with pCD-M were fractionated into nuclear membrane, soluble nuclear, mitochondrial, and cytoplasmic fractions using differential centrifugation. Briefly, three 10-cm plates were transfected with pCD-M. The following day, cells were scraped from the plate and resuspended in 5 volumes of mitochondrial isolation buffer (220 mM mannitol, 68 mM sucrose, 10 mM HEPES, pH 7.4, 10 mM KCl, 1 mM EGTA, 1 mM EDTA, 1 mM MgCl2). Cells were incubated for 5 min on ice and then Dounce homogenized. Homogenate was centrifuged for 10 min at 1,000 × g. The supernatant was further centrifuged for 10 min at 10,000 × g, providing the mitochondrial pellet and cytoplasmic fraction (supernatant). The previous pellet was resuspended with RIPA lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris, pH 8.0, 1 mM phenylmethylsulfonyl [PMS]) and incubated on ice for 15 min. Finally, the lysate was centrifuged at 10,000 × g for 10 min to provide the nucleoplasm and nuclear membrane extracts. Both final pellet fractions were washed three times with the buffer used in the previous step.
Immunoblotting.
Cell lysate was separated by SDS-polyacrylamide gel electrophoresis (PAGE) as previously described (69). Briefly, samples were run on 12.5% gel. Proteins then were transferred to an Immobilon P polyvinylidene difluoride (PVDF) membrane and blocked in 1% BSA in TBST. Membranes were incubated overnight with primary antibody (1:1,000 in 1% BSA in TBST) at 4°C, washed in TBST, and then incubated with secondary antibody conjugated with horseradish peroxidase (HRP) in 1% BSA in TBST (for 1 h at room temperature). Membranes were washed in TBST and incubated with enhanced-chemiluminescence reagent (Pierce, Rockford, IL) for 2 min and then visualized using a ChemiDoc-It2 510 imager (UVP).
Real-time qPCR.
EPC cells transfected with various plasmids or infected with the viruses indicated above were subjected to RNA isolation using TRIzol (Invitrogen) according to the manufacturer's protocol. Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI) was used to reverse transcribe 1 μg of isolated RNA. Reverse transcription reactions were carried out by incubating 1 μg of RNA with 100 ng of random hexamer primer and water to a total volume of 7 μl at 70°C for 10 min. For RT-qPCR studies with recombinant viruses, 1 ng of in vitro-transcribed GFP RNA also was added to the reaction mixture to serve as an internal control for normalization, since virus infection itself shuts down endogenous RNA synthesis. Reaction mixtures were briefly cooled to 4°C before addition of 13 μl of an M-MLV–reverse transcriptase mixture (4 μl of 5× M-MLV–reverse transcriptase reaction buffer, 2 μl of 5 mM deoxynucleoside triphosphates [dNTPs; Invitrogen], 0.5 μl M-MLV–reverse transcriptase [Promega], and water to 13 μl). Samples then were incubated for 1 h at 42°C. cDNA samples were diluted 1:10 with water and then subjected to qPCR using GFP, EPC β-actin, VHSV M, Mx-1, and EPC IFN primers (Table 1). RT-qPCR was performed using 5 μl of 2× Radiant green from a Lo-ROX qPCR kit (Alkali Scientific, Pompano Beach, FL), 1 μl of diluted cDNA, 50 ng of each primer, and water to a total volume of 10 μl. Reactions and data collection were performed with a Bio-Rad C1000 real-time thermocycler for 3 min of initial denaturation at 95°C, followed by 40 cycles of 15 s at 95°C for denaturation and 30 s at 60°C for elongation. Readings were taken at the end of each elongation step. Threshold numbers were obtained by an automated single point threshold within the log-linear range. Samples were normalized to EPC β-actin or GFP, and relative gene expression levels were calculated using the 2−ΔΔCT method, where CT is threshold cycle.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed as previously described with the following modifications (70). Briefly, 107 cells were cross-linked with 1% formaldehyde for 10 min and then quenched with 125 mM glycine for 5 min at room temperature. Nuclei were prepared in cell lysis buffer (5 mM Tris-HCl, pH 8, 85 mM KCl, 0.5% NP-40, 0.5 mM PMSF, 1× protease inhibitor cocktail [PIC; Thermo Scientific]) on ice for 10 min and sonicated to yield chromatin fragments (200 to 700 bp). Immunoprecipitations were performed overnight at 4°C using 1 μg of anti-RNAP IIa A304-405A (Bethyl Laboratories, Montgomery, TX) or IgG and then incubated with protein A-agarose (EMD Millipore, Billerica, MA), which was preequilibrated with sonicated herring sperm DNA and BSA. Immunoprecipitated material was washed extensively, and the cross-links were reversed. DNA from the eluted chromatin was purified with a PCR purification kit by following the manufacturer's instructions (Qiagen). Differences in DNA enrichment for ChIP samples were determined by qPCR using 4% of the precipitated sample DNA and 1% of the input DNA. The primers used for ChIP assay are listed in Table 1.
