Abstract
Thymidylate synthase (TSase) is responsible for synthesizing the sole de novo source of dTMP in all organisms. TSase is a drug target and as such it is well studied both in terms of structure and reaction mechanism. Cysteine 146 in E. coli TSase is universally conserved because it serves as the nucleophile in the enzyme mechanism. Here we use the C146S mutation to probe the role of the sulfur atom in early events in the catalytic cycle beyond serving as the nucleophile. Surprisingly, the single atom substitution severely decreases substrate binding affinity, and the unfavorable ΔΔGbind° is comprised of roughly equal enthalpic and entropic components at 25 °C. Chemical shifts in the free and dUMP-bound states show the mutation causes perturbations throughout TSase, including regions important for complex stability, in agreement with a less favorable enthalpy change. We measured the NMR methyl symmetry axis order parameter (S2axis), a proxy for conformational entropy, for TSase at all vertices of the dUMP binding/C146S mutation thermodynamic cycle and found that the calculated TΔΔS°conf is of similar sign and magnitude as the calorimetric TΔΔS°. Further, we ascribed minor resonances in wild-type-dUMP spectra to a state with a covalent bond between the Sγ of C146 and C6 of dUMP, and find S2axis values are unaffected by covalent bond formation, indicating this reaction step is neutral with respect to ΔS°conf. Lastly, the C146S mutation enabled us to measure cofactor analog binding by ITC without the confounding heat signature of covalent bond formation. Raltitrexed binds free and singly bound TSase with similar affinities, yet the two binding events have different enthalpy changes, providing further evidence of communication between the two active sites.
Introduction
Thymidylate synthase (TSase) catalyzes the conversion of dUMP to dTMP using the cofactor, 5,10-methylenetetrahydrofolate (CH2H4Fol), as both a methylene and hydride donor. Without TSase activity, cells are starved of one of the four DNA nucleotides and die a “thymineless death”. Thus TSase is an attractive target for drugs treating proliferative diseases. Indeed, for nearly 40 years, clinicians have prescribed thymidylate synthase inhibitors to combat an array of malignancies (1, 2). As a drug target and model system for understanding catalyzed hydride transfer, TSase has received significant attention from investigators using steady-state kinetics (3), pre-steady state kinetics (4), kinetic isotope effect studies (4-8), QM-MM simulations (9-13), and x-ray crystallography (14) to understand inhibitor binding and the reaction mechanism. In addition, TSase is a homodimer and a half-the-sites reactive enzyme (15, 16), indicating that the two active sites communicate with each other and thus making the enzyme an excellent model system for probing allostery in homo-oligomeric proteins.
In the early stages of the reaction, TSase uses a universally conserved cysteine (C146 in E. coli TSase) as a nucleophile to attack the C-6 position of the substrate dUMP (17). Previous studies suggest the C146 side-chain function is limited to its role as a nucleophile based on the following: 1) Mutation of C146 to serine causes a severe (∼5,000-fold) reduction in kcat, but only modest increases in Km (∼2-fold) for both substrate and cofactor (18). 2) KD measurements show C146S appears to bind dUMP ∼2-fold more weakly than the wild-type (18). 3) The C146S mutation results in no change in TSase fluorescence spectra (19), protease susceptibility, or dimer stability (20), whereas other C146 substitutions (e.g. C146W) or mutations to nearby residues (e.g. H147) do manifest in changes to these parameters. However, the equilibrium binding studies found only one molecule of dUMP binds to each dimer of wild-type and C146S TSase (18), which is not in agreement with crystal structures of the wild-type-dUMP complex (21) or our rigorous ITC measurements of the wild-type-dUMP interaction (22), both showing unequivocally that the two active sites bind substrate.
Here we revisit the C146S mutation and exploit it as a useful reagent that allowed us to probe multiple aspects of TSase function. To resolve the discord in stoichiometry between our wild-type dUMP-binding ITC data and the previously published binding data on C146S, and to ask what role, if any, the sulfur atom plays in early events of the catalytic cycle beyond serving as the nucleophile, we used ITC to measure dUMP binding to the C146S mutant of E. coli TSase. We find a stoichiometry of two dUMP molecules per C146S dimer, which is in agreement with both wild-type crystal structures (21) and our wild-type binding data (22). However, this subtle, single atom mutant binds dUMP nearly 20-fold more weakly than the wild-type enzyme with the unfavorable ΔΔG°bind apportioned roughly evenly between enthalpic and entropic components. The geometry of the wild-type dUMP crystal structure indicates this binding defect does not result from loss of a hydrogen bond between C146 and dUMP. Rather, a comparison of backbone NMR chemical shifts between wild-type and C146S TSase in the free and dUMP bound states shows the mutation results in wide-spread perturbation beyond the immediate vicinity of the substitution, including both phosphate binding loops and the dimer interface. Further, methyl symmetry axis NMR order parameters (S2axis), which have been shown to be a proxy for conformational entropy (23-26), indicate differences in S°conf in both the free and dUMP bound states of C146S TSase that account for the differences we observe in calorimetric entropies. In addition, because S146 is severely limited in its ability to form a covalent bond with substrate, we used the C146S mutation to deduce that weak resonances in wild-type-dUMP NMR spectra report on a minor state in which C146 is covalently bound to dUMP. S2axis values for the covalently bound state are indistinguishable from those in the non-covalent complex indicating that bond formation is neutral with respect to ΔS°conf. Lastly, we took advantage of the covalent bond defect of C146S to measure binding of the cofactor analog drug, Raltitrexed, to the dUMP binary complex by ITC without the confounding heat signature of covalent bond formation. These data show that cofactor analog binds free and singly bound TSase-dUMP with minimal cooperativity. However, the active sites clearly communicate with each other based on differences in ΔH° for the two binding events. This observation is in accord with measurements of chemical shifts in singly bound states showing that binding in one active site is sensed by the other (22),53.
Experimental Procedures
Materials
The C146S TSase mutation was made using the QuikChange procedure. TSase for ITC experiments was expressed and purified as described (22). The multiple NMR approaches used herein required different isotopic labeling schemes; therefore details on sample growth and preparation will be described in the relevant NMR experimental sections. TSase concentration was determined using Σ280 = 103,820 L•mol−1•cm−1. dUMP (Σ262 = 9,660 L•mol−1•cm−1) and 5F-dUMP (Σ262 = 9,660 L•mol−1•cm−1) were purchased from Sigma, CH2H4-Fol (Σ290 = 32,000 L•mol−1•cm−1) was from Merck and Cie (Switzerland), and Raltitrexed was from LKT laboratories. The concentration of Raltitrexed was determined using the PULCON (27) NMR approach using L-Tyr and L-Trp as concentration standards.
Isothermal titration calorimetry
ITC experiments to measure dUMP and Raltitrexed binding to C146S TSase were conducted on an Auto-iTC200 instrument. For dUMP binding, the enzyme concentration in the cell was 696 μM (dimer) and the dUMP concentration was 13.92 mM in the syringe. For Raltitrexed titrations into the C146S-dUMP complex, 50 μM C146S TSase (dimer) and 20 mM dUMP were present in the cell, and 1 mM Raltitrexed plus 20 mM dUMP were in the syringe. Buffer conditions for all ITC experiments were 25 mM NaPO4, 1 mM EDTA, 0.01% NaN3, 1 mM TCEP-HCl, pH 7.5. dUMP titrations were performed at 25 °C, and Raltitrexed titrations were performed at 5, 15, and 25 °C. Raw thermograms were integrated in Origin v. 7 to generate isotherms that were fit as previously described (22) using in-house MATLAB scripts. Briefly, our approach implements a general two-site model based on the binding polynomial (28) that includes cell concentration as a fitted parameter to allow for small errors in enzyme concentration and/or an active enzyme fraction of less than one.
