Abstract
Introduction
Human induced pluripotent stem cells (iPSCs) are a promising source of endothelial cells (iPSC-ECs) for engineering three-dimensional (3D) vascularized cardiac tissues. To mimic cardiac microvasculature, in which capillaries are oriented in parallel, we hypothesized that endothelial differentiation of iPSCs within topographically aligned 3D scaffolds would be a facile one-step approach to generate iPSC-ECs as well as induce aligned vascular organization.
Methods
Human iPSCs underwent endothelial differentiation within electrospun 3D polycaprolactone (PCL) scaffolds having either randomly oriented or parallel-aligned microfibers. Using gene, protein, and endothelial functional assays, endothelial differentiation was compared between conventional two-dimensional (2D) films and 3D scaffolds having either randomly oriented or aligned microfibers. Furthermore, the role of parallel-aligned microfiber patterning on the organization of vessel-like networks was assessed.
Results
The cells in both the randomly oriented and aligned 3D scaffolds demonstrated an 11-fold upregulation in gene expression of the endothelial phenotypic marker, CD31, compared to cells on 2D films. This upregulation corresponded to >3-fold increase in CD31 protein expression in 3D scaffolds, compared to 2D films. Concomitantly, other endothelial phenotypic markers including CD144 and endothelial nitric oxide synthase also showed significant transcriptional upregulation in 3D scaffolds by >7-fold, compared to 2D films. Nitric oxide production, which is characteristic of endothelial function, was produced 4-fold more abundantly in 3D scaffolds, compared to on 2D PCL films. Within aligned scaffolds, the iPSC-ECs displayed parallel-aligned vascular-like networks with 70% longer branch length, compared to cells in randomly oriented scaffolds, suggesting that fiber topography modulates vascular network-like formation and patterning.
Conclusion
Together, these results demonstrate that a 3D scaffold structure promotes endothelial differentiation, compared to 2D substrates, and that aligned topographical patterning induces anisotropic vascular network-like organization.
Electronic supplementary material
The online version of this article (doi:10.1007/s12195-017-0502-y) contains supplementary material, which is available to authorized users.
Keywords: Endothelial cell, Three-dimensional scaffolds, Topography, Anisotropy, Tissue engineering, Vascularization, Tissue engineering, Induced pluripotent stem cell
Introduction
Three-dimensional (3D) engineered tissues are a promising solution to the shortage of donor myocardium for transplantation. With the advent of human induced pluripotent stem cell (iPSC) technology,50 the ability to derive patient-specific cardiovascular lineages for tissue engineering is no longer a limitation. However, a major remaining roadblock to engineering scalable 3D myocardial tissue constructs is the lack of effective strategies to transport nutrients to cells.30 Physiologically, cells reside within 100–200 μm from blood vessels in order to survive, due to the diffusion limit of oxygen through biological tissues.27 Moreover, the microvascular architecture of the myocardium is well-organized and anisotropic, in which capillaries lined by vascular endothelial cells (ECs) are oriented in parallel along the direction of the muscle fibers.19 Accordingly, the ability to derive ECs from human iPSCs and organize them into ordered vascular networks is important for developing vascularized engineered cardiac tissues.
Stem cell specification and tissue morphogenesis are largely dependent on spatiotemporally coordinated signaling mechanisms16,25 and architectural organization.15 Cells use mechanosensitive mechanisms to sense their surroundings, and are often highly influenced developmentally and functionally by their extracellular microenvironment.13 Since vascular patterning during embryonic development takes place within a 3D environment, 3D scaffolds may confer more physiologically relevant spatial cues to ECs, compared to conventional 2D substrates.34 Moreover, since topographical cues from scaffolds have been shown to modulate the orientation, organization and fate commitment of cells,14 their strategic incorporation into scaffold design can serve as an additional modulator of cellular organization. However, the effect of 3D scaffold structure and aligned topographical patterning on endothelial differentiation and organization is largely unknown. Aside from what has been previously shown of endothelial differentiation within 3D fibrin hydrogels,57 very little is known of how 3D scaffolds and/or aligned topographical cues influence this process.
A common strategy to create 3D vascularized engineered tissues is to differentiate human iPSCs into iPSC-derived ECs (iPSC-ECs) on a 2D platform using soluble factors, followed by embedding the cells into 3D scaffolds to induce vascular network organization.33,58 We hypothesized that endothelial differentiation directly within topographically aligned 3D scaffolds may be a facile one-step approach to generate iPSC-ECs with aligned network-like organization that mimics myocardial microvasculature.
Here, we investigated the endothelial differentiation process and vascular network-like formation of iPSC-ECs in topographically parallel-aligned 3D microfibrous scaffolds. We demonstrate that both randomly oriented and aligned 3D scaffolds promote more endothelial differentiation, compared to 2D films. However, aligned 3D scaffolds direct the formation of parallel-oriented vascular network-like structures, producing significantly longer branches than that on randomly oriented scaffolds. These findings have important implications in engineering cardiac tissues with ordered microvasculature.
Materials and Methods
Polymer Preparation and Electrospinning
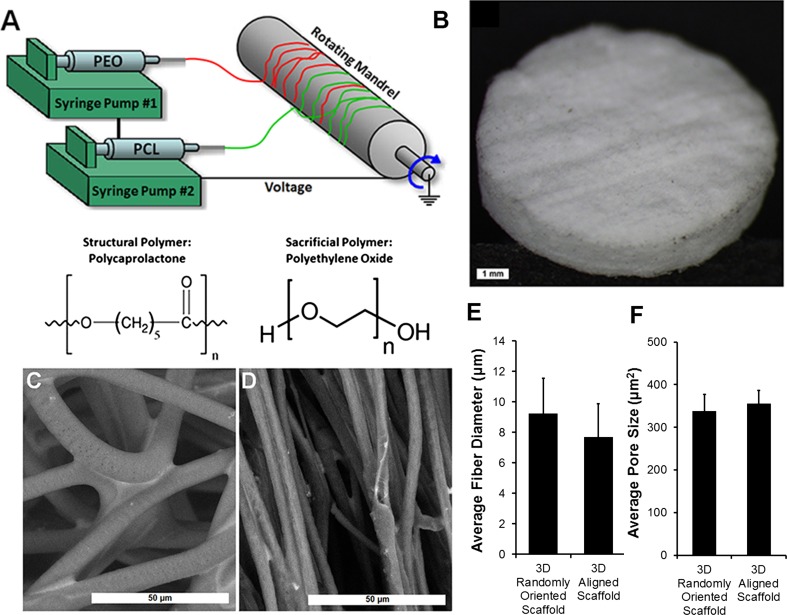
3D porous microfibrous scaffolds with either randomly oriented or parallel-aligned fiber orientation were fabricated by electrospinning of polycaprolactone (PCL) scaffolds (Suppl Fig. 1). Polyethylene oxide (PEO) was concurrently electrospun with PCL as a sacrificial polymer to further increase the porosity of the scaffolds. To prepare 3D scaffolds of random fiber orientation by electrospinning, a 30% solution of PCL (Sigma Aldrich, Mn 80,000) was dissolved in chloroform and loaded into a syringe pump (New Era Pump Systems Inc.). PEO (18%, Sigma Aldrich, Mv 100,000) was dissolved in chloroform and loaded into a separate syringe. PCL and PEO were simultaneously electrospun from separate spinnerets at 2.5 mL/h using a commercial electrospinner (Nanospinner 24-XP, Inovenso Ltd.) at a voltage of 30.0 kV, mandrel speed of 200 RPM, and a distance from spinneret to mandrel of 100 mm (Fig. 1a). After electrospinning, the scaffold sheet was dessicated for 24 h. The 3D scaffold of aligned fiber orientation was generated from 3D random scaffolds by heat-based mechanical stretching.24 Briefly, scaffolds strips (0.5 × 1 × 0.04 inches) were stretched uniaxially to 300% of original length using a 1 kg weight for 1 h at 55 °C. After fabrication of both random and aligned fiber scaffold strips, circular scaffold discs were cut using a 6 mm biopsy punch (Medex Supply), and then scaffold discs (6 mm in scaffold diameter, 0.8 mm in thickness) were submerged in ddH2O at 37 °C overnight, with agitation, to remove the PEO. The following day, scaffold discs were incubated in 70% ethanol with agitation at room temperature for 2 h for disinfection. After ethanol disinfection, the scaffold discs were washed three times in sterile ddH2O, and stored in sterile ddH2O until ready for cell seeding. As a basis for comparison, 2D PCL films were prepared by preparing a 1% (w/v) solution in chloroform. The PCL solution was then poured onto 18 mm round glass coverslips (Fisher Scientific) and allowed to cure for 24 h prior to ethanol disinfection and cell seeding.
Figure 1.
Fabrication and characterization of electrospun 3D PCL scaffolds. (a) Schematic of concurrent electrospinning of PCL and PEO; (b) Macroscopic view of 3D scaffold; (c–d) SEM images of randomly oriented (c) and aligned (d) scaffold structure; (e) Quantification of average microfiber diameter (n = 3); (f) Quantification of average pore size (n = 3). Scale bar: 1 mm (b); 50 µm (c–d).
