Significance
The endoplasmic reticulum (ER) serves as a platform for the packaging of most secretory proteins into conserved coat protein complex II (COPII)-coated transport carriers destined for ER–Golgi intermediate compartments (ERGIC) in animal cells. In this work, we demonstrate that Trk-fused gene (TFG), a protein implicated in multiple neurodegenerative diseases and oncogenesis, functions in this pathway by interacting directly with the COPII protein Sec23. Specifically, we show that TFG outcompetes interactions between the inner and outer layers of the COPII coat, indicating that TFG promotes the uncoating process after transport carriers undergo scission from the ER. Moreover, we demonstrate that TFG simultaneously captures and concentrates COPII transport carriers at the ER/ERGIC interface to enable the rapid movement of secretory cargoes to the ERGIC.
Keywords: COPII, Trk-fused gene, tether, coat disassembly, endoplasmic reticulum
Abstract
The conserved coat protein complex II (COPII) mediates the initial steps of secretory protein trafficking by assembling onto subdomains of the endoplasmic reticulum (ER) in two layers to generate cargo-laden transport carriers that ultimately fuse with an adjacent ER–Golgi intermediate compartment (ERGIC). Here, we demonstrate that Trk-fused gene (TFG) binds directly to the inner layer of the COPII coat. Specifically, the TFG C terminus interacts with Sec23 through a shared interface with the outer COPII coat and the cargo receptor Tango1/cTAGE5. Our findings indicate that TFG binding to Sec23 outcompetes these other associations in a concentration-dependent manner and ultimately promotes outer coat dissociation. Additionally, we demonstrate that TFG tethers vesicles harboring the inner COPII coat, which contributes to their clustering between the ER and ERGIC in cells. Together, our studies define a mechanism by which COPII transport carriers are retained locally at the ER/ERGIC interface after outer coat disassembly, which is a prerequisite for fusion with ERGIC membranes.
In most metazoan systems, cargoes transported from the endoplasmic reticulum (ER) must traverse a vesicular-tubular cluster of membranes known as the “ER–Golgi intermediate compartment” (ERGIC) en route to the Golgi apparatus (1–5). This process involves the function of two distinct coat complexes (6–9). The conserved coat protein complex II (COPII) assembles at ER subdomains and mediates transport to the ERGIC (5, 10–12), whereas the COPI complex directs transport from the ERGIC to the Golgi as well as retrograde transport to the ER (5, 8, 13–15). In both cases, the coats define the architecture of the transport carriers but must eventually disassemble to enable membrane fusion at the target compartment (16, 17). In the case of COPI, which minimally consists of seven core subunits and an ADP ribosylation factor (Arf)-type GTPase (18, 19), uncoating is promoted by association with target membrane tethers such as the Dsl1 complex, which interferes with COPI subunit interactions, and stimulation of GTP hydrolysis on Arf by members of the Arf guanine nucleotide-activating protein (GAP) (ArfGAP) family (20–23).
In contrast, mechanisms that direct COPII uncoating have been more controversial. The COPII coat consists of two layers, an inner adaptor layer composed of the GTPase Sar1 and Sec23-Sec24 heterodimers and an outer lattice-like cage made up of Sec13-Sec31 heterotetramers (7, 24, 25). Initial studies suggested that GTP hydrolysis on Sar1, which is activated by the inner coat GAP Sec23 and further stimulated by the outer coat protein Sec31, plays an important role in vesicle uncoating (10, 26–29). However, subsequent in vitro studies contended that Sar1 is rapidly removed from COPII carriers immediately following their scission at the ER (14, 30). More recently, phosphorylation of COPII subunits at ERGIC and Golgi membranes has been suggested to promote uncoating, but in vitro studies have failed to support this idea, and the consequences of posttranslational modifications on COPII coat stability remain unclear (14, 31–33). Related to COPI tethering by the Dsl1 complex, the transport protein particle (TRAPP) complex tethers COPII transport carriers at the ERGIC and cis-Golgi (14, 20, 30, 34). However, unlike the function of Dsl1, biochemical studies suggest that TRAPP associates with Sec23 on a site that overlaps with its binding domain for Sar1, which is not predicted to disrupt inner or outer coat integrity. Moreover, recent work suggests that TRAPP actually promotes outer COPII coat assembly during the formation of transport carriers (35). Thus, it remains unknown how the COPII coat disassembles before fusion at the ERGIC.
Several additional regulators of COPII-mediated trafficking have also been described recently, some of which directly impact coat stability. Members of the Sec16 family function at ER subdomains to inhibit Sar1 GTPase activity and facilitate COPII coat assembly (36–38). In a related manner, the cargo receptors Tango1 and cTAGE5 associate directly with Sec23 at the ER to promote the formation of elongated COPII-coated tubules (39, 40). Ultimately, outer coat assembly has been suggested to outcompete Tango1/cTAGE5 and Sec16 interactions with the inner coat to enable fission of transport carriers in a manner dependent on Sar1 GTP hydrolysis (40, 41).
Following release from the ER, COPII transport carriers are restricted to the ER/ERGIC interface (42, 43). Our previous studies indicated that this distribution is regulated by Trk-fused gene (TFG), a homo-oligomeric protein complex that assembles into a meshwork and colocalizes with COPII carriers in cells (44, 45). Here, we demonstrate that TFG associates directly with Sec23 and exhibits the ability to outcompete interactions between layers of the COPII coat. Our data are most consistent with a model in which TFG promotes the release of the outer COPII coat and restricts diffusion of carriers harboring the inner coat until tethering occurs at ERGIC membranes. In doing so, TFG functions to maintain the integrity of the early secretory pathway.
Results
TFG Facilitates the Export of Conventional Cargoes from the ER.
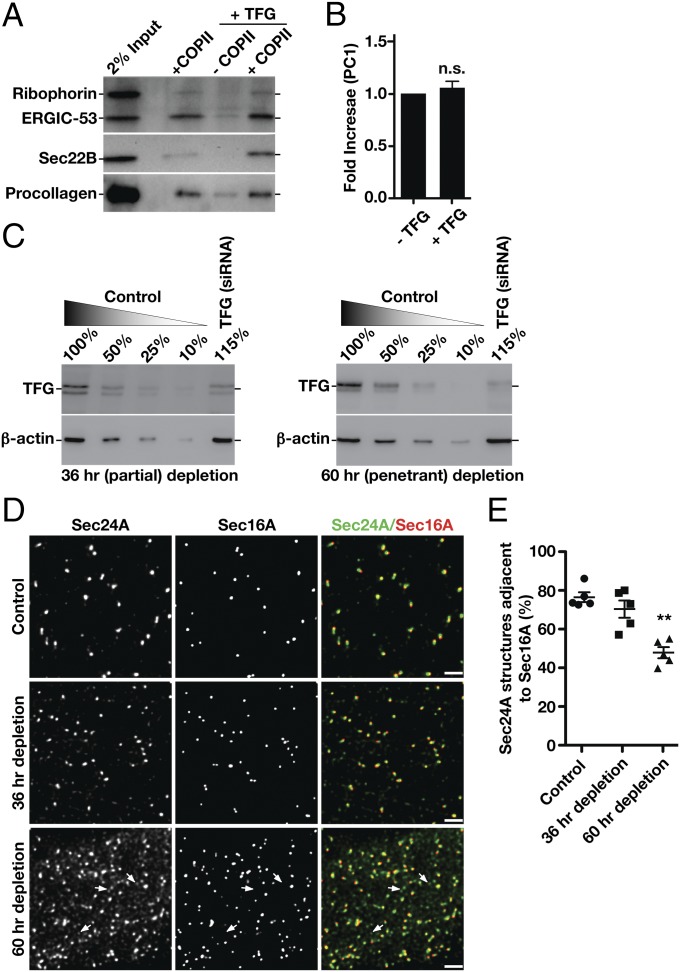
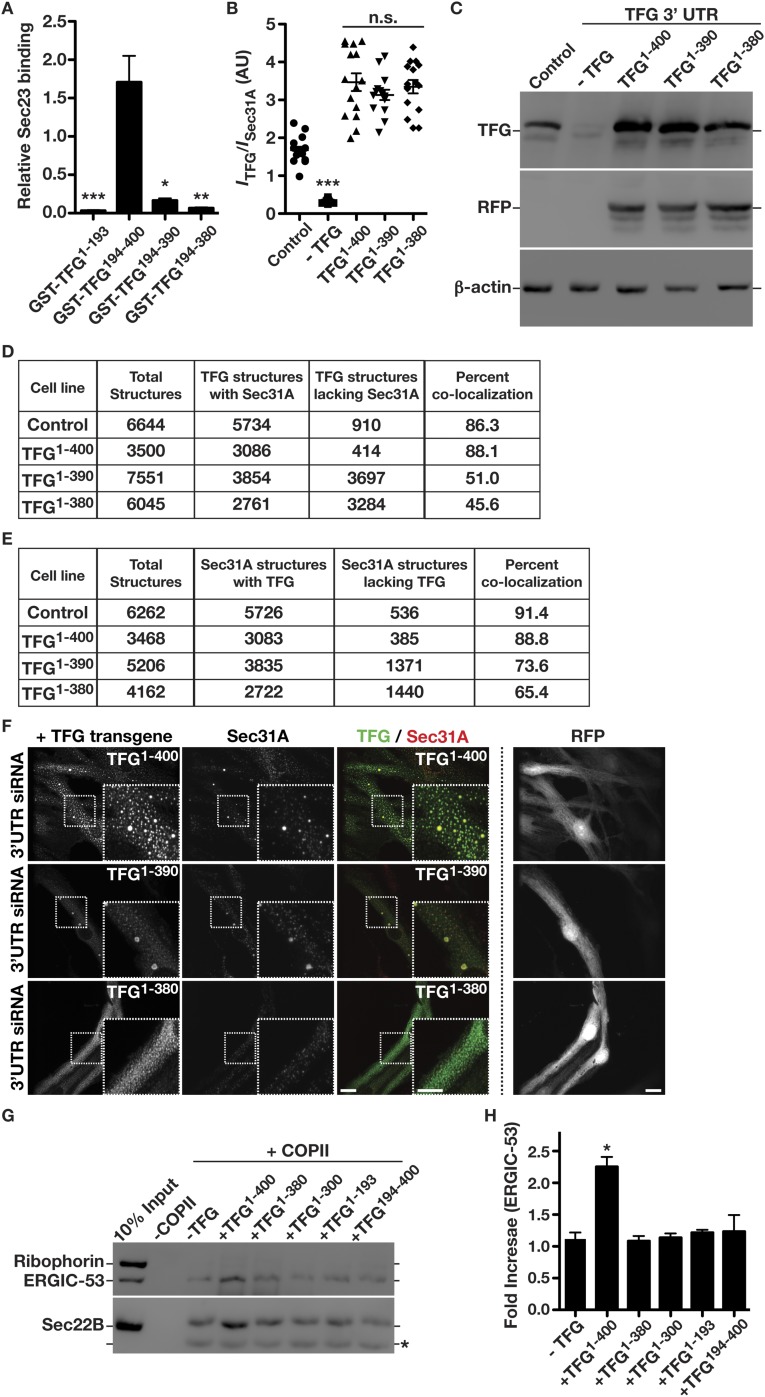
We demonstrated previously that TFG plays a conserved role in regulating the export of diverse cargoes, including SNAREs, Golgi enzymes, and other biosynthetic transmembrane proteins of all sizes, from the ER (44, 45). Additionally, we found that penetrant depletion of TFG disrupts early secretory pathway integrity, leading to the accumulation of inner and outer COPII-coated transport carriers away from the ER/ERGIC interface (44, 46). A recent report similarly demonstrated that TFG promotes organization of ER exit sites but suggested a specialized function for TFG in the export of large cargoes, such as procollagens (47). To test directly the contribution of TFG to COPII-dependent cargo transport, we used a reconstituted system in which permeabilized cells lacking cytosol were supplemented with purified recombinant components (10, 11, 48). As described previously, the addition of COPII subunits was sufficient to promote the formation of transport carriers harboring small, native cargoes, including the lectin ERGIC-53 and the R-SNARE Sec22B, but not ribophorin, an ER resident protein (Fig. 1A and Fig. S1A). The addition of TFG further stimulated the release of ERGIC-53– and Sec22B-containing transport carriers (Fig. S1B) but exhibited a negligible effect on procollagen (PC1) secretion (Fig. 1 A and B) (49), indicating that the role of TFG in the early secretory pathway is unlikely to be specific for large cargoes.
Fig. 1.
TFG regulates the transport of conventional COPII carriers. (A) Representative immunoblot of recovered COPII transport carriers isolated following budding reactions performed in the presence or absence of TFG using antibodies directed against ribophorin (Top), ERGIC-53 (Top), Sec22B (Middle), and procollagen (Bottom). (B) Quantification of the relative amounts of procollagen present in COPII transport carriers, comparing budding reactions performed in the presence and absence of TFG (n = 3). Unlike the addition of cytosol (49), recombinant TFG fails to stimulate the formation of COPII carriers containing procollagen. n.s., not significant. (C) Representative immunoblots conducted using TFG antibodies of extracts from cells that were partially depleted (Left) or more fully depleted (Right) of TFG. Immunoblotting for actin was used as a load control to quantify levels of TFG depletion. (D) Control cells (mock transfected) and cells depleted of TFG for 36 h (partial depletion) or 60 h (penetrant depletion) were immunostained using antibodies directed against Sec24A and Sec16A and were imaged using STED microscopy. Representative deconvolved images are shown. Arrows highlight Sec24A-positive structures that do not stain with Sec16A antibodies (Bottom). (Scale bars, 4 μm.) (E) Quantification of the percentage of Sec24A-labeled structures that are juxtaposed to Sec16A under the conditions specified. Error bars represent mean ± SEM; n = at least 10 different cells per condition. **P < 0.01 (penetrant depletion compared with control), calculated using a paired t test.
Fig. S1.