NMR resonance assignments
Wild type TSase ILV methyl resonance assignment experiments were performed on a 500 μM (dimer) sample of U-[2H, 13C, 15N] methyl protonated [Ile(13C, δ1only), Leu(13CH3,12CD3), Val(13CH3,12CD3)] labeled TSase in 99.8% D2O NMR buffer (150 mM NaCl, 25 mM NaPO4, 1 mM EDTA, 0.01% NaN3, pD 7.1). We were able to make most assignments using HMCM(CG)CBCA and HMCM(CGCBCA)CO experiments acquired on a 700 MHz Avance III spectrometer equipped with a TCI CryoProbe, using non-uniform sampling (NUS) acquisition strategy covering 45% and 35% of the F1F2 matrices respectively. Assignments were completed with the aid of a [13C–F1,13C–F2]-edited NOESY with a 70 ms mixing time acquired on a 500 μM (dimer) sample of U-[2H, 15N] methyl protonated [Ile(13C, δ1only), Leu(13CH3,13CH3), Val(13CH3,13CH3)] labeled TSase in 99.8% D2O NMR buffer. This dataset was also acquired at 700 MHz, using NUS covering 45% of the F1F2 matrix. Methyl ILV resonances for the substrate-bound complex were made via a dUMP titration monitored by a 1H-13C HSQC and a [13C–F1,13C–F2]-edited NOESY as described above for the apo enzyme. To assign amide and ILV methyl assignments for free and dUMP-bound C146S, we made assignments based on nearest neighbors in wild-type spectra. NMR spectra for resonance assignments, pH dependence, and spin relaxation were acquired at 25 °C, processed with nmrPipe (29), and peak picking and integration were done in NMRViewJ (30). The three dimensional resonance assignment spectra acquired with NUS were reconstructed using iterative soft thresholding (31).
NMR chemical shift perturbation and spin relaxation
Chemical shift perturbation (CSP) based on TROSY 1H-15N HSQCs was calculated by:
15N R2 relaxation rates were measured in the Hahn-echo mode for 500 μM (dimer) samples of U-[2H, 15N] free, dUMP-bound, and 5F-dUMP+CH2H4-Fol-bound TSase using the pulse sequence described by Bax and co-workers (32). Two “planes” were collected, one at 600 MHz (Avance III, TCI CryoProbe) with a relaxation delays of zero and 15 ms, and the other at 850 MHz (Avance III, TCI CryoProbe), with relaxation delays of zero and 12 ms. R2 values at each field were calculated at each field using where T is the difference in relaxation times between the two planes (i.e. 15 ms at 600 MHz), and Io and I are the resonance intensities in the zero and T, planes respectively. Because Rex is proportional to the square of the magnetic field, residues with exchange were identified by taking the difference in R2 rates between the two fields. We then used boxplots to identify outliers with ΔR2 values greater than Q1 (quartile1) plus the Q3-Q1 interquartile range. This corresponds to values greater than or equal to two standard deviations from the mean.
To measure the tumbling times of the free and dUMP-bound TSase, we used TROSY versions of 15N R1, R1ρ, and {1H}-15N heteronuclear NOE pulse sequences (33) collected on 500 μM (dimer) samples of U-[2H, 15N] TSase at a single magnetic field (600 MHz, Avance III, TCI CryoProbe) as described (8). Global tumbling times of free and dUMP-bound TSase were 32.3 and 31.5 ns/rad, respectively. The global tumbling time of C146S-dUMP TSase was assumed to be the same its wild-type counterpart. This proved to be accurate given that the difference in methyl symmetry axis order parameters (S2axis, see below) between the wild-type and C146S complexes cluster around zero (Figure 5C). Due to limited sample, we used a less concentrated sample of free C146S TSase (370 μM) as compared to free wild-type TSase (500 μM). The lower viscosity of this more dilute sample required a 5% reduction in tumbling time (from 39.91 ns to 37.71 ns/rad in D2O) in order for the differences in order parameters to be clustered at zero (Figure 5B).
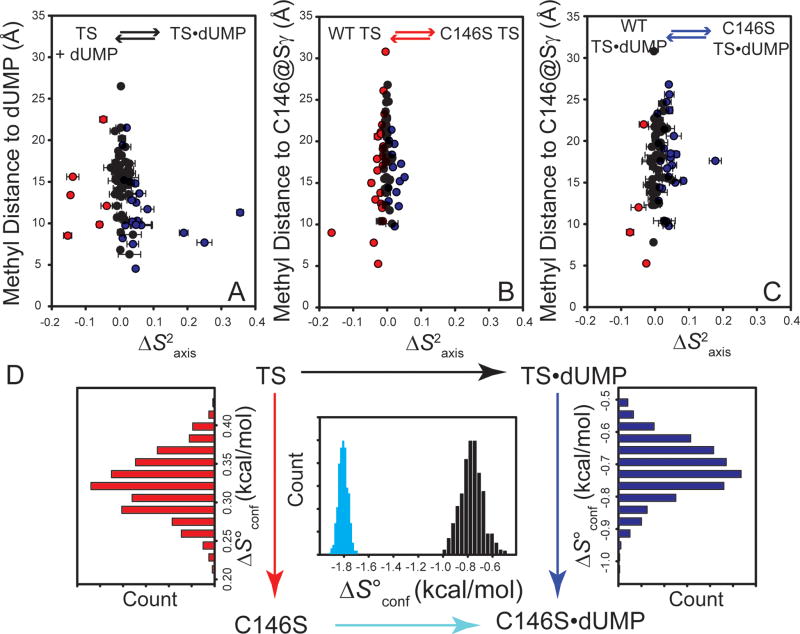
Figure 5.
Change in methyl S2axis and conformational entropy for wild-type and C146S dUMP binding. (A) ILV methyl ΔS2axis for wild-type dUMP binding plotted as a function of distance from a pseudo atom placed at the average methyl proton position, and the nearest dUMP atom. Methyl groups becoming significantly more rigid upon binding are blue, methyl groups becoming significantly more flexible are red, and methyl groups with no significant change are black. Significance criterion is ΔS2axis must be greater than 2σ. ΔS2axis associated with the wild-type to C146S mutation in the free and dUMP-bound states are shown in Panels B and C, respectively. (D) The conformational entropy meter (Experimental Procedures) was used to convert ΔS2axis to ΔS°conf for the different legs of the thermodynamic cycle. The entropy meter shows that ΔS°conf for mutant binding is ∼ 1 kcal/mol (298 K) less favorable than for the wild-type. The histograms for the vertical legs show this difference originates both from increased dynamics in the free state and decreased dynamics in the bound state. Histograms take into account noise in the S2axis measurements (See Experimental Procedures).
ILV methyl S2axis values were measured using the intra-methyl 1H-1H dipolar cross-correlated spin relaxation approach of Kay and co-workers (34) on 500 μM (dimer) samples of U-[2H, 15N] methyl protonated [Ile(13C, δ1only), Leu(13CH3,12CD3), Val(13CH3,12CD3)] labeled TSase in 99.8% D2O NMR buffer. To minimize the relaxation effects of nearby protons, the labeling scheme in which only one of the LV methyl groups is protonated was used, and experiments were performed in D2O buffer. To account for the increased viscosity in D2O buffer, global tumbling times measured in H2O (see above) were simply multiplied by the D20/H2O viscosity ratio of 1.235 (35). The time dependent build-up of triple quantum coherences (36) (Ib) and the bi-exponential decay of 1H magnetization (Ia) were measured in an interleaved manner at 850 MHz. Relaxation delays were: 0.3, 0.6, 0.8, 1.2, 1.6, 2.4, 3.2, 4.0, and 5.6 ms, with underlined values acquired in duplicate for the purposes of error estimation. The cross relaxation rate, η, was determined by fitting the ratio of the intensities in the two experiments to the following:
where T is the relaxation delay, and δ is related to the relaxation contribution from external 1H spins (37). Errors in η were estimated using Monte Carlo methods bounded by noise in the intensity measurements as estimated from duplicate points. The relative error was then propagated to S2axis, which is calculated by:
where μo is the vacuum permittivity constant, P2(x) = 1/2(3x2 − 1), θaxis,HH is the angle between the methyl 3-fold axis and a vector connecting a pair of methyl 1H nuclei (90°)(37), γH is the gyromagnetic ratio of a proton spin, τc is the global tumbling time, and rHH is the distance between pairs of methyl protons (1.813 Å) (34, 38).
