Abstract
Background
The monitoring of vectors is one of the key surveillance measures to assess the risk of arbovirus transmission and the success of control strategies in endemic regions. The recent re-emergence of Zika virus (ZIKV) in the tropics, including Singapore, emphasizes the need to develop cost-effective, rapid and accurate assays to monitor the virus spread by mosquitoes. As ZIKV infections largely remain asymptomatic, early detection of ZIKV in the field-caught mosquitoes enables timely implementation of appropriate mosquito control measures.
Results
We developed a rapid, sensitive and specific real-time reverse transcription polymerase chain reaction (rRT-PCR) assay for the detection of ZIKV in field-caught mosquitoes. The primers and PCR cycling conditions were optimized to minimize non-specific amplification due to cross-reactivity with the genomic material of Aedes aegypti, Aedes albopictus, Culex quinquefasciatus, Culex tritaeniorhynchus, Culex sitiens and Anopheles sinensis, as well as accompanying microbiota. The performance of the assay was further evaluated with a panel of flaviviruses and alphaviruses as well as in field-caught Ae. aegypti mosquitoes confirmed to be positive for ZIKV. As compared to a probe-based assay, the newly developed assay demonstrated 100% specificity and comparable detection sensitivity for ZIKV in mosquitoes.
Conclusions
Being a SYBR Green-based method, the newly-developed assay is cost-effective and easy to adapt, thus is applicable to large-scale vector surveillance activities in endemic countries, including those with limited resources and expertise. The amplicon size (119 bp) also allows sequencing to confirm the virus type. The primers flank relatively conserved regions of ZIKV genome, so that, the assay is able to detect genetically diverse ZIKV strains. Our findings, therefore, testify the potential use of the newly-developed assay in vector surveillance programmes for ZIKV in endemic regions.
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2373-4) contains supplementary material, which is available to authorized users.
Keywords: Zika virus, RT-PCR, Vector, Diagnostics, Surveillance
Background
Zika virus (ZIKV) is an arbovirus that was first isolated in 1947 from a rhesus monkey in Uganda and was subsequently identified in Aedes africanus mosquitoes in 1948 [1]. Although the first authentic ZIKV infection in humans was described in early 1960s [2], ZIKV remained dormant until 2007 when it re-emerged in Micronesia [3]. The re-emergence has caused a new wave of epidemics in Africa, Asia, islands of the tropical Pacific Ocean and the Americas [4]. Despite being associated with complications such as microcephaly and Guillain-Barré syndrome, a large proportion of ZIKV infections are asymptomatic and self-limiting [4], thereby, is silently transmitted and underreported. ZIKV is transmitted primarily through the bites of Aedes mosquitoes, which also transmit dengue (DENV) and chikungunya viruses [5]. As Aedes mosquitoes are widely distributed, there is an imminent threat of recurrent ZIKV epidemics, especially in the tropics and subtropics.
Singapore reported the first indigenous outbreak of ZIKV in August 2016 [6]. Due to the widespread presence of Aedes mosquitoes, high human density and lack of population exposure to ZIKV, there was a substantial threat of an island wide epidemic and sustained transmission. Vector control is the cornerstone in controlling ZIKV transmission, and the National Environment Agency, Singapore stepped up the vector surveillance and control activities to avert further virus spread in the country. One of the main components of the vector surveillance activities was to screen vector populations for the presence of ZIKV in order to monitor the risk of sustained transmission and to prioritize resource allocation for vector control operations.
The laboratory detection of ZIKV mainly relies on the detection of virus RNA by molecular methods such as polymerase chain reaction (PCR) and related technologies; e.g. loop mediated isothermal amplification [7–11]. The majority of available assays are probe-based real-time reverse transcription PCR (rRT-PCR) methods. While probe-based rRT-PCR protocols tend to be more sensitive and specific than SYBR Green-based protocols, the latter is more cost-effective and easier to adopt from published primer sequences. This makes SYBR Green-based methods one of the preferred choices for PCR-based mass screening purposes in disease surveillance programmes, especially in resource-poor settings that also lack expertise in assay design and optimisation. Moreover, SYBR Green-based methods allow the amplification of longer PCR fragments than probe-based protocols, thus enable sequencing a reasonable length of genomic fragments to affirm the pathogen/s, especially in non-endemic regions. Nevertheless, only a handful of SYBR Green-based RT-PCR methods have been published for the detection of ZIKV RNA [8, 11].