Depletion of TFG disrupts early secretory pathway organization and the secretion of conventional cargoes. (A) Representative immunoblot of recovered COPII transport carriers isolated following budding reactions performed in the presence or absence of TFG using antibodies directed against ERGIC-53 (Upper), ribophorin (Upper), and Sec22B (Lower). An asterisk highlights a nonspecific band recognized by the Sec22B antibody. (B) Quantification of the relative amounts of ERGIC-53 present in COPII transport carriers, comparing budding reactions performed in the presence or absence of TFG (n = 3). *P < 0.05, calculated using a paired t test. (C) Relative fluorescence intensity (I) of Sec24A juxtaposed to Sec16A-labeled structures in cells depleted of TFG to varying extents. *P < 0.05 (compared with control), calculated using a paired t test. (D) Control cells and cells depleted of TFG for 36 or 60 h were fixed and stained using antibodies directed against TFG and were imaged using confocal microscopy. Images shown are representative of at least 30 individual cells analyzed for each condition. Nonspecific staining within the nucleus is observed following penetrant depletion of TFG (60 h time point). (Scale bar, 10 μm.) (E) Control cells and cells depleted of TFG for 36 or 60 h were fixed and stained using antibodies directed against Sec16A and ERGIC-53 and were imaged using STED microscopy. (Scale bars, 4 μm; Inset, 0.5 μm.) Images shown are representative of at least 30 individual cells analyzed for each condition. (F) Quantification of the percentage of Sec16A-labeled structures that are juxtaposed to Sec24A under the conditions specified. (G) Localization of various integral membrane proteins within the C. elegans germline in control and TFG-depleted animals. Arrows highlight the aberrant accumulation of cargoes in the perinuclear ER. Images shown are representative of at least six different animals analyzed for each condition. (Scale bars, 5 μm.) (H) Sequence traces obtained from heterozygous animals (following TOPO cloning) harboring a 44-bp deletion mutation in the first coding exon of rat TFG. The deleted sequence is underlined in the wild-type trace (Upper), and nonnative amino acids encoded following a frame shift are highlighted in red.
We therefore questioned whether the modest effects of TFG inhibition on the export of small artificial cargoes reported recently (47) could be a consequence of insufficient protein depletion. To address this possibility, we first defined conditions under which TFG was partially (∼44%) and more fully (∼77%) depleted (Fig. 1C). We then examined the distribution of COPII carriers relative to Sec16A, a marker of ER subdomains at which COPII coat assembly occurs (50). When TFG was partially depleted, COPII structures that were modestly reduced in intensity remained juxtaposed to sites marked by Sec16A (Fig. 1D and Fig. S1C), consistent with recently published findings (47). However, upon more penetrant inhibition of TFG, as determined by immunofluorescence studies (Fig. S1D), we observed a dramatic increase in the number of COPII and ERGIC-53 labeled structures, which were no longer juxtaposed to Sec16A (Fig. 1 D and E and Fig. S1E). Notably, under all conditions, Sec16A-labeled sites continued to exhibit juxtaposed Sec24A, indicating that ER subdomains continue to produce COPII transport carriers in the absence of TFG (Fig. S1F).
Since human tissue-culture cells undergo apoptosis rapidly after penetrant depletion of TFG (44), we turned to the Caenorhabditis elegans germline as a model system, where we previously demonstrated an ability to achieve greater than 95% inhibition of TFG expression (45). Examination of both large (E-cadherin; >3,000 amino acids) and small (eight different transmembrane SNAREs and the minimal transmembrane domain of mannosidase II) cargoes demonstrated a uniform defect in secretion from the ER following TFG depletion (Fig. S1G). Taken together, these data indicate that TFG plays a general role in the normal trafficking of conventional COPII carriers.
Dysregulation of COPII function in rodent models results in varying phenotypes ranging from early embryo lethality to more modest impacts on physiology due to redundancies in subunit expression (51, 52). If TFG functioned only in the secretion of large cargoes such as procollagens, we would predict that its deletion phenotype would be similar to that of Tango1-knockout animals, which exhibit several developmental abnormalities but are born in normal Mendelian frequencies (53). Using CRISPR-mediated genome editing, we isolated heterozygous rats harboring a 44-bp deletion within the first coding exon of TFG, which results in an early frame shift and produces only the first 21 amino acids of native TFG (Fig. S1H). After outcrossing one of the founders six times, we mated heterozygous animals and genotyped 114 progeny that were born (from five distinct mating pairs). In total, we obtained 39 wild-type animals, 75 heterozygous animals, and no homozygous-null animals (Table S1). We also performed timed pregnancies and isolated embryos at E9.5 and E14. Genotyping revealed that none of the recovered embryos were homozygous-null (Table S1). Together, these data suggest that loss of TFG results in early embryo lethality in rats before E9.5, which is more consistent with a role for TFG in general protein secretion.
Table S1.
Genotypes of progeny from mating heterozygous (TFG+/−) pairs of animals
| Genotype | |||
| Stage of development | +/+ | +/− | −/− |
| Newborn | 39 | 75 | 0 |
| Embryo E9.5 | 5 | 12 | 0 |
| Embryo E14 | 7 | 15 | 0 |
For newborn analysis, five mating pairs were analyzed (two rounds of mating). For embryonic analysis, two timed matings were conducted at each time point, and embryos were harvested from pregnant dams at either E9.5 or E14.
TFG Associates Directly with the Inner COPII Coat Protein Sec23.
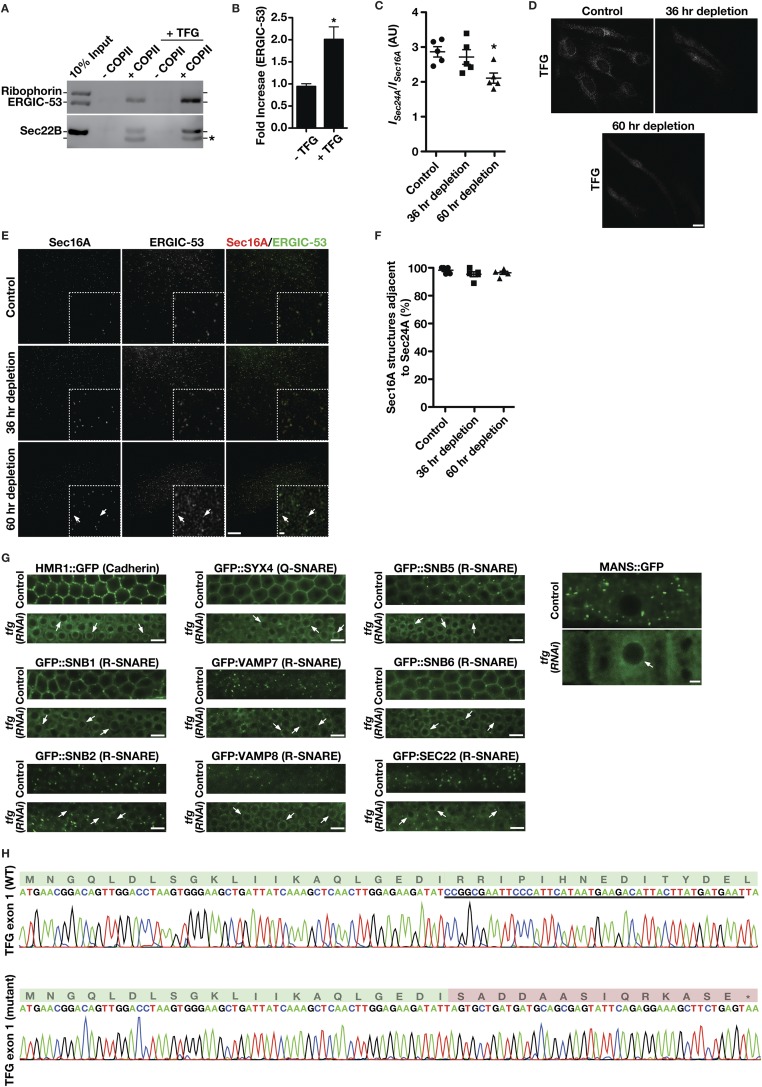
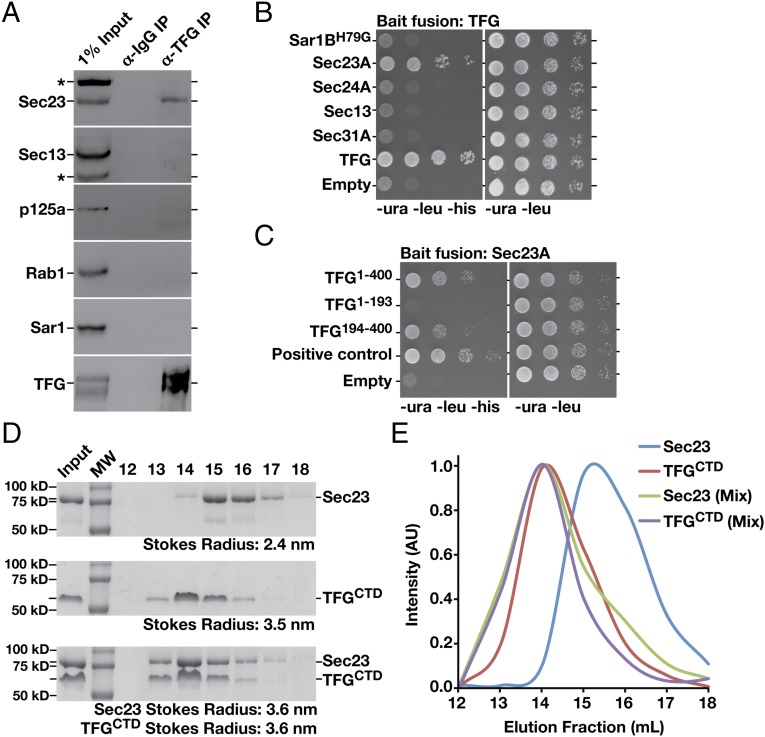
To investigate potential mechanisms by which TFG regulates COPII-mediated protein transport, we conducted a series of immunoprecipitations using TFG antibodies and freshly prepared rat liver cytosol and two distinct human cell lines (RPE1 and HeLa). These studies revealed specific interactions with Sec23 but not other components of the COPII machinery (Fig. 2A and Table S2). Additionally, we conducted a directed yeast two-hybrid screen to identify TFG-interacting partners in the early secretory pathway (Table S3). This approach again revealed a specific association between TFG and the inner COPII coat protein Sec23 (Fig. 2B). Consistent with these findings, recombinant GST-tagged TFG, but not GST alone, was also capable of recovering native Sec23 from rat liver cytosol (Fig. S2A).
Fig. 2.
TFG interacts directly with the inner COPII coat subunit Sec23. (A) Immunoprecipitations using IgG or antibodies directed against TFG were conducted using rat liver cytosol, separated by SDS/PAGE, and immunoblotted using the indicated antibodies (n = 3 in each condition). Asterisks highlight nonspecific bands recognized by Sec23 and Sec13 antibodies. (B) Yeast coexpressing plasmids encoding TFG (bait fusion) and several unique prey constructs were plated (10-fold dilutions, left to right) on either selective (−Ura, −Leu, −His) or histidine-supplemented medium for 48 h (n = 3). The GTPase-deficient form of Sar1B (H79G) was used in these experiments. (C) Yeast coexpressing plasmids encoding Sec23A (bait fusion) and prey constructs encoding distinct regions of TFG were plated as described in A (n = 3). Coexpression of bait and prey constructs encoding TFG fusions was used as a positive control, and an empty prey construct was used for a negative control. (D) Purified C. elegans Sec23 and TFG C-terminal domain (CTD), amino acids 196–486, were separated individually and as a mixture by gel filtration chromatography, and specific fractions were analyzed by SDS/PAGE followed by Coomassie staining (n = 3 in each condition). Stokes radii were calculated based on the elution profile of known standards. MW, molecular weight marker. (E) Retention times of C. elegans TFG (amino acids 196–486) and full-length Sec23 (individually and as a mixture) were plotted based on densitometry after gel filtration chromatography and SDS/PAGE analysis of fractions. AU, arbitrary units.
Table S2.
TFG-interacting proteins identified by solution mass spectrometry after immunoprecipitation from HeLa and RPE1 cell extracts
| Protein | Unique peptides identified |
| M.NGQLDLSGK.L | |
| K.AQLGEDIR.R | |
| R.GKLLSNDEVTIK.Y | |
| K.LTLFVNGQPR.P | |
| R.PLESSQVK.Y | |
| R.LLDSLEPPGEPGPSTNIPENDTVDGR.E | |
| TFG | G.PSTNIPENDTVDGREEK.S |
| K.QSTQVMAASMSAFDPLK.N | |
| M.SAFDPLKNQDEINK.N | |
| K.NVMSAFGLTDDQVSGPPSAPAEDR.S | |
| R.SGTPDSIASSSSAAHPPG.V | |
| P.PPSGPNPYAR.N | |
| F.GQGYTQPGPGYR.- | |
| R.PAQPQEHPFASSR.F | |
| Sec23B | R.YINTEHGGSQAR.F |
| K.AGYQDMPEYENFK.H |
Antibodies directed against TFG were used in immunoprecipitation studies, and peptides corresponding to proteins involved in the early secretory pathway that were identified by mass spectrometry analysis are shown. Immunoprecipitation using IgG failed to identify peptides corresponding to either TFG or Sec23.
Table S3.
Yeast two-hybrid screen to identify TFG-interacting proteins
| Prey fusion | UniProt ID | Interaction |
| TFG | Q92734 | ++ |
| TRAPPC3 | O43617 | — |
| REEP1 | Q9H902 | — |
| Atlastin-1 | Q8WXF7 | — |
| Rab1A | P62820 | — |
| Sar1B | Q9Y6B6 | — |
| Sec23A | Q15436 | ++ |
| Sec24A | O95486 | — |
| Sec13 | P55735 | — |
| Sec31A | O94979 | — |
| Sec16B | Q96JE7 | — |
A TFG bait fusion was cotransformed into yeast expressing the specified prey constructs and assayed for growth on selective medium lacking histidine. ++, strong interaction; —, no interaction.
Fig. S2.