Calculation of ΔS°conf based on ΔS2axis using the entropy meter
We used the entropy meter calibrated (25) by Wand and co-workers to convert changes in S2axis to changes in S°conf. Briefly, ΔS°conf was calculated based on the following:
where T is the temperature in Kelvin, is the entropy meter slope (m=−0.0018 kcal−1 mol−1 K−1 ΣNχ−1), Δ<S2axis> is the change in average methyl order parameters common to the end states in the ΔS°conf calculation, and ΣNχ is the sum of the methyl side-chain χ angles associated with the probes used in the Δ<S2axis> calculation. We estimated the uncertainty in ΔS°conf using Monte Carlo simulations bounded by the errors of the constituent values in the Δ<S2axis> calculation. We note that there is also uncertainty in the entropy meter itself as the data points used to calibrate the meter are linear (25), but not perfectly so. The magnitude of this uncertainty is not explicitly stated in the entropy meter report, so we do not include it here. Nonetheless the precision of experimental S2axis measurements place high confidence in the <S2axis> associated with each of the states.
Results and Discussion
C146S mutation significantly weakens substrate binding
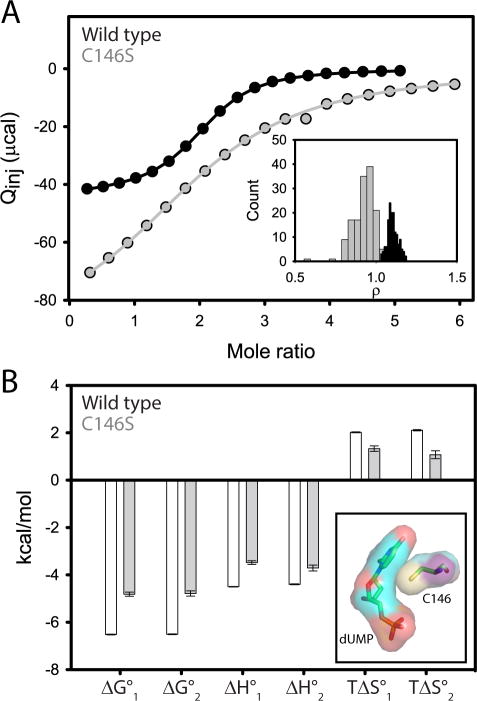
We used ITC to probe the effect of the C146S mutation on substrate binding in phosphate buffer at 25 °C. As with the wild-type interaction (22), the C146S ITC data were fit to a general two-site model based on the binding polynomial (28), modified to allow for a correction of enzyme concentration or active enzyme fraction (See Experimental Procedures). Substrate binds with similar affinities to both the free and singly bound forms of C146S TSase as evidenced by a cooperativity constant, ρ, very close to one (0.9 ± 0.1, Figure 1A inset and Table S1), which is similar to the wild-type interaction (22). Despite the lack of binding cooperativity, there is communication between the two sites because ΔH° and TΔS° are slightly different for the two binding events (Table S1). This is reminiscent of the wild-type enzyme-dUMP interaction(22), which also lacks binding cooperativity but possesses “silent allostery” (39) in the form of differences in ΔH° and TΔS°. Surprisingly, this subtle mutation results in a 1.7 kcal/mol (18-fold) penalty with respect to formation of the dUMP complex (Figure 1 and Table S1). The ΔΔG°bind is essentially split evenly between enthalpic and entropic components, with ΔΔH° accounting for 1 and 0.7 kcal/mol of the difference in binding to free and singly-bound forms, respectively. Similarly, TΔΔS° accounts for −0.7 and −1 kcal/mol (Figure 1B and Table S1) of the difference in the first and second binding events. The sulfur atom does pack against the dUMP sugar and base ring (Figure 1B, inset), which could provide some additional stability in the wild-type relative to the C146S complex. We point out that differences to direct contacts are likely limited to changes in London dispersion forces since it is clear from the x-ray model (pdbid 1BID) that the geometries of C146 and dUMP do not favor a SH-π hydrogen bond (40).
Figure 1.
dUMP binding to wild-type and C146S TSase by ITC at 25 °C. Wild-type data were presented previously 22. (A) ITC isotherms for wild-type (black) and C146S (gray) binding to dUMP with fitted lines shown. Note the dimeric TSase concentrations in the cell were 206 μM and 696 μM for wild-type and C146S TSase respectively. The inset shows the cooperativity factor, ρ, which is the ratio of the intrinsic association constants for binding to the free and singly bound enzymes. (B) ITC-derived thermodynamic parameters for the two dUMP binding events. Note the less favorable ΔG°bind is the net of less favorable enthalpic and entropic components. The inset shows the surfaces of dUMP and C146 from the wild-type dUMP-complex x-ray model (pdbid 1BID).
Small population of Wild-type TSase-dUMP complex has covalent bond between C146 and substrate, but its disruption is not the source of the C146S binding defect
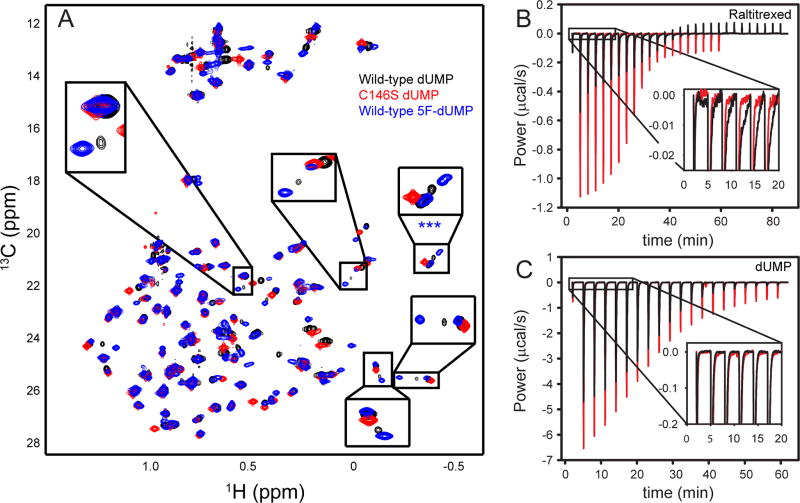
Given the evidence that TSase can form a covalent bond between the sulfur atom of C146 and C6 of dUMP even in the absence of cofactor (41), and the bond cannot form in the C146S mutant, an important question is whether this defect is responsible for the apparent weaker binding affinity. To consider this, we turned to 1H-13C ILV methyl HSQC spectra of the wild-type dUMP-bound complex, in which we detect a minor state that is likely the covalent complex based on the following: 1) The minor resonances are not present in C146S-dUMP spectra and the single set of resonances in the mutant spectrum overlap with the major state in the wild-type spectrum (Figure 2A). 2) In wild-type dUMP NMR titrations, the free and bound major states are in fast exchange on the NMR timescale, while the free state and bound minor-state are in slow exchange, as is expected for a non-covalent complex with modest binding affinity (17 μM (22)) and a longer-lived, covalent complex, respectively. 3) The population of the minor state is significantly increased in the substrate analog 5F-dUMP complex (Figure 2A), in agreement with stabilization of the covalently bound adduct by the strength of the C5-F bond and destabilization of the aromatic ring system by the electronegativity of fluorine. 4) The putative covalent complex resonances of the dUMP and 5F-dUMP complexes are close to each other, but the 5F-dUMP covalent peaks are always shifted further from their non-covalent pairs than is the case for the dUMP complex (Figure 2A), consistent with the relative strengths of the two covalent bonds. Collectively, these data provide strong evidence that the minor state observed in spectra of the wild-type-dUMP complex reflects a population with a covalent bond between C146@Sγ and dUMP@C6.
Figure 2.
A fraction of the TSase-dUMP complex contains a covalent bond between C146@Sγ and dUMP@C6. (A) ILV methyl 1H-13C HSQC spectra of the wild-type dUMP complex (black), C146S-dUMP complex (red), and wild-type 5F-dUMP complex (blue). Expanded boxes contain resonances that report on a minor state with a covalent bond between C146 and dUMP (see text). Note the minor state is more highly populated in the 5F-dUMP complex and absent in the C146S complex. Note also that the C146S-dUMP complex resonances overlay with the major state resonances of the wild-type-dUMP complex. (B) ITC thermograms for Raltitrexed binding to the wild-type and C146S-dUMP complexes in black and red respectively. The expanded region shows a slow exothermic component in the wild-type, but not the mutant thermogram, that is therefore likely to report on covalent bond formation. (C) ITC thermograms for wild-type and C146S dUMP binding are colored similarly to panel B. Expanded baseline shows ITC does not detect covalent bond formation in either of these titrations.
