Abstract
RNA thermometers regulate expression of some genes involved in virulence of pathogenic bacteria such as Yersinia, Neisseria, and Salmonella. They often function through temperature-dependent conformational changes that alter accessibility of the ribosome-binding site. The 5′-untranslated region (UTR) of the htrA mRNA from Salmonella enterica contains a very short RNA thermometer. We have systematically characterized the structure and dynamics of this thermometer at single-nucleotide resolution using SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) assays. Our results confirm that the htrA thermometer adopts the predicted hairpin conformation at low temperatures, with conformational change occurring over a physiological temperature regime. Detailed SHAPE melting curves for individual nucleotides suggest that the thermometer unfolds in a cooperative fashion, with nucleotides from both upper and lower portions of the stem gaining flexibility at a common transition temperature. Intriguingly, analysis of an extended htrA 5′ UTR sequence revealed not only the presence of the RNA thermometer, but also an additional, stable upstream structure. We generated and analyzed point mutants of the htrA thermometer, revealing elements that modulate its stability, allowing the hairpin to melt under the slightly elevated temperatures experienced during the infection of a warm-blooded host. This work sheds light on structure–function relationships in htrA and related thermometers, and it also illustrates the utility of SHAPE assays for detailed study of RNA thermometer systems.
Keywords: SHAPE, structure probing, RNA thermometer, htrA
INTRODUCTION
RNA thermometers are structural elements that affect expression of the mRNAs in which they reside in direct response to temperature change (Kortmann and Narberhaus 2012; Shapiro and Cowen 2012; Grosso-Becera et al. 2015). These elements do not appear to require protein cofactors for proper function, and they can control expression of reporter genes in heterologous systems. RNA thermometers commonly occur in the 5′-untranslated region (UTR) of bacterial mRNAs that encode proteins involved in the heat shock response or in pathogenic virulence. In these cases, they adopt temperature-sensitive structures that prevent ribosomal binding to the mRNA at low temperatures. These structures melt open under higher temperatures, allowing translational initiation. Control of gene expression via structural transitions in an RNA element allows rapid response to changing environmental conditions, augmenting regulation on the transcriptional level. RNA thermometers have also been implicated in several eukaryotic systems (Meyer et al. 2011; Wan et al. 2012). Furthermore, given the low requirements for sequence conservation of these elements, vast numbers of novel RNA thermometers likely await discovery.
Some natural and artificial thermometers form a single hairpin that occludes the ribosome-binding site (RBS) (Neupert et al. 2008; Kortmann et al. 2011; Kouse et al. 2013), but many natural thermometers are significantly more complex (Morita et al. 1999; Kortmann and Narberhaus 2012). The reasons for this variability are not well understood. Many RNA thermometers contain noncanonical base pairs, bulges, and other structural features thought to modulate their stability, which allows response over a physiological temperature range (Kortmann and Narberhaus 2012). Detailed molecular-level characterization of the effect of temperature on the structure and dynamics of these important regulators is critical for full understanding of their biological functions.
Here, we have utilized selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Merino et al. 2005) assays for a detailed characterization of the structure and behavior of the htrA RNA thermometer from Salmonella enterica. SHAPE assays provide a quantitative method for simultaneous characterization of the backbone flexibility of each nucleotide in a complex RNA molecule (Gherghe et al. 2008b; Weeks and Mauger 2011). Critical for detailed study of RNA thermometers, SHAPE assays produce reliable and comparable results at a wide range of incubation temperatures (Wilkinson et al. 2005, 2006). As such, SHAPE has been used for detailed studies of temperature-dependent structural transitions in tRNA molecules (Wilkinson et al. 2005; Strulson et al. 2014), and in one recent study of an RNA thermometer, the CssA thermometer from Neisseria meningitidis (Barnwal et al. 2016).
The htrA thermometer is classified as a fourU thermometer (Waldminghaus et al. 2007), because it is predicted to form a hairpin with a stretch of four uridine nucleotides opposing the Shine–Dalgarno (SD) sequence (Fig. 1A; Klinkert et al. 2012). Other fourU RNA thermometers have been identified in pathogenic bacteria, such as in the 5′ UTRs of the agsA mRNA of S. enterica (Waldminghaus et al. 2007) and the shuA mRNA of Shigella dysenteriae (Kouse et al. 2013), as well as in the intergenic region of the lcrF mRNA of Yersinia pseudotuberculosis (Böhme et al. 2012). The agsA gene encodes a heat shock protein, while shuA and lcrF encode virulence factors. The sequence of the htrA RNA thermometer is strictly conserved over a wide range of S. enterica strains, including those responsible for typhoid fever and gastroenteritis (Supplemental Fig. 1A). In contrast, it is not present in the 5′ UTR sequences from the htrA genes of many other organisms, such as Helicobacter pylori and humans.
FIGURE 1.
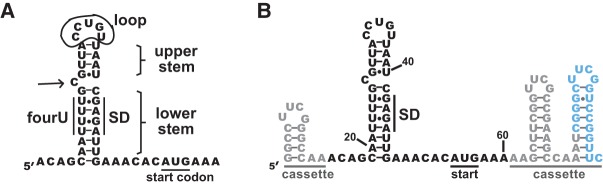
Predicted secondary structure of the htrA RNA thermometer from Salmonella enterica. (A) The 5′ UTR of the htrA mRNA is shown from the standard transcriptional start site to one codon past the AUG start codon. The fourU element and the Shine–Dalgarno (SD) sequence are labeled. As defined in the text, the lower stem and upper stem nucleotides are labeled, and hairpin loop nucleotides are circled. The single-nucleotide bulge on the 5′ side of the stem is indicated with an arrow. The structure was predicted using mfold 3.0 (Zuker 2003), as shown in Klinkert et al. (2012). (B) The putative structure of the htrA thermometer embedded in the SHAPE structure cassette (Wilkinson et al. 2006; Vachon and Conn 2012). The primer-binding site is shown in blue, and the remainder of the cassette regions are shown in gray. Numbering is from the 5′ end of the SHAPE cassette; this numbering is used throughout the manuscript. The structure shown was predicted using RNAstructure (Reuter and Mathews 2010) at 37°C.
The S. enterica htrA thermometer is among the shortest known natural RNA thermometers, completely contained in the 40-nucleotide (nt) 5′ UTR of the mRNA for the protein HtrA (high temperature requirement A) (Klinkert et al. 2012). HtrA is a periplasmic serine protease reported to degrade aggregated or misfolded proteins (Pallen and Wren 1997). In some species, including E. coli, it is required for survival at high temperature (>42°C) (Lipinska et al. 1989). In Salmonella species, it increases viability at elevated temperatures, but it is not essential for survival (Johnson et al. 1991; Mo et al. 2006). However, HtrA is critical for the pathogenic virulence of a number of bacterial species, including Salmonella (Pallen and Wren 1997). For example, S. enterica subsp. enterica serovar Typhimurium (S. Typhimurium) strains lacking or bearing mutant HtrA show reduced infectivity of mice (Mo et al. 2006; Lewis et al. 2009), as well as reduced survival in murine macrophages (Bäumler et al. 1994). HtrA is also required for persistence of Salmonella strains in chicken eggs, a major source of human infection (Raspoet et al. 2014). Numerous attenuated strains of S. enterica used for live vaccines have been made through mutation or deletion of the htrA gene (Dunstan et al. 1998; Lowe et al. 1999). Greater understanding of the determinants for thermometer structure and function could thus provide insight into the virulence of these pathogenic bacteria.
In this work, we sought to understand the htrA thermometer from S. enterica in greater depth through the combination of biochemical and biological assays. We used SHAPE assays to investigate the structure and dynamics of the htrA RNA thermometer on a single-nucleotide level. Assays were conducted under a wide range of temperatures, including those experienced under conditions of standard growth and of infection, allowing construction of individual melting curves for nucleotides throughout the thermometer stem. In order to assess the contribution of particular nucleotides, we produced a panel of mutant sequences, and their structure and inducibility were also analyzed via SHAPE and reporter gene assays. The results further illustrate the utility of SHAPE assays for the study of thermometer systems, reveal key features for htrA temperature response in the physiological regime, and provide insight into the melting behavior of this thermometer.