One of the main limitations in using the available methods for the vector surveillance is that they have primarily been designed for ZIKV detection in human biological samples [9, 12, 13]. The molecular assays optimised for human biological samples may not necessarily perform in a similar manner in field-caught mosquito samples, potentially due to complex nature of inherent and microbiota genomic composition that may affect the assay specificity [14]. None of the methods described thus far, except for that of Faye et al. [10], has compared the assay performance in field-caught mosquitoes. In order to support ZIKV surveillance in mosquito populations, we developed a SYBR Green-based RT-PCR assay optimized for the detection of ZIKV in mosquitoes and compared its performance with one of the probe-based assays [9] in field-caught mosquitoes.
Methods
Designing of primers
Whole genome sequences of all genotypes of ZIKV were retrieved from the GenBank nucleotide database and aligned using the BioEdit software [15]. The alignment was used to map already-published ZIKV-specific primers [9] and to design new primers in adjoining upstream and downstream conserved regions overlapping the membrane and envelope genes (Table 1).
Table 1.
Details of the primer sequences and combinations evaluated for the assay development
| Primer name | Sequence (5′-3′) | Genome positiona |
|---|---|---|
| Forward primers | ||
| ZIKV-Fb | TTGGTCATGATACTGCTGATTGC | 941–963 |
| ZIKV-F2 | TCAACGAGCCAAAAAGTCAT | 917–936 |
| ZIKV-F3 | ATCAGGTGCATAGGAGTCAG | 977–996 |
| Reverse primers | ||
| ZIKV-Rb | CCTTCCACAAAGTCCCTATTGC | 996–1016 |
| ZIKV-R3 | CAGGTCCCACCTGACATGC | 1017–1035 |
| ZIKV-R4 | CAACGACTGTCCGAAGCCATG | 1177–1197 |
aGenome positions were calculated from the first base of the 5′ non-translated region of the ZIKV reference sequence in GenBank nucleotide database (Accession no. NC012532.1)
bPrimers previously used for a probe-based rRT-PCR protocol published by Lanciotti et al. [9]
Viruses
ZIKV strains ATCC® VR-84 (GenBank: NC012532.1) and PLCal_ZV (GenBank: KF993678) provided by Public Health Agency of Canada [16] were used to prepare viral stocks. Viral stocks were prepared by inoculating each isolate into Vero cells (African Green Monkey Kidney) in M199 growth medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum, 1% L-glutamine solution and 100 mM Penicillin/Streptomycin (Sigma-Aldrich, St. Louis, MO, USA). After five days of propagation, viral titres were quantified by using the plaque assay performed on BHK-21 cells as described elsewhere [17].
Extraction of viral RNA and mosquito-derived RNA
Viral RNA was extracted from known titres of viral stocks by using the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. All primers were initially optimised using ATCC® VR-84 RNA for the amplification efficiency and annealing temperature.
Virus stocks of both ATCC® VR-84 and PLCal_ZV strains were 9-fold diluted to obtain virus titres from 104 to 10−4 PFU/ml. Pools (five mosquitoes per pool) of laboratory-strains of Aedes aegypti, Aedes albopictus and Culex quinquefasciatus mosquitoes in 1× phosphate-buffered saline were spiked with serial dilutions of viruses. In order to evaluate the primer cross-reactivity with RNA derived from wild mosquitoes, we included field-caught Ae. aegypti, Ae. albopictus, Culex tritaeniorhynchus, Culex sitiens and Anopheles sinensis adult mosquitoes that were screened negative for flaviviruses, using a pan-flavivirus PCR assay [18]. These samples were not spiked with ZIKV RNA before RNA extraction. Spiked and non-spiked mosquito samples were homogenized in a mixer mill (Retsch, Haan, Germany) as described before [19]. RNA was extracted from 140 μl of each homogenate by using the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. The quantification of RNA extracted from non-spiked field-caught mosquitoes is given in Additional file 1: Table S1. The amount of mosquito-derived RNA varied based on the species as well as tissues used (whole mosquitoes versus head and thorax only) and reached levels similar to RNA extracted from virus cultures (> 1000 ng/ml). Forty units of RNaseOUT (Life Technologies, Carlsbad, CA, USA) were added to each elute to minimize RNA degradation. RNA extracted from non-spiked samples was mixed with an equal volume of ZIKV (PLCal_ZV strain, 104 PFU/ml) RNA to test for the cross-reactivity of primers in the presence of genomic material derived from mosquitoes.