Having identified resonances associated with the covalently-bound nucleotide, we measured the fraction of this state based on resonance intensities in the wild-type spectrum (19%, see Figure 2A), which, together with the previously measured covalent bond breakage rate (H5 exchange with solvent at similar pH and temperature with a rate of 0.01 min−1) (41), allows us to calculate the forward rate of covalent bond formation (0.0019 min−1), which is negligible over the course of the sixty minute ITC experiment. This conclusion is further supported by comparing the wild-type and C146S ITC thermograms for cofactor analog binding to the dUMP complex (Figure 2B), and for dUMP binding (Figure 2C). Cofactor and cofactor analog binding to the substrate complex is known to induce covalent bond formation between C146 and C6 of the nucleotide (42, 43) and we observe a slow exothermic event in wild-type ITC thermograms that is absent in C146S counterparts (Figure 2B). We therefore attribute this slow exothermic signal to covalent bond formation. By contrast, thermograms of wild-type and C146S dUMP binding are indistinguishable (Figure 2C), consistent with the assertion that covalent bond formation in the wild-type dUMP complex is not yet formed during our ITC experiments and thus is not the basis for the observed weaker binding.
C146 side-chain is likely predominantly protonated at neutral pH in free and dUMP-bound TSase
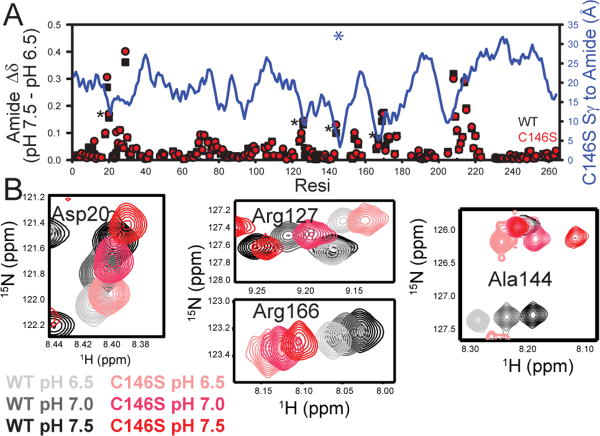
We also considered the possibility that if C146 is deprotonated, the C146S mutation could represent more than a single atom substitution as it would also remove a negative charge. Previous calculations have estimated the C146 side chain has a pKa of 6.7 in the closed ternary complex (10), but to our knowledge, this has not been confirmed experimentally and the protonation state of C146 in neither the free nor the nucleotide-bound states has been reported. Given that our ITC and NMR experiments were collected at pH 7.5, we looked for evidence of different titration behavior of apo TSase by examining a pH titration by TROSY 1H-15N HSQC spectra. Our range was limited to pH 6.5-pH 7.5 due to enzyme stability issues at the lower end of this range, and disruption of the dimer interface under basic conditions, but we would expect to see changes in the titrations if C146 has a pKa near 6.7. Figure 3A shows the change in amide chemical shifts between pH 6.5 and pH 7.5 for the wild-type and C146S enzymes. From this plot it is clear that the mutation has little effect on the titration behavior as sensed by the available amide probes. Further, titration trajectories of the four residues closest to the site of mutation, D20, R127, A144, and R166 are not affected (Figure 3B). Given that the C146 sulfur atom is less than 4 Å from the guanidinium group of R166 and this side-chain stabilizes the anionic form of the C146 side-chain after concerted hydride transfer and C146-dUMP bond breakage (13), we would expect the pH sensitivity of the R166 amide to be affected by a nearby charge change. We point out that it is not necessary to also look at differences in dUMP complex pH titrations because we have previously shown by ITC that dUMP binding is associated with minimal change in protonation state (loss of less than 0.1 proton per binding event) (22). Therefore dUMP binding does not cause any significant pKa shifts in the experimental pH range. However, we raise the caveat that these are indirect measurements. Ideally, we would measure the pH dependence of C146 chemical shifts directly, and look for changes of specific magnitude and direction (44) that would implicate titration of C146 itself. We are unfortunately limited to the approach described here because C146 resonances are broadened away in both the free and dUMP-bound forms (see below). Nonetheless, based on the available data we assert that the protonated form of the C146 side-chain predominates at pH 7.5 in free and dUMP-bound TSase, and therefore differences in binding (above), structure (below), and dynamics (below) are not due to a charge change.
Figure 3.
pH dependence of free wild-type and C146S TSase amide chemical shifts. (A) Difference in apo enzyme chemical shift at two pH values (pH 7.5 vs. 6.5) for the wild-type and C146S apo enzymes in black and red respectively. The blue line plot with the right-hand y-axis shows the amide distance to the site of the mutation and the blue asterisk marks the spot of the C146S mutation. (B) Spectra for residues that have pH dependent behavior and are closest to the mutation site. Wild-type titration series is in gray to black and mutant in pink to red. These residues are marked with asterisks in Panel A.
Chemical shift changes reveal widespread effects of C146S mutation in free and substrate-bound TSase
The sulfur atom of C146 packs against the dUMP base ring in wild-type complex x-ray model (Figure 1B, inset), which may contribute favorably to the binding free energy. However, it is unclear whether perturbation of this interaction is solely responsible for the observed thermodynamic differences reported above. Given the large and unexpected change in binding affinity, we turned to NMR to probe the range of the mutation's effect. Several lines of evidence show differences in the C146-containing active site loop among multiple states of the wild-type enzyme cycle, and these differences are not evident in the corresponding x-ray models. First, we were unable to locate resonances corresponding to C146-F149 in the free enzyme, and C146-H147 in the dUMP complex in TROSY triple resonance experiments, indicating chemical exchange broadening by motions on the μs-ms timescale. This conclusion is supported by Hahn-Echo R2 measurements (32), showing chemical exchange (Rex) in F150 in the free enzyme and F149-F150 in the dUMP complex; these residues flank the aforementioned “invisible” amino acids (Figure S1A&B). Chemical exchange of this region could be hinted at in the free enzyme by the x-ray model (pdbid 2FTQ) in which the side chains of C146 and H147 have two conformations. The first conformation of C146 can be superimposed with C146 in the dUMP (21) and 5F-dUMP-CH2H4-Fol (43) complexes, and places the side-chain near the guanidinium group of R166 (Figure S1D). The second pose leaves the side chain more solvent exposed as it is rotated away from R166. The dUMP complex x-ray model shows the active site loop in a single conformation, so dUMP binding may lower the population of second conformations below the threshold required to visualize in electron density maps, or some dynamic process not detected by x-ray crystallography is responsible for the Rex observed in both the free and bound states. By contrast, we were able to visualize triple resonance spin-systems for the entire loop, including C146, in the closed ternary complex with the substrate analog, 5F-dUMP, and the biological cofactor, CH2H4Fol (45). Hahn-Echo measurements on this complex are in agreement as we observe no Rex in the C146-loop (Figure S1C). Collectively, these data show that the C146 loop is switching between multiple conformations even in the presence of substrate. Cofactor binding, with its associated conformational change (14), is required to quench these motions. This picture is inconsistent with a single conformation in which the C146 side-chain is anchored in place by the London dispersion forces depicted by the dUMP complex crystal structure (Figure 1B, inset).
