CryoEM structure of MxB tubes at 4.6 Å resolution reveals novel interfaces responsible for assembly and anti–HIV-1 activity.
Abstract
Human dynamin–like, interferon-induced myxovirus resistance 2 (Mx2 or MxB) is a potent HIV-1 inhibitor. Antiviral activity requires both the amino-terminal region of MxB and protein oligomerization, each of which has eluded structural determination due to difficulties in protein preparation. We report that maltose binding protein–fused, full-length wild-type MxB purifies as oligomers and further self-assembles into helical arrays in physiological salt. Guanosine triphosphate (GTP), but not guanosine diphosphate, binding results in array disassembly, whereas subsequent GTP hydrolysis allows its reformation. Using cryo-electron microscopy (cryoEM), we determined the MxB assembly structure at 4.6 Å resolution, representing the first near-atomic resolution structure in the mammalian dynamin superfamily. The structure revealed previously described and novel MxB assembly interfaces. Mutational analyses demonstrated a critical role for one of the novel interfaces in HIV-1 restriction.
INTRODUCTION
Myxovirus resistance (Mx) proteins are important restriction factors in the interferon response against viruses (1). There are two isoforms in mammals, grouped into Mx1 (MxA)–like and Mx2 (MxB)–like proteins, based on evolutionary analysis (2). Although human MxA has been well documented to restrict a broad range of viruses, including influenza, vesicular stomatitis, and Thogoto (3), the antiviral activity of human MxB has only been recently discovered, decades after its initial identification (4) and characterization (5). Studies have shown strong inhibition of HIV-1 replication by ectopic (6–8) or endogenous expression (8) of MxB. MxB primarily targets the viral core after cell entry and after reverse transcription (9). In certain cell types, this interaction is dependent on the HIV-1 capsid protein (CA) host factor cyclophilin A (8, 10). MxB also blocks nuclear import of preintegration complexes and proviral integration (6, 11). Several primate MxB proteins have since been reported to show species-dependent variation in inhibiting the infection of lentiviruses (13) and transmitted/founder virus strains (10) have also been found to escape inhibition by MxB, suggesting an active selective pressure on HIV-1 evolution.
Mx proteins belong to the dynamin superfamily of large guanosine triphosphatases (GTPases) (fig. S1), which share a core structure comprising a GTPase, a bundle signaling element (BSE), and a stalk domain (1). The structural homology among these proteins is exemplified in the crystal structures of human MxA (14) and human MxB lacking its N-terminal region (NTR) (Δ1–83) (15), where individual domains are practically superimposable (root mean square deviation of 0.8 to 1.1 Å) (15). Dynamin family proteins share common properties of self-assembling into ordered helical arrays and exhibiting guanosine triphosphate (GTP)–dependent assembly or disassembly (16). Although these properties, along with the L4 loop that confers antiviral specificity (17), were found to be important for MxA function, the L4 loop (11, 18) and GTPase activity (6, 7, 11) appear to be dispensable for MxB anti–HIV-1 activity. Instead, the antiviral activity of MxB requires its capsid-binding NTR (6) and the ability to oligomerize (18, 19). However, no structural information is available for either the protein oligomers or the NTR, owing to difficulties with protein preparation of wild-type MxB. Current knowledge of protein oligomerization and assembly of the dynamin family is largely based on the cryo-electron microscopy (cryoEM) structure of dynamin–ΔPRD (deleted proline-rich domain) tubes at 12 Å resolution (20). Therefore, we sought to obtain essential structural information of MxB oligomers and to dissect the specific interfaces responsible for the protein’s anti–HIV-1 activity.
RESULTS AND DISCUSSION
Purification of full-length wild-type MxB
To obtain full-length wild-type MxB, we expressed and purified an N-terminal maltose binding protein (MBP) fusion protein from mammalian cells. MBP-MxB is purified as oligomers when eluted from a Sephacryl S-500 HR gel filtration column following amylose affinity chromatography (Fig. 1A) such that a gradient of different oligomeric species was observed (Fig. 1, B to E). Single-particle analysis and two-dimensional (2D) classification of the negatively stained EM images from the main peak fraction (Fig. 1C) revealed that these MxB oligomers have various extents of packing (Fig. 1F), indicating that the sample was too heterogeneous for further structural analysis. Like other dynamin family members (16), MxB spontaneously assembled into highly ordered long helical tubes at 150 mM NaCl (Fig. 2A), even at low protein concentrations (0.05 mg/ml; 0.4 μM). Immunogold labeling localized the MBP fusion tag to the outer surface of the tube (Fig. 2B), suggesting that the MxB NTR is oriented toward its outer circumference. The helical assembly was not induced by the MBP tag because removal of the tag did not affect tube formation and instead induced tube bundling/aggregation (fig. S2).
Fig. 1. Full-length wild-type MxB purified as oligomers.
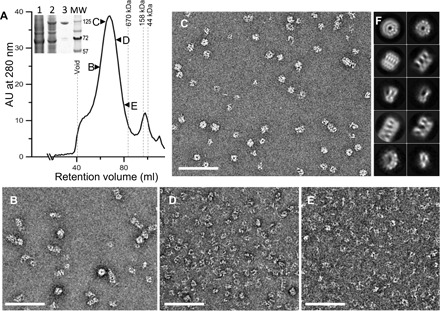
(A) Purification of MBP-MxB from Expi293F cells by amylose affinity chromatography, followed by gel filtration through a Sephacryl S-500 HR column. The fractions indicated by arrowheads were visualized by negative stain EM. Inset, Coomassie-stained SDS–polyacrylamide gel electrophoresis (PAGE) gel of untransfected cells (1), transfected cells (2), and elution from amylose resin (3). Molecular weight (MW) markers are shown in kilodaltons (kDa). AU, absorbance units. (B to E) Representative negative stain micrographs of the respective fractions indicated in (A). (F) 2D class averages of the negatively stained MxB sample from fraction “C.” Scale bars, 200 nm.
Fig. 2. GTP binding induces depolymerization of the helical MxB assembly.