Cell Culture and Endothelial Differentiation
The human iPSC line (obtained from Dr. Joshua Knowles, MD, PhD) was generated from reprogramming of healthy human adult peripheral blood mononuclear cells using Sendai virus-mediated transduction of Sox2, Oct3/4, KLF4, and c-myc.7 The iPSCs were expanded on tissue culture polystyrene (TCPS) dishes coated with geltrex (Sigma) in Essential 8 medium (Gibco). Endothelial differentiation was carried out using a protocol modified from Patsch et al. 45 The iPSCs were seeded into randomly oriented or aligned scaffolds at 250,000 cells/cm2 in Essential 8 medium. After 6 h, during which cells have attached to the scaffolds, endothelial differentiation was initiated using a differentiation media composed of Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12, Gibco), supplemented with 1X B27 (Gibco), 1X N27 (Gibco), 5 µM Wnt agonist CHIR-99021 (Selleckchem) and 25 ng/mL bone morphogenetic protein-4 (BMP-4, Peprotech). The cells were maintained in this media formulation for 3 days, after which the media was replaced with StemPro-34 media (Gibco), supplemented with 50 ng/mL vascular endothelial growth factor-A (VEGF-A,Peprotech) and 5 µM forskolin (LC Labs). Cells were then cultured in this media for 2 days, and subsequently prepared for analysis. As controls for 3D scaffolds, iPSCs were seeded onto 2D PCL films at a seeding density of 50,000 cells/cm2 and differentiated using the same method as that of 3D scaffolds.
Immunofluorescence Staining of Endothelial Differentiation
Immunofluorescence staining was performed to assess endothelial differentiation based on the expression of the phenotypic marker, CD31, using established procedures.22,44 In brief, at 1, 3, and 5 days of differentiation, the cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences), permeabilized in 0.1% Triton X-100 (Sigma-Aldrich), and then blocked with 5% normal goat serum (Cell Signaling Technology). The samples were then incubated with primary antibodies mouse anti-human CD31 (Dako), sheep anti-human CD31 antibody (R&D Systems) or mouse anti-human paxillin (BD Transduction). Afterwards, the cells were washed in phosphate buffered saline (PBS) before the application of AlexaFluor 488- or AlexaFluor 594-conjugated secondary antibodies (Thermo Fisher Scientific) for 2 h. Next, the samples were washed in PBS, and total nuclei were counterstained using either Hoechst 33342 or TO-PRO-3 (Thermo Fisher Scientific) nuclear dyes. Samples were then stored at 4 °C protected from light until ready for imaging using a laser scanning confocal microscope (LSM 710, Zeiss). Confocal 3D z-stacks were acquired with a z-stack thickness of 300–400 µm. Using ImageJ (version 1.48, NIH), confocal microscopy depicting maximum projections were generated from the z-stacks. Each maximum projection image depicting CD31 was converted to grayscale, and the mean gray value was computed using ImageJ. In the corresponding image stained using TO-PRO-3 for nuclei, total nuclei were counted using the particle counting tool. Measurements of CD31 mean intensity were extracted and then normalized by total cell nuclei. For each treatment group at each time point, three independent samples were assessed.
Analysis of Cell Penetration Depth
After 5 days of differentiation within the 3D scaffolds, the depth of cell penetration was analyzed histologically. The scaffold samples containing cells were embedded in optimal cutting temperature compound (OCT, Sakura Finetek) and then cryosectioned transversely at 50 µm thickness and then mounted onto glass slides. The cell nuclei within the sections were counterstained with TO-PRO-3. The mounted sections were subsequently imaged with a confocal microscope. The depth of cellular infiltration into the scaffold was quantified using ImageJ using the line measurement tool. For each treatment group, five sections per sample were analyzed, and the data was averaged among three independent samples.
Morphological Assessment of Differentiated iPSC-ECs Seeded in 3D Scaffolds
To better visualize the morphology of iPSC-ECs within 3D randomly oriented or aligned scaffolds, iPSC-ECs were generated using a high efficiency differentiation protocol modified from previous literature38,54 and then seeded onto the 3D scaffolds. In brief, iPSCs cultured on tissue culture dishes were incubated with CHIR-99021 (5 µM), BMP-4 (25 ng/mL), B27 supplement, and N2 supplement. After 3 days, the cells were dissociated with HyQtase (Fisher Scientific) and plated back onto tissue culture dishes (3.3 × 104 cells/cm2) in StemPro-34 media (Gibco), containing forskolin (5 µM), VEGF-A (50 ng/mL), and polyvinyl alcohol (2 mg/mL, Sigma). After 7 days, the cells were washed twice with PBS, and cultured in endothelial growth media (EGM-2MV, Lonza) supplemented with additional VEGF (100 ng/mL) for 7 more days. The differentiated iPSC-ECs were then seeded into randomly oriented or aligned scaffolds for 2 days before fixation in 4% paraformaldehyde and immunofluorescence staining for CD31 (Dako) as described above.
Scanning Electron Microscopy (SEM)
For variable pressure SEM (VP-SEM) analysis, samples were fixed for 24 h with 4% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2), rinsed in the same buffer and post-fixed for 1 h with 1% aqueous OsO4 to enhance contrast of cellular material. Samples were mounted fully hydrated onto 10 mm cup-shaped stubs custom-fitted for the Deben cold-stage (Deben Ltd, Suffolk), and temperature was gradually decreased during evacuation. VP-SEM was carried out with a Hitachi S-3400 N VP-SEM (Hitachi High Technologies) operated at 15 kV and 50–60 Pa, using Backscattered Electron detection and cold-stage (−25 °C) control of hydration.
Field emission SEM (FESEM) analysis was carried out according to previous methods.44 In brief, samples were fixed with 4% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2), and post-fixed with 1% aqueous OsO4, followed by dehydration in an ascending ethanol series. Samples were then critical point dried with liquid CO2 in a Tousimis Autosamdri-815B apparatus (Tousimis), mounted onto double-sided copper tape on 15 mm aluminum stubs (Electron Microscopy Sciences), and sputter-coated with 50 Å of gold–palladium using a Denton Desk II Sputter Coater (Denton Vacuum). Visualization was performed with a Zeiss Sigma FESEM (Carl Zeiss Microscopy) operated at 2–5 kV using a combination of InLens and SE2 (Everhart–Thornley) secondary electron detectors. Images were captured in TIFF using store resolution 2048 × 1536 pixels and a line averaging noise reduction algorithm.
Scaffold attributes were quantified using ImageJ. The line measurement tool was used to measure fiber diameter, with a minimum of ten measurements made per SEM image, with a total of ten images per sample. Pore sizes were quantified by thresholding using a high-pass filter to detect the void spaces between fibrils. For each treatment group, three independent samples per treatment group were assessed.
Reflectance Microscopy Imaging of Scaffold Topography
Complementary to SEM imaging, reflectance microscopy was employed to visualize scaffold structure using a laser scanning confocal microscope (LSM 710, Zeiss). Two-dimensional Fast Fourier Transform (2D FFT) analysis was performed using an ImageJ plugin to generate frequency plots and alignment histograms that depict fiber orientation (n = 3).3
Quantitative Polymerase Chain Reaction
Gene expression analysis was conducted using RT-PCR of cDNA synthesized from purified RNA. The GeneJET RNA purification kit (Thermo-Fisher Scientific) was used to isolate and purify total RNA, according to the manufacturer’s instructions. The concentration of total RNA was measured using a UV–Vis Spectrophotometer (NanoDrop 2000, Thermo Scientific), and cDNA was synthesized from total RNA using the SuperScript II First-Strand cDNA Synthesis kit (Thermo Fisher Scientific) following the manufacturer’s instructions and synthesized in a compact thermal cycler (T100 Thermal Cycler, Bio-Rad). Real time PCR was carried out using Taqman primers (Thermo Fisher Scientific) for CD31, CD144, VEGF-A, von Willebrand factor (vWF), endothelial nitric oxide synthase (eNOS), and GAPDH using a 7300 Real-Time PCR System (Applied Biosystems) with the following thermal profile: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s/cycle, and then 60 °C for 1 min. The data were quantified by the ΔΔCt method,40 normalized to GAPDH housekeeping gene, and then expressed as relative fold changes (n = 3).
Nitric Oxide Assay and Quantification
Endothelial cell function was assessed via quantification of nitric oxide production using the fluorescent probe, 4-amino-5-methylamino-2′,7′-difluorofluorescein (DAF-FM) diacetate (Thermo Fisher Scientific).32 Samples were incubated with 5 μM solution of DAF-FM in DMSO for 40 min at 37 °C and then washed in PBS to remove excess probe. After an additional 30 min of incubation in EGM-2MV endothelial growth medium (Lonza) to allow for complete de-esterification of intracellular diacetates, the living cells were then imaged using a confocal microscope (Zeiss LSM710). 3D z-stacks were acquired, with a z-stack thickness of 300–400 µm, and a 1 µm optical section thickness. DAF-FM intensity was captured and quantified using ImageJ by thresholding image stacks based on hue, saturation and pixel value. The measurement of DAF-FM integrated density was extracted, and the values were normalized by total cell number based on Hoechst staining. For each sample, 5 images at 10× magnification were analyzed (n = 3).
Analysis of Vascular Network-Like Formation
Vascular network organization metrics were obtained using Imaris 8.3 (Bitplane) equipped with the Filament Tracer tool. Using whole z-stack confocal microscopy images, filament objects were added and segmented using the “autopath no loops” algorithm setting. The average filament length and number of branch points were quantified from image stacks of CD31 expression as measures of vascular branch length and complexity (n = 5).