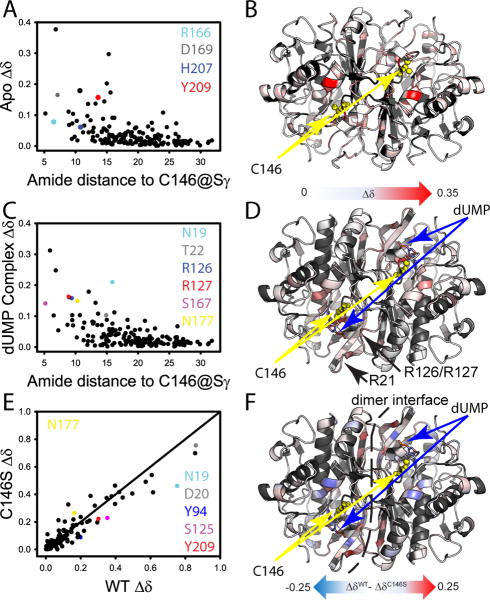
Comparison of wild-type and C146S TROSY 1H-15N HSQCs of free and dUMP-bound TSase (Figure S2) shows the mutation causes extensive changes to the enzyme. In the free state, several sites important for dUMP complex formation are perturbed: Arg166, Asp169, His207, and Tyr209 all make contacts to dUMP (21) and show significant chemical shift perturbations (CSPs) due to C146S mutation (Figure 4A). These effects are not completely unexpected as the amides are relatively close to the mutation site, with Tyr209 being the most distal amide at ∼14 Å away (Figure 4A). While chemical shift perturbation generally dampens with distance, the effects of this substitution are felt throughout the free enzyme with significant changes observed at sites greater than 25 Å from residue 146 (Figure 4A&B). The mutation is likewise “felt” in the dUMP complex. (Note wild-type and C146S-dUMP complex comparisons are based on the major, non-covalent state (see above)). A similar pattern of distance-dependent dampening is observed in the complex in that there is widespread CSP at distal sites with effects reaching beyond 25 Å (Figure 4C&D). Further, the mutation perturbs several residues important for substrate binding in the dUMP complex. For example, the Arg21 and Arg126 loops provide the basic residues that contact the dUMP phosphate in the complex, and both of these loops are affected by the mutation in the dUMP-bound state (Figure 4C&D). This is also true of S167 and N177, which make hydrogen bonds to the dUMP phosphate and pyrimidine ring respectively (Figure 4C&D).
Figure 4.
Effect of C146S mutation on free and dUMP-bound TSase from NMR amide chemical shifts. (A)Mutational CSPs (See Experimental Procedures) in the apo-enzyme are plotted vs the distance to the site of mutation. Residues important to dUMP binding and discussed in the text are highlighted in color. (B)CSPs from panel A are mapped onto the apo-enzyme structure with the site of mutation in yellow. (C) Mutational CSPs in the dUMP complex are plotted vs the distance to the site of mutation. Residues important to dUMP binding and discussed in the text are highlighted in color. (D) CSPs from panel D are mapped onto the dUMP-complex structure with the site of mutation and substrate highlighted. The CSP scales in Panels B&D are the same. The phosphate binding arginine loops are highlighted in this panel. (E) The CSPs for wild-type dUMP binding are plotted against the CSPs for C146S dUMP binding. Important residues having different CSPs associated with complex formation are highlighted in color. (F) The difference in CSP is mapped onto the dUMP complex structure. The model is annotated to show significant differences in the binding site and dimer interface.
Given the observed differences in thermodynamics of binding, it is important to ask how the C146S mutation affects the change in chemical shift in going from the free to bound state. While we show above that the mutant free and bound forms are different from the wild-type counterparts, it is possible that parallel changes in the two end states could indicate similarity in the binding process. Figure 4E shows this is not the case. While there is generally a linear relationship between the wild-type and C146S dUMP binding CSPs, there are large and widespread deviations from the line of unit slope. Interestingly, there are outliers both above and below the line, but the residues showing the largest CSPs all experience a smaller change for the mutant binding process than for the wild-type (Figure 4E). Several regions important for dUMP binding, including both phosphate binding loops, Y94, N177, and Y209 all have different changes in chemical shift upon dUMP binding (Figure 4E), indicating functional changes in the binding process. Lastly, it is noteworthy that the beta sheet comprising the dimer interface is also significantly affected, with the mutation causing both increases and decreases in CSP for dUMP binding (Figure 4F). These data are consistent with the hypothesis that the C146S mutation affects ΔH°bind by subtly rearranging hydrogen bond strengths at the interface and the strength of the interactions between the phosphate binding loops and dUMP. Taken together, the CSPs show this subtle mutation has widespread effects on the conformational ensembles of the free enzyme and the dUMP complex, which leads to changes in the process, and hence the thermodynamics of dUMP complex formation.
Differences in conformational entropy partially underlie differences in substrate binding affinity
Weaker dUMP binding by the C146S mutant is the result of both less favorable enthalpy and entropy changes (Figure 1B) relative to the wild-type. Given that the mutation is essentially isosteric, changes in solvent entropy are not likely responsible for the modest change in TΔS° of ∼ 1 kcal/mol per binding event (Figure 1B and Table S1). Instead, we focused our attention on the mutation's effect on conformational entropy, ΔS°conf. It has recently been shown that the methyl symmetry axis order parameter, S2axis, is an excellent proxy for conformational entropy (23, 25, 26, 46-48), so as a means to determine the mutational effect on ΔS°conf, we measured S2axis for ILV (Ile, Leu, and Val) methyl groups within the free and dUMP-bound states of wild-type and C146S TSase using the intra-methyl 1H-1H dipolar cross-correlated spin relaxation approach of Kay and co-workers (34).
Because the ps-ns dynamics of various states along the wild-type reaction coordinate have not yet been probed, we first determined the S2axis for the free and dUMP-bound wild-type enzyme (Table S2). As with effects of ligand binding in other systems (49-53), dUMP binding elicits a heterogeneous dynamic response, in that there are both decreases and increases in flexibility (Figure 5A, Figure S3). It is important to note that dUMP binds to an essentially pre-formed active site and binding is not accompanied by significant conformational change(14), indicating the following changes in S2axis and hence S°conf occur within a single conformational well. There are no methyl groups making direct contact with dUMP in the complex as L143@CD1 makes the closest approach to the ligand (4.6 Å). However, in general, the largest dynamic perturbations do tend to be close to dUMP and involve rigidification upon binding (Figure 5A, Figure S3), including L7, V11, L170, and L208. There are two probes in the C-terminus (V262 and I 264) and S2axis values of ∼0.1 in both the free and dUMP-bound point at significant flexibility on the ps-ns timescale consistent with an open conformation (Tables S2). However, the C-terminus is sensitive to binding as the probes exhibit small but significant decreases in flexibility upon complex formation (Figure S3). The unfavorable ΔS°conf associated with rigidification is partially offset by methyl groups that become more flexible upon binding (Figure 5A). I129 is noteworthy as it has the largest change in S2axis among probes in this class and it resides at the dimer interface (Figure S3). When thinking about this change, it is important to consider that the apo and dUMP states are symmetrical species with a single set of resonances (excluding the small fraction of covalently bound dUMP, see above), so we are measuring the change in S2axis associated with binding both dUMP molecules to a homodimer. In the case of I129, its methyl group is actually closer to dUMP in the opposite subunit, emphasizing that it is unclear to what extent the changes report upon effects associated with binding to the local subunit, the distal subunit, or both. We are currently using a mixed-dimer strategy 53 to understand how single ligand binding affects dynamics in the both the local and distal subunits.
Due to the high sensitivity of these experiments, we were also able to obtain precise measurements of S2axis for the minor population of TSase covalently bound to dUMP. We unambiguously assigned and measured relaxation for 19 resonances in both the major and minor states, and surprisingly, none of these probes have significantly different dynamics in the non-covalent and covalent complexes (Figure S4). This was unexpected given that we (54-60) and others (23, 47) have shown that S2axis is highly sensitive to even the most subtle of perturbations (e.g. even the C146S mutation, see below). To our knowledge, covalent bond formation with dUMP is a rare example of a perturbation that is silent at the level of S2axis. However, we point out that while this perturbation is sufficient to cause wide-spread chemical shift changes (Figure 2A), it is more subtle than a typical covalent bond since the TSase catalytic chemistry demands this covalent bond is weak (12, 13), which is supported by its low population (see above). Further, reversibility in the binary complex (41) and diffuse electron density in the ternary complex with 5F-dUMP and CH2H4-Fol (43) also support the idea of a metastable bond. Nonetheless, the data presented here show that formation of the catalytically relevant covalent bond in the binary complex is neutral with respect to S2axis and therefore ΔS°conf.
