Abstract
Riboswitches are noncoding mRNA elements that control gene expression by altering their structure upon metabolite binding. Although riboswitch crystal structures provide detailed information about RNA–ligand interactions, little knowledge has been gathered to understand how riboswitches modulate gene expression. Here, we study the molecular recognition mechanism of the S-adenosylmethionine SAM-I riboswitch by characterizing the formation of a helical stacking interaction involving the ligand-binding process. We show that ligand binding is intimately linked to the formation of the helical stacking, which is dependent on the presence of three conserved purine residues that are flanked by stacked helices. We also find that these residues are important for the formation of a crucial long-range base pair formed upon SAM binding. Together, our results lend strong support to a critical role for helical stacking in the folding pathway and suggest a particularly important function in the formation of the long-range base pair.
Keywords: Bacillus subtilis, RNA structure, gene regulation, S-adenosylmethionine, riboswitch
INTRODUCTION
RNA-mediated genetic regulatory mechanisms are involved in a wide range of physiological processes (Waters and Storz 2009). Among RNA regulators, riboswitches are highly structured elements located in untranslated regions of mRNAs that modulate gene expression upon metabolite binding (Serganov and Nudler 2013). Riboswitches have been shown to act at various genetic levels such as transcription termination, translation initiation, mRNA decay, and splicing (Breaker 2006; Serganov and Nudler 2013) and are involved in the control of genes usually associated with the biosynthesis or transport of target metabolites (Edwards et al. 2007; Schwalbe et al. 2007; Roth and Breaker 2009). Ligand binding to riboswitches usually results in the genetic repression of adjacent genes, but a few cases have been reported where ligand binding positively regulates gene expression (Barrick and Breaker 2007). Riboswitches are composed of two modular regions corresponding to the aptamer domain and the expression platform. For each riboswitch class, the aptamer domain is involved in the specific recognition of a cellular metabolite and is highly conserved at both sequence and structure levels (Serganov and Nudler 2013). In contrast, the expression platform does not show any significant conservation in sequence or structure and controls gene expression upon structural reorganization. Transcriptional pause sites have been recently shown in riboswitch expression platforms to be important for RNA folding and metabolite sensing (Perdrizet et al. 2012; Chauvier et al. 2017). Additionally, transcriptional intermediates corresponding to differently elongated riboswitch transcripts have been characterized, which revealed that riboswitches may selectively fold into different structures along the transcriptional landscape (Watters et al. 2016; Chauvier et al. 2017; Helmling et al. 2017), indicating a sequential folding process (Pan and Sosnick 2006).
S-adenosylmethionine (SAM)-sensing riboswitches are involved in the control of the metabolism, biosynthesis, and transport of cysteine, methionine, and SAM (Wang and Breaker 2008; Garst et al. 2011). SAM riboswitches are unique since at least five structurally different riboswitch families specifically recognize SAM (Wang and Breaker 2008). The Bacillus subtilis metI SAM-I riboswitch acts through a mechanism controlling premature transcription termination (McDaniel et al. 2003; Winkler et al. 2003). In the absence of SAM, the riboswitch adopts a conformation characterized by the presence of an antiterminator (AT), allowing transcription of the downstream-regulated gene (Fig. 1A; McDaniel et al. 2003; Winkler et al. 2003). However, SAM binding to the aptamer favors the formation of the anti-antiterminator (P1 stem), which promotes the folding of the transcription terminator (T) required for premature transcription termination (Fig. 1A). The SAM-I aptamer is organized around a four-way junction composed of four stems (P1–P4) that are joined by unpaired nucleotides (Fig. 1B,C). The aptamer architecture is established by two sets of coaxially stacked helices, one of them comprising stems P1 and P4 and the other involving stems P2 and P3 (Fig. 1C; Montange and Batey 2006; Lu et al. 2010). The coaxial units are oriented relatively to each other to provide the P1–P3 close juxtaposition necessary for the formation of the ligand-binding site (Fig. 1C). Previous studies have determined that the SAM-I aptamer adopts a two-step folding pathway selectively induced by Mg2+ and SAM cofactors (Fig. 1D; Stoddard et al. 2010; Heppell et al. 2011). While the binding of Mg2+ ions prefolds the aptamer to form a pseudoknot interaction as well as the P1–P3 close juxtaposition (FMg), the addition of SAM results in several discrete structural rearrangements such as the P1–P4 stacking interaction (Fig. 1D), all of which lead to the formation of the native state (FNS). However, the precise mechanism by which the ligand-binding site is assembled and how the P1–P4 stacking interaction is involved during the aptamer folding process still remains to be established.
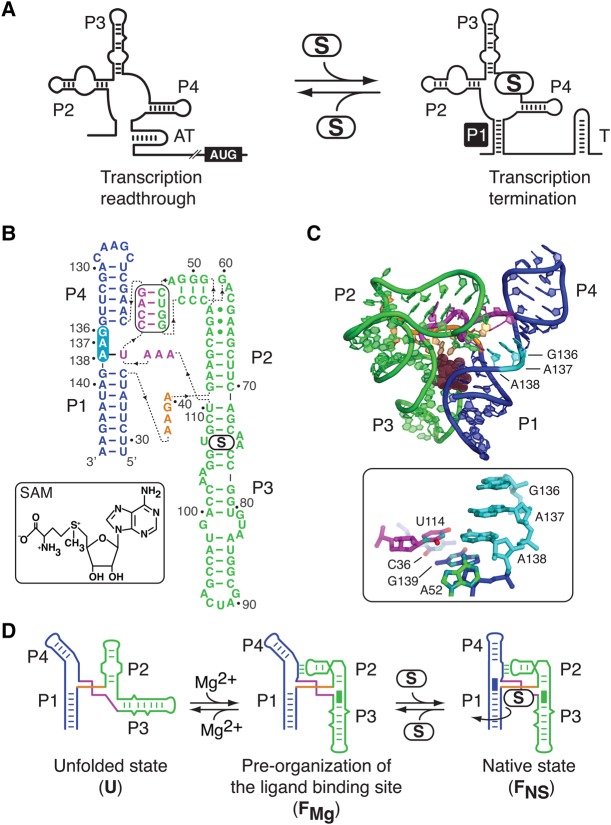
FIGURE 1.
Structure and folding of the SAM-I riboswitch. (A) Schematic representation of the SAM-I riboswitch transcriptional control. In the absence of SAM (S), the folding of an antiterminator structure (AT) permits transcription of the adjacent coding region (AUG). However, upon SAM binding, the formation of the P1 stem (black square) favors the adoption of a transcription terminator (T), leading to premature transcription termination and genetic repression. (B) Secondary structure of the B. subtilis yitJ aptamer based on crystal structures (Montange and Batey 2006; Lu et al. 2010). The P1–P4 and the P2–P3 stacking units are represented in blue and green, respectively. The pseudoknot involving the P2 stem–loop and the J3/4 region is boxed. Nucleotides 136–138 are highlighted in cyan. Dotted lines indicate the polarity of the sequence. (C) The three-dimensional structure of the yitJ SAM-I riboswitch (Montange and Batey 2006). The ligand is represented as spheres. Note the location and the stacked configuration of residues 136–138 located between stems P1 and P4. The local environment of the U114–A138 base pair is boxed. Nitrogen and oxygen atoms of nucleobases are depicted in cyan and red, respectively. (D) The SAM-I folding pathway consists of the unfolded (U), the Mg2+-induced (FMg), and the native state (FNS) structures (Heppell et al. 2011). Formation of the FMg state comprises the P2–P3 helical stacking (green rectangle), the pseudoknot interaction, and the P1–P3 close juxtaposition. The binding of SAM (S) leads to the FNS state characterized by the P1–P4 helical stacking (blue rectangle) and the rotation of the P1 stem (curved arrow). The disposition of the U state is arbitrary, as no information is known regarding the exact position of each helical unit.
Herein, we characterized the ligand recognition mechanism of the SAM-I riboswitch. Using a combination of biochemical and biophysical assays, we investigated the formation of the P1–P4 stacking interaction that is observed upon ligand binding. We found that the stacking interaction is dependent on a region (J1/4 region) that is located at the interface of stems P1 and P4 containing three conserved purine residues. Although the J1/4 region is not located in the expression platform, the structural properties of J1/4 could be important to modulate gene expression by directing the riboswitch ligand-sensing mechanism. By introducing multiple pyrimidines in the J1/4 region, we observed that both ligand binding and helical stacking formation are highly inhibited, indicating that both processes are intimately linked. Our results also show that the formation of a previously observed long-range base pair (Montange and Batey 2006; Lu et al. 2010) is crucial for the adoption of the P1–P4 helical stacking and for ligand binding. In contrast, none of the mutations disrupting the P1–P4 helical stacking were found to perturb the adoption of the FMg state, suggesting that the conserved purine residues are not significantly involved in the formation of the FMg structural conformer. Together, our results lend strong support to a critical role for the P1–P4 stacking interaction in the folding pathway, suggesting a particularly important function for the conserved purine residues of the J1/4 region in the formation of the ligand binding site.