Statistical Analysis
All data are expressed as mean ± standard deviation. Statistical analysis was performed by the Student’s t test for comparison of two groups or one-way analysis of variance (ANOVA) with Holm’s adjustment for multiple comparisons. For comparison of the same treatment group at three time points, a repeated measures ANOVA with Holm’s adjustment was performed. Statistical significance was accepted at p < 0.05.
Results
Characterization of 3D Microfibrous Scaffold
The randomly oriented fibrous polymer scaffolds were fabricated by co-electrospinning of the PCL structural polymer with PEO sacrificial polymer (Fig. 1a). To generate parallel-aligned fibrous scaffolds, the resultant scaffolds were further mechanically stretched uniaxially by 300% of original length. After thorough removal of PEO, the remaining PCL scaffold was approximately 800 µm in thickness (Fig. 1b). The scaffolds were imaged using VP-SEM to capture the geometrical architecture of scaffolds in the wet state. Quantitative characterization of PCL scaffolds by VP-SEM imaging revealed a mean fiber diameter of 9.2 ± 0.7 µm on the randomly oriented scaffolds (Figs. 1c and 1e). Mechanical stretching to create aligned fibrous scaffolds resulted in a mean fiber diameter of 7.6 ± 0.3 µm (Figs. 1d and 1e), which was not significantly different from the mean fiber diameter of randomly oriented scaffolds. In addition, the average pore area between the randomly oriented and aligned fiber configurations was similar (randomly oriented scaffold 338 ± 39 µm2, aligned scaffold: 356 ± 32 µm2, Fig. 1f). The randomly oriented and aligned scaffold configurations were used for all subsequent assays to compare the effect of three-dimensionality and fiber topography on endothelial differentiation and vascular network-like formation.
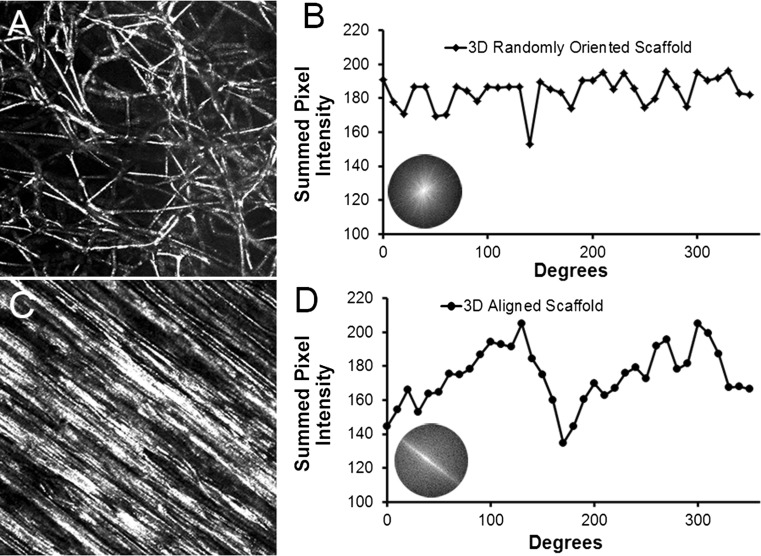
To quantify the degree of parallel fiber alignment (anisotropy), we performed 2D FFT analysis using the reflectance microscopy images to generate frequency plots and alignment histograms (Fig. 2). A frequency plot (Figs. 2b and 2d insets) is composed of grayscale pixels distributed about the center. For images that lack parallel fiber alignment, the frequency plot consists of white pixels distributed symmetrically about the center, in which pixels close to the center represent pixels in the source reflectance microscopy image that do not vary notably in intensity with adjacent pixels in the image.3 In contrast, for images that contain parallel fiber alignment, the frequency plot contains pixels distant from the center and concentrated along the axis of anisotropy.3 In comparing the frequency plots between aligned and randomly oriented scaffolds, the aligned scaffold showed distinctive characteristics of anisotropy based on the pattern of white pixels along the axis of fiber alignment (Fig. 2d inset), whereas the randomly oriented scaffold showed characteristics of isotropy in which the white pixels were generally distributed about the center (Fig. 2b inset).
Figure 2.
Characterization of 3D scaffolds using 2D Fast Fourier Transform (FFT) analysis. (a–b) Representative reflectance microscopy image of randomly oriented scaffold (a) and corresponding alignment histogram (b); (c–d) Representative reflectance microscopy image of an aligned scaffold (c) and corresponding alignment histogram (d). Insets in B and D depict frequency plots.
The frequency plots were also displayed quantitatively as frequency alignment histograms that depict the principal angle of orientation within 360° of space (Figs. 2b and 2d). In brief, the frequency alignment histogram depicts pixel intensities summed along the radius for each angle of circular projection (0–360°).3 Notably, images showing parallel fiber alignment have histograms with preferential arrayed pixels along the axis of fiber orientation, producing a prominent peak within the 0–180° followed by a second peak 180° later due to symmetry. As shown in Figs. 2b and 2d, the aligned scaffold was characterized by two distinctive peaks, whereas the randomly oriented scaffold was not characterized by any distinctive peaks. These results show that the aligned microfibrous scaffolds were similar to the randomly oriented 3D scaffolds in fiber diameter and pore size, with the notable difference of parallel-aligned fiber organization.
Cell Viability in 3D Scaffolds
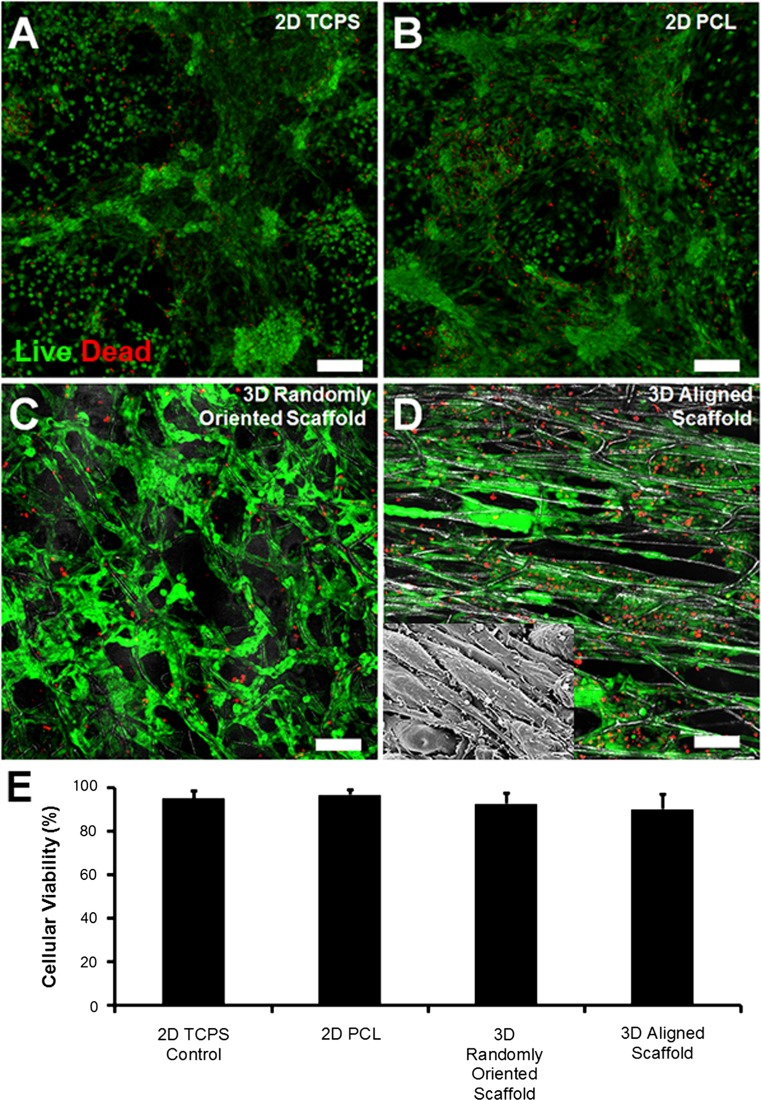
The biocompatibility of PCL scaffolds and their ability to support cellular viability were assessed using vital fluorescent dyes. After 5 days of differentiation, the majority of the cells in both the randomly oriented and aligned fibrous scaffolds showed similar levels of viability that were not significantly different from that on 2D PCL films or TCPS (2D TCPS: 96 ± 3%; 2D-PCL: 96 ± 3%; 3D-random oriented scaffold: 93 ± 5%; and 3D aligned scaffold: 91 ± 8%, (Figs. 3a–3e). These results demonstrated high viability of cells cultured in all PCL conditions and comparable viability to conventional TCPS. Therefore, there did not appear to be cytotoxic effects associated with PCL scaffolds in 2D or 3D configurations.
Figure 3.
Cell viability in 3D micofibrous scaffolds after 5 days of differentiation using Live/Dead cytotoxicity vital dyes. Representative fluorescent images depicting viable (green) and dead (red) cells are shown in samples consisting of 2D tissue culture polystyrene (TCPS) (a), 2D PCL (b), 3D randomly oriented scaffold (c), or 3D aligned scaffold (d). Depicted in the inset is the cellular morphology after 5 days of differentiation as imaged by SEM (e). Quantification of cell viability (n = 3). Scale bar: 100 µm.