RESULTS
Structure and temperature response of the htrA thermometer
The sequence upstream of the htrA coding region is quite conserved among S. enterica strains (Supplemental Fig. 1B). Several studies have mapped transcriptional start sites (TSSs) for htrA mRNA from S. Typhimurium (Lewis et al. 2009; Kröger et al. 2013; Srikumar et al. 2015). In each study, three distinct TSSs were found, producing a 40-nt 5′ UTR, a 47-nt 5′ UTR, and a much longer 215-nt or 449-nt 5′ UTR. The longer transcripts begin within the coding sequence for the upstream gene, deoxyguanosinetriphosphate triphosphohydrolase (dgt). Transcripts with 215-nt and 47-nt long 5′ UTRs were initially reported to be produced constitutively, but at low levels, with slight induction upon heat shock (Lewis et al. 2009). In that work, the transcript with a 40-nt 5′ UTR was only observed under stress conditions. More recent RNA-seq and dRNA-seq studies on S. Typhimurium 4/74 have shown that both of the short transcripts are expressed under a variety of growth and stress conditions, whereas transcripts with a 449-nt long 5′ UTR are expressed at very low levels under normal growth conditions, but induced under a number of stress conditions (Kröger et al. 2013; Srikumar et al. 2015). Relative contributions of each promoter to the overall transcript level varied with the particular growth or stress condition. However, transcription from all three sites was up-regulated upon infection of murine macrophages, with higher production of the transcripts with the two shorter 5′ UTRs (Srikumar et al. 2015). As the 40-nt 5′ UTR entirely contains the thermometer sequence, we have primarily focused on this variant.
Computational predictions show that the hairpin of the S. enterica htrA thermometer is interrupted by a single-nucleotide bulge (Fig. 1A; Zuker 2003; Klinkert et al. 2012). The lower stem of the hairpin includes the four uridine nucleotides of the fourU motif paired with the Shine–Dalgarno sequence, and the upper stem is capped with an unusual 5-nt loop. First, we experimentally validated the computational prediction using SHAPE assays. For this, the entire 40-nt 5′ UTR plus the first two codons of the coding region were embedded in a set of stable hairpins known as a structure cassette (Fig. 1B; Wilkinson et al. 2006; Vachon and Conn 2012). The structure cassette provides a binding site for primer extension and prevents information loss from the 5′ and 3′ ends of the RNA of interest, allowing all nucleotides of the RNA thermometer to be simultaneously assessed. The structure cassette also provided us a means for scaling and normalizing the SHAPE reactivities between experiments conducted at different temperatures, as its hairpins were not observed to melt over the temperature range studied. Computational predictions using mfold (Zuker 2003) and RNAstructure (Reuter and Mathews 2010) suggested that the cassette sequence would not affect the secondary structure of the embedded thermometer RNA.
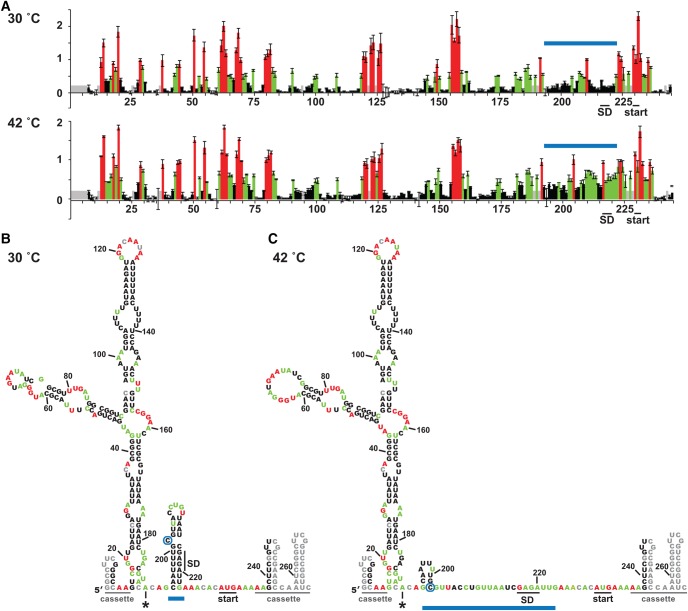
SHAPE reactions were performed in the presence of 2 mM MgCl2 in order to mimic the physiological concentration of free Mg2+ in bacteria (Alatossava et al. 1985; Froschauer et al. 2004; Kolisek et al. 2008). We used the commercially available SHAPE reagent N-methylisatoic anhydride (NMIA) (Merino et al. 2005). This acylating agent reacts preferentially with flexible 2′-hydroxyl groups in the RNA backbone. Primer extension products were resolved via capillary electrophoresis, and the results were analyzed using QuShape (Karabiber et al. 2013). At 30°C, representing standard growth conditions, the SHAPE results for the Salmonella htrA RNA thermometer were consistent with the predicted structure (Fig. 2). Low, but nonzero, reactivity (indicated in black) was observed throughout the putative stem region, while moderate (green) to high (red) reactivity was generally observed in the regions corresponding to the hairpin loop and to the flanking nucleotides. As has been previously observed for a small percentage of unpaired nucleotides (Jones et al. 2008; Wilkinson et al. 2009), a few flanking nucleotides showed low reactivity in our SHAPE experiments. Moderate reactivity was also observed for the bulged C nucleotide, as well as for one U nucleotide in the upper stem. Overall, the reactivity levels observed for the stem nucleotides were higher than the mean values observed for internally paired nucleotides (Wilkinson et al. 2009), suggestive of mild instability of the thermometer stem even at 30°C.
FIGURE 2.
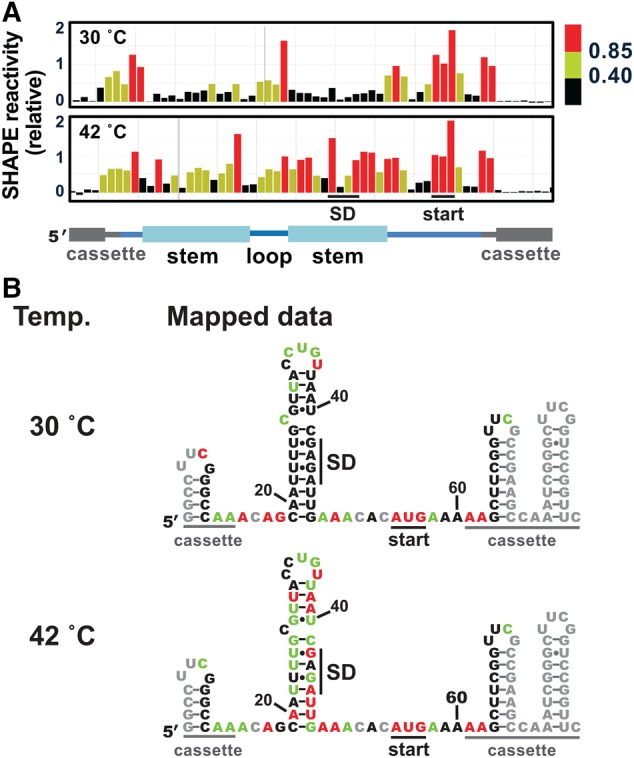
The wild-type htrA RNA thermometer adopts a hairpin conformation at low temperature. (A) Representative SHAPE results at 30°C (top) and at 42°C (bottom) for the wild-type htrA RNA thermometer embedded in the structure cassette shown in Figure 1B. Data were processed using QuShape (Karabiber et al. 2013), and results mapped against a cartoon of the thermometer secondary structure, including cassette regions. Nucleotides with low SHAPE reactivity (<0.40), medium SHAPE reactivity (0.40–0.85), and high SHAPE reactivity (≥0.85) are shown in black, green, and red, respectively (Karabiber et al. 2013). In all cases, representative results were prepared in QuShape. (B) Average SHAPE results at 30°C (top) and 42°C (bottom) mapped on the putative secondary structure of the htrA thermometer embedded in the structure cassette (Fig. 1B). Coloring is as in part A. No data were collected for the nucleotides shown in gray, which were either too close to the RNA ends or showed atypically high signals in the SHAPE (−) reaction. Mapped values are the average of six (30°C) or four (42°C) independent experiments (Supplemental Fig. 4A).
