Abstract
Splicing in S. cerevisiae has been shown to proceed cotranscriptionally, but the nature of the coupling remains a subject of debate. Here, we examine the effect of nineteen complex–related splicing factor Prp45 (a homolog of SNW1/SKIP) on cotranscriptional splicing. RNA-sequencing and RT-qPCR showed elevated pre-mRNA levels but only limited reduction of spliced mRNAs in cells expressing C-terminally truncated Prp45, Prp45(1–169). Assays with a series of reporters containing the AMA1 intron with regulatable splicing confirmed decreased splicing efficiency and showed the leakage of unspliced RNAs in prp45(1–169) cells. We also measured pre-mRNA accumulation of the meiotic MER2 gene, which depends on the expression of Mer1 factor for splicing. prp45(1–169) cells accumulated approximately threefold higher levels of MER2 pre-mRNA than WT cells only when splicing was induced. To monitor cotranscriptional splicing, we determined the presence of early spliceosome assembly factors and snRNP complexes along the ECM33 and ACT1 genes. We found that prp45(1–169) hampered the cotranscriptional recruitment of U2 and, to a larger extent, U5 and NTC, while the U1 profile was unaffected. The recruitment of Prp45(1–169) was impaired similarly to U5 snRNP and NTC. Our results imply that Prp45 is required for timely formation of complex A, prior to stable physical association of U5/NTC with the emerging pre-mRNA substrate. We suggest that Prp45 facilitates conformational rearrangements and/or contacts that couple U1 snRNP-recognition to downstream assembly events.
Keywords: cotranscriptional splicing, spliceosome assembly, nineteen complex, Prp8, RES complex, chromatin immunoprecipitation
INTRODUCTION
Introns are removed from protein-coding transcripts based on the combination of splicing sequences and inputs from other complexes involved in gene expression, all of which need to be interpreted during the splicing process. The spliceosome in budding yeast consists of five conserved U-rich small nuclear RNAs (snRNAs) and almost 100 proteins; the overall architecture as well as most of the splicing factors is conserved in higher eukaryotes (Bessonov et al. 2008; Fabrizio et al. 2009). These snRNAs form the active site of the spliceosome and base pair with transcripts, directly contributing to intron recognition and splicing catalysis (Valadkhan and Jaladat 2010; Hang et al. 2015). Proteins have structural, enzymatic, and regulatory functions (Jurica and Moore 2003; Will and Lührmann 2011) and also support a dynamic interaction network which links splicing with all other stages of gene expression (Alexander and Beggs 2010; Bentley 2014). However, the functioning of this network and its role in gene expression regulation are still far from understood.
The spliceosome assembles de novo on each intron in a stepwise fashion. Proper execution of the rearrangements required to reach the catalytically competent spliceosome depends on DExD/H ATPases, which couple ATP hydrolysis with structural alterations. Early in assembly, U1 snRNP binds the 5′ splice site (5′ss), forming the commitment complex 1 (CC1) and is then joined by Msl5 (branchpoint binding protein; BBP), which recognizes the branch site (Ruby and Abelson 1988; Berglund et al. 1997). Subsequently, Msl5 and its heterodimer partner Mud2 are displaced in favor of U2 snRNP, after which U4/U6.U5 tri-snRNP is incorporated into the splicing complex. U1 snRNP is then exchanged for U6 and U5 snRNPs at 5′ss (see Wang and Burge 2008 and references therein). A complex of Prp19-associated factors (nineteen complex; NTC) together with “NTC-related” factors (Prp17, Prp45, and Prp46, among others) are incorporated with tri-snRNP (Fabrizio et al. 2009), i.e., before or during unwinding of U4 from U6 snRNA, and remain part of the spliceosome through its cycle. In yeast, NTC is required for the stable association of U5/U6 with the spliceosome after U1 and U4 are released (Chan et al. 2003). NTC helps to facilitate proper base-pairing interactions between pre-mRNA and specific parts of U6 and U5 snRNAs in the active spliceosome (Chan and Cheng 2005). Apart from splicing, the complex plays roles in transcription elongation, mRNA export, and DNA repair (for review, see Chanarat and Sträßer 2013).
In this work, we examine the role of the NTC-related factor Prp45 of Saccharomyces cerevisiae in cotranscriptional splicing. The gene product corresponding to Prp45 is present throughout eukaryotes (SNW1 in Schizosaccharomyces pombe, SKIP in human; Dahl et al. 1998; Albers et al. 2003), with the remarkable exception of Cyanidioschyzon merolae, an alga with an extremely reduced spliceosome that lacks both U1 snRNP and NTC components (Stark et al. 2015). SKIP has been shown to be part of the CDC5-related NTC during splicing (Ajuh et al. 2000; Makarov et al. 2002). In early studies, SKIP was reported in many interactions with transcription factors and coregulators (for review, see Folk et al. 2004). More recent evidence suggests that SKIP participates in the functioning of positive transcription elongation factor complex P-TEFb (Brès et al. 2005, 2009).
High-resolution structural information on spliceosomes has recently become available, including the conformation and contacting partners for part of Prp45 itself (Yan et al. 2015, 2016, 2017; Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016; Bertram et al. 2017; Fica et al. 2017). The structures of S. cerevisiae, S. pombe, and human spliceosomes from stages Bact, C, C*, and ILS all contain parts of Prp45/SNW1/SKIP and reveal a strikingly extended chain spanning the whole spliceosome structure (Yan et al. 2015, 2016; Bertram et al. 2017). The arrangement observed for Prp45 is in accordance with the intrinsically disordered character predicted for parts of Prp45 (Korneta and Bujnicki 2012); similar extended chain-interfaces are observed for other proteins containing intrinsically disordered regions, such as the CDK inhibitors of the CIP/KIP family (Das et al. 2016). The protein may be capable of mediating interactions with various partners during stages of spliceosome assembly, aiding their effective contacts or conformational changes. The N-terminal part of Prp45 is shorter than it is in S. pombe or metazoan counterparts (S. cerevisiae lacks the G-rich box amino acids 63–79 of the S. pombe structure) and is traceable in the structure beginning at amino acid 30. The protein is then resolved (excluding several gaps) through the conserved SNW domain and helical region up to amino acid 350 of the C-terminal part (in the Bact complex) (Rauhut et al. 2016; Yan et al. 2016).
Previously, we reported that a truncation of Prp45 weakened the association of second-step helicase Prp22 with spliceosome complexes and affected the fidelity of the 3′ splice site choice. In addition to NTC components, we further identified several second-step factors to be synthetically lethal with prp45(1–169) (Gahura et al. 2009). Here, we demonstrate using RNA-seq and splicing reporters that prp45(1–169) leads to splicing-dependent pre-mRNA accumulation. ChIP analysis of snRNP recruitment in prp45(1–169) cells revealed the delay of U2, U5, and NTC, implying that splicing in these cells is completed post-transcriptionally. We suggest that Prp45 is important for timely (i.e., cotranscriptional) U2 snRNP association as well as the subsequent stages of spliceosome formation. The effect of the PRP45 allele on U2 resembles the “early roles” reported for PRP8 and PRP28 (Price et al. 2014) in that it occurs before the stable association of the respective splicing factor with pre-mRNA (Ohi et al. 2002).