RESULTS
Importance of the J1/4 region for the SAM-I riboswitch transcription regulation activity
According to available crystal structures (Montange and Batey 2006; Lu et al. 2010), the J1/4 single strand region of the SAM-I riboswitch contains three conserved purine residues stacked between helices P1 and P4 (Fig. 1B,C). Sequence alignments have shown that the J1/4 consensus sequence corresponds to R136–R137–A138, where R is a purine (Barrick and Breaker 2007). Although crystal structures show that G136 and A137 are only involved in stacking interactions, A138 also participates in the formation of a long-range Watson–Crick base pair with U114 (Fig. 1B,C, see box). In addition, A138 is involved via its 2′-OH with the N1 position of residue A52 that is located in the P2 stem–loop (Fig. 1C, see box; Montange and Batey 2006).
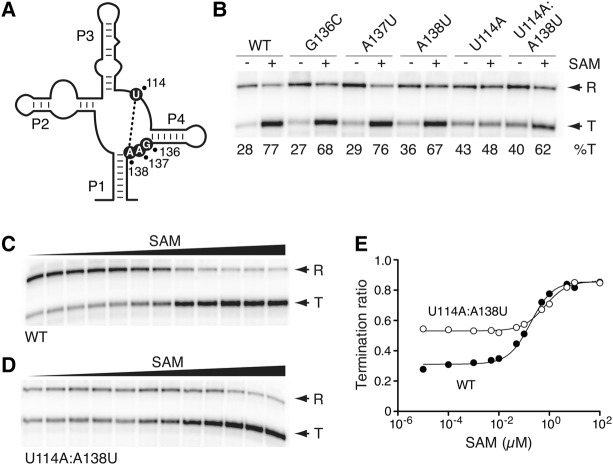
To establish the importance of J1/4 nucleotides for riboswitch regulatory activity, we used single-round in vitro transcription assays as a function of SAM concentration (McDaniel et al. 2003, 2005; Heppell et al. 2011). We used a DNA template containing the B. subtilis glyQS promoter fused to the metI SAM-I riboswitch and a 63-nt sequence located downstream from the transcription terminator. While transcription readthrough occurs in the absence of SAM, the addition of ligand favors premature transcription termination (McDaniel et al. 2003, 2005; Heppell et al. 2011). When transcribing the wild-type riboswitch in the absence of SAM under single-round conditions, a proportion of 28% transcription termination was observed showing a low fraction of transcription termination in the absence of ligand (Fig. 2B). However, the addition of 1 µM ligand resulted in higher transcription termination (77%), in agreement with the riboswitch promoting transcription termination when bound to SAM (McDaniel et al. 2003, 2005; Heppell et al. 2011). We next introduced mutations in the J1/4 region and assessed their effect on riboswitch activity by measuring the extent of transcription termination. Each mutation was designed to correspond to the Watson–Crick complementary residue of the studied position. We obtained a different reaction profile for each tested position. For example, although the introduction of mutations G136C or A137U did not strongly perturb SAM-induced transcription termination (Fig. 2B), the riboswitch activity was modestly affected when introducing an A138U mutation since termination was altered both in the absence and presence of SAM (36% and 67% termination, respectively).
FIGURE 2.
The importance of the J1/4 region for the SAM-I riboswitch transcriptional activity. (A) Schematic representing the secondary structure of the SAM-I riboswitch aptamer. The location of residues 114, 136, 137, and 138 is shown using black circles. The tertiary interaction occurring between positions 114 and 138 is shown by a dotted line. (B) Single-round in vitro transcriptions performed in the absence (−) or in the presence of 1 µM SAM for the wild-type and selected J1/4 riboswitch mutants. Readthrough (R) and prematurely terminated (T) transcripts are indicated on the right and percentages of termination (%T) are shown below the gel. (C,D) Single-round in vitro transcriptions performed as a function of SAM concentration for the wild-type (C) and the U114A:A138U (D) riboswitch mutant. Terminated and readthrough products are indicated on the right. SAM concentrations correspond to 10 pM, 100 pM, 1 nM, 5 nM, 10 nM, 50 nM, 100 nM, 500 nM, 1 µM, 5 µM, 10 µM, and 100 µM. (E) Quantification analysis of single-round in vitro transcriptions shown in C and D. The relative termination ratios are reported for the wild-type (black circles) and the U114A:A138U (white circles) riboswitch mutant.
Since crystal structures show that A138 is involved in a long-range Watson–Crick base pair with residue U114 (Fig. 2A; Montange and Batey 2006; Lu et al. 2010), we assessed the importance of the base pair U114–A138 for riboswitch activity by using the U114A mutant. We found that transcription termination ratios obtained both in the absence and presence of SAM were very close (43% vs. 48%, respectively), indicating that the riboswitch regulatory activity was strongly perturbed (Fig. 2B). To verify that this loss of activity resulted from the disruption of the U114–A138 base pair or from the identity change of U114, we made an additional construct restoring the base pair interaction (Fig. 2B, U114A:A138U). We observed that the ligand-dependent transcription termination was partially recovered in this context (40% and 62% in the absence and in the presence of SAM, respectively), indicating that the identity is important, but not crucial for riboswitch activity. To further understand the influence of the U114–A138 base pair on riboswitch activity, we performed in vitro transcriptions as a function of ligand concentration. By fitting the extent of transcription termination using a simple two-state model (Blouin and Lafontaine 2007; Heppell et al. 2011), we determined the SAM concentration corresponding to half of the variation in transcription termination (referred to as T50), effectively revealing the propensity of the riboswitch to perform ligand binding in a transcriptional context. As expected, when performing transcription reactions using the wild-type riboswitch, we observed that the proportion of prematurely terminated mRNA increased over the range of SAM concentration (Fig. 2C). Fitting the data to the model gave a T50 value of ∼183 nM (Fig. 2E), in good agreement with our previous study (Heppell et al. 2011). When performing the same experiment using the U114A:A138U mutant, a T50 value of ∼673 nM was obtained (Fig. 2D,E), indicating that the riboswitch mutant requires ∼3.7 times more ligand to efficiently achieve premature transcription termination. Although our results show that the nature of the base pair U114–A138 is not critical for riboswitch activity, they suggest its importance for ligand sensing in a transcriptional context.
The FMg state relies on the pseudoknot interaction but not on the J1/4 region
To decipher the influence of J1/4 residues on riboswitch activity, we used the fluorescence resonance energy transfer (FRET) technique to monitor the folding of the SAM-I aptamer. FRET is a powerful method to study conformational changes of macromolecules in solution (Lilley 2009). By using FRET assays, we determined the relative extent of energy transfer (EFRET) occurring between a pair of donor and acceptor fluorophores attached at defined positions. We used fluorescein and Cy3 since these fluorophores are well characterized (Norman et al. 2000). The efficiency of transfer is inversely proportional to the sixth power of the distance between both fluorophores and is defined by a Förster distance (R0) of 56 Å for the fluorescein–Cy3 pair (Norman et al. 2000). Variations in FRET efficiency can be interpreted as distance changes occurring between fluorophores, which is useful to deduce RNA global changes and to understand associated folding pathways (Bassi et al. 1997; Lafontaine et al. 2002; Lemay et al. 2006; Lilley 2009; Blouin et al. 2011; Heppell et al. 2011; Wood et al. 2012; Kim et al. 2014; Rinaldi et al. 2016).