Sec23 and the TFG C terminus form a stable heterodimer in solution. (A) GST or a GST fusion to full-length human TFG was immobilized on glutathione resin and incubated with rat liver cytosol. The eluates were separated by SDS/PAGE and immunoblotted using antibodies directed against Sec23 (n = 3). An asterisk highlights a nonspecific band recognized by Sec23 antibodies in rat liver cytosol. (B–D) Representative light-scattering profiles (Left) and Coomassie-stained SDS/PAGE gels (Right) following size-exclusion chromatography of purified C. elegans Sec23 (B), the TFG C terminus (amino acids 196–486) (C), and a mixture of the proteins (D). (E) Summary of the measured molecular masses of Sec23, TFG, and the Sec23-TFG complex, compared with the expected massed predicted by amino acid composition (n = 3, each condition). (F) Sequence alignment of human and zebrafish TFG, with identical amino acids highlighted in green. (G) Sequence alignment of human, zebrafish, and C. elegans TFG (the C-terminal peptide necessary for Sec23 binding), with identical amino acids highlighted in yellow.
To delineate the region of TFG necessary for its association with Sec23, we again used a yeast two-hybrid approach. Our findings indicated that the C-terminal half of TFG (amino acids 194–400), which we demonstrated previously to be intrinsically disordered (44), was necessary and sufficient to bind to Sec23 (Fig. 2C). To determine whether the interaction was direct, we purified recombinant forms of Sec23 and the TFG C terminus and examined their elution profiles separately and in combination following gel filtration chromatography. Individually, Sec23 exhibited a Stokes radius of 2.4 nm, whereas the TFG C terminus exhibited a Stokes radius of 3.5 nm (Fig. 2D). In contrast, when the proteins were mixed, they coeluted with an average Stokes radius of 3.6 nm, indicating that they formed a complex (Fig. 2 D and E). To determine the molecular mass of the complex, we used a combination of size-exclusion chromatography and multiangle light scattering (Fig. S2 B–E). These data demonstrated that Sec23 and the TFG C terminus form a 127.8 (±1.4)-kDa complex, consistent with a 1:1 association (Fig. S2E).
The Localization of TFG Is Governed by Its Association with COPII-Coated Transport Carriers.
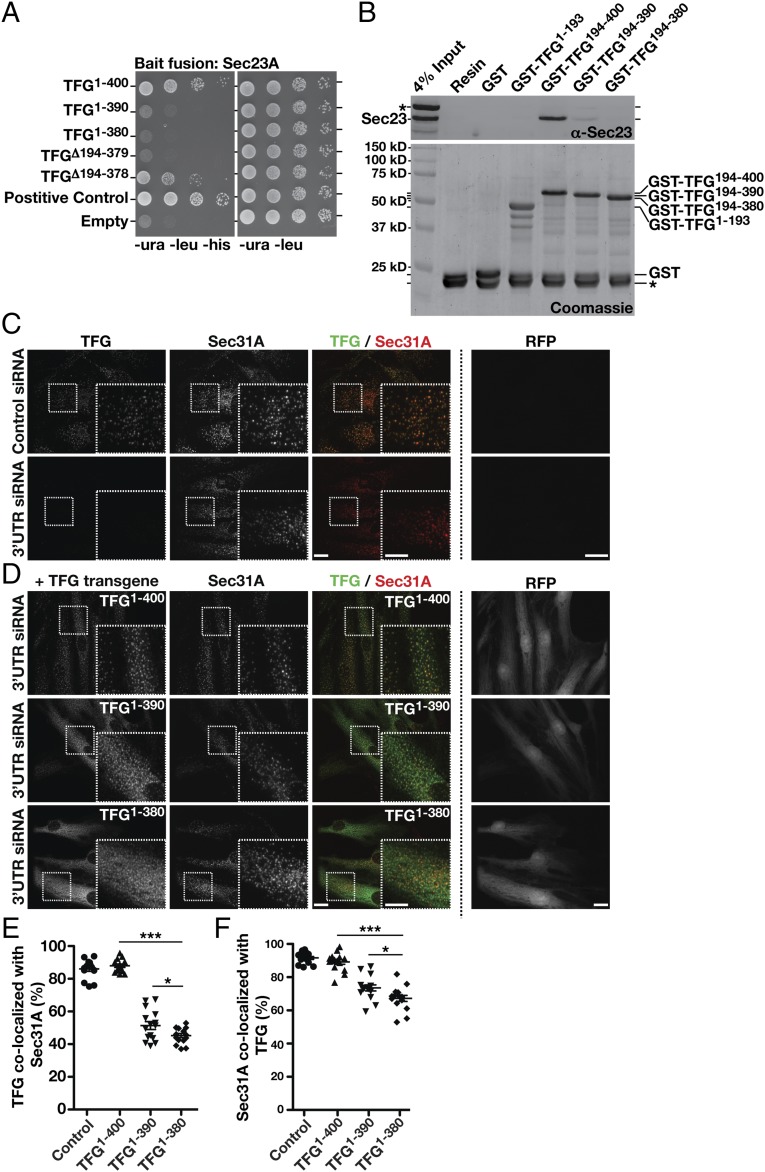
The C-terminal portion of TFG is poorly conserved across metazoans based on amino acid alignment algorithms (Fig. S2F). However, its overall amino acid content varies relatively little from C. elegans (≈30% proline, 17% glycine, 13% glutamine, 10% alanine, and 8% serine) to humans (≈20% proline, 10% glycine, 21% glutamine, 11% alanine, and 9% serine). To determine the specific region of TFG necessary for binding to Sec23, we began truncating TFG from its C-terminal end and measuring the impact on Sec23 binding using the yeast two-hybrid system. Removal of as little as 10 amino acids from the C terminus of TFG had a dramatic impact on Sec23 binding, and deletion of 20 amino acids reduced binding to background levels (Fig. 3A). These findings were confirmed using recombinant forms of TFG in pull-down experiments from rat liver cytosol (Fig. 3B and Fig. S3A). By contrast, deletion of the entire C-terminal half of TFG with the exception of the last 22 amino acids (i.e., deletion of amino acids 194–378) preserved its ability to interact with Sec23 (Fig. 3A). These data define the minimal domain of TFG that is necessary for its association with Sec23 (Fig. S2G), although its proline-rich domain (PRD) may also contribute to binding, based on previous work examining PRDs from Sec31 and Tango1/cTAGE5 (40).
Fig. 3.
An interaction with Sec23 is necessary for TFG localization at the ER/ERGIC interface. (A) Yeast coexpressing plasmids encoding Sec23A (bait fusion) and prey constructs encoding distinct regions of TFG were plated as described in Fig. 2A (n = 3). (B) Immobilized GST or GST fused to the C terminus of human TFG (amino acids 194–400) was incubated with rat liver cytosol, washed extensively, and eluted using glutathione. Recovered samples (15% total) were immunoblotted using Sec23 antibodies (Upper) or were Coomassie stained (Lower) following SDS/PAGE (n = 3, each condition). An asterisk highlights native rat glutathione S-transferases, which bind to the resin under all conditions. (C and D) Control cells (mock transfected) and cells depleted of TFG in the presence of doxycycline (3 ng/mL) to drive expression of various forms of TFG were immunostained using antibodies directed against TFG (green) and Sec31A (red) and were imaged using confocal microscopy. RFP fluorescence intensity was used to determine relative expression levels of each transgene. Images shown are projections of 3D datasets (4 μm in z). (Scale bars, 15 μm.) Higher-magnification views of the boxed regions are also shown in the lower right portion of each panel. (Inset scale bars, 5 μm.) Images shown are representative of at least 30 individual cells analyzed for each condition. (E and F) Quantification of the percentage of TFG-labeled structures that colocalize with Sec31A (E) and the percentage of Sec31A-labeled structures that colocalize with TFG (F) under the conditions specified. Error bars represent mean ± SEM; n = at least 15 different cells per condition. ***P < 0.001 [cells expressing full-length TFG compared with cells expressing truncated TFG (amino acids 1–390 and 1–380)], calculated using a paired t test. *P < 0.05 (comparing cells expressing each truncated form of TFG), calculated using a paired t test.
Fig. S3.
The TFG C terminus directs its localization in cells. (A) Quantification of relative Sec23 binding to various GST-TFG fusion proteins. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with 4% load), calculated using a paired t test. (B) Relative fluorescence intensities of TFG and Sec31A were calculated under control conditions, TFG-depleted conditions, and conditions in which endogenous TFG is depleted and an exogenous isoform is expressed. ***P < 0.001 (compared with control), calculated using a paired t test; n.s., not significant. (C) Extracts from cells expressing various forms of TFG, following depletion of endogenous TFG, were immunoblotted using TFG (Top), RFP (Middle), and actin (load control) (Bottom) antibodies (n = 3). (D and E) Total number of TFG-labeled structures that colocalize with Sec31A (D) and the total number of Sec31A-labeled structures that colocalize with TFG (E) under control conditions and conditions under which endogenous TFG is depleted and an exogenous isoform is expressed. (F, Left) Inducible genome-edited cell lines depleted of endogenous TFG and treated with doxycycline to overexpress exogenous full-length TFG (amino acids 1–400) (Top), truncated TFG lacking the last 10 residues (amino acids 1–390) (Middle), and truncated TFG lacking the last 20 residues (amino acids 1–380) (Bottom) were fixed and stained using antibodies directed against TFG and Sec31A. (Right) RFP expression also induced by doxycycline is shown in each case. Images shown are representative of at least 30 individual cells analyzed for each condition. (Scale bar, 10 μm; Inset, 5 μm.) (G) Representative immunoblot of recovered COPII transport carriers isolated following budding reactions (n = 3) performed in the presence or absence of various TFG isoforms using antibodies directed against ERGIC-53 (Upper), ribophorin (Upper), and Sec22B (Lower). (H) Quantification of the relative amounts of ERGIC-53 present in COPII transport carriers, comparing budding reactions performed in the presence or absence of various isoforms of TFG. *P < 0.05 (compared with control), calculated using a paired t test.
We previously demonstrated that TFG localizes to the ER/ERGIC interface together with COPII-coated transport carriers (44, 45). To determine whether its interaction with Sec23 is necessary, we depleted the endogenous form of TFG using a siRNA targeting its 3′ UTR and replaced it with various TFG transgenes that we predict would alter Sec23 binding in cells (Fig. 3 C and D). To maintain tight control over relative transgene expression (Fig. S3 B and C), we inserted a single copy of each at the adeno-associated virus integration site 1 (AAVS1) safe harbor locus using TALEN-mediated genome editing. All transgenes were placed under control of the Tet-On system and were followed by an internal ribosome entry site (IRES) RFP cassette to monitor expression levels quantitatively by fluorescence microscopy (Fig. 3D and Fig. S3C). Replacement of endogenous TFG with a full-length, untagged transgene did not affect its colocalization with COPII carriers (Fig. 3D). In contrast, transgenes lacking either 10 or 20 amino acids from the TFG C terminus exhibited reduced colocalization with COPII-labeled structures (Fig. 3 D–F and Fig. S3 D and E). Additionally, overexpression of TFG was shown previously to generate large, potentially phase-separated regions of the cytoplasm capable of titrating COPII transport carriers (44). However, overexpression of truncated TFG (amino acids 1–380) abolished this ability (Fig. S3F). Truncated isoforms of TFG were also incapable of promoting COPII carrier budding in vitro, unlike the full-length protein (Fig. S3 F and G). Together, these data are consistent with the idea that an interaction with Sec23 is necessary for TFG to assemble at the ER/ERGIC interface and facilitate COPII-mediated trafficking.
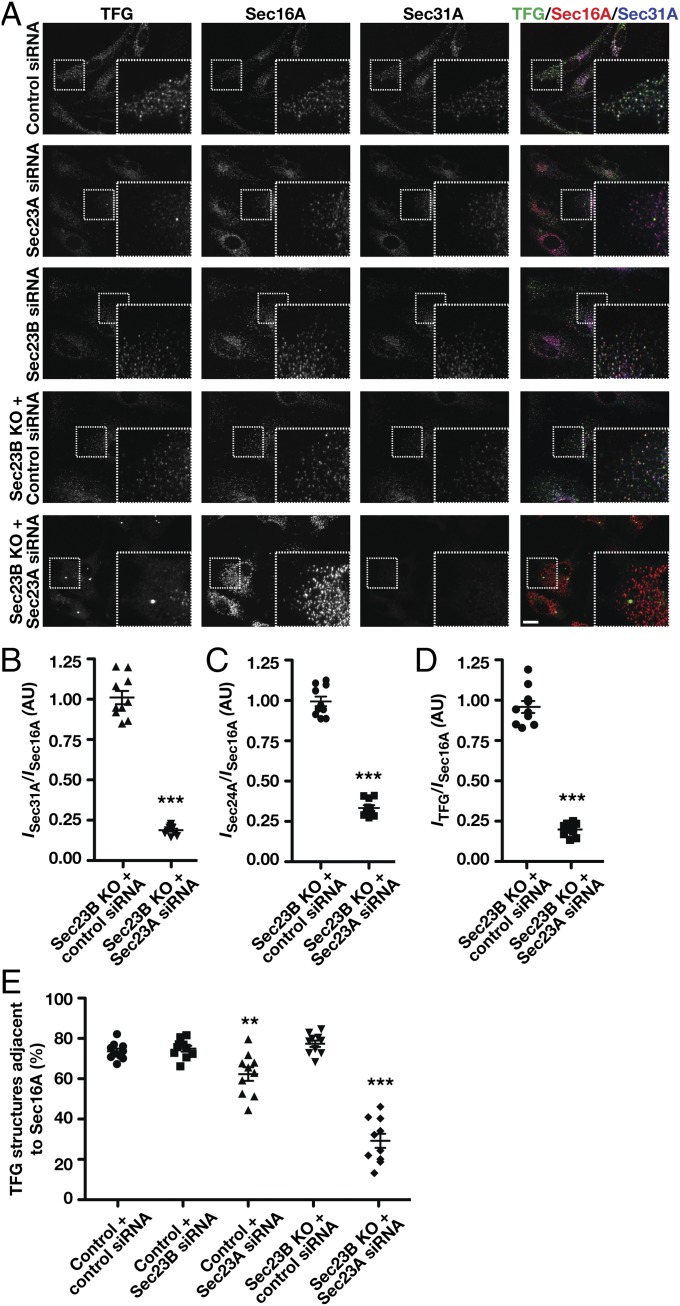
To confirm a role for COPII transport carriers in directing TFG distribution, we examined the impact of inhibiting inner COPII coat assembly. In mammals, two largely redundant isoforms of Sec23 are expressed (Sec23A and Sec23B). Using CRISPR-mediated genome editing, we generated three independent RPE1 cell lines lacking the Sec23B isoform (Fig. 4A and Fig. S4 A–E), which enabled us to achieve a dramatic reduction in COPII accumulation at ER subdomains upon siRNA-mediated depletion of Sec23A (Fig. 4 B and C and Fig. S4F). Under these conditions, we found that TFG was distributed diffusely throughout the cytoplasm and largely failed to accumulate near Sec16A-labeled subdomains at the ER (Fig. 4 A, D, and E and Fig. S4 A–C, F, and G), even though Sec16A levels were dramatically elevated (Fig. S4F). These data indicate that Sec23 is the major binding partner of TFG in the early secretory pathway, although other COPII-associated factors may also contribute to its recruitment (45), and that the presence of COPII transport carriers is required for TFG assembly at the ER/ERGIC interface.