Statistics.
Data management, analysis, and graphing were done in Microsoft Excel 2016. Analysis was performed by two-tailed, unpaired Student t tests. Graphs represent the statistical means ± standard errors of the means (SEM).
ACKNOWLEDGMENTS
This work benefitted from critical comments provided by anonymous reviewers. We also thank Shelby Powell, Tyler Williams, Tyler Popil, and Jacob Blandford for technical assistance with some of the experiments described herein.
This study was funded by NSF grant DBI-1354806/1354684 (D.W.L, C.A.S., V.N.V.) and USDA/ARS CRIS project number 5090-31320-004-00D under specific cooperative agreement number 58-3655-9-748 (D.W.L., C.A.S.).
The views contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Government. Mention of trade names, proprietary products, or specific equipment does not constitute a guarantee or warranty by the USDA and does not imply its approval to the exclusion of other products that may be suitable.
Footnotes
This is PMEL contribution number 4623.
REFERENCES
- 1.Janeway CA. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symp Quant Biol 54:1–13. doi: 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA Jr, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 3.Loo YM, Gale M Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 5.Lin R, Mamane Y, Hiscott J. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol 19:2465–2474. doi: 10.1128/MCB.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J 17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 9.Stark GR. 2007. How cells respond to interferons revisited: from early history to current complexity. Cytokine Growth Factor Rev 18:419–423. doi: 10.1016/j.cytogfr.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galabov AS. 1981. Induction and characterization of tortoise interferon. Methods Enzymol 78:196–208. doi: 10.1016/0076-6879(81)78118-7. [DOI] [PubMed] [Google Scholar]
- 11.Altmann SM, Mellon MT, Distel DL, Kim CH. 2003. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J Virol 77:1992–2002. doi: 10.1128/JVI.77.3.1992-2002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz U, Kaspers B, Staeheli P. 2004. The interferon system of non-mammalian vertebrates. Dev Comp Immunol 28:499–508. doi: 10.1016/j.dci.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Zou J, Tafalla C, Truckle J, Secombes CJ. 2007. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J Immunol 179:3859–3871. doi: 10.4049/jimmunol.179.6.3859. [DOI] [PubMed] [Google Scholar]
- 14.Sun B, Robertsen B, Wang Z, Liu B. 2009. Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev Comp Immunol 33:547–558. doi: 10.1016/j.dci.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Zou J, Gorgoglione B, Taylor NGH, Summathed T, Lee P-T, Panigrahi A, Genet C, Chen Y-M, Chen T-Y, Ul Hassan M, Mughal SM, Boudinot P, Secombes CJ. 2014. Salmonids have an extraordinary complex type I IFN system: characterization of the IFN locus in rainbow trout Oncorhynchus mykiss reveals two novel IFN subgroups. J Immunol 193:2273–2286. doi: 10.4049/jimmunol.1301796. [DOI] [PubMed] [Google Scholar]
- 16.Chang M, Collet B, Nie P, Lester K, Campbell S, Secombes CJ, Zou J. 2011. Expression and functional characterization of the RIG-I-like receptors MDA5 and LGP2 in rainbow trout (Oncorhynchus mykiss). J Virol 85:8403–8412. doi: 10.1128/JVI.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biacchesi S, LeBerre M, Lamoureux A, Louise Y, Lauret E, Boudinot P, Bremont M. 2009. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol 83:7815–7827. doi: 10.1128/JVI.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verrier ER, Langevin C, Benmansour A, Boudinot P. 2011. Early antiviral response and virus-induced genes in fish. Dev Comp Immunol 35:1204–1214. doi: 10.1016/j.dci.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 19.World Organisation for Animal Health. 2016. Manual of diagnostic tests for aquatic animals. World Organisation for Animal Health, Paris, France. [Google Scholar]