Depth of Cell Penetration into 3D Scaffolds
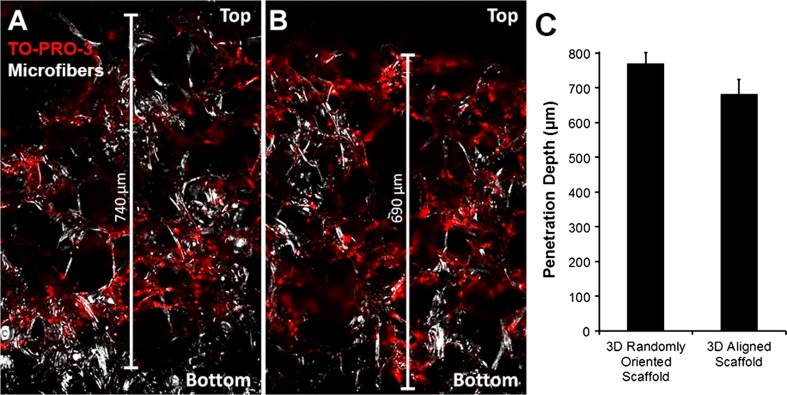
In order to determine the penetration depth of differentiating iPSCs, the cell-seeded aligned or randomly oriented scaffolds were cryosectioned transversely, and then stained with TO-PRO-3 nuclear dye for fluorescence visualization of cells. Cryosections were fluorescently visualized using a laser scanning confocal microscope. In both randomly oriented and aligned 3D PCL scaffolds, the cells were able to infiltrate nearly through the entire depth of the scaffolds after 5 days of culture in endothelial differentiation media (Figs. 4a and 4b). The average depth of penetration for randomly oriented scaffolds was 770 ± 14 µm, corresponding to 683 ± 18 µm for aligned scaffolds (Fig. 4c). The total scaffold thickness was ~800 µm. These results demonstrated that the geometrical architecture of both random and aligned PCL scaffolds enabled deep cellular penetration.
Figure 4.
Analysis of the depth of cellular penetration into 3D scaffolds. TO-PRO-3 (red) nuclear labeling of cells in transverse sections of randomly oriented (a) or aligned scaffolds (b); (c) Quantification of penetration depth (n = 3). Top side refers to the air-facing side of the scaffold. Corresponding microfibers are visualized using reflectance microscopy (white). Scale bar: 100 µm.
Phenotypic and Functional Analysis of Endothelial Differentiation
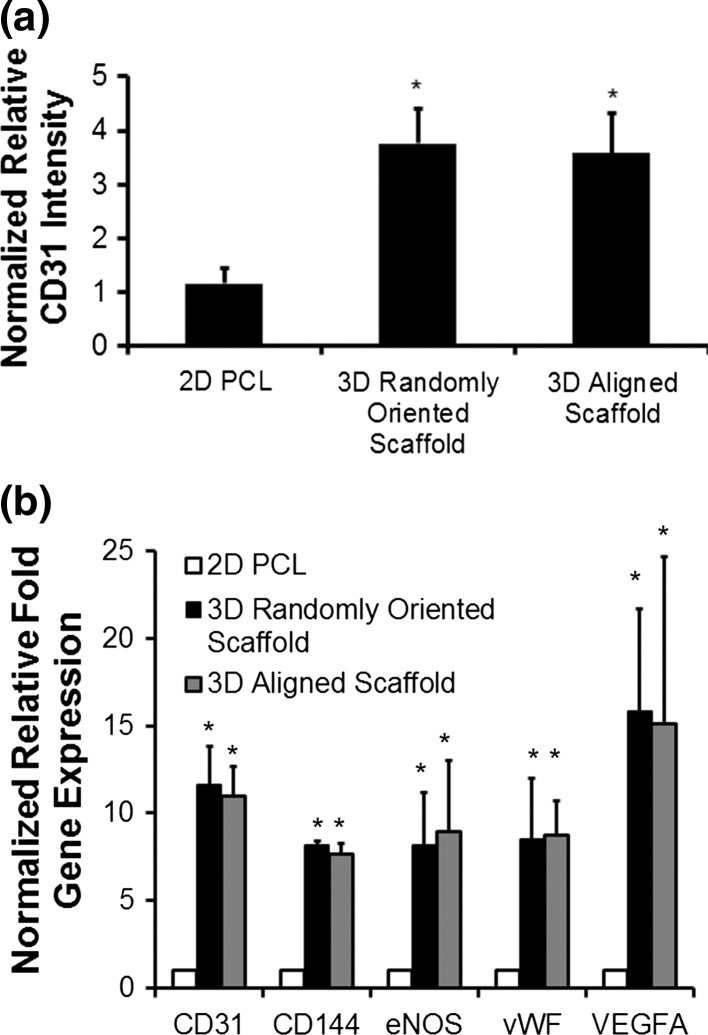
After confirmation of cellular viability and adequate depth penetration of iPSCs in 3D microfibrous scaffolds, the capacity of iPSCs to undergo endothelial differentiation was subsequently investigated. After 5 days of differentiation in either randomly oriented or aligned fibrous scaffold configurations, as well as on 2D PCL film control samples, the cells were fixed and immunofluorescently stained for the endothelial cell adhesion marker, CD31. Confocal stacked images were acquired using a laser scanning confocal microscope, and the CD31 intensity was normalized to total nuclei and then expressed relative to the 2D PCL treatment group. The normalized relative intensity of CD31 was significantly higher in the 3D scaffolds (randomly oriented scaffold: 3.8 ± 0.6 fold higher; aligned scaffold: 3.6 ± 0.7 fold higher), compared to 2D PCL films (p < 0.05) (Fig. 5a).This enhancement in CD31 protein expression in 3D environments suggested that, regardless of fiber topography, 3D porous scaffolds induced more endothelial differentiation than 2D substrates. To explore the temporal response of endothelial differentiation in 3D scaffolds, normalized relative CD31 protein expression measured between 1 through 5 days of differentiation revealed the direct relationship of increasing CD31 protein expression with time (Suppl Fig. 2). However, there was no significant difference in expression between the randomly oriented scaffolds and aligned scaffolds.
Figure 5.
Quantification of endothelial differentiation after 5 days of differentiation. (a) Comparison of normalized relative CD31 intensity between 2D PCL and 3D randomly oriented or aligned scaffolds (n = 3); (b) Normalized relative fold change in gene expression for endothelial-related genes (n = 3). * indicates p < 0.05 (n = 3).
To verify these results derived from protein expression, we further compared the gene expression of endothelial phenotypic markers for cells that were differentiated on either 2D films or the 3D scaffolds for 5 days (Fig. 5b). In agreement with the CD31 protein expression results, the qPCR analysis demonstrated that gene expression of CD31 was significantly upregulated when differentiated within 3D scaffolds (randomly oriented scaffold: 11.6 ± 1.3 fold change; aligned scaffold: 11.0 ± 0.98 fold change), than when compared to 2D films (p < 0.05). Similarly, CD144/VE-cadherin, an endothelial specific adhesion molecule located at junctions between ECs, was also significantly increased by >7-fold in 3D scaffolds (p < 0.05). A similar induction of >8-fold in 3D scaffolds was shown for von Wille brand Factor (vWF), a plasma glycoprotein that mediates platelet adhesion to endothelial cell surface. Since ECs are known to secrete paracrine factors such as VEGF-A, we also assessed the expression of this angiogenic growth factor and showed a >15-fold upregulation of VEGF-A in 3D scaffolds, compared to 2D films (p < 0.05). An additional gene we assessed was eNOS, an endothelial-specific enzyme that catalyzes the production of nitric oxide that confers vasorelaxation in blood vessels, and eNOS was also significantly increased in 3D scaffolds (p < 0.05) However, there was no significant difference in gene expression between randomly oriented and aligned fibrous scaffolds. These results suggested that the 3D scaffolds promoted greater endothelial differentiation, when compared to 2D substrates, regardless of fiber topographical orientation.