Next we measured the ILV methyl order parameters for free and dUMP-bound C146S TSase. Overall, when compared to dUMP binding (Figure 5A), the mutation has a smaller effect on S2axis in the free state (Figure 5B and Figure S5). There are significant changes throughout the molecule, which is consistent with the widespread effect of the mutation on chemical shift. While there are some probes that become more rigid in the free state, increased flexibility is the overall trend, with I129 showing a large decrease in S2axis relative to the wild-type (Figure S5). Interestingly, the direction and magnitude of this change are very similar to what is observed in wild-type enzyme upon dUMP binding (Figure S3). However, the mutation does not generally recapitulate the dynamic response of dUMP as several diagnostic probes change in opposite directions upon binding in the wild-type and mutation (e.g. L27, L170 in Figures S3 vs. S5). The C146S mutation generally results in rigidification in the dUMP complex, as compared to the wild-type counterpart, with the largest changes observed in I55, I112, V185, and V200 (Figure 5C and Figure S6).
By taking the average change in S2axis, (Δ<S2axis>) and scaling it by the number of side-chain dihedral angles associated with the methyl probes (ΣNχ), we can use the entropy meter (See Experimental Procedures) to determine ΔS°conf for the legs of dUMP binding/C146S mutation thermodynamic cycle and gain insight into how the mutation affects ΔS°conf for dUMP binding (Figure 5D). In the free state, the mutation results in a favorable ΔS°conf of 0.32 ± 0.03 kcal/mol at 298 K. However, the mutation has the opposite effect in the bound state with the mutation giving ΔS°conf of −0.71 ± 0.09 kcal/mol (Figure 5D). This gives a net TΔΔS°conf of −1.03 ± 0.09 kcal/mol (Figure 5D) for C146S-dUMP binding relative to the wild-type, which is on the order of what we observe for the change in calorimetric entropy (Figure 1B and Table S1). These results are therefore consistent with the assertion that changes in S°conf in both the free and dUMP-bound states underlie the observed differences in calorimetric entropy. We note that these calculations are for a single binding event and that we therefore implicitly assume that Δ<S2axis> is the same for dUMP binding to the free enzyme and the singly bound forms. We therefore use a single subunit to count Nχ and scale Δ<S2axis>. This is a reasonable approximation because the ΔS°bind for the two binding events (Table S1) are very similar at 298.15 K, which is the temperature used for the ITC and NMR comparisons. We are currently performing relaxation experiments using mixed TSase dimers with only a single binding competent active site to test this assumption. Lastly, we note that if the average change in order parameter is scaled according to the number of probes in the dimer, we arrive at a total TΔΔS°conf of −2.08 ± 0.18 kcal/mol associated with binding both dUMP molecules. This agrees well with the sum of calorimetric TΔΔS° values for both binding events (−1.7 kcal/mol; see Figure 1B and Table S1).
Both subunits of C146S-substrate complex bind cofactor analogs with similar affinity
Previously we used NMR to show the 5F-dUMP-CH2H4-Fol “diligand” binds to free and singly bound TSase with similar affinities (22). The drawbacks to this approach are: 1) 5F-dUMP makes a covalent bond with both TSase and CH2H4-Fol (hence the term “diligand”) rendering the interaction so tight that we could only measure the relative binding affinities of the two sites, and 2) The diligand probes a composite of both the substrate and cofactor binding steps, and we would gain more knowledge by isolating the two binding events. Unfortunately cofactor analog binding to the wild-type substrate complex is accompanied by covalent bond formation between C146 and the substrate (42, 43), and the heat of fusion confounds analysis of ITC thermograms (e.g. Figure 2B). ITC is by far our method of choice for measuring binding in multi-ligand systems because we can unambiguously measure thermodynamic parameters for discrete binding events, and the C146S mutant allows us to isolate the cofactor-analog binding step using ITC without the effects of covalent bond formation. While we show above that the mutation does affect substrate binding, it does not affect the structure of the closed, ternary complex (backbone RMSD = 0.129 Å between the wild-type diligand complex and the C146S product complex (61)), thus C146S is a useful tool to measure cofactor binding.
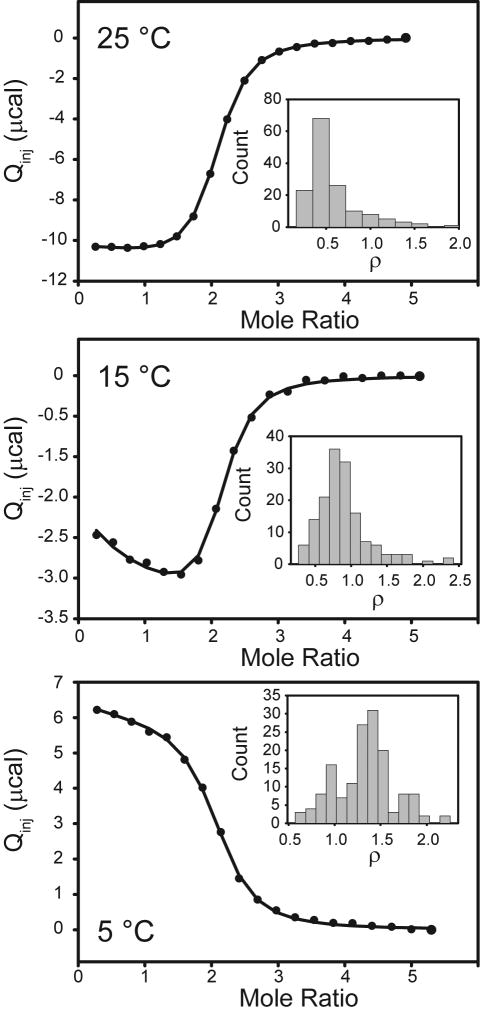
To probe the cofactor binding step, we injected cofactor analog (Raltitrexed) into the pre-formed C146S-dUMP complex at 5, 15, and 25°C (Figure 6). Raltitrexed binds to the dUMP complex with a KD of ∼0.5 μM at 25 °C (Table S3), which is significantly tighter than dUMP binding (16 μM, Table S1). As with binding to substrate, cofactor analog binds to the free and singly bound dimers with similar affinity with ρ values hinting at slight negative cooperativity at 25 °C (ρ=0.5 ± 0.4, Table S3) or slight positive cooperativity at 5 °C (ρ=1.3 ± 0.3). It should be emphasized that a ρ value of 0.5 indicates a two-fold difference in binding affinities, so the magnitude of cooperativity is indeed small. In addition, ρ-value histograms of Monte Carlo simulations show that the noise in the data are generally consistent with 0.5 ≤ ρ ≤ 2.0 (Figure 6, insets). However, the binding sites are not equivalent as evidenced by the clear difference in ΔH° for the two binding events. The difference in ΔH° is small (maximum of 0.6 kcal/mol, Table S3), but it is robust as the early slopes in the isotherms at 5 and 15 °C clearly show a mixture of multiple processes with different enthalpies (Figure 6). Now, it is clear that E. coli TSase is not significantly cooperative in either the substrate or cofactor binding steps. However, several lines of evidence show that the effects of substrate or cofactor binding are indeed communicated to the second subunit based on the following: 1) Changes in in enthalpy (and entropy) are not equivalent for the two binding events. 2) We observe quartets of resonances for a subset of amides at intermediate points in diligand NMR titrations (22, 62). Two of the resonances correspond to the free and doubly bound enzymes. The other two are attributable to the free and bound subunits of the singly bound state, indicating that binding of the first diligand binding event is communicated to the second subunit. 3) Studies in which we used mixed dimers to isolate the singly bound dUMP state and measure the chemical shifts of all microstates also show crosstalk between binding and non-binding subunits (62). Thus the structure and/or dynamics of the second subunit are affected by binding the first dUMP, cofactor, or diligand molecules. We are currently performing experiments designed to understand the nature of this communication.
Figure 6.
Cofactor analog binding to the C146S-dUMP complex by ITC. Raltitrexed was titrated into the pre-formed nucleotide complex at 25 °C (Top), 15 °C (Middle), and 5 °C (Bottom). The inset histograms plot the cooperativity parameter, ρ, which is a ratio of the two intrinsic association constants for binding to the free and singly bound enzyme.