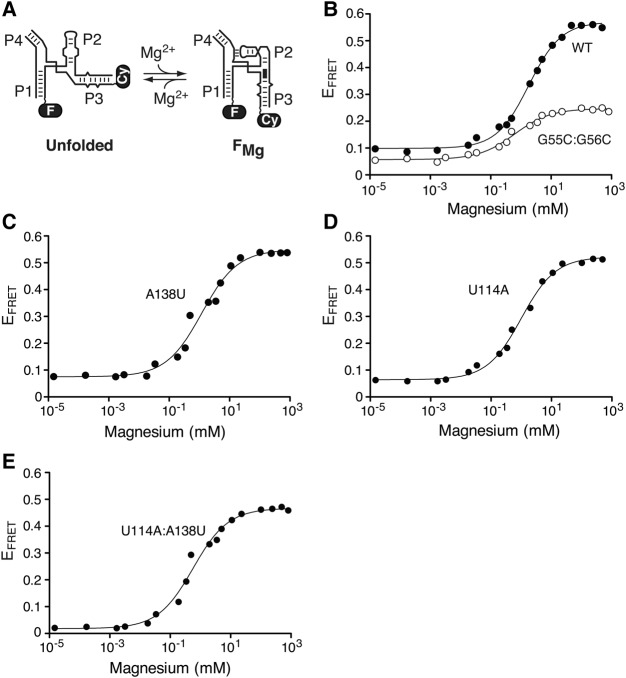
Using this approach, we monitored the formation of the FMg state by using a construct of the SAM-I aptamer labeled with fluorescein and Cy3 on P1 and P3 stems, respectively (Fig. 3A). Representative data for the wild-type aptamer indicated that a large increase in EFRET is obtained when raising the Mg2+ concentration (Fig. 3B). These data show that the binding of Mg2+ ions is important for the close juxtaposition occurring between stems P1 and P3 that is found in the FMg state (Heppell et al. 2011), and which is important for the formation of the ligand-binding site (Montange and Batey 2006; Lu et al. 2010). By fitting FRET data to a model describing an all-or-none conformational transition induced by the binding of Mg2+ ions, we determined the Mg2+ concentration ([Mg2+]1/2) at which the transition is 50% complete. The P1–P3 transition occurs in the low mM range and is characterized by a value of [Mg2+]1/2 = 1.6 ± 0.1 mM (Fig. 3B; Supplemental Table S1). This value is similar to what is typically obtained for other junction-containing riboswitches (Blouin and Lafontaine 2007; Baird and Ferre-D'Amare 2010; Wood et al. 2012) and ribozymes (Bassi et al. 1997; Zhao et al. 2000; Lafontaine et al. 2002; Penedo et al. 2004; Pereira et al. 2008). The P1–P3 folding transition was determined to be highly dependent on the formation of the pseudoknot interaction since the introduction of mutations (G55C:G56C) destabilizing the interaction severely perturbed the transition by reducing the [Mg2+]1/2 value to 0.71 ± 0.09 mM (Supplemental Table S1). The severity of the destabilizing effect was also apparent from the P1–P3 transition not adopting the WT folded state even at saturating Mg2+ concentrations, as observed from the low EFRET value (∼0.25) of the mutant (Fig. 3B). These results suggest that the close juxtaposition of P1–P3 requires both the binding of Mg2+ ions and the formation of the pseudoknot interaction, consistent with our previous findings (Heppell et al. 2011).
FIGURE 3.
The sequence of the J1/4 region is not important for the formation of the FMg state. (A) The Mg2+-dependent folding process of the SAM-I aptamer is shown with fluorescein (F) and Cy3 (Cy) dyes located on stems P1 and P3, respectively. The Mg2+-dependent P2-P3 helical stacking is shown by a black rectangle. (B) Folding transitions of the wild-type (black circles) and G55C:G56C aptamer mutant (white circles) as a function of Mg2+ concentration. Experimental data were fitted (lines) in each case by nonlinear regression to a simple two-state model where the binding of metal ions induces a structural change. (C,D) Folding transitions of the A138U (C) and U114A (D) mutant aptamers as a function of the Mg2+ concentration. Both A138U and U114A mutations are predicted to disrupt the A138:U114 long-range Watson–Crick base pairing. (E) Folding transitions of the U114A:A138U mutant aptamer as a function of Mg2+ concentration. The U114A:A138U aptamer is predicted to allow the formation of the A138-U114 long-range Watson–Crick base pair.
We studied the sequence requirements of the J1/4 region for the adoption of the FMg conformation. In each experiment, we measured the FRET vector 1–3 as a function of the Mg2+ concentration, and all calculated [Mg2+]1/2 values are reported in Supplemental Table S1. When introducing mutations at position 136 (G136C) or 137 (A137U), no significant variation of the [Mg2+]1/2 value was observed for the P1–P3 folding transition (Supplemental Table S1), suggesting that these residues are not critically involved in the formation of the FMg state. We also investigated the importance of the U114–A138 long-range base pair by altering each nucleotide for its respective Watson–Crick complement (A138U and U114A). For both A138U (Fig. 3C) and U114A (Fig. 3D), although the [Mg2+]1/2 value was found to decrease to ∼1 mM (Supplemental Table S1), the FRET transition exhibited an EFRET value similar to the WT aptamer (Fig. 3B) at a saturating ion concentration. In contrast to the G55C:G56C mutant, these results suggest that both A138U and U114A mutants are able to undergo the P1–P3 folding transition in the presence of sufficient Mg2+ ions. The simultaneous introduction of both mutations (Fig. 3E, U114A:A138U) further decreased the [Mg2+]1/2 value to 0.66 ± 0.09 mM (Supplemental Table S1), suggesting a combined detrimental effect of U114A and A138U. However, the presence of a saturating magnesium concentration resulted in a P1–P3 folded state (EFRET ∼0.45) close to the WT aptamer (EFRET ∼0.55). Together, these results show that the close juxtaposition of helices P1 and P3 does not rely on the identity of J1/4 residues or the U114–A138 base pair, but rather on the formation of the pseudoknot interaction.
The U114–A138 base pair is important for the formation of the FNS state
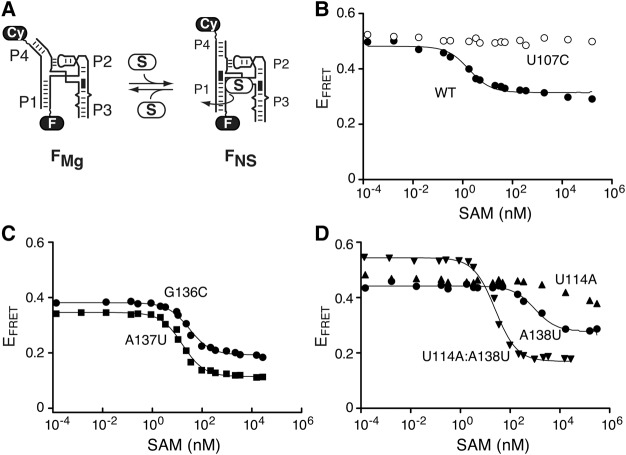
We previously showed that ligand binding to the SAM-I aptamer leads to the formation of the FNS state, which includes several structural changes such as the coaxial stacking of helices P1 and P4 (Fig. 1D; Heppell et al. 2011). The P1–P4 structural transition was monitored by, respectively, incorporating fluorescein and Cy3 in stems P1 and P4 and by measuring FRET as a function of SAM concentration (Fig. 4A). By using this approach, it is possible to estimate the dissociation constant (Kd) for the formation of the aptamer–ligand complex. When monitoring the P1–P4 folding process in the context of the wild-type aptamer, we found that the EFRET decreased from ∼0.5 to ∼0.3 (Fig. 4B) and that the transition was characterized by a value of Kd = 1.8 ± 0.4 nM (Supplemental Table S2). This value is in good agreement with previously established affinities for different aptamer variants (Winkler et al. 2003; Montange et al. 2010; Heppell et al. 2011). As expected, no SAM-dependent transition was detected when using a U107C mutant preventing ligand binding (Fig. 4B), consistent with SAM binding being crucial for the FRET transition (Heppell et al. 2011). For all tested constructs, estimated Kd values are reported in Supplemental Table S2.
FIGURE 4.
The U114:A138 long-range base pair is crucial for the formation of the FNS state. (A) The SAM-induced folding process of the aptamer is shown with fluorescein (F) and Cy3 (Cy) dyes located on stems P1 and P4, respectively. The formation of the helical stacking is shown by a black rectangle located at the interface of both P1 and P4 stems. The curved arrow represents the helical twist that is observed upon SAM binding. (B) Folding transitions of the wild-type (black circles) and U107C mutant (white circles) aptamers as a function of the SAM concentration. The lack of significant change prevented the analysis of the U107C mutant. (C) Folding transitions of the G136C (circles) and A137U (squares) mutant aptamers as a function of the SAM concentration. (D) Folding transitions of the U114A (up-pointing triangles), A138U (circles), and U114A:A138U (down-pointing triangles) as a function of the SAM concentration. Note that U114A was not analyzed due to the lack of significant FRET change.