Fig. 4.
COPII transport carriers are required for TFG to assemble at the ER/ERGIC interface. (A) Control cells and genome-edited cells lacking Sec23B were transfected with the indicated siRNA and immunostained using antibodies directed against TFG (green), Sec16A (red), and Sec31A (blue) and were imaged using confocal microscopy. Images shown are projections of 3D datasets (4 μm in z). (Scale bar, 15 μm.) Higher-magnification views of the boxed regions are also shown in the lower right portion of each panel. (Inset scale bar, 5 μm.) Images shown are representative of at least 30 individual cells analyzed for each condition. (B–D) Fluorescence intensity (I) of Sec31A (B), Sec24A (C), or TFG (D) relative to juxtaposed Sec16A-labeled structures in cells lacking Sec23B following delivery of control siRNAs or siRNAs directed against Sec23A. ***P < 0.001 (compared with control), calculated using a paired t test. (E) Quantification of the percentage of TFG-labeled structures that are juxtaposed to Sec16A under specified conditions. Error bars represent mean ± SEM; n = at least 10 different cells per condition. **P < 0.01, ***P < 0.001 (compared with control), calculated using a paired t test.
Fig. S4.
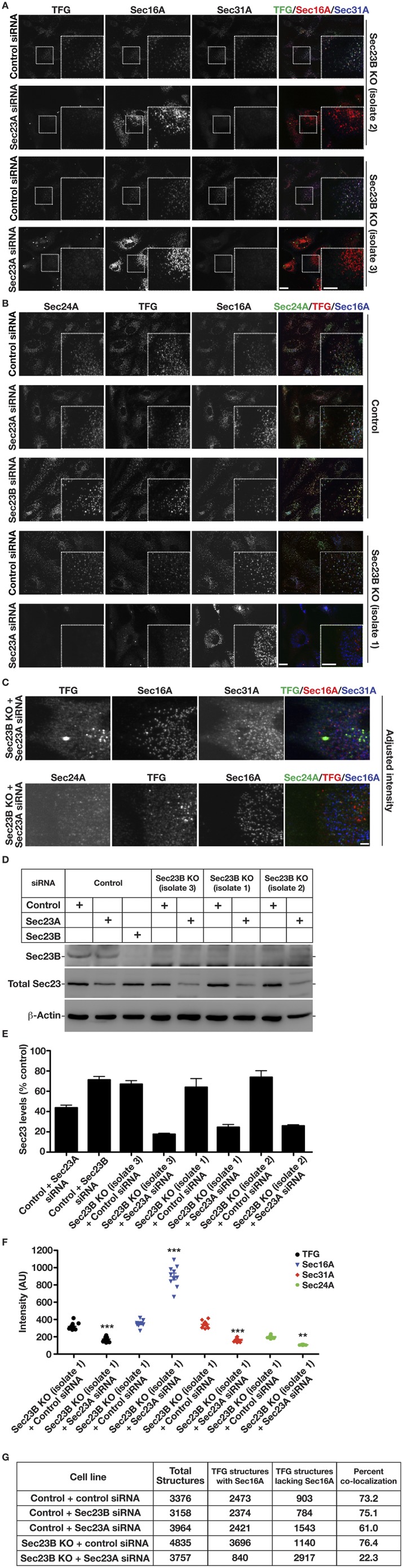
Sec23 directs TFG localization in cells. (A) Independently isolated cell lines harboring deletion mutations in Sec23B were treated with control siRNAs or with siRNAs targeting Sec23A and were fixed and stained using antibodies against TFG (green), Sec16A (red), and Sec31A (blue). (Scale bar, 10 μm.) Higher-magnification views of the boxed regions are shown in the lower right portion of each panel. (Inset scale bar, 5 μm.) Images shown are representative of at least 30 individual cells analyzed for each condition. (B) Control cells and cells harboring a deletion mutation in Sec23B were treated with control siRNAs or with siRNAs targeting Sec23A and were fixed and stained using antibodies against TFG (red), Sec24A (green), and Sec16A (blue). (Scale bar, 10 μm.) Higher-magnification views of the boxed regions are shown in the lower right portion of each panel. (Inset scale bar, 5 μm.) Images shown are representative of at least 30 individual cells analyzed for each condition. (C) Genome-edited cells lacking Sec23B and depleted of Sec23A were fixed and stained using antibodies directed against TFG (green), Sec16A (red), and Sec31A (blue). The intensity of TFG and Sec24A staining was adjusted to highlight their lack of colocalization with Sec31A and Sec16A. (Scale bar, 5 μm.) (D) Extracts from control cells and cell lines lacking Sec23B, following treatment with various siRNAs, were immunoblotted (n = 3) using antibodies directed against Sec23B specifically (Top), both Sec23 isoforms (Middle), and actin (load control) (Bottom). (E) Quantification of Sec23 depletion, based on immunoblotting studies conducted in D. (F) Quantification of the relative fluorescence intensities of TFG (black), Sec16A (blue), Sec31A (red), and Sec24A (green) in cells lacking Sec23B and treated with control siRNAs or siRNAs targeting Sec23A. **P < 0.01, ***P < 0.001 (compared with control), calculated using a paired t test. (G) Total number of TFG-labeled structures that are juxtaposed to Sec16A and the total number of Sec16A-labeled structures that are juxtaposed to TFG under control conditions and conditions under which Sec23 expression is reduced.
TFG Competes with the Outer COPII Coat for Inner Coat Binding.
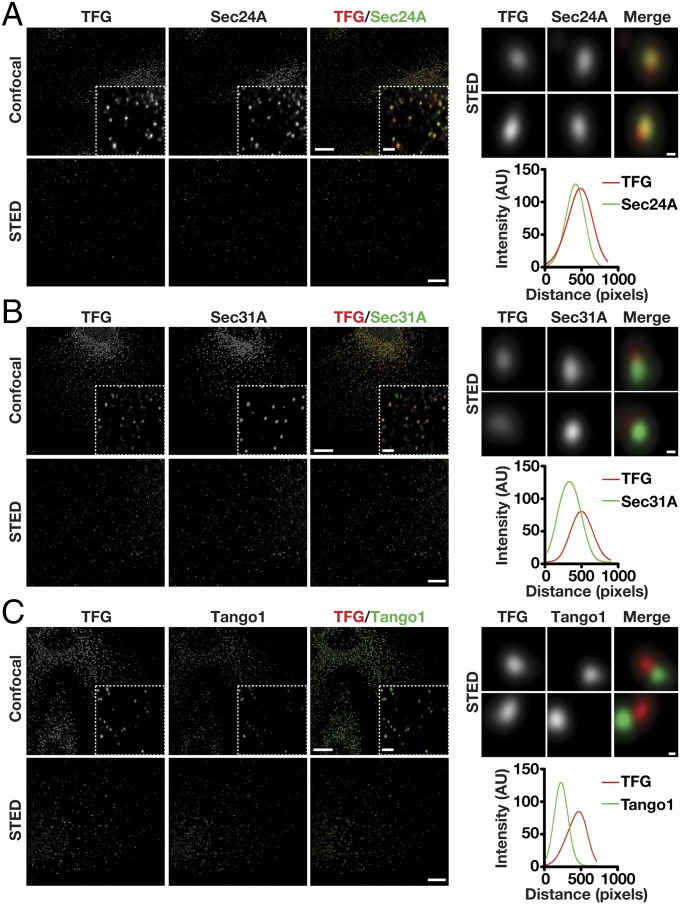
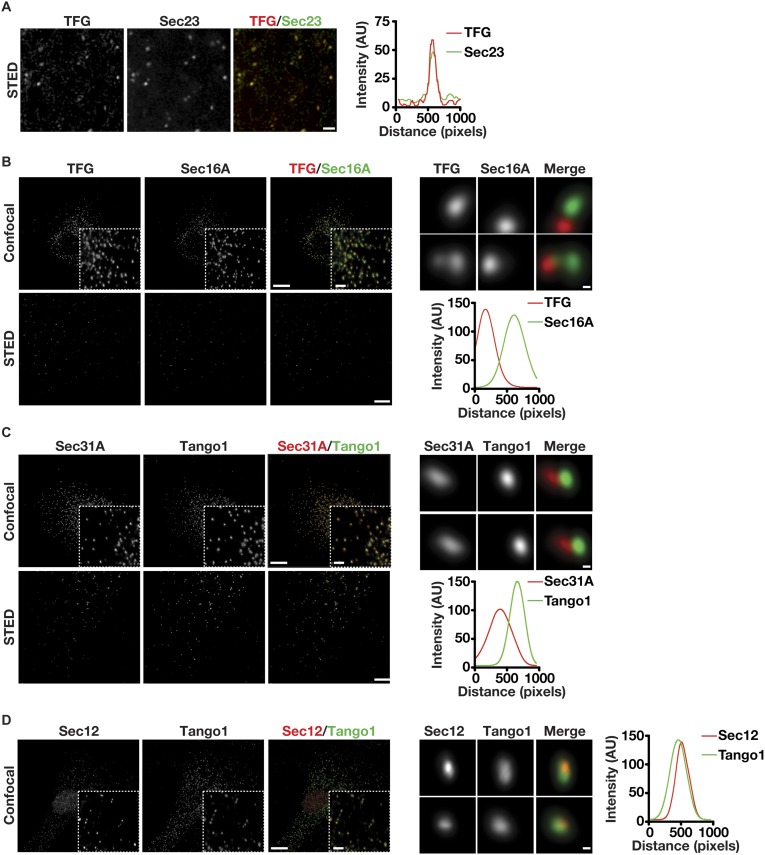
Based on our studies and previous work from others, Sec23 appears to integrate the actions of multiple regulators of COPII-mediated transport via its ability to associate with numerous factors including Sar1, Sec24, Sec31, Sec16, TRAPPC3, p125A, members of the TANGO1 cargo receptor family, and, now, TFG (10, 14, 36, 39, 54–56). Some of these interactions have been shown to be competitive, suggesting that unique associations with Sec23 occur sequentially as transport carriers form, undergo scission from the ER, and tether to ERGIC membranes (14, 40). We used confocal and stimulated emission depletion (STED) microscopy to define the relative distribution of TFG with several of these factors, based on the availability of validated antibodies. Consistent with our previous work, TFG was juxtaposed to Sec16, a marker of ER subdomains that produce COPII transport carriers, and colocalized with Sec23, Sec24, and Sec31, coat proteins on the carriers that accumulate at the ER/ERGIC interface under steady-state conditions (Fig. 5 A and B and Fig. S5 A and B). Notably, based on the analysis of numerous points of colocalization, TFG exhibited more extensive overlap with inner COPII subunits than with outer COPII subunits (Fig. 5 A and B and Fig. S5A). Tango1 colocalized with the Sar1 exchange factor Sec12 on ER subdomains and was juxtaposed to TFG, consistent with the idea that Tango1 associates with COPII carriers as they initially form but fails to be incorporated into the carriers when they leave the ER (Fig. 5C and Fig. S5 C and D). Together, our data suggest that TFG binds to COPII transport carriers subsequent to their scission from ER subdomains.
Fig. 5.
TFG and the inner COPII subunit Sec24A exhibit overlapping distributions. Control RPE1 cells were fixed and stained with antibodies directed against TFG and Sec24A (A), Sec31A (B), or Tango1 (C) and were imaged using confocal and STED microscopy. [Scale bars, 10 μm (confocal) and 4 μm (STED).] Higher-magnification views (confocal) are shown in the lower right portion of each panel in the upper rows. (Inset scale bars, 2 μm.) Higher-magnification views (STED) are also shown for two sites harboring TFG (Right), and a representative linescan measurement is shown to highlight the relative distributions of labeled proteins. (Scale bars, 200 nm.) Images shown are representative of at least 30 individual cells analyzed for each condition.
Fig. S5.
Organization of the early secretory pathway components TFG, Sec16A, COPII subunits, and Tango1. (A, Left) Control C. elegans embryos were fixed and stained using antibodies directed against Sec23 and TFG and were imaged using STED microscopy (n = 6 embryos). (Scale bar, 1 μm.) (Right) A representative linescan measurement is shown to highlight the relative distributions of TFG and Sec23. (B–D, Left) Control RPE1 cells were fixed and stained with antibodies directed against TFG and Sec16A (B), Sec31A and Tango1 (C), or Sec12 and Tango1 (D) and were imaged using confocal and STED microscopy. (Scale bars, 10 μm for confocal imaging; 4 μm for STED microscopy.) Higher-magnification views (confocal) are shown in the lower right portion in the top row of each panel. (Inset scale bar, 2 μm.) (Right) Higher-magnification views (STED) are also shown for sites labeled with the specified antibodies, and a representative linescan measurement is shown to highlight the relative distributions of labeled proteins. (Scale bars, 200 nm.) Images shown are representative of at least 30 individual cells analyzed for each condition.
