Abstract
The five human RecQ helicases participate in multiple processes required to maintain genome integrity. Of these, the disease-linked RecQ4 is the least studied because it poses many technical challenges. We previously demonstrated that the yeast Hrq1 helicase displays similar functions to RecQ4 in vivo, and here, we report the biochemical and structural characterization of these enzymes. In vitro, Hrq1 and RecQ4 are DNA-stimulated ATPases and robust helicases. Further, these activities were sensitive to DNA sequence and structure, with the helicases preferentially unwinding D-loops. Consistent with their roles at telomeres, telomeric repeat sequence DNA also stimulated binding and unwinding by these enzymes. Finally, electron microscopy revealed that Hrq1 and RecQ4 share similar structural features. These results solidify Hrq1 as a true RecQ4 homolog and position it as the premier model to determine how RecQ4 mutations lead to genomic instability and disease.
INTRODUCTION
The human genome encodes five RecQ family helicases (RecQ1, BLM, WRN, RecQ4 and RecQ5), three of which are linked to genetic disorders when mutated: BLM, WRN and RecQ4 (1). The BLM and WRN proteins have been extensively investigated and found to serve important roles in multiple nuclear processes. Understanding the functions of RecQ4 is also extremely important because mutations in RecQ4 are linked to three different human diseases (2–6), and RecQ4 is the only human RecQ known to impact both nuclear and mitochondrial genome integrity (7).
However, RecQ4 is much less well studied than the other RecQs. This is partially due to experimental difficulties with human RecQ4 in vivo. The metazoan RecQ4 helicases are evolutionary chimeras created by the apparent fusion of an essential DNA replication initiation factor in lower eukaryotes, Sld2 (8), to the N-terminus of a RecQ helicase (Figure 1A) (9). The vast majority of recq4 clinical alleles map to the helicase-like portion of RecQ4 (2), presumably because mutations in the Sld2-like domain would be lethal. It is difficult to study the effects of these clinical mutations in the context of the full-length protein in vivo due to pleiotropic defects in DNA replication and the variable effects of different clinical alleles in various cell lines (10,11). RecQ4 also presents challenges to biochemical investigations because it is difficult to purify in large amounts (12,13), and its helicase activity is masked by a strong DNA annealing activity present in the Sld2-like N-terminus (12,14). The Saccharomyces cerevisiae Sld2 protein likewise displays DNA annealing activity in vitro (15). These technical challenges have made it difficult to ascertain any mechanistic details of RecQ4 in disease prevention. Therefore, investigators have searched for a more experimentally amenable RecQ4 homolog in a simpler model system to study the fundamental mechanisms of RecQ4 sub-family helicases in the maintenance of genome stability.
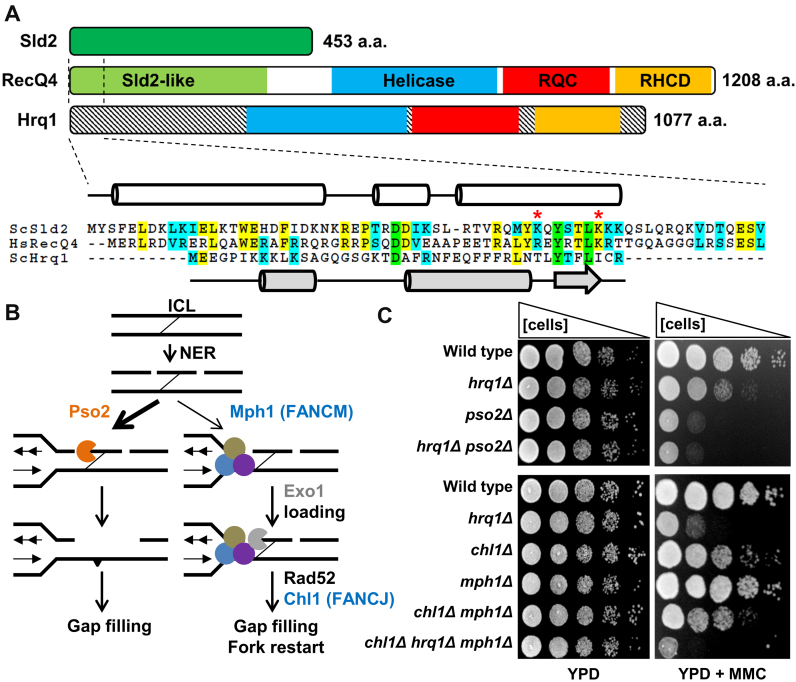
Figure 1.
Hrq1 functions in the Pso2 pathway during ICL repair. (A) Domain schematics for human RecQ4, S. cerevisiae Hrq1, and S. cerevisiae Sld2 showing a similar predicted domain organization between the two helicases. The demarcation of the Sld2-like domain, helicase domain, RecQ C-terminal (RQC) domain, and the RecQ4p/Hrq1p-conserved domain (RHCD) are based on (16,18). Note that the N-termini of these helicases are not conserved, at least in part due to the presence of a Sld2-like domain in the N-terminus of RecQ4. The highly conserved N-terminal 65 amino acids (aa) of RecQ4 and 67 aa of Sld2 are aligned below, along with the non-conserved N-terminus of Hrq1. Green highlighting denotes completely conserved residues in all three sequences, yellow denotes identical residues in two of the sequences, and blue indicates similarity between two sequences. The known secondary structure (cylinders = α-helices and lines = random coils) of the first 50 aa of RecQ4 (47) is shown in white, and the predicted secondary structure (broad arrow = β-sheet) of the first 43 aa of Hrq1 is in gray. Hs, Homo sapiens and Sc, Saccharomyces cerevisiae. Two residues implicated in origin DNA binding by Sld2 (67) are marked with red asterisks. (B) The two ICL repair pathways in S. cerevisiae (adapted from (68)). ICL repair is initiated by NER, and then the lesion-containing DNA is preferentially processed by the Pso2-dependent pathway (left). The Pso2 nuclease (orange) degrades one strand of the crosslinked DNA, and the lesion is eventually healed by gap filling. In the absence of Pso2, an alternative FA-like pathway (right) can repair the ICL. This requires the FANCM homolog Mph1 (blue) and other factors (tan and purple circles) that recruit the ExoI nuclease (gray) and other downstream factors such as Rad52 and (likely) the FANCJ homolog Chl1. (C) The deletion of HRQ1 is epistatic to pso2Δ but not chl1Δ mph1Δ. Serial dilutions of saturated overnight cultures of the indicated strains were spotted onto plates containing rich medium (YPD, left) or rich medium supplemented with MMC (right). The Pso2 pathway experiment (top) used 50 μg/ml MMC and the FA-like pathway experiment (bottom) used 100 μg/ml MMC.