We found the P1–P4 structural transition to be quite sensitive to mutations introduced in the J1/4 region. While the SAM binding affinity was affected by ∼10-fold for replacements at positions G136 and A137 (Fig. 4C; Supplemental Table S2), the affinity was reduced by more than 500-fold for the A138U mutant (Fig. 4D; Supplemental Table S2). Moreover, the folding transition was severely perturbed when introducing the U114A mutation (Fig. 4D), for which a reliable affinity constant could not be calculated. In contrast, when both U114A and A138U mutations were introduced simultaneously (U114A:A138U mutant), a Kd value of 25 ± 1 nM was obtained corresponding to an ∼14-fold difference with the wild-type (Fig. 4D; Supplemental Table S2). These data clearly suggest that the presence of a base pairing between positions 114 and 138 is crucial for the P1–P4 structural transition, which is in contrast to its lack of importance for the formation of the FMg conformer (Fig. 3). Furthermore, because there is a good correlation between P1–P4 structural transition and the formation of the 114–138 long-range base pair, our results suggest that the 114–138 base pair is important for riboswitch ligand binding.
Given that FRET assays monitor the formation of the P1–P4 folding transition, it is possible that mutations strongly affecting the ligand-dependent P1–P4 helical stacking interaction (e.g., A138U) could still allow SAM binding but not P1–P4 stacking, effectively uncoupling ligand binding from helical stacking. We tested this hypothesis by using a competition assay with a wild-type aptamer having a 2-aminopurine (2AP) at position 138 that exhibits strong fluorescence quenching upon SAM binding (Fig. 5A; Heppell and Lafontaine 2008; Heppell et al. 2009, 2011). In this binding assay, the SAM affinity of a nonfluorescent competitor aptamer is assessed by incubating it in the presence of the fluorescent construct (Fig. 5B). The fluorescence signal is thus directly related to the binding affinity of competitors and can be normalized to compare the affinity of various SAM binding aptamers. Control experiments showed that incubation of the fluorescent aptamer with SAM (Fig. 5A) resulted in low 2AP fluorescence emission (Fig. 5C, “–”). However, the addition of an excess of the nonfluorescent wild-type aptamer (Fig. 5B) resulted in high 2AP fluorescence signal (Fig. 5C, WT), indicating that 2AP fluorescence results from SAM sequestration by the competing nonfluorescent wild-type aptamer. When experiments were done using competing aptamers such as U114A and A138U, no 2AP fluorescence emission was detected, showing that both aptamers did not efficiently bind SAM (Fig. 5C). These results are in agreement with FRET data showing that both aptamers exhibit severely reduced binding affinity toward SAM (at least 500-fold reduction, Fig. 4D). In contrast, the use of the U114A:A138U competing aptamer resulted in significant 2AP fluorescence emission (Fig. 5C), consistent with its ability to sequester SAM (Fig. 4D). Together, these results indicate that the adoption of the P1–P4 helical stack is intimately associated with SAM binding, and consequently, that the formation of the U114–A138 long-range base pair is crucial for ligand binding.
FIGURE 5.
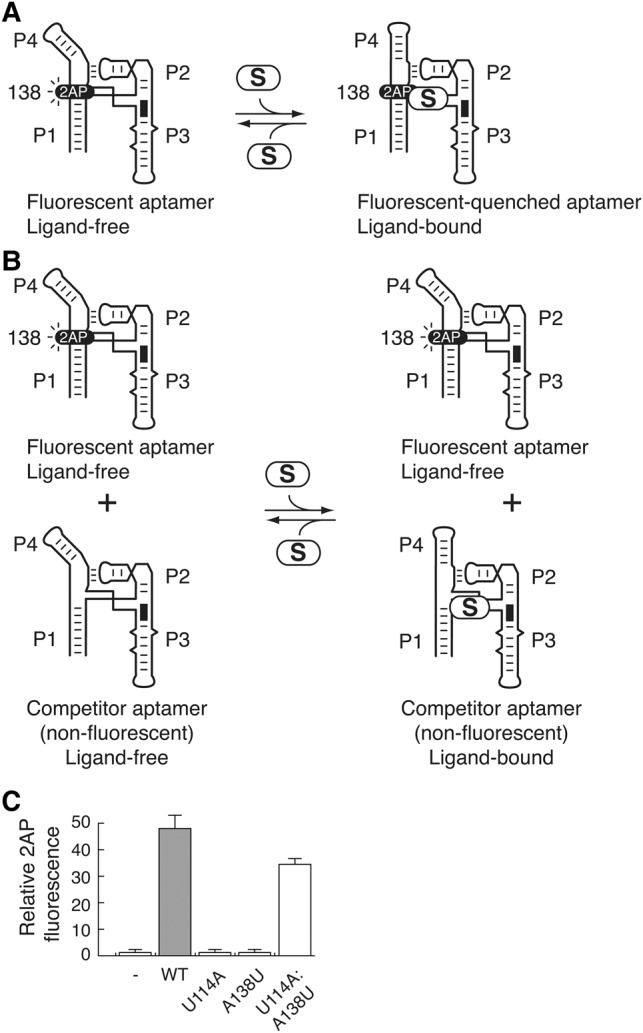
U114A and A138U mutants do not efficiently perform SAM binding. (A) Schematic representing the fluorescence emission of a 2-aminopurine (2AP) fluorophore at position 138 of the SAM aptamer. A wild-type aptamer containing a 2AP at position 138 is used as a fluorescent reporter that is quenched upon SAM binding. In the presence of Mg2+, the aptamer is folded in a conformation allowing 2AP fluorescence. However, SAM (S) binding to the aptamer results in the P1–P4 helical stacking and 2AP fluorescence quenching. (B) Schematic representing the competition assay in which the fluorescent aptamer is incubated in the presence of a nonfluorescent competitor aptamer. The binding of SAM by the competitor allows the fluorescent aptamer to remain free from SAM binding, thus leading to a 2AP fluorescence increase. (C) Relative binding (%) is reported as a function of competitor aptamer. In the absence of any competitor (−), very little fluorescence can be detected due to the efficient 2AP fluorescence quenching that occurs through SAM binding. The relative binding is shown for the wild-type (WT), U114A, A138U, and U114A:A138U aptamer competitors.
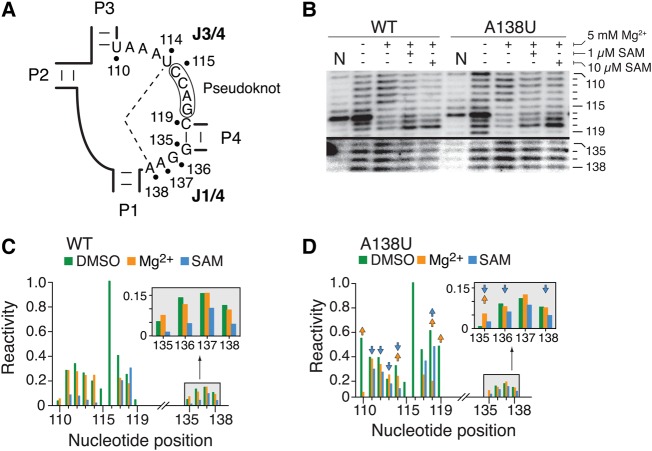
SHAPE analysis of the SAM-I A138U aptamer variant
To characterize the folding of the J1/4 region (Fig. 6A), we performed selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Merino et al. 2005) both on the wild-type and A138U mutant, the latter exhibiting a strong reduction in the formation of the P1–P4 helical stacking (Fig. 4D). SHAPE enables users to extract information about the relative flexibility of RNA linkages, which can effectively be used to study riboswitch conformational changes (Lu et al. 2010; Steen et al. 2010; Stoddard et al. 2010; Hennelly and Sanbonmatsu 2011; Heppell et al. 2011). This assay is particularly relevant to detect nucleotide flexibility against constrained RNA regions, where 2′-OH groups in flexible parts are more susceptible to react with the electrophile N-methylisatoic anhydride (NMIA).
FIGURE 6.
