Abstract
Interplay between DNA repair of the oxidatively modified base 8-oxo-7,8-dihydroguanine (OG) and transcriptional activation has been documented in mammalian genes. Previously, we synthesized OG into the VEGF potential G-quadruplex sequence (PQS) in the coding strand of a luciferase promoter to identify that base excision repair (BER) unmasked the G-quadruplex (G4) fold for gene activation. In the present work, OG was site-specifically synthesized into a luciferase reporter plasmid to follow the time-dependent expression in mammalian cells when OG in the VEGF PQS context was located in the coding vs template strands of the luciferase promoter. Removal of OG from the coding strand by OG glycosylase-1 (OGG1)-mediated BER upregulated transcription. When OG was in the template strand in the VEGF PQS context, transcription was downregulated by a BER-independent process. The time course changes in transcription show that repair in the template strand was more efficient than repair in the coding strand. Promoters were synthesized with an OG:A base pair that requires repair on both strands to yield a canonical G:C base pair. By monitoring the up/down luciferase expression, we followed the timing of repair of an OG:A base pair occurring on both strands in mammalian cells in which one lesion resides in a G-quadruplex loop and one in a potential i-motif. Depending on the strand in which OG resides, coding vs template, this modification is an up/downregulator of transcription that couples DNA repair with transcriptional regulation.
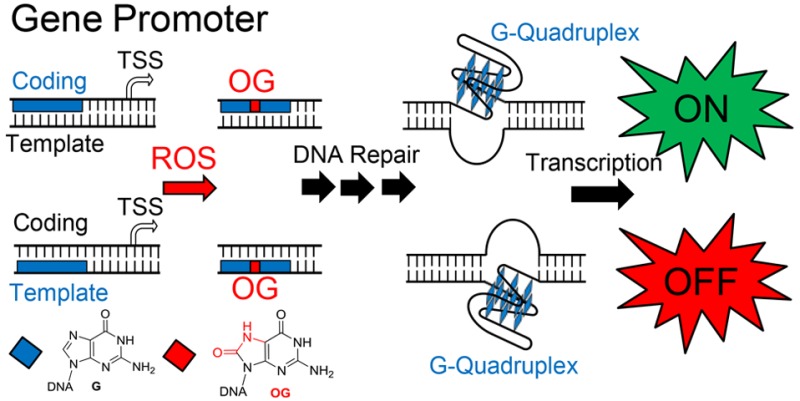
Oxidatively modified sites in DNA, such as the two-electron oxidation of guanine (G) to 8-oxo-7,8-dihydroguanine (OG, Scheme 1A), are targets for DNA repair.1,2 Recent reports have demonstrated an interplay between DNA repair and transcriptional regulation when OG resides in a gene promoter.3−9 However, many details regarding the process are poorly understood, providing opportunities for further inquiry. Herein, chemical synthesis has provided the ability to prepare plasmids with a site-specific OG modification in the promoter of a luciferase gene followed by transfection into mammalian cells to probe the coupling of DNA repair with transcriptional regulation. The OG-containing reporters allowed examination into the strand impact (i.e., nontranscribed or “coding” vs template), sequence context effects, and base pair partner impact (i.e., OG:C vs OG:A) on transcription. In the present studies, we discovered that the strand in which OG resides in a PQS context modulates the up/downregulation and time course of gene expression. The results provide further evidence for the modification of G to OG in DNA poising a gene for up- or down-expression depending on the context. These studies add support to a growing body of evidence and discussions that OG is an epigenetic-like modification to DNA.3,4,10−15
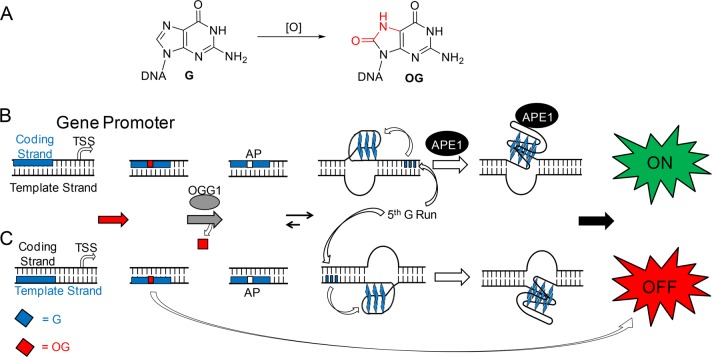
Scheme 1. Oxidation of G to OG in the VEGF Promoter PQS Can Turn Transcription On or Off.
(A) Scheme for oxidation of G to OG. (B) Oxidation of the VEGF PQS in the coding strand turns transcription on. (C) Oxidation of the VEGF PQS in the template strand turns transcription off.
Eukaryotic genes consist of a promoter, 5′-UTR, coding region comprised of exons and introns, and a 3′-UTR. Previous studies analyzed how introduction of the OG lesion site specifically in the coding region of a gene impacts transcription.7−9 When OG is in the template strand of a coding region paired with C in the opposite strand, initiation of base excision repair (BER) to yield an abasic site (AP) stalls progression of the transcription elongation complex, resulting in downregulation of mRNA synthesis.16 Alternatively, if OG is detected by the sensor protein CSB that initiates transcription-coupled nucleotide excision repair (TC-NER), mRNA synthesis is also downregulated.17,18 When OG is in the template strand in a coding region, the dominant repair pathway is TC-NER in mammals.19 In the absence of OG repair, the elongation complex can bypass the modification to yield the background level of mRNA.8,17 On the other hand, when OG is located in the coding strand of an exon paired with C in the opposite strand, OG is preferentially repaired by BER.7,17 Initiation of BER to yield a strand break stalls the transcription elongation complex leading to downregulation of mRNA synthesis7 or facilitates mRNA synthesis in G-rich sequence contexts capable of R loop formation.20 In the absence of BER, OG in the coding strand does not impact transcription. Local sequence differences influence the impact OG has on transcription,21 and the promoter strength can influence the magnitude by which OG in a coding region alters transcription.22 Key findings from these studies include a strand bias in the magnitude of change in transcription by OG and a strand dependency in the preferred DNA repair pathway utilized in mammalian cells for removal of OG. However, studies addressing how OG incorporated site specifically alters transcription in other gene regions, such as promoters, are limited. This is a fundamentally different question because OG in a promoter impacts the transcriptional preinitiation complex that can lead to coupling of DNA repair and initiation of transcription; in contrast, OG in a coding region impedes progression of the transcriptional elongation complex leading to stalling or truncated transcription.
