Abstract
MicroRNAs (miRNAs) play major roles in various biological processes and have been implicated in the pathogenesis and malignant progression of glioblastoma multiforme (GBM). The aim of this study was to assess the predictive values of miRNAs for overall survival (OS) of patients with GBM. MiRNA expression profiles and clinical information of 563 GBM patients were obtained from the Cancer Genome Atlas. The most significantly altered miRNAs were identified and miRNA expression profiles were performed, through principal component analysis, the least absolute shrinkage and selection operator method. The survival analysis was performed using the Cox regression models. Additionally, receiver operating characteristic (ROC) analysis was used to assess the performance of survival prediction. We used the bioinformatics tools to establish the miRNA signature for biological relevance assessment. A linear prognostic model of three miRNAs was developed and the patients were divided into high risk and low risk groups based this model. The area under the ROC curve (AUC) for the three miRNA signature predicting 5-year survival was 0.894 (95%CI, 0.789-1.000) in the testing set and0.841 (95%CI, 0.689-0.993) in all GBM patients. High risk patients had significantly shorter OS than patients with low risk (P< 0.001). The results from this study support a three miRNA signature for outcome prediction of GBM. These results provided a new prospect for prognostic biomarker of GBM.
Keywords: glioblastoma multiforme, microRNA, prognosis, overall survival, TCGA
INTRODUCTION
Glioblastoma multiforme (GBM), a World Health Organization grade IV glioma, is one of the most common and aggressive primary malignancies in adults. Patients with GBM have a poor prognosis despite advances in diagnostic and therapeutic approaches and recent considerable research efforts, with a mean survival rate of approximately 3.3 % at 2 years and 1.2 % at 3 years as well as a median overall survival (OS) time of 12 to 17 months [1–3]. Besides typical genetic alterations, aberrant epigenetic mechanisms, such as DNA methylation, histone modifications, chromatin remodeling, or altered noncoding RNA expression (e.g, microRNAs), have been implicated in the pathogenesis and malignant progression of GBM [4]. Therefore, these findings have inspired the new discovery of molecularly targeted therapies as a novel approach for GBM treatment.
MicroRNAs (miRNAs) are small non-coding RNAs of 18-25 nucleotides in length that regulate gene expression by base-pairing with the 3’-untranslated region (3’-UTR) of mRNA targets, resulting in mRNA degradation and/or inhibition of mRNA translation [5]. MiRNAs play major roles in many biological processes such as cellular proliferation, apoptosis, migration, and differentiation [6–8]. To date, at least 253 and 95 miRNAs were found to be significantly upregulated and downregulated in GBM, respectively [9]. Some miRNAs have been implicated in glioblastoma development, progression and prognosis [10–12]. However, they do not necessarily reveal the similar results, due perhaps to molecular heterogeneity in tumors [13]. Moreover, there are some inconsistencies between studies due to variations in the approaches for selecting miRNAs, study design features such as small sample size, a heterogeneous admixture of cancer types, various lengths of follow-up, and data analysis without full adjustment for important prognostic parameters and treatments as well as type of array platform utilized, and the choice of control tissue. All these issues may be important sources of heterogeneity of the results reported and may bias the estimated outcomes.
By characterizing genetic and epigenetic alterations, and the expression of cancer genomes, the Cancer Genome Atlas (TCGA) project has provided a comprehensive way to understand various cancers. Prognostic miRNA signature has shown a predictive value for OS of patients with breast cancer [14, 15], colon cancer [16] and lung cancer [17] using TCGA datasets, while the similar prediction of prognosis has not performed in GBM. Since the extensive TCGA study also demonstrated a large number of miRNA expression data for GBM patients, to take advantage of utilizing this specific resource we aimed in the current study to assess the predictive value of specific miRNA signature for OS of patients with GBM from an existing large dataset of TCGA.