Collection of field mosquitoes in a ZIKV cluster
Adult mosquitoes were caught using Gravitraps [20] between epidemiological week (EW) 34 of 2016 and EW12 of 2017 to monitor ZIKV transmission in the first zika fever cluster in Singapore [6]. The field-caught mosquitoes were first identified to species level before being excised individually into two parts: abdomen and head/thorax. One to five abdomens were pooled before the extraction of RNA using the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The mosquito pools were screened for ZIKV by using a probe-base rRT-PCR assay as described elsewhere [9]. The head and thorax samples of mosquitoes in positive abdomen pools were screened individually by using the same rRT-PCR assay to determine the extact location of virus transmission.
SYBR green-based real time RT-PCR assay
One-step rRT-PCR assay was carried out using QuantiTect SYBR Green RT-PCR Kit (Qiagen, Hilden, Germany) in a 20 μl reaction consisting of 5 μl of extracted ZIKV RNA and 1 μl of each primer mix (Table 2) in a LightCycler® 2.0 (Roche Diagnostics GmbH, Mannheim, Germany) machine. The following thermal profile was used: reverse transcription at 50 °C for 20 min and inactivation at 95 °C for 15 min, followed by 40 cycles of 94 °C for 15 s, 52–55 °C for 30 s and 72 °C for 30 s. The exact annealing temperatures used for each primer pair is given in Table 2. A melting curve analysis was performed at 65 °C for 15 s followed by a cooling step at 37 °C for 20 s to verify the accuracy of amplified products. The amplicon sizes were confirmed by 2% agarose gel electrophoresis. A standard curve for the quantification of viral titres was obtained by plotting quantification cycle (Cq) values against known virus titres for each dilution. The amplification efficiency and coefficient of determination (R2) were calculated using Microsoft Excel (Microsoft, Redmond, WA, USA).
Table 2.
Primer combinations evaluated during the assay development
| Primer combination | Amplification success | Product size (bp) |
|---|---|---|
| F + R | Successful amplification; annealing at 55 °C | 76 |
| F + R3 | Successful amplification; annealing at 55 °C | 95 |
| F + R4 | Successful amplification; annealing at 55 °C | 257 |
| F2 + R | Successful amplification; annealing at 52 °C | 100 |
| F2 + R3 | Successful amplification; annealing at 52 °C | 119 |
| F2 + R4 | Successful amplification; annealing at 52 °C | 281 |
| F3 + R | Failed amplification | 40 |
| F3 + R3 | Failed amplification | 59 |
| F3 + R4 | Successful amplification; annealing at 52 °C | 221 |
Performance parameters of the assay
The specificity of the assay for ZIKV was determined by using RNA extracted from a panel of flaviviruses; DENV-1 (GenBank: KM403575), DENV-2 (GenBank: KR779784), DENV-3 (GenBank: KR779787), DENV-4 (GenBank: KR779790), Japanese encephalitis virus (Nakayama Strain, courtesy of Dr. Y.C. Chan), West Nile virus, Kunjin virus, yellow fever virus (17D vaccine strain), and alphaviruses; Ross River virus, Sindbis virus and chikungunya virus (GenBank: KY883764).
Results
Assay design and optimisation
The published [9] and newly designed primers were permutated to evaluate the performance of primer mixes that generate different amplicon sizes. Of nine primer mixes tested, only seven pairs were able to yield successful amplification (Table 2). We further evaluated the sensitivity and specificity of four different primer pairs (ZIKV-F + R3, F + R4, F2 + R3, and F3 + R4) that amplified a range of amplicon sizes (76–257 bp). Longer amplicon sizes were preferred because of their suitability for sequencing to determine virus lineages.