RESULTS
Cells expressing truncated Prp45 show decreased splicing efficiency
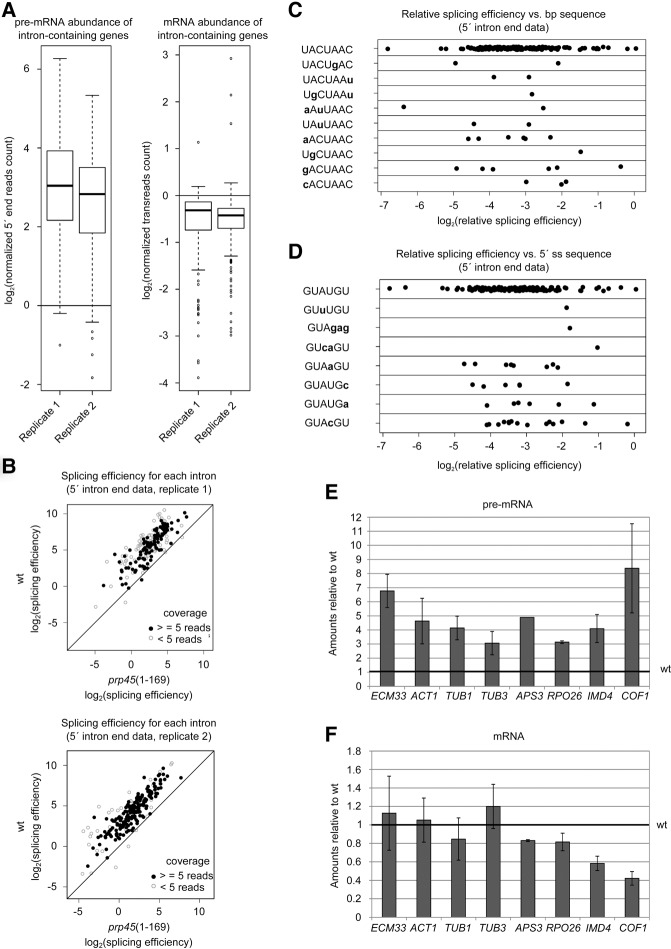
We found previously that prp45(1–169) cells expressing a splicing reporter containing an actin intron generate the reporter mRNA at levels similar to wild-type (WT) cells (Gahura et al. 2009). Only when the reporter contained suboptimal splicing signals (5′ss, 3′ss, or branch site) limiting either the first or second step (Konarska et al. 2006) was mRNA production impaired. A part of the splicing phenotype of Prp45(1–169) may be rescued by coexpression of the missing C-terminal Prp45 region in trans (Gahura et al. 2009). To characterize the extent of the splicing defects caused by Prp45 truncation, we analyzed the levels of unspliced and spliced transcripts in prp45(1–169) cells of a BY4741 genetic background using RNA-seq of two biological replicates. We obtained data (≥5 reads covering the 5′ss) for the 175 and 157 introns in WT, and the 232 and 228 introns in prp45(1–169) cells. In comparison to WT, pre-mRNA was accumulated in the mutant strain with a median of enrichment of about 7–8× (Fig. 1A, left panel). The change of mRNA levels was much less pronounced; the median of the decline in the mutant strain was about 0.7–0.8× (Fig. 1A, right panel). For example, the normalized mRNA and pre-mRNA read counts for the ECM33 gene were 1764 and 9 in the WT strain, respectively, while in the mutant strain the counts were 1588 (mRNA) and 80 (pre-mRNA).
FIGURE 1.
Impairment of Prp45 function leads to pre-mRNA accumulation. (A–C) Transcriptome analysis of prp45(1–169) cells in comparison with WT in two biological replicates. (A) pre-mRNA is accumulated in the prp45 mutant (left panel), while the mRNA levels of intron-containing genes are affected only to a limited extent (right panel). The bottom and top of the box represent the second and third quartiles, respectively; the line represents the median. Whiskers extend up to 1.5× of the interquartile range. (B) The overall relative splicing efficiency is impaired in prp45(1–169) cells. However, this drop in relative splicing efficiency does not show any tendency with respect to the branch point (C) or 5′ss sequence (D). Graphs C and D show results from one biological replicate. Ratios of mutant/WT splicing efficiencies are plotted. (E,F) RT-qPCR quantification of splicing of selected genes. Spliced (E) and unspliced (F) transcripts were quantified by RT-qPCR and normalized to TOM22. Data are expressed relative to WT levels. Error bars represent standard deviations (SD) of at least three biological experiments.
To quantify splicing efficiency, we first calculated the ratio of the transreads (i.e., reads spanning an exon–exon junction) and intronic reads spanning the first intron nucleotide (Převorovský et al. 2016) and then plotted these values against each other. This analysis showed that splicing efficiency is markedly lower in the prp45(1–169) cells than in the WT strain (Fig. 1B). Moreover, splicing efficiency in the prp45 mutant was impaired to a similar extent regardless of the branch site (Fig. 1C) or 5′ss (Fig. 1D) sequences, demonstrating the universal effect of the mutation irrespective of the defining intronic cis-acting features. There was no correlation between relative splicing efficiency in the mutant and branch point to 3′ss distance, overall intron length, transcript abundance (transreads), or ORF length (data not shown). Apparently, longer pre-mRNA-mediated interactions of the (pre)spliceosome with RNA polymerase II (RNA Pol II; on genes with long introns) or more RNA Pol II complexes (on highly expressed genes) were not able to compensate for the deficient assembly process. To verify the RNA-seq results, we measured the levels of unspliced precursors and spliced mRNAs of several endogenous genes by RT-qPCR. We confirmed a three- to sixfold accumulation of all pre-mRNAs tested in prp45(1–169) cells (Fig. 1E,F).
Our data set also contained transreads corresponding to unannotated alternatively spliced RNAs as reported previously (Volanakis et al. 2013; Kawashima et al. 2014; Schreiber et al. 2015). In agreement with the earlier reports, these isoforms represented a minor proportion of spliced RNAs of the particular gene (typically <5%). We detected 60 such previously published alternative splice isoforms, 36 of which were less abundant in the prp45 mutant in comparison to WT (0.5–0.1-fold change; data not shown). It seems that these low-frequency alternative splicing events are more sensitive to prp45(1–169).
Prp45(1–169) causes increased leakage of unspliced reporters
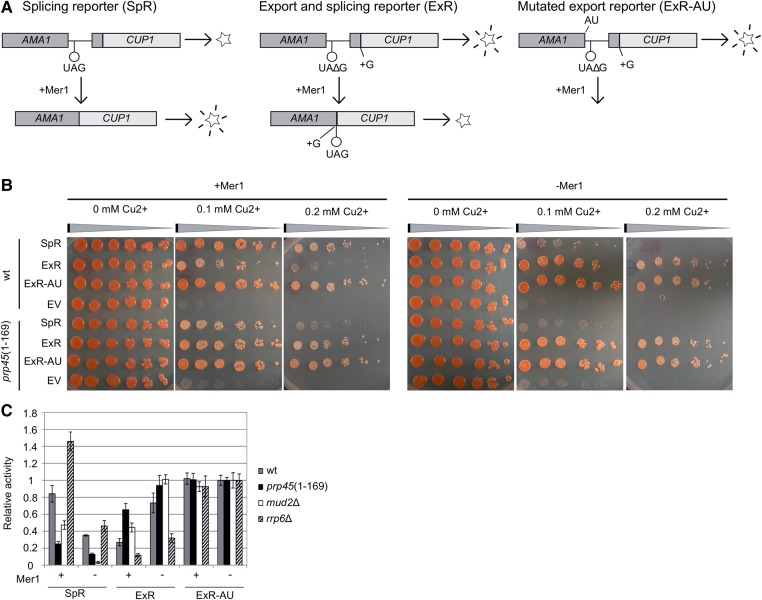
To monitor pre-mRNA accumulation under conditions of regulated splicing, we used a set of AMA1-CUP1-based reporter constructs expressed in cup1Δ cells (Fig. 2A). AMA1, together with the MER2/REC107, MER3/HFM1, and SPO22 genes, is spliced efficiently only in meiotic Mer1-expressing cells. Mer1 binds to the intron's splicing enhancer (Spingola and Ares 2000) and facilitates the recruitment of U1 snRNP, Mud2, and U2 snRNP to pre-mRNA (Spingola et al. 2004; Balzer and Henry 2008). In vegetative cells, the splicing of AMA1 intron-containing reporters can be activated by ectopic expression of Mer1. The reporters provide complementary information: (i) The splicing reporter (SpR) yields functional Cup1 protein only when spliced; (ii) the export reporter (ExR) is translated to Cup1 only when unspliced; and (iii) the control ExR-AU reporter, which has a modified 5′ss and cannot be spliced, yields Cup1 irrespective of changes in splicing. We also used modified reporters, which enable the CUP1 sequence to be replaced by the lacZ gene encoding β-galactosidase, to obtain an independent and quantifiable readout of the system (Fig. 2C).