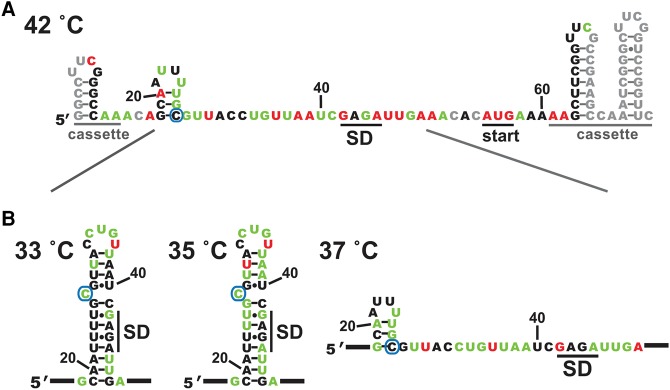
We next investigated the response of the RNA thermometer to temperature. SHAPE assays were performed at 42°C, representing the temperature experienced under heat-shock conditions or upon Salmonella infection of chickens (Raspoet et al. 2014). For the majority of the htrA stem nucleotides, SHAPE reactivity increased considerably from 30°C to 42°C, while only minor changes were observed to nucleotides in the flanking regions or in the hairpin loop (Fig. 2). These trends were independent of the cassette sequence and SHAPE reagent used (Supplemental Fig. 2). Interestingly, reactivity decreased dramatically for three nucleotides: the final U in the hairpin loop, the single-nucleotide bulge, and the G immediately preceding the thermometer stem. Overall, the SHAPE results suggested that the htrA hairpin no longer forms at 42°C, consistent with the model of a structural transition which occurs upon infection of a warm-blooded host.
In order to better understand this structural transition, we next investigated the htrA thermometer over a broad range of temperatures bracketing the physiological regime. SHAPE reactions were conducted at ∼3°C intervals from 20°C to 62°C (Fig. 3), allowing a detailed assessment of the behavior of each individual nucleotide position of the thermometer with respect to temperature. At the lowest temperatures (20°C and 24°C), stem nucleotides displayed even lower reactivity than at 30°C, while only minor changes were observed to nucleotides in the flanking regions or in the hairpin loop (Fig. 3A; Supplemental Fig. 2). These results confirm the mild instability proposed above for the thermometer hairpin at 30°C, which could allow leaky expression of the encoded protein at this temperature. At 37°C, reactivity of the stem nucleotides was intermediate between that observed at 30°C and 42°C.
FIGURE 3.
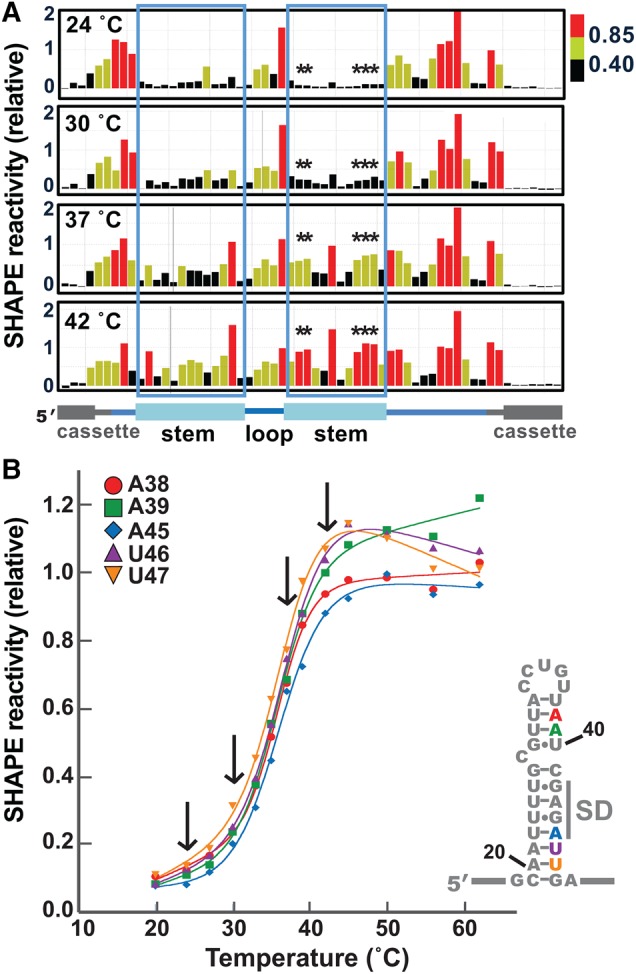
The htrA thermometer appears to open cooperatively upon increasing temperature. (A) Representative SHAPE data for the htrA thermometer, comparing 24°C, 30°C, 37°C, and 42°C. Coloring is as in Figure 2. The regions corresponding to the 5′ and 3′ sides of thermometer stem are boxed, and nucleotides graphed in part B are marked with asterisks. (B) SHAPE melting curves for selected nucleotides in the 3′ side of the stem, including A38, A39, A45, U46, and U47. Arrows indicate the temperatures shown in part A. Each data point is the average of two to six independent experiments. Error bars are not shown, in the interest of clarity (see Supplemental Fig. 3 for representative curves with error).
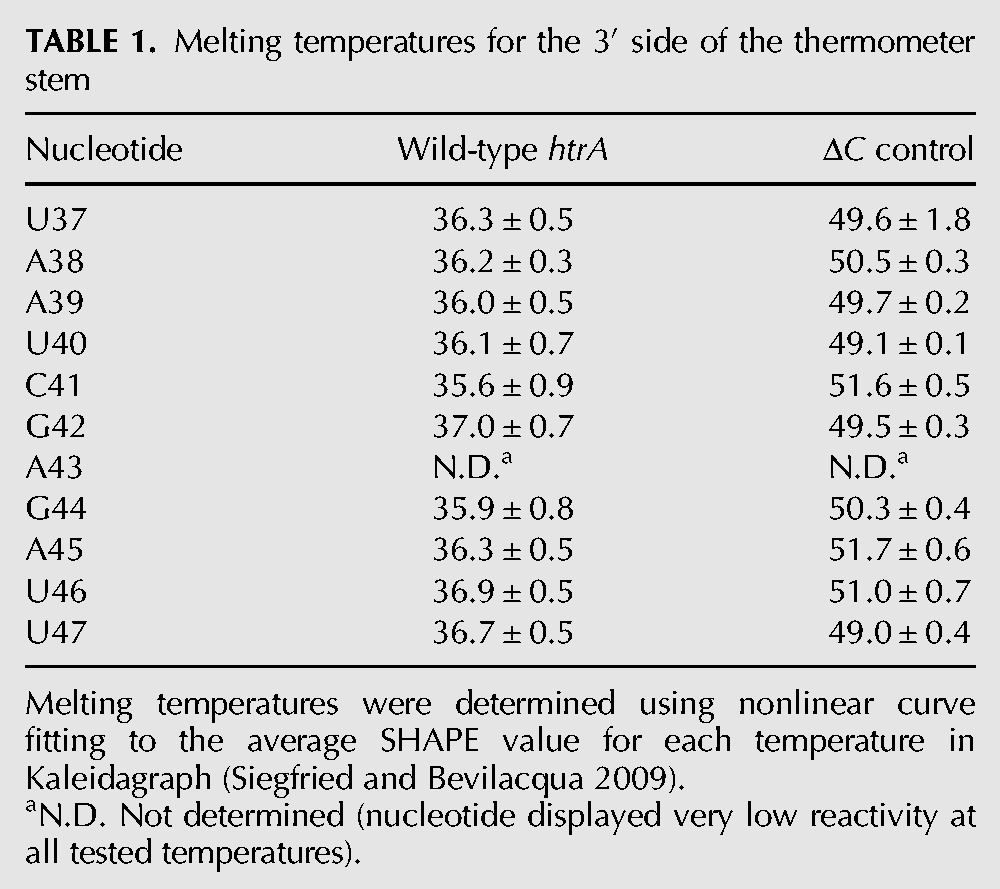
Therefore, we used the SHAPE results acquired at each temperature to construct melting curves for individual nucleotides throughout the RNA molecule (Fig. 3B). Overall, SHAPE reactivity increased in a sigmoidal fashion for the nucleotides in the thermometer stem, with the exception of the bulged C and its flanking nucleotides. Nonlinear curve fitting was performed for each nucleotide in the 3′ side of the thermometer stem individually (Siegfried and Bevilacqua 2009), revealing a common melting temperature of ∼36°C (Table 1). These results suggest that the upper and lower stem melt simultaneously, consistent with a cooperative melting transition for the entire thermometer. The individual nucleotide melting curves of nucleotides in the 5′ side of the thermometer stem also suggest thermometer melting between 35°C and 37°C (data not shown).