To determine the assay specificity in the presence of mosquito RNA, the four different primer pairs (ZIKV-F + R3, F + R4, F2 + R3, and F3 + R4) were first tested with RNA extracted from mosquito pools spiked with ZIKV (Table 3) and pools that were not spiked with ZIKV RNA (Additional file 2: Table S2). This step also determined any background signal generated due to primer cross-reactivity with mosquito-derived RNA. Among the four combinations tested, ZIKV-F + R4 and ZIKV-F3 + R4 primer pairs demonstrated substantial cross-reactivity (Additional file 3: Figure S1a, b). Therefore, ZIKV-F + R3 (Additional file 3: Figure S1c) and ZIKV-F2 + R3 (Fig. 1a) combinations were shortlisted for further evaluation of the cross-reactivity and specificity in a panel of flavivirus and alphavirus RNA as well as with mosquito-derived RNA. None of the eight flaviviruses tested gave positive results for both primer pairs (Fig. 1b; Additional file 3: Figure S1d). However, two alphaviruses, namely Sindbis virus (Tm = 80.07, Cq = 30.24) and Ross River virus (Tm = 80.02, Cq = 27.09) gave positive signals for ZIKA-F + R3 (Additional file 3: Figure S1d), but not for the ZIKV-F2 + R3 primer mix (Fig. 1b). Therefore, ZIKV-F2 + R3 primer pair was selected for the further evaluation.
Table 3.
Comparison of the sensitivity of ZIKV detection between a probe-based rRT-PCR assay and the SYBR Green-based assay utilising the primer combination F2 + R3
| ZIKV strainb | Mosquito species | Detection threshold of virus titre (PFU/ml) | |
|---|---|---|---|
| Probe-based rRT-PCRa | SYBR Green-based rRT-PCR | ||
| ATCC® VR-84 | Aedes aegypti | 100 | 10 |
| Aedes albopictus | 10 | 1 | |
| Culex quinquefasciatus | 10 | 100 | |
| PLCal_ZV | Aedes aegypti | 1 | 1 |
| Aedes albopictus | 0.1 | 0.1 | |
| Culex quinquefasciatus | 10 | 10 | |
aThe assay protocol has previously been described by Lanciotti et al. [9]
bZIKV strain ATCC® VR-84 is the prototype strain isolated in Uganda in 1947 (NCBI accession No. NC012532.1). The strain belonging to African genotype. PLCal_ZV strain was isolated in a Canadian traveller from Thailand in 2013 (NCBI accession No. KF993678). PLCal_ZV belongs to the Asian genotype
Fig. 1.
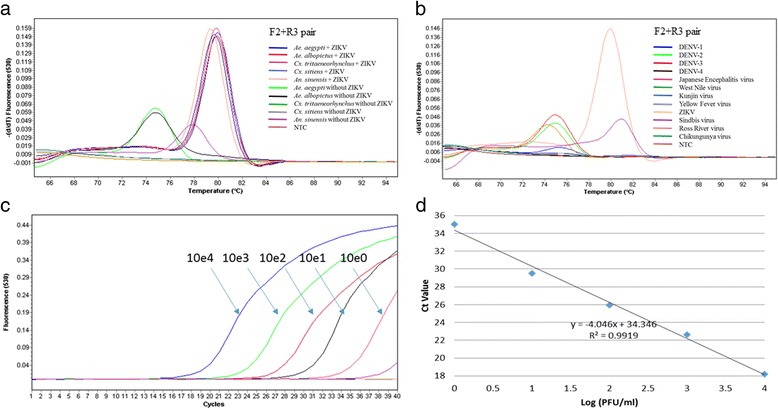
Performance of the rRT-PCR assay utilising the primer combination F2 + R3. a Specificity of the assay in the presence of mosquito RNA. The mosquito RNA was spiked with a 5-fold dilution (102–10-2 PFU/ml) of ZIKV RNA, to determine any primer cross-reactivity with mosquito-derived genomic material. b Specificity of the assay in a panel of flaviviruses and alphaviruses. The melting peak for ZIKV ranged between 80.3–80.4 °C, while that of primer dimers ranged between 73.0–75.8 °C. c Amplification curves for 10-fold serial dilutions (104–10-4 PFU/ml) of ZIKV. d Standard curve for the quantification of ZIKV infectious virus particles. The curve was generated by plotting the quantification cycle (Cq) values against the log10 of infectious virus particles (PFU/ml). Only the detection range is shown. RNA from ZIKV isolate PLCal_ZV was used in the analysis
Evaluation of sensitivity
In the absence of mosquito RNA, the detection sensitivity of the assay ranged between 104 and 100 PFU/ml (Fig. 1c). The Cq values of ZIKV-F2 + R3 primer pair showed a linear curve (R 2 = 0.99) for the detection of ZIKV (Fig. 1d) and the standard curve generated allows quantification of infectious virus particles based on Cq values. In order to determine the ZIKV detection sensitivity in the presence of mosquito RNA, we used a 5-fold serial dilution (102–10-2 PFU/ml) of the total RNA extracted from pools of mosquitoes spiked with ZIKV (104 PFU/ml). The sensitivity of detection of ZIKV RNA in the presence of Ae. aegypti, Ae. albopictus and Cx. quinquefasciatus RNA was 10, 1, and 100 PFU/ml, respectively, indicating at least one log drop of sensitivity in the presence of mosquito RNA (Table 3). Of two reference strains tested, the assay generated better detection sensitivity for the Asian genotype of ZIKV (PLCal_ZV) (Table 3), which is attributable to the recent emergence of ZIKV in the Americas and Asia [4]. Overall, the detection sensitivity of the newly developed assay for ZIKV in mosquitoes was comparable to that of a probe-based assay described previously [9] (Table 3).