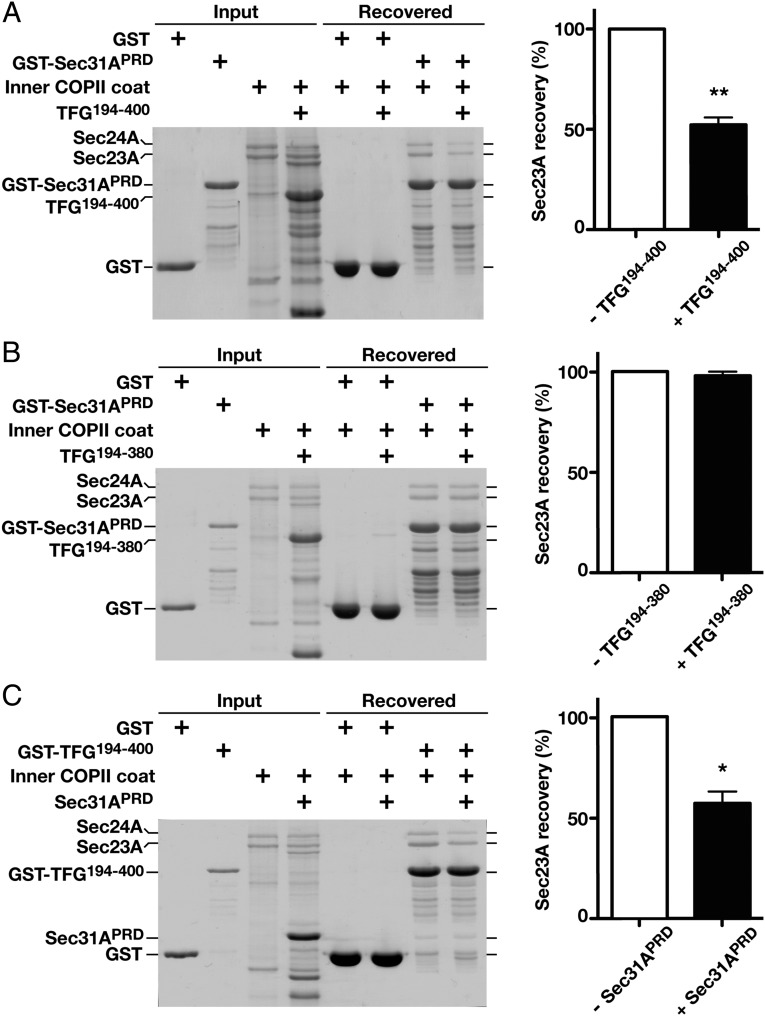
Both Sec31 and TANGO1/cTAGE5 exploit PRDs to associate with a common motif on Sec23 (40). Given the presence of a PRD in TFG, we investigated the possibility that it interacts with Sec23 in a manner that would compete with Sec31 and/or TANGO1. We first purified a recombinant form of Sec31 fused to GST, which included its PRD and a neighboring fragment previously shown to stimulate Sec23-mediated GTP hydrolysis on Sar1 (29), and immobilized it on glutathione agarose. In contrast to GST alone, Sec23-Sec24 heterodimers were retained on beads harboring GST-Sec31 (Fig. 6A). We then repeated the assay in the presence of either a twofold or fivefold molar excess of the soluble recombinant TFG C terminus and found that Sec23-Sec24 retention by GST-Sec31 was significantly reduced in both cases (Fig. 6A and Fig. S6A). These data suggest that TFG disrupts the association between the inner and outer layers of the COPII coat. In contrast, a truncated form of TFG lacking the final 20 residues implicated in Sec23 binding failed to compete with Sec31 (Fig. 6B). In parallel, we also tested the ability of the TFG C terminus to compete with the Tango1 PRD for Sec23 binding. Again, we found that TFG could successfully outcompete Tango1 in retaining Sec23-Sec24 heterodimers, consistent with current models indicating that Tango1 is efficiently removed from COPII transport carriers upon their scission from the ER (Fig. S6B).
Fig. 6.
TFG outcompetes Sec31A for Sec23A binding. (A, Left) Immobilized GST-Sec31A (PRD) was incubated with recombinant Sar1B-Sec23A-Sec24A in the presence or absence of the C terminus of human TFG (amino acids 194–400; 1:1:5 ratio), eluted using sample buffer, and separated by SDS/PAGE followed by Coomassie staining. (Right) The amount of Sec23A recovered in each case is quantified (n = 3) relative to GST-Sec31A. (B, Left) Immobilized GST-Sec31A (PRD) was incubated with recombinant Sar1B-Sec23A-Sec24A in the presence or absence of a truncated form of the TFG C terminus (amino acids 194–380; 1:1:5 ratio), eluted using sample buffer, and separated by SDS/PAGE followed by Coomassie staining. (Right) The amount of Sec23A recovered in each case is quantified (n = 3), relative to GST-Sec31A). (C, Left) Immobilized GST-TFG (amino acids 194–400) was incubated with recombinant Sar1B-Sec23A-Sec24A in the presence or absence of Sec31A (PRD; 1:1:5 ratio), eluted using sample buffer, and separated by SDS/PAGE followed by Coomassie staining. (Right) The amount of Sec23A recovered in each case is quantified (n = 3) relative to GST-TFG. In all cases, error bars represent mean ± SEM. **P < 0.01; *P < 0.05, calculated using a paired t test.
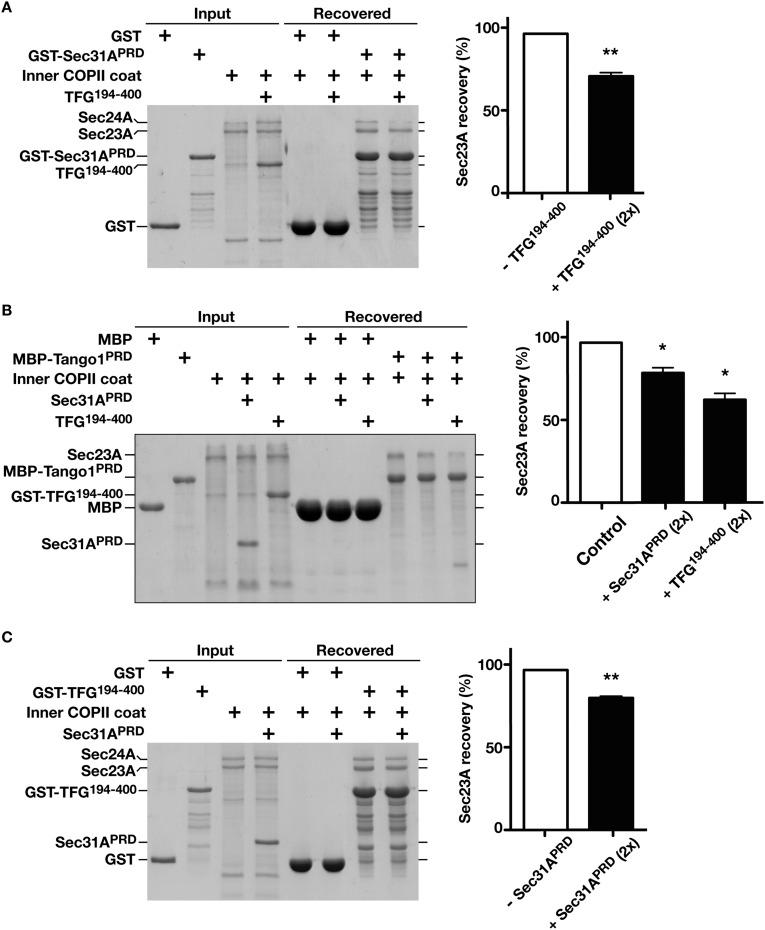
Fig. S6.
TFG competes with Sec31A and Tango1 for Sec23 association. (A, Left) Immobilized GST-Sec31A (PRD) was incubated with recombinant Sar1B-Sec23A-Sec24A in a 1:1:2 ratio in the presence or absence of the C terminus of human TFG (amino acids 194–400), eluted using sample buffer, and separated by SDS/PAGE followed by Coomassie staining. (Right) The amount of Sec23A recovered in each case is quantified (n = 3) relative to GST-Sec31A. (B, Left) Immobilized MBP-Tango1 (amino acids 1,800–1,907) was incubated with recombinant Sar1B-Sec23A-Sec24A in the presence or absence of Sec31A (PRD) (1:1:2 ratio) or TFG (amino acids 194–400) (1:1:2 ratio), eluted using sample buffer, and separated by SDS/PAGE followed by Coomassie staining. (Right) The amount of Sec23A recovered in each case is quantified (n = 3) relative to MBP-Tango1. (C, Left) Immobilized GST-TFG (amino acids 194–400) was incubated with recombinant Sar1B-Sec23A-Sec24A in the presence or absence of Sec31A (PRD) (1:1:2 ratio), eluted using sample buffer, and separated by SDS/PAGE followed by Coomassie staining. (Right) The amount of Sec23A recovered in each case is quantified (n = 3) relative to GST-TFG. In all cases, error bars represent the mean ± SEM. **P < 0.01; *P < 0.05, calculated using a paired t test.
To determine the relative affinities of TFG and Sec31 for Sec23, we performed a reciprocal competition assay in which a GST fusion to the TFG C terminus was immobilized on glutathione agarose. Sec23-Sec24 heterodimers bound to resin containing GST-TFG but not to GST alone (Fig. 6C). The addition of a twofold and fivefold molar excess of soluble Sec31 resulted in decreased Sec23-Sec24 retention on the resin, suggesting that Sec31 can similarly compete with TFG for Sec23 binding (Fig. 6C and Fig. S6C). However, based on the high concentration of TFG that accumulates at the ER/ERGIC interface (44, 45) combined with its ability to form octameric complexes (44) with high avidity for Sec23, our findings are consistent with a model in which TFG actively facilitates dissociation of the outer COPII coat.
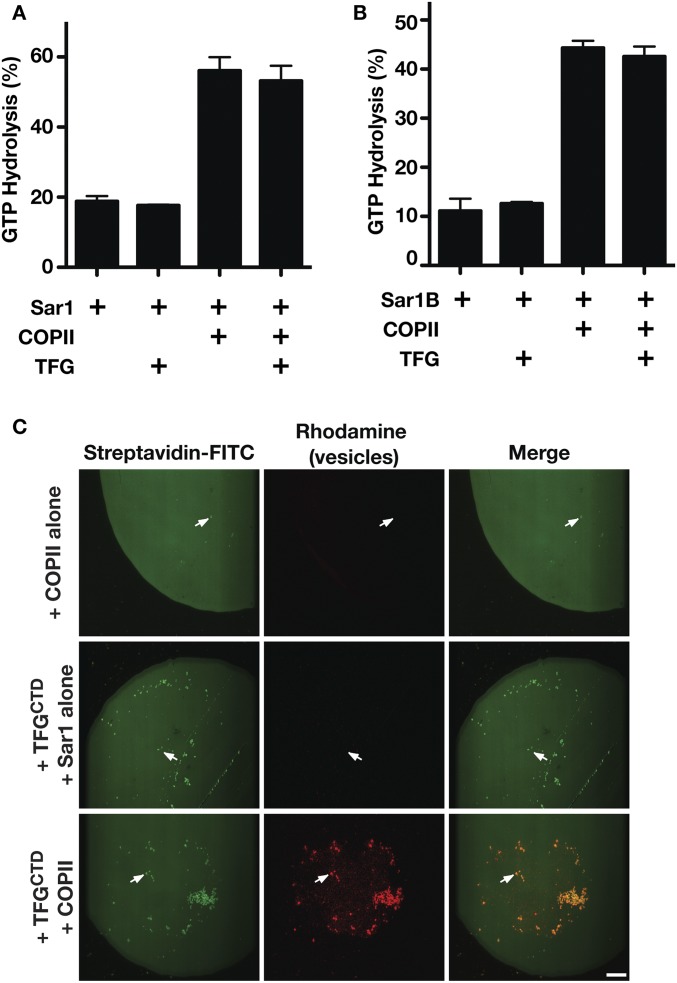
Sec31 was previously demonstrated to stimulate Sec23-mediated GTP hydrolysis on Sar1 (27). Since TFG and Sec31 exhibit an overlapping binding site on Sec23, we examined whether TFG also plays a role in modulating Sec23 activity, which may promote inner coat disassembly. We used two distinct assays to monitor GTP hydrolysis on Sar1 following the addition of Sec23-Sec24 in the presence or absence of TFG. In both cases, TFG failed to modulate Sec23 GAP activity (Fig. S7A). These data suggest that TFG is unlikely to facilitate inner COPII coat disassembly at the ER/ERGIC interface.
Fig. S7.
The TFG C terminus does not promote Sec23-mediated GAP activity on Sar1 but is capable of tethering COPII-coated liposomes. (A) C. elegans TFG (amino acids 196–486) was incubated with Sar1 loaded with GTP in the presence or absence of Sec23-Sec24, and the extent of GTP hydrolysis was measured (n = 3). (B) Human TFG (amino acids 194–400) was incubated with Sar1B loaded with GTP in the presence or absence of Sec23A-Sec24A, and the extent of GTP hydrolysis was measured (n = 3). (C) Microcontact printing was used to generate biotinylated surfaces to which FITC-streptavidin was bound. Incubations in the presence or absence of the C. elegans TFG C terminus (amino acids 196–486) fused to the streptavidin-binding peptide and liposomes containing rhodamine-PE that were coated either with Sar1GTP and Sec23 (COPII) or with Sar1GTP alone were conducted. Surfaces were inverted onto glass coverslips and imaged using confocal microscopy (n = 3). Arrows highlight streptavidin-labeled structures, which only localize with rhodamine-labeled vesicles in the presence of TFG and COPII. (Scale bar, 25 μm.)
The TFG C Terminus Is Sufficient to Capture COPII-Coated Carriers.