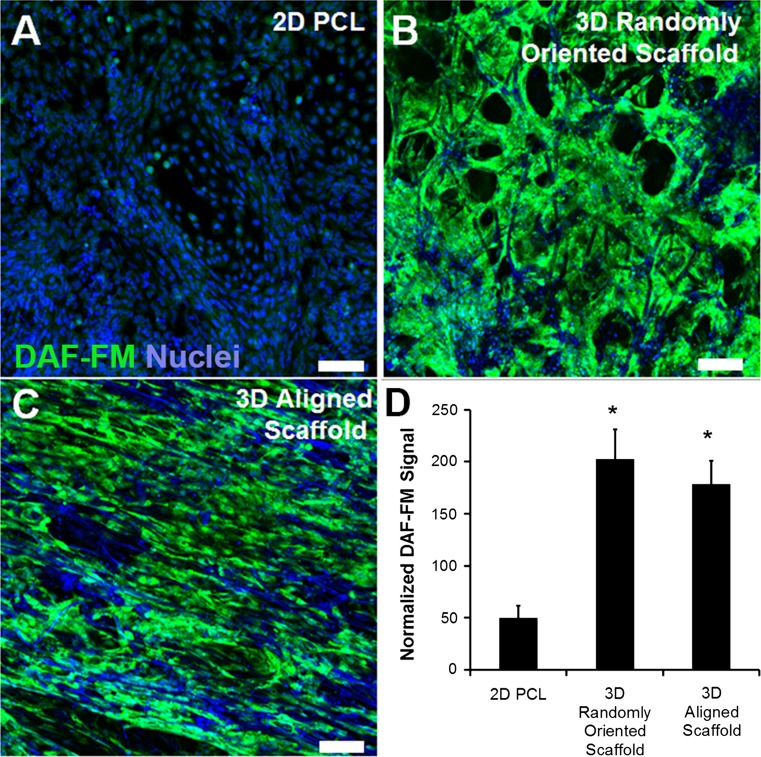
Since the ability to release nitric oxide is an important characteristic of functional ECs, to further corroborate the results derived from gene and protein analysis, the functional capacity of iPSC-ECs to produce nitric oxide was quantified using DAF-FM, a nitric oxide fluorescent probe. After normalization of fluorescent signal to cell area, the DAF-FM signal showed approximately a 4-fold increase in 3D randomly oriented scaffolds and approximately a 3.6-fold increase in 3D aligned scaffolds, compared to 2D films (p < 0.05, Fig. 6). These results indicate that this characteristic functionality of ECs is enhanced in three dimensions, compared to two dimensions, although there was no significant difference between randomly oriented vs. aligned scaffolds.
Figure 6.
Nitric oxide production in 3D microfibrous scaffolds after 5 days of differentiation. (a–c) Representative confocal images of nitric oxide fluorescent probe (green) are depicted in the 2D PCL film (a), 3D randomly oriented scaffold (b), and 3D aligned scaffold (c); (d) Quantification of normalized fluorescence signal (n = 3). Total nuclei were labeled using Hoechst 33342 (blue). * Denotes statistically significant relationship, compared to 2D PCL group (p < 0.05). Scale bar = 100 µm.
Vascular Network-Like Organization
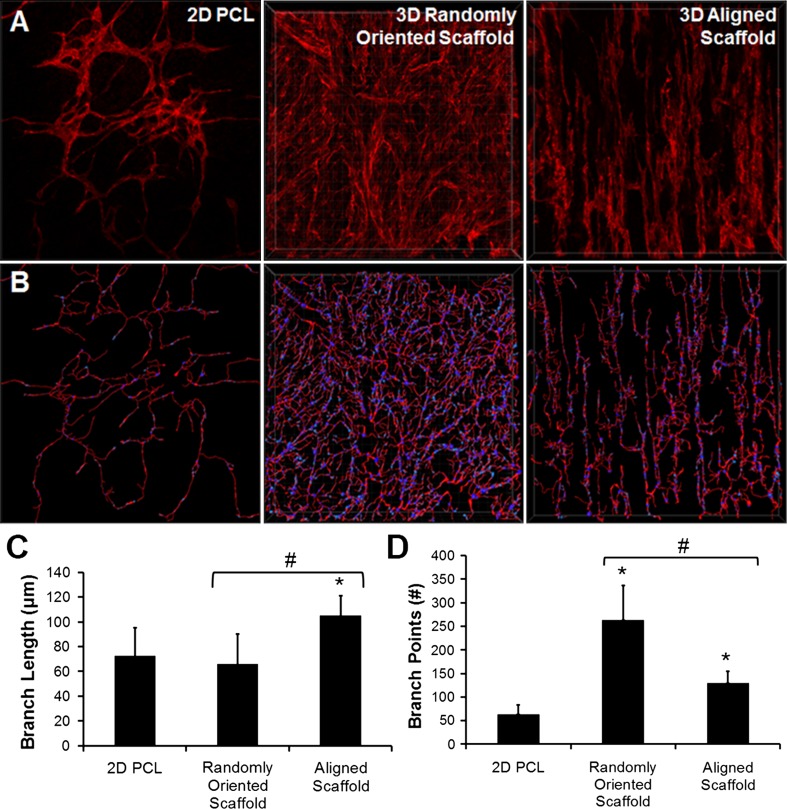
In the aligned fibrous scaffolds, the CD31-expressing iPSC-ECs organized themselves into vascular network-like structures that were oriented anisotropically along the direction of the fibers (Figs. 5a and 7a, Supplementary Video 1). In contrast, in the randomly oriented scaffolds, the CD31-expressing iPSC-ECs were more disorganized in their formation of vascular network-like structures (Figs. 5a and 7a, Supplementary Video 2).
Figure 7.
Vascular network-like formation in 3D microfibrous scaffolds. (a) 3D stacked confocal images of CD31 staining in 2D PCL film, 3D randomly oriented scaffold, and 3D aligned scaffold. (b) Transformation of CD31 expression into skeletonized filaments; (c–d) Quantification of branch length (c) and branch points (d). * indicates statistically significant relationship to 2D PCL film, and # indicates statistically significant comparison between 3D groups. p < 0.05 (n = 5).
To further characterize the vascular network-like formation, we analyzed the geometrical features by recreating 3D configurations of the vascular structures using Imaris software (Figs. 7a and 7b). From the 3D images, we quantified the branch length and number of nodes within the tube-like networks (Figs. 7c and 7d) as indicators of vascular organization and complexity. The branch length was significantly higher in the aligned fibrous scaffolds (105.0 ± 16.2 µm), compared to in the randomly oriented scaffolds (65.9 ± 24.1 µm) and 2D PCL scaffolds (72.5 ± 22.6 µm), which was consistent with longer and well-organized vascular-like structures in the aligned scaffolds (p < 0.05). Conversely, the number of nodes was higher in the randomly oriented scaffold (263.4 ± 72.9), compared to the aligned fibrous scaffolds (129.8 ± 24.5) and 2D PCL films (63.8 ± 18.9), which was consistent with the aligned scaffolds producing parallel-aligned network-like structures with fewer intersecting branches (p < 0.05). Together, these results demonstrated that aligned fibrous scaffolds could organize the vascular network-like structures anisotropically, whereas randomly oriented scaffolds produced more disorganized vascular network-like structures.
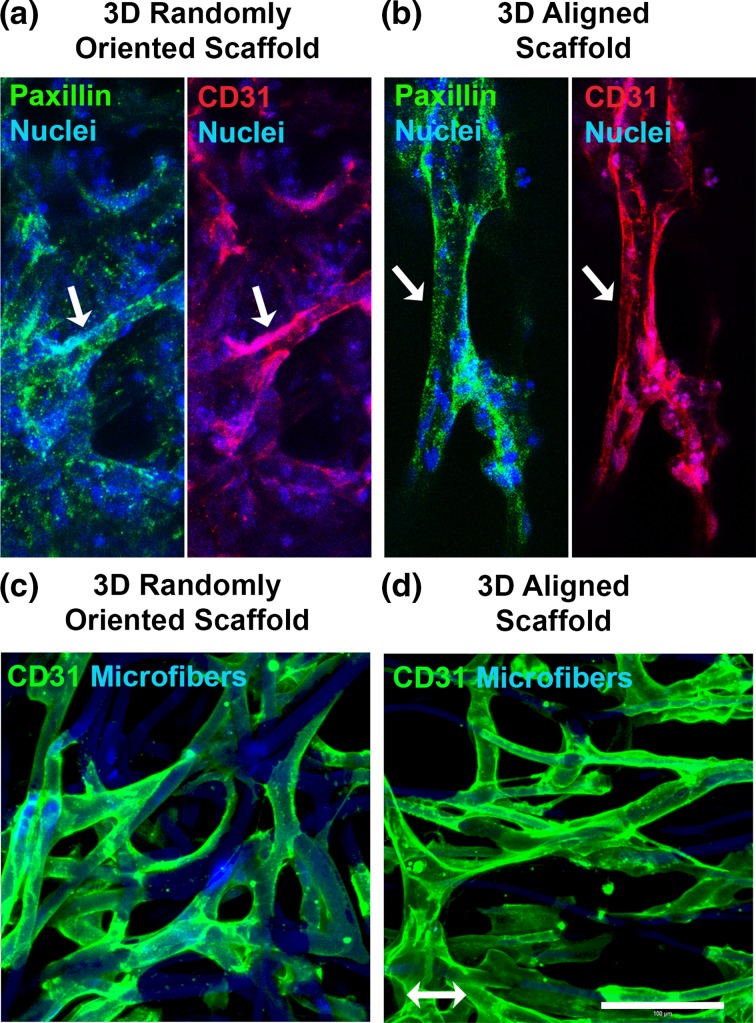
To better understand the mechanism by which iPSC-ECs formed aligned network-like structures in the aligned nanofibrillar scaffolds, we examined the focal adhesions formed by the cells to the microfibers. As shown in Figs. 8a and 8b, the CD31+ iPSC-ECs showed abundant expression of paxillin, a focal adhesion adapter protein that mediates cellular attachment to extracellular matrix (ECM) proteins.56 In both the randomly oriented and aligned scaffolds, the punctate assembly of paxillin was observed throughout the iPSC-ECs, especially along the primary axis of the elongated cell body.