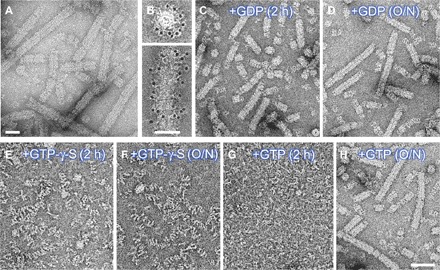
(A) MBP-MxB self-assembled into helical structures at 150 mM NaCl. (B) Gold labeling of MBP shows that MBP is located on the surface of assembled MBP-MxB tubes. End-on view (top) and side view (bottom) are shown. (C to H) Representative micrographs of MBP-MxB in the presence of 1 mM GDP (C and D), GTP-γ-S (E and F), and GTP (G and H), incubated at room temperature for 2 hours (C, E, and G) or overnight (O/N) (D, F, and H). Scale bars, 50 nm (A and B) and 100 nm (H). Panels (C) to (H) are of the same scale.
Effect of GTP binding on MxB helical assembly
MxB was previously shown to have GTPase activity in immunoprecipitates (5). We determined that purified MxB hydrolyzes GTP with the GTPase activity (kobs) comparable to the basal GTPase hydrolysis rates of other dynamin-like proteins (fig. S3) (16). The addition of GTP (Fig. 2G and fig. S4B) or nonhydrolyzable GTP analogs, such as guanosine 5′-O-(3′-thiotriphosphate) (GTP-γ-S) or guanosine-5′-[(β,γ)-methylene] triphosphate (GMP-PCP) (Fig. 2, E and F), to the MxB tubes completely disrupted them, whereas the addition of guanosine diphosphate (GDP) (Fig. 2, C and D) or GTP without MgCl2 (fig. S4A) had no effect. Overnight treatment with GTP (Fig. 2H), but not with GMP-PCP or GTP-γ-S (Fig. 2F), resulted in MxB tube reassembly. These results suggest that GTP binding, but not hydrolysis, is sufficient to exert conformational changes that disrupt the MxB helical assembly, wherein upon hydrolysis, MxB reverts back to the assembly-competent conformation. It should be noted that although GTP-γ-S or GMP-PCP binding disassembles the MxB tubes or depolymerizes the MxA rings (21), binding of these GTP analogs, on the contrary, promotes helical assembly of other dynamin family members such as human dynamin 1 (22), yeast Dnm1 (23), and human Drp1 (24).
CryoEM structure of the MxB assembly
The assembled MxB tubes were highly ordered with an inner and outer diameter of 55 and 275 Å, respectively (Fig. 3A and fig. S5). There was an undifferentiated density out to 360 Å, as shown in one of the 2D classes (Fig. 3A, between white and blue dashed lines), presumably corresponding to the MxB NTR and MBP tag. The Fourier transforms of the MxB tubes indicated that they belong to a one-start helical family of (−6, 1) (fig. S5), a right-handed helix (fig. S6) with a rise of 8.237 Å and a twist of 58.4°. Using cryoEM and real-space helical reconstruction (25), we determined the 3D density map of the MxB helical assembly at 4.6 Å resolution (Fig. 3B and fig. S7). The local resolution of the density map varies (Fig. 3B); α-helical turns (fig. S8, D to G) and some bulky side-chain densities (fig. S8, D and G) are resolved at the inner core (stalk and BSE), whereas the GTPase domain appears to be more flexible. The NTR, together with the MBP tag, is not resolved, probably because of their flexibility. Initial rigid-body docking of individual domains from the crystal structure of the NTR-truncated MxB dimer [Protein Data Bank (PDB) ID: 4WHJ] (15) resulted in a reasonable overall fit, although it revealed substantial deviations, particularly at the first stalk helix Sα1 and unaccounted extra helical density (fig. S8A, arrows). This helical region is part of the L4 loop, which conveys antiviral specificity for MxA-like proteins (17). It was previously thought to be completely unstructured and was not observed in either the MxA (26) or MxB (15) crystal structure.
Fig. 3. CryoEM structure of the MxB helical assembly.
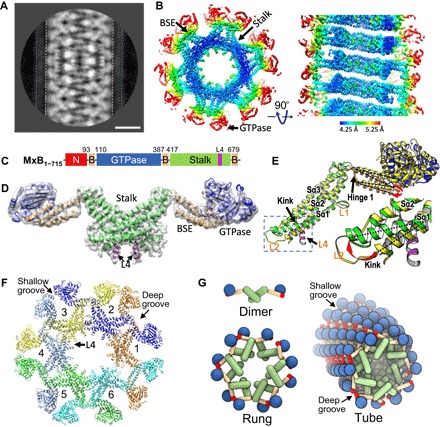
(A) 2D class average of MBP-MxB helical segments from cryoEM images. Dashed white and blue lines indicate the outer surfaces of the MxB density and MBP density, respectively. Scale bar, 10 nm. (B) CryoEM reconstruction of an MxB tube shown in surface rendering, contoured at 4.5σ, and viewed parallel (left) and perpendicular (right) to the tube axis. Color indicates the local resolution of the density map ranging from 4.25 Å (blue) to 5.25 Å (red). The locations of the domains are indicated. (C) Domain structure of MxB. (D) Density map (contoured at 3σ) of an MxB dimeric assembly unit overlaid with a real-space refinement model of the MxB dimer. Domains are color-coded as in (C). Arrows point to the new L4 helix (purple). (E) Superposition of the cryoEM model and the crystal structure of MxB monomer (PDB ID: 4WHJ) (yellow). Arrows indicate the differences between the two structures: hinge 1, kinked extended stalk helix Sα1 and the new helix in L4. Inset, enlarged view of the boxed region. Dashed lines and red arrows show the change in helix orientation. (F) Atomic model of the MxB assembly in one rung, comprising six MxB dimers (1 to 6), each uniquely colored. Surface grooves are marked. (G) Schematic of the MxB helical assembly, hierarchically through three levels: a dimer, a rung, and a tube.