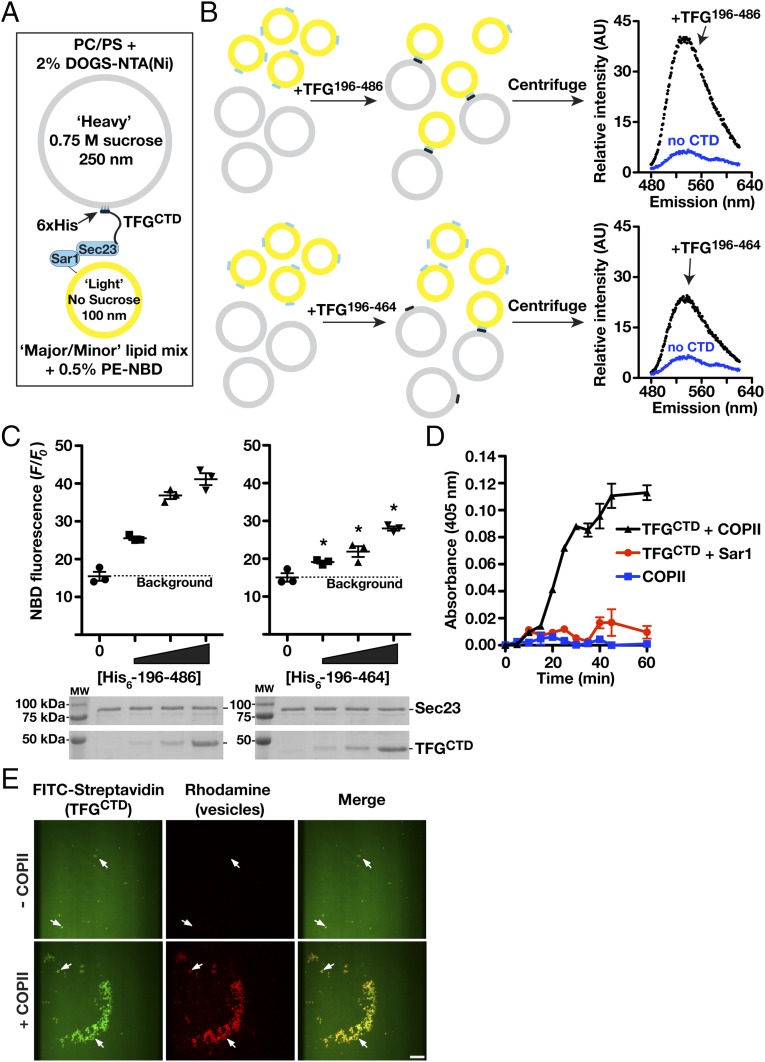
In the absence of TFG, transport carriers harboring both the inner and outer layers of the COPII coat accumulate throughout cells (also see Fig. 1) (44). These data suggest that TFG functions both to promote outer coat dissociation and to retain COPII carriers at the ER/ERGIC interface. To directly test whether TFG is capable of capturing COPII-coated vesicles, we developed an in vitro assay using two types of synthetic liposomes (Fig. 7A) (57). One set of “heavy” liposomes (∼250 nm in diameter) was generated in the presence of sucrose to enable facile sedimentation upon low-speed centrifugation and contained lipids that bind polyhistidine-tagged proteins (DOGS-NTA-Ni) with high affinity. Another set of “light” liposomes (∼100 nm in diameter) was generated using a mixture of lipids, which enable assembly of the inner COPII coat. A polyhistidine-tagged form of the recombinant TFG C terminus was bound to the heavy liposomes and mixed with COPII-coated light liposomes (Fig. 7B). Upon sedimentation specifically in the presence of TFG, we recovered both types of liposomes, indicating that the TFG C terminus was sufficient to bind inner COPII-coated vesicles (Fig. 7C). Consistent with these findings, the presence of the TFG C terminus on vesicles was sufficient to promote the aggregation of COPII-coated vesicles, as determined by spectrophotometric measurements (Fig. 7D). Importantly, a truncated version of the TFG C terminus lacking the last 20 amino acids necessary for efficient Sec23 binding exhibited a reduced ability to mediate cosedimentation of COPII-coated light liposomes (Fig. 7 B and C).
Fig. 7.
TFG tethers COPII-coated liposomes. (A) Diagram depicting the liposome-tethering assay highlighting the presence of C. elegans TFG (amino acids 196–486, harboring a polyhistidine tag) on heavy liposomes containing DOGS-NTA(Ni) (gray) and COPII components (C. elegans Sar1GTP and Sec23) bound to light liposomes (yellow). (B, Left) Light liposomes harboring COPII (Sar1GTP, 2 μM; Sec23, 250 nM) were mixed with heavy liposomes (1:1 ratio, 1 mM total lipids), bound to varying amounts (0, 10, 30, and 100 nM) of full-length TFG C terminus (amino acids 196–486) or truncated TFG C terminus (amino acids 196–464), and centrifuged. (Right) The liposome pellet was resuspended with buffer containing 0.2% Triton X-100, and nitrobenzoxadiazole (NBD) fluorescence was quantified to determine the degree of tethering. AU, arbitrary units. (C, Upper) Quantification of NBD fluorescence (F) obtained from sedimented liposomes in the presence of different concentrations of the full-length or truncated TFG C terminus (graphical plots; n = 3). *P < 0.05 (compared with control), calculated using a paired t test. (Lower) The presence of Sec23 (10% total) and TFG (100% total) with sedimented liposomes was confirmed by SDS/PAGE analysis followed by Coomassie staining. (D) Liposome aggregation was measured (absorbance: 405 nm) over time. Light liposomes harboring Sar1GTP alone or Sar1GTP with Sec23 (COPII) were incubated with heavy liposomes in the presence or absence of TFG (30 nM). Reactions (n = 3) were maintained at 4 °C, and samples were taken at the indicated time points. (E) Microcontact printing was used to generate biotinylated surfaces to which FITC-streptavidin was bound. The C. elegans TFG C terminus (amino acids 196–486) fused to the streptavidin-binding peptide was incubated with these regions and subsequently exposed to liposomes containing rhodamine-PE, which were coated with either Sar1GTP and Sec23 or Sar1GTP alone. Surfaces were inverted onto glass coverslips and imaged using confocal microscopy (n = 3). Arrows indicate TFG clusters that recruit COPII-coated liposomes. (Scale bar, 25 μm.)
To confirm a role for TFG in capturing COPII transport carriers, we generated clusters of its C terminus on glass coverslips using a microcontact printing approach (58, 59). The TFG-enriched domains were exposed to synthetic liposomes containing rhodamine-labeled lipids and were imaged using confocal microscopy. Only when the synthetic liposomes were precoated with the inner COPII proteins were they retained on the artificial TFG microdomains (Fig. 7E and Fig. S7B). Together, our data support a model in which TFG facilitates outer COPII coat dissociation but local retention of inner COPII-coated transport carriers at the ER/ERGIC interface.
Discussion
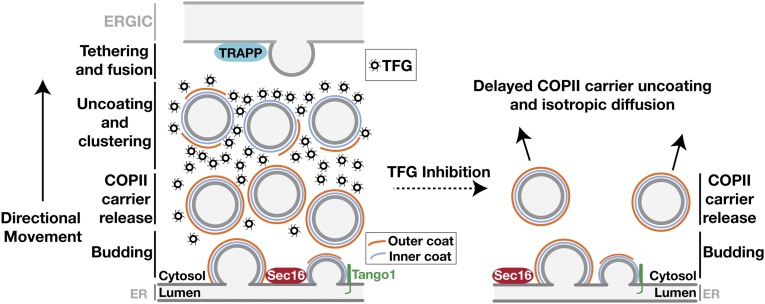
In general, current models depicting the ER/ERGIC interface fail to account for the spatial and temporal relationships between COPII coat disassembly and the local retention of transport carriers before target membrane fusion (14, 30, 34, 47, 60). Based largely on work conducted in yeast, which lack ERGIC membranes, a stepwise transport mechanism has been proposed that relies on sequential interactions between the inner COPII coat protein Sec23 and three distinct binding partners: Sar1, TRAPPC3, and casein kinase 1δ (14, 61). However, precisely how transport carriers traverse the 300- to 500-nm distance between the ER and ERGIC has remained unclear (43). Our data support a concerted tethering model in which TFG promotes outer COPII coat dissociation after transport carriers leave the ER. Simultaneously, TFG functions to retain inner COPII-coated carriers at the ER/ERGIC interface. By concentrating transport carriers with only partial coats, TFG generates a platform that facilitates TRAPP complex-mediated homotypic fusion and nascent ERGIC formation (Fig. 8).
Fig. 8.
Model depicting the organization of the early secretory pathway in the presence and absence of TFG. Initiation of COPII carrier formation occurs at ER subdomains harboring Sec16A and Tango1-like cargo receptors, which interact directly with Sec23. Full assembly of the outer COPII cage interferes with these interactions and facilitates the release of the transport carrier into the ER/ERGIC interface, where TFG competes with Sec31 for Sec23 binding. The outer coat is ultimately displaced by TFG, which clusters inner COPII-coated transport carriers until they tether and fuse with adjacent ERGIC membranes. In the absence of TFG, transport carriers harboring both the inner and outer layers of the COPII coat continue to form but diffuse away from the ER/ERGIC interface, slowing secretory transport of conventional COPII-dependent cargoes.
The minimal COPII machinery required to generate cargo-laden, ER-derived transport carriers has been defined for several years, but regulators of this pathway continue to emerge, several of which are expressed only in metazoan organisms, which have evolved more complex systems to control cargo efflux in response to environmental and developmental cues (62, 63). Biochemical and genetic screens in human cells, Drosophila, and C. elegans have identified several factors that play important roles at the ER in cargo selection, exit site organization, and modulating secretory capacity, but components that facilitate movement of COPII-coated carriers between the ER and ERGIC have been more challenging to define (55, 64–67). Microtubules and other cytoskeletal elements are absent within this region, but, nevertheless, most COPII-coated carriers cluster within the ER/ERGIC interface, suggesting the presence of an underlying meshwork that links the organelles to form an integrated secretory unit (44, 68). In Drosophila, Tango1 plays an integral role in organizing the early secretory pathway by forming micron-sized ring structures that encircle COPII-coated carriers and the cis-Golgi (55). Upon Tango1 inhibition, exit sites on the ER become uncoupled from the Golgi, and both conventional and unconventional (large cargo) secretion are negatively impacted. Depletion of TFG in C. elegans similarly blocks general secretion and uncouples exit sites on the ER from ERGIC membranes in human cells (44, 45). Notably, whereas mammals express both Tango1 and TFG, Drosophila lack a TFG ortholog, and C. elegans lack Tango1-like receptors, suggesting that these components evolved separately to regulate early secretory pathway organization in unique ways (69). Consistent with this idea, Tango1 and TFG are spatially restricted from one another in human cells (Fig. 5C). Current evidence suggests that Tango1 acts early to organize exit sites on the ER and enable procollagen export (40, 70, 71), whereas TFG functions to initiate dissociation of the outer COPII coat and restrict diffusion of inner COPII-coated carriers at the ER/ERGIC interface.
A recent study suggested that TFG acts specifically to regulate the transport of large cargoes (47). However, our findings are inconsistent with this idea. In particular, it is difficult to reconcile how the association between TFG and Sec23 would be restricted to large transport carriers. Additionally, early embryo lethality observed upon TFG knockout in rodents is inconsistent with the more subtle impacts of deleting receptors known to regulate the secretion of large cargoes (53, 55). In contrast to our studies, the prior report failed to use immunofluorescence to determine the degree to which TFG was depleted in cells, raising the distinct possibility that TFG was only partially reduced (47). In this scenario, the specific impact on collagen secretion may have been indirectly caused by disruption of exit site organization as a result of long-term partial depletion of TFG, which could disproportionately affect the export of large cargoes over others.
The ability of TFG to tether inner COPII-coated carriers is highly reminiscent of the functions proposed for synapsin and synuclein in synaptic vesicle clustering (72–76). All three proteins self-associate and harbor disordered regions composed largely of proline, glutamine, glycine, and serine, which form relatively weak but multivalent interactions with vesicle-associated proteins (77–79). In particular, TFG assembles into octameric ring structures (44), with intrinsically disordered C-terminal domains that interact directly with Sec23 on COPII carriers. By tethering neighboring transport carriers, TFG may facilitate a liquid-phase separation at the ER/ERGIC interface (80). Consistent with this idea, blocking COPII transport disrupts the ability of TFG to assemble at exit sites on the ER. Through phase separation, TFG would establish a nonmembranous compartment, which promotes the tight clustering of transport carriers. Synaptic vesicles are readily exchanged in and out of liquid-phase clusters, allowing unimpeded fusion with the plasma membrane when appropriately stimulated (81). Similarly, upon inner coat disassembly, COPII transport carriers would escape the TFG liquid phase and fuse homotypically or heterotypically with juxtaposed ERGIC membranes, potentially in a manner dependent on the TRAPP complex (30). Based on previous work, TRAPPC3 and casein kinase 1δ outcompete Sar1 for Sec23 binding, which may promote inner COPII coat disassembly in vivo and expose SNAREs necessary for membrane fusion (14).
Materials and Methods
All rodent experiments were conducted in the Sprague–Dawley background (Envigo) and were conducted after approval by the University of Wisconsin–Madison Institutional Animal Care and Use Committee.
Recombinant Protein Expression and Purification and Yeast Two-Hybrid Studies.
Recombinant proteins were purified following expression in BL21 (E. coli T1R, Sigma) (82) or baculovirus-mediated expression in Sf9 cells (83). Protein purification was conducted using glutathione agarose beads (for GST fusions) or nickel-affinity resin (for polyhistidine-tagged proteins), followed by ion exchange chromatography and gel filtration. For Sar1 isoforms, the GST tag was removed using PreScission protease in the presence of 500 μM GDP overnight to facilitate nucleotide exchange. Expression and purification of other COPII proteins and TFG were carried out as described previously (44).
For yeast two-hybrid studies, all bait and prey constructs were sequence verified. Yeast transformed with the indicated bait and target plasmids were grown in medium lacking uracil and leucine overnight at 30 °C with shaking, diluted to an OD600 of 0.25 in medium lacking amino acids, and spot-plated onto selective medium. Plates were incubated at 30 °C for 48 h and then were imaged.
Synthetic Liposome Generation, Immunoblotting, and Immunoprecipitations.