Previous experiments to establish G oxidation to OG in gene promoters impacts transcription proceeded by inducing oxidative stress in cells and then utilizing a low-resolution sequencing method (e.g., ChIP-qPCR or immunofluorescence) to determine the possible presence of OG in a specific promoter.4,6,23 These approaches cannot uniquely identify OG from the >20 G oxidation products characterized.24,25 The possible cellular formation of OG in the promoter regions of the TNFα,4VEGF,26BCL2,6 and SIRT1(5) genes was observed in tandem with a ∼3-fold upregulation of transcription; further, these reports suggest a coupling of BER with transcriptional activation. Our studies on this topic used reporter plasmids with OG site-specifically synthesized into a gene promoter to unambiguously verify the impact on transcription, and to begin mapping the protein and DNA structure switching pathways involved in the activation process.3 Specifically, we synthesized OG into the regulatory VEGF promoter potential G-quadruplex sequence (PQS) in the coding strand to inspect how the modified base altered transcription of a reporter gene. We found the OG-modified system after 48 h transfection into mammalian cells induced transcription by ∼3-fold relative to an unmodified control.3 Additionally, the BER proteins OGG1 and APE1 were essential for gene activation, as was the ability of the sequence to adopt a G-quadruplex (G4) structure. Mechanistically, OGG1-mediated release of OG to yield a helix-destabilizing AP provides the thermodynamic drive for the PQS to switch structures to a G4 fold. This structure switching ability is possible because the VEGF PQS possesses a fifth G run (aka, “spare tire”) that replaces the damaged run by extruding it out of the G4 core and establishing a stable topology (Scheme 1B).27 The G4 fold with an AP site in a long loop allows APE1 to bind, but it stalls the endonuclease activity providing a context for APE1 to interact with transcriptional activating factors.5,28,29 Stalled APE1 activity resulting in the transcriptional change was supported by experiments with chemicals that inhibit the activity of APE1 and modified substrates that are bound but inefficiently cleaved by APE1.3 Our results demonstrate that OG in the coding strand of the VEGF promoter PQS induces transcription via adoption of a G4 fold, stalling the BER process for recruitment of activating factors.
In the present report, we took the next steps to explore what happens if the VEGF PQS containing OG is located in the template strand instead of the coding strand of the promoter. When flipping the OG and PQS to the template strand, we found transcription was significantly attenuated (Scheme 1C). This observation led to further studies to begin to understand the strand bias in DNA repair and determine how different base pairs of OG (OG:C vs OG:A) impact transcription. In the present experiments, the time-course transcription profiles were monitored to gauge the relative differences of OG repair and gene activation in three different mammalian cell lines. The results demonstrate that OG in the context of a PQS in a gene promoter can alter the transcriptional state of a gene by guiding DNA repair interactions with transcriptional machinery.
Results and Disscussion
Time-Dependent Profiles for OG-Containing Promoters
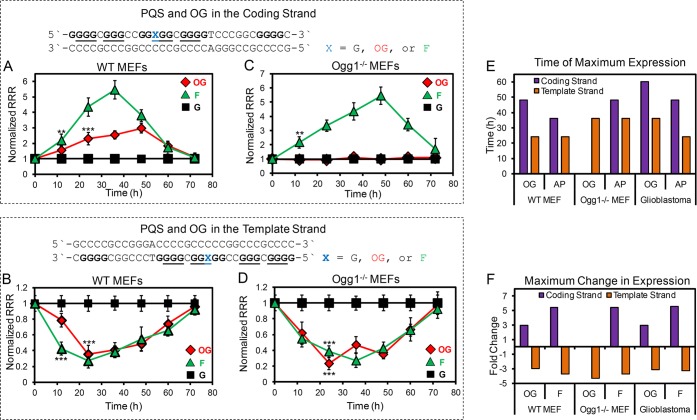
The site-specifically modified reporter plasmids were synthesized and verified following methods previously described by our laboratory.3,30 The G nucleotide modified in the VEGF PQS was previously found to be at a site sensitive to oxidation by inflammation-derived CO3•– to yield OG as a product.27 The modified reporter plasmid possesses two luciferase genes among which the Renilla luciferase gene had the chemically modified promoter while the firefly luciferase gene was not modified so as to be used as an internal standard for the quantitative studies presented. Transfection of the OG-modified plasmids into wild-type mouse embryonic fibroblasts (WT MEFs), Ogg1 knocked out MEFs (Ogg1–/– MEFs), and human glioblastoma (U87 MG) cells was conducted. Determination of the relative response ratios (RRR) is described in the Methods section. A time-course analysis from 12–72 h to determine the rate of Renilla luciferase expression when OG and the PQS were located in the coding strand (Figure 1A) or template strand (Figure 1B) relative to a control plasmid without the OG modification were first conducted in the WT MEF and glioblastoma (Figure S1) cells. In the coding strand, the presence of OG in the VEGF PQS (Figure 1A, red diamonds) showed a time-dependent increase in expression relative to cells transfected with the all-G VEGF PQS-containing plasmid utilized as a control to determine background levels of transcription (Figure 1A, black square). Expression was nearly 3-fold greater than the control experiments at 48 or 60 h post-transcription for the WT MEF or glioblastoma cells, respectively. These observations are consistent with our previous work.3 In contrast, placement of OG in the VEGF PQS in the template strand resulted in a time-dependent decrease in Renilla expression relative to the all-G control experiments (Figure 1B). The expression was maximally suppressed by nearly 70% at 24 and 36 h post transfection in the WT MEF and glioblastoma cells (Figure S1), respectively. These observations identify a strand dependency in the up- or downregulation of transcription induced by OG in the context of the promoter VEGF PQS.
Figure 1.