RESULTS
Selection of miRNAs with prognostic value
According to inclusive criteria, a total of 563 GBM patients were finally enrolled in this study. The principal component analysis included the selected 315 miRNAs with component score coefficient matrix ≥0.4 or ≤0.4 from 470 miRNAs. After the least absolute shrinkage and selection operator (LASSO) analysis, 315 miRNAs were further reduced to 9 potential predictors (Figure 1A and 1B), including hsa-miR-148a, hsa-miR-175p, hsa-miR-222, hsa-miR-302d, hsa-miR-487b, hsa-miR-608, hsa-miR-646, hsa-miR-649, and hsa-miR-675. For subsequent analysis, we randomly divided the total patients into the training set (n=282) and testing set (n=281) respectively. The mean age of patients in the training and testing groups was 58.0 and 57.9 years old, respectively. There was no significant differences of patient's age and survival time between the two sets (P> 0.05).
Figure 1. MiRNA selection using the least absolute shrinkage and selection operator (LASSO) binary logistic regression model.
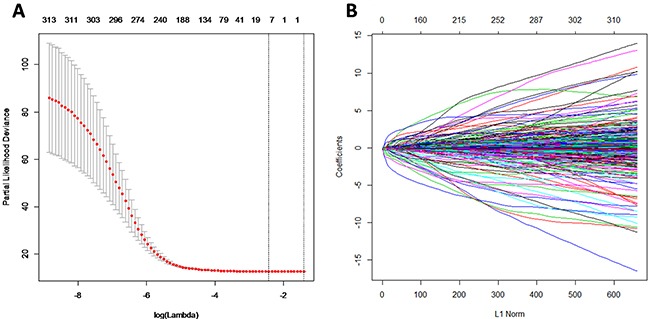
(A) Tuning parameter (λ) selection in the LASSO model. The area under the receiver operating characteristic (AUC) curve was plotted versus log (λ). (B) LASSO coefficient profiles.
Multivariable logistic regression analyses, including following clinical candidate predictors (e.g. age, gender) and the 9 selected miRNAs (e.g., hsa-miR-148a, hsa-miR-175p, hsa-miR-222, hsa-miR-302d, hsa-miR-487b, hsa-miR-608, hsa-miR-646, hsa-miR-649, and hsa-miR-675 miRNAs), were used to evaluate the contribution of each miRNA as independent prognostic factor of patient survival in the testing set. The backward stepwise method identified four best predictors including hsa-miR-222, hsa-miR-302d, hsa-miR-646, and age (Table 1).
Table 1. Multivariate Cox proportional hazards analysis.
| Variables | Coefficient | P-value | HRs | 95%CI | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Hsa-miR-222 | 0.112 | 0.028 | 1.119 | 1.012 | 1.237 |
| Hsa-miR-302d | -3.671 | 0.036 | 0.025 | 0.001 | 0.784 |
| Hsa-miR-646 | -2.971 | 0.044 | 0.051 | 0.003 | 0.917 |
| Age | .023 | 0.000 | 1.023 | 1.013 | 1.033 |
HR, hazard ratio; CI, confidence interval.
MiRNA prognostic model
With coefficients from Cox regression analysis, the prognostic model was built and prognostic score was calculated as follows: Prognostic-score = (0.112×expression level of hsa-miR-222) + (-3.671×expression level of hsa-miR-302d) + (-2.971×expression level of hsa-miR-646) + (0.023×age). The miRNAs expression level was as the log2 reads per million of total aligned miRNA reads. The prognostic-score showed a great prediction of prognostic capacity for 5-year survival in GBM with an area under the curve (AUC) of 0.841 (95%CI, 0.689-0.993) in the training set (Figure 2A), an AUC of 0.894 (95%CI, 0.789-1.000) in the testing set, (Figure 2B), and an AUC of 0.854 (95%CI, 0.744-0.964) in all GBM patients (Figure 2C), respectively.
Figure 2. The ROC curves for the three microRNA signature in TCGA GBM cohort.