We modeled the L4 helix de novo using Rosetta into the MxB dimer structure and carried out molecular dynamics flexible fitting (MDFF), followed by real-space refinement to obtain an all-atom cryoEM structure model of the MxB assembly (Fig. 3, D and F, fig. S8, and movie S1). The resulting model displays a good match to the experimental density (Fig. 3D and fig. S8, C to G). In the cryoEM structure, the entire MxB assembly is made up of MxB dimer units. Six dimers go hand in hand, interlocking with each other through the stalk and BSE domains to form one rung, where the sixth dimer comes around to interact with the first dimer, forming the one-start right-handed helix (Fig. 3, F and G, and movie S2). The tube surface displays a shallow groove, where the GTPase domains cluster, and a deep groove, where the NTRs are presumably located (Fig. 3, F and G). The MxB dimer in the assembly is substantially different from the crystal dimer (PDB ID: 4WHJ), displaying (i) a highly kinked and extended stalk Sα1 helix C terminus (Sα1c), which effectively displaces the base of Sα1c by 27°, (ii) a shift in the domain orientations about hinge 1 between the BSE and the stalk by 22°, and (iii) a well-ordered L4 loop helix (Fig. 3E). As a result, these new features led to the formation of completely new assembly interfaces, which are distinct from those previously inferred from crystal contacts (table S1) (discussed in detail below). There is no swapped dimer present, as previously thought for the dynamin tube (20).
Novel MxB assembly interfaces
The cryoEM structure revealed three levels of assembly interfaces that are likely shared by members of the dynamin superfamily of GTPases: dimer interface [interface 2; following the MxA convention (26)], oligomer interfaces (interfaces 1 and 3), and helical (higher-order) assembly interface (interface 4) (Figs. 3G and 4, A and B). The dimer interface (interface 2) is essentially the same, as seen in the MxB crystal structure (fig. S9A). However, the other three interfaces are novel in the cryoEM structure, as described in detail below.
Fig. 4. MxB intermolecular assembly interfaces and their role in the MxB assembly and HIV-1 inhibition.
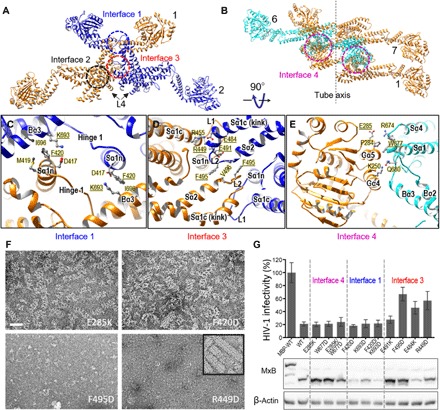
(A) Intermolecular interfaces in an MxB oligomer: the canonical MxB dimer interface, interface 2 (black circle), and the lateral interfaces that link adjacent MxB dimers, interfaces 1 (blue circle) and 3 (red circle). The same color scheme is used as in Fig. 3F. (B) Interface 4 (magenta circles), the vertical interface between adjacent rungs. MxB dimer 7 starts the next rung in the helix, colored the same as dimer 1. (C to E) Expanded views of the intermolecular interfaces: interface 1 (C), interface 3 (D), and interface 4 (E). Specific residues at the interfaces are labeled, along with the secondary structures. Underlined amino acids were subjected to mutational analysis. (F) Effects of interface mutations on the MxB assembly: E285K (interface 4), F420D (interface 1), F495D, and R449D (interface 3). Negatively stained images of purified interface mutant proteins under helical assembly conditions are shown. Inset, wild-type MxB. Scale bar, 50 nm. (G) Effects of interface mutations on MxB anti–HIV-1 activity (top) (mean ± SD for minimally n = 3 independent experiments) and Western blot analysis of MxB protein expression (bottom). MBP-tagged wild-type (WT) MxB, which does not inhibit HIV-1, was used as a control.
Interface 1 is formed by the symmetric interaction of the tip of the stalk domain (Sα1n) of one dimer and the BSE domain (Bα3) from another, mediated by a salt bridge (D417-K693) and hydrophobic contacts (F420, M419, and I696) (Fig. 4C and fig. S9B). This interface is markedly different from the putative interface 1 (fig. S10A and table S1) gleaned from the crystallographic symmetries of MxA and MxB structures, which shows stacking of the stalk domains (Sα1n and Sα4) of the dimers to form a linear array (14, 15).
Interface 3 is completely novel and composed of the new extended base of kinked Sα1c and its connecting loops at the either end of Sα1c, L1 and L2 (Fig. 4D and fig. S9C). Conserved hydrophobic L2 residues (F495 and V496) from adjacent dimers form a symmetric interaction (Fig. 4D, blue and orange, and fig. S9C). Additional salt bridges, R449-E491 between L1 and L2 from neighboring dimers as well as R455-E484 between L1 and the base of Sα1c, further stabilize the interface (Fig. 4D and fig. S9C). Because of the kinking of Sα1c and consequential positioning of its base and L2 (Fig. 3E and movie S3), these interactions are absent from the previous crystal structures (fig. S10B). However, a kink in Sα1c was observed in a crystal structure of tetrameric human dynamin 3 (fig. S11C, black arrow) where interface contacts in L2 similar to MxB were also observed (fig. S11E) (27), further emphasizing the critical role of this new interface in MxB oligomerization (movie S4).
Interface 4 is formed between the GTPase domain of dimer 1 and the stalk–hinge 1 region of dimer 6, as it completes a full rung (Fig. 4, B and E). Our structure lacks evidence for GTPase domain dimerization, as proposed previously for dynamin helical assembly (20), supporting stalk-driven oligomerization. The GTP hydrolysis–deficient MxB mutant, T151A, was shown to retain the ability to inhibit HIV-1 (6, 11) without affecting GTP binding (28). The T151A alteration does not affect the MxB helical assembly (fig. S12A); however, unlike wild-type MxB, GTP or GTP-γ-S binding has a marginal effect on MxB T151A assemblies (fig. S12). The residues involved in interface 4 include P284, E285, and K250 from the GTPase domain of dimer 1 and R674, W677, and Q680 from the stalk–hinge 1 region of dimer 6, mediating charge interactions and hydrophobic contacts (Fig. 4E and fig. S9D). This is the only assembly interface involving the GTPase domain and is the interface responsible for helical assembly by stabilizing rung stacking (Figs. 3G and 4B). Thus, it explains our observations that the MxB tubes are disrupted into oligomers upon GTP binding (Fig. 2, E and G). Low-salt conditions, which promote helical assembly of MxB (Fig. 2A) and other dynamin family members (29), likely facilitate hydrophobic interactions at interface 4.