FIGURE 2.
Splicing/export reporters indicate a defect before the first transesterification in prp45(1–169) cells. (A) Principle of assay using the AMA1 intron-splicing/export reporter substrates (Scherrer and Spingola 2006). AMA1 is a meiotic gene; its splicing in vegetative cells depends on the presence of Mer1, a first-step accessory factor. The reporters SpR and ExR contain STOP codons in different positions; their transcripts can be translated to functional proteins (CUP1 or β-galactosidase) only when spliced (SpR) or not spliced (ExR). The control ExR-AU reporter with mutated 5′ss (GU to AU) is not recognized by the spliceosome and serves as a control to exclude export-specific defects. (B) CUP1-based reporters suggest that prp45(1–169) cells are impaired before the first splicing step. Mid-log cells (cup1Δ background) expressing the indicated reporter plasmid in the absence or presence of Mer1 were harvested and spotted in eightfold dilution series on media containing increasing concentrations of copper ions. “EV” cells bear an empty vector. Compare the growth of SpR/ExR-expressing cells between prp45(1–169) and WT strains in the presence of Mer1 (reporter intron is recognized by the spliceosome) and in its absence. (C) ONPG assay. Mid-log cells expressing the indicated reporter plasmid in the absence or presence of Mer1 were harvested and β-galactosidase activity was measured by ONPG assay. Values were normalized to the activity measured for the ExR-AU reporter in the respective strains in the absence of Mer1. Error bars represent SD of at least three independent experiments.
The spot-tests shown in Figure 2B display the differences in reporter-conferred Cu2+-resistance between prp45(1–169) and WT cells in the presence or absence of Mer1. prp45(1–169) cells show impaired splicing in comparison to WT, which is documented in the complementary readouts of SpR and ExR reporters in Mer1-expressing cells. No differences between mutant and WT were observed in the ExR-AU expressing cells, ruling out unrelated splicing defects. The lacZ reporters confirmed the results of CUP1 reporters in prp45(1–169) cells (Fig. 2C). The comparison of the prp45(1–169) mutant with cells that lack Mud2, a protein involved in the early stages of spliceosome assembly (Abovich et al. 1994), revealed a similar pattern of effects. In contrast, the deletion of the nuclear exosome component RRP6 had an opposite impact on the reporter readouts. The splicing-dependent accumulation of unspliced ExR in prp45(1–169) cells is in agreement with a defect preceding the first splicing step. A defect affecting only the second step, i.e., the accumulation of intron–exon 2 lariat, would not influence the ExR signal.
To support the data obtained on reporter genes by the measurement of an endogenous gene, we monitored the splicing of MER2 by RT-qPCR. We compared MER2 pre-mRNA and mRNA levels in WT and prp45(1–169) in the absence or presence of Mer1. Splicing proceeded efficiently only in the presence of the splicing activator Mer1, as revealed by high mRNA levels in both strains (Fig. 3). However, the MER2 pre-mRNA level dropped to about one half in the presence of Mer1 in WT cells, but remained unchanged in prp45(1–169) cells. We conclude that the unspliced MER2 accumulated in prp45(1–169) cells, in comparison to WT, only under conditions when the splicing was active. These observations demonstrate that pre-mRNA accumulation in prp45(1–169) cells is associated with active splicing.
FIGURE 3.
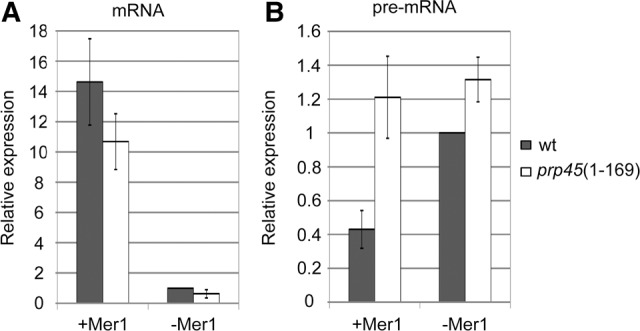
prp45(1–169) cells accumulate an excess of MER2 pre-mRNA only when splicing is active. Mer1, a meiotic factor indispensable for MER2 splicing, was ectopically expressed in vegetative cells. The levels of MER2 mRNA (A) and pre-mRNA (B) were quantified by RT-qPCR. prp45(1–169) cells accumulated more MER2 precursors than WT cells only when Mer1 was present (see pre-mRNA levels, B) and splicing was active (see mRNA levels, A). Error bars represent SD of three biological experiments.
Truncation of Prp45 decreases its recruitment to intron-containing genes and disrupts cotranscriptional splicing
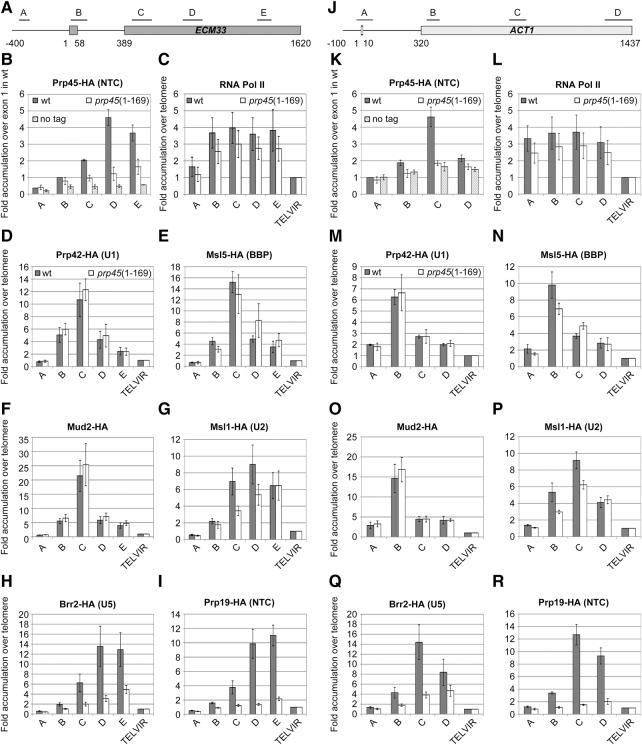
ChIP assays of tagged spliceosomal components have been successfully used to prove the cotranscriptional character of spliceosome recruitment and splicing in both yeast and mammalian systems (Görnemann et al. 2005; Tardiff and Rosbash 2006; Price et al. 2014). The ChIP signals along the gene can be interpreted as the “time” of the stable association of a particular splicing component (Kotovic et al. 2003; Görnemann et al. 2005). Such experiments as well as single-molecule studies also provide evidence in favor of the stepwise spliceosome assembly model (Hoskins et al. 2011; Carillo Oesterreich et al. 2016). To address the association of Prp45 with intron-containing genes, we prepared two strains expressing either C-terminally HA-tagged Prp45 or Prp45(1–169) and performed ChIP-qPCR analysis using an anti-HA antibody. We designed primer pairs to span ECM33 and ACT1 genes with PCR amplicons positioned along the genes (Fig. 4A,J). Prp45 was detected with a maximum signal in the middle of exon 2 (Fig. 4B,K). In contrast, the Prp45(1–169) signal was dramatically lower in all Prp45-positive amplicons, with the tendency to grow gradually toward the 3′ end of genes.