Liposomes were generated as described previously (84); detailed compositions can be found in SI Materials and Methods. Immunoblotting of extracts was performed as described previously (85) using antibodies described in SI Materials and Methods. For immunoprecipitation studies, antibodies were covalently linked to protein A beads (Bio-Rad). Rat liver cytosol (25 mg protein; ∼1 mL) or cell-line extracts were added to the beads and incubated at 4 °C for 1 h. The beads were washed, and bound proteins were eluted in 100 mM glycine (pH 2.6). Following TCA precipitation, samples were examined by solution mass spectrometry or were separated by SDS/PAGE for immunoblot analysis (45).
CRISPR-Mediated Genome Editing, TALEN Expression System, and siRNA-Mediated Depletion.
A guide RNA (gRNA) (a gift from David Ginsburg, University of Michigan, Ann Arbor, MI) directed to exon 1 of Sec23B together with Cas9-GFP were transfected into RPE1 cells using FuGENE HD (Promega). Cells were single-cell sorted using FACS, and clones were examined by immunoblot analysis. Sanger sequencing was used to confirm the presence of mutations that led to frameshifts and truncation of Sec23B. To generate clonal doxycycline-inducible cell lines to express TFG transgenes, we used a TALEN-mediated targeting system, which incorporated the transgenes at the AAVS1 locus (86). A puromycin-resistant donor cassette encoding various forms of TFG followed by an IRES and a cDNA encoding RFP was used in all cases. Depletion studies were carried out using siRNAs described in SI Materials and Methods.
Immunofluorescence, Imaging Studies, and Antibodies.
For all immunofluorescence experiments, RPE1 cells were grown as described previously (44) and were processed as detailed in SI Materials and Methods. Confocal imaging was conducted on a Nikon Eclipse TiE swept-field confocal microscope using a 60× objective and a CoolSNAP HQ2 CCD camera. Acquisition parameters were controlled by Nikon Elements software, and image analysis was conducted using Imaris software. Immunofluorescence of fixed cells was performed as described previously (87). Briefly, 20–30 Z sections at 0.2-μm steps were acquired (depending on sample thickness). Colocalization and spot intensity analysis were performed using Imaris software. Specified areas of cells located distally from the perinuclear Golgi were selected, and thresholds for size and intensity were set for each channel respectively, as described in SI Materials and Methods. STED microscopy was preformed on a Leica TCS SP8 3× superresolution microscope. Images were collected using a 775-nm depletion laser for all dyes used (Alexa 555, Alexa 594, and Alexa 660) and were processed using Huygens deconvolution software. The FWHM of the STED point-spread function was determined to be 90 nm, based on the imaging of diffraction-limited fluorescent beads.
For all doxycycline-inducible expression studies, wild-type or the indicated TALEN-edited RPE1 cells were plated on glass coverslips and treated with doxycycline (3 ng/mL) and either control or TFG 3′ UTR siRNA (10 pmol per well). For all Sec23A-knockdown experiments, either wild-type or Sec23B-knockout cell lines were plated onto glass coverslips at 7.5% confluency and were treated with the appropriate Sec23 siRNAs (10 pmol total per well) for 96 h. C. elegans strains used in this study were derived from the Bristol strain N2. With the exception of the strain expressing HMR-1::GFP from its endogenous locus (88), all transgenic animals were created by biolistic transformation (89), and the transgenes used were under the control of PIE1 regulatory sequences to enable germline expression (90, 91).
GST Pull-Down Experiments, COPII Budding Reactions, and Liposome-Tethering Assays.
Fifty micrograms of the indicated GST-fusion protein was incubated with 50 μL of fresh glutathione resin in 50 mM Hepes (pH 7.6), 100 mM NaCl, 1 mM MgCl2, and 1 mM DTT for immobilization. The resin was washed and incubated with 1.5 mg of protein from cleared rat liver cytosol, prepared as described previously (92), at 4 °C for 1 h. After extensive washing, bound proteins were eluted in 1× PBS (pH 8.0) and 30 mM reduced glutathione to a final volume of 900 μL and were TCA precipitated. The precipitate was resuspended in sample buffer and separated by SDS/PAGE. All GST competition experiments were conducted as described previously (40) and are detailed in SI Materials and Methods.
In vitro budding reactions to analyze the formation of large transport carriers harboring procollagen were conducted as described previously (49). Light liposomes for tethering reactions were extruded through a 0.05-μm filter and were clarified by centrifugation at 16,163 × g for 15 min. Heavy liposomes were reconstituted with Sar1 buffer supplemented with 0.75 M sucrose and were extruded through a 0.4-μm filter. Liposome-tethering assays and liposome-aggregation experiments (determined by measuring absorbance at 405 nm) were based on previous work (57, 93) and are described in SI Materials and Methods.
SI Materials and Methods
Rodent Studies, C. elegans Depletions, and siRNA-Mediated Depletion Studies.
All rodent experiments were conducted in the Sprague–Dawley background (Envigo) and were conducted after approval by the University of Wisconsin–Madison Institutional Animal Care and Use Committee. To generate rats harboring a frameshift mutation in TFG, purified Cas9 and an in vitro-transcribed gRNA (5′-ATTATGAATGGGAATTCGC-3′) were coinjected into embryos, which were implanted into foster mothers. Viable pups were genotyped by Sanger sequencing, and founders harboring appropriate mutations were outcrossed to control animals (Envigo) before mating between heterozygous animals. For timed pregnancies, pairings were arranged for 6 h, and females were subsequently checked for vaginal plugs to confirm mating.
For RNAi experiments in C. elegans, early L4-stage hermaphrodites were fed dsRNA targeting TFG for 48 h at 20 °C. For imaging, live transgenic animals were immobilized as described previously (91). All images shown are representative of experiments conducted more than three times independently.
For siRNA-mediated depletions, the following siRNAs were transfected by using Lipofectamine RNAiMAX (Invitrogen): 3′ UTR TFG: 5′-CCAAAAGACUCCAGUACUA-3′; exon 1 TFG: 5′-ACUUCUGAGUAAUGAUGAA-3′; Sec23A-1: 5′-GUUAUGCUGGUAUAUCUGA-3′; Sec23A-2: 5′-GCAUAAUGCUCCAAUUCCU-3′; Sec23B-1; 5′-CACGUUACAUCAACACGGA-3′; Sec23B-2: 5′-CACUAUGAGAUGCUUGCUA-3′. Cells were analyzed at the time points indicated after transfection of siRNAs, and depletions were confirmed by immunoblotting cell extracts and/or immunostaining.
COPII Budding Reactions.
Budding reactions were conducted as described previously (10). To make semi-intact cells, two 10-cm dishes of RPE1 cells at ∼80% confluency were collected by trypsinization, centrifuged in complete medium at 600 rcf for 5 min, and permeabilized. For the conventional budding reaction, wild-type Sar1A (1 μg) was mixed with Sec23A/Sec24D (1 μg), Sec13/Sec31A (1 μg), and GTP (300 μM) in B-88 buffer (100 μL final volume). Semi-intact cells (OD600 = 0.2) were added, and the reaction was incubated at 30 °C for 1 h. After incubation, the reaction was centrifuged at 14,000 rcf for 25 min to sediment the semi-intact cells, and 75 μL of the supernatant was transferred to an ultracentrifuge tube and centrifuged at 150,000 rcf for 20 min. The supernatant was removed, and COPII vesicles were collected into sample buffer. Samples were separated by SDS/PAGE, and the indicated proteins were detected using immunoblot analysis.
Immunofluorescence and Immunoblotting Studies.
All images shown are representative of experiments conducted at least three times independently and include the analysis of at least 10 fields of cells for each condition unless otherwise noted. Cells were grown on glass coverslips before fixation using paraformaldehyde (4%) or cold methanol (20 min at −20 °C). Cells were permeabilized using PBS + 0.5% Triton X-100 for 10 min and blocked using antibody dilution buffer [AbDil: 50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 3% BSA] for 1 h at 25 °C. Coverslips were incubated with the indicated primary antibody diluted in AbDil overnight at 4 °C. Slides were washed and subsequently incubated with the indicated secondary antibody for 45 min at room temperature in the dark before mounting. Staining using commercially available antibodies directed against Sec16A (Bethyl Laboratories), Sec24a (Santa Cruz), ERGIC-53 (Enzo Life Sciences), and Tango1 (Sigma) was preceded by fixation using cold methanol, as these antibodies were largely incompatible with paraformaldehyde fixation.
Immunoblotting was conducted using antibodies directed against Sec23 (Thermo Scientific), TFG (LifeSpan BioSciences), Sec13 (Proteintech), p125a (Novus Biologicals), Rab1A (Cell Signaling Technology), β-actin (Sigma-Aldrich), ERGIC-53 (Santa Cruz), and Sar1, Sec23B, and Sec22B (gifts of the R.S. laboratory).
Confocal and STED Image Analysis.
The lower limits of intensity thresholds were defined by staining with secondary antibodies alone. Structure size threshold was set to 0.7 μm for all image quantifications, unless otherwise noted below. To quantify whether two markers were juxtaposed or overlapped, distance thresholds between 0.5 and 0.6 μm were used. The number of colocalized spots was divided by the total spots counted [colocalized/(colocalized + nonlocalized)] to determine the percentage of colocalization. For colocalization studies between Sec16A and Sec24A (STED imaging), the minimum diameter of structures was set at 0.25 μm, with intensities of at least 5,000 arbitrary units (AU) (Sec16A) and 2,500 AU (Sec24A). Structures were considered juxtaposed if their centroids were less than 0.3 μm apart. For colocalization studies between Sec31A and TFG (confocal imaging), the minimum diameter of structures was set at 0.7 μm, with intensities of at least 225 AU (Sec31A) and 265 AU (TFG). Structures were considered colocalized if their centroids were less than 0.5 μm apart. For colocalization studies between Sec16 and TFG (confocal imaging), the minimum diameter of structures was set at 0.7 μm, with intensities of at least 275 AU (Sec16A) and 265 AU (TFG). Structures were considered juxtaposed if they were less than 0.5 μm apart (from their centroids).
GST Competition Experiments.
A saturating amount of GST or an indicated GST fusion protein (>100 μg) was incubated with 8 μL of glutathione resin (16 μL slurry) and mixed for 1 h at 4 °C in 20 mM Hepes (pH 7.6), 150 mM KOAc, 1 mM MgCl2, 1 mM DTT, and 0.2% Triton X-100. Saturated resin was washed twice with 1 mL of wash buffer to remove unbound protein. Competition experiments were carried out in 600-μL reactions (fivefold molar excess conditions) or 300-μL reactions (twofold molar excess conditions). Reactions were mixed at 4 °C for 1 h, after which the resin was washed extensively to remove unbound protein. Sample buffer was used to elute the resin, and eluted proteins were separated by SDS/PAGE.
Size-Exclusion Chromatography and Multiangle Light Scattering.
Size-exclusion chromatography was conducted using a S200 gel filtration column. The Stokes radius of each protein or protein complex was calculated from its elution volume based on the elution profiles of characterized standards (45). For mass determination studies, samples (500 μL) were separated on a Wyatt WTC-030S5 column coupled to a Wyatt miniDAWN TREOS three-angle light-scattering detector (82).
Synthetic Liposome Characterization and Composition.
Liposomes were analyzed by dynamic light scattering (DLS) using a Wyatt DynaPro NanoStar DLS instrument to determine their average diameters (84). Phospholipids (Avanti Polar Lipids) in chloroform were mixed to generate lipid mixtures, including the major/minor mix (in lipid, mol%): 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 50; 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 21; 1,2-Dioleoyl-sn-glycero-3-phosphoserine (DOPS), 8; 1,2-Dioleoyl-sn-glycero-3-phosphate (DOPA), 5; phosphatidylinositol (PI), 9; phosphatidylinositol 4-phosphate (PI4P), 2.2; phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], 0.8; diacylglycerol (DAG), 2; 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-PE), 0.5; and supplemented with 20% cholesterol, which was shown previously to facilitate maximal binding of Sar1 isoforms (11). Heavy liposomes used in tethering reactions were generated using a phospholipid mixture containing (in lipid, mol%): 1,2- DOPC, 88; DOPS, 10; 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (DGS-NTA), 2. Lipids were dried, resuspended in 50 mM Hepes (pH 7.6), 100 mM NaCl, 1 mM MgCl2, and 1 mM DTT, and subjected to extrusion through a nitrocellulose filter (Whatman) of a desired pore size.
Liposome-Tethering Assay.
Sar1GDP (2 μM), GTP (300 μM), and Sec23 (0.25 μM), where indicated, were mixed at 4 °C in Sar1 buffer along with the indicated TFG C-terminal domain fragment. Light and heavy liposomes (0.5 mM) were mixed into the reaction in a 1:1 ratio to initiate tethering. The reaction was carried out at 4 °C for 1 h and then was centrifuged at 4 °C at 14,800 rcf for 15 min to sediment heavy liposomes. After sedimentation, the supernatant was removed, and the recovered liposomes were resuspended in Sar1 buffer supplemented with 0.2% Triton X-100 to a total of 600 μL. Total NBD fluorescence was measured using a spectrofluorometer (excitation: 460 nm; emission scan: 480–620 nm), and emission was plotted to calculate the area under the curve (AUC).
For liposome-aggregation assays, reactions were maintained on ice, and 4-μL samples were taken at the indicated time points for measurement using a nanoVette microliter cell (Beckman Coulter). Biotin array slides were generated as described previously (58, 59). FITC-labeled streptavidin (100 μg) (Life Technologies) was incubated with each biotin array slide for 1 h at room temperature. The slide was washed five times with 1 mL Sar1 buffer. Then 100 μg of TFG (resides 196–486, fused to an N-terminal streptavidin-binding peptide) was incubated with the biotin slide for 1 h. The slide was again washed five times with 1 mL Sar1 buffer and submerged in 200 μL of Sar1 buffer. Fifty microliters of the reaction (final concentrations: liposomes, 500 μM; Sar1GTP, 2 μM; Sec23, 250 nM where indicated) were added to the slide to initiate tethering. The reaction was allowed to incubate for 1 h at room temperature. Slides were washed with 1 mL buffer and then were inverted on a 32 × 32 mm glass coverslip and imaged.
