Abstract
Individual bacteria and shifts in microbiome composition are associated with human disease, including cancer. To unravel the connections underlying oral bacterial dysbiosis and oral squamous cell carcinoma (OSCC), cancer lesion samples and anatomically matched normal samples were obtained from the same patients. We then profiled the bacteria within OSCC lesion surface samples at the species level using next-generation sequencing to comprehensively investigate bacterial community composition and functional genes in these samples. Significantly greater bacterial diversity was observed in the cancer samples than in the normal samples. Compared with previous studies, we identified many more taxa demonstrating remarkably different distributions between the groups. In particular, a group of periodontitis-correlated taxa, including Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella and Parvimonas, was significantly enriched in OSCC samples. Additionally, several operational taxonomic units (OTUs) associated with Fusobacterium were highly involved in OSCC and demonstrated good diagnostic power. Our study revealed drastic changes in surface bacterial communities of OSCC. The findings enrich knowledge of the association between oral bacterial communities and oral cancer.
Introduction
The human body is inhabited by over 100 trillion microbial cells living in symbiosis with their host1. Bacteria at certain body sites have long been believed to be involved in immune modulation, disease development, and health maintenance. The term microbiome was coined to describe “the collective genomes and gene products of all microbes residing within an organism”2. With the advent of high-throughput, next-generation sequencing (NGS), there has been a surge of interest in studying the human microbiome in the context of disease. Recent studies have demonstrated the importance of the gut microbiota in digestion, fat storage, angiogenesis, immune system development and responses, resistance to colonization, epithelial architecture3,4, and dysbiosis, which is believed to contribute to the pathogenesis of local and systemic diseases, including inflammatory bowel disease, diabetes, and colorectal cancer5. Located at the beginning of the aerodigestive tract, approximately 700 prokaryote species have been detected in the human oral cavity. These species belong to 185 genera and 12 phyla, of which approximately 54% are officially named, 14% unnamed (but cultivated) and 32% known only as uncultivated phylotypes6. This oral bacterial flora plays an essential role in maintaining a normal oral physiological environment and is associated with host health7. In contrast to traditional views, recent analyses suggest the involvement of a consortium of microbes, rather than a single species, as causing disease8, a phenomenon that has been well characterized for periodontal diseases9.
Oral cancer, primarily oral squamous cell carcinoma (OSCC) deriving from the oral mucosa, is a disease that arises from both host genetics and environmental factors; tobacco and alcohol consumption, betel quid chewing, and human papillomavirus infection are well-known risk factors10. The incidence of oral cancer is increasing, and this disease continues to be a major global health problem. Furthermore, approximately 15% of oral cancer cases cannot be attributed to the aforementioned major risk factors, resulting in the need to explore other potential risk factors11. A plethora of bacteria, the proverbial bacterial biofilm, coat each surface of the oral cavity12, and groups inhabiting the mucosal surface might constitute the bulk of the tumor microenvironment. To date, various microbes and changes in different bacteria have been associated with several types of cancer13. Cancer-associated changes in the oral microbiome have been assessed in several early studies employing culture-based or molecular techniques14–19, but a consensus has not been reached due to the limited number of strains/clones that it is feasible to test. However, the emergence of NGS allows microbial communities to be profiled at an unprecedented depth and coverage.
To date, several studies have employed NGS to assess bacterial profiles associated with OSCC. Pushalkar et al. evaluated the diversity and relative abundance of bacteria in the saliva of subjects with OSCC; however, only three OSCC cases and two healthy controls were included20. Later, a larger-scale study analyzed swabs of lesion surfaces and contra-lateral normal mucosae from 18 OSCC patients, and significant decreases in the abundances of the genera Firmicutes and Actinobacteria were observed in cancer samples21. In another report, saliva bacterial communities in six OSCC patients were elucidated by performing pyrosequencing, and paired taxa within the family Enterobacteriaceae together with the genus Oribacterium were suggested to distinguish OSCC samples from oropharyngeal squamous cell carcinoma (OPSCC) and normal samples22. Regardless, the studies above failed to provide bacterial composition at the species level, even though specific species or even strains are usually involved in disease. Al-hebshi successively profiled bacterial communities within 23 OSCC tissue samples from Yemeni patients at the species level, providing the first epidemiological evidence for associations of Fusobacterium nucleatum and Pseudomonas aeruginosa with OSCC23,24. Given the limited number of OSCC samples included, the significance of the findings from these studies is unclear. Accordingly, more studies are warranted to validate these results.
In the current study, cancer lesion samples and matched controls were procured from 40 Chinese subjects with OSCC. Bacterial profiles within the samples were characterized at the species level. Shifts in bacterial composition and gene functions associated with OSCC were described and analyzed. In particular, we detected a group of periodontitis-related taxa that was significantly enriched in OSCC samples. Our findings may contribute to further clarification of the connection between OSCC and oral bacteria.