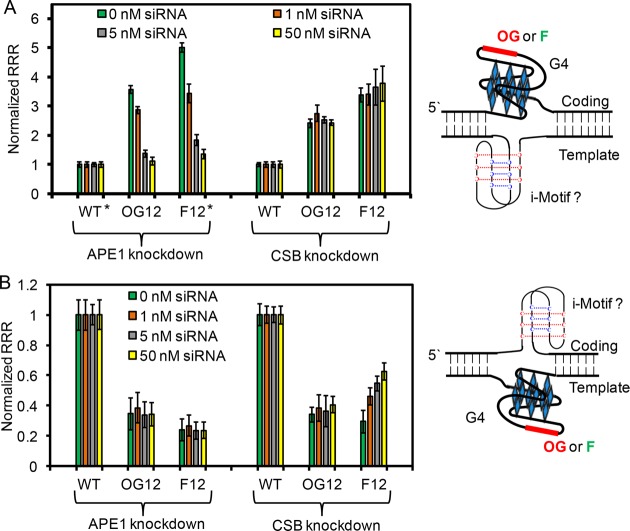
Time-dependent studies of the impact of G, OG, or an AP analog, F, on transcription when the sites of interest were located in the promoter VEGF PQS in either the coding or template strand of the promoter for the Renilla luciferase gene. Studies for the modification and PQS in the coding strand (A) or template strand (B) of the promoter for the Renilla luciferase gene reporter plasmid transfected in WT MEFs. Studies for the modification and PQS in the coding strand (C) or template strand (D) of the promoter for the Renilla luciferase gene reporter plasmid transfected in Ogg1–/– MEFs. (E) Time of maximal change in gene expression when OG or F were located in the gene promoter for the MEF and glioblastoma cell lines. (F) Maximum changes in expression levels observed when OG or F was located in the gene promoter relative to the native sequence control in all three cell lines. Determination of the RRR is described in the Methods section of the Supporting Information. On the basis of a Student’s t test, the time in which significance at **P < 0.01 or ***P < 0.001 was first observed is marked on each plot.
Our studies and those of others found that BER of OG initiated by the DNA glycosylase Ogg1 was the gatekeeper for setting off a cascade of events for transcriptional induction when operating in a gene promoter with the VEGF PQS.3−6 The studies in our laboratory found this phenomenon occurred when OG was located in the coding strand of the promoter.3 Therefore, time-dependent studies with the OG-modified plasmids in the two different strand orientations were conducted in Ogg1–/– MEFs to determine the importance of Ogg1 activity. Two interesting observations were made in these Ogg1-knockout cell experiments. First, when OG in the PQS was in the coding strand of the promoter, no increase in Renilla expression relative to the control was observed over the 72-h analysis. This result supports Ogg1 being essential for transcriptional induction when OG is present in the VEGF PQS context in the coding strand. From the DNA repair perspective, this null result in the knockout cells suggests that Ogg1-mediated BER is the dominant process for removal of OG in the coding strand of a gene promoter. Second, when OG was in the template strand, a 3-fold reduction in gene expression between WT and Ogg1–/– MEFS was observed (Figure 1B and D). Additionally, the time profile for the transcriptional change with OG in the template strand was similar between the WT and Ogg1–/– MEFS. This similar result in both MEF cell types means that OG in the template strand of a gene promoter is not predominantly repaired by BER and is likely corrected by TC-NER.19 The observation of a strand bias in the dominant DNA repair pathway in a gene promoter is consistent with the bias observed in gene coding regions.16−19 Additional experiments to understand the strand bias in repair are described below.
Time-Dependent Profiles for Promoters with Abasic Site Analogs
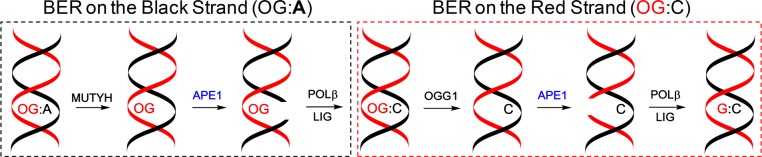
Base excision repair initiated by OGG1 in mammals removes OG when base paired with C. In the cellular context, the product of OG release by OGG1 has been proposed as either an AP resulting from monofunctional activity (i.e., glycosylase reaction only)7 or a nick in the backbone resulting from bifunctional activity (i.e., glycosylase and β-lyase reactions).31 Regardless of the product identity, the AP or nick are substrates for APE1 to yield a gap at the modification site.32,33 The gap is then filled with the correct nucleotide by POLβ and returned back to the duplex state via LIGIII-mediated ligation (Scheme 2).1,2 When DNA modifications are repaired by TC-NER, a multiprotein complex finds the lesion by the sensor protein CSB and then catalyzes the releases of a 25–30-mer single strand surrounding the site.19
Scheme 2. BER Pathway for Removal of OG from a Duplex DNA When Base Paired with A (Left) or C (Right).
First, to better understand the steps following release of OG by BER, an AP analog F was synthesized into the position of interest in either the coding or template strand in order to follow the time-course change in gene expression. In the WT cells (Figure 1A,B, and S1), the presence of F led to a 6-fold increase in transcription in the coding strand that was nearly 2-fold greater than OG in the same context; on the other hand, when F was in the template strand, it led to a 3-fold decrease in transcription with a similar magnitude as OG. In the Ogg1–/– MEFs, the presence of F in the coding strand allowed bypass of Ogg1, leading to a 6-fold increase in transcription at 48 h post-transfection (Figure 1C). This is consistent with our previous results and others that the AP in a structured context (i.e., G4 or hairpin) facilitates upregulation of transcription by an APE1-mediated process.3,5 Last, the presence of F in the template strand in the PQS context continued to give a similar decrease in transcription between the WT and Ogg1–/– MEFs with a similar time profile (Figure 1D). Results with the F-containing reporter plasmids further support the strand bias in up- or downregulation of transcription by modifications in the context of the VEGF PQS. The results identify that BER on the coding strand is a mechanism for DNA repair leading to gene activation; in contrast, the same modifications on the template strand are not predominantly repaired by BER, resulting in a decrease in gene expression.