TABLE 1.
Melting temperatures for the 3′ side of the thermometer stem
RNAstructure was then used to refine the thermometer structure at each temperature, incorporating the experimentally determined SHAPE reactivity values as a pseudo-free energy term in the calculations (Deigan et al. 2009; Reuter and Mathews 2010). Incorporation of SHAPE reactivity can greatly improve the accuracy of structure prediction. This is particularly true for calculations performed at temperatures other than 37°C, as such calculations require extrapolation of thermodynamic parameters that were originally measured at 37°C. In the case of the htrA thermometer, the refined structure at 30°C is the same as predicted in the absence of experimental restraints. Without experimental restraints, the lowest energy structure predicted for 42°C by RNAstructure alone still contains the thermometer stem. In contrast, incorporation of experimental results confirms that this stem no longer folds at 42°C (Fig. 4A). At this temperature, the SHAPE data support the presence of an alternate hairpin that involves both the fourU motif and the C from the single-nucleotide bulge, leaving the SD sequence exposed and available for ribosomal interactions. Comparison of the lowest energy structure at each temperature from 20°C–62°C reveals that the conformational transition between folded and unfolded thermometer stem occurs between 35°C and 37°C (Fig. 4B), consistent with the experimental melting curves, as well as with a biological role in pathogenic virulence. Interestingly, the alternate hairpin mentioned above appears to be present at temperatures from 37°C to 45°C (data not shown).
FIGURE 4.
Incorporation of SHAPE constraints into structural predictions supports a cooperative transition. (A) SHAPE-constrained secondary structure of the htrA thermometer at 42°C. Shown is the lowest energy structure predicted using RNAstructure, with incorporation of SHAPE values as pseudo-free energy restraints. The C that forms the single-nucleotide bulge in the folded thermometer is circled. Diagonal lines indicate the region expanded in B below. Coloring as in Figure 2. (B) SHAPE-constrained secondary structure predicted at temperatures bracketing the structural transition (33°C, 35°C, 37°C), focusing in on the thermometer hairpin sequence. No structural change was predicted in the flanking cassette regions over a temperature range from 20°C to 56°C. SHAPE-constrained secondary structures reveal that the alternative stem seen here at 37°C and 42°C melts out between 45°C and 50°C.
Analysis of a complex, extended 5′ UTR for the htrA mRNA
As previously mentioned, transcription initiation in S. enterica can also occur from promoters located far upstream of the htrA thermometer sequence (Lewis et al. 2009; Kröger et al. 2013; Srikumar et al. 2015). In order to assess whether thermometer structure or temperature response would be altered in the context of this extended upstream sequence, we transcribed the full 215-nt htrAp1 5′ UTR in the context of the structure cassette and subjected this RNA to SHAPE assays. The activated SHAPE reagent 1M7 (1-methyl-7-nitroisatoic anhydride) (Mortimer and Weeks 2007; Turner et al. 2013) was used for these experiments, because its shorter half-life reduced the incubation times required, which proved valuable for minimizing degradation of this much longer RNA. Qualitatively similar results were observed upon use of NMIA, although the greater background signal hindered quantitative analysis.
Overall, our SHAPE results revealed that the htrA thermometer behaved very similarly in the context of the extended 5′ UTR as it did as an isolated element (Fig. 5). At 30°C, SHAPE reactivity patterns for the thermometer region were consistent with the presence of the thermometer stem (Fig. 5A,B), which presumably blocks the Shine–Dalgarno sequence from ribosome accessibility. At the elevated temperature (42°C), SHAPE reactivity again dramatically increased for nucleotides of the upper stem, as well as for those in the 3′ side of the lower stem (Fig. 5A,C). Incorporation of the 42°C experimental data into structure predictions confirmed that the thermometer stem is not folded under these conditions. Instead, the 5′ side of the lower stem again appears to form the alternate upstream hairpin seen in the shorter RNA (Fig. 4), exposing the Shine–Dalgarno sequence for ribosomal interaction.
FIGURE 5.
The RNA thermometer forms in the context of an extended htrA 5′ UTR. (A) Average SHAPE results at 30°C (top) and at 42°C (bottom) for the wild-type htrA RNA thermometer in the context of the extended 5′ UTR produced from transcription at the upstream promoter htrAp1 (Lewis et al. 2009). Error bars show the standard deviation from three independent experiments. Coloring is as in Figure 2, with the thermometer hairpin indicated with a blue bar. (B,C) SHAPE-constrained secondary structures of the htrAp1 5′ UTR at 30°C (B) and 42°C (C). Structure prediction was performed with RNAstructure, as in Figure 4. Shown is the lowest energy SHAPE-constrained secondary structure predicted at each temperature. The bulged C is circled, and the asterisk denotes the transcriptional start site for the standard htrA mRNA.
The additional sequence present in the extended 5′ UTR showed patterns of high and low SHAPE reactivity consistent with a structured RNA. The lowest energy SHAPE-derived secondary structures indicated that the additional sequence upstream of the thermometer is quite structured at both temperatures, with minimal change between 30°C and 42°C (Fig. 5B,C). The consistency of the results between the two temperatures for the upstream region also provides an internal control for the method, as it clearly highlights differences in the behavior of the thermometer region from that of these other structured RNA elements.
Determinants for thermometer function
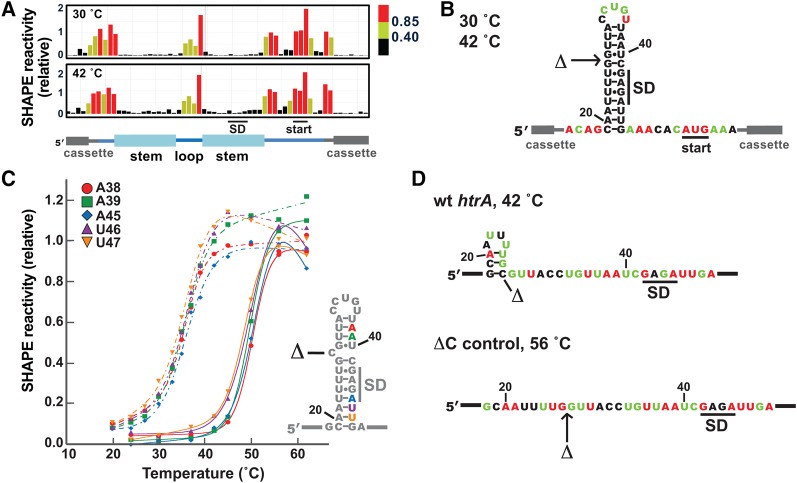
In order to correlate thermometer melting with particular structural features, we investigated several changes to the htrA thermometer sequence. First, we probed the role of the cytosine bulge in thermometer structure and behavior. Removal of this nucleotide was previously shown to greatly reduce translation under heat-shock conditions in reporter gene assays (Klinkert et al. 2012). We therefore generated an RNA construct lacking the bulged nucleotide (ΔC control) and performed SHAPE analysis on it (Fig. 6A,B). At 30°C, the ΔC control showed extremely low reactivity throughout the hairpin stem, while displaying similar reactivity to the wild-type RNA in the hairpin loop and flanking unstructured sequences. Reactivity levels increased only slightly at the elevated temperature of 42°C, indicating that the hairpin remained closed at this temperature. In fact, reactivity levels for the stem nucleotides of the ΔC control RNA at 42°C were comparable or lower than levels for the corresponding nucleotides of the wild-type RNA at 30°C.