Results
Overall structure of bacterial communities across samples
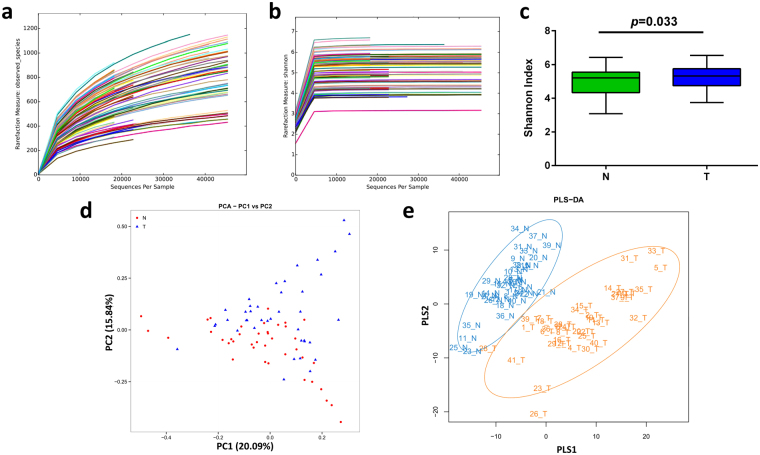
In the current study, 80 samples were sequenced using an Illumina MiSeq system, and a total of 5,075,391 raw sequences were generated. After quality trimming and chimera checking, 4,075,169 high-quality sequences with an average length of 391 bp were recovered for downstream analysis, with an average of 50,940 reads (ranging from 19,353 to 117,244 reads) per sample. After alignment in the HOMD, unique representative sequences were classified into 2,334 operational taxonomic units (OTUs) at a 97% similarity level, from which 11 phyla, 130 genera and 389 species were detected. Good’s estimator of coverage was 99.26%, indicating the 16 S rRNA sequences identified in this study likely represent the majority of bacterial sequences present in the samples. Different indexes (Shannon, Simpson, Abundance-based Coverage Estimator (ACE), and Chao 1) were employed to estimate the α-diversity of the bacterial community. Although rarefaction curves of numbers of observed OTUs per sample suggested new phylotypes would be expected with additional sequencing (Fig. 1a), the rarefaction curves for the Shannon diversity index for each sample reached plateaus, indicating that the majority of the diversity was already procured (Fig. 1b). As revealed by the Shannon diversity index (Fig. 1c), the diversity of the bacterial community in the cancer samples was significantly increased compared with that of the corresponding clinically normal control samples. A similar trend was observed when employing other diversity indexes (Simpson, ACE, Chao 1), although without statistical significance (Supplementary Fig. S1).
Figure 1.
Comparison of oral microbiota structures in the N and T groups. (a) Rarefaction analysis of bacterial 16 S rRNA gene sequences was performed to evaluate whether further sequencing would likely detect additional taxa, indicated by a plateau. Different colors represent different samples. (b) Shannon index curves were constructed to evaluate the numbers of samples likely required to identify additional taxa, indicated by a plateau. Different colors represent different samples. (c) Box plots depict differences in bacterial diversity between the N and T groups according to the Shannon index. (d) PCA at the OTU level. (e) Partial least square discriminant score plot of oral microbiota between the N and T groups. N, clinical normal samples; T, oral cancer samples.
To evaluate the extent of the similarity of the bacterial communities, unweighted UniFrac Principal Component Analysis (PCA) at the OTU level was employed and indicated no obvious separation between groups (Fig. 1d). Then, Partial Least Squares Discriminant Analysis (PLS-DA), a supervised analysis suitable for high-dimensional data, was performed (Fig. 1e). The bacterial communities in the cancer samples and the matched controls clustered separately, suggesting the overall structures of the bacterial communities in the groups were significantly different. Spots representing the cancer samples presented more dispersed distribution patterns than those of the controls, aligning with the increased level of bacterial diversity found in the cancer samples. A nonparametric multivariate analysis of variance (Adonis) and an analysis of similarities (ANOSIM) based on UniFrac distances were performed, and the calculated P values (P = 0.002 for Adonis, P = 0.001 for ANOSIM) further demonstrated the remarkable differences between the bacterial communities in the groups.
Common and distinct bacterial taxa in the analyzed groups
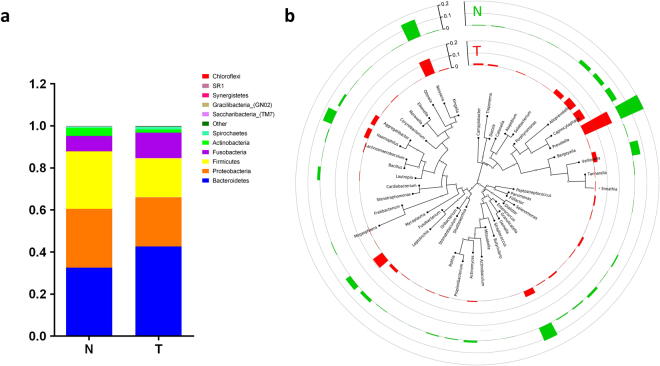