Cell Line and Modification Differences in the Transcription Profiles
On the basis of the time to reach maximal Renilla expression, the peak of repair coupled with transcriptional modulation could be estimated and compared. The peak change in expression for either OG or F was 12 h earlier in WT MEF compared to glioblastoma cells, independent of the strand in which the modification resided (Figure 1E). Inspection of the strand bias for achieving the maximum expression change found modifications in the template strand peaked 12 h earlier than observed for modifications in the coding strand for both wild-type cells. These observations point out that OG or F modifications on the template strand are acted upon faster than those in the coding strand of the promoter region to induce a change in transcription. Comparisons between the WT and Ogg1–/– MEFs for the F modification in either strand found the time of maximal change in expression was observed 12 h earlier in the WT cells. Because OG did not change expression in the knockout cell line when found in the coding strand, a comparison could not be made. On the template strand, the change resulting from OG was observed ∼12 h earlier in the WT MEFs than in the Ogg1–/– MEFs. If DNA repair initiation is the rate-limiting step leading to the change in gene expression, these observations support repair, leading to a change in transcription that is more efficient on the template strand relative to the coding strand. The concept of greater repair efficiency on the template strand has been reported for modifications in gene coding regions that interfere with the transcriptional elongation complex;19 additionally, recent high-throughput sequencing studies conclude that mutations from OG are more likely when located in the coding strand as a result of less efficient DNA repair.34 Modifications on the template strand of a promoter impact the transcriptional preinitiation complex, while that is not the case for modifications in the coding strand.35 This difference likely leads to the more efficient activity observed on the template strand.
The maximum change in expression for each modification in each cell line was then compared. In the template strand, OG or F in all three cell lines led to nearly a 3-fold reduction in transcription (Figure 1F). This observation identifies that modifications to the template strand that impact transcription initiation result in gene suppression. In the coding strand, OG in the VEGF PQS context furnished a 3-fold increase in transcription in WT MEF and glioblastoma cells, while F in the same context and cells yielded a 6-fold increase in transcription. The additional increase in expression observed with the AP analog compared to OG could result from either the OGG1 release of OG not yielding an intact AP or rather the glycosylase being bifunctional and generating a strand break. Perhaps this difference is not reproduced with the synthetic plasmids, or cellular APE1 operates more slowly on the AP analog (i.e., F) than on an authentic AP that would be formed by monofunctional activity on OG. This is possible because the F analog is more stable to APE1 cleavage than an authentic AP.36 The present results cannot rule out either possibility.
The PQS Context Facilitates a Greater Change in Transcription
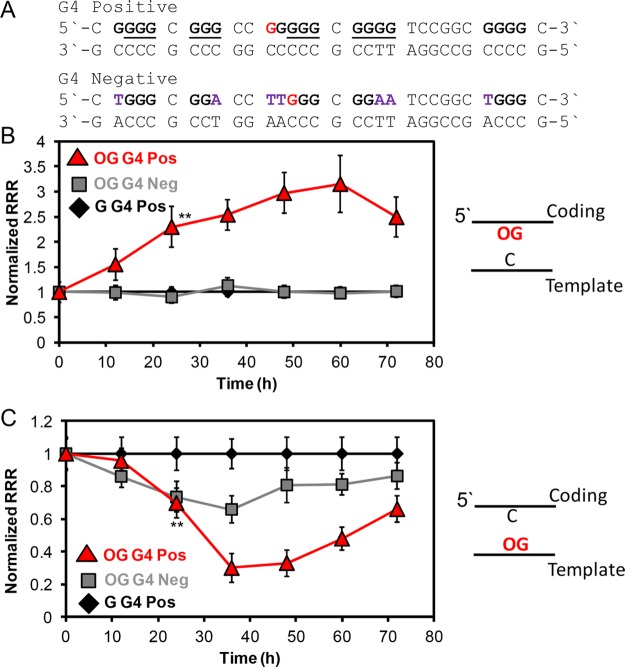
Formation of G4 structures by PQSs in coding vs template strands was proposed to alter the direction in which transcription is modulated (i.e., G4 folds are up- or downregulatory sequences).37 The data so far identify that OG or F in the coding or template strand in the context of a PQS modulates transcription; however, these results cannot conclude whether a G4 fold was involved in the change in transcription observed. Therefore, time-course studies to compare OG in the context of the VEGF PQS vs a sequence not capable of G4 formation were conducted in glioblastoma cells (Figure 2A). The VEGF PQS is bound by three equivalents of the SP1 transcription factor,38 and therefore, the G4 null sequence retained the SP1 transcription factor consensus sequence to ensure the studies only analyzed G4 formation. When OG was placed in the context of a G4-null sequence in the coding strand, no induction of transcription was observed up to 72 h (Figure 2B). In contrast, OG in a G4-null sequence in the template strand caused a nearly 30% reduction in transcription after 36 h, less than the ∼70% reduction in transcription observed for OG in the VEGF PQS context but still significant (Figure 2C). The presence of OG in the template strand must impact loading of the transcriptional preinitiation complex; however, when OG can facilitate G4 formation, the reduction in transcription was much greater. These results do not provide unequivocal support for G4 formation in the cellular context; however, they do advance experimental data for G-rich PQSs to guide cellular processes under oxidative stress conditions with the strong possibility of G4 formation.
Figure 2.
Impact of OG on transcription when located in the context of a G4 positive or negative sequence found in the coding or template strand of the Renilla luciferase reporter gene. (A) The sequences for the G4 positive and negative strands studied. (B) Time-dependent studies for OG in the coding strand or (C) template strand of the reporter plasmid. The reporter plasmids were analyzed in glioblastoma cells. On the basis of a Student’s t test, the time in which significance at **P < 0.01 was first observed is marked on each plot.