NMIA probing of the wild-type and A138U aptamers. (A) Schematic representing the J1/4 and J3/4 regions of the SAM-I aptamer. The long-range U114–A138 base pair is shown by a dashed line. (B) SHAPE modifications of the wild-type and A138U SAM-I aptamers. Reactions were performed in the absence or presence of Mg2+ or SAM. Only regions corresponding to J3/4 (110–119 nt) and J1/4 (135–138 nt) are shown. Control reactions were done in which DMSO was used instead of NMIA (N). The complete gel is shown in Supplemental Figure S1A. (C,D) NMIA chemical reactivity (normalized units) of the wild-type (C) and A138U (D) aptamer according to nucleotide positions. Reactions were performed in the absence or in the presence of 5 mM Mg2+ (Mg2+), or in the presence of 5 mM Mg2+ and 1 µM SAM (SAM). Only regions corresponding to J3/4 (110–119 nt) and J1/4 (135–138 nt) are shown. Arrows represent a folding effect in the A138U mutant that is at least 1.8-fold smaller (downward pointing) or greater (upward pointing) than the value observed in the context of the wild-type aptamer. The insets represent a magnification of the 135–138 nt region. Complete quantifications are shown in Supplemental Figure S1B.
Representative SHAPE profiles of J1/4 and J3/4 regions for the wild-type aptamer are shown in Figure 6B. The complete gel and quantification are shown in Supplemental Figure S1A,B, respectively. When the wild-type aptamer was subjected to NMIA in the presence of Mg2+, a strong protection was observed in the J3/4 region at nucleotides 115–117, which are involved in the formation of the pseudoknot interaction (Fig. 6B,C). As expected from the magnesium-induced folding (Lu et al. 2010; Stoddard et al. 2010; Heppell et al. 2011), changes in NMIA reactivity were also observed in other residues located throughout the aptamer domain (Supplemental Fig. S1A). These changes are located in the L2 loop, the kink-turn motif and stems P3 and P4 (Supplemental Fig. S2A). Furthermore, the addition of 1 µM or 10 µM SAM resulted in strong protections in the P3 stem (Supplemental Fig. S2A) and in the region J3/4 (110–114 nt) (Fig. 6B,C). We also detected smaller but significant protections in the J1/4 region (Fig. 6B,C, positions 135–138), indicating that these residues become more constrained upon SAM binding. Overall, our SHAPE results are in agreement with the bulk of available chemical probing data showing that both Mg2+ and SAM cofactors promote the folding of the aptamer domain in the presence of Mg2+ and SAM, respectively (Winkler et al. 2003; McDaniel et al. 2005; Lu et al. 2010; Stoddard et al. 2010; Heppell et al. 2011).
SHAPE experiments were also done using the A138U mutant exhibiting a strongly altered SAM-dependent P1–P4 stacking (Fig. 6B). SHAPE reactivity showed several variations (at least 1.8-fold change) when comparing the Mg2+-induced folding of the A138U mutant to the wild-type aptamer (see arrows in Fig. 6D; Supplemental Figs. S1B, S2B). For example, we observed that U110 exhibited an increased NMIA protection in the presence of Mg2+ ions, suggesting that the A71–U110 base pair exhibits increased Mg2+-induced changes in the context of the A138U mutant when compared to the wild type (Supplemental Fig. S2C). A significant difference of Mg2+ reactivity was also observed for the C119–G135 base pair (Supplemental Fig. S2C). Given that both base pairs showing differences in NMIA reactivity are located in proximity to the aptamer core junction, it suggests that the A138U mutation perturbs the local structure of the aptamer core domain (Supplemental Fig. S2C). SHAPE reactions performed in the presence of 1 µM SAM revealed a reaction pattern different from the wild-type aptamer (Fig. 6B,D). For instance, lower NMIA protections induced by SAM were observed for regions J3/4 (positions 110–114) and J1/4 (positions 135, 136, and 138) (Fig. 6D; Supplemental Fig. S2C). Additional differences were also detected in the P3 stem region (Supplemental Fig. S2C, residues 79–82), consistent with the lower affinity of the A138U aptamer for SAM. However, when performing SHAPE assays in the presence of 10 µM SAM, an overall SHAPE profile very similar to the wild-type in the presence of SAM was obtained (Fig. 6B; Supplemental Fig. S1A), consistent with the A138U aptamer binding SAM at high concentrations (Fig. 4D).
Multiple uracils in the J1/4 region strongly alters riboswitch folding and activity
According to our results, as long as the U114–A138 base pair is preserved, sequence changes in the J1/4 region do not strongly impair the adoption of the P1–P4 helical stacking (Fig. 4; Supplemental Table S2). Given that conserved purines of the J1/4 region are stacked between stems P1 and P4 (Fig. 1C; Montange and Batey 2006; Lu et al. 2010), we speculated that the phylogenetic preference for purines (Winkler et al. 2003) could be related to their high stacking propensity (Patel et al. 1987; Woodson and Crothers 1988; Kalnik et al. 1989; Nikonowicz et al. 1990; Rosen et al. 1992; Rosen and Patel 1993). We determined the tolerance of J1/4 to the presence of pyrimidines by analyzing additional naturally occurring J1/4 sequences containing pyrimidine residues (see Supplemental Table S2 for description of mutants). When needed, the identity of U114 was altered to preserve the long-range Watson–Crick base pair with residue 138.
Using FRET assays to monitor the formation of the P1–P4 stacking, we first analyzed the effect of naturally occurring J1/4 sequences containing cytosines at various positions. When monitoring the P1–P4 helical stacking process with a construct containing a cytosine at position 137, thus corresponding to a J1/4 sequence of 5′-GCA-3′ (Supplemental Table S2), we measured a SAM affinity of 21 ± 1 nM, suggesting that the identity of A137 is not highly important for the P1–P4 stacking interaction. Similarly, when the A137C mutation was introduced in a different J1/4 sequence, we obtained a comparable SAM-binding affinity (ACA sequence, Kd = 19 ± 1 nM) (Supplemental Table S2). We also analyzed the effect of a cytosine at position A138 with an aptamer containing an AAC J1/4 sequence (Supplemental Table S2). In this case, a U114G mutation was introduced to preserve the 114–138 long-range base pair. Using this construct, we observed a ligand-binding affinity of 15 ± 4 nM (Supplemental Table S2), indicating that an A138C substitution is not detrimental for the P1–P4 stacking transition. A J1/4 sequence containing multiple cytosines (CCA) yielded results very similar to those obtained with single cytosine-containing mutants (Supplemental Table S2, Kd = 26 ± 1 nM), consistent with the P1–P4 stacking transition not being perturbed by the presence of cytosines in the J1/4 region.
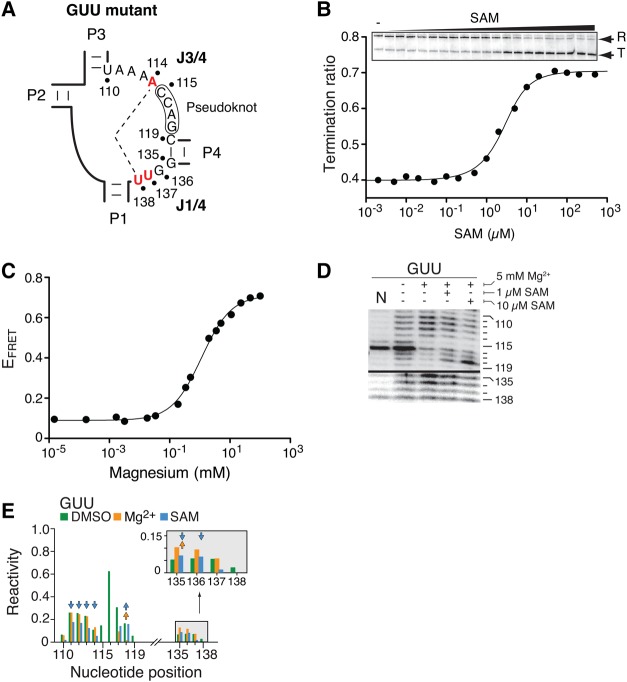
A similar analysis was also performed by selecting natural J1/4 sequences containing uracils. Similar to sequences containing cytosines, aptamers containing a single uracil at position 136 (UAA sequence) or 137 (GUA sequence) exhibited binding affinities in the low nM range (Supplemental Table S2), suggesting that ligand binding is not strongly altered. In contrast, a natural sequence containing both A137U and A138U mutations (Fig. 7A, GUU sequence) showed an ∼300-fold reduction in ligand-binding sensing (Supplemental Table S2). These results indicate that a cumulative disrupting effect is observed when both A137U and A138U mutations are simultaneously introduced. Using single-round transcription experiments, we next analyzed the ability of the GUU variant to prematurely terminate transcription as a function of SAM concentration. As expected, we found that the riboswitch activity was significantly affected given that the T50 value was increased to ∼1.3 µM (Fig. 7B), which is approximately sevenfold higher than what was obtained for the wild-type riboswitch (Fig. 2C). We also studied the Mg2+-induced P1–P3 folding transition of the GUU variant by following the P1–P3 vector using FRET assays (Fig. 7C). The magnesium titration revealed that the conformational change was not significantly perturbed compared to the wild-type ([Mg2+]1/2 = 1.1 mM), consistent with our data showing that J1/4 mutations do not affect the P1–P3 folding transition (Fig. 3). We next performed SHAPE experiments to characterize the structure of the GUU aptamer variant (Fig. 7D,E). Similar to the A138U mutant, Mg2+-dependent protections located in the P3 stem (positions 81 and 82) were less pronounced in the GUU variant when compared to the wild-type aptamer (Supplemental Figs. S3A,B, S4A). Furthermore, SAM-dependent NMIA protections in the P3 stem and J3/4 region (positions 111–114 and 118) were decreased in the context of the GUU aptamer when compared to the wild-type (Fig. 7D,E; Supplemental Figs. S3, S4). Such decreased variations in residue protection were also detected in the J1/4 region for positions 135 and 136, consistent with the reduced ability of the mutant to perform SAM binding (Supplemental Table S2). Reactions performed with 10 µM SAM resulted in a pattern consistent with ligand binding (Fig. 7D,E), as observed for the A138U mutant. Overall, SHAPE reactivity of the GUU aptamer mutant showed that both J1/4 and J3/4 regions have smaller SAM-dependent changes when compared to the wild-type (Supplemental Fig. S4B), which is similar to what was obtained for the A138U mutant (Supplemental Fig. S2C). However, in contrast to both the wild-type and A138U, a very low reactivity for positions 137 and 138 in the GUU aptamer was observed in all tested conditions, suggesting that the presence of both A137U and A138U mutations lead to a decrease in NMIA reactivity (Fig. 7D). Together, our SHAPE results suggest that the J1/4 GUU sequence does not strongly alter the Mg2+-induced conformation, but rather affects the SAM-dependent folding step of the aptamer.