Functional importance of MxB interface 3 in assembly and antiviral activity
With detailed knowledge of all interfaces responsible for assembly, we tested which of these interfaces is relevant to the MxB function. Because the dimer interface was previously well characterized for its important role in MxB antiviral function (15, 18, 19), we focused on the three new assembly interfaces: interfaces 1, 3, and 4. Representative residues at these interfaces were substituted to evaluate their effects on the ability of MxB to form oligomers or assemble into tubes and to restrict HIV-1 in cultured cells. Mutations across these interfaces—F420D in interface 1, F495D and R449D in interface 3, and E285K in interface 4—diminished MxB tube formation (Fig. 4F). Although interface 1 and 4 mutants retained the ability to form oligomers greater than a dimer, only interface 3 mutants (F495D and R449D) failed to oligomerize (Fig. 4F). Coincidentally, only interface 3 mutations (F495D, R449D, and E484K) appreciably affected the anti–HIV-1 activity of MxB (Fig. 4G). Substitution of R455, which forms a salt bridge with E484 in our structure (Fig. 4D and fig. S9C), was previously found to disrupt HIV-1 restriction by MxB (table S1) (11, 18). Correlating the biochemical results with infection data strongly suggests that nonhelical MxB oligomers greater than a dimer are the active anti–HIV-1 species and that the new interface 3 is critical for MxB oligomerization and antiviral function. Given the prevailing GTP concentrations in the cell (~0.47 mM) (30), MxB is likely GTP-bound and not helically assembled. Although the structure of the MxB NTR remains elusive, our near-atomic resolution cryoEM structure of assembled, wild-type MxB allows for hypothetical models of possible MxB-capsid interactions, wherein an MxB tetramer could potentially recognize the mature capsid lattice, thus precluding the need for GTPase activity and interface 4 for MxB anti–HIV-1 activity (31, 32). Further, the high-resolution structure of a member of dynamin superfamily and the detailed molecular interactions responsible for GTP-dependent assembly or disassembly provide a structural framework for the function of other dynamin family GTPases. Dynamins generally catalyze GTP hydrolysis through the dimerization of GTPase domains (16), and in the context of helical assembly, this dimerization interface (GG interface) is thought to form between rungs (33). The GG interface is completely absent in the MxB helical assembly, and the conformation of the BSE domains of the helical MxB relative to the GTPase domain may represent the “closed” conformation, similar to that observed in the GDP-bound or apo crystal structures of dynamin proteins (fig. S13) (16). We speculate that the trans-dimerization of the GTPase domains upon GTP binding of MxB is sensed by the BSE domains, causing them to transition to the open conformation, which effectively disrupts the helical assemblies.
MATERIALS AND METHODS
Plasmid construction
Cloning vectors containing the gene for the full-length wild-type human MxB (UniProt ID: P20592-1) and the MBP tag were gifts from J. Ahn (Pittsburgh Center for HIV Protein Interactions, University of Pittsburgh School of Medicine). The expression vector pcDNA3.1(+) was obtained from Life Technologies (Invitrogen). The MxB gene and the MBP tag were amplified by polymerase chain reaction (PCR) and then subcloned, using the NEBuilder HiFi Assembly kit (New England Biolabs Inc.), into pcDNA3.1(+) that had been linearized by the restriction enzymes Eco RV and Xba I. The resulting insert, designated as MBP-MxB-H6, has a leading Kozak sequence, an N-terminal MBP tag, followed by a human rhinovirus 3C protease cut site, the full-length wild-type MxB, and a C-terminal hexahistidine tag (H6). The plasmid used for the infectivity assay was generated by transferring the PCR construct for MBP-MxB-H6 or MxB-hemagglutinin from pcDNA3.1(+) to pIRES2–enhanced green fluorescent protein (eGFP) (34).The sequences of the inserts were confirmed by DNA sequencing (Genewiz Inc.).
Sequence alignment
Amino acid sequences of Mx and dynamin proteins were obtained from UniProt using the indicated IDs and were then aligned using MUSCLE (35). The alignment was visualized using ESPript3 (36).
Expression and purification of MxB
Recombinant MxB was transiently expressed in mammalian cells using the Expi293 Expression kit from Life Technologies (Invitrogen). Suspension-adapted Expi293F cells were grown in Expi293 Expression Medium to a density of 3.5 × 106 to 4 × 106 cells/ml and a viability of >95% 24 hours before transfection. Plasmid DNA and ExpiFectamine reagent (Invitrogen) were diluted in Opti-MEM I Reduced Serum (Invitrogen) into separate tubes, incubated for 5 min at room temperature, and then mixed together for 25 min. Cells were transfected with the DNA-ExpiFectamine complex at a DNA/transfection reagent/cell culture volume ratio of 30 μg/1.5 ml/30 ml and to a final cell density of 2.9 × 106 cells/ml. Cells were then incubated at 37°C and 125-rpm agitation with 8% CO2 in air. After 18 hours of incubation, 150 μl of transfection enhancer 1 and 1.5 ml of transfection enhancer 2 for every 30 ml of cells were added into the suspension. Twenty-four hours after the addition of the enhancers, cells were harvested by centrifugation at low speed (100g). Cells were washed once with cold phosphate-buffered saline, and the cell pellet was flash-frozen and stored at −80°C for later use.
The thawed cell pellet was resuspended in buffer A [50 mM Hepes-KOH (pH 8), 250 mM NaCl, and 5% glycerol] supplemented with detergents (1% Tween 20 and 0.3% NP-40), deoxyribonuclease I (50 μg/ml; Sigma-Aldrich) in the presence of 5 mM MgCl2, 10 mM β-mercaptoethanol, and a cocktail of protease inhibitors (Roche). After 1 hour of rotation at 4°C, the lysate was homogenized by 15 strokes in an ice-cold, tight-fitting Dounce homogenizer. The homogenate was then centrifuged at 21,000g at 4°C for 30 min. After centrifugation, the supernatant was collected and mixed with 1 ml of amylose agarose resin (New England Biolabs Inc.) (per 50 ml of cell suspension) pre-equilibrated with buffer A. The mixture was incubated with rotation at 4°C for 1 hour and then transferred to a column to flow through. The resin was washed with 50× resin volume of buffer A. To elute the recombinant protein, the resin was incubated, in batch, with buffer A containing 50 mM maltose for 15 min at 4°C, and then, the flow through was collected as elution. The purified protein was detected by Western blot using antibodies against the MBP tag (Abcam) and MxB (N-17) (Santa Cruz Biotechnology) and HisProbe–horseradish peroxidase (HRP) (Thermo Fisher Scientific) for the hexahistidine tag.