Figure 8.
iPSC-EC morphology and focal adhesion assembly in 3D microfibrous scaffolds. (a–b) Confocal microscopy images depict paxillin expression in iPSC-ECs within 3D randomly oriented (a) or aligned (b) microfibrous scaffolds. CD31 (red), paxillin (green), and nuclei (blue). Arrows point to a representative CD31+ iPSC-EC. (c–d) Confocal microscopy images of iPSC-ECs cultured within randomly oriented or aligned scaffold depict elongated cellular morphology and wrapping of cell bodies around microfibers. The iPSC-ECs were visualized by CD31 (green) expression. The microfibers were visualized by autofluorescence (blue) near the 480 nm emission wavelength. Arrow denotes bulk microfiber alignment direction. Scale bar: 100 µm.
To better visualize the morphology of iPSC-ECs in the absence of other cell types, we seeded iPSC-ECs directly into the randomly oriented or aligned microfibrous scaffolds for 2 days and then visualized the cellular morphology based on CD31 expression (Figs. 8c and 8d). Owing to the autofluorescence of the microfibers near 490 nm wavelength, the iPSC-ECs could be visualized relative to the spatial geometries of the microfibers. The aligned scaffolds appeared to adopt an aligned organization by wrapping their cell bodies along the anisotropically patterned microfibers, resulting in an aligned organization of the cells (Fig. 8d). On the randomly oriented scaffolds, however, the cells wrapped their cell bodies around the randomly arranged microfibers, resulting in non-organized iPSC-ECs (Fig. 8c). These findings suggest that the iPSC-ECs differentiated within 3D scaffolds could sense the topographical cues of the scaffolds, leading to global arrangement of the iPSC-ECs along the direction of the aligned microfibers.
Discussion
The progress and promise of tissue engineering to produce patient-specific, functional and implantable tissues and organs hinges on the ability to safely and efficiently generate purified cell types of interest, grow them in biocompatible materials that mimic specific in vivo microenvironments, and ultimately provide these cell-seeded engineered constructs with functional vasculature. There are a variety of biophysical and biochemical cues that contribute to stem cell differentiation, and the identification of specific microenvironmental cues that direct lineage specification can facilitate this process. In this study, we investigated the combined effect of three dimensionality and topographical architecture on endothelial differentiation and organization of iPSC-ECs into aligned vascular-like networks. Our findings suggest that aside from the presentation of defined soluble factors in growth media, the 3D environment and topography also contribute to enhanced endothelial differentiation efficiency and control of vascular network-like formation. The salient findings from this study are: (1) 3D porous scaffolds augment endothelial differentiation, capacity (Figs. 5 and 6); and (2) anisotropic topography enhances the branch length frequency in the formation of vascular network-like structures (Fig. 7).
It is well recognized from in vivo physiology that an optimal balance of microenvironmental cues is required to support proper tissue development and function.28 To mimic vascular development in the embryo, a number of endothelial differentiation protocols employ soluble growth factors and/or extracellular matrix (ECM) proteins. Blancas et al. reported a chemically defined protocol using growth factors, including VEGF-A and BMP-4, along with ECMs such as fibronectin.6 Wu et al. employed not only soluble inducing factors of CHIR-99021and VEGF-A, but also the anti-absorptive agent polyvinyl alcohol to preferentially detach non-endothelial lineages, resulting in high yields of iPSC-ECs.54 Whereas high-efficiency differentiation protocols such as that from Wu et al. 54 might mask the potential benefit of 3D scaffolds, in this study we employed a differentiation protocol45 that, in our hands, yielded ~20% efficiency of CD31+ iPSC-ECs in conventional tissue culture dishes, and thereby allow for the possibility of observing an enhancement in differentiation in 3D scaffolds.
To date, there have been reports that 3D environments promote stem cell differentiation into ECs,57 cardiomyocytes,8 fibroblasts,47 smooth muscle cells,39,53 and other lineages.9,31,41,46,52 Our finding that 3D biomaterials are potent modulators of endothelial specification concurs with a study by Zhang et al. that demonstrated a significant improvement of endothelial differentiation in 3D fibrin (45% efficiency), compared to 2D (10% efficiency) substrates.57 We believe that the present study is the first report of endothelial differentiation of iPSCs within 3D synthetic polymer scaffolds.
The mechanism by which 3D structure influences cell fate specification is largely unknown, but is actively investigated.4,34 Since cells exist in 3D environments in vivo, they may respond differentially in vitro to 3D environments, compared to 2D environments, in which the degrees of freedom for motility and intercellular communication are reduced. In the context of angiogenesis, the sprouting of blood vessels is a phenomenon that requires 3D microenvironments to achieve higher orders of complexity.36 This complexity is largely a result of distinctions in cell adhesion patterns, tissue-specific mechanotransduction, and release kinetics of paracrine factors.4 One comprehensive genome-wide study comparing iPSC-ECs grown in 2D or 3D substrates revealed that iPSC-ECs developed in 2D adopt a more proliferative phenotype characterized by increased focal adhesion kinase (FAK)–extracellular signal-regulated kinase (ERK) activity, in which inhibition of ERK signaling increased the expression of vasculature development genes.58 Therefore, it is possible that the difference in signal transduction dynamics between integrins and cell adhesion kinases in 2D vs. 3D contributes highly to endothelial differentiation capacity and cellular organization.
Although prior studies have reported the importance of topographical cues for reprogramming efficiency in 2D substrates,14,49 we show that anisotropy of microfibrous 3D scaffolds not only enhanced endothelial differentiation, compared to 2D substrates, but also modulated the organization of the resulting CD31+ iPSC-ECs. The vascular-like structures in the aligned scaffolds were distinctively organized along the direction of the microfibers, whereas in the randomly oriented scaffolds the vascular network-like structures were highly disorganized (Fig. 7). The aligned microfibrous scaffolds provided biophysical cues that enabled the iPSC-ECs to form focal adhesions and wrap around the microfibers (Fig. 8). Our findings concur with the literature that demonstrates the role of aligned nano- or microfibers in modulating stem cell differentiation and/or cellular organization. For example, randomly oriented nanofibers induced significantly higher differentiation of embryonic stem cells (ESCs) into astrocytes than on aligned nanofibers.55 However, the ESC-derived astrocytes formed longer neurite outgrowth on aligned scaffolds, when compared to randomly oriented scaffolds. Another group showed that embryoid bodies seeded onto aligned scaffolds produced longer extensions and neuron outgrowth along the axis of the nanofibers, concomitant with upregulation of mature neural genes, compared to cells cultured on randomly oriented scaffolds.1
Although, the effects of aligned microfibrous 3D scaffolds on endothelial differentiation is largely unknown, we have previously demonstrated that aligned nanofibers not only induce iPSC-EC alignment, but also augment their survival and angiogenic function in ischemic tissues.23,43 The mechanism by which scaffold anisotropy modulates differentiation and cellular reorganization is not well-understood, but likely mediated by cell-ECM interactions. We previously showed that aligned nanofibrous scaffolds significantly upregulated the gene expression of integrin α1 subunit in primary human ECs, when compared to randomly oriented scaffolds Further work in this area will elucidate the signaling pathways by which anisotropic scaffolds modulate differentiation and cellular function.
Numerous technologies exist to fabricate aligned scaffolds, including mechanical loading,48 microfluidics,35,37 magnetic fields,20 and electrospinning.21,24 The advantages of electrospinning over other techniques include the ability to fabricate 3D scaffolds with scalable size and tunable fiber diameters. We created electrospun scaffolds composed of PCL owing to its biocompatibility and controllable mechanical properties.11 Furthermore, concurrent electrospinning with PEO as a sacrificial polymer further increased the porosity of the resulting scaffold.29
To characterize the endothelial identity of the iPSC-ECs generated within the 3D scaffolds, nitric oxide production served as an important functional assessment of endothelial identity.10,12 As a vasodilator and anti-atherogenic molecule, nitric oxide is an important gas that is released by ECs to maintain cardiovascular homeostasis.17,42 For this reason, nitric oxide production was selected as a functional assessment of endothelial identity. A 3-fold increase in the production of NO suggested a strong improvement in endothelial function in 3D scaffolds, compared to 2D films. However, to substantiate the significant increase in VEGF-A gene expression, other functional assays including proteomic analysis of secreted VEGF-A and other growth factors is warranted.
Given the stark differences in cellular organization on randomly oriented scaffolds compared to aligned scaffolds, these two kinds of scaffolds may be beneficial for different tissue engineering applications, depending on the tissue type of interest. The aligned scaffolds may be beneficial for engineering cardiac19 and skeletal muscle,5 which consist of parallel-aligned blood vessels interspersed among muscle fibers. In contrast, the randomly oriented scaffold may be well-suited for mimicking the disorganized vasculature of the dermis.51 Therefore, the topography of the scaffold should be strongly considered in the development of engineered tissues.
As with most in vitro culture systems, there are some limitations in this study that warrant further investigation. For example, basal and inducible permeability is differentially regulated across the vascular tree based on the EC phenotype and abundance of tight junctions.2 Moreover, in response to cues from the microenvironment, ECs from arteries and veins present epigenetic, morphological and functional differences,2 including those that control cell motility and thrombosis.18 Furthermore, the vascular network-like structures in this study were not developed to the point where they could support the perfusion of liquid through them. Incorporation of a bioreactor system may help sustain the survival of the cells to allow for further vascular maturation.26 In addition, endothelial progenitor cells also express CD31 and may be present within the population of iPSC-ECs.38 Since the iPSC differentiation often leads to a heterogeneous population of differentiating cells, future studies to identify the non-endothelial populations in the scaffolds are warranted.