siRNA Knockdown Studies to Probe the Repair Pathways
The results up to this point conclude that the removal of OG from the coding strand within a promoter is achieved by the BER pathway; in contrast, the dominant DNA repair pathway for removal of OG in the template strand appears not to be BER. To better understand the strand dependency in repair of OG, a series of siRNA knockdown experiments to probe BER or TC-NER proteins were conducted 24 h prior to transfection of the reporter plasmids in glioblastoma cells. The cellular reporter plasmids were then incubated for 48 h before determining the luciferase expression levels. The Ogg1–/– MEF experiments identified the role of Ogg1 in the strand dependency of coupling DNA repair with transcription. Therefore, the first siRNA knockdown studies were commenced with APE1-specific siRNAs in glioblastoma cells transfected with OG or F modified reporter plasmids in the coding strand (Figure 3A). The knockdown studies found that as the siRNA concentration was increased from 1–50 nM for OG- or F-containing plasmids, the level of Renilla luciferase expression decreased with the titration series. This finding further supports that BER with APE1 activity is the dominant repair pathway for these lesions in the coding strand, and APE1 is essential for coupling DNA repair with transcriptional activation. In the second study, the TC-NER lesion sensor protein CSB (aka ERCC6) was knocked down by siRNAs. For the cases of OG or F in the coding strand of the plasmid, no impact on Renilla expression was observed as the CSB-specific siRNAs were titrated into the cell culture from 1–50 nM, suggesting that TC-NER is not a major DNA repair pathway for OG or F located in the coding strand of a gene promoter. This observation is consistent with studies conducted on these modifications located in the coding strand of a gene coding region.8
Figure 3.
siRNA knockdown studies of APE1 or CSB during transfection of OG or F containing reporter plasmids to determine the strand dependency in the major DNA repair pathway observed. (A) Studies conducted when OG or F were located in the coding strand in the VEGF PQS context. (B) Studies conducted when OG or F were located in the template strand in the VEGF PQS context. All transfection experiments were conducted in human glioblastoma cells by transfecting the siRNAs 24 h prior to transfection of the reporter plasmids. Luciferase expression was measured 48 h after transfection of the plasmids. *These data were previously reported by our laboratory and are provided for comparative purposes.3
The same siRNA knockdown studies were then conducted on glioblastoma cells transfected with OG or F modifications in the template strand in the context of the VEGF PQS in the promoter of the reporter gene. Knockdown of APE1 with siRNAs yielded an insignificant impact on Renilla expression when OG or F was present in the promoter (Figure 3B). Knockdown of CSB with siRNAs generated a significant change in Renilla expression when F was present in the template strand, and the impact was siRNA dose dependent; however, Renilla expression remained similar when OG was in the template strand throughout the CSB siRNA titration study. These results suggest TC-NER is the DNA repair pathway functional for AP in the template strand, a finding consistent with DNA repair of AP in the template strand of gene coding regions.39 However, knocking down key BER or TC-NER proteins did not have an effect when OG was in the template strand. One of two possibilities may occur with OG in the template strand. (1) By knocking down either BER or TC-NER, this may activate the other pathway to repair OG, or (2) repair of OG in the template strand of a gene promoter is achieved by an alternative mechanism using other damage sensor proteins. The possibility of an alternative repair pathway has been proposed for repair of the template strand OG located in a gene promoter.18 Maher et al. recently found that an additional unknown factor is required for BER of a base lesion,40 and therefore, the siRNA knockdown studies may have failed to target the correct sensor protein. The present results cannot add further support for either claim. It is interesting that the siRNA studies with the AP analog did not mirror the results found with OG. This difference may result from the AP analog driving the PQS to the G4 state more so than OG, which likely drives the mechanism of repair in the cell. Future inquiry is needed to begin to address these differences.
Repair of an OG:A Base Pair Up/Downregulates Transcription
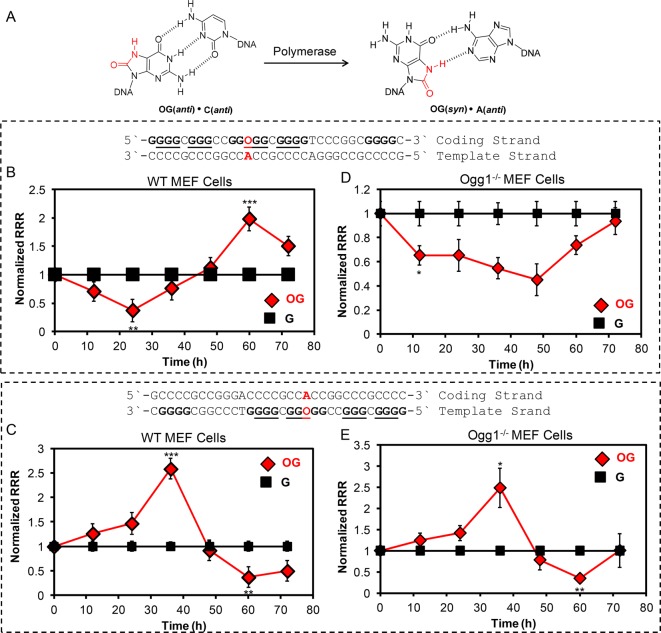
The experiments establish that repair of OG in the context of the VEGF PQS in the coding strand leads to an increase in gene expression, while repair of OG in the same context but in the template strand leads to a decrease in transcription (Scheme 1B and C). This is the case when OG is base-paired with C. Polymerase bypass of OG in a template strand has a high probability of base paring with A as a consequence of OG favoring the syn conformation allowing an OG:A Hoogsteen base pair to form (Figure 4A).1 Repair of an OG:A base pair is initiated by MUTYH-mediated removal of A to yield an AP that is a substrate for APE1, POLβ, and LIGIII to yield an OG:C base pair (Scheme 2).1 Repair of the OG:C base pair follows the mechanism described above using OGG1, APE1, POLβ, and LIGIII to yield a G:C base pair. Repair of an OG:A base pair occurs on both strands in a defined order. Therefore, we were intrigued to see if we could monitor the repair processes of an OG:A base pair by following the time-course expression of Renilla luciferase allowing the up- to downregulation or down- to upregulation of Renilla expression to be monitored depending on the strand in which OG resides.
Figure 4.
Base pairs of OG with A or C and the impact an OG:A base pair has on transcription in WT- or Ogg1–/–-MEF cells when located in the VEGF PQS in a luciferase reporter gene. (A) Structures for the OG:C and OG:A base pairs. Expression observed in WT MEFs when OG is in either the coding (B) or template (C) strand base paired with A. Expression observed in Ogg1–/– MEFs when OG is in either the coding (D) or template (E) strand base paired with A. On the basis of a Student’s t test, the time in which significance at *P < 0.05, **P < 0.01, or ***P < 0.001 was first observed is marked on each plot.