FIGURE 7.
The introduction of multiple pyrimidines strongly disrupts ligand binding and riboswitch regulation. (A) Schematic representing the GUU SAM-I aptamer exhibiting multiple nucleotide substitutions in the core domain. Mutated nucleotides U114A, A137U, and A138U are shown in red. The long-range base pair is preserved due to the Watson–Crick complementarity of both U114A and A138U mutations. (B) Single-round in vitro transcriptions performed as a function of SAM concentrations for the GUU riboswitch mutant. Terminated and readthrough products are indicated on the right. SAM concentrations correspond to 0 nM (−), 1 nM, 2 nM, 5 nM, 10 nM, 20 nM, 50 nM, 100 nM, 200 nM, 500 nM, 1 µM, 2 µM, 5 µM, 10 µM, 20 µM, 50 µM, 100 µM, 200 µM, 500 µM, 1 mM, and 2 mM. (C) Folding transitions of the GUU aptamer mutant as a function of the Mg2+ concentration. The FRET efficiency is increasing as expected from the close juxtaposition of stems P1 and P3. (D) SHAPE modifications of the GUU SAM-I aptamer. Reactions were performed in the absence or presence of Mg2+ or SAM. Only regions corresponding to J3/4 (110–119 nt) and J1/4 (135–138 nt) are shown. A control reaction was done in which DMSO was used instead of NMIA (N). The complete gel is shown in Supplemental Figure S3A. (E) NMIA chemical reactivity (normalized units) of the GUU aptamer mutant according to nucleotide positions. Reactions were performed in the absence (DMSO) or in the presence of 5 mM Mg2+ (Mg2+), or in the presence of 5 mM Mg2+ and 1 µM SAM (SAM). Only regions corresponding to J3/4 (110–119 nt) and J1/4 (135–138 nt) are shown. The inset represents a magnification of the 135–138 nt region. Arrows represent a folding effect in the A138U mutant that is at least 1.8-fold smaller (downward pointing) or greater (upward pointing) than the value observed in the context of the wild-type aptamer. The complete quantification is shown in Supplemental Figure S3B.
DISCUSSION
The outcome of riboswitch gene regulation ultimately relies on the riboswitch folding pathway, which is intimately involved in the sensing of target cellular metabolites. In our previous study of the SAM-I riboswitch (Heppell et al. 2011), we found that the folding pathway of the aptamer domain follows a two-step hierarchical process selectively induced by metal ions (FMg) and ligand (FNS), each of them being important for riboswitch regulatory control. The adoption of the FMg state is characterized by the formation of a pseudoknot interaction and coaxial stacking involving stems P2 and P3. In addition, the FMg state comprises the close juxtaposition of stems P1 and P3 that is important for the formation of the ligand-binding site. Although there is still no clear mechanism explaining how stems P1 and P3 are brought together into close juxtaposition, it is likely that the pseudoknot is used as a lever to position the P2–P3 coaxial unit in a conformation locating both P1 and P3 stems close to each other (Fig. 1D; Heppell et al. 2011). This model is supported by 2-aminopurine fluorescence and FRET data showing that the pseudoknot is important for both P2–P3 helical stacking and P1–P3 close juxtaposition, respectively (Fig. 3B; Heppell et al. 2011). A similar structural arrangement has previously been described for the VS ribozyme in which helices 2 and 5 are juxtaposed to form a cleft that is required for the docking of the stem–loop substrate (Hiley and Collins 2001; Lafontaine et al. 2002; Lipfert et al. 2008; Suslov et al. 2015). In the case of the VS ribozyme, the close juxtaposition is induced by the binding of Mg2+ ions that occurs in the low millimolar range, suggesting that the VS ribozyme and the SAM-I aptamer fold within a similar range of Mg2+ concentration (Stoddard et al. 2010; Heppell et al. 2011). Furthermore, SAXS analysis revealed that the P1 and P3 stems of the SAM-I riboswitch exhibit conformational heterogeneity in the presence of Mg2+, which was suggested to be important for SAM sensing (Stoddard et al. 2010). This conformational heterogeneity was directly observed in sm-FRET experiments where fast dynamics could be observed between U and FMg in the low Mg2+ millimolar range (Heppell et al. 2011).
Ligand binding to the SAM-I aptamer results in the formation of the FNS state (Heppell et al. 2011) characterized by the coaxial stacking of stems P1 and P4 as well as a helical rotation of the P1 stem (Fig. 1D). Crystal structures show that the single strand J1/4 region constituted of three purines is stacked between stems P1 and P4 (Montange and Batey 2006; Lu et al. 2010). Because phylogenetic data indicate that the J1/4 region exhibits a marked preference for purine residues (Barrick and Breaker 2007), known to have high stacking propensity (Patel et al. 1987; Woodson and Crothers 1988; Kalnik et al. 1989; Nikonowicz et al. 1990; Rosen et al. 1992; Rosen and Patel 1993), it suggests that J1/4 purine residues are important to contribute to the stacking interaction between stems P1 and P4. Moreover, the recently demonstrated importance of the P4 stem for ligand binding and transcription control (Heppell et al. 2011) suggests that the P1–P4 stacking interaction is a fundamental folding process that is intimately involved in the riboswitch regulatory mechanism.