Conclusion
In summary, we demonstrated that 3D microfibrous scaffolds enhanced the endothelial differentiation capacity of human iPSCs, compared to 2D substrates. Furthermore, parallel-aligned scaffolds induced spatially aligned vascular network-like structures that better mimic the organization of cardiac microvasculature. These findings suggest that conventional 2D cell culture systems may be limited in achieving endothelial differentiation and/or vessel-like structures that are physiologically observed in 3D microenvironments. Thus, these findings have important implications in the design of vascularized engineered tissues, in which 3D structure may enhance the differentiation efficiency of stem cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Video 1. 3D reconstructed view of CD31 (red) and total nuclei (green) in aligned microfibrous scaffolds, based on confocal microscopy. Supplementary material 1 (AVI 244517 kb)
Supplementary Video 2. 3D reconstructed view of CD31 (red) and total nuclei (green) in randomly oriented microfibrous scaffolds, based on confocal microscopy. Supplementary material 3 (AVI 242789 kb)
Acknowledgments
We thank Joshua Knowles, MD, Ph.D. and Ivan Carcamo-Orive, Ph.D., for technical assistance in endothelial differentiation. This study was supported by grants to NFH from the US National Institutes of Health (R00HL098688, R01HL127113, and R21EB020235), Merit Review Award (1I01BX002310) from the Department of Veterans Affairs Biomedical Laboratory Research and Development, the Stanford Women and Sex Differences in Medicine Center, the Stanford Child Health Research Institute. NFH was also supported by a McCormick Gabilan fellowship. MW was supported by a diversity supplement through the US National Institutes of Health (R01HL127113). In addition, this study was supported in part by a grant from US National Institutes of Health (NCATS-CTSA, UL1 TR001085). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Conflict of interest
The authors (Joseph J. Kim, Luqia Hou, Guang Yang, Nicholas P. Mezak, Maureen Wanjare, Lydia M. Joubert, and Ngan F. Huang) declare that they have no conflicts of interest.
Ethical Approval
All human subjects research were carried out with informed consent in accordance with institutional guidelines and approved by the Institutional Review Board at Stanford University. No animal studies were carried out by the authors for this article.
Author Contributions
J.J.K. and N.F.H. designed the experiments. J.J.K, L.H., G.Y., N.P.M, M.W. and L.M.J. carried out the experiments. J.J.K., L.H., G.Y., and N.F.H. analyzed and interpreted the data. J.J.K., L.H., G.Y., L.M.J. and N.F.H. wrote the manuscript.
Footnotes
This article is part of the 2017 CMBE Young Innovators special issue.
Ngan F. Huang is an Assistant Professor in the Department of Cardiothoracic Surgery at Stanford University and Principal Investigator at the Veterans Affairs Palo Alto Health Care System. Dr. Huang completed her BS in Chemical Engineering from the Massachusetts Institute of Technology under the research guidance of Dr. Robert Langer. She then received her MS and Ph.D. in Bioengineering from the University of California Berkeley & University of California San Francisco Joint Program in Bioengineering under the mentorship of Dr. Song Li. Prior to joining the faculty, she was a postdoctoral scholar in the Division of Cardiovascular Medicine at Stanford University under the guidance of Dr. John Cooke. Her laboratory investigates the interactions between stem cells and the extracellular matrix microenvironment for engineering cardiovascular tissues to treat cardiovascular and musculoskeletal diseases. Dr. Huang has authored over 60 publications and patents, including reports in Nat Med, PNAS, and Nano Lett. She has received numerous honors, including a NIH K99/R00 Career Development Award, Fellow of the American Heart Association, a Young Investigator award from the Society for Vascular Medicine, and a Rising Star award at the CMBE-BMES conference. Her research is funded by the NIH, Department of Defense, and Department of Veteran Affairs.
Joseph J. Kim, Luqia Hou, and Guang Yang have contributed equally to this work.
References
- 1.Abbasi N, Hashemi SM, Salehi M, Jahani H, Mowla SJ, Soleimani M, Hosseinkhani H. Influence of oriented nanofibrous pcl scaffolds on quantitative gene expression during neural differentiation of mouse embryonic stem cells. J Biomed Mater Res A. 2016;104:155–164. doi: 10.1002/jbm.a.35551. [DOI] [PubMed] [Google Scholar]
- 2.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 3.Ayres CE, Jha BS, Meredith H, Bowman JR, Bowlin GL, Henderson SC, Simpson DG. Measuring fiber alignment in electrospun scaffolds: a user’s guide to the 2d fast fourier transform approach. J Biomater Sci Polym Ed. 2008;19:603–621. doi: 10.1163/156856208784089643. [DOI] [PubMed] [Google Scholar]
- 4.Baker BM, Chen CS. Deconstructing the third dimension: how 3d culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bearden SE, Segal SS. Neurovascular alignment in adult mouse skeletal muscles. Microcirculation. 2005;12:161–167. doi: 10.1080/10739680590904964. [DOI] [PubMed] [Google Scholar]
- 6.Blancas AA, Wong LE, Glaser DE, McCloskey KE. Specialized tip/stalk-like and phalanx-like endothelial cells from embryonic stem cells. Stem Cells Dev. 2013;22:1398–1407. doi: 10.1089/scd.2012.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D’Souza SL, Knowles JW, Patel A, Papatsenko D, Abbasi F, Reaven GM, Whalen S, Lee P, Shahbazi M, Henrion MY, Zhu K, Wang S, Roussos P, Schadt EE, Pandey G, Chang R, Quertermous T, Lemischka I. Analysis of transcriptional variability in a large human ipsc library reveals genetic and non-genetic determinants of heterogeneity. Cell stem cell. 2016 doi: 10.1016/j.stem.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Zeng D, Ding L, Li XL, Liu XT, Li WJ, Wei T, Yan S, Xie JH, Wei L, Zheng QS. Three-dimensional poly-(epsilon-caprolactone) nanofibrous scaffolds directly promote the cardiomyocyte differentiation of murine-induced pluripotent stem cells through wnt/beta-catenin signaling. BMC Cell Biol. 2015;16:22. doi: 10.1186/s12860-015-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chitrangi S, Nair P, Khanna A. Three-dimensional polymer scaffolds for enhanced differentiation of human mesenchymal stem cells to hepatocyte-like cells: a comparative study. J Tissue Eng Regen Med. 2016 doi: 10.1002/term.2136. [DOI] [PubMed] [Google Scholar]
- 10.Cooke JP. Flow, no, and atherogenesis. Proc Natl Acad Sci USA. 2003;100:768–770. doi: 10.1073/pnas.0430082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dash TK, Konkimalla VB. Poly-є-caprolactone based formulations for drug delivery and tissue engineering: a review. J Controll Release. 2012;158:15–33. doi: 10.1016/j.jconrel.2011.09.064. [DOI] [PubMed] [Google Scholar]
- 12.Davignon J, Ganz P. Role of endothelial dysfunction in atherosclerosis. Circulation. 2004;109:27–32. doi: 10.1161/01.CIR.0000115644.35804.8B. [DOI] [PubMed] [Google Scholar]
- 13.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 14.Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 16.Festuccia N, Osorno R, Wilson V, Chambers I. The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states. Curr Opin Genet Dev. 2013;23:504–511. doi: 10.1016/j.gde.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–376. doi: 10.1038/288373a0. [DOI] [PubMed] [Google Scholar]
- 18.Geenen IL, Molin DG, van den Akker NM, Jeukens F, Spronk HM, Schurink GW, Post MJ. Endothelial cells (ecs) for vascular tissue engineering: venous ecs are less thrombogenic than arterial ecs. J Tissue Eng Regen Med. 2015;9:564–576. doi: 10.1002/term.1642. [DOI] [PubMed] [Google Scholar]
- 19.Greenbaum RA, Ho SY, Gibson DG, Becker AE, Anderson RH. Left ventricular fibre architecture in man. Br Heart J. 1981;45:248–263. doi: 10.1136/hrt.45.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo C, Kaufman LJ. Flow and magnetic field induced collagen alignment. Biomaterials. 2007;28:1105–1114. doi: 10.1016/j.biomaterials.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 21.He W, Yong T, Ma ZW, Inai R, Teo WE, Ramakrishna S. Biodegradable polymer nanofiber mesh to maintain functions of endothelial cells. Tissue Eng. 2006;12:2457–2466. doi: 10.1089/ten.2006.12.2457. [DOI] [PubMed] [Google Scholar]