Conclusion
In this study we used the C146S mutation of E. coli TSase to probe the function of this catalytically critical amino acid beyond its role as the substrate attacking nucleophile. The mutant proved to be a valuable reagent that revealed a number of previously unknown aspects of TSase function. First, by comparing the pH dependence of wild-type and mutant NMR spectra, we are able to assert that the major form of the C146 side-chain is protonated in free and dUMP-bound TSase at neutral pH. In addition, the mutation was instrumental in allowing us to deduce that ∼20% of the wild-type TSase-dUMP complex contains a covalent bond between the C146 side-chain and C146, which is not seen in crystal structures. Further and surprisingly, this single atom substitution causes a 20-fold reduction in dUMP binding affinity, indicating the side-chain sulfur atom provides significant stability to the complex. The binding defect is composed of roughly equal enthalpic and entropic components. Chemical shift perturbations show wide-spread effects of the mutation in multiple regions important for dUMP binding with the most notable changes involving the Arg21 and Arg126 phosphate binding loops, which may account for the less favorable enthalpy change. Thus, the C146 side-chain contacts the dUMP base, as is apparent in X-ray crystal structures, but these solution studies show the sulfur atom also influences the environment of multiple other key dUMP recognition elements. Further, by using an entropy meter, which converts changes in methyl symmetry axis NMR order parameters (S2axis) into conformation entropy, we show that the sign and magnitude of the calculated ΔΔS°conf (wild-type dUMP binding vs. C146S dUMP binding) are in agreement with the change in the overall calorimetric entropy. Taken together, the cysteine side chain plays several key roles in stabilizing the substrate complex, which is the first step in the reaction coordinate. We also highlight that this is the first report of the role of conformational entropy in the wild-type reaction coordinate, in which binding of each dUMP molecule is opposed by ∼1 kcal/mol of ΔS°conf. Lastly, we used the C146S mutation to measure cofactor analog, Raltitrexed, binding to the TSase-dUMP complex by ITC without the confounding heat signal associated with covalent bond formation. We show this analog binds to the free and singly bound subunits with very similar affinity (within two-fold), yet the ΔH (and ΔS) associated with the two binding events are different, which is consistent with the communication between binding sites observed previously by ITC and NMR.
Supplementary Material
Acknowledgments
Funding: This work is supported by a National Institutes of Health (NIH) grant, GM083059, to A. L. L.
We would like to thank Dr. Vitali Tugarinov for the pulse sequence used to measure intramethyl 1H-1H dipolar cross-correlated relaxation rates, and some of the pulse sequences used to assign ILV methyl groups (HMCM(CG)CBCA and HMCM(CGCBCA)CO). We also thank Dr. Scott Robson and Prof. Gerhard Wagner for sharing the istHMS software used for spectrum reconstruction. Lastly, we thank Professor Amnon Kohen, Zahidul Islam, and Bradley Falk for helpful discussions.
Footnotes
Supporting Information Available: Additional figures mentioned in the text, tables containing thermodynamic data for dUMP and Raltitrexed binding, and a table with raw S2axis values.
References
- 1.Danenberg PV. Thymidylate synthetase - a target enzyme in cancer chemotherapy. Biochimica et biophysica acta. 1977;473:73–92. doi: 10.1016/0304-419x(77)90001-4. [DOI] [PubMed] [Google Scholar]
- 2.Mathews CK. Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat Rev Cancer. 2015;15:528–539. doi: 10.1038/nrc3981. [DOI] [PubMed] [Google Scholar]
- 3.Fantz C, Shaw D, Jennings W, Forsthoefel A, Kitchens M, Phan J, Minor W, Lebioda L, Berger FG, Spencer HT. Drug-resistant variants of Escherichia coli thymidylate synthase: effects of substitutions at Pro-254. Molecular pharmacology. 2000;57:359–366. [PubMed] [Google Scholar]
- 4.Spencer HT, Villafranca JE, Appleman JR. Kinetic scheme for thymidylate synthase from Escherichia coli: determination from measurements of ligand binding, primary and secondary isotope effects, and pre-steady-state catalysis. Biochemistry. 1997;36:4212–4222. doi: 10.1021/bi961794q. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal N, Hong B, Mihai C, Kohen A. Vibrationally enhanced hydrogen tunneling in the Escherichia coli thymidylate synthase catalyzed reaction. Biochemistry. 2004;43:1998–2006. doi: 10.1021/bi036124g. [DOI] [PubMed] [Google Scholar]
- 6.Hong B, Haddad M, Maley F, Jensen JH, Kohen A. Hydride transfer versus hydrogen radical transfer in thymidylate synthase. J Am Chem Soc. 2006;128:5636–5637. doi: 10.1021/ja060196o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong B, Maley F, Kohen A. Role of Y94 in proton and hydride transfers catalyzed by thymidylate synthase. Biochemistry. 2007;46:14188–14197. doi: 10.1021/bi701363s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Sapienza PJ, Abeysinghe T, Luzum C, Lee AL, Finer-Moore JS, Stroud RM, Kohen A. Mg2+ binds to the surface of thymidylate synthase and affects hydride transfer at the interior active site. J Am Chem Soc. 2013;135:7583–7592. doi: 10.1021/ja400761x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanaan N, Ferrer S, Marti S, Garcia-Viloca M, Kohen A, Moliner V. Temperature dependence of the kinetic isotope effects in thymidylate synthase. A theoretical study. J Am Chem Soc. 2011;133:6692–6702. doi: 10.1021/ja1114369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanaan N, Marti S, Moliner V, Kohen A. A quantum mechanics/molecular mechanics study of the catalytic mechanism of the thymidylate synthase. Biochemistry. 2007;46:3704–3713. doi: 10.1021/bi061953y. [DOI] [PubMed] [Google Scholar]
- 11.Kanaan N, Marti S, Moliner V, Kohen A. QM/MM study of thymidylate synthase: enzymatic motions and the temperature dependence of the rate limiting step. The journal of physical chemistry A. 2009;113:2176–2182. doi: 10.1021/jp810548d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swiderek K, Kohen A, Moliner V. The influence of active site conformations on the hydride transfer step of the thymidylate synthase reaction mechanism. Physical chemistry chemical physics: PCCP. 2015;17:30793–30804. doi: 10.1039/c5cp01239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Ferrer S, Moliner V, Kohen A. QM/MM calculations suggest a novel intermediate following the proton abstraction catalyzed by thymidylate synthase. Biochemistry. 2013;52:2348–2358. doi: 10.1021/bi400267q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroud RM, Finer-Moore JS. Conformational dynamics along an enzymatic reaction pathway: thymidylate synthase, “the movie”. Biochemistry. 2003;42:239–247. doi: 10.1021/bi020598i. [DOI] [PubMed] [Google Scholar]
- 15.Maley F, Pedersen-Lane J, Changchien L. Complete restoration of activity to inactive mutants of Escherichia coli thymidylate synthase: evidence that E. coli thymidylate synthase is a half-the-sites activity enzyme. Biochemistry. 1995;34:1469–1474. doi: 10.1021/bi00005a001. [DOI] [PubMed] [Google Scholar]
- 16.Johnson EF, Hinz W, Atreya CE, Maley F, Anderson KS. Mechanistic characterization of Toxoplasma gondii thymidylate synthase (TS-DHFR)-dihydrofolate reductase. Evidence for a TS intermediate and TS half-sites reactivity. The Journal of biological chemistry. 2002;277:43126–43136. doi: 10.1074/jbc.M206523200. [DOI] [PubMed] [Google Scholar]
- 17.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 18.Dev IK, Yates BB, Leong J, Dallas WS. Functional role of cysteine-146 in Escherichia coli thymidylate synthase. Proc Natl Acad Sci U S A. 1988;85:1472–1476. doi: 10.1073/pnas.85.5.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaPat-Polasko L, Maley GF, Maley F. Properties of bacteriophage T4 thymidylate synthase following mutagenic changes in the active site and folate binding region. Biochemistry. 1990;29:9561–9572. doi: 10.1021/bi00493a010. [DOI] [PubMed] [Google Scholar]