- 20.Einer-Jensen K, Ahrens P, Forsberg R, Lorenzen N. 2004. Evolution of the fish rhabdovirus viral haemorrhagic septicaemia virus. J Gen Virol 85:1167–1179. doi: 10.1099/vir.0.79820-0. [DOI] [PubMed] [Google Scholar]
- 21.Schutze H, Mundt E, Mettenleiter TC. 1999. Complete genomic sequence of viral hemorrhagic septicemia virus, a fish rhabdovirus. Virus Genes 19:59–65. doi: 10.1023/A:1008140707132. [DOI] [PubMed] [Google Scholar]
- 22.Ammayappan A, Vakharia VN. 2009. Molecular characterization of the Great Lakes viral hemorrhagic septicemia virus (VHSV) isolate from USA. Virol J 6:171. doi: 10.1186/1743-422X-6-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snow M, Cunningham CO, Melvin WT, Kurath G. 1999. Analysis of the nucleoprotein gene identifies distinct lineages of viral haemorrhagic septicaemia virus within the European marine environment. Virus Res 63:35–44. doi: 10.1016/S0168-1702(99)00056-8. [DOI] [PubMed] [Google Scholar]
- 24.Pierce LR, Stepien CA. 2012. Evolution and biogeography of an emerging quasispecies: diversity patterns of the fish viral hemorrhagic septicemia virus (VHSv). Mol Phylogenet Evol 63:327–341. doi: 10.1016/j.ympev.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 25.Stone DM, Ferguson HW, Tyson PA, Savage J, Wood G, Dodge MJ, Woolford G, Dixon PF, Feist SW, Way K. 2008. The first report of viral haemorrhagic septicaemia in farmed rainbow trout, Oncorhynchus mykiss (Walbaum), in the United Kingdom. J Fish Dis 31:775–784. doi: 10.1111/j.1365-2761.2008.00951.x. [DOI] [PubMed] [Google Scholar]
- 26.Toplak I, Hostnik P, Rihtaric D, Olesen NJ, Skall HF, Jencic V. 2010. First isolation and genotyping of viruses from recent outbreaks of viral haemorrhagic septicaemia (VHS) in Slovenia. Dis Aquat Organ 92:21–29. doi: 10.3354/dao02251. [DOI] [PubMed] [Google Scholar]
- 27.Elsayed E, Faisal M, Thomas M, Whelan G, Batts W, Winton J. 2006. Isolation of viral haemorrhagic septicaemia virus from muskellunge, Esox masquinongy (Mitchill), in Lake St Clair, Michigan, USA reveals a new sublineage of the North American genotype. J Fish Dis 29:611–619. doi: 10.1111/j.1365-2761.2006.00755.x. [DOI] [PubMed] [Google Scholar]
- 28.Lumsden JS, Morrison B, Yason C, Russell S, Young K, Yazdanpanah A, Huber P, Al-Hussinee L, Stone D, Way K. 2007. Mortality event in freshwater drum Aplodinotus grunniens from Lake Ontario, Canada, associated with viral haemorrhagic septicemia virus, type IV. Dis Aquat Organ 76:99–111. doi: 10.3354/dao076099. [DOI] [PubMed] [Google Scholar]
- 29.Groocock GH, Getchell RG, Wooster GA, Britt KL, Batts WN, Winton JR, Casey RN, Casey JW, Bowser PR. 2007. Detection of viral hemorrhagic septicemia in round gobies in New York State (USA) waters of Lake Ontario and the St. Lawrence River. Dis Aquat Organ 76:187–192. doi: 10.3354/dao076187. [DOI] [PubMed] [Google Scholar]
- 30.Gagne N, Mackinnon AM, Boston L, Souter B, Cook-Versloot M, Griffiths S, Olivier G. 2007. Isolation of viral haemorrhagic septicaemia virus from mummichog, stickleback, striped bass and brown trout in eastern Canada. J Fish Dis 30:213–223. doi: 10.1111/j.1365-2761.2007.00802.x. [DOI] [PubMed] [Google Scholar]
- 31.Thompson TM, Batts WN, Faisal M, Bowser P, Casey JW, Phillips K, Garver KA, Winton J, Kurath G. 2011. Emergence of viral hemorrhagic septicemia virus in the North American Great Lakes region is associated with low viral genetic diversity. Dis Aquat Organ 96:29–43. doi: 10.3354/dao02362. [DOI] [PubMed] [Google Scholar]
- 32.Cornwell ER, Eckerlin GE, Getchell RG, Groocock GH, Thompson TM, Batts WN, Casey RN, Kurath G, Winton JR, Bowser PR. 2011. Detection of viral hemorrhagic septicemia virus by quantitative reverse transcription polymerase chain reaction from two fish species at two sites in Lake Superior. J Aquat Anim Health 23:207–217. doi: 10.1080/08997659.2011.644411. [DOI] [PubMed] [Google Scholar]