GTPase assay
The GTPase function of MxB was assessed using a continuous NADH (reduced form of nicotinamide adenine dinucleotide)–coupled assay (37). The reaction mixture was prepared to achieve the following final concentrations in the assay solution: 50 mM Hepes-KOH (pH 7), 150 mM NaCl, 10 mM MgCl2, 2 mM dithiothreitol (DTT), 4 mM phosphenolpyruvate, 0.35 mM NADH, 25 U of pyruvate kinase/lactate dehydrogenase, and 1 mM GTP. MxB was added just before measurement. The decrease in the NADH absorbance at 340 nm was monitored in a 96-well plate using a Synergy H1 Hybrid Reader (BioTek) at 37°C. The rate of NADH oxidation was measured and used to calculate the kobs of MxB. NADH oxidation was also monitored in the absence of protein using buffer only as control. The experimental values were normalized to correct for background. Results are representative of three independent measurements.
Mass spectrometry analysis
MxB was digested overnight at 37°C with sequencing grade trypsin in the presence of 1 M urea. Digested protein (100 pmol) was captured on a Phenomenex Aeris C18 column (2.1 × 50 mm) (Phenomenex) and eluted with a 0 to 65% gradient of acetonitrile at 400 μl/min. Lock mass–corrected mass spectra were acquired in MSE (all-ion) mode using a Waters Q-ToF Premier, and peptides were identified using ProteinLynx Global Server.
Electron microscopy
Sample preparation
To prepare samples for initial screening by negative stain, the elution from the amylose resin was immediately filtered through a HiPrep Sephacryl S-500 HR (GE Healthcare) in buffer A with an additional 2 mM DTT. To induce the formation of the long tubes, the elution from the amylose resin was diluted in assembly buffer [20 mM Hepes-KOH (pH 7), 150 mM NaCl, 1 mM MgCl2, 2 mM EGTA, and 2 mM DTT] at 0.5 to 1 mg/ml, and then incubated for the indicated period of time.
Negative stain EM
Aliquots (3 μl) from the gel filtration samples or the helical assembly were adsorbed to a glow-discharged, 400-mesh, carbon-coated copper grid and stained with fresh uranyl formate (2%). Images were recorded on a TF20 electron microscope (FEI) equipped with a field-emission gun at the indicated magnification on a 4k × 4k Gatan UltraScan charge-coupled device camera (Gatan).
Immunogold labeling
Immunogold labeling, modified from the study by Mears et al. (38), was performed to determine the orientation of the NTR of MxB in its helical form. Samples containing the MxB tubes were prepared and applied to a grid, as described above. The grid was successively floated on the following solutions: (i) twice with blocking buffer [bovine serum albumin (BSA;10 mg/ml) in oligomerization buffer] for 5 min, (ii) with blocking buffer containing primary antibody against MBP tag (Abcam) (1:250 dilution) for 1 hour, (iii) twice with blocking buffer for 5 min, and (iv) with blocking buffer containing a 5-mm gold-labeled secondary antibody (Ted Pella) (1:250 dilution) for 1 hour. All incubations were carried out at 4°C, and the grid was washed once with blocking buffer and twice with oligomerization buffer before staining with uranyl formate.
CryoEM
Three microliters of the MxB tubes (0.5 mg/ml) was applied on the carbon side of glow-discharged holey R2/1 Quantifoil grids (Quantifoil Micro Tools GmbH), manually blotted from the backside, and then plunge-frozen in liquid ethane using a homemade manual plunger. Images were collected under low-dose conditions (~40 e−/Å2 total) using a Polara 300-kV microscope with a field-emission gun and an FEI Falcon II detector. Movies (~1000, each with seven frames) were manually collected using SerialEM (39) at a nominal magnification of ×98,000 (1.147 Å/pixel), with under-focus values ranging from 1.5 to 3.5 μm.
Image processing and helical reconstruction
Movie frames were aligned using UCSF (University of California, San Francisco) MotionCorr v2.1 (40), and the resulting motion-corrected sums were used for contrast transfer function estimation using Gctf v0.50 (41). Micrographs were then sorted on the basis of image quality, and ~630 micrographs were used for subsequent helical reconstruction using RELION 2.0 beta (25), which is a software package in development that integrates a helical processing workflow. Helical segments were boxed using EMAN 2.0 helixboxer.py (42), and diffraction patterns from individual tubes or 2D class averages generated using Spring (43) were used to estimate the helical parameters. A small data set with 3486 segments (500-pixel box size) was used in the helical processing workflow in RELION 2.0 beta to refine the helical parameters, which converged to a rotation angle of 58.4° and a rise of 8.25 Å. Using the previously generated 2D classes as templates, helical filaments were automatically picked from the full data set, resulting in 51,553 segments (450-pixel box size, 90.41% overlap between neighboring boxes, and interbox distance six times the helical rise). The segments were then analyzed by 2D classification, and 64 of 100 classes showing clear structural details were then selected for further processing (44,955 segments correspond to ~270,000 subunits). The first round of refinement, using a featureless cylinder (236 pixels in diameter and low-pass–filtered to 30 Å), resulted in a map with a resolution of 6.9 Å. For the next round of refinement, the previous reconstruction was used as an initial reference and converged to a map with 6.6 Å overall resolution, which was improved to 5.0 Å after postprocessing (sharpening and application of soft-edged mask). A final round of refinement was performed using polished particles, which gave a map with 5.3 Å resolution. Postprocessing by soft-edge masking and B-factor sharpening resulted in a map with 4.6 Å resolution. The final map was then low-pass–filtered, according to the local resolution estimated using RELION 2.0. Further 3D classification did not reveal distinguishable reconstructions, suggesting that there was no mixing of helical symmetry in the data set. Attempts to resolve the outer portion of the tube by 3D classification using cylindrical masks (150 to 360 Å in diameter) did not show any improvement to the features of the outer portion of the map.