As a first step, we determined if repair in the complementary strand (i.e., potential i-motif sequence, PIMS) to the VEGF PQS could facilitate a change in gene expression. Plasmids were prepared placing OG in the loop of the VEGF PIMS with C opposite in either the coding or template strand of the promoter for Renilla luciferase. The time-course expression of Renilla was monitored from 12–48 h post-transfection in glioblastoma cells to find that when OG was repaired in the i-motif context residing in the coding strand, Renilla expression increased relative to the control (Figure S2); in contrast, when OG was repaired in the i-motif context in the template strand, Renilla expression decreased relative to the control (Figure S2). These results show that we can monitor in both strands the repair of an OG:A base pair. Also, these results demonstrate that the PQS is not the only structured sequence context that can lead to a change in gene expression when DNA repair is coupled to transcription.3,5
The plasmids containing OG in either the coding or template strand paired with A were transfected into the WT MEF, Ogg1–/– MEF, or glioblastoma cells to follow the time-course expression profiles. When OG was in the coding strand in the PQS context and A was in the template strand in the PIMS context, studies in WT MEFs showed Renilla expression first decreased by nearly 50% up to 24 h while Mutyh repaired A in the template strand (Figure 4B). After 24 h, Renilla expression increased to be 2-fold greater at 60 h than the control while the OG was being repaired on the coding strand (Figure 4B). Next, the OG:A base pair and contexts were flipped to place OG in the template strand and A in the coding strand, and the opposite profile was observed (Figure 4C). Repair of A on the coding strand led first to a 2-fold increase in expression up to 36 h, followed by repair of OG on the template strand and a decrease in expression (Figure 4C). Similar changes for monitoring the OG:A base pair repair in the coding or template strand were observed in glioblastoma cells (Figure S3). In the final study, we placed the OG:A base pair containing plasmids in the Ogg1–/– MEF cells that could initiate repair but not complete the task. Placement of OG in the VEGF PQS context in the coding strand and A opposite in the template strand yielded a decrease in expression of 60% at 48 h (Figure 4D). Because OG in the coding strand does not change expression in Ogg1–/– MEFs, the expression returned to the level observed in the all-G control study. Finally, placement of OG in the template strand and A in the coding strand furnished an increase in expression of more than 2-fold up to 36 h, and then a decrease of 50% less than the control was observed at 60 h (Figure 4E). These observations are consistent with OG in the template strand without Ogg1-mediated BER decreasing transcription (Figure 1D). The ability to synthesize OG into a reporter plasmid in the OG:A base pair context allowed us to monitor the complete repair processes of returning this promutagenic base pair back to the correct G:C base pair.
Oxidiatively Modified Promoter PQSs Up/Downregulate Transcription
Synthetic manipulation of reporter plasmids to install DNA modifications with single-nucleotide precision provided the opportunity to study the impact on gene expression of G oxidation to OG in the context of a gene promoter. The case in which OG was located in the VEGF PQS context in the coding strand of a gene promoter initiated BER, leading to gene induction by nearly 3-fold (Figures 1A and S1). The activation process occurs by BER site specifically introducing an AP where the OG was located to unmask the G4 fold for binding by APE1 (Scheme 1B). Although the BER glycosylase OGG1 operated on OG in a duplex context, the resulting AP site is highly destabilizing to the duplex, permitting an equilibrium shift in favor of the G4 fold in which the AP resides in an unstructured loop. Next, we propose that AP recruits APE1 to bind but not cleave in the G4 context because the conformation of the lesion site is inappropriate for phosphodiester hydrolysis (Scheme 1B). In support of our proposed mechanism, a previous study found the activity of APE1 is highly attenuated when an AP resides in the loop of a G4 fold.41
Of the many roles for APE1 in the cell, another is to function as a transcriptional activator by interacting with activator proteins.15,29 We reported on this finding and have now extended our studies to include the time-course analysis of gene induction, strand dependency, and OG base-pairing dependency on the gene modulation process (Figures 1–4). We have found that OG or an AP analog, F, in the coding strand was repaired to induce transcription 12 h faster in MEF cells than glioblastoma cells (Figure 1E). When Ogg1–/– MEFs were studied, coding strand OG was not repaired, while F produced a slower rate of repair and gene induction than observed in the WT MEFs (Figure 1E). This observation suggests the involvement of OGG1 in repair of substrates outside its scope to increase the efficiency of other DNA repair proteins. In vitro kinetic studies have found OGG1 and APE1 stimulate each other’s activity,42,43 and the present findings support an interaction between these two BER proteins in vivo. The key finding with respect to DNA repair is that BER is the dominant pathway for removal of OG or F in the coding strand of a promoter element (Figures 1C and 3A). Depending on the sequence context (i.e., PQS or PIMS), the repair process can facilitate gene induction, as described below.
The presence of OG or F in the template strand of a gene promoter in the VEGF PQS context led to a decrease in the transcriptional output of the gene. These findings conclude that OG or F in this context results in transcriptional repression, and as described below, this finding was independent of sequence context. The repair process stimulated by F appears to be activation of TC-NER via CSB (Figures 1D and 3B), while the present results are inconclusive with respect to the major repair mechanism for OG in the template strand of a promoter. Nevertheless, OG is a downregulator of transcription when located in the template strand, which in the PQS context may facilitate G4 formation to further downregulate mRNA synthesis (Scheme 1C). Comparing the time-course analysis of OG or F in the coding vs template strands, we found the gene modulation process via DNA repair to return to background levels more quickly in the template strand than the coding strand. Our studies with synthetic reporter plasmids provide direct evidence for the claim that DNA repair is more efficient in template strands relative to coding strands.34 Thus, oxidation of G in a promoter PQS context will provide a faster change in transcription than oxidation of the same context on the coding strand.