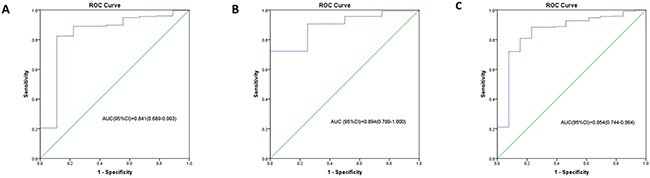
The ROC curve for predicting 5-year survival in GBM with an AUC of 0.841 (95%CI, 0.689-0.993) in the training set (A), an AUC of 0.894 (95%CI, 0.789-1.000) in the testing set (B), and an AUC of 0.854 (95%CI, 0.744-0.964) in all GBM patients (C), respectively.
Based on ROC curve for predicting 5-year survival in the testing set, we categorized the samples into two groups as a high risk and a low risk group using the best cutoff point of prognostic scores with optimum sensitivity and specificity. The cutoff point was -36.5428 with 90.6% sensitivity and 75.0% specificity. Compared with the patients with a low risk score, the patients with a high risk score in the TCGA GBM cohort had a significantly shorter OS (P < 0.001) (Figure 3).
Figure 3. Kaplan Meier curves for the three microRNA signature in TCGA GBM cohort.
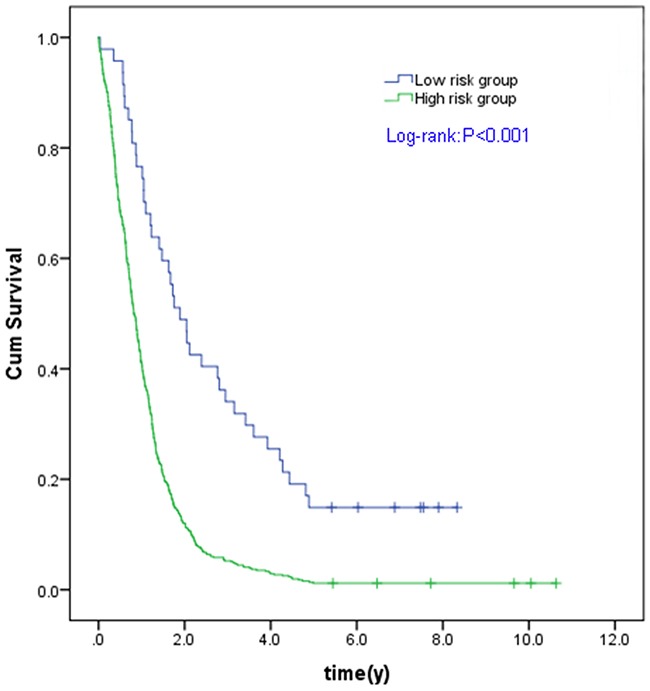
Individual patient was scored according to the three miRNAs signature. The Kaplan-Meier curves for GBM risk groups obtained from the TCGA cohort divided by the cut-off point. The OS of high risk group is significantly lower than that of low risk group in all GBM patients (C). The P values of the log-rank tests are <0.001.
Target prediction and functional enrichment of the three miRNA signature in GBM
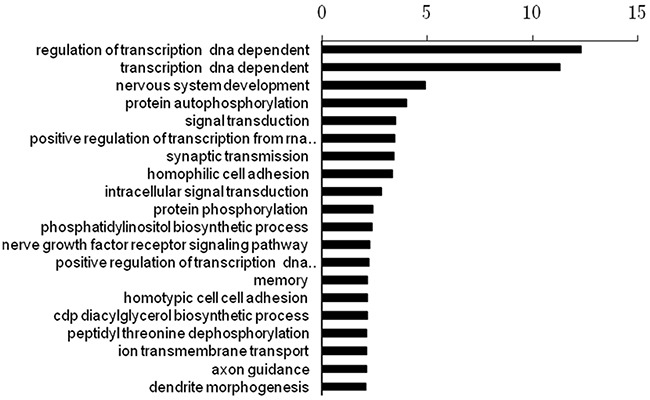
The numbers of target genes of the three miRNAs were 9017, 15613, and 9625, which were predicted by the database of miRwalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). We then performed a functional enrichment analysis to elucidate the biological function of target genes of the three miRNA signature. A total of 645 pathways were enriched. The top 20 enriched functional analysis from Gene ontology (GO) analysis was shown in Figure 4. The top enriched biological process was regulation of transcription. The top enriched pathway was the Axon guidance pathway.