Infectivity assay
Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), and streptomycin (0.1 mg/ml). For virus production, cells were plated 1 day before transfection. Single-round HIV-1 harboring the gene for firefly luciferase (HIV-Luc) was generated by cotransfecting HEK293T with pNLX.Luc.(R−).ΔAvrII and pCG-VSV-G (44). The virus yield was assessed by HIV-1 CA p24 enzyme-linked immunosorbent assay (ABL Inc.).
Before infection, HEK293T cells were transiently transfected with parental pIRES2-eGFP vector or derivatives expressing MxB. At 24 hours after transfection, GFP-positive cells selected by fluorescence-activated cell sorting were infected in duplicate with HIV-Luc (0.1 pg of p24 per cell) in the presence of polybrene (4 μg/ml). Forty-eight hours after infection, cells were lysed, and luciferase activity was determined as described (34, 44). Luciferase values were normalized to the level of total proteins in cell lysates, as determined using a bicinchoninic acid (BCA) assay (Pierce).
Western blotting
Cells were lysed in 50 mM tris-HCl (pH 8), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS, and total protein concentration was determined using the BCA assay. Samples (10 μg of total protein) were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and reacted with goat polyclonal antibody to MxB (N-17), followed by HRP-conjugated secondary bovine anti-goat antibody (Santa Cruz Biotechnology). As an internal control, HRP-conjugated antibody to β-actin (Sigma-Aldrich) was used. Membranes were developed using the ECL Prime reagent (Amersham Biosciences) and imaged with a ChemiDoc MP imager (Bio-Rad).
De novo structure modeling of the L4 helix (residues 579 to 598)
Structure modeling of the L4 helix, which was apparent in the cryoEM density map but was missing in the crystal structure (PDB ID: 4WHJ), was modeled using Rosetta (45). Fragments containing sequence 3-mers and 9-mers were generated using the Rosetta server, using the MxB wild-type sequence (46). One thousand candidate folds were generated using the TopologyBroker protocol in RosettaScripts (47). The structural variability of the predicted folds was modest because all the predicted structures corresponded to an extended α helix. Using the Talaris2014 potential (48), the lowest energy fold was selected for further refinement, as explained in the following sections.
Molecular dynamics flexible fitting
The x-ray–derived structure of the MxB dimer (PDB ID: 4WHJ) was used as the initial model. Missing loops were modeled using Modeller v9.17 (49). In addition, residues 487 to 490, which contain four alanine mutations in the structure (PDB: 4WHJ), were reverted back to the wild-type sequence. For the initial modeling of full-length MxB, the L4 helix was omitted, and residues 577 and 626 were treated as C-terminal and N-terminal, respectively, to cap the ends. Secondary structural assignments used as structural restraints (ss-restraints) during the MDFF protocol (50) were derived using a secondary structure prediction program, DSSP (51). To preserve the cis/trans conformations present in the initial model (PDB ID: 4WHJ), cis-peptide restraints were also used. The model was then subjected to MDFF, with the backbone atoms coupled to the experimental density, with a coupling constant ramping from 0.05 to 0.15 over 5 ns, resulting in a cross-correlation between the map and the structure of 0.85. Because of the ss-restraints, the fitting of the stalk helix Sα1c into the experimental density was poor; therefore, the ss-restraints for residues 472 to 474 were manually removed. Subsequently, the model was subjected to MDFF, with a coupling constant ramping from 0.05 to 0.15 over 5 ns, resulting in a cross-correlation of 0.86. All molecular dynamics simulations were performed using NAMD 2.10 (52), with an integration time step of 2 fs, bonded interactions computed every time step, and electrostatics updated every 4 fs. Particle mesh Ewald was used for long-range electrostatics with a grid size of 1 Å. The CHARMM36 force field (53) and the TIP3P water model (54) were used in all simulations.
Iterative refinement of the MxB dimer by Rosetta and MDFF
The MDFF-derived MxB dimer model was further refined using Rosetta by following a similar procedure as the one developed by Lindert and McCammon (55). In particular, the MxB model was refined using the CartesianSampler available in RosettaScripts. For this purpose, the de novo model of L4 helix was docked into the experimental density using UCSF Chimera (56). First, a hybrid model of dimeric MxB containing the docked L4 helix and the MDFF-derived MxB dimer was obtained using Modeller (49). The hybrid model was then subjected to further refinement using the CartesianSampler in RosettaScripts (47). The model, which consists of the MDFF-derived MxB dimer and the Rosetta-derived model for residues 579 to 598, was further refined using MDFF, with a density coupling ramping from 0.05 to 0.15 over 5 ns.
Modeling of the MxB helical assembly
Using UCSF Chimera, starting from the Rosetta-MDFF MxB dimer, an entire MxB helical assembly was constructed by using a helical symmetry with a rise of 8.327 Å and an angle of 58.4°. The helical model was refined using MDFF with symmetry restraints (57). In addition, fitting of L1 and L2 into the density was further improved by using the interactive MDFF protocol with a coupling between the backbone atoms and a density of 0.1 (50). The interactive MDFF protocol manually guides the backbone of the loops into the experimental density. The final cross-correlation between the helical model and the density map was 0.92.
Molecular dynamics simulations and analysis
An equilibrium molecular dynamics simulation of the entire MxB tube was performed starting from the refined helical MDFF-Rosetta model. The molecular dynamics simulations were performed using NAMD 2.10 (52), with an integration time step of 2 fs, bonded interactions computed every time step, and electrostatics updated every 4 fs. Particle mesh Ewald was used for long-range electrostatics with a grid size of 1 Å. The CHARMM36 force field (53) and the TIP3P (54) water model were used in all simulations. Analysis of salt bridges, hydrogen bonds, and hydrophobic contacts was performed in VMD (visual molecular dynamics) (58) and averaged over all interfaces of the tube. A probability score was assigned to each contact, based on the occupancy of the contact in the assembled structure during the molecular dynamics simulation.