Base pairs between OG and C or A determine the initial steps of DNA repair (Scheme 2). By following Renilla luciferase expression with synthetic reporters, the repair process of each base pair context of OG could be monitored (Figures 1A–D and 4B,C). When OG was base paired with C in the VEGF PQS context, gene expression was either enhanced or repressed depending on the strand in which the OG and PQS resided. Interestingly, when OG was base paired with A, transcription was enhanced and then repressed, and the order of the events depended on the strand in which OG resided (Figures 4B and C). This experiment is not a biologically relevant one; it is highly unlikely OG base paired with A will drive gene expression changes during oxidative stress, because A is only inserted opposite OG during polymerase extension. However, the unique pattern of gene expression modulation (i.e., up/down or down/up) provides an opportunity to monitor DNA repair of an OG:A base pair in the cellular context. In addition, this experiment provided evidence that both the VEGF G4 and the i-motif sequences are capable of folding under identical cellular conditions because the induction of gene expression is only observed when the lesions are present in folded secondary structures of the coding strand.
The ability of the VEGF PQS context in which OG or F was housed to possibly adopt a G4 fold was found to be critical for driving the direction of the gene modulation process (Figures 1A–D). In the coding strand, repair initiation of OG to an AP provides a drive for the sequence to shift structures from a duplex to a G4 fold to guide induction of transcription (Scheme 1B). The present data, in tandem with our previous studies,3 supports APE1 as the central BER protein for gene induction. Many reports have found APE1 interacts with activating transcription factors, such as HIF-1α, AP1, or NF-κB and others for transcriptional regulation.15,29 Future cellular and genome level studies are needed to better understand the choreography of G4 formation and protein interactions that regulate transcription. On the other hand, OG or F in the VEGF PQS context in the template strand led to a greater repression of transcription (Scheme 1C). This observation is consistent with studies finding G4 folds block the progression of polymerases on template strands to stall biological processes.44,45 Therefore, OG or F can function as up/downregulatory modifications in gene promoters on the basis of the strand and sequence context of the modification.
Bioinformatic studies suggest that PQSs are nearly equally distributed in the coding and template strands of gene promoters.46 Thus, under conditions of oxidative stress that effect G oxidation to OG, the impact on transcription would yield nearly equal up or down transcriptional regulation globally if other nucleosomal or protein factors are not considered. Further studies into different PQSs in plasmid-based systems and experiments on the genome level are needed to clarify the sequence requirements and nucleosome impact for G oxidation to drive transcription. The existence of G4 folds in the nucleus has been subject to much debate; recent G4 ChIP-Seq studies found ∼10 000 G4s folded in human skin cells under normal growth conditions.47 In light of the present studies, the population of folded G4s may change as a result of oxidative stress. Future studies on the genome level will guide a better understanding of whether promoter PQSs are sensors of oxidative stress by direct oxidation of G nucleotides in the PQS context to modulate transcription in response to the stress. Last, this study provides evidence for the oxidative DNA modification OG functioning as a regulatory mark to up- or downregulate transcription, leading us and many others to hypothesize OG could be an epigenetic-like DNA modification.3,4,10−13
Acknowledgments
We thank the National Cancer Institute for financial support of the project (R01 CA090689). The oligonucleotides and Sanger sequencing were provided by the University of Utah Health Sciences Core facilities that are supported in part by a National Cancer Institute Cancer Center Support grant (P30 CA042014).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.7b00636.
Additional materials and methods information, time-course analyses for luciferase expression in glioblastoma cells of a wild-type G-quadruplex, OG in an i-motif, and an OG-A base pair with accompanying relevant references (PDF)
Author Contributions
A.M.F. and C.J.B. conceived the research and wrote the manuscript. A.M.F., J.Z., and Y.D. conducted the experiments. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- David S. S.; O’Shea V. L.; Kundu S. (2007) Base-excision repair of oxidative DNA damage. Nature 447, 941–950. 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S. S. (2014) Base excision repair: A critical player in many games. DNA Repair 19, 14–26. 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. M.; Ding Y.; Burrows C. J. (2017) Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 114, 2604–2609. 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L.; Zhu B.; Hao W.; Zeng X.; Vlahopoulos S. A.; Hazra T. K.; Hegde M. L.; Radak Z.; Bacsi A.; Brasier A. R.; Ba X.; Boldogh I. (2016) Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J. Biol. Chem. 291, 25553–25566. 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniali G.; Lirussi L.; D’Ambrosio C.; Dal Piaz F.; Vascotto C.; Casarano E.; Marasco D.; Scaloni A.; Fogolari F.; Tell G. (2014) SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell 25, 532–547. 10.1091/mbc.E13-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B.; Ombra M. N.; Bertoni A.; Cuozzo C.; Sacchetti S.; Sasso A.; Chiariotti L.; Malorni A.; Abbondanza C.; Avvedimento E. V. (2008) DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206. 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Allgayer J.; Kitsera N.; Bartelt S.; Epe B.; Khobta A. (2016) Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 44, 7267–7280. 10.1093/nar/gkw473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornaletti S.; Maeda L. S.; Kolodner R. D.; Hanawalt P. C. (2004) Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair 3, 483–494. 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Kathe S. D.; Shen G. P.; Wallace S. S. (2004) Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J. Biol. Chem. 279, 18511–18520. 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- Fleming A. M.; Burrows C. J. (2017) 8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis. DNA Repair 56, 75–83. 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifermann M.; Epe B. (2017) Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark?. Free Radical Biol. Med. 107, 258–265. 10.1016/j.freeradbiomed.2016.11.018. [DOI] [PubMed] [Google Scholar]