The bacterial communities in the cancer lesions and the controls were analyzed at different taxonomic levels (Fig. 2 and Supplementary Fig. S2). Bacteroidetes, Proteobacteria, Firmicutes, Fusobacteria, Actinobacteria, the top five most abundant phyla, together comprised 98.62% of all sequences (Fig. 2a). Bacteroidetes was the most abundant phylum, accounting for 37.6% of sequences. In contrast, the abundances of the other detected phyla, including Synergistetes, SR1, and Chloroflexi, were less than 0.1%. At the genus level, Prevotella, Neisseria, Streptococcus, Fusobacterium, and Haemophilus were the five most abundant genera, comprising 22.46%, 13.67%, 8.17%, 6.95%, and 5.74% of sequences, respectively (Fig. 2b). Of all genera detected, 18 taxa were found in all samples; in addition to the five genera mentioned above, these included Capnocytophaga, Veillonella, Alloprevotella, Porphyromonas, Leptotrichia, Aggregatibacter, Selenomonas, Campylobacter, Granulicatella, Actinomyces, Gemella, Lachnoanaerobaculum, and Bergeyella. The shared genera collectively represented over 80.0% of all detected sequences. At the species level, an average of approximately 200 species were detected per sample. The relative abundances of eleven species each surpassed 2%; specifically, Neisseria flavescens, Prevotella melaninogenica, Fusobacterium periodonticum, Streptococcus oralis, Prevotella intermedia, Veillonella atypica, Haemophilus parahaemolyticus, Porphyromonas sp. _oral_taxon_279, Capnocytophaga leadbetteri, Alloprevotella sp. _oral_taxon_473 and Haemophilus parainfluenzae together accounted for a total of 53.8% of sequences (Supplementary Fig. S2). Neisseria flavescens was the most abundant species in both groups, with a relative abundance of 10.73% in cancer lesions and 12.19% in normal controls. Despite significant inter-individual variation, 14 species were detected across all samples, including Actinomyces odontolyticus, Prevotella melaninogenica, Prevotella scopos, Capnocytophaga gingivalis, Gemella sanguinis, Granulicatella adiacens, Streptococcus oralis, Streptococcus salivarius, Lachnoanaerobaculum umeaense, Veillonella atypica, Fusobacterium periodonticum, Neisseria elongata, Neisseria flavescens, and Haemophilus parainfluenzae, and constituted the oral mucosal core bacteriome of the OSCC patients.
Figure 2.
Composition of bacterial communities across samples at the phylum and genus levels. (a) Relative abundance of bacterial phyla among the N and T groups. (b) Classification tree of the 50 most abundant genera across all samples. The outer cycle colored bars represent the relative abundances of taxa in each group. N, clinical normal samples; T, oral cancer samples.
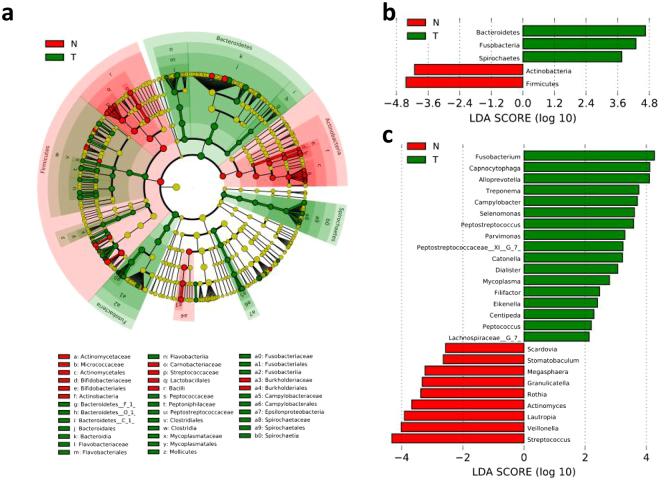
The bacterial composition of the cancer samples varied from that of the controls. There were 6 phyla and 68 genera with higher abundances in the cancer samples than in the controls. To identify the distinguishing taxa within the groups, the linear discriminant analysis (LDA) effect size (LEfSe) method was implemented (Fig. 3). At the phylum level, Spirochaetes, Fusobacteria, and Bacteroidetes were significantly enriched in diseased samples, while Firmicutes and Actinobacteria were significantly decreased (Fig. 3b). At the genus level, 17 taxa exhibited significantly higher abundances in the cancer samples than in the controls, including Mycoplasma, Treponema, Campylobacter, Eikenella, Centipeda, Lachnospiraceae_G_7, Alloprevotella, Fusobacterium, Selenomonas, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella, Parvimonas, Capnocytophaga, and Peptostreptococcaceae_XI_G_7. The taxa Megasphaera, Stomatobaculum, Granulicatella, Lautropia, Veillonella, Streptococcus, Scardovia, Rothia, and Actinomyces were remarkably prevalent in the controls (Fig. 3c). Prevotella and Neisseria were the predominant genera in both groups: there was no significant difference in distribution between the two groups (Fig. 2b). At the species level, 39 species were significantly increased, while 28 species were significantly decreased in diseased samples compared to those levels in the controls (Supplementary Fig. S3).
Figure 3.
Distinct taxa identified in the N and T groups using LEfSe analysis. (a) Cladogram constructed using the LEfSe method to indicate the phylogenetic distribution of bacteria that were remarkably enriched in the N and T groups. (b) LDA scores showed significant bacterial differences within groups at the phylum level.(c) LDA scores showed significant bacterial differences within groups at the genus level. N, clinical normal samples; T, oral cancer samples.