Figure 4. The top 20 enriched functional analysis from Gene ontology (GO) analysis.
DISCUSSION
In this study, we found that 3 selected miRNA signature may serve as predictor of GBM patient survival, providing novel insights into the significance of miRNAs as molecular markers in predicting the prognosis of GBM patients.
MiRNAs are regulatory nucleic acids that modulate the epigenetic state and gene expression of cells from both the tumor and its microenvironment, thereby influencing tumor cell proliferation, differentiation, survival and invasion. Alterations of miRNA expression have now been identified to be correlated with diagnosis, prognosis, and response to therapy for a range of human malignancies, including lung cancer [18, 19], breast cancer [20, 21], ovarian cancer [22] and prostate cancers [23]. Since the first two studies on investigation of miRNA expression profiles in GBM [24, 25], numerous miRNA-mediated mechanisms have been found to be implicated in the pathogenesis and malignant progression of GBM [26–28]. Dysregulation of miRNA expression level and functionality has been detected to be associated with GBM [9], although the functional properties might remain controversial or fully unknown. The pattern of miRNA expression has become a recognized tool besides other gene expression profiling for stratifying GBM patients into different groups [29]. In a previous study, a ten-miRNA expression signature was identified as survival predictors in a relatively smal sample size of 222 GBM patients [30].
Some miRNAs associated with longer or shorter survival have been identified [31]. Moreover, many of these prognostic miRNAs have validated targets and functional characteristics. In the present study, we have identified a three microRNA signature, consisting of hsa-miR-222, hsa-miR-302d and hsa-miR-646, which were validated as an independent predictor for GBM patients’ survival. The AUC of the ROC curve for the three microRNA signature predicting 5-year survival was 0.894 in the testing set and 0.854 in the total dataset, confirming a good performance for predicting survival of GBM. Among the three miRNAs, overexpression of hsa-miR-222 has been associated with poor prognosis as previously reported, and the increased expression of miR-222-3p, a target of PTEN, inhibited PTEN in glioma [32, 33]. MiR-222 is also detected to target O6-methylguanine methyltransferase (MGMT) mRNA. Chronic miR-222-mediated MGMT downregulation might render cells unable to repair genetic damage, leading to poor GBM prognosis [34]. Furthermore, miR-222 targeted an important cell cycle regulator of p27/KIP1 [35, 36], supporting a critical role of miR-222 in the regulation of the cell cycle, proliferation, and invasion. Suppression of miR-222 upregulated p27/ KIP1, increased the number of cells in G0/G1 stage, reduced G1/S progression, and induced apoptosis. MiR-222 also modulated the IFN-α signaling pathway including regulation of STAT1/STAT2 phosphorylation and nuclear localization [37]. An inverse relationship between pro-apoptotic PUMA gene and miR-222 expression has been demonstrated in glioma tissues [38]. MiR-222has also been shown to target intracellular adhesion molecule 1 (ICAM-1), where expression of miR-222 was inversely correlated to ICAM-1 expression in GBM tumors [39]. Microarray analysis of gene expression after inhibition of miR-221/miR-222 in GBM showed 158 differentially expressed genes involved in cell metabolism, cytoskeletal organization, and molecular signaling [37]. In a separate bioinformatics analysis, miR-221/miR-222 regulated about 70 common target genes [40]. These various targets explain the widespread regulation of miR-222 in GBM function.
On the contrary, low expression of hsa-miR-302d and hsa-miR-646 was associated with poor prognosis in patients with GBM in the current study. The miR-302 members were down-regulated in P-glycoprotein (P-gp)-overexpressing breast cancer cell lines [41], and downregulated BCRP expression to increase chemosensitivity of breast cancer cells [42]. MiR-302dmay also control proliferation and cell survival of human adipose tissue-derived mesenchymal stem cells through different targets [43]. MiR-646 is downregulated in many human cancers, and functions as a tumor suppressor. Overexpression of miR-646 inhibited lung cancer cell proliferation and metastasis, and acted as a predictor of OS [44]. Downregulation of miR-646 was also associated with progression of osteosarcoma, and played a key role in suppression of tumor in osteosarcoma [45–46]. Another study demonstrated that miR-646 may play an important role in the development and tumor metastasis of clear cell renal carcinoma [47]. However, to the best of our knowledge, this is the first to report an association of hsa-miR-302d and hsa-miR-646 with GBM.