Evaluation of the assay in ZIKV infected field caught mosquitoes
To determine the applicability of the SYBR Green rRT-PCR assay for ZIKV surveillance in field-caught mosquitoes during disease outbreaks, we screened a panel of Ae. aegypti mosquitoes collected from the first local ZIKV outbreak cluster [6] (Table 4). The panel included seven head/thorax samples that were confirmed to be positive for ZIKV by the probe-based assay described by Lanciotti et al. [9]. These ZIKV-positive mosquitoes were collected as part of the vector surveillance activities carried out during the outbreak. We also included a similar number of ZIKV-negative mosquito RNA samples to determine the specificity. The results, summarized in Table 4, showed that the SYBR Green assay was 100% specific and produced either an equal or better sensitivity profile as compared to the probe-based assay. The results also testified the absence of cross-reactivity with the genomic material of field-caught mosquitoes (Additional file 4: Figure S2).
Table 4.
Evaluation of SYBR Green-based rRT-PCR assay performance in ZIKV infected field caught mosquitoes confirmed by a probe-based rRT-PCR assay
| Sample IDb | Probe-based rRT-PCRa | SYBR Green-based rRT-PCR | ||
|---|---|---|---|---|
| Cq value | Result | Cq value (Tm) | Result | |
| HT55 | 21.06 | Positive | 17.57 (80.38) | Positive |
| HT66 | 19.54 | Positive | 15.23 (80.24) | Positive |
| HT67 | 28.77 | Positive | 26.06 (80.16) | Positive |
| HT68 | 29.19 | Positive | 26.98 (80.22) | Positive |
| HT74 | 17.09 | Positive | 13.74 (80.42) | Positive |
| HT81 | 19.66 | Positive | 16.33 (80.1) | Positive |
| HT83 | 33.23 | Positive | 35 (80.04) | Positive |
| Ae. albopictus | Not available | Negative | Not available | Negative |
| Ae. albopictus | Not available | Negative | Not available | Negative |
| Culex spp. | Not available | Negative | Not available | Negative |
| Culex spp. | Not available | Negative | Not available | Negative |
| Ae. aegypti | Not available | Negative | Not available | Negative |
| Ae. aegypti | Not available | Negative | Not available | Negative |
| Ae. aegypti | Not available | Negative | Not available | Negative |
| ZIKV positive control (ATCC® VR-84) | 16.77 | Positive | 13.84 (79.88) | Positive |
| Negative control | Not available | Negative | Not available | Negative |
Abbreviations: C q quantification cycle, T m melting temperature
aThe assay protocol has previously been described by Lanciotti et al. [9]
bSample IDs from HT55 to HT83 are Ae. aegypti specimens collected from a zika fever cluster during the first outbreak of ZIKV in Singapore in August 2016 [6]. They were confirmed to be ZIKV-positive by the probe-based rRT-PCR assay. ZIKV negative mosquito specimens collected from the field, including the same cluster, are included for the comparison of specificity
Discussion
Screening of both human and vector populations for the presence of pathogens is equally important to vector-borne disease surveillance programmes. First, the data obtained from vector populations enables the confirmation of autochthonous transmission, in the event of emergence/re-emergence of non-endemic pathogens. The data also helps to monitor the subsequent expansion of pathogen transmission. The longitudinal monitoring of vector populations, especially in disease clusters, signals whether the transmission is sustained or interrupted, and thereby the successfulness of control measures in place.