Co-occurrence network analysis and function predictions
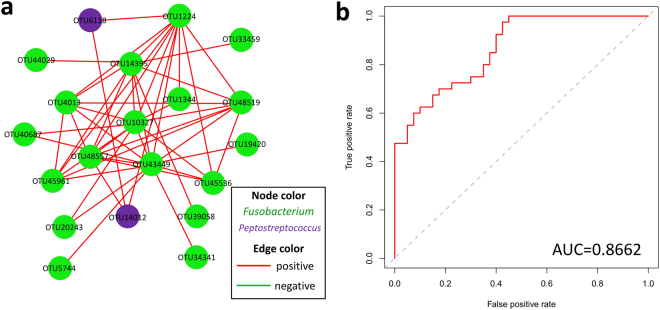
To predict the ecological relationships across different bacterial communities, spatial Pearson’s correlations between bacterial species were visualized and then analyzed (Supplementary Fig. S4). In the network, pairwise relationships were represented by edges connecting two nodes. There were 1,574 associations among 306 nodes in the controls and 1,794 associations among 366 nodes in the lesions at the OTU level, indicating increased network complexity in the latter group. The genera Prevotella and Neisseria comprised the two densest clusters in both groups. The hub OTU with the most associations in the clusters, OTU50445 (belonging to Prevotella melaninogenica), primarily represented the genus Prevotella in both groups, while the hub OTUs in the clusters primarily representing the genus Neisseria were OTU52958 in the controls and OTU46085 in the cancer samples, both of which belonged to Neisseria flavescens. There was a highly connected bacterial cluster of Fusobacterium comprising OTUs (OTU48557, 45961, 43449, 14395, 1224, 10327, and 4013) that were heavily involved in the bacterial ecology structure of the diseased samples compared with the controls, suggesting that Fusobacterium may play a critical role in the development of oral cancer (Fig. 4a). The predictive power of the seven described OTUs was further assessed by constructing a receiver operating characteristic (ROC) curve, a function of the true-positive rate (TPR or sensitivity) and false-positive rate (FPR or 1-specificity). The area under the ROC curve (AUC) reached 0.866, indicating good diagnostic performance (Fig. 4b).
Figure 4.
Co-occurrence network of Fusobacterium comprising OTUs and the diagnostic power of selected OTUs. (a) Each node represents an OTU colored for its genus-level phylotypes, and each edge represents a significant co-occurrence relationship colored according to its association (red: positive, green: negative). (b) ROC curves for selected Fusobacterium OTUs were constructed to predict diagnostic power.
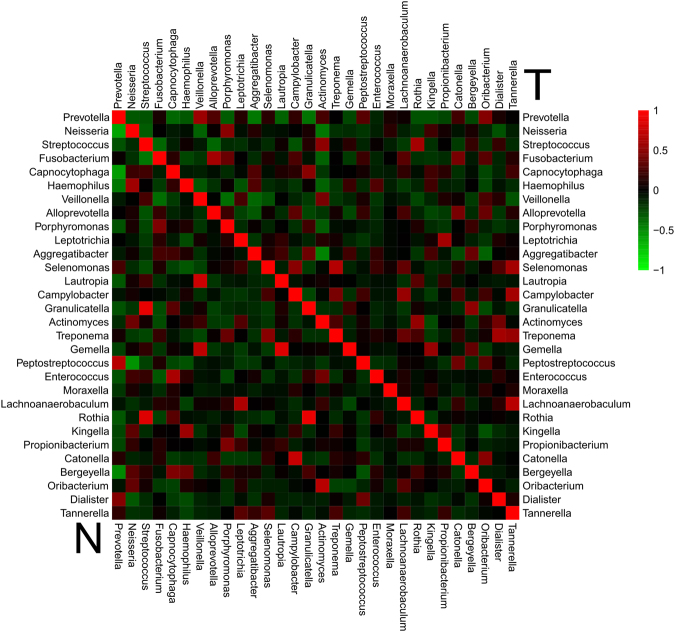
The co-occurrence patterns of the 30 most abundant bacterial genera in each group were investigated in detail (Fig. 5). In matched normal controls, Tannerella and Lachnoanaerobaculum were the most positively correlated (ρ = 0.681), whereas Actinomyces and Aggregatibacter were the most negatively correlated (ρ = −0.462). In the cancer samples, the most positively correlated genera were Rothia and Granulicatella (ρ = 0.903), and Neisseria and Prevotella were the most negatively correlated (ρ = −0.618). Additionally, opposing co-occurrence patterns at the genus and species level were observed. For example, the genera Actinomyces and Neisseria correlated positively (ρ = 0.383) in the lesions but were negatively correlated (ρ = −0.429) in normal controls. Prevotella pallens and Prevotella intermedia demonstrated negative correlations (ρ = −0.222) in lesions but positively correlated in the control group (ρ = 0. 342) (Supplementary Fig. S5).
Figure 5.
Co-occurrence and co-exclusion analysis of bacterial genera. Pearson correlations among the top 30 most abundant bacterial genera were calculated and analyzed; groups are shown on the left and right. Correlation values ranged from −1.00 (green) to 1.00 (red).