FIGURE 6.
The single-nucleotide bulge has a profound effect on thermometer behavior. (A) Representative SHAPE results at 30°C (top) and 42°C (bottom) for the closed control RNA lacking the C bulge (ΔC control). (B) SHAPE results mapped on the predicted secondary structure of the hairpin in the ΔC control RNA. The position of the deleted nucleotide is indicated with an arrow. (C) SHAPE melting curves comparing the melting behavior of the ΔC control RNA with the wild-type thermometer. Solid lines show melting curves for selected nucleotides in the 3′ side of the ΔC control stem (A38, A39, A45, U46, and U47, with numbering as in the wild-type sequence). Each data point is the average of two independent experiments. Error bars are not shown in the interests of clarity (see Supplemental Fig. 3 for representative curves with error). Melting curves from the corresponding nucleotides for the wild-type RNA (from Fig. 3B) are shown with dashed lines. (D) Comparison of the wild-type thermometer at 42°C and the ΔC control at 56°C. Shown are the lowest energy SHAPE-derived secondary structures, with nucleotides color-coded by the average SHAPE values that were incorporated into structure predictions. The C nucleotide that is removed to make the ΔC control is marked; note that its absence precludes formation of the alternate hairpin. SHAPE values used represent the average of two to four experiments.
SHAPE reactions were then conducted at a wider range of temperatures (24°C–62°C) to allow a more detailed examination of the effect of the bulged nucleotide on thermometer melting and dynamics (Fig. 6C). SHAPE reactivity values were virtually identical at 24°C, 30°C, and 37°C, suggesting that the hairpin remains firmly closed under this temperature range. For the majority of the stem nucleotides in the ΔC control, SHAPE reactivity increased dramatically from 45°C to 56°C, while only minor changes were observed to nucleotides in the flanking regions or in the hairpin loop. Several nucleotides in the stem region of the ΔC control proved unreactive even at the highest temperatures (Fig. 6D). In general, these were the same nucleotides that were unreactive at high temperatures in the wild-type sequence.
The SHAPE results acquired at each temperature were again used to construct melting curves for individual nucleotides throughout the RNA molecule. SHAPE reactivity also increased in a sigmoidal fashion for the stem nucleotides of the ΔC control. Representative melting curves of stem nucleotides from throughout the 3′ side of the hairpin stem of the ΔC control were compared with the corresponding nucleotides in the wild-type thermometer (Fig. 6C). Nonlinear curve fitting indicated a melting temperature of ∼50°C, representing an increase of 14°C from that of the wild-type sequence (Table 1). Similar results were obtained when all data points for the 3′ side of the hairpin stem were normalized, then treated as a single data set (data not shown), again suggesting a cooperative transition between open and closed forms of the RNA, as might be expected for this simple, unbroken stem. These results also confirmed the importance of the single-nucleotide bulge for maintaining the thermometer melting temperature in the physiological range.
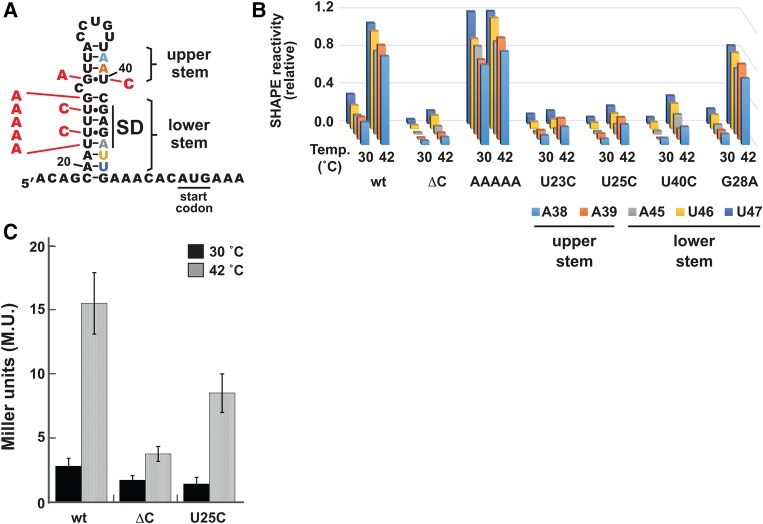
We next assessed a series of mutations that directly impacted base-pairing in the thermometer stem (Fig. 7A). The first mutation was chosen to disrupt the lower stem of the thermometer, replacing the fourU motif opposite the SD sequence, as well as the subsequent G, with a series of five A nucleotides (AAAAA mutant). As expected, SHAPE assays on the AAAAA mutant revealed high reactivity for nucleotides in the lower stem at 30°C (Fig. 7B; Supplemental Fig. 4C). Reactivity levels were also high for nucleotides in the upper stem, consistent with destabilization of the entire thermometer hairpin by this mutation.
FIGURE 7.
Structural determinants for thermometer formation. (A) Cartoon of mutant hairpins studied. Mutations are shown in red against the secondary structure of the wild-type htrA thermometer. The AAAAA mutation replaces the four U nucleotides as well as the G just prior to the bulge with A nucleotides. All other mutations are single-point mutations. Stem nucleotides shown in part B are color coded accordingly. (B) SHAPE results comparing the wild-type thermometer (wt), the closed control thermometer (ΔC), and the RNAs with stem mutations (AAAAA, U23C, U25C, U40C, and G28A). In each case, RNA was probed in the context of the standard SHAPE cassette shown in Figure 2. SHAPE results are displayed for the 3′ stem nucleotides A38, A39, A45, U46, and U47. Full reactivity profiles at 30°C and 42°C are shown in Supplemental Figure 4. (C) Reporter gene assay results for the wild-type thermometer and selected mutants. In each case, the thermometer sequence provided the SD sequence for expression of heat-stable β-galactosidase from the pBAD2-bgaB plasmid (Klinkert et al. 2012). Shown are the averages ± error at the 95% confidence level for 18 (wild-type htrA), 17 (ΔC), or 10 (U25C) independent experiments.
Further mutations were chosen to test the impact of strengthening either the upper or lower stem of the thermometer hairpin. This hairpin includes three noncanonical U•G or G•U base pairs. Two U•G base pairs are found in the lower stem, forming interactions between the fourU motif and the G nucleotides of the SD sequence. These U•G base pairs are not consecutive, unlike in the canonical fourU thermometer from the agsA gene (Waldminghaus et al. 2007). A final G•U base pair is located in the upper stem, immediately above the cytosine bulge. To understand the role that these base pairs play in thermometer structure and stability, we generated and analyzed several point mutations that introduced canonical Watson–Crick base pairs in these positions (Fig. 7A).
The two mutations to the fourU region each replaced one of the two U•G base pairs with a C–G pair. In each case, the point mutation was made to the nucleotide on the 5′ side of the thermometer stem (U23C and U25C), so as to not disrupt the SD sequence itself. These mutations are predicted to stabilize the RNA thermometer. SHAPE assays revealed that these mutations had the expected effect, as little to no reactivity was observed for stem nucleotides even at 42°C. The reduction in reactivity was observed not only for nucleotides in the lower stem, but also for nucleotides in the upper stem, supporting a cooperative melting transition for the entire hairpin. However, introduction of neither C–G base pair had as profound an effect as the outright removal of the C bulge (Fig. 7B; Supplemental Fig. 4), highlighting the importance of the single-nucleotide bulge for the structural transition.
We then performed reporter assays in order to correlate changes in thermometer structure and stability with effects on thermometer function in bacterial cells. The reporter gene was the bgaB gene, which produces a heat-stable β-galactosidase from Bacillus stearothermophilus, with thermometer variants embedded in its 5′ UTR (Klinkert et al. 2012). This reporter system has been validated for study of RNA thermometers, including htrA (Klinkert et al. 2012). We thus prepared and tested a mutated version of the reporter plasmid, replacing the final U of the fourU motif with C (U25C), comparing its behavior to that of the wild-type thermometer and the ΔC control (Fig. 7C). As expected, the reporter gene bearing the wild-type thermometer showed low expression at 30°C, with a substantial increase in expression at the permissive temperature of 42°C. In contrast, the ΔC control led to low expression at both temperatures. These results were consistent with previous observations (Klinkert et al. 2012), as well as with our SHAPE studies above. The mutant U25C element led to reduced expression at both temperatures compared to the wild-type thermometer, suggesting that it more effectively sequestered the SD sequence from interactions with ribosomal rRNA. However, it did not have as pronounced an effect as the ΔC control at 42°C, again consistent with our SHAPE studies.