Pathogen-specific molecular assays are unarguably applicable to both human and vector samples. However, as compared to human biological samples, vector specimens tend to generate higher “background genetic noise” due to the presence of host as well as other related organisms in the microbiome. The potential “cross-talk” between the primers and non-targeted template sites may influence the sensitivity, specificity and efficiency of PCR amplification. Therefore, assays optimized for human samples may need to be modified, re-developed and evaluated before being used for pathogen surveillance in vector populations. This is notably applicable to assays with detection mechanisms that tend to be less specific. For example, SYBR Green intercalates with any double-stranded DNA product and emits a positive signal. Consequently, non-specific amplifications may be captured as false positives. This is in contrast to probe-based assays that require the template annealing of a specific, short genomic sequence to generate a positive signal, thereby maintaining a high level of specificity. Therefore, SYBR Green rRT-PCR assays need extra diligence in primer designing and protocol optimization to achieve comparable performance to probe-based assays [21, 22]. As demonstrated in the present study, we tested several combinations of primers that were either adapted from a previously published protocol [9] or newly-designed within the same conserved region of ZIKV genome. Many of them generated non-specific, background noise that was difficult to be differentiated from a true signal in the SYBR Green assay format. However, ZIKV-F2 + R3 primer combination exhibited negligible level of cross-reactivity with a panel of different mosquito species, alphaviruses and flaviviruses, while maintaining detection sensitivity comparable to a known probe-based assay for ZIKV [9].
Despite the common drawback of being less-specific than probe-based assays, SYBR Green technology is widely used as a cheap alternative for the rRT-PCR-based pathogen detection [21]. The cost benefit of SYBR Green approach justifies its applicability in mass screening exercises, such as surveillance programmes. The newly-developed SYBR Green assay showed 100% sensitivity and specificity in a panel of ZIKV-positive mosquitoes collected from a disease cluster, demonstrating its suitability for vector surveillance programmes. Importantly, the assay demonstrated higher sensitivity of detection for the Asian genotype of ZIKV, which is attributed to the recent emergence of zika fever in the Americas and Asia [4]. Even though the assay was optimized by using two ZIKV strains of both Asian and African genotypes, the primers flank relatively conserved regions of ZIKV genome, indicating the universal applicability of our assay for the detection of genetically diverse ZIKV strains. Our findings, therefore, support the potential use of newly-developed assay for the screening of field-caught mosquitoes as part of ZIKV surveillance programmes in endemic regions.
Conclusions
Recent emergence and inter-continental spread of ZIKV remind us of the need for cost-effective, specific, sensitive and technically less-demanding molecular assays to monitor human and vector populations in vulnerable regions. Rapid detection of ZIKV in the field collected mosquitoes assists in optimizing resource allocation for vector control and monitoring the control success. Because ZIKV co-circulates in regions where both dengue and chikungunya viruses are endemic, molecular assays that perform optimally among human and vector specimens play an important role not only in an epidemiological perspective, but also in clinical management, because the infections due to these viruses are often clinically indistinguishable.