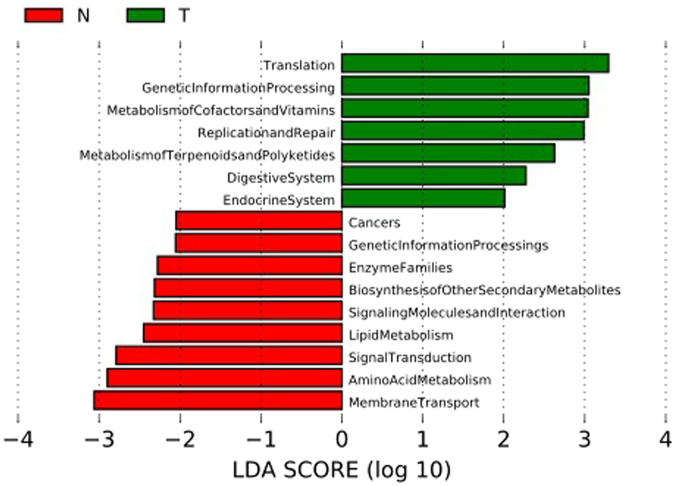
Finally, the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) algorithm was employed to predict bacterial functions in the two groups. The LEfSe outputs showed a series of metabolic pathways presenting significantly different distributions in each group (Fig. 6). Pathways related to Genetic Information Processing were remarkably enriched in cancer lesions.
Figure 6.
LDA scores predict gene function enriched in different groups using PICRUSt. N, clinical normal samples; T, oral cancer samples.
Discussion
Interest in a possible relationship between bacteria and different stages of cancer development has been increasing since the classification of Helicobacter pylori as a definite carcinogen by the World Health Organization25. Although microorganisms have been implicated in 15.4% of human malignancies26, there is a paucity of knowledge regarding the role of bacteria in the progression of OSCC. Because bacterial composition at the species level within OSCC samples has rarely been reported, we attempted to add knowledge regarding this aspect.
Compared with the pyrosequencing applied in previous studies20–23, Illumina Miseq is a popular established platform of NGS due to its reduced run times and lower cost of reagents, with an overall error rate below 1%, and the 2 × 300 bp read length is flexible for sequencing of small genomes27,28. However, the different sequencing regions (V4-V520, V421, V3-V522, V1-V323,24) targeted between studies, along with differences in sample types, selection of control tissues, and number of samples included, may produce inconsistent results. In this study, the V4-V5 region was chosen for sequencing, as it is proposed to be an ideal target for bacteria in 16 S rRNA-based analyses29. In the current study, we obtained many more raw sequences and OTUs than those obtained in earlier studies21,23, making it possible to comprehensively profile bacterial structure in cancer lesions. Then, we compared the complexity of the bacterial communities present at mucosal sites in both groups, and our results unraveled critical yet pronounced bacterial characteristics associated with disease. Different from previous reports in which tissue biopsies or swabs were analyzed17,19,21,24, greater phylogenetic diversity was observed in the OSCC lesion surface in our study, as suggested by rarefaction curves for the Shannon diversity index. With regard to saliva samples, Guerrero-Preston reported a significant loss in richness and diversity of oral bacterial species in oral cancer patients compared to controls22, whereas another study revealed much greater diversity of bacterial communities in OSCC samples30. Moreover, Adonis and ANOSIM analyses further corroborated the significant differences between bacterial communities in oral cancer lesions and those in matched controls. Thus, oral bacteria dysbiosis appears to be present during OSCC development.
Overall, five of the most abundant phyla detected in our study were consistent with those found in previous studies, specifically, Bacteroidetes, Proteobacteria, Firmicutes, Fusobacteria, and Actinobacteria, while the less abundant phyla detected in different surveys varied. Although Firmicutes was the most abundant phylum in previous studies20–22, Bacteroidetes was detected as the most abundant phylum in the current study. Of the highly abundant genera in this report, Fusobacterium, Neisseria, Prevotella, Streptococcus, Leptotrichia, Veillonella and Capnocytophaga were previously shown to be the most abundant genera in samples from periodontitis patients31. As opposed to Streptococcus, which was predominant in Pushalkar’s study17, Prevotella was the most abundant genus across all samples in our analysis, accounting for 22.5% of abundance, which is comparable to previous findings21. The use of HOMD enabled the assignment of many more OTUs at the species level. Neisseria flavescens and Fusobacterium periodonticum are consistently predominant in cancer lesions23.