Supplementary Material
Acknowledgments
We thank P. E. Prevelige Jr. for the mass spectrometry analysis, T. Brosenitsch for reading the manuscript, and X. Fu, G. Zhao, C. Liu, M. DeLucia, M. Ford, and T. Krzysiak for the technical assistance. Funding: This work was supported by the NIH (GM082251, GM085043, GM104601, GM067887, OD019995, and AI039394), the Wellcome Trust Investigator Award (206422/Z/17/Z), and the UK Medical Research Council (MC_UP_A025_1013). Molecular dynamics simulations were performed on the Blue Waters Computer, financed by the NSF (OCI 07-25070 and ACI-1238993). Author contributions: F.J.D.A. and P.Z. designed the research. F.J.D.A. expressed and purified the MxB, carried out the biochemical analysis and EM imaging, prepared the samples for cryoEM, and collected the cryoEM data. F.J.D.A., S.H., S.H.W.S., and P.Z. carried out the cryoEM image analysis and 3D reconstruction. J.R.P. and K.S. performed the molecular dynamics simulations and real-space refinement and, with F.J.D.A., built the atomic model of the MxB assembly. F.J.D.A., J.R.P., and P.Z. analyzed the model and designed the mutations. F.J.D.A. carried out the mutagenesis. S.J. and A.N.E. performed the viral infectivity assays. F.J.D.A. and P.Z. wrote the manuscript, with support from all the authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The cryoEM density map of wild-type MxB has been deposited in the Electron Microscopy Data Bank under the accession code EMD-8577. The resulting atomic model has been deposited in the PDB under the accession code PDB-5UOT.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/9/e1701264/DC1
table S1. Effects of MxB mutations at intermolecular interfaces 1, 3, and 4.
fig. S1. Sequence alignment of Mx and dynamin proteins.
fig. S2. MxB assembles into helical tubes with and without the MBP tag.
fig. S3. GTPase activity of MBP-MxB at different protein and salt concentrations.
fig. S4. MBP-MxB tubes in the presence of GTP without or with MgCl2.
fig. S5. CryoEM of the MBP-MxB assembly.
fig. S6. Handedness of the MxB helical assembly map.
fig. S7. Gold-standard Fourier shell correlation curve of the MBP-MxB density map.
fig. S8. MDFF and real-space refinement of the MxB helical assembly model.
fig. S9. Enlarged views of the intermolecular interfaces in the MxB assembly.
fig. S10. Comparison of interfaces between cryoEM and crystal structures.
fig. S11. Comparison of L1 and L2 contacts of MxB and dynamin 3.
fig. S12. Interface 4 mutations relative to GTP-binding site.
fig. S13. Comparison of MxB and MxA GTPase-BSE domains.
movie S1. Molecular dynamics flexible fitting of MxB into the cryoEM map.
movie S2. MxB assembles into a helical array.
movie S3. Sα1c helix kinks from linear to tubular array.
movie S4. Formation of Interface 3 during helical assembly.
REFERENCES AND NOTES
- 1.Haller O., Staeheli P., Schwemmle M., Kochs G., Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 23, 154–163 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P. S., Young J. M., Emerman M., Malik H. S., Evolutionary analyses suggest a function of MxB immunity proteins beyond lentivirus restriction. PLOS Pathog. 11, e1005304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhelst J., Hulpiau P., Saelens X., Mx proteins: Antiviral gatekeepers that restrain the uninvited. Microbiol. Mol. Biol. Rev. 77, 551–566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebi M., Fäh J., Hurt N., Samuel C. E., Thomis D., Bazzigher L., Pavlovic J., Haller O., Staeheli P., cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9, 5062–5072 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melén K., Keskinen P., Ronni T., Sareneva T., Lounatmaa K., Julkunen I., Human MxB protein, an interferon-α-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 271, 23478–23486 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Kane M., Yadav S. S., Bitzegeio J., Kutluay S. B., Zang T., Wilson S. J., Schoggins J. W., Rice C. M., Yamashita M., Hatziioannou T., Bieniasz P. D., MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goujon C., Moncorgé O., Bauby H., Doyle T., Ward C. C., Schaller T., Hué S., Barclay W. S., Schulz R., Malim M. H., Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502, 559–562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z., Pan Q., Ding S., Qian J., Xu F., Zhou J., Cen S., Guo F., Liang C., The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14, 398–410 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Fricke T., White T. E., Schulte B., de Souza Aranha Vieira D. A., Dharan A., Campbell E. M., Brandariz-Nuñez A., Diaz-Griffero F., MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11, 68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z., Pan Q., Liang Z., Qiao W., Cen S., Liang C., The highly polymorphic cyclophilin A-binding loop in HIV-1 capsid modulates viral resistance to MxB. Retrovirology 12, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matreyek K. A., Wang W., Serrao E., Singh P. K., Levin H. L., Engelman A., Host and viral determinants for MxB restriction of HIV-1 infection. Retrovirology 11, 90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busnadiego I., Kane M., Rihn S. J., Preugschas H. F., Hughes J., Blanco-Melo D., Strouvelle V. P., Zang T. M., Willett B. J., Boutell C., Bieniasz P. D., Wilson S. J., Host and viral determinants of Mx2 antiretroviral activity. J. Virol. 88, 7738–7752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W., Guo H., Ma M., Markham R., Yu X.-F., Accumulation of MxB/Mx2-resistant HIV-1 capsid variants during expansion of the HIV-1 epidemic in human populations. EBioMedicine 8, 230–236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao S., von der Malsburg A., Dick A., Faelber K., Schröder G. F., Haller O., Kochs G., Daumke O., Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35, 514–525 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Fribourgh J. L., Nguyen H. C., Matreyek K. A., Alvarez F. J. D., Summers B. J., Dewdney T. G., Aiken C., Zhang P., Engelman A., Xiong Y., Structural insight into HIV-1 restriction by MxB. Cell Host Microbe 16, 627–638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daumke O., Praefcke G. J. K., Invited review: Mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers 105, 580–593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell P. S., Patzina C., Emerman M., Haller O., Malik H. S., Kochs G., Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe 12, 598–604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dicks M. D. J., Goujon C., Pollpeter D., Betancor G., Apolonia L., Bergeron J. R. C., Malim M. H., Oligomerization requirements for MX2-mediated suppression of HIV-1 infection. J. Virol. 90, 22–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buffone C., Schulte B., Opp S., Diaz-Griffero F., Contribution of MxB oligomerization to HIV-1 capsid binding and restriction. J. Virol. 