Additional files
Quantification of RNA extracted from field-caught mosquitoes. (DOCX 13 kb)
Cross-reactivity of different primer combinations in the presence/absence of RNA derived from Aedes spp., Culex spp. and Anopheles spp. mosquitoes. (DOCX 15 kb)
Melting curve analysis demonstrating the non-specific amplification by primer combinations F + R3, F + R4 and F3 + R4 in the presence of mosquito-derived RNA and specificity of F + R3 in a panel of flavivirus and alphavirus RNA. a. Melting peak analysis for the F + R4 primer pair b. Melting peak analysis for the F3 + R4 primer pair c. Melting peak analysis for the F + R3 primer pair. d. Melting peak analysis for the F + R3 primer pairs within a panel of flavivirus and alphavirus RNA. Abbreviation: NTC, Negative control. (TIFF 2225 kb)
Melting peak analysis for the rRT-PCR assay performance in field-caught ZIKV-infected mosquitoes. The evaluation is based on the primer combination F2 + R3. The melting peak for ZIKV infected mosquitoes (1–7) and ZIKV non-infected mosquito species (8–14) falls within the same range as the positive control (ATCC® VR-84). (TIFF 679 kb)
Acknowledgements
We thank colleagues at Environmental Health Institute, especially Dr. Li-Kiang Tan, for providing virus stocks, and Applied Entomology team for coordinating the field collection of mosquitoes and laboratory investigations.
Funding
This study was supported by the National Environmental Agency, Singapore. The funding sources of this study had no role in the study design, data collection, data analysis, data interpretation, writing of the report, or in the decision to submit the paper for publication.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional files. The whole genome sequence of the chikungunya virus isolate used in the assay evaluation was deposited in the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) under the accession number KY883764.
Abbreviations
- DENV
Dengue virus
- PFU
Plaque forming units
- rRT-PCR
real-time reverse transcription polymerase chain reaction
- ZIKV
Zika virus
Authors’ contributions
HCH conceptualised and coordinated the assay design. WPT designed, optimized the assay and performed evaluations. CSC supervised the entomological surveillance and morphological identification. WPT, GY, GL and SNC conducted the screening of field-caught mosquitoes for ZIKV. WPT, HCH, CSC and NLC wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2373-4) contains supplementary material, which is available to authorized users.
Contributor Information
Wei-Ping Tien, Email: tien_wei_ping@nea.gov.sg.
Gareth Lim, Email: gareth_lim@nea.gov.sg.
Gladys Yeo, Email: gladys_yeo@nea.gov.sg.
Suzanna Nicole Chiang, Email: suzanna_chiang@nea.gov.sg.
Chee-Seng Chong, Email: chong_chee_seng@nea.gov.sg.
Lee-Ching Ng, Email: ng_lee_ching@nea.gov.sg.
Hapuarachchige Chanditha Hapuarachchi, Email: chanditha_hapuarachchi@nea.gov.sg, Email: chandith@yahoo.com.
References
- 1.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Simpson DI. Zika virus infection in man. Trans R Soc Trop Med Hyg. 1964;58:335–338. doi: 10.1016/0035-9203(64)90201-9. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 4.Musso D, Gubler DJ. Zika virus. Clin Microbiol Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vorou R. Zika virus, vectors, reservoirs, amplifying hosts, and their potential to spread worldwide: what we know and what we should investigate urgently. Int J Infect Dis. 2016;48:85–90. doi: 10.1016/j.ijid.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Group SZS Outbreak of Zika virus infection in Singapore: an epidemiological, entomological, virological, and clinical analysis. Lancet Infect Dis. 2017;17:813–821. doi: 10.1016/S1473-3099(17)30249-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Yin F, Bi Y, Cheng G, Li J, Hou L, et al. Rapid and sensitive detection of Zika virus by reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2016;238:86–93. doi: 10.1016/j.jviromet.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Xu MY, Liu SQ, Deng CL, Zhang QY, Zhang B. Detection of Zika virus by SYBR green one-step real-time RT-PCR. J Virol Methods. 2016;236:93–97. doi: 10.1016/j.jviromet.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–9. [DOI] [PMC free article] [PubMed]