Compared with other studies, our study identified larger numbers of distinguishing taxa at each level using the LEfSe method. At the phylum level, Firmicutes and Actinobacteria presented with the same patterns reported by Schmidt21 and were remarkably decreased in cancer lesions, while significant increases in Fusobacteria was also observed, consistently24. In line with the previous studies, genera Streptococcus and Rothia were significantly decreased in cancer lesions21,24. Intriguingly, the majority of these significantly enriched genera in lesions are involved in periodontal disease, including Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella, and Parvimonas 31. Consistent with previous findings, remarkable enrichment of Peptostreptococcus and Parvimonas was observed in cancer samples17,22. Additionally, Veillonella was significantly decreased in cancer lesions, a finding that was previously reported in 73% of oral cancer patients after treatment17, indicating Veillonella correlates with a healthy status. Of the distinguishing species identified across the groups, forty species were highly abundant in cancer lesions, including Porphyromonas endodontalis, Filifactor alocis and Dialister pneumosintes, which are newly recognized periodontal pathogens32. Of all oral bacteria, Porphyromonas gingivalis and Fusobacterium nucleatum might possess the greatest potential to be correlated with oral cancer, as both have been implicated in pancreatic and colorectal cancers. Recently, a report by Gallimidi indicated P. gingivalis and F. nucleatum promote oral cancer progression via direct interactions with oral epithelial cells through Toll-like receptors33. However, P. gingivalis did not differ in abundance between groups. Fusobacterium, comprising the species periodonticum, naviforme, and nucleatum_subsp, was significantly enriched in lesions, accounting for 8.33%, 0.103%, and 0.297% of sequences in the cancer group, respectively. In another study, F. periodonticum, F. naviforme, and F. nucleatum_subsp were reported to account for 4.08%, 0.01% and 11.67% of sequences in cancer samples, respectively24. Thus, the different prevalences of Fusobacterium species detected in OSCC samples between studies may largely be due to differences in sample types, races and geographic regions of the subjects recruited. Further evidence is needed to verify these findings. A higher abundance of several Treponema species was observed in cancer lesions. T. denticola, a member of the periodontal “red complex” involved in pancreatic cancer34, was not included. In the literature, Bacteroides fragilis has been linked to colon cancer35, but it was not observed in our study, although it was detected in OSCC tissues in another report23. Capnocytophaga levels were significantly higher in the saliva of lung cancer patients36 than in healthy controls, and Capnocytophaga gingivalis was previously suggested to be a potential salivary biomarker of oral cancer37. In this study, C. gingivalis was detected at higher levels in control samples without any significance, while C. leadbetteri and C. sp_oral_taxon_902 were remarkably overabundant in lesions. Members of the genus Selenomonas have been repeatedly associated with periodontal disease, although the Selenomonas species detected in this study did not correlate with known diseases38. Several species of Peptostreptococcus and Parvimonas were extensively enriched in cancer samples, including Peptostreptococcus stomatis and Parvimonas micra, both of which are reportedly related to colorectal cancer39. Eikenella corrodens, a fastidious gram-negative facultative anaerobic bacillus, was also detected in another study24. The genus Eikenella is significantly overrepresented in colorectal cancer5 and is associated with HPV-negative head and neck squamous cell carcinoma samples22. Given its documented history of pathogenicity, further investigation of the potential role of E. corrodens in the etiology of OSCC is warranted. In our design, paired lesion and control samples were procured from one individual, eliminating inter-individual variation. Therefore, even slight differences in the bacterial profiles between groups may be closely correlated with OSCC. Although several of the distinguishing taxa were present in relatively tiny proportions, their role in the development of OSCC should not be ignored.
Bacteria coexist in complex interaction webs, and interactions within these webs affect the species involved, while perturbations may contribute to disease40. As revealed in our network analysis, bacterial communities in OSCC samples presented with more complex webs depicting ecological relationships, consistent with the extensive bacterial diversity detected in the samples. The genera Prevotella and Neisseria clustered, forming two of the densest interaction webs in both groups. Prevotella and Neisseria may play key roles in maintaining the stability of the oral bacterial community across samples. Conversely, an association network centered around Fusobacterium arose in the cancer group, indicating that the genus Fusobacterium was implicated in the development of OSCC in accordance with its significant increase in the cancer group. Fusobacterium tends to co-adhere with other species in oral biofilms by forming bridges between early and late colonizers41,42. Thus, it was reasonable to infer a critical role for Fusobacterium in increasing OSCC bacterial diversity. Further evaluation of the role of Fusobacterium in OSCC may require more study. It was observed that the same paired taxa showed absolute opponent relationships within the groups, implicating that some drastic changes in the bacterial symbiotic relationships occurred during the oral carcinogenesis.
Functions of bacterial communities were inferred using PICRUSt based on 16 S rRNA sequence information, and differences between controls and diseased samples were further analyzed using the LEfSe method. Overall, the most abundant gene categories were replication and repair, membrane transport, amino acid metabolism, carbohydrate metabolism, and translation, likely reflecting the fundamental requirements for bacterial life in the oral mucosal habitat. As illustrated above, several gene functions exhibited remarkably different distributions within the groups. Notably, among the functions that were significantly decreased in cancer lesions, those related to membrane transport, amino acid metabolism, signal transduction, and lipid metabolism were also under-represented in periodontitis samples43. Periodontitis is suggested to be an independent risk factor for OSCC44, and bacterially induced chronic inflammation has been anticipated as being involved in oral carcinogenesis45. The findings observed in our study, such as several predicted functions in OSCC presenting same patterns as periodontitis and periodontitis-related taxa being significantly enriched in OSCC, indicate the pro-inflammatory potential of the bacterial communities of OSCC samples, which is consistent with a recent study24. Nonetheless, to obtain further details regarding the changes in gene functions of bacterial communities presenting in lesions, whole-metagenome sequencing is warranted in future studies.
In conclusion, bacterial dysbiosis was observed within OSCC surface lesion samples in our study, with drastic changes in bacterial composition and bacterial gene functions compared to controls. In particular, a group of periodontitis-correlated taxa, including Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella and Parvimonas, was found to be significantly enriched in OSCC samples. In addition, several OTUs belonging to Fusobacterium were inferred to be heavily involved in OSCC and demonstrated good diagnostic power. The oral microbiota towards OSCC actually is considered like comorbidity factor. According to the current design and observation, it is difficult to determine whether bacterial dysbiosis changed the local microenvironment and then drove carcinogenesis or cancerization in bacterial habitats allowed bacteria suitable for a tumor microenvironment to thrive, resulting in shifts in bacterial communities. More investigation is needed.