FIGURE 4.
prp45(1–169) delays cotranscriptional spliceosome assembly. ChIP-qPCR analysis of cotranscriptional recruitment of spliceosomal components was performed on the ECM33 and ACT1 genes (A,J; positions of PCR amplicons are indicated). Cells expressing HA-tagged Prp45 or Prp45(1–169) were used for ChIP-qPCR with the anti-HA antibody. Signals were normalized to input values and expressed as the fold enrichment relative to the signal from exon 1 in the Prp45-HA strain. The strain without the tagged protein served as a negative control (B,K). RNA Pol II ChIP signals were obtained using the anti-Rpb3 antibody. Cotranscriptional spliceosome assembly was monitored via the recruitment of HA-tagged components of commitment complex 1 using HA-tagged Prp42 (D,M), Msl5 (E,N), and Mud2 (F,O). Subsequent stages of spliceosome formation were followed using HA-tagged members of U2 snRNP (Msl1; G,P), U5 snRNP (Brr2; H,Q), and NTC (Prp19; I,R). Signals were normalized to input values and expressed as the fold enrichment relative to the signal obtained from the telomeric region (TELVIR; graphs C–I and L–R). Error bars represent SD from at least four independent experiments.
We used the HA-tagged splicing factors Prp42, Msl5, Mud2, Msl1, Brr2, and Prp19 to monitor the presence of U1 snRNP, BBP, Mud2, U2 snRNP, U5 snRNP, and NTC, respectively, along the ECM33 and ACT1 genes. The profiles of U1 snRNP and Msl5/Mud2 showed maxima at the beginning of exon 2, which is in agreement with their roles in the early phases of spliceosome assembly (CC2). The profiles of U2 and U5 snRNPs peaked afterward. The levels of NTC were highest in the second half of exon 2. These data are in agreement with previously reported ChIP analyses (Görnemann et al. 2005; Lacadie and Rosbash 2005). prp45(1–169) mutant cells did not show any alterations of either U1 snRNP (Fig. 4D,M) or Mud2 recruitment (Fig. 4F,O). The profile of Msl5 was also similar to WT; we observed only a slightly higher signal from the middle of exon 2 in prp45 mutants (Fig. 4E,N). In contrast, prp45(1–169) hampered the cotranscriptional recruitment of U2 snRNP (Fig. 4G,P), the U5 snRNP component Brr2 (Fig. 4H,Q), and the NTC member Prp19 (Fig. 4I,R). The profile of Prp19 resembled the behavior of Prp45 and showed a similar decrease of recruitment in prp45(1–169) cells (Fig. 4B,K). ChIP on DBP2 gave principally the same data but of lower resolution, because of the short second exon of this gene (data not shown). We also measured RNA Pol II ChIP to complement the spliceosome recruitment data. RNA Pol II was evenly distributed along the genes with slightly lower signals in mutant cells; we did not observe any accumulation of the RNA Pol II signal on the tested intron-containing genes in the prp45 mutant. Our ChIP results show that Prp45 affects U2 snRNP recruitment and subsequent spliceosome formation events, which supports the conclusion that this factor plays a role in the early stages of spliceosome assembly.
DISCUSSION
Extensive truncation of Prp45 has a mild splicing phenotype in yeast
The RNA-seq data analyzed in this paper document that the prp45(1–169) allele resulted in a seven- to eightfold increase of pre-mRNA levels, whereas most of the corresponding mRNAs were only subtly affected. When comparing the relative splicing efficiencies of genes with noncanonical splice sites against the changes induced by the prp45 mutation, we did not observe any splice site sequence-specific effects (Fig. 1C,D). We examined pre-mRNA and mRNA levels of AMA1-based splicing and export reporters, which depend on Mer1 for 5′ss recognition and splicing. In these assays, which measure the proportion of spliced and leaked (unspliced) pre-mRNAs relative to WT, prp45(1–169) and mud2Δ mutants showed a similar defect, namely a decreased proportion of mRNA-rendered versus leaked pre-mRNA molecules (Fig. 2C). In a separate experiment, we analyzed the pre-mRNA and mRNA levels of the endogenous MER2 gene. prp45(1–169) cells accumulated a relative excess of pre-mRNA only when splicing was active (Fig. 3B).
The phenotypes of mutant PRP45, i.e., elevated pre-mRNA levels, decreased splicing efficiency of first-step limiting substrates (Gahura et al. 2009), and impaired nuclear retention of pre-mRNA, are similar to factors involved in the early stages of prespliceosome formation, such as cap-binding complex (CBC) or Mud2 (Colot et al. 1996; Rain and Legrain 1997). MSL5 (BBP) mutants also affect the splicing of nonconsensus introns and cause leakage of pre-mRNA into the cytoplasm, albeit to a much larger extent than prp45(1–169) (Rutz and Séraphin 2000). The early role of the NTC-related component PRP45 is also supported by the findings of yeast two-hybrid interactions between Snw1, the Prp45/SKIP ortholog in S. pombe, and U2AF23, as well as human SKIP and both U2AF65/35 subunits (the major interaction being with U2AF65) (Ambrozková et al. 2001; Chen et al. 2011). The U2AF65-related protein of yeast, Mud2, has been found to interact with the NTC component Clf1 (Chung et al. 1999). Intriguingly, none of the MS analyses of purified staged spliceosomal complexes have presented an assembly where the U2AF subunits and the NTC-related component Prp45/SKIP were found together (Jurica et al. 2002; Makarov et al. 2002; Ohi et al. 2002; Fabrizio et al. 2009).
The early effects of Prp45 on the association of U2 snRNP with pre-mRNA
We used HA-tagged versions of yeast splicing factors and Prp45 to gain information on the impact of its truncation on cotranscriptional splicing. Prp45(1–169) severely delayed cotranscriptional recruitment of a subset of splicing subcomplexes to ACT1 and ECM33 pre-mRNAs. Because catalysis can occur after the stable association of tri-snRNP and NTC, our results suggest that splicing in prp45(1–169) cells proceeds only after RNA Pol II reaches the end of the gene. The transcriptome analysis [performed with poly(A)-enriched RNA] shows that splicing is eventually accomplished, albeit post-transcriptionally and with lower efficiency (higher steady-state levels of pre-mRNA). Notably, the ACT1 gene, where we observed the delay, shows no second-step splicing defect in prp45(1–169) cells using ACT1 reporter-based primer extension assays (Gahura et al. 2009).