Here, we have characterized the influence of the J1/4 region and the U114–A138 base pair on the folding and activity of the SAM-I riboswitch. Remarkably, our FRET results have shown that while the formation of the FMg state is not sensitive to the identity of J1/4 residues, its formation is highly dependent on the pseudoknot structure (Supplemental Table S1). In contrast, we found that the global conformation of the FNS state is sensitive to changes introduced in the J1/4 region. Although replacement of G136 and A137 did not significantly alter the transition of the P1–P4 FRET vector (Fig. 4C), the introduction of either U114A or A138U mutation dramatically perturbed the ligand-dependent folding of the aptamer (Fig. 4D). However, efficient ligand binding was obtained by the simultaneous introduction of both U114A and U138U mutations (Figs. 4D, 5C), which re-enables the formation of the Watson–Crick base pair. The ∼14-fold decrease in binding affinity observed for the compensatory mutant compared to the wild-type aptamer (Supplemental Table S2) also suggests that the identity of the base pair is important for ligand binding. Since the 2′OH of A138 is involved with A52, it is likely that the identity of A138 is important to support this tertiary interaction. Furthermore, the presence of an A138U mutation could possibly stabilize the P1 stem by forming an extra base pair with A37, which is consistent with a higher transcription termination ratio in the absence of ligand (Fig. 2B). The importance of A138 is in agreement with the natural base pair representation showing that ∼81% of natural representatives exhibit the U114–A138 base pair. In this context, the composition of the J1/4 sequence is primarily constituted by AAA (28%), GAA (18%), and AGA (14%) (Supplemental Table S3), indicating that a homopurine adenine tract is the most represented sequence. The second most represented base pair is the A114–U138 combination (∼5%), which is associated with J1/4 sequences corresponding to AAU (1.6%), CAU (1.3%) and GAU (<1%) (Supplemental Table S3), showing a lower degree of preference for purine residues. Other base pair configurations are also possible but are only predicted in lower proportion, suggesting that they are not optimal for riboswitch activity or that they are tuned for particular growth conditions. Surprisingly, the C114:G138 variant is not naturally occurring, suggesting that additional evolutionary pressures are involved. A study performed by Henkin and coworkers previously reported the natural variability of SAM riboswitches found in B. subtilis (Tomsic et al. 2008). Among the 11 SAM representatives, only the metK variant was found to be unresponsive to SAM when assessed by in vitro transcription assays. Visual inspection of metK secondary structure reveals that a possible A114:C138 combination is present in the core region. This combination could explain why the riboswitch is not SAM-responsive like other natural representatives. In this regard, the composition of the long-range base pair could modulate gene expression in a similar manner to the P4 stem that was recently shown to influence the riboswitch response (Heppell et al. 2011).
From available crystal structures (Montange and Batey 2006; Lu et al. 2010), no clear mechanism can be deduced describing the underlying molecular process implying the stacking of the P1–P4 helical domain, the role of the U114–A138 base pair and SAM recognition. Nevertheless, several lines of evidence suggest that the base pair is intimately linked to ligand binding and P1–P4 helical stacking. First, we showed here using 2AP competition assays that the presence of the 114:138 base pair is essential for SAM binding (Fig. 5C). Second, FRET assays indicated a strong link between base pair formation and the ligand-dependent P1–P4 helical stacking (Fig. 4D). Third, SHAPE data showed that disruption of the base pair in the A138U mutant requires an increased SAM concentration to obtain a folding pattern similar to the wild type (Supplemental Fig. S1A). Finally, in vitro transcription assays revealed that the ligand-dependent riboswitch activity is more efficient in the presence of the base pair (Fig. 2B; Lu et al. 2010). Therefore, based on our study, we postulate a ligand-binding mechanism where SAM recognition by the aptamer involves the formation of the U114–A138 base pair, which in turn allows the stacking of both P1 and P4 stems by virtue of J1/4 stacking interactions. Although our data do not exclude that the U114–A138 base pair is formed prior to SAM binding, the fact that the J1/4 region is reorganized and stabilized upon SAM binding, as revealed by standard and temperature-dependent SHAPE experiments (Lu et al. 2010; Stoddard et al. 2010; Heppell et al. 2011), suggests that the base pair is formed only when the aptamer is bound to SAM. This is supported by FRET assays showing that the FMg conformer is not affected when disrupting the base pair interaction by individually mutating residues U114 or A138 (Supplemental Table S1). Since no direct interaction between the bound SAM and the U114–A138 base pair is observed in crystal structures (Montange and Batey 2006; Lu et al. 2010), the formation of the base pair likely results from the reorganization of the aptamer core region upon SAM binding (Lu et al. 2010; Stoddard et al. 2010; Heppell et al. 2011). Whether the ligand-dependent formation of the base pair is conformationally captured or induced-fit is not currently known and will require further single-molecule FRET assays to shed light on the underlying molecular mechanism (Haller et al. 2011; Heppell et al. 2011). Furthermore, the relationship between J1/4 folding and the recently observed P1 helical rotation will need to be addressed since no clear mechanism is currently known regarding the ligand-dependent rotation of the P1 stem (Heppell et al. 2011).
While FRET experiments indicated that the P1–P3 close juxtaposition is not affected when altering the J1/4 sequence, SHAPE assays showed that the A138U mutation does not only affect ligand binding, but also the Mg2+-induced folding of the aptamer core region (Fig. 6B; Supplemental Fig. S2C, base pairs A71–U110 and C119–G135). Although such a mutation impairs ligand binding (Figs. 4D, 5C, 6D), a productive RNA–ligand complex can still be obtained given that higher ligand concentrations allow ligand binding and riboswitch activity as observed using in vitro transcription (Fig. 2B), FRET (Fig. 4D), and SHAPE assays (Fig. 6B; Supplemental Fig. S1A). These results suggest that the aptamer core is conserved not only to allow efficient binding, but also to avoid the presence of mutations that could trap the aptamer in an inefficient conformation for ligand binding, as previously seen for purine riboswitches (Delfosse et al. 2010; Tremblay et al. 2011). Importantly, this deleterious effect on ligand binding is more pronounced when introducing additional uracils (Fig. 7; Supplemental Table S2, GUU and UUU mutants). According to SHAPE data (Figs. 6B, 7D), no strong protections are observed for J1/4 residues in the presence of SAM for the A138U and GUU mutants. The lack of strong protection could be partly explained by the fact that there is a general lower SHAPE reactivity in the J1/4 region of the wild-type, making less straightforward the structural interpretation about the J1/4 region. Such a low reactivity is also observed in the A138U and GUU mutants. However, lower NMIA reactivity has previously been observed for pyrimidine residues (Wilkinson et al. 2009), suggesting that the absence of strong NMIA protections is a consequence of a lower NMIA reactivity toward uracils contained in the J1/4 region. Moreover, the modulation of transcription termination by both riboswitch mutants in the presence of SAM suggests that the U114–A138 base pair is formed in both mutants (Figs. 2B, 7B). Thus, our results suggest that the presence of multiple uracils likely destabilizes the J1/4 stacking interaction, which would result in the inefficient P1–P4 helical stacking and ligand-binding activity. Clearly, more work will be required to elucidate the exact molecular mechanism involving the U114–A138 base pair interaction and its role in ligand-binding recognition.
In conclusion, our study provides strong evidence that the ligand-induced P1–P4 helical stacking interaction is very important for the folding and thus the regulatory activity of the SAM-I riboswitch. Therefore, this work further expands the repertoire of mechanisms that riboswitches use to recognize target metabolite and to regulate gene expression. Given that pairwise helical stacking interactions are found across several riboswitches (Serganov and Nudler 2013), we expect that ligand-induced stacking interaction is a widespread mechanism used by riboswitches to stabilize their ligand-bound form.
MATERIALS AND METHODS
Single-round in vitro transcription assays
DNA templates for transcriptions were obtained as previously described (Blouin et al. 2011; Heppell et al. 2011). Sequences for all templates are provided in the Supplemental Information. Briefly, the promoter sequence of glyQS was fused to a B. subtilis metI variant to produce a transcription start site 20 nucleotides (nt) upstream of the aptamer domain. The PCR product was designed to initiate transcription with a dinucleotide GpC with a halt at position +17 nt achieved by omission of CTP. The readthrough transcript is 252-nt long and includes 41 nt after the AUG start codon. However, SAM binding to the riboswitch results in premature transcription termination resulting in a shorter transcript of ∼185 nt.
FRET analysis
Wild-type constructs used for FRET analysis have been described previously (Heppell et al. 2011). Sequences for constructs are provided in the Supplemental Information. Mutations performed for aptamer variants are indicated in the text. Aptamer molecules were assembled from a combination of T7 RNAP transcripts (Milligan et al. 1987) and synthetic RNA strands coupled with fluorescein or Cy3 dyes. FRET data were collected at 10°C using 10 pmol of fluorescent aptamers in 90 mM Tris–borate pH 8.3 and 100 mM KCl and analyzed as previously described (Blouin et al. 2011; Heppell et al. 2011). Magnesium ions and SAM were titrated over a range of concentrations as indicated in the text. The Mg2+-dependent proportion of folded aptamer was fitted to a simple two-state model assuming an all-or-none conformational transition (Lemay et al. 2006). The SAM-induced folding process was analyzed by fitting the FRET transition using a stoichiometric-binding model (Turner et al. 2005). Experiments were performed at least three times and exhibited very similar uncertainties (<5%). Reported errors are the standard uncertainties of the data from the best-fit theoretical curves. The standard measurement uncertainty is approximated by the SD of the points from the fitted curve (Flannery et al. 1992; Rist and Marino 2001).