- 10.Faye O, Diallo D, Diallo M, Weidmann M, Sall AA. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol J. 2013;10:311. doi: 10.1186/1743-422X-10-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charrel RN, Leparc-Goffart I, Pas S, de Lamballerie X, Koopmans M, Reusken C. Background review for diagnostic test development for Zika virus infection. Bull World Health Organ. 2016;94:574–84D. doi: 10.2471/BLT.16.171207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faye O, Dupressoir A, Weidmann M, Ndiaye M, Alpha SA. One-step RT-PCR for detection of Zika virus. J Clin Virol. 2008;43:96–101. doi: 10.1016/j.jcv.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Gourinat AC, O'Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis. 2015;21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro MG, Nogueira RM, Filippis AM, Ferreira AA, Lima Mda R, Faria NR, et al. Dengue virus type 4 in Niteroi, Rio de Janeiro: the role of molecular techniques in laboratory diagnosis and entomological surveillance. Mem Inst Oswaldo Cruz. 2012;107:940–945. doi: 10.1590/S0074-02762012000700017. [DOI] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acid Symp Ser. 1999;41:95–98. [Google Scholar]
- 16.Fonseca K, Meatherall B, Zarra D, Drebot M, MacDonald J, Pabbaraju K, et al. First case of Zika virus infection in a returning Canadian traveler. Am J Trop Med Hyg. 2014;91:1035–1038. doi: 10.4269/ajtmh.14-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan LK, Lam S, Low SL, Tan FH, Ng LC, Teo D. Evaluation of pathogen reduction systems to inactivate dengue and chikungunya viruses in apheresis platelets suspended in plasma. Adv Infect Dis. 2013;3:1–9.
- 18.Yang CF, Chen CF, Su CL, Teng HJ, Lu LC, Lin C, et al. Screening of mosquitoes using SYBR green I-based real-time RT-PCR with group-specific primers for detection of flaviviruses and alphaviruses in Taiwan. J Virol Methods. 2010;168:147–51. [DOI] [PubMed]
- 19.Kek R, Hapuarachchi HC, Chung CY, Humaidi MB, Razak MA, Chiang S, et al. Feeding host range of Aedes albopictus (Diptera: Culicidae) demonstrates its opportunistic host-seeking behavior in rural Singapore. J Med Entomol. 2014;51:880–884. doi: 10.1603/ME13213. [DOI] [PubMed] [Google Scholar]
- 20.Lee C, Vythilingam I, Chong CS, Abdul Razak MA, Tan CH, Liew C, et al. C. Gravitraps for management of dengue clusters in Singapore. Am J Trop Med Hyg. 2013;88:888–892. doi: 10.4269/ajtmh.12-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tajadini M, Panjehpour M, Javanmard SH. Comparison of SYBR green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv Biomed Res. 2014;3:85. doi: 10.4103/2277-9175.127998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao H, Shockey JM. Comparison of TaqMan and SYBR green qPCR methods for quantitative gene expression in tung tree tissues. J Agric Food Chem. 2012;60:12296–12303. doi: 10.1021/jf304690e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of RNA extracted from field-caught mosquitoes. (DOCX 13 kb)
Cross-reactivity of different primer combinations in the presence/absence of RNA derived from Aedes spp., Culex spp. and Anopheles spp. mosquitoes. (DOCX 15 kb)
Melting curve analysis demonstrating the non-specific amplification by primer combinations F + R3, F + R4 and F3 + R4 in the presence of mosquito-derived RNA and specificity of F + R3 in a panel of flavivirus and alphavirus RNA. a. Melting peak analysis for the F + R4 primer pair b. Melting peak analysis for the F3 + R4 primer pair c. Melting peak analysis for the F + R3 primer pair. d. Melting peak analysis for the F + R3 primer pairs within a panel of flavivirus and alphavirus RNA. Abbreviation: NTC, Negative control. (TIFF 2225 kb)
Melting peak analysis for the rRT-PCR assay performance in field-caught ZIKV-infected mosquitoes. The evaluation is based on the primer combination F2 + R3. The melting peak for ZIKV infected mosquitoes (1–7) and ZIKV non-infected mosquito species (8–14) falls within the same range as the positive control (ATCC® VR-84). (TIFF 679 kb)
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its additional files. The whole genome sequence of the chikungunya virus isolate used in the assay evaluation was deposited in the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) under the accession number KY883764.