- 33.Kim R, Faisal M. 2010. Comparative susceptibility of representative Great Lakes fish species to the North American viral hemorrhagic septicemia virus sublineage IVb. Dis Aquat Organ 91:23–34. doi: 10.3354/dao02217. [DOI] [PubMed] [Google Scholar]
- 34.Kim R, Faisal M. 2011. Emergence and resurgence of the viral hemorrhagic septicemia virus (Novirhabdovirus, Rhabdoviridae, Mononegavirales). J Adv Res 2:9–23. doi: 10.1016/j.jare.2010.05.007. [DOI] [Google Scholar]
- 35.Ammayappan A, Vakharia VN. 2011. Nonvirion protein of novirhabdovirus suppresses apoptosis at the early stage of virus infection. J Virol 85:8393–8402. doi: 10.1128/JVI.00597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi MK, Moon CH, Ko MS, Lee U-H, Cho WJ, Cha SJ, Do JW, Heo GJ, Jeong SG, Hahm YS. 2011. A nuclear localization of the infectious haematopoietic necrosis virus NV protein is necessary for optimal viral growth. PLoS One 6:e22362. doi: 10.1371/journal.pone.0022362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MS, Kim KH. 2013. The role of viral hemorrhagic septicemia virus (VHSV) NV gene in TNF-alpha- and VHSV infection-mediated NF-kappaB activation. Fish Shellfish Immunol 34:1315–1319. doi: 10.1016/j.fsi.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 38.Chiou PP, Kim CH, Ormonde P, Leong JA. 2000. Infectious hematopoietic necrosis virus matrix protein inhibits host-directed gene expression and induces morphological changes of apoptosis in cell cultures. J Virol 74:7619–7627. doi: 10.1128/JVI.74.16.7619-7627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stepien CA, Pierce LR, Leaman DW, Niner MD, Shepherd BS. 2015. Gene diversification of an emerging pathogen: a decade of mutation in a novel fish viral hemorrhagic septicemia (VHS) substrain since its first appearance in the Laurentian Great Lakes. PLoS One 10:e0135146. doi: 10.1371/journal.pone.0135146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan H, Yoza BK, Lyles DS. 1998. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology 251:383–392. doi: 10.1006/viro.1998.9413. [DOI] [PubMed] [Google Scholar]
- 41.Tornoe CW, Christensen C, Meldal M. 2002. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper (i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem 67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 42.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. 2002. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl 41:2596–2599. doi:. [DOI] [PubMed] [Google Scholar]
- 43.Dixon P, Feist S, Kehoe E, Parry L, Stone D, Way K. 1997. Isolation of viral haemorrhagic septicaemia virus from Atlantic herring Clupea harengus from the English Channel. Dis Aquat Organ 30:81–89. doi: 10.3354/dao030081. [DOI] [Google Scholar]
- 44.Ahmed M, Lyles DS. 1998. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J Virol 72:8413–8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell 17:93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 46.Her LS, Lund E, Dahlberg JE. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 47.Petersen JM, Her LS, Varvel V, Lund E, Dahlberg JE. 2000. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol Cell Biol 20:8590–8601. doi: 10.1128/MCB.20.22.8590-8601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Kobbe C, van Deursen JM, Rodrigues JP, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo-Fonseca M, Izaurralde E. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell 6:1243–1252. doi: 10.1016/S1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 49.Corden JL. 1990. Tails of RNA polymerase II. Trends Biochem Sci 15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 50.O'Brien T, Hardin S, Greenleaf A, Lis JT. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 51.Bensaude O, Bonnet F, Casse C, Dubois MF, Nguyen VT, Palancade B. 1999. Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD). Biochem Cell Biol 77:249–255. doi: 10.1139/o99-047. [DOI] [PubMed] [Google Scholar]