SHAPE analysis
SHAPE reactions were performed as previously described (Blouin et al. 2011; Heppell et al. 2011). Sequences for aptamers are provided in the Supplemental Information. Briefly, each reaction was prepared using 1 pmol of aptamer incubated in the absence or presence of cofactors as indicated. RNA molecules were incubated for 80 min at 37°C in N-methylisatoic anhydride (NMIA) dissolved in DMSO. Reverse transcription was used to detect modified positions. Individual band intensities were integrated using SAFA (Das et al. 2005).
2-Aminopurine (2AP) fluorescence
Sequences for all fluorescent aptamers are provided in the Supplemental Information. Data were collected using a Quanta Master fluorimeter at 15°C in a reaction buffer containing 50 mM Tris–HCl (pH 8.3), 25 mM NaCl, 10 mM MgCl2. Competition assays were performed as previously described (Heppell et al. 2011) and were done using a concentration 1.5 µM aptamer labeled with 2AP, 0.8 µM SAM, and 1.5 µM of the competing nonfluorescent aptamer in 50 mM Tris–HCl (pH 8.3), 25 mM NaCl and 10 mM MgCl2. Reagents were incubated for a duration of 15 min prior to measurements. 2AP fluorescence was recorded at least three times and all exhibited very similar uncertainties (<5%).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Carlos Penedo, Alain Lavigueur, and members of the laboratory for discussion and critical reading of the manuscript. This work was supported by the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada. D.A.L. was supported through a Fonds de Recherche Santé Québec Senior Scholar.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061796.117.
REFERENCES
- Baird NJ, Ferre-D'Amare AR. 2010. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi GS, Murchie AI, Walter F, Clegg RM, Lilley DM. 1997. Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study. EMBO J 16: 7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Lafontaine DA. 2007. A loop-loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13: 1256–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Chinnappan R, Lafontaine DA. 2011. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res 39: 3373–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2006. Riboswitches and the RNA world. In The RNA world, 3rd ed (ed. Cech TR, et al.), Vol. 4, pp. 1–15. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Chauvier A, Picard-Jean F, Berger-Dancause JC, Bastet L, Naghdi MR, Dubé A, Turcotte P, Perreault J, Lafontaine DA. 2017. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat Commun 8: 13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. 2005. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfosse V, Bouchard P, Bonneau E, Dagenais P, Lemay JF, Lafontaine DA, Legault P. 2010. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res 38: 2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Klein DJ, Ferre-D'Amare AR. 2007. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol 17: 273–279. [DOI] [PubMed] [Google Scholar]
- Flannery BP, Teukolsky SA, Vetterling WT. 1992. Numerical recipes in Fortran, 2nd ed Cambridge University Press, Cambridge, UK. [Google Scholar]
- Garst AD, Edwards AL, Batey RT. 2011. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol 3: a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. 2011. Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7: 393–400. [DOI] [PubMed] [Google Scholar]
- Helmling C, Wacker A, Wolfinger MT, Hofacker IL, Hengesbach M, Fürtig B, Schwalbe H. 2017. NMR structural profiling of transcriptional intermediates reveals riboswitch regulation by metastable RNA conformations. J Am Chem Soc 139: 2647–2656. [DOI] [PubMed] [Google Scholar]
- Hennelly SP, Sanbonmatsu KY. 2011. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res 39: 2416–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppell B, Lafontaine DA. 2008. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry 47: 1490–1499. [DOI] [PubMed] [Google Scholar]
- Heppell B, Mulhbacher J, Penedo JC, Lafontaine DA. 2009. Application of fluorescent measurements for characterization of riboswitch-ligand interactions. Methods Mol Biol 540: 25–37. [DOI] [PubMed] [Google Scholar]
- Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA. 2011. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol 7: 384–392. [DOI] [PubMed] [Google Scholar]
- Hiley SL, Collins RA. 2001. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J 20: 5461–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalnik MW, Norman DG, Swann PF, Patel DJ. 1989. Conformation of adenosine bulge-containing deoxytridecanucleotide duplexes in solution. Extra adenosine stacks into duplex independent of flanking sequence and temperature. J Biol Chem 264: 3702–3712. [PubMed] [Google Scholar]
- Kim H, Abeysirigunawarden SC, Chen K, Mayerle M, Ragunathan K, Luthey-Schulten Z, Ha T, Woodson SA. 2014. Protein-guided RNA dynamics during early ribosome assembly. Nature 506: 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DM. 2002. The global structure of the VS ribozyme. EMBO J 21: 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. 2006. Folding of the adenine riboswitch. Chem Biol 13: 857–868. [DOI] [PubMed] [Google Scholar]
- Lilley DM. 2009. The structure and folding of branched RNA analyzed by fluorescence resonance energy transfer. Methods Enzymol 469: 159–187. [DOI] [PubMed] [Google Scholar]
- Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM. 2008. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16: 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A. 2010. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J Mol Biol 404: 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. 2003. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci 100: 3083–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Henkin TM. 2005. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol Microbiol 57: 1008–1021. [DOI] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231. [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT. 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Montange RK, Mondragon E, van Tyne D, Garst AD, Ceres P, Batey RT. 2010. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol 396: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonowicz EP, Meadows RP, Gorenstein DG. 1990. NMR structural refinement of an extrahelical adenosine tridecamer d(CGCAGAAT TCGCG)2 via a hybrid relaxation matrix procedure. Biochemistry 29: 4193–4204. [DOI] [PubMed] [Google Scholar]
- Norman DG, Grainger RJ, Uhrin D, Lilley DM. 2000. Location of cyanine-3 on double-stranded DNA: importance for fluorescence resonance energy transfer studies. Biochemistry 39: 6317–6324. [DOI] [PubMed] [Google Scholar]
- Pan T, Sosnick T. 2006. RNA folding during transcription. Annu Rev Biophys Biomol Struct 35: 161–175. [DOI] [PubMed] [Google Scholar]
- Patel DJ, Shapiro L, Hare D. 1987. Nuclear magnetic resonance and distance geometry studies of DNA structures in solution. Annu Rev Biophys Biophys Chem 16: 423–454. [DOI] [PubMed] [Google Scholar]
- Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DM. 2004. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10: 880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdrizet GA II, Artsimovitch I, Furman R, Sosnick TR, Pan T. 2012. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc Natl Acad Sci 109: 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MJ, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. 2008. Single VS ribozyme molecules reveal dynamic and hierarchical folding toward catalysis. J Mol Biol 382: 496–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi AJ, Lund PE, Blanco MR, Walter NG. 2016. The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts. Nat Commun 7: 8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rist M, Marino J. 2001. Association of an RNA kissing complex analyzed using 2-aminopurine fluorescence. Nucleic Acids Res 29: 2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MA, Patel DJ. 1993. Conformational differences between bulged pyrimidines (C-C) and purines (A-A, I-I) at the branch point of three-stranded DNA junctions. Biochemistry 32: 6563–6575. [DOI] [PubMed] [Google Scholar]
- Rosen MA, Live D, Patel DJ. 1992. Comparative NMR study of An-bulge loops in DNA duplexes: intrahelical stacking of A, A-A, and A-A-A bulge loops. Biochemistry 31: 4004–4014. [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR. 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78: 305–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. 2007. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl 46: 1212–1219. [DOI] [PubMed] [Google Scholar]
- Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen KA, Malhotra A, Weeks KM. 2010. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease. J Am Chem Soc 132: 9940–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. 2010. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18: 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslov NB, DasGupta S, Huang H, Fuller JR, Lilley DMJ, Rice PA, Piccirilli JA. 2015. Crystal structure of the Varkud satellite ribozyme. Nat Chem Biol 11: 840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. 2008. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol 190: 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay R, Lemay JF, Blouin S, Mulhbacher J, Bonneau E, Legault P, Dupont P, Penedo JC, Lafontaine DA. 2011. Constitutive regulatory activity of an evolutionary-excluded riboswitch variant. J Biol Chem 286: 27406–27415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DM. 2005. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA 11: 1192–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Breaker RR. 2008. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol 86: 157–168. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, Lucks JB. 2016. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol 23: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. 2009. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA 15: 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707. [DOI] [PubMed] [Google Scholar]
- Wood S, Ferre-D'Amare AR, Rueda D. 2012. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem Biol 7: 920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA, Crothers DM. 1988. Structural model for an oligonucleotide containing a bulged guanosine by NMR and energy minimization. Biochemistry 27: 3130–3141. [DOI] [PubMed] [Google Scholar]
- Zhao ZY, Wilson TJ, Maxwell K, Lilley DM. 2000. The folding of the hairpin ribozyme: dependence on the loops and the junction. RNA 6: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.