Composition analyses of purified splicing complexes using MS indicated that Prp45 was present as part of the NTC-containing spliceosome (i.e., B complex and later stages) (Jurica et al. 2002; Makarov et al. 2002), but not at earlier points of the splicing cycle (CC2 or A complex) (Hartmuth et al. 2002). However, the truncation of Prp45 affected the spliceosome before tri-snRNP, and NTC are observed to be stably associated with pre-mRNA. The purification schemes used in these MS studies may have precluded the isolation of transiently associating components, including highly flexible Prp45. For example, recent in vitro analysis of human spliceosomes stalled because a mutation in Prp28/DHX8 identified a complex that contained U1, U2, and loosely associated tri-snRNP (called pre-B complex) (Boesler et al. 2016). This particle, which apparently precedes the formation of B complex, evaded identification previously.
Early effects on assembly, i.e., effects preceding the stage when a protein becomes detectable as part of the (pre)spliceosome, have been reported for Prp8, Prp28 (Li et al. 2013; Price et al. 2014), as well as the NTC component Clf1 (Chung et al. 2002; Wang et al. 2003), among others. The mutant form of U5 snRNP-associated DEAD-box ATPase Prp28, Prp28-1, hampers the formation of CC2 complex and subsequent U2 recruitment. Prp28-1 also leads to the delayed dissociation of U1. Importantly, mutations in the bromodomain-like domain of Prp8 are able to suppress both these effects of Prp28-1 (Price et al. 2014). These results argue that Prp8 must gain access to Prp28 during its ATP-independent role in CC2 formation and/or to U1 snRNP (Li et al. 2013). U5 and Prp8 have been repeatedly found crosslinked to 5′ss during the early phase of splicing, both in yeast (Newman et al. 1995) and in Metazoa (Wyatt et al. 1992; Ast and Weiner 1997). The interaction of Prp8 with 5′ss requires U4 and U6, which suggests that the interacting entity is in the form of tri-snRNP (Maroney et al. 2000). The defect in the cotranscriptional recruitment of U2 snRNP caused by Prp45 truncation bears resemblance to the early effects of Prp8 and Prp28. Intriguingly, Prp45 is found in many contacts with Prp8 (see below).
The dissociation of U1 is uncoupled from downstream binding events in prp45(1–169) cells
The profile of U1 was identical for WT and mutant, while the stable association of U2, U5, and NTC was delayed in prp45(1–169) cells (U2<U5<NTC). This implies that U1 displacement is not coordinated with tri-snRNP binding (U6–5′ss pairing) (Lacadie and Rosbash 2005; Tardiff and Rosbash 2006). Information in the literature suggests that U1 recruitment and dissociation are dependent on many factors, including later-acting ones, such as Prp28 or Prp8 (Price et al. 2014). In cotranscriptional assays, the interaction of U1 with pre-mRNA was extended toward the 3′ end of exon 2 in deletion mutants of the cap-binding complex (CBC) (Görnemann et al. 2005) or after U2 snRNA depletion (Tardiff and Rosbash 2006). In contrast, the dissociation of Msl5/Mud2 has not been reported to be directly coupled to U2 binding. Temperature-sensitive mutations of Msl5, which block the formation of CC2 in vitro at nonpermissive temperatures, do not affect prespliceosome assembly (U2 association) (Rutz and Séraphin 1999, 2000). The observed lack of U1 stabilization on exon 2 in the context of reduced U2 binding in prp45(1–169) cells suggests that Prp45 contributes to the coupling of U1 dissociation to subsequent binding events.
U1 snRNP occupancy may decrease too early for the delayed U2/tri-snRNP to bind efficiently. Splicing can be accomplished without U1 only in genes with specific cis elements (Crispino et al. 1996). Overexpression of SR proteins can compensate for the depletion of U1 in HeLa cells (Crispino et al. 1994). Intriguingly, the organism that lacks the Prp45 homolog, Cyanidioschyzon merolae, has a reduced splicing system without U1 snRNP, NTC, and RES proteins (see also below; Stark et al. 2015). Splicing in this reduced setting probably proceeds through the formation of the U2/tri-snRNP complex without the U1 recognition event.
Truncated Prp45 lacks parts that contact U2 components
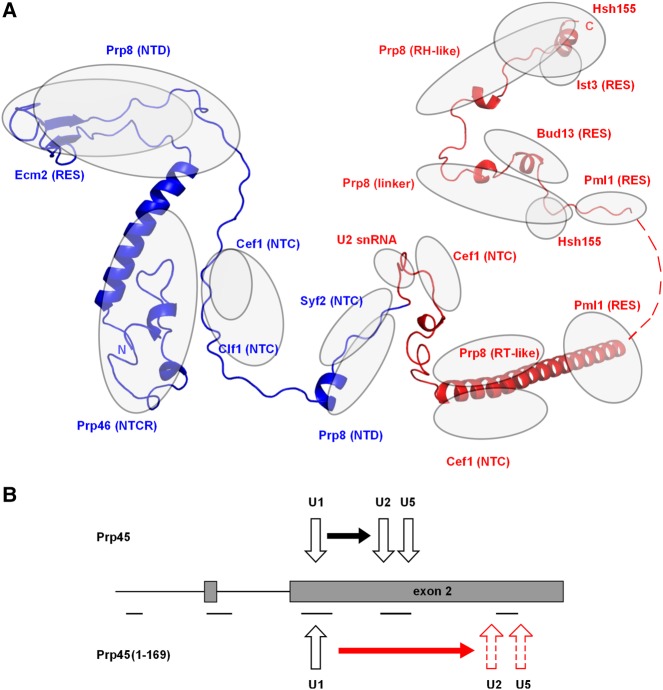
Prp45 is an essential protein and its extensive truncation, which lacks the conserved SNWKN motif as well as the C-terminal 210 amino acids of the protein, is compatible with growth (Gahura et al. 2009). According to available spliceosome structures of S. cerevisiae, S. pombe, and H. sapiens (Yan et al. 2015, 2016; Bertram et al. 2017), Prp45/Snw1/SKIP interacts with a number of proteins across the spliceosome, with contacts to Prp8 occurring along the entire length of Prp45/Snw1/SKIP (Fig. 5A). The NTD domain of Prp8 is bound through the conserved DPL and SNWKN motifs of Prp45 (amino acids 120–122 and 170–174, respectively), and the RT domain is contacted by the C-terminal helix of Prp45. The structure of the S. cerevisiae Bact complex (unlike other available spliceosome structures) also contains structural data for part of the C terminus of Prp45 (amino acids 303–350). In the Bact spliceosome, amino acids 303–350 of Prp45 contact with the linker domain and the RH domain of Prp8 (the amino acids 303–320 and 329–350 of Prp45, respectively). Interestingly, the RH domain changes its position with respect to other domains of Prp8 during Bact–C–C* transitions (Bertram et al. 2017). The contacts and position of the C terminus of Prp45 (amino acids 329–350) stabilized by the RH domain are thus likely to be stage-specific.
FIGURE 5.
Truncation of Prp45 opens a gap between U1 dissociation and U2/U5 snRNP recruitment. (A) Scheme of Prp45 conformation and contacts within the spliceosome. The ribbon diagram combines information from the cryo-EM structure of the S. cerevisiae Bact (5GM6) and human C* (5MQF) complexes (Yan et al. 2016; Bertram et al. 2017). Amino acids missing in the Prp45(1–169) mutant are labeled red (i.e., 170–379). Regions of close contact for Prp45 and proteins/RNA (i.e., encompassing <4 Å intermolecular distances) are indicated by gray ovals. (B) Scheme summarizing the effect of the prp45(1–169) allele on cotranscriptional splicing. The amplicons used for ChIP analysis are indicated by lines under the gene diagram.