Methods
Subject recruitment and sample collection
Subjects with OSCC with a median age of 62 (60% male and 40% female) were recruited from the Department of Oral and Maxillofacial-Head and Neck Oncology of the Ninth People’s Hospital (Shanghai, China). All subjects consented to clinical examination and sampling. Subjects did not have detectable periodontal inflammation, visible carious lesions, oral mucosal diseases, or any severe systemic disorders (such as diabetes, immune compromise, or genetic diseases). Moreover, they had not received treatment for OSCC or taken antibiotics at least two weeks prior to sampling. This study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital affiliated with the Shanghai Jiao Tong University School of Medicine. Written informed consent was obtained from all subjects. All experiments were performed in accordance with the approved guidelines.
According to a well-defined clinical protocol, swabs of oral lesions and anatomically matched normal sites were collected. Subjects were prevented from drinking and eating for at least 2 h before sampling. All samples were transported to the laboratory on ice within 2 h of collection and were stored at −80 °C before subsequent processing.
DNA extraction
Metagenomic DNA was individually extracted from swabs using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quantity and quality of the isolated DNA was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and by performing agarose gel electrophoresis, respectively. DNA samples were frozen at −20 °C for further analysis.
Illumina sequencing and bioinformatics analysis of 16 S rRNA gene amplicons
Gene sequencing of 16 S rRNA was conducted on an Illumina MiSeq platform according to a previously described protocol46. PCR amplifications were performed with the 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 926 R(5′-CCGTCAATTYYTTTRAGTTT-3′) primers, which targeted the V4-V5 hypervariable region of the 16 S rRNA gene. DNA was amplified following a previously described protocol47. Pairs of reads from the original DNA fragments were merged using fast length adjustment of short reads (FLASH) software48, and sequences were analyzed using quantitative insights into microbial ecology (QIIME) software49. Sequences were assigned to OTUs at 97% similarity; a representative sequence was selected for each OTU, and the RDP classifier was employed to assign taxonomic data to each representative sequence.
Representative sequences were assigned at different taxonomic levels (from phylum to species) to the Human Oral Microbiome Database following the Bayesian approach with a 97% cutoff value. Bacterial diversity was determined by performing a sampling-based OTU analysis and was displayed as a rarefaction curve. Bacterial richness and diversity across samples were assessed using the following α indexes, which were estimated at a distance of 3%: Chao 1, ACE, Simpson, Shannon and Good’s coverage50. Student’s t-test was used to compare bacterial diversity. GraphPad Prism V.6.0 (San Diego, CA, USA) was used for graph preparation. PCA using unweighted UniFrac distance metrics was carried out, and the R package was used to visualize interactions among bacterial communities in different samples. In addition, PLS-DA, nonparametric analysis of Adonis distance matrices and ANOSIM were also performed to compare bacterial composition between samples51. LEfSE (http://huttenhower.sph.harvard.edu/galaxy/) was employed to identify distinguishing taxa between the two groups at multiple levels and to visualize the results using taxonomic bar charts and cladograms52. Network structures in the bacterial communities of the samples were defined by the Molecular Ecological Network Analysis Pipeline53 and visualized using Cytoscape54. A ROC curve was constructed to determine the diagnostic values of Fusobacterium OTUs for OSCC. Co-occurrence patterns for the 30 most abundant taxonomic groups across samples were explored by calculating Pearson correlation coefficients. The results were clustered and visualized using the MeV package55. Functional compositions of the bacterial communities were predicted using PICRUSt according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) dataset56.
Data Availability
The sequencing data from this study have been deposited in the GenBank Sequence Read Archive under accession number SRP097643.
Electronic supplementary material
Acknowledgements
This research was financially supported by Shanghai Summit & Plateau Disciplines. We acknowledge Shanghai Personal Biotechnology Co., Ltd. for their kind help with gene sequencing and bioinformatics analysis.
Author Contributions
Conception and design of the experiments: J.L., Z.H. and B.H. Performed the experiments: H.Z., M.C. and S.R. Data processing and analysis: H.Z. and Z.H. Volunteer recruitment and sample collection: H.Z., C.Z., M.C. and X.Y. Paper writing: H.Z. Revision of the manuscript: J.L., C.Z., B.H., Z.H., M.C. and H.Z.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Hongsen Zhao and Min Chu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11779-9
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bin Hu, Email: hub1093@2m9h.net.
Chenping Zhang, Email: zhang.chenping@hotmail.com.
Jingping Liang, Email: liangjpdentist@126.com.