- 22.Huang NF, Fleissner F, Sun J, Cooke JP. Role of nitric oxide signaling in endothelial differentiation of embryonic stem cells. Stem Cells Dev. 2010;19:1617–1626. doi: 10.1089/scd.2009.0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang NF, Okogbaa J, Lee JC, Jha A, Zaitseva T, Paukshto M, Sun J, Fuller GG, Cooke JP. The modulation of endothelial cell morphology, function, and survival using anisotropic nanofibrillar collagen scaffolds. Biomaterials. 2013;34:4038–4047. doi: 10.1016/j.biomaterials.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang NF, Patel S, Thakar RG, Wu J, Hsiao BS, Chu B, Lee RJ, Li S. Myotube assembly on nanofibrous and micropatterned polymers. Nano Lett. 2006;6:537–542. doi: 10.1021/nl060060o. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 26.Jeong SI, Kim SY, Cho SK, Chong MS, Kim KS, Kim H, Lee SB, Lee YM. Tissue-engineered vascular grafts composed of marine collagen and plga fibers using pulsatile perfusion bioreactors. Biomaterials. 2007;28:1115–1122. doi: 10.1016/j.biomaterials.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Kannan RY, Salacinski HJ, Sales K, Butler P, Seifalian AM. The roles of tissue engineering and vascularisation in the development of micro-vascular networks: a review. Biomaterials. 2005;26:1857–1875. doi: 10.1016/j.biomaterials.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Keung AJ, Kumar S, Schaffer DV. Presentation counts: microenvironmental regulation of stem cells by biophysical and material cues. Ann Rev Cell Dev Biol. 2010;26:533–556. doi: 10.1146/annurev-cellbio-100109-104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khorshidi S, Solouk A, Mirzadeh H, Mazinani S, Lagaron JM, Sharifi S, Ramakrishna S. A review of key challenges of electrospun scaffolds for tissue-engineering applications. J Tissue Eng Regen Med. 2016;10:715–738. doi: 10.1002/term.1978. [DOI] [PubMed] [Google Scholar]
- 30.Kim JJ, Hou L, Huang NF. Vascularization of three-dimensional engineered tissues for regenerative medicine applications. Acta Biomater. 2016 doi: 10.1016/j.actbio.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3d culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kojima H, Urano Y, Kikuchi K, Higuchi T, Hirata Y, Nagano T. Fluorescent indicators for imaging nitric oxide production. Angew Chem Int Ed Engl. 1999;38:3209–3212. doi: 10.1002/(SICI)1521-3773(19991102)38:21<3209::AID-ANIE3209>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Kusuma S, Shen YI, Hanjaya-Putra D, Mali P, Cheng L, Gerecht S. Self-organized vascular networks from human pluripotent stem cells in a synthetic matrix. Proc Natl Acad Sci USA. 2013;110:12601–12606. doi: 10.1073/pnas.1306562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kutys ML, Chen CS. Forces and mechanotransduction in 3d vascular biology. Curr Opin Cell Biol. 2016;42:73–79. doi: 10.1016/j.ceb.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanfer B, Freudenberg U, Zimmermann R, Stamov D, Korber V, Werner C. Aligned fibrillar collagen matrices obtained by shear flow deposition. Biomaterials. 2008;29:3888–3895. doi: 10.1016/j.biomaterials.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Laschke MW, Harder Y, Amon M, Martin I, Farhadi J, Ring A, Torio-Padron N, Schramm R, Rucker M, Junker D, Haufel JM, Carvalho C, Heberer M, Germann G, Vollmar B, Menger MD. Angiogenesis in tissue engineering: Breathing life into constructed tissue substitutes. Tissue Eng. 2006;12:2093–2104. doi: 10.1089/ten.2006.12.2093. [DOI] [PubMed] [Google Scholar]
- 37.Lee P, Lin R, Moon J, Lee LP. Microfluidic alignment of collagen fibers for in vitro cell culture. Biomed Microdevices. 2006;8:35–41. doi: 10.1007/s10544-006-6380-z. [DOI] [PubMed] [Google Scholar]
- 38.Lian X, Bao X, Al-Ahmad A, Liu J, Wu Y, Dong W, Dunn KK, Shusta EV, Palecek SP. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of wnt signaling. Stem cell Rep. 2014;3:804–816. doi: 10.1016/j.stemcr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin S, Mequanint K. Activation of transcription factor gax and concomitant downregulation of il-1beta and erk1/2 modulate vascular smooth muscle cell phenotype in 3d fibrous scaffolds. Tissue Eng Part A. 2015;21:2356–2365. doi: 10.1089/ten.tea.2015.0153. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Maldonado M, Ico G, Low K, Luu RJ, Nam J. Enhanced lineage-specific differentiation efficiency of human induced pluripotent stem cells by engineering colony dimensionality using electrospun scaffolds. Adv Healthc Mater. 2016;5:1408–1412. doi: 10.1002/adhm.201600141. [DOI] [PubMed] [Google Scholar]
- 42.Moncada S, Higgs A. The l-arginine-nitric oxide pathway. N Engl J Med. 1993;329:2002–2012. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama KH, Hong G, Lee JC, Patel J, Edwards B, Zaitseva TS, Paukshto MV, Dai H, Cooke JP, Woo YJ, Huang NF. Aligned-braided nanofibrillar scaffold with endothelial cells enhances arteriogenesis. ACS Nano. 2015;9:6900–6908. doi: 10.1021/acsnano.5b00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayama KH, Joshi PA, Lai ES, Gujar P, Joubert LM, Chen B, Huang NF. Bilayered vascular graft derived from human induced pluripotent stem cells with biomimetic structure and function. Regen Med. 2015;10:745–755. doi: 10.2217/rme.15.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O’Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K, Xia Y, Florido MH, He W, Pan W, Prummer M, Warren CR, Jakob-Roetne R, Certa U, Jagasia R, Freskgard PO, Adatto I, Kling D, Huang P, Zon LI, Chaikof EL, Gerszten RE, Graf M, Iacone R, Cowan CA. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul A, Manoharan V, Krafft D, Assmann A, Uquillas JA, Shin SR, Hasan A, Hussain MA, Memic A, Gaharwar AK, Khademhosseini A. Nanoengineered biomimetic hydrogels for guiding human stem cell osteogenesis in three dimensional microenvironments. J Mater Chem B Mater Biol Med. 2016;4:3544–3554. doi: 10.1039/C5TB02745D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothan HA, Djordjevic I, Bahrani H, Paydar M, Ibrahim F, Abd Rahmanh N, Yusof R. Three-dimensional culture environment increases the efficacy of platelet rich plasma releasate in prompting skin fibroblast differentiation and extracellular matrix formation. Int J Med Sci. 2014;11:1029–1038. doi: 10.7150/ijms.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellaro TL, Hildebrand D, Lu Q, Vyavahare N, Scott M, Sacks MS. Effects of collagen fiber orientation on the response of biologically derived soft tissue biomaterials to cyclic loading. J Biomed Mater Res A. 2007;80:194–205. doi: 10.1002/jbm.a.30871. [DOI] [PubMed] [Google Scholar]
- 49.Sia J, Yu P, Srivastava D, Li S. Effect of biophysical cues on reprogramming to cardiomyocytes. Biomaterials. 2016;103:1–11. doi: 10.1016/j.biomaterials.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Wang XN, McGovern N, Gunawan M, Richardson C, Windebank M, Siah TW, Lim HY, Fink K, Li JL, Ng LG, Ginhoux F, Angeli V, Collin M, Haniffa M. A three-dimensional atlas of human dermal leukocytes, lymphatics, and blood vessels. J Investig Dermatol. 2014;134:965–974. doi: 10.1038/jid.2013.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang XF, Song Y, Liu YS, Sun YC, Wang YG, Wang Y, Lyu PJ. Osteogenic differentiation of three-dimensional bioprinted constructs consisting of human adipose-derived stem cells in vitro and in vivo. PLoS One. 2016;11:e0157214. doi: 10.1371/journal.pone.0157214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wingate K, Bonani W, Tan Y, Bryant SJ, Tan W. Compressive elasticity of three-dimensional nanofiber matrix directs mesenchymal stem cell differentiation to vascular cells with endothelial or smooth muscle cell markers. Acta Biomater. 2012;8:1440–1449. doi: 10.1016/j.actbio.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu YT, Yu IS, Tsai KJ, Shih CY, Hwang SM, Su IJ, Chiang PM. Defining minimum essential factors to derive highly pure human endothelial cells from ips/es cells in an animal substance-free system. Sci Rep. 2015;5:9718. doi: 10.1038/srep09718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie J, Willerth SM, Li X, Macewan MR, Rader A, Sakiyama-Elbert SE, Xia Y. The differentiation of embryonic stem cells seeded on electrospun nanofibers into neural lineages. Biomaterials. 2009;30:354–362. doi: 10.1016/j.biomaterials.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- 57.Zhang S, Dutton JR, Su L, Zhang J, Ye L. The influence of a spatiotemporal 3d environment on endothelial cell differentiation of human induced pluripotent stem cells. Biomaterials. 2014;35:3786–3793. doi: 10.1016/j.biomaterials.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Schwartz MP, Hou Z, Bai Y, Ardalani H, Swanson S, Steill J, Ruotti V, Elwell A, Nguyen BK, Bolin J, Stewart R, Thomson JA, Murphy WL. A genome-wide analysis of human pluripotent stem cell-derived endothelial cells in 2d or 3d culture. Stem Cell Rep. 2017;8:907–918. doi: 10.1016/j.stemcr.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. 3D reconstructed view of CD31 (red) and total nuclei (green) in aligned microfibrous scaffolds, based on confocal microscopy. Supplementary material 1 (AVI 244517 kb)
Supplementary Video 2. 3D reconstructed view of CD31 (red) and total nuclei (green) in randomly oriented microfibrous scaffolds, based on confocal microscopy. Supplementary material 3 (AVI 242789 kb)