In order to further address whether the upper and lower stems melt cooperatively, we investigated the effect of mutation of the G•U base pair in the upper stem on the temperature response of the lower stem. We generated and analyzed two point mutations of the upper G•U base pair (Fig. 7A). The first mutation swapped this G•U base pair for a G–C base pair, through substitution of the U nucleotide immediately prior to the SD sequence with a C. SHAPE assays revealed that this mutation (U40C) had a similar effect as the U → C mutations in the lower stem (Fig. 7B; Supplemental Fig. 4), dramatically reducing SHAPE reactivity of nucleotides throughout the upper and lower portions of the stem at both 30°C and 42°C. These results contrast with the effect of replacing the G of the upper G•U base pair with an A nucleotide, creating instead an A–U base pair (G28A). Here, we observed a decreased reactivity for the stem nucleotides at 30°C, when compared with the wild-type hairpin (Fig. 7B; Supplemental Fig. 4). However, the mutant sequence still appears to be unfolded at 42°C, based on reactivity of nucleotides both in the upper and lower portion in the stem (Fig. 7B; Supplemental Fig. 4). It is critical to note that for both of these G•U mutants, alteration of a nucleotide in the upper stem affected the reactivity of nucleotides from the lower stem, as well. These results suggest that the upper and lower stems share a common melting transition, again supporting cooperative melting behavior of the entire thermometer hairpin.
DISCUSSION
RNA thermometers provide a rapid and potentially reversible method to regulate gene expression in response to changing environmental conditions. Many questions remain regarding the range of structural features that will lead to thermometer behavior (Kortmann and Narberhaus 2012). Here, we performed an in-depth study of the S. enterica htrA thermometer, originally identified on the basis of computational prediction and reporter gene assays (Klinkert et al. 2012). The htrA thermometer is located in the 5′ UTR of the gene encoding the protein HtrA (high-temperature requirement A) in a wide range of S. enterica strains, including those responsible for typhoid fever and gastroenteritis (Supplemental Fig. 1A). Although HtrA is essential for response to heat shock in E. coli, the S. enterica version is intrinsically involved in pathogenic virulence (Mo et al. 2006).
Here, we used SHAPE analysis and mutagenesis to investigate the structure, dynamics, and temperature response of the htrA thermometer. These assays confirmed the predicted hairpin fold of the thermometer at 30°C, while indicating exposure of the Shine–Dalgarno sequence at 42°C (Figs. 2, 4). We then went on to investigate the mechanism of thermometer melting. We initially hypothesized that the bulged C might provide the nucleation point for a gradual melting of the entire stem with increasing temperature. This would parallel what has been observed with RNA thermometers such as the ROSE1 element from Bradyrhizobium japonicum, the cyanobacterial hsp17 thermometer, and the CssA thermometer from Neisseria meningitidis, for which thermometer melting occurs progressively from a destabilizing element upon temperature increase (Chowdhury et al. 2006; Wagner et al. 2015; Barnwal et al. 2016). An alternative hypothesis was that the bulge would decouple the upper and lower stem into two independently folding units, with distinct melting transitions based on their individual stability.
Contrary to these initial hypotheses, analysis of SHAPE melting curves for individual nucleotides suggest that the thermometer unfolds in a concerted and cooperative fashion, with nucleotides in both the upper and lower stem gaining flexibility at a common transition temperature (∼36°C) (Table 1; Fig. 3). Incorporation of SHAPE results into structure–prediction algorithms also supports a concerted melting transition in the physiological regime, again with a melting temperature of ∼36°C (Fig. 4). SHAPE assays performed on stem mutants of the htrA thermometer further support communication between the upper and lower stems, despite the intervening bulge. Mutations which strengthened base-pairing in the upper stem also increased the melting temperature of the lower stem, and vice versa (Fig. 7). A cooperative melting transition is also observed in the agsA fourU thermometer from S. enterica (Rinnenthal et al. 2010), despite other differences between the structures of these two thermometers. It is tempting to speculate that the combination of the single-nucleotide bulge and the cooperativity displayed between the stems of the htrA thermometer allows for tuning of the temperature response of the thermometer in response to the body temperature of the host.
Removal of the bulged nucleotide is predicted to stabilize the htrA hairpin stem dramatically (ΔGfold, wt: −5.8 kcal/mol versus ΔGfold, ΔC: −9.6 kcal/mol, as calculated in RNAstructure [Reuter and Mathews 2010]). Increased stability is experimentally evident through a shift in the melting temperature for nucleotides throughout the hairpin stem. Comparison of individual melting curves suggest a shift of ∼14°C (Table 1; Fig. 6C), such that little to no melting is observed over the temperature range required for thermometer response. Intermediate reactivities are observed when stabilizing mutations are introduced to the stem itself, even in the presence of the single-nucleotide bulge (Fig. 7A,B). These results agree with those from reporter gene assays, where a mutant thermometer with a stabilized stem (U25C) directs levels of gene expression intermediate between the wild-type thermometer and the ΔC control (Fig. 7C).
Intriguingly, several nucleotides from the wild-type thermometer showed decreased reactivity upon temperature increase. One of these was the U nucleotide that forms the final nucleotide of the thermometer hairpin loop, which has significantly higher reactivity than any other nucleotide in the thermometer hairpin at 30°C (Fig. 2A). Some instances of extreme reactivity arise from constriction of the nucleotide sugar into a defined conformation that enhances the nucleophilicity of the 2′-hydroxyl group, for instance, by favoring a 2′-endo conformation of the ribose group (Gherghe et al. 2008a; Mortimer and Weeks 2009). If this is the situation for the final loop nucleotide, the decrease in reactivity at higher temperatures could be explained by thermometer melting, which would release it from the reactive conformation.
A similar effect might be responsible for the behavior of the bulged C nucleotide, which also displays decreased reactivity at increased temperatures. At low temperatures (below 30°C), it is the most reactive nucleotide in the stem portion of the thermometer hairpin, whereas under heat shock or infection conditions, it becomes one of the least reactive stem nucleotides (Fig. 3A). Reactivity levels again increase under the highest temperatures studied (data not shown). However, higher reactivity levels at low temperatures might simply be a result of greater flexibility of this nucleotide in the single-nucleotide bulge than in the alternate hairpin. Formation of the alternate hairpin also explains the decrease in reactivity observed for the G that precedes the standard thermometer stem (Figs. 3, 4), as well as the difference in reactivity of the 3′ versus the 5′ side of the lower stem, which we observe in the contexts of both the short and the long 5′ UTRs at 42°C. Further work will address the importance of this alternate hairpin, as well as the identity of the bulged nucleotide and its flanking base pairs, on thermometer behavior and function.
SHAPE assays are readily adapted for study of longer RNA sequences. We thus used SHAPE assays to investigate an extended htrA 5′ UTR (∼215 nt) produced by S. enterica (Lewis et al. 2009). Our results reveal that thermometer structure and behavior is similar in the context of this longer RNA as it is in the standard 5′ UTR (Fig. 5), with sequestration of the SD sequence at 30°C, but exposure at 42°C. The presence of structured elements upstream of the thermometer at both temperatures is suggestive of additional function for this region, potentially allowing for combinatorial regulation of HtrA expression by both temperature and a yet-unknown signal. Such combinatorial control has been produced in genes containing tandem riboswitches (Serganov and Nudler 2013) or artificial riboswitch/thermometer hybrids (Roßmanith and Narberhaus 2016), for example. The additional upstream sequence is highly conserved among Salmonella strains (Supplemental Fig. 1B), suggesting a possible common role for this structure in regulation.