With respect to other proteins, Prp45(1–169) extensively contacts Prp46 (wrapping around its globular fold) and Slt11/Ecm2. In addition, contacts also occur with Cef1, Clf1, Cwc15, and Syf2, all of which are NTC components. The portion of Prp45 that is ablated (amino acids 170–379) has closer interactions with Prp8, Syf2, and Cef1, and, in addition, contacts the SF3b scaffold Hsh155 and the RES proteins Ist3, Cwc26/Bud13, and Pml1 (see Yan et al. 2017). Truncated Prp45 ends only N-terminally at the highly conserved SNWKN motif (amino acids 170–174), thus lacking a contact which is present in both S. cerevisiae and S. pombe structures between this motif and the U-rich loop of U2 snRNA of the Bact complex (Yan et al. 2015, 2016). Remarkably, despite the loss of this interaction, the ablation of the SNWKN motif is compatible with splicing. The more extensive C-terminal truncation of prp45(1–131) is viable, but the cells are already severely growth-impaired (K Abrhámová, unpubl.). The contacts of the Prp45 chain between amino acids 131 and 169 include Prp46, Cef1, and Clf1. N-terminal truncations (implicating the lack of Prp46-binding interface parts) are also viable, provided, however, that the C-terminal part is retained (Martínková et al. 2002; Albers et al. 2003).
Because of its intrinsically disordered character and nonglobular nature, it is likely that Prp45 acts in a complex with its partners, not as a free protein. The truncated form may exist in a complex with Prp8 and NTC, similar to the WT protein. However, based on the structural information from the Bact complex, the mutant should be deficient in mediating contacts or regulatory inputs to U2 snRNP and the RES complex (Fig. 5A).
After assembly, Prp45 is present in the spliceosome through the whole splicing cycle (Albers et al. 2003). Using chemical crosslinking, Prp45 has been found to be in the vicinity of intronic RNA in the C complex, where it overlaps the contacts of the RES complex subunit: Snu17, Hsh155, and Prp8 (Schneider et al. 2015). In the human C* complex, the C-terminal helix of SKIP contacts with Prp22 helicase (DHX8) (Bertram et al. 2017). Accordingly, biochemical evidence implicates Prp45 in the recruitment of Prp22 (Gahura et al. 2009). SNW1/SKIP thus not only spans the whole spliceosomal core, but also connects it with effector enzymes positioned at the periphery—with Prp22 helicase via its C-terminal helix (Bertram et al. 2017) and PPIL1 isomerase through its N terminus (Skružný et al. 2001; Xu et al. 2006).
Post-transcriptional splicing is compatible with growth in S. cerevisiae
Data from single-molecule intron-tracking experiments document that the speeds of splicing and transcription are tuned with respect to each other, so that splicing occurs in close vicinity of the RNA Pol II holocomplex (Carillo Oesterreich et al. 2016). This physical proximity may be important for the availability of some splicing factors/regulators. Increasing the distance between the two complexes also changes the circumstances, which are important for splice site choice. For example, more alternative splice sites or regulatory cis sequences can compete (as they emerge from the RNA Pol II exit channel) (Carillo Oesterreich et al. 2016 and references therein). Truncation of Prp45 hinders spliceosome assembly so that splicing can no longer keep pace with transcribing RNA Pol II. Nevertheless, the defect is still compatible with viability at 30°C. While S. cerevisiae cells tolerate extensive truncations of Prp45, S. pombe cells do not [Snw1(1–280) does not complement SNW1 deletion; data not shown]. It is possible that S. pombe is more dependent on the coordination of transcription and splicing, as it has less conserved splicing sequences, more alternative splice sites, and more Metazoan-like organization of its intronome (Wood et al. 2002; Kuhn and Käufer 2003; Carillo Oesterreich et al. 2016).
We conclude that the C-terminal part of Prp45 is important for both U2 and tri-snRNP association kinetics, perhaps enabling them to attain an association-competent state (Fig. 5B). These results thus add Prp45 to the list of spliceosomal components that have been shown to affect spliceosome assembly “from afar,” even though not yet incorporated into the prespliceosome–pre-mRNA complex (Price et al. 2014). This suggests that the interactions that must be considered in order to better explain splice site recognition and spliceosome assembly encompass not only the firmly bound snRNP particles but also the surrounding compartment with transiently interacting components.
MATERIALS AND METHODS
Yeast strains, plasmids, and media
Yeast strains are listed in Table 1. The strains were grown in synthetic (SD) or YPAD media. The PRP45 truncation was generated using an integration cassette obtained by PCR with pFA6-3HA-NatMX6 (Van Driessche et al. 2005) or pFA6-3HA-kanMX6 (Longtine et al. 1998) as a template, or by replacing the PRP45 coding sequence for amino acids 170–379 using the delitto perfetto method (Storici and Resnick 2006).
TABLE 1.
Strains used in this study
Plasmids R1070 (Mer1-expressing plasmid), R1130 (empty vector), SpR-AMA1-CUP1, ExR-AMA1-CUP1, ExR-AU-AMA1-CUP1, SpR-AMA1-lacZ, and ExR-AMA1-lacZ were generously gifted to us by Marc Spingola (Engebrecht et al. 1991; Scherrer and Spingola 2006). Plasmid ExR-AU-AMA1-lacZ was prepared from ExR-AU-AMA1-CUP1 by replacing the CUP1 gene using the KpnI fragment encoding lacZ.
Nucleic acid extraction for transcriptome analysis
Yeast cultures were grown at 30°C in a YPAD medium. Cells (8 mL) were harvested by centrifugation (1000g, 3 min, room temperature) during the exponential growth phase. Cell pellets were stored at −80°C. The pellets were thawed on ice and then 400 µL of extraction buffer (0.1 M Tris–HCl pH 7.5, 1 mM EDTA, 0.1 M LiCl, 1% SDS), 400 µL glass beads, and 400 µL of acid phenol (pH 4.3):chloroform:isoamyl alcohol (25:24:1) solution were added. Cells were disrupted using FastPrep-24 (MP Biomedicals), lysates were spun, and the total RNA was precipitated from the aqueous layer using 96% EtOH. One hundred and fifty micrograms of RNA was taken and adjusted with DEPC-treated water to 175 µL of the total volume. To remove DNA, 20 µL of the DNase buffer and 5 µL of DNase from a MasterPure Yeast RNA Purification Kit (Epicentre Biotechnologies) were added; further steps were performed according to the manufacturer's instructions. After rinsing in 70% EtOH, 500 µL of 70% EtOH was added to the RNA pellets and stored at −20°C.
Transcriptome sequencing
Poly(A)-enrichment and strand-specific RNA-seq library preparation and sequencing (Illumina, 100 nt, paired-end) were carried out by BGI, Hong Kong. The sequencing data are available from the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5149.
RNA-seq data analysis
The quality of sequencing reads was verified with fastQC 0.11.4 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the S. cerevisiae genome r64 using HISAT 2.0.3-beta (Kim et al. 2015). Nonuniquely mapping reads (MAPQ < 10) were filtered out using SAMtools 1.3.1 (Li et al. 2009). Analysis of differential gene expression was performed using R (www.r-project.org/) and the Bioconductor package DESeq2 (Love et al. 2014; Huber et al. 2015).
Splicing efficiency for each intron was calculated as described previously (Převorovský et al. 2016). Briefly, transreads (reads spanning exon–exon junctions, reflecting the abundance of spliced mRNA) were extracted using regtools 0.2.0 (https://regtools.readthedocs.io) and quantified for each junction. The number of reads spanning the most 5′-terminal base of each intron (reflecting unspliced pre-mRNA abundance) was determined using BEDTools 2.25.0 (Quinlan 2014). Splicing efficiency (Fig. 1B) was then calculated as the ratio of transreads (mRNA) to intron-end reads (pre-mRNA).