GTPase Assay.
The Promega GTPase assay (GTPase-Glo, V7681) was used to measure GTP hydrolysis as described previously (84). Briefly, the indicated reaction (Sar1GDP, 0.5 μM; Sec23-Sec24, 200 nM; TFG, 1 μM) was mixed with liposomes (major minor mix; 500 μM final concentration) and added to a 96-well plate. The reaction was initiated upon the addition of GTP (5 μM).
Acknowledgments
We thank Alma Seitova and Ashley Hutchinson for technical support in baculovirus expression, Pietro De Camilli for useful discussions, and David Melville for help with technical aspects of the COPII budding reaction. This work was supported in part by NIH Grants GM110567 (to A.A.) and P41 GM103533 (to J.R.Y.), American Cancer Society Grant 123268-RSG-12-139-01-CSM (to A.A.), Brain Research Foundation Grant BRFSG-2015-03 (to A.A.), the University of Wisconsin Carbone Cancer Center Grant P30 CA014520, and the University of Wisconsin Institute for Clinical and Translational Research Grant UL1TR000427. The Structural Genomics Consortium (SGC) is a registered charity (no. 1097737) that receives funds from numerous sources (www.thesgc.org/about/partners).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709120114/-/DCSupplemental.
References
- 1.Hobman TC, Zhao B, Chan H, Farquhar MG. Immunoisolation and characterization of a subdomain of the endoplasmic reticulum that concentrates proteins involved in COPII vesicle biogenesis. Mol Biol Cell. 1998;9:1265–1278. doi: 10.1091/mbc.9.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venditti R, Wilson C, De Matteis MA. Exiting the ER: What we know and what we don’t. Trends Cell Biol. 2014;24:9–18. doi: 10.1016/j.tcb.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appenzeller-Herzog C, Hauri H-P. The ER-Golgi intermediate compartment (ERGIC): In search of its identity and function. J Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Menárguez JA, Geuze HJ, Slot JW, Klumperman J. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 1999;98:81–90. doi: 10.1016/S0092-8674(00)80608-X. [DOI] [PubMed] [Google Scholar]
- 6.Antonny B, Gounon P, Schekman R, Orci L. Self-assembly of minimal COPII cages. EMBO Rep. 2003;4:419–424. doi: 10.1038/sj.embor.embor812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zanetti G, et al. The structure of the COPII transport-vesicle coat assembled on membranes. eLife. 2013;2:e00951. doi: 10.7554/eLife.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aridor M, Bannykh SI, Rowe T, Balch WE. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- 10.Barlowe C, et al. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 11.Matsuoka K, et al. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 12.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 13.Orci L, et al. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 14.Lord C, et al. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473:181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lord C, Ferro-Novick S, Miller EA. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the golgi. Cold Spring Harb Perspect Biol. 2013;5:a013367. doi: 10.1101/cshperspect.a013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowe T, et al. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J Cell Biol. 1996;135:895–911. doi: 10.1083/jcb.135.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato K, Nakano A. Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat Struct Mol Biol. 2005;12:167–174. doi: 10.1038/nsmb893. [DOI] [PubMed] [Google Scholar]
- 18.Kuge O, et al. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodonova SO, et al. VESICULAR TRANSPORT. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science. 2015;349:195–198. doi: 10.1126/science.aab1121. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 22.Tanigawa G, et al. Hydrolysis of bound GTP by ARF protein triggers uncoating of Golgi-derived COP-coated vesicles. J Cell Biol. 1993;123:1365–1371. doi: 10.1083/jcb.123.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zink S, Wenzel D, Wurm CA, Schmitt HD. A link between ER tethering and COP-I vesicle uncoating. Dev Cell. 2009;17:403–416. doi: 10.1016/j.devcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- 25.Stagg SM, et al. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allan BB, et al. Stage-specific assays to study biosynthetic cargo selection and role of SNAREs in export from the endoplasmic reticulum and delivery to the Golgi. Methods. 2000;20:411–416. doi: 10.1006/meth.2000.0954. [DOI] [PubMed] [Google Scholar]
- 27.Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- 28.Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 29.Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai H, et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 31.Koreishi M, Yu S, Oda M, Honjo Y, Satoh A. CK2 phosphorylates Sec31 and regulates ER-To-Golgi trafficking. PLoS One. 2013;8:e54382. doi: 10.1371/journal.pone.0054382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin L, et al. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharpe LJ, Luu W, Brown AJ. Akt phosphorylates Sec24: New clues into the regulation of ER-to-Golgi trafficking. Traffic. 2011;12:19–27. doi: 10.1111/j.1600-0854.2010.01133.x. [DOI] [PubMed] [Google Scholar]
- 34.Yu S, et al. mBet3p is required for homotypic COPII vesicle tethering in mammalian cells. J Cell Biol. 2006;174:359–368. doi: 10.1083/jcb.200603044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao S, et al. Mammalian TRAPPIII complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci Rep. 2017;7:43207. doi: 10.1038/srep43207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Supek F, Madden DT, Hamamoto S, Orci L, Schekman R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J Cell Biol. 2002;158:1029–1038. doi: 10.1083/jcb.200207053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yorimitsu T, Sato K. Insights into structural and regulatory roles of Sec16 in COPII vesicle formation at ER exit sites. Mol Biol Cell. 2012;23:2930–2942. doi: 10.1091/mbc.E12-05-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kung LF, et al. Sec24p and Sec16p cooperate to regulate the GTP cycle of the COPII coat. EMBO J. 2012;31:1014–1027. doi: 10.1038/emboj.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito K, et al. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 2009;136:891–902. doi: 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 40.Ma W, Goldberg J. TANGO1/cTAGE5 receptor as a polyvalent template for assembly of large COPII coats. Proc Natl Acad Sci USA. 2016;113:10061–10066. doi: 10.1073/pnas.1605916113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salama NR, Chuang JS, Schekman RW. Sec31 encodes an essential component of the COPII coat required for transport vesicle budding from the endoplasmic reticulum. Mol Biol Cell. 1997;8:205–217. doi: 10.1091/mbc.8.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeuschner D, et al. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat Cell Biol. 2006;8:377–383. doi: 10.1038/ncb1371. [DOI] [PubMed] [Google Scholar]
- 44.Johnson A, et al. TFG clusters COPII-coated transport carriers and promotes early secretory pathway organization. EMBO J. 2015;34:811–827. doi: 10.15252/embj.201489032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witte K, et al. TFG-1 function in protein secretion and oncogenesis. Nat Cell Biol. 2011;13:550–558. doi: 10.1038/ncb2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beetz C, et al. Inhibition of TFG function causes hereditary axon degeneration by impairing endoplasmic reticulum structure. Proc Natl Acad Sci USA. 2013;110:5091–5096. doi: 10.1073/pnas.1217197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCaughey J, et al. TFG promotes organization of transitional ER and efficient collagen secretion. Cell Rep. 2016;15:1648–1659. doi: 10.1016/j.celrep.2016.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindler AJ, Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc Natl Acad Sci USA. 2009;106:17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorur A, et al. COPII-coated membranes function as transport carriers of intracellular procollagen I. J Cell Biol. 2017;216:1745–1759. doi: 10.1083/jcb.201702135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X-W, et al. SEC24A deficiency lowers plasma cholesterol through reduced PCSK9 secretion. eLife. 2013;2:e00444. doi: 10.7554/eLife.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baines AC, Adams EJ, Zhang B, Ginsburg D. Disruption of the Sec24d gene results in early embryonic lethality in the mouse. PLoS One. 2013;8:e61114. doi: 10.1371/journal.pone.0061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson DG, et al. Global defects in collagen secretion in a Mia3/TANGO1 knockout mouse. J Cell Biol. 2011;193:935–951. doi: 10.1083/jcb.201007162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tani K, Mizoguchi T, Iwamatsu A, Hatsuzawa K, Tagaya M. p125 is a novel mammalian Sec23p-interacting protein with structural similarity to phospholipid-modifying proteins. J Biol Chem. 1999;274:20505–20512. doi: 10.1074/jbc.274.29.20505. [DOI] [PubMed] [Google Scholar]
- 55.Liu M, et al. Tango1 spatially organizes ER exit sites to control ER export. J Cell Biol. 2017;216:1035–1049. doi: 10.1083/jcb.201611088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lees JA, et al. Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science. 2017;355:eaah6171. doi: 10.1126/science.aah6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knight GT, Sha J, Ashton RS. Micropatterned, clickable culture substrates enable in situ spatiotemporal control of human PSC-derived neural tissue morphology. Chem Commun (Camb) 2015;51:5238–5241. doi: 10.1039/c4cc08665a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sha J, Lippmann ES, McNulty J, Ma Y, Ashton RS. Sequential nucleophilic substitutions permit orthogonal click functionalization of multicomponent PEG brushes. Biomacromolecules. 2013;14:3294–3303. doi: 10.1021/bm400900r. [DOI] [PubMed] [Google Scholar]
- 60.Hughes H, et al. Organisation of human ER-exit sites: Requirements for the localisation of Sec16 to transitional ER. J Cell Sci. 2009;122:2924–2934. doi: 10.1242/jcs.044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menon S, et al. mBET3 is required for the organization of the TRAPP complexes. Biochem Biophys Res Commun. 2006;350:669–677. doi: 10.1016/j.bbrc.2006.09.096. [DOI] [PubMed] [Google Scholar]
- 62.Saito K, Katada T. Mechanisms for exporting large-sized cargoes from the endoplasmic reticulum. Cell Mol Life Sci. 2015;72:3709–3720. doi: 10.1007/s00018-015-1952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scharaw S, et al. The endosomal transcriptional regulator RNF11 integrates degradation and transport of EGFR. J Cell Biol. 2016;215:543–558. doi: 10.1083/jcb.201601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simpson JC, et al. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat Cell Biol. 2012;14:764–774. doi: 10.1038/ncb2510. [DOI] [PubMed] [Google Scholar]
- 65.Bard F, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 66.Green RA, et al. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wendler F, et al. A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 2010;29:304–314. doi: 10.1038/emboj.2009.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Presley JF, et al. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 69.Glick BS. New insights into protein secretion: TANGO1 runs rings around the COPII coat. J Cell Biol. 2017;216:859–861. doi: 10.1083/jcb.201701142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santos AJM, Raote I, Scarpa M, Brouwers N, Malhotra V. TANGO1 recruits ERGIC membranes to the endoplasmic reticulum for procollagen export. eLife. 2015;4:e10982. doi: 10.7554/eLife.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maeda M, Katada T, Saito K. TANGO1 recruits Sec16 to coordinately organize ER exit sites for efficient secretion. J Cell Biol. 2017;216:1731–1743. doi: 10.1083/jcb.201703084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gitler D, et al. Different presynaptic roles of synapsins at excitatory and inhibitory synapses. J Neurosci. 2004;24:11368–11380. doi: 10.1523/JNEUROSCI.3795-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milovanovic D, De Camilli P. Synaptic vesicle clusters at synapses: A distinct liquid phase? Neuron. 2017;93:995–1002. doi: 10.1016/j.neuron.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walter H, Brooks DE. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 1995;361:135–139. doi: 10.1016/0014-5793(95)00159-7. [DOI] [PubMed] [Google Scholar]
- 75.Wang L, et al. α-synuclein multimers cluster synaptic vesicles and attenuate recycling. Curr Biol. 2014;24:2319–2326. doi: 10.1016/j.cub.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shupliakov O, Haucke V, Pechstein A. How synapsin I may cluster synaptic vesicles. Semin Cell Dev Biol. 2011;22:393–399. doi: 10.1016/j.semcdb.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 77.Clayton DF, George JM. The synucleins: A family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci. 1998;21:249–254. doi: 10.1016/s0166-2236(97)01213-7. [DOI] [PubMed] [Google Scholar]
- 78.Burré J, et al. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benfenati F, Bähler M, Jahn R, Greengard P. Interactions of synapsin I with small synaptic vesicles: Distinct sites in synapsin I bind to vesicle phospholipids and vesicle proteins. J Cell Biol. 1989;108:1863–1872. doi: 10.1083/jcb.108.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Malinovska L, Kroschwald S, Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim Biophys Acta. 2013;1834:918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 81.Rizzoli SO, Betz WJ. The structural organization of the readily releasable pool of synaptic vesicles. Science. 2004;303:2037–2039. doi: 10.1126/science.1094682. [DOI] [PubMed] [Google Scholar]
- 82.Schuh AL, et al. The VPS-20 subunit of the endosomal sorting complex ESCRT-III exhibits an open conformation in the absence of upstream activation. Biochem J. 2015;466:625–637. doi: 10.1042/BJ20141202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Espenshade PJ, Li W-P, Yabe D. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc Natl Acad Sci USA. 2002;99:11694–11699. doi: 10.1073/pnas.182412799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanna MG, et al. Sar1 GTPase activity is regulated by membrane curvature. J Biol Chem. 2016;291:1014–1027. doi: 10.1074/jbc.M115.672287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Audhya A, et al. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Z, Wu E, Qian Z, Wu W-S. A multicolor panel of TALE-KRAB based transcriptional repressor vectors enabling knockdown of multiple gene targets. Sci Rep. 2014;4:7338. doi: 10.1038/srep07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178:43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marston DJ, et al. MRCK-1 drives apical constriction in C. elegans by linking developmental patterning to force generation. Curr Biol. 2016;26:2079–2089. doi: 10.1016/j.cub.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang L, Johnson A, Hanna M, Audhya A. Eps15 membrane-binding and -bending activity acts redundantly with Fcho1 during clathrin-mediated endocytosis. Mol Biol Cell. 2016;27:2675–2687. doi: 10.1091/mbc.E16-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weissman JT, Aridor M, Balch WE. Purification and properties of rat liver Sec23-Sec24 complex. Methods Enzymol. 2001;329:431–438. doi: 10.1016/s0076-6879(01)29104-6. [DOI] [PubMed] [Google Scholar]
- 93.Hui E, et al. Mechanism and function of synaptotagmin-mediated membrane apposition. Nat Struct Mol Biol. 2011;18:813–821. doi: 10.1038/nsmb.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]