The carcinogenic process of GBM is a multi-step process driven by a series of genetic and epigenetic alterations, resulting in the progression from a normal cell to a cancer cell. Through the enrichment and function analysis, we found that the target genes of prognostic miRNA signature may participate in many important biological processes, including regulation of transcription, nervous system development, and protein autophosphorylation.
In conclusion, we have identified a three-miRNA integrated signature that can predict the survival outcome of GBM patients. Our findings may help researchers in understanding GBM cell death and survival, developing targeted therapy, identifying high-risk GBM patients for better treatment, and warranting future evaluation of these biomarkers in patients with GBM. Modulating miRNA expression hold strong potential in therapeutic strategies and development of new molecular targeted treatment. Further understanding of molecular miRNA signatures would lead to increased survival and improved quality of life for GBM patients.
MATERIALS AND METHODS
Patient cohort and miRNA data
The level 3 miRNA expression profiles (level 3 data) with clinical information for GBM patients were obtained from TCGA data portal (May 2016, https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). Both the miRNA expression data and clinical data are open-access. To avoid the impact of unrelated causes of death, the cases with less than 1-month OS and death from other diseases or accidents were excluded in this study.
Identification of miRNAs with prognostic value in GBM
MiRNA expression data was normalized by using the R/ Bioconductor package edgeR, which is designed for digital gene expression data [48]. The principal component analysis and LASSO method, which is suitable for the regression of high-dimensional data [49], were adopted for selection of miRNAs. The selected miRNAs were those with component score coefficient matrix ≥0.4 or ≤0.4.
Multivariable logistic regression analysis, including following clinical candidate predictors: age, gender, and selected miRNAs, was used to evaluate the contribution of each clinical variable and miRNA as independent prognostic factors of patient's survival in the testing set. The miRNA expression level was as the log2 reads per million of total aligned miRNA reads. The backward stepwise method was employed to select the best predictors. All analyses were performed using R (Packages: survival, survivalROC, boot, and superpc).
Definition of prognostic model and ROC curve
The linear prognostic model was developed based on coefficient from Cox regression analysis. We used the linear miRNA prognostic model obtained from the training set to calculate a prognostic score for each patient. The prognostic performance was measured using time-dependent receiver operating characteristic (ROC) curves. Since the majority of events occurred before 60 months, the ability of models to predict outcome around 60 months was assessed.
In the ROC curve of the testing set for predicting 5-year survival, we chose the prognostic scores with optimum sensitivity and specificity as the cutoff values, to divide the patients into the high risk or low risk group. Kaplan-Meier curves were used to estimate the survival for patients within the two risk groups. All analyses were performed using R (Packages: survival, survivalROC, boot, and superpc). For all analyses, statistical significance was set at p <0.05, and all tests were two-sided.
Target prediction and enrichment analysis
The target genes of miRNAs were predicted by miRWalk (www.umm.uni-heidelberg.de), which offers a comprehensive data of possible miRNA targets. The pathway enrichment analysis was conducted with the GeneTrail gene set enrichment tool. The results were considered significant when P value was less than 0.05 after FDR corrected.
Abbreviations
- GBM
glioblastoma multiforme
- AUC
area under the curve
- GO
gene ontology
- LASSO
least absolute shrinkage and selection operator
- miRNA
MicroRNA
- OS
overall survival
- ROC
receiver operating characteristic
- TCGA
the Cancer Genome Atlas.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This work was supported by National Natural Science Foundation of China (No. 81402461, 81471709, 81373092); Subject Chief Scientist of Shanghai, Science and Technology Commission of Shanghai Municipality (grant 13XD1402400); and the Fundamental Research Funds for the Central Universities, HUST (No. 2014TS057).