In the comparisons of pre-mRNA and mRNA abundance of intron-containing genes (Fig. 1A), sequencing read counts were first normalized to account for different sequencing library sizes (i.e., sample sequencing depth). To this end, a group of control intron-less genes was selected, which showed unchanged expression between WT and the prp45(1–169) mutant according to DESeq2 analyses (n = 178 control genes). The median of the mutant/WT mRNA abundance ratios of these control genes was then used to normalize the respective read counts from the mutant to the WT in each biological replicate. Relative pre-mRNA and mRNA abundance for the WT and mutant were then analyzed by comparing the normalized 5′ intron-end read counts and transread counts, respectively.
RT-qPCR
Total RNA was extracted using the MasterPure Yeast RNA Purification Kit (Epicentre Biotechnologies) according to the manufacturer's protocol. Complementary DNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific), random hexamer primers, and 2 µg of RNA as a template. qPCR was performed on a LightCycler 480 II (Roche). Each reaction (total volume of 12.5 µL) consisted of 6.25 µL of MESA GREEN qPCR MasterMix Plus for SYBR Assay, no ROX (Eurogentec), 0.3 mM primers, and 0.05 µL of cDNA. Primer sequences are summarized in Table 2. Each sample was run in triplicate. Results were calculated either using the ΔΔCt method (Livak and Schmittgen 2001) or using a calibration curve calculated from standards included in each run.
TABLE 2.
Primers used in this study

ONPG assay
Cells were cultivated to the middle exponential phase (corresponding to OD 0.5–0.8) in the appropriate media and the equivalent of 4 OD was harvested by centrifugation. The cell pellet was washed with Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4 [pH 7.0], 10 mM KCl, 1 mM MgSO4) and resuspended in the Z-buffer to a final volume of 1 mL. Samples were transferred on ice and 3.4 µL of β-mercaptoethanol was added. The cell suspension (300 µL) was mixed with glass beads and lysed using FastPrep-24 (MP Biomedicals; speed 6; four cycles; cycle duration 15 sec; 5 min on ice between cycles). After lysis, 300 µL of Z-buffer was added and samples were mixed by vortexing. Samples (250 µL each) were incubated with 350 µL of Z-buffer and 80 µL of freshly made ortho-nitrophenyl-β-galactoside (ONPG) solution (4 mg/mL) in Z-buffer at 30°C for 60 min in a shaker. The reaction was then stopped by adding 250 µL of 1 M Na2CO3; samples were centrifuged, and supernatants collected. Absorbance was measured at 420 and 550 nm using the spectrophotometer UV Mini 1240 (Shimadzu). Miller units of activity were calculated using the formula MU = 1000 × [(A420 − 1.75 × A550)]/(T × V × OD600), where OD600 is the optical density of the cell suspension after centrifugation, washing and volume adjustment to 1 mL, T is the incubation time in minutes, and V is the volume of the cell suspension used for the enzymatic reaction in milliliters.
Chromatin immunoprecipitation
Cells were grown in 50 mL YPAD or SD media to OD 0.6 and fixed by shaking for 30 min in 1% formaldehyde (Sigma-Aldrich; 1% solution in medium) at room temperature. Formaldehyde was quenched with 0.125 M glycine for 10 min. Cells were harvested by centrifugation and washed with deionized water; the cell pellet was stored at −80°C. The pellet was thawed in 270 µL of LB buffer (50 mM HEPES pH 7.6, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.1 mM Na-DOC, protease inhibitor cocktail [Serva]), mixed with glass beads and lysed using FastPrep-24 (MP Biomedicals; speed 5.5; three cycles; cycle duration 13 sec; 5 min on ice between cycles). The disrupted cells were collected by centrifugation; the pellet was washed with 600 µL of LB buffer and resuspended in 270 µL of the same buffer. Chromatin was fragmented to ∼500 bp by sonication (Bioruptor sonicator, Diagenode; “high” settings; 15 cycles; 30 sec on followed by 30 sec off; 2°C water bath). Samples were centrifuged and the supernatant collected. Protein concentration was measured by UV absorption at 280 nm using a Nanodrop 2000 (Thermo Scientific) and adjusted to the same level with the LB buffer. Samples were cleared for 1 h with 20 µL of protein A sepharose (GE Healthcare; blocked with 0.5 mg/mL BSA). An aliquot of the sonicated sample was saved as an input; 240 µL was mixed with 10 µL of an anti-HA (HA.11, Covance) or anti-Rbp3 (1Y26[1Y27], Santa Cruz Biotechnology) antibody. A 1-h preincubation was followed by a 3-h incubation with 40 µL of protein A sepharose. All sepharose incubations were carried out on a rotator in a cold room. Sepharose beads were washed six times with IP buffer (50 mM Tris–HCl pH 7.5, 1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40). For decrosslinking and DNA isolation, 100 µL of 10% Chelex (Bio-Rad) was added to the beads, mixed by vortexing, and incubated for 10 min at 95°C. Samples were then cooled down and incubated with 1 µL of proteinase K (20 mg/mL) for 30 min at 55°C on a shaker. Proteinase K was inactivated by incubating for 10 min at 95°C. Samples were centrifuged and the DNA containing the supernatant was collected. The slurry was washed once again with 60 µL of deionized water, centrifuged, and the supernatants pooled. Input DNA was isolated using the same protocol. qPCR analysis was performed as described above with 20 ng of DNA as a template. Primer sequences are summarized in Table 2.
ACKNOWLEDGMENTS
We thank K. Neugebauer for the strains with HA-tagged splicing factors and M. Spingola's laboratory for the AMA1-based reporters. This work was supported by Charles University grants GAUK (Grantová Agentura Univerzity Karlovy) 441711, UNCE (Univerzita Karlova) 204013, SVV244-260435, and PRIMUS/MED/26, and by the European Social Fund Project no. CZ.1.07/2.3.00/30.0022.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061986.117.