- Moore S. P.; Toomire K. J.; Strauss P. R. (2013) DNA modifications repaired by base excision repair are epigenetic. DNA Repair 12, 1152–1158. 10.1016/j.dnarep.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Zarakowska E.; Gackowski D.; Foksinski M.; Olinski R. (2014) Are 8-oxoguanine (8-oxoGua) and 5-hydroxymethyluracil (5-hmUra) oxidatively damaged DNA bases or transcription (epigenetic) marks?. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 764–765, 58–63. 10.1016/j.mrgentox.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Park J.; Park J. W.; Oh H.; Maria F. S.; Kang J.; Tian X. (2016) Gene-specific assessment of guanine oxidation as an epigenetic modulator for cardiac specification of mouse embryonic stem cells. PLoS One 11, e0155792. 10.1371/journal.pone.0155792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniali G.; Malfatti M. C.; Tell G. (2017) Unveiling the non-repair face of the base excision repair pathway in RNA processing: A missing link between DNA repair and gene expression?. DNA Repair 56, 65–74. 10.1016/j.dnarep.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Kitsera N.; Stathis D.; Luhnsdorf B.; Muller H.; Carell T.; Epe B.; Khobta A. (2011) 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 39, 5926–5934. 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen E.; Kwon K.; Coin F.; Egly J. M.; Klungland A. (2004) Transcription activities at 8-oxoG lesions in DNA. DNA Repair 3, 1457–1468. 10.1016/j.dnarep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Le Page F.; Klungland A.; Barnes D. E.; Sarasin A.; Boiteux S. (2000) Transcription coupled repair of 8-oxoguanine in murine cells: the Ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl. Acad. Sci. U. S. A. 97, 8397–8402. 10.1073/pnas.140137297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P. C.; Spivak G. (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970. 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Roy D.; Zhang Z.; Lu Z.; Hsieh C. L.; Lieber M. R. (2010) Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 30, 146–159. 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allgayer J.; Kitsera N.; von der Lippen C.; Epe B.; Khobta A. (2013) Modulation of base excision repair of 8-oxoguanine by the nucleotide sequence. Nucleic Acids Res. 41, 8559–8571. 10.1093/nar/gkt620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastoriza-Gallego M.; Armier J.; Sarasin A. (2007) Transcription through 8-oxoguanine in DNA repair-proficient and Csb(−)/Ogg1(−) DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis 22, 343–351. 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- Clark D. W.; Phang T.; Edwards M. G.; Geraci M. W.; Gillespie M. N. (2012) Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radical Biol. Med. 53, 51–59. 10.1016/j.freeradbiomed.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. M.; Burrows C. J. (2017) Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radical Biol. Med. 107, 35–52. 10.1016/j.freeradbiomed.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J.; Wagner J. R.; Shafirovich V.; Geacintov N. E. (2014) One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int. J. Radiat. Biol. 90, 423–432. 10.3109/09553002.2013.877176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastukh V.; Roberts J. T.; Clark D. W.; Bardwell G. C.; Patel M.; Al-Mehdi A. B.; Borchert G. M.; Gillespie M. N. (2015) An oxidative DNA ″damage″ and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol. 309, 1367–1375. 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. M.; Zhou J.; Wallace S. S.; Burrows C. J. (2015) A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these ″spare tires″ have an evolved function?. ACS Cent. Sci. 1, 226–233. 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G.; Quadrifoglio F.; Tiribelli C.; Kelley M. R. (2009) The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid. Redox Signaling 11, 601–620. 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Wilson D. M. 3rd. (2014) Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signaling 20, 678–707. 10.1089/ars.2013.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J.; Fleming A. M.; Burrows C. J. (2016) Sequencing of DNA lesions facilitated by site-specific excision via base excision repair DNA glycosylases yielding ligatable gaps. J. Am. Chem. Soc. 138, 491–494. 10.1021/jacs.5b11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimoff R.; Kosmatchev O.; Kirchner A.; Pfaffeneder T.; Spada F.; Brantl V.; Muller M.; Carell T. (2017) 5-Formyl- and 5-carboxydeoxycytidines do not cause accumulation of harmful repair intermediates in stem cells. J. Am. Chem. Soc. 139, 10359–10364. 10.1021/jacs.7b04131. [DOI] [PubMed] [Google Scholar]
- Parsons J. L.; Dianova I. I.; Dianov G. L. (2004) APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 32, 3531–3536. 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. M. 3rd (2005) Ape1 abasic endonuclease activity is regulated by magnesium and potassium concentrations and is robust on alternative DNA structures. J. Mol. Biol. 345, 1003–1014. 10.1016/j.jmb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Haradhvala N. J.; Polak P.; Stojanov P.; Covington K. R.; Shinbrot E.; Hess J. M.; Rheinbay E.; Kim J.; Maruvka Y. E.; Braunstein L. Z.; Kamburov A.; Hanawalt P. C.; Wheeler D. A.; Koren A.; Lawrence M. S.; Getz G. (2016) Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 164, 538–549. 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S.; Bernecky C.; Cramer P. (2015) Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143. 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- Schermerhorn K. M.; Delaney S. (2013) Transient-state kinetics of apurinic/apyrimidinic (AP) endonuclease 1 acting on an authentic AP site and commonly used substrate analogs: the effect of diverse metal ions and base mismatches. Biochemistry 52, 7669–7677. 10.1021/bi401218r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M. L.; Paeschke K.; Zakian V. A. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780. 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer G.; Cramer T.; Suske G.; Kemmner W.; Wiedenmann B.; Hocker M. (2003) Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J. Biol. Chem. 278, 8190–8198. 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- Kim N.; Jinks-Robertson S. (2010) Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 30, 3206–3215. 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher R. L.; Marsden C. G.; Averill A. M.; Wallace S. S.; Sweasy J. B.; Pederson D. S. (2017) Human cells contain a factor that facilitates the DNA glycosylase-mediated excision of oxidized bases from occluded sites in nucleosomes. DNA Repair 57, 91–97. 10.1016/j.dnarep.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxson C.; Hayner J. N.; Beckett J.; Bloom L. B.; Tornaletti S. (2014) Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res. 42, 7708–7719. 10.1093/nar/gku417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko V. S.; Nevinsky G. A.; Zharkov D. O. (2007) Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair 6, 317–328. 10.1016/j.dnarep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Esadze A.; Rodriguez G.; Cravens S. L.; Stivers J. T. (2017) AP-endonuclease 1 accelerates turnover of human 8-oxoguanine DNA glycosylase by preventing retrograde binding to the abasic-site product. Biochemistry 56, 1974–1986. 10.1021/acs.biochem.7b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. (2015) G4-associated human diseases. EMBO Rep. 16, 910–922. 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P.; Reams C.; Simpson L. J.; Sale J. E. (2010) Epigenetic instability due to defective replication of structured DNA. Mol. Cell 40, 703–713. 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413. 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel-Hertsch R.; Beraldi D.; Lensing S. V.; Marsico G.; Zyner K.; Parry A.; Di Antonio M.; Pike J.; Kimura H.; Narita M.; Tannahill D.; Balasubramanian S. (2016) G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 48, 1267–1272. 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.