In conclusion, we have used SHAPE assays to investigate the structure and dynamics of the htrA RNA thermometer from S. enterica. This thermometer forms a hairpin which sequesters the Shine–Dalgarno sequence under normal growth temperatures, but which melts open at moderately elevated temperatures, such as those experienced during infection. SHAPE analysis allowed a detailed and quantitative analysis of the behavior of each nucleotide of the htrA thermometer with respect to temperature. The presence of a single-nucleotide bulge was found both to promote stem flexibility at normal growth temperatures and also to shift the melting temperature of the hairpin into the relevant physiological range. The stems flanking this bulge were found to melt in a concerted fashion, rather than progressively from the single-nucleotide bulge. U•G base pairs involving the Shine–Dalgarno sequence and a G•U base pair in the upper stem were all found to modulate the melting temperature. It is tempting to speculate that these elements work in concert, tuning the thermometer behavior to allow leaky expression under standard growth conditions, but a differential response upon infection, depending on the body temperature of the warm-blooded host infected. Further investigation will address the effect of the identity of the bulge nucleotide and surrounding base pairs on thermometer structure and behavior, focusing specifically on the role of the alternate hairpin formed under intermediate temperatures.
MATERIALS AND METHODS
RNA production
DNA templates for the standard RNAs used for SHAPE were generated via extension of partially overlapping DNA oligonucleotides (Supplemental Table 1, Integrated DNA Technologies, IDT) using the Klenow fragment of DNA polymerase I (Life Technologies). These templates embedded the htrA or variant RNAs within 5′ and 3′ cassette sequences to allow analysis of the full RNA of interest (Wilkinson et al. 2006; Vachon and Conn 2012). Templates were amplified by PCR using Platinum pfx polymerase (Life Technologies), then cleaned using the Qiagen PCR Kit. In some cases, dsDNA produced from Klenow reactions was directly used as the transcription template, without prior amplification. Template sequences were verified at the Plant-Microbe Genomics Facility at The Ohio State University (OSU PMGF). RNAs were produced via in vitro transcription using the T7 polymerase HiScribe High Yield Quick Kit (New England Biolabs) or with recombinant T7 RNA polymerase produced in-house. RNAs were purified from denaturing urea polyacrylamide gels (10%) and extracted from the gel using passive diffusion. RNA concentrations were quantitated using a Tecan F200 Infinite Spectrophotometer equipped with a NanoQuant plate. RNA homogeneity was assessed using analytical urea polyacrylamide gels (20%) stained with 1× SYBR Gold (Life Technologies).
Construction of the extended 5′ htrA UTR
The DNA corresponding to the full-length 215-nt UTR plus the first two codons of the coding region, preceded by the T7 RNA polymerase promoter and flanked by restriction enzyme sites and SHAPE cassette sequences (Supplemental Table 1), was ordered as a gBlock DNA from IDT. The sequence used corresponds to that of S. Enteritidis and S. Paratyphi B; it differs from that of S. Typhimurium by 1 nt (Supplemental Fig. 1B). Following the cloning protocol from IDT, this DNA was digested with BamHI and EcoRI and ligated into the corresponding sites of the pUC19 plasmid (New England Biolabs, NEB) using the NEB Quick Ligase Kit. DNA sequences were verified at the OSU PMGF. Double-stranded DNA templates were prepared via PCR amplification (Supplemental Table 1). RNAs were transcribed from these dsDNA templates as above, then purified from 8% denaturing polyacrylamide gels. RNA homogeneity was assessed using analytical urea polyacrylamide gels (8%) stained with 1× SYBR Gold.
SHAPE experiments
SHAPE experiments were conducted as described previously (Wilkinson et al. 2006; McGinnis et al. 2009), but with minor modifications as follows. In short, 8–12 pmol of purified RNA were denatured at 95°C in water, then snap-cooled on ice to favor intramolecular folding. One-fifth volume of 5× SHAPE folding mix (250 mM HEPES, pH 8.0, 500 mM potassium acetate, and 10 mM MgCl2) was added, making the final concentrations in the reaction 50 mM HEPES, pH 8.0, 100 mM potassium acetate, and 2 mM MgCl2. RNAs were allowed to equilibrate at 30°C for 15 min, then at the desired assay temperature for 20 min. Then 3–4.5 pmol of RNA (27 µL) was transferred to prewarmed tubes containing 3 µL of DMSO (− reaction) or SHAPE reagent in DMSO (+ reaction). Final concentration of SHAPE reagent was 13 mM for N-methylisatoic anhydride (NMIA, Life Technologies) or 10 mM for 1-methyl-7-nitroisatoic anhydride (1M7, gift of Manuel Ares, Jr.). Reactions proceeded for at least five times the hydrolysis half-life (t1/2) of the reagent at the desired temperature (Wilkinson et al. 2006), followed by ethanol precipitation in the presence of 20 µg glycogen.
For primer extension, pellets were resuspended in 10.5 µL H2O. To this was added 2.5 µL of a 0.6 µM 5′ 6-FAM labeled DNA primer complementary to the 3′ end of the SHAPE cassette. Annealing occurred at 65°C for 5 min, then 37°C for 5 min. Primer extension was carried out in the presence of 50 mM Tris–HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, 5 mM DTT, 0.5 mM of each dNTP, and 200 units of SuperScript III (Thermo Fisher). Extension reactions were incubated at 45°C for 1 min, 52°C for 25 min, then 65°C for 10 min. Reactions were then quenched with the addition of 4 µL of a solution of 2 M sodium acetate (unbuffered) and 40 mM EDTA. Sequencing reactions were performed similarly, but with unmodified RNA, a DNA primer bearing a 5′ VIC fluorophore, and addition of ddATP or ddTTP (0.5 mM final concentration) to the primer extension reaction. Each SHAPE reaction (+ or − SHAPE reagent) was mixed with an aliquot of sequencing reaction, followed by ethanol precipitation. Pellets were washed 2–3 times with 500–750 µL 70% ethanol to remove trace salts, then air-dried. Pellets were resuspended in 9.5 µL Hi-Di Formamide and 0.5 µL size standards, then resolved via capillary electrophoresis on an Applied Biosystems, Inc. 3730 DNA Analyzer (OSU PMGF). Inspection of electropherograms indicated that SHAPE reactions were carried out under single-hit conditions in all cases, as assessed by the presence of significant amounts of the full-length product. Electropherograms were processed using QuShape (Karabiber et al. 2013). QuShape uses model-free normalization with 0 signifying no reactivity and 1 indicating the average reactivity of a highly reactive nucleotide. Manual adjustments were made to the suggested scaling parameter and percent outliers in order to scale the + trace to the − trace (focusing on the stem nucleotides of the cassette, which remain unreactive over this temperature range) and to normalize the reactivity of a highly reactive, unstructured nucleotide (A61) for each temperature. Nucleotides were binned by normalized SHAPE reactivity, with values that are <0.40, 0.40–0.85, and ≥0.85 indicating low, moderate, and high SHAPE reactivity, respectively (Karabiber et al. 2013).
Melting curves were constructed using reactivity values from SHAPE experiments conducted at a range of temperatures from 20–62°C (for wild-type htrA) or 24–62°C (for the ΔC control). For melting curves, NMIA was used as the SHAPE reagent. Each data point was the average value from two to six independent experiments. Melting temperatures were obtained by nonlinear curve fitting using Kaleidagraph (Siegfried and Bevilacqua 2009). Structure prediction was performed with RNAstructure (Deigan et al. 2009), using the average SHAPE reactivities as pseudo-free energy change constraints (slope of 1.8 kcal/mol and y-intercept of −0.6 kcal/mol).