- 20.Saxl RL, Maley GF, Hauer CR, Maccoll R, Changchien L, Maley F. Significance of mutations on the structural perturbation of thymidylate synthase: implications for their involvement in subunit exchange. Protein Sci. 2007;16:1439–1448. doi: 10.1110/ps.062509807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stout TJ, Sage CR, Stroud RM. The additivity of substrate fragments in enzyme ligand binding. Structure. 1998;6:839–848. doi: 10.1016/S0969-2126(98)00086-0. [DOI] [PubMed] [Google Scholar]
- 22.Sapienza PJ, Falk BT, Lee AL. Bacterial Thymidylate Synthase Binds Two Molecules of Substrate and Cofactor without Cooperativity. J Am Chem Soc. 2015;137:14260–14263. doi: 10.1021/jacs.5b10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igumenova TI, Frederick KK, Wand AJ. Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution. Chemical reviews. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasinath V, Sharp KA, Wand AJ. Microscopic insights into the NMR relaxation based protein conformational entropy meter. J Am Chem Soc. 2013;135:15092–15100. doi: 10.1021/ja405200u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wider G, Dreier L. Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc. 2006;128:2571–2576. doi: 10.1021/ja055336t. [DOI] [PubMed] [Google Scholar]
- 28.Freire E, Schon A, Velazquez-Campoy A. Isothermal titration calorimetry: general formalism using binding polynomials. Methods Enzymol. 2009;455:127–155. doi: 10.1016/S0076-6879(08)04205-5. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in molecular biology. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 31.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. Journal of biomolecular NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakomek NA, Kaufman JD, Stahl SJ, Louis JM, Grishaev A, Wingfield PT, Bax A. Internal dynamics of the homotrimeric HIV-1 viral coat protein gp41 on multiple time scales. Angewandte Chemie. 2013;52:3911–3915. doi: 10.1002/anie.201207266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakomek NA, Ying J, Bax A. Measurement of (1)(5)N relaxation rates in perdeuterated proteins by TROSY-based methods. Journal of biomolecular NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tugarinov V, Kay LE. Relaxation rates of degenerate 1H transitions in methyl groups of proteins as reporters of side-chain dynamics. J Am Chem Soc. 2006;128:7299–7308. doi: 10.1021/ja060817d. [DOI] [PubMed] [Google Scholar]
- 35.Cho CH, Urquidi J, Singh S, Robinson GW. Thermal offset viscosities of liquid H2O, D2O, and T2O. Journal of Physical Chemistry B. 1999;103:1991–1994. [Google Scholar]
- 36.Sun H, Kay LE, Tugarinov V. An optimized relaxation-based coherence transfer NMR experiment for the measurement of side-chain order in methyl-protonated, highly deuterated proteins. The journal of physical chemistry B. 2011;115:14878–14884. doi: 10.1021/jp209049k. [DOI] [PubMed] [Google Scholar]
- 37.Tugarinov V, Sprangers R, Kay LE. Probing side-chain dynamics in the proteasome by relaxation violated coherence transfer NMR spectroscopy. J Am Chem Soc. 2007;129:1743–1750. doi: 10.1021/ja067827z. [DOI] [PubMed] [Google Scholar]
- 38.Ishima R, Petkova AP, Louis JM, Torchia DA. Comparison of methyl rotation axis order parameters derived from model-free analyses of (2)H and (13)C longitudinal and transverse relaxation rates measured in the same protein sample. J Am Chem Soc. 2001;123:6164–6171. doi: 10.1021/ja0104711. [DOI] [PubMed] [Google Scholar]
- 39.Fisher HF. Detecting “silent” allosteric coupling. Methods in molecular biology. 2012;796:71–96. doi: 10.1007/978-1-61779-334-9_5. [DOI] [PubMed] [Google Scholar]
- 40.Duan GL, Smith VH, Weaver DF. Characterization of aromatic-thiol pi-type hydrogen bonding and phenylalanine-cysteine side chain interactions through ab initio calculations and protein database analyses. Mol Phys. 2001;99:1689–1699. [Google Scholar]
- 41.Pogolotti AL, Jr, Weill C, Santi DV. Thymidylate synthetase catalyzed exchange of tritiumfrom [5-3H]-2′-deoxyuridylate for protons of water. Biochemistry. 1979;18:2794–2798. doi: 10.1021/bi00580a016. [DOI] [PubMed] [Google Scholar]
- 42.Rutenber EE, Stroud RM. Binding of the anticancer drug ZD1694 to E. coli thymidylate synthase: assessing specificity and affinity. Structure. 1996;4:1317–1324. doi: 10.1016/s0969-2126(96)00139-6. [DOI] [PubMed] [Google Scholar]
- 43.Hyatt DC, Maley F, Montfort WR. Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate. Biochemistry. 1997;36:4585–4594. doi: 10.1021/bi962936j. [DOI] [PubMed] [Google Scholar]
- 44.Platzer G, Okon M, McIntosh LP. pH-dependent random coil (1)H, (13)C, and (15)N chemical shifts of the ionizable amino acids: a guide for protein pK a measurements. Journal of biomolecular NMR. 2014;60:109–129. doi: 10.1007/s10858-014-9862-y. [DOI] [PubMed] [Google Scholar]
- 45.Sapienza PJ, Lee AL. Backbone and ILV methyl resonance assignments of E. coli thymidylate synthase bound to cofactor and a nucleotide analogue. Biomolecular NMR assignments. 2014;8:195–199. doi: 10.1007/s12104-013-9482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp KA, O'Brien E, Kasinath V, Wand AJ. On the relationship between NMR-derived amide order parameters and protein backbone entropy changes. Proteins. 2015;83:922–930. doi: 10.1002/prot.24789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 48.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci U S A. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. Journal of molecular biology. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Law AB, Fuentes EJ, Lee AL. Conservation of side-chain dynamics within a protein family. J Am Chem Soc. 2009;131:6322–6323. doi: 10.1021/ja809915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee AL, Kinnear SA, Wand AJ. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nature structural biology. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- 52.Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: modulation of dynamics within a structural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sapienza PJ, Mauldin RV, Lee AL. Multi-timescale dynamics study of FKBP12 along the rapamycin-mTOR binding coordinate. Journal of molecular biology. 2010;405:378–394. doi: 10.1016/j.jmb.2010.10.037. [DOI] [PubMed] [Google Scholar]
- 54.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clarkson MW, Lee AL. Long-range dynamic effects of point mutations propagate through side chains in the serine protease inhibitor eglin c. Biochemistry. 2004;43:12448–12458. doi: 10.1021/bi0494424. [DOI] [PubMed] [Google Scholar]
- 56.Hu H, Clarkson MW, Hermans J, Lee AL. Increased rigidity of eglin c at acidic pH: evidence from NMR spin relaxation and MD simulations. Biochemistry. 2003;42:13856–13868. doi: 10.1021/bi035015z. [DOI] [PubMed] [Google Scholar]
- 57.Whitley MJ, Lee AL. Exploring the role of structure and dynamics in the function of chymotrypsin inhibitor 2. Proteins. 2011;79:916–924. doi: 10.1002/prot.22930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitley MJ, Zhang J, Lee AL. Hydrophobic core mutations in CI2 globally perturb fast side-chain dynamics similarly without regard to position. Biochemistry. 2008;47:8566–8576. doi: 10.1021/bi8007966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL. Evaluation of energetic and dynamic coupling networks in a PDZ domain protein. Journal of molecular biology. 2006;364:337–351. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 60.McDonald LR, Whitley MJ, Boyer JA, Lee AL. Colocalization of fast and slow timescale dynamics in the allosteric signaling protein CheY. Journal of molecular biology. 2013;425:2372–2381. doi: 10.1016/j.jmb.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fauman EB, Rutenber EE, Maley GF, Maley F, Stroud RM. Water-mediated substrate/product discrimination: the product complex of thymidylate synthase at 1.83 A. Biochemistry. 1994;33:1502–1511. doi: 10.1021/bi00172a029. [DOI] [PubMed] [Google Scholar]
- 62.Falk BT, Sapienza PJ, Lee AL. Chemical shift imprint of intersubunit communication in a symmetric homodimer. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1604748113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.