REFERENCES
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Crespo I, Vital AL, Gonzalez-Tablas M, Patino Mdel C, Otero A, Lopes MC, de Oliveira C, Domingues P, Orfao A, Tabernero MD. Molecular and genomic alterations in glioblastoma multiforme. Am J Pathol. 2015;185:1820–33. doi: 10.1016/j.ajpath.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–71. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 7.Cheng HY, Obrietan K. Revealing a role of microRNAs in the regulation of the biological clock. Cell Cycle. 2007;6:3034–5. doi: 10.4161/cc.6.24.5106. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol. 2013;47:131–44. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, Califano A. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriksen M, Johnsen KB, Olesen P, Pilgaard L, Duroux M. MicroRNA expression signatures and their correlation with clinicopathological features in glioblastoma multiforme. Neuromolecular Med. 2014;16:565–77. doi: 10.1007/s12017-014-8309-7. [DOI] [PubMed] [Google Scholar]
- 12.Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C, Zhang H, James CD, Sznajder JI, et al. MiR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29:732–45. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong J, Bing Z, Su Y, Deng D, Peng X. An integrated mRNA and microRNA expression signature for glioblastoma multiforme prognosis. PLoS One. 2014;9:e98419. doi: 10.1371/journal.pone.0098419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci U S A. 2013;110:7413–7. doi: 10.1073/pnas.1304977110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JT, Wang F, Chapin W, Huang RS. Identification of microRNAs as breast cancer prognosis markers through the cancer genome atlas. PLoS One. 2016;11:e0168284. doi: 10.1371/journal.pone.0168284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Zhao J, Zhang R. Four microRNAs signature for survival prognosis in colon cancer using TCGA data. Sci Rep. 2016;6:38306. doi: 10.1038/srep38306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X, Wu Y, Yu W, Li H. Identification of a seven-miRNA signature as prognostic biomarker for lung squamous cell carcinoma. Oncotarget. 2016;7:81670–9. doi: 10.18632/oncotarget.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eder M, Scherr M. MicroRNA and lung cancer. N Engl J Med. 2005;352:2446–8. doi: 10.1056/NEJMcibr051201. [DOI] [PubMed] [Google Scholar]
- 19.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Dvinge H, Git A, Gräf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, Chin SF, Provenzano E, Turashvili G, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–82. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 21.Cuiffo BG, Campagne A, Bell GW, Lembo A, Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, El-Ashry D, Hematti P, Polyak K, et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell. 2014;15:762–74. doi: 10.1016/j.stem.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–35. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 25.Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 26.Liu N, Zhang L, Wang Z, Cheng Y, Zhang P, Wang X, Wen W, Yang H, Liu H, Jin W, Zhang Y, Tu Y. MicroRNA-101 inhibits proliferation, migration and invasion of human glioblastoma by targeting SOX9. Oncotarget. 2016;8:19244–54. doi: 10.18632/oncotarget.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C, Zhang H, James CD, Sznajder JI, et al. MiR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29:732–45. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiou GY, Chien CS, Wang ML, Chen MT, Yang YP, Yu YL, Chien Y, Chang YC, Shen CC, Chio CC, Lu KH, Ma HI, Chen KH, et al. Epigenetic regulation of the miR142-3p/interleukin-6 circuit in glioblastoma. Mol Cell. 2013;52:693–706. doi: 10.1016/j.molcel.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by microRNAs. Cancer Res. 2011;71:3387–99. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henriksen M, Johnsen KB, Andersen HH, Pilgaard L, Duroux M. MicroRNA expression signatures determine prognosis and survival in glioblastoma multiforme--a systematic overview. Mol Neurobiol. 2014;50:896–913. doi: 10.1007/s12035-014-8668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Guo F, Wang P, Hong S, Zhang C. miR-221/222 confers radioresistance in glioblastoma cells through activating Akt independent of PTEN status. Curr Mol Med. 2014;14:185–95. doi: 10.2174/1566524013666131203103147. [DOI] [PubMed] [Google Scholar]