89, 3285–3294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chappie J. S., Mears J. A., Fang S., Leonard M., Schmid S. L., Milligan R. A., Hinshaw J. E., Dyda F., A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell 147, 209–222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nigg P. E., Pavlovic J., Oligomerization and GTP-binding requirements of MxA for viral target recognition and antiviral activity against influenza A virus. J. Biol. Chem. 290, 29893–29906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P., Hinshaw J. E., Three-dimensional reconstruction of dynamin in the constricted state. Nat. Cell Biol. 3, 922–926 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., Nunnari J., Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon Y., Pitts K. R., McNiven M. A., Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12, 2894–2905 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao S., von der Malsburg A., Paeschke S., Behlke J., Haller O., Kochs G., Daumke O., Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Reubold T. F., Faelber K., Plattner N., Posor Y., Ketel K., Curth U., Schlegel J., Anand R., Manstein D. J., Noé F., Haucke V., Daumke O., Eschenburg S., Crystal structure of the dynamin tetramer. Nature 525, 404–408 (2015). [DOI] [PubMed] [Google Scholar]
- 28.King M. C., Raposo G., Lemmon M. A., Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl. Acad. Sci. U.S.A. 101, 8957–8962 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Praefcke G. J. K., McMahon H. T., The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Traut T. W., Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Kong J., Ma M., He S., Qin X., Mx oligomer: A novel capsid pattern sensor? Future Microbiol. 11, 1047–1055 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Zhao G., Perilla J. R., Yufenyuy E. L., Meng X., Chen B., Ning J., Ahn J., Gronenborn A. M., Schulten K., Aiken C., Zhang P., Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundborger A. C., Fang S., Heymann J. A., Ray P., Chappie J. S., Hinshaw J. E., A dynamin mutant defines a superconstricted prefission state. Cell Rep. 8, 734–742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shun M.-C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A., LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21, 1767–1778 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingerman E., Nunnari J., A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 404, 611–619 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Mears J. A., Ray P., Hinshaw J. E., A corkscrew model for dynamin constriction. Structure 15, 1190–1202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Li X., Mooney P., Zheng S., Booth C. R., Braunfeld M. B., Gubbens S., Agard D. A., Cheng Y., Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Desfosses A., Ciuffa R., Gutsche I., Sachse C., SPRING—An image processing package for single-particle based helical reconstruction from electron cryomicrographs. J. Struct. Biol. 185, 15–26 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Koh Y., Wu X., Ferris A. L., Matreyek K. A., Smith S. J., Lee K., KewalRamani V. N., Hughes S. H., Engelman A., Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J. Virol. 87, 648–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leaver-Fay A., Tyka M., Lewis S. M., Lange O. F., Thompson J., Jacak R., Kaufman K., Renfrew P. D., Smith C. A., Sheffler W., Davis I. W., Cooper S., Treuille A., Mandell D. J., Richter F., Ban Y.-E. A., Fleishman S. J., Corn J. E., Kim D. E., Lyskov S., Berrondo M., Mentzer S., Popović Z., Havranek J. J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J. J., Kuhlman B., Baker D., Bradley P., ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chivian D., Kim D. E., Malmström L., Bradley P., Robertson T., Murphy P., Strauss C. E. M., Bonneau R., Rohl C. A., Baker D., Automated prediction of CASP-5 structures using the Robetta server. Proteins 53, 524–533 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Fleishman S. J., Leaver-Fay A., Corn J. E., Strauch E.-M., Khare S. D., Koga N., Ashworth J., Murphy P., Richter F., Lemmon G., Meiler J., Baker D., RosettaScripts: A scripting language interface to the Rosetta macromolecular modeling suite. PLOS ONE 6, e20161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Meara M. J., Leaver-Fay A., Tyka M. D., Stein A., Houlihan K., DiMaio F., Bradley P., Kortemme T., Baker D., Snoeyink J., Kuhlman B., Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J. Chem. Theory Comput. 11, 609–622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.B. Webb, A. Sali, Comparative protein structure modeling using MODELLER, in Current Protocols in Bioinformatics (John Wiley & Sons Inc., 2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trabuco L. G., Schreiner E., Gumbart J., Hsin J., Villa E., Schulten K., Applications of the molecular dynamics flexible fitting method. J. Struct. Biol. 173, 420–427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 52.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Best R. B., Zhu X., Shim J., Lopes P. E. M., Mittal J., Feig M., Mackerell A. D. Jr, Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 55.Lindert S., McCammon J. A., Improved cryoEM-guided iterative molecular dynamics–Rosetta protein structure refinement protocol for high precision protein structure prediction. J. Chem. Theory Comput. 11, 1337–1346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Chan K.-Y., Gumbart J., McGreevy R., Watermeyer J. M., Sewell B. T., Schulten K., Symmetry-restrained flexible fitting for symmetric EM maps. Structure 19, 1211–1218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/9/e1701264/DC1
table S1. Effects of MxB mutations at intermolecular interfaces 1, 3, and 4.
fig. S1. Sequence alignment of Mx and dynamin proteins.
fig. S2. MxB assembles into helical tubes with and without the MBP tag.
fig. S3. GTPase activity of MBP-MxB at different protein and salt concentrations.
fig. S4. MBP-MxB tubes in the presence of GTP without or with MgCl2.
fig. S5. CryoEM of the MBP-MxB assembly.
fig. S6. Handedness of the MxB helical assembly map.
fig. S7. Gold-standard Fourier shell correlation curve of the MBP-MxB density map.
fig. S8. MDFF and real-space refinement of the MxB helical assembly model.
fig. S9. Enlarged views of the intermolecular interfaces in the MxB assembly.
fig. S10. Comparison of interfaces between cryoEM and crystal structures.
fig. S11. Comparison of L1 and L2 contacts of MxB and dynamin 3.
fig. S12. Interface 4 mutations relative to GTP-binding site.
fig. S13. Comparison of MxB and MxA GTPase-BSE domains.
movie S1. Molecular dynamics flexible fitting of MxB into the cryoEM map.
movie S2. MxB assembles into a helical array.
movie S3. Sα1c helix kinks from linear to tubular array.
movie S4. Formation of Interface 3 during helical assembly.