Reporter gene assays
Mutants were prepared using the Q5 Site-Directed Mutagenesis Kit (NEB), according to the manufacturer's protocol. The plasmid pBAD2-bgaB-htrAp3 (gift of Franz Narberhaus) was used to prepare pBAD2-bgaB-htrAp3-U25C. Mutagenic primers were designed using NEBaseChanger and produced by IDT, and PCR reactions used an annealing temperature of 60°C. Sequences were verified at the OSU PMGF. NEB5α cells (NEB) bearing the reporter plasmid (pBAD2-bgaB-htrAp3, pBAD2-bgaB-htrAp3-ΔC [gift of Franz Narberhaus], or pBAD2-bgaB-htrAp3-U25C) were grown overnight at 37°C in the presence of 100 µg/mL carbenicillin. One milliliter of overnight culture was then added to 25 mL LB broth with 50 µg/mL carbenicillin. When cultures reached an OD600 of 0.5, control aliquots were removed and incubated at 30°C. Arabinose was then added to 0.01% in order to induce transcription of the reporter gene mRNA, and 8 mL aliquots of each culture were shifted to prewarmed tubes at 30°C and 42°C. After a further 30-min incubation, 0.5 mL aliquots were removed for analysis. The β-galactosidase assay was conducted as by Miller (Miller 1972), with the modification that the enzymatic assay was conducted for 30 min at 42°C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Manuel Ares, Jr., University of California, Santa Cruz, for the gift of the SHAPE reagent 1M7 and Franz Narberhaus, Ruhr Universität Bochum, Germany, for the gift of the pBAD2-bgaB-htrAp3 and pBAD2-bgaB-htrAp3-ΔC plasmids. We acknowledge Hilary Cornell, Casey Cempre, Megan Brooks, and Patrick Dunn for technical assistance, and we thank Michael Zianni and Anthony McCoy of the Plant-Microbe Genomics Facility at The Ohio State University for their contributions to this work. We also thank Joan Steitz and Philip Bevilacqua for their helpful comments on the manuscript. Financial support was provided by Denison University, the Reid and Polly Anderson Endowment (E.K.C., J.K.F., K.A.U., and Y.A.N.), the Denison University Research Foundation (R.M.M.-F.), and a Cottrell College Science Award from the Research Corporation for Science Advancement to R.M.M.-F. (ID #23326).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062299.117.
REFERENCES
- Alatossava T, Jütte H, Kuhn A, Kellenberger E. 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol 162: 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwal RP, Loh E, Godin KS, Yip J, Lavender H, Tang CM, Varani G. 2016. Structure and mechanism of a molecular rheostat, an RNA thermometer that modulates immune evasion by Neisseria meningitidis. Nucleic Acids Res 44: 9426–9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Kusters JG, Stojiljkovic I, Heffron F. 1994. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun 62: 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, et al. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog 8: e1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Maris C, Allain FHT, Narberhaus F. 2006. Molecular basis for temperature sensing by an RNA thermometer. EMBO J 25: 2487–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci 106: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan SJ, Simmons CP, Strugnell RA. 1998. Comparison of the abilities of different attenuated Salmonella typhimurium strains to elicit humoral immune responses against a heterologous antigen. Infect Immun 66: 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. 2004. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett 237: 49–55. [DOI] [PubMed] [Google Scholar]
- Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. 2008a. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc 130: 8884–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. 2008b. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc 130: 12244–12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso-Becera MV, Servín-González L, Soberón-Chávez G. 2015. RNA structures are involved in the thermoregulation of bacterial virulence-associated traits. Trends Microbiol 23: 509–518. [DOI] [PubMed] [Google Scholar]
- Johnson K, Charles I, Dougan G, Pickard D, O'Gaora P, Costa G, Ali T, Miller I, Hormaeche C. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol 5: 401–407. [DOI] [PubMed] [Google Scholar]
- Jones CN, Wilkinson KA, Hung KT, Weeks KM, Spremulli LL. 2008. Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA 14: 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabiber F, McGinnis JL, Favorov OV, Weeks KM. 2013. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B, Cimdins A, Gaubig LC, Roßmanith J, Aschke-Sonnenborn U, Narberhaus F. 2012. Thermogenetic tools to monitor temperature-dependent gene expression in bacteria. J Biotechnol 160: 55–63. [DOI] [PubMed] [Google Scholar]
- Kolisek M, Launay P, Beck A, Sponder G, Serafini N, Brenkus M, Froschauer EM, Martens H, Fleig A, Schweigel M. 2008. SLC41A1 is a novel mammalian Mg2+ carrier. J Biol Chem 283: 16235–16247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 10: 255–265. [DOI] [PubMed] [Google Scholar]
- Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F. 2011. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res 39: 2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouse AB, Righetti F, Kortmann J, Narberhaus F, Murphy ER. 2013. RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS One 8: e63781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, et al. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14: 683–695. [DOI] [PubMed] [Google Scholar]
- Lewis C, Skovierova H, Rowley G, Rezuchova B, Homerova D, Stevenson A, Spencer J, Farn J, Kormanec J, Roberts M. 2009. Salmonella enterica Serovar Typhimurium HtrA: regulation of expression and role of the chaperone and protease activities during infection. Microbiology 155: 873–881. [DOI] [PubMed] [Google Scholar]
- Lipinska B, Fayet O, Baird L, Georgopoulos C. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol 171: 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DC, Savidge TC, Pickard D, Eckmann L, Kagnoff MF, Dougan G, Chatfield SN. 1999. Characterization of candidate live oral Salmonella typhi vaccine strains harboring defined mutations in aroA, aroC, and htrA. Infect Immun 67: 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis JL, Duncan CDS, Weeks KM. 2009. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol 468: 67–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231. [DOI] [PubMed] [Google Scholar]
- Meyer M, Plass M, Pérez-Valle J, Eyras E, Vilardell J. 2011. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell 43: 1033–1039. [DOI] [PubMed] [Google Scholar]
- Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. [Google Scholar]
- Mo E, Peters SE, Willers C, Maskell DJ, Charles IG. 2006. Single, double and triple mutants of Salmonella enterica serovar Typhimurium degP (htrA), degQ (hhoA) and degS (hhoB) have diverse phenotypes on exposure to elevated temperature and their growth in vivo is attenuated to different extents. Microb Pathog 41: 174–182. [DOI] [PubMed] [Google Scholar]
- Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. 1999. Translational induction of heat shock transcription factor σ32: evidence for a built-in RNA thermosensor. Genes Dev 13: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. 2007. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 129: 4144–4145. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. 2009. C2′-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc Natl Acad Sci 106: 15622–15627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. 2008. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res 36: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW. 1997. The HtrA family of serine proteases. Mol Microbiol 26: 209–221. [DOI] [PubMed] [Google Scholar]
- Raspoet R, Shearer N, Appia-Ayme C, Haesebrouck F, Ducatelle R, Thompson A, Van Immerseel F. 2014. A genome-wide screen identifies Salmonella Enteritidis lipopolysaccharide biosynthesis and the HtrA heat shock protein as crucial factors involved in egg white persistence at chicken body temperature. Poult Sci 93: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. 2010. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res 38: 3834–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roßmanith J, Narberhaus F. 2016. Exploring the modular nature of riboswitches and RNA thermometers. Nucleic Acids Res 44: 5410–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Cowen LE. 2012. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. MBio 3: e00238-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried NA, Bevilacqua PC. 2009. Thinking inside the box: designing, implementing, and interpreting thermodynamic cycles to dissect cooperativity in RNA and DNA folding. Methods Enzymol 455: 365–393. [DOI] [PubMed] [Google Scholar]
- Srikumar S, Kröger C, Hébrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron ADS, Hokamp K, Hinton JCD. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11: e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strulson CA, Boyer JA, Whitman EE, Bevilacqua PC. 2014. Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA 20: 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R, Shefer K, Ares M Jr. 2013. Safer one-pot synthesis of the ‘SHAPE’ reagent 1-methyl-7-nitroisatoic anhydride (1m7). RNA 19: 1857–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachon VK, Conn GL. 2012. Plasmid template design and in vitro transcription of short RNAs within a “structure cassette” for structure probing experiments. Methods Mol Biol 941: 157–169. [DOI] [PubMed] [Google Scholar]
- Wagner D, Rinnenthal J, Narberhaus F, Schwalbe H. 2015. Mechanistic insights into temperature-dependent regulation of the simple cyanobacterial hsp17 RNA thermometer at base-pair resolution. Nucleic Acids Res 43: 5572–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. 2007. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 65: 413–424. [DOI] [PubMed] [Google Scholar]
- Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, Makino DL, Nutter RC, Segal E, Chang HY. 2012. Genome-wide measurement of RNA folding energies. Mol Cell 48: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM, Mauger DM. 2011. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res 44: 1280–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc 127: 4659–4667. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. 2009. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA 15: 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.