- 33.Tokudome T, Sasaki A, Tsuji M, Udaka Y, Oyamada H, Tsuchiya H, Oguchi K. Reduced PTEN expression and overexpression of miR-17-5p,-19a-3p, -19b-3p, -21-5p, -130b-3p, -221-3p and -222-3p by glioblastoma stem-like cells following irradiation. Oncol Lett. 2015;10:2269–72. doi: 10.3892/ol.2015.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quintavalle C, Mangani D, Roscigno G, Romano G, Diaz-Lagares A, Iaboni M, Donnarumma E, Fiore D, De Marinis P, Soini Y, Esteller M, Condorelli G. MiR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS One. 2013;8:e74466. doi: 10.1371/journal.pone.0074466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2009;8:2685. doi: 10.4161/cc.8.17.9489. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Kang C, You Y, Pu P, Yang W, Zhao P, Wang G, Zhang A, Jia Z, Han L, Jiang H. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol. 2009;34:1653–60. doi: 10.3892/ijo_00000296. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Han L, Zhang A, Yang W, Zhou X, Pu P, Du Y, Zeng H, Kang C. Global changes of mRNA expression reveals an increased activity of the interferon-induced signal transducer and activator of transcription (STAT) pathway by repression of miR-221/222 in glioblastoma U251 cells. Int J Oncol. 2010;36:1503–12. doi: 10.3892/ijo_00000637. [DOI] [PubMed] [Google Scholar]
- 38.Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ, Kang CS. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda R, Kohanbash G, Sasaki K, Fujita M, Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze MT, Khan SA, Sobol RW, Okada H. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci U S A. 2009;106:10746–51. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, Zhong Y, You Y, Pu P, Kang C. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913–20. doi: 10.3892/ijo_00000570. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L, Wang Y, Jiang L, He M, Bai X, Yu L, Wei M. MiR-302a/b/c/d cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein(P-gp) by targeting MAP/ERK kinase kinase 1 (MEKK1) J Exp Clin Cancer Res. 2016;35:25. doi: 10.1186/s13046-016-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Zhao L, Xiao Q, Jiang L, He M, Bai X, Ma M, Jiao X, Wei M. miR-302a/b/c/d cooperatively inhibit BCRP expression to increase drug sensitivity in breast cancer cells. Gynecol Oncol. 2016;141:592–601. doi: 10.1016/j.ygyno.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 43.Kim JY, Shin KK, Lee AL, Kim YS, Park HJ, Park YK, Bae YC, Jung JS. MicroRNA-302 induces proliferation and inhibits oxidant-induced cell death in human adipose tissue-derived mesenchymal stem cells. Cell Death Dis. 2014;5:e1385. doi: 10.1038/cddis.2014.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Y, Chen Y, Ma D, Ji Z, Cao F, Chen Z, Ning Y, Bai C. miR-646 is a key negative regulator of EGFR pathway in lung cancer. Exp Lung Res. 2016;42:286–95. doi: 10.1080/01902148.2016.1207726. [DOI] [PubMed] [Google Scholar]
- 45.Azam AT, Bahador R, Hesarikia H, Shakeri M, Yeganeh A. Downregulation of microRNA-217 and microRNA-646 acts as potential predictor biomarkers in progression, metastasis, and unfavorable prognosis of human osteosarcoma. Tumour Biol. 2016;37:5769–73. doi: 10.1007/s13277-015-3821-4. [DOI] [PubMed] [Google Scholar]
- 46.Sun XH, Geng XL, Zhang J, Zhang C. miRNA-646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2) Tumour Biol. 2015;36:2127–34. doi: 10.1007/s13277-014-2822-z. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Liu M, Feng Y, Xu YF, Huang YF, Che JP, Wang GC, Yao XD, Zheng JH. Downregulated miR-646 in clear cell renal carcinoma correlated with tumour metastasis by targeting the nin one binding protein (NOB1) Br J Cancer. 2014;111:1188–200. doi: 10.1038/bjc.2014.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauerbrei W, Royston P, Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med. 2007;26:5512–28. doi: 10.1002/sim.3148. [DOI] [PubMed] [Google Scholar]