- 52.Komarnitsky P, Cho EJ, Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ammayappan A, Kurath G, Thompson TM, Vakharia VN. 2011. A reverse genetics system for the Great Lakes strain of viral hemorrhagic septicemia virus: the NV gene is required for pathogenicity. Mar Biotechnol (NY) 13:672–683. doi: 10.1007/s10126-010-9329-4. [DOI] [PubMed] [Google Scholar]
- 54.Holland JJ, Peterson JA. 1964. Nucleic acid and protein synthesis during poliovirus infection of human cells. J Mol Biol 8:556–575. doi: 10.1016/S0022-2836(64)80011-5. [DOI] [PubMed] [Google Scholar]
- 55.Wertz GW, Youngner JS. 1970. Interferon production and inhibition of host synthesis in cells infected with vesicular stomatitis virus. J Virol 6:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferran MC, Lucas-Lenard JM. 1997. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol 71:371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmed M, McKenzie MO, Puckett S, Hojnacki M, Poliquin L, Lyles DS. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J Virol 77:4646–4657. doi: 10.1128/JVI.77.8.4646-4657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clinton GM, Little SP, Hagen FS, Huang AS. 1978. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell 15:1455–1462. doi: 10.1016/0092-8674(78)90069-7. [DOI] [PubMed] [Google Scholar]
- 59.Dancho B, McKenzie MO, Connor JH, Lyles DS. 2009. Vesicular stomatitis virus matrix protein mutations that affect association with host membranes and viral nucleocapsids. J Biol Chem 284:4500–4509. doi: 10.1074/jbc.M808136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mebatsion T, Weiland F, Conzelmann KK. 1999. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol 73:242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Black BL, Rhodes RB, McKenzie M, Lyles DS. 1993. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J Virol 67:4814–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaudier M, Gaudin Y, Knossow M. 2002. Crystal structure of vesicular stomatitis virus matrix protein. EMBO J 21:2886–2892. doi: 10.1093/emboj/cdf284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graham SC, Assenberg R, Delmas O, Verma A, Gholami A, Talbi C, Owens RJ, Stuart DL, Grimes JM, Bourhy H. 2008. Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathog 4:e1000251. doi: 10.1371/journal.ppat.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glodowski DR, Petersen JM, Dahlberg JE. 2002. Complex nuclear localization signals in the matrix protein of vesicular stomatitis virus. J Biol Chem 277:46864–46870. doi: 10.1074/jbc.M208576200. [DOI] [PubMed] [Google Scholar]
- 66.Ogden JR, Pal R, Wagner RR. 1986. Mapping regions of the matrix protein of vesicular stomatitis virus which bind to ribonucleocapsids, liposomes and monoclonal antibodies. J Virol 58:860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Connor JH, Lyles DS. 2005. Inhibition of host and viral translation during vesicular stomatitis virus infection. J Biol Chem 280:13512–13519. doi: 10.1074/jbc.M501156200. [DOI] [PubMed] [Google Scholar]
- 68.Ammayappan A, Lapatra SE, Vakharia VN. 2010. A vaccinia-virus-free reverse genetics system for infectious hematopoietic necrosis virus. J Virol Methods 167:132–139. doi: 10.1016/j.jviromet.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 69.Lai SH, Jiang GB, Yao JH, Li W, Han BJ, Zhang C, Zeng CC, Liu YJ. 2015. Cytotoxic activity, DNA damage, cellular uptake, apoptosis and Western blot analysis of ruthenium(II) polypyridyl complex against human lung decarcinoma A549 cell. J Inorg Biochem 152:1–9. doi: 10.1016/j.jinorgbio.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 70.Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci U S A 95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]