REFERENCES
- Abovich N, Liao XC, Rosbash M. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev 8: 843–854. [DOI] [PubMed] [Google Scholar]
- Ajuh P, Kuster B, Panov K, Zomerdijk JCBM, Mann M, Lamond AI. 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J 19: 6569–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers M, Diment ANN, Muraru M, Russell CS, Beggs JD. 2003. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander R, Beggs JD. 2010. Cross-talk in transcription, splicing and chromatin: who makes the first call? Biochem Soc Trans 38: 1251–1256. [DOI] [PubMed] [Google Scholar]
- Ambrozková M, Půta F, Fuková I, Skruzný M, Brábek J, Folk P. 2001. The fission yeast ortholog of the coregulator SKIP interacts with the small subunit of U2AF. Biochem Biophys Res Commun 284: 1148–1154. [DOI] [PubMed] [Google Scholar]
- Ast G, Weiner AM. 1997. A novel U1/U5 interaction indicates proximity between U1 and U5 snRNAs during an early step of mRNA splicing. RNA 3: 371–381. [PMC free article] [PubMed] [Google Scholar]
- Balzer RJ, Henry MF. 2008. Snu56p is required for Mer1p-activated meiotic splicing. Mol Cell Biol 28: 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Liu W-T, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. [DOI] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850. [DOI] [PubMed] [Google Scholar]
- Boesler C, Rigo N, Anokhina M, Tauchert MJ, Agafonov DE, Kastner B, Urlaub H, Ficner R, Will CL, Lührmann R. 2016. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat Commun 7: 11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Gomes N, Pickle L, Jones KA. 2005. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev 19: 1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Yoshida T, Pickle L, Jones KA. 2009. SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol Cell 36: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillo Oesterreich FC, Herzel L, Straube K, Hujer K, Howard J, Neugebauer KM. 2016. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell 165: 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Cheng SC. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199. [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. [DOI] [PubMed] [Google Scholar]
- Chanarat S, Sträßer K. 2013. Splicing and beyond: the many faces of the Prp19 complex. Biochim Biophys Acta 1833: 2126–2134. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang L, Jones KA. 2011. SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev 25: 701–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Mclean MR, Rymond BC. 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Zhou Z, Huddleston KA, Harrison DA, Reed R, Coleman TA, Rymond BC. 2002. Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biochim Biophys Acta 1576: 287–297. [DOI] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev 10: 1699–1708. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Blencowe BJ, Sharp PA. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265: 1866–1869. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Mermoud JE, Lamond AI, Sharp PA. 1996. Cis-acting elements distinct from the 5′ splice site promote U1-independent pre-mRNA splicing. RNA 2: 664–673. [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Wani B, Hayman MJ. 1998. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene 16: 1579–1586. [DOI] [PubMed] [Google Scholar]
- Das RK, Huang Y, Phillips AH, Kriwacki RW, Pappu RV. 2016. Cryptic sequence features within the disordered protein p27Kip1 regulate cell cycle signaling. Proc Natl Acad Sci 113: 5616–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J, Voelkel-Meiman K, Roeder GS. 1991. Meiosis-specific RNA splicing in yeast. Cell 66: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608. [DOI] [PubMed] [Google Scholar]
- Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai X-C, Newman AJ, Nagai K. 2017. Structure of a spliceosome remodelled for exon ligation. Nature 542: 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk P, Půta F, Skruzný M. 2004. Transcriptional coregulator SNW/SKIP: the concealed tie of dissimilar pathways. Cell Mol Life Sci 61: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahura O, Abrhámová K, Skruzný M, Valentová A, Munzarová V, Folk P, Půta F. 2009. Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J Cell Biochem 106: 139–151. [DOI] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Serebriiskii I, Finley RL, Kolonin MG, Gyuris J, Brent R. 1998. Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Prot Sci 82: 20.1.1–20.1.35. [Google Scholar]
- Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63. [DOI] [PubMed] [Google Scholar]
- Hang J, Wan R, Yan C. 2015. Structural basis of pre-mRNA splicing. Science 349: 1191–1198. [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Lührmann R. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci 99: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. 2011. Ordered and dynamic assembly of single spliceosomes. Science 331: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12: 5–14. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Douglass S, Gabunilas J, Pellegrini M, Chanfreau GF. 2014. Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae. PLoS Genet 10: e1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC. 2006. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell 21: 543–553. [DOI] [PubMed] [Google Scholar]
- Korneta I, Bujnicki JM. 2012. Intrinsic disorder in the human spliceosomal proteome. PLoS Comput Biol 8: e1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM. 2003. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol 23: 5768–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, Käufer NF. 2003. Pre-mRNA splicing in Schizosaccharomyces pombe: regulatory role of a kinase conserved from fission yeast to mammals. Curr Genet 42: 241–251. [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M. 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75. [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. 1993. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang W, Xu T, Ramsey J, Zhang L, Hill R, Hansen KC, Hesselberth JR, Zhao R. 2013. Comprehensive in vivo RNA-binding site analyses reveal a role of Prp8 in spliceosomal assembly. Nucleic Acids Res 41: 3805–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Longtine SM, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle RJ. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Romfo CM, Nilsen TW. 2000. Functional recognition of the 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol Cell 6: 317–328. [DOI] [PubMed] [Google Scholar]
- Martínková K, Lebduška P, Skružný M, Folk P, Půta F. 2002. Functional mapping of Saccharomyces cerevisiae Prp45 identifies the SNW domain as essential for viability. J Biochem 132: 557–563. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Teigelkamp S, Beggs JD. 1995. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1: 968–980. [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol 22: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Převorovský M, Hálová M, Abrhámová K, Libus J, Folk P. 2016. Workflow for genome-wide determination of pre-mRNA splicing efficiency from yeast RNA-seq data. Biomed Res Int 2016: 4783841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AM, Görnemann J, Guthrie C, Brow DA. 2014. An unanticipated early function of DEAD-box ATPase Prp28 during commitment to splicing is modulated by U5 snRNP protein Prp8. RNA 20: 46–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain J, Legrain P. 1997. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J 16: 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee C, Urlaub H, Kastner B. 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Abelson J. 1988. An early hierarchic role of P1 small nuclear ribonucleoprotein in spliceosome assembly. Science 242: 1028–1035. [DOI] [PubMed] [Google Scholar]
- Rutz B, Séraphin B. 1999. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5: 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B, Séraphin B. 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J 19: 1873–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer FW, Spingola M. 2006. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA 12: 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Agafonov DE, Schmitzová J, Hartmuth K, Fabrizio P, Lührmann R. 2015. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3′ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet 11: e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber K, Csaba G, Haslbeck M, Zimmer R. 2015. Alternative splicing in next generation sequencing data of Saccharomyces cerevisiae. PLoS One 10: e0140487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skružný M, Ambrozková M, Fuková I, Martínková K, Blahůšková A, Hamplová L, Půta F, Folk P. 2001. Cyclophilins of a novel subfamily interact with SNW/SKIP coregulator in Dictyostelium discoideum and Schizosaccharomyces pombe. Biochim Biophys Acta 1521: 146–151. [DOI] [PubMed] [Google Scholar]
- Spingola M, Ares M. 2000. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol Cell 6: 329–338. [DOI] [PubMed] [Google Scholar]
- Spingola M, Armisen J, Ares M. 2004. Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res 32: 1242–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MR, Dunn EA, Dunn WSC, Grisdale CJ, Daniele AR, Halstead MRG, Fast NM, Rader SD. 2015. Dramatically reduced spliceosome in Cyanidioschyzon merolae. Proc Natl Acad Sci 112: E1191–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Resnick MA. 2006. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol 409: 329–345. [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Rosbash M. 2006. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA 12: 968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Jaladat Y. 2010. The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics 10: 4128–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J. 2005. Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast 22: 1061–1068. [DOI] [PubMed] [Google Scholar]
- Volanakis A, Passoni M, Hector RD, Shah S, Kilchert C, Granneman S, Vasiljeva L. 2013. Spliceosome-mediated decay (SMD) regulates expression of nonintronic genes in budding yeast. Genes Dev 27: 2025–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hobbs K, Lynn B, Rymond BC. 2003. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J Biol Chem 278: 7875–7883. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Wyatt JR, Sontheimer EJ, Steitz JA. 1992. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev 6: 2542–2553. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 27: 275–288. [DOI] [PubMed] [Google Scholar]
- Xu C, Zhang J, Huang X, Sun J, Xu Y, Tang Y, Wu J, Shi Y, Huang Q, Zhang Q. 2006. Solution structure of human peptidyl prolyl isomerase-like protein 1 and insights into its interaction with SKIP. J Biol Chem 281: 15900–15908. [DOI] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CCL, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Huang G, Shi Y. 2016. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353: 904–911. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2017. Structure of a yeast step II catalytically activated spliceosome. Science 355: 149–155. [DOI] [PubMed] [Google Scholar]