References
- 1.Kudo Y, et al. Oral environment and cancer. Genes Environ. 2016;38:13. doi: 10.1186/s41021-016-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 4.Burcelin R. Gut microbiota and immune crosstalk in metabolic disease. Mol Metab. 2016;5:771–781. doi: 10.1016/j.molmet.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allali I, et al. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes. 2015;6:161–172. doi: 10.1080/19490976.2015.1039223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gholizadeh P, et al. Role of oral microbiome on oral cancers, a review. Biomed Pharmacother. 2016;84:552–558. doi: 10.1016/j.biopha.2016.09.082. [DOI] [PubMed] [Google Scholar]
- 8.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, et al. A Filifactor alocis-centered co-occurrence group associates with periodontitis across different oral habitats. Sci Rep. 2015;5:9053. doi: 10.1038/srep09053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng JH, Iyer NG, Tan MH, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: A global study. Head Neck. 2017;39:297–304. doi: 10.1002/hed.24589. [DOI] [PubMed] [Google Scholar]
- 11.Perera M, Al-Hebshi NN, Speicher DJ, Perera I, Johnson NW. Emerging role of bacteria in oral carcinogenesis: a review with special reference to perio-pathogenic bacteria. J Oral Microbiol. 2016;8:32762. doi: 10.3402/jom.v8.32762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35:249–255. doi: 10.1093/carcin/bgt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy KN, Sonkodi I, Szoke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral Oncol. 1998;34:304–308. doi: 10.1016/S1368-8375(98)80012-2. [DOI] [PubMed] [Google Scholar]
- 15.Hooper SJ, et al. Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol. 2006;44:1719–1725. doi: 10.1128/JCM.44.5.1719-1725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper SJ, et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol. 2007;56:1651–1659. doi: 10.1099/jmm.0.46918-0. [DOI] [PubMed] [Google Scholar]
- 17.Pushalkar S, et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12:144. doi: 10.1186/1471-2180-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bebek G, et al. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum Mol Genet. 2012;21:1557–1565. doi: 10.1093/hmg/ddr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, H. et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. 9, 14 (2017). [DOI] [PMC free article] [PubMed]
- 20.Pushalkar S, et al. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol. 2011;61:269–277. doi: 10.1111/j.1574-695X.2010.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt BL, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One. 2014;9:e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerrero-Preston, R. et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, Human Papilloma Virus infection and surgical treatment. Oncotarget (2016). [DOI] [PMC free article] [PubMed]
- 23.Al-Hebshi NN, Nasher AT, Idris AM, Chen T. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples. J Oral Microbiol. 2015;7:28934. doi: 10.3402/jom.v7.28934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Hebshi, N.N. et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. 7, 1834 (2017). [DOI] [PMC free article] [PubMed]
- 25.Khatoon J, Rai RP, Prasad KN. Role of Helicobacter pylori in gastric cancer: Updates. World J Gastrointest Oncol. 2016;8:147–158. doi: 10.4251/wjgo.v8.i2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plummer M, et al. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609–616. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D-L, Jiang X, Wu QL, Zhou N-Y. Intragenomic Heterogeneity of 16S rRNA Genes Causes Overestimation of Prokaryotic Diversity. Applied and Environmental Microbiology. 2013;79:5962–5969. doi: 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X, Zhang Q, Hua H, Chen F. Changes in the salivary microbiota of oral leukoplakia and oral cancer. Oral Oncol. 2016;56:e6–8. doi: 10.1016/j.oraloncology.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. Isme j. 2014;8:1879–1891. doi: 10.1038/ismej.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goncalves C, et al. Association of three putative periodontal pathogens with chronic periodontitis in Brazilian subjects. J Appl Oral Sci. 2016;24:181–185. doi: 10.1590/1678-775720150445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binder Gallimidi A, et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6:22613–22623. doi: 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogrendik M. Oral bacteria in pancreatic cancer: mutagenesis of the p53 tumour suppressor gene. Int J Clin Exp Pathol. 2015;8:11835–11836. [PMC free article] [PubMed] [Google Scholar]
- 35.Toprak NU, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 36.Yan X, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111–3122. [PMC free article] [PubMed] [Google Scholar]
- 37.Mager DL, et al. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagpal D, Prakash S, Bhat KG, Singh G. Detection and comparison of Selenomonas sputigena in subgingival biofilms in chronic and aggressive periodontitis patients. J Indian Soc Periodontol. 2016;20:286–291. doi: 10.4103/0972-124X.181247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 40.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 41.Nobbs AH, Jenkinson HF, Jakubovics NS. Stick to your gums: mechanisms of oral microbial adherence. J Dent Res. 2011;90:1271–1278. doi: 10.1177/0022034511399096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abed J, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, et al. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci Rep. 2013;3:1843. doi: 10.1038/srep01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tezal M, et al. Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:2406–2412. doi: 10.1158/1055-9965.EPI-09-0334. [DOI] [PubMed] [Google Scholar]
- 45.Hooper SJ, Wilson MJ, Crean SJ. Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head Neck. 2009;31:1228–1239. doi: 10.1002/hed.21140. [DOI] [PubMed] [Google Scholar]
- 46.Zhang G, Wei L, Chang CC, Zhang Y, Wei D. Molecular Biological Methods in Environmental Engineering. Water Environ Res. 2016;88:930–953. doi: 10.2175/106143016X14696400494371. [DOI] [PubMed] [Google Scholar]
- 47.Xiao C, Ran S, Huang Z, Liang J. Bacterial Diversity and Community Structure of Supragingival Plaques in Adults with Dental Health or Caries Revealed by 16S Pyrosequencing. Front Microbiol. 2016;7:1145. doi: 10.3389/fmicb.2016.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warton DI, Wright ST, Wang Y. Distance-based multivariate analyses confound location and dispersion effects. Methods in Ecology and Evolution. 2012;3:89–101. doi: 10.1111/j.2041-210X.2011.00127.x. [DOI] [Google Scholar]
- 52.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng Y, et al. Molecular ecological network analyses. BMC Bioinformatics. 2012;13:113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mar JC, Matigian NA, Quackenbush J, Wells CA. attract: A method for identifying core pathways that define cellular phenotypes. PLoS One. 2011;6:e25445. doi: 10.1371/journal.pone.0025445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data from this study have been deposited in the GenBank Sequence Read Archive under accession number SRP097643.