The discovery of Hrq1, a putative RecQ4 homolog in fungi (16), held the promise to circumvent many of the issues of working with human RecQ4 itself. First, it is not involved in DNA replication because fungal Sld2 exists as a separate protein rather than a fusion to the helicase (Figure 1A) (16). Additionally, cells deleted for HRQ1 grow like wild-type under normal conditions (13). Therefore, mutational analysis of Hrq1 can be used to study the disease-linked repair function(s) of RecQ4 sub-family helicases without compromising DNA replication. Second, recombinant Hrq1 can be purified in larger quantities than RecQ4 (13), facilitating the in vitro characterization of its biochemical activities. However, due to low sequence conservation and the scarcity of information on these helicases in the literature, it is necessary to establish that Hrq1 is a functional homolog of RecQ4 (i.e. that it performs the same functions in genome maintenance as RecQ4) rather than just a homolog at the primary sequence level. This fact is typified by the second S. cerevisiae RecQ family helicase, Sgs1. Its protein sequence is most homologous to RecQ1 (16), but Sgs1 is a functional homolog of BLM (reviewed in (17)).
At the primary sequence level, S. cerevisiae Hrq1 and human RecQ4 contain a degenerate RecQ C-terminal (RQC) domain that is otherwise highly conserved in all studied RecQ family helicases (16,18). As opposed to all other RecQs, Hrq1 and RecQ4 sub-family helicases all also contain a characteristic RecQ4p/Hrq1p-conserved domain (RHCD). In vivo, our previous work further supports our hypothesis that Hrq1 is functionally homologous to RecQ4. Indeed, we demonstrated that Hrq1 plays an important catalytic role in DNA inter-strand crosslink (ICL) repair (13), similar to that of RecQ4 in humans (11). Further, it was recently shown that RecQ4 functions in telomere maintenance (19), and we found that Hrq1 inhibits telomere addition to DNA double-strand breaks, limits telomere lengthening in cells lacking the telomerase inhibitor Pif1, and is needed for the Type I telomerase-independent telomere maintenance pathway in S. cerevisiae (13). Unfortunately, the mechanistic role of Hrq1 in these pathways is unclear because relatively little biochemical work with Hrq1 has been performed (13,20).
Here, we extended our investigation of the role of Hrq1 in ICL repair and performed a thorough biochemical comparison of Hrq1 and RecQ4 to examine the extent of their functional similarities. We found that Hrq1 and RecQ4 are both DNA-stimulated ATPases that display high binding affinity for the same DNA structures, with a strong preference for D-loops and G-quadruplex (G4) DNA. The biochemical activities of Hrq1 and RecQ4 were also stimulated by telomeric repeat sequence DNA. This is consistent with the known and hypothesized roles of Hrq1 and RecQ4 in DNA recombination, repair, and telomere maintenance (13,21,22). Finally, transmission electron microscopy (TEM) and single-particle 2D image averaging revealed that Hrq1 and RecQ4 also possess similar tertiary structures. Thus, we report the first direct biochemical and structural comparison of Hrq1 and RecQ4, establishing Hrq1 as a RecQ4 homolog and an excellent experimental model to interrogate the roles of human RecQ4 in maintaining genome integrity.
MATERIALS AND METHODS
Online methods
Nucleotides, oligonucleotides and other reagents
32P-ATP was purchased from PerkinElmer (Waltham, MA, USA), and unlabeled ATP was from GE Healthcare (Little Chalfont, UK) or DOT Scientific (Burton, MI, USA). The oligonucleotides used in this work were synthesized by IDT (Coralville, IA, USA) and are listed in Supplemental Table S1. All restriction enzymes were from New England Biolabs (Ipswich, MA, USA).
Protein sequence analysis
The default settings of the suite of tools available on Biology Workbench (http://workbench.sdsc.edu) were used to analyse the RecQ4, Hrq1, and Sld2 sequences. To generate the alignment in Figure 1A, the N-terminal domains of RecQ4 (aa 1–464) and Hrq1 (aa 1–279) were aligned with full-length Sld2 (453 aa) using CLUSTALW (23) and formatted and shaded using BOXSHADE (v.3.3.1). The secondary structure of Hrq1 was predicted using GOR4 tool (24).
Mitomycin C (MMC) sensitivity assay
The strains listed in Figures 1 and 2 were tested for mitomycin C sensitivity essentially as described (13). Briefly, the cells were grown overnight in 1% yeast extract, 2% peptone and 2% dextrose (YPD) medium at 30°C with aeration, diluted to an optical density at 660 nm of 1 with sterile water, and then 10-fold serially diluted likewise to 10−4. Then, 5 μl of each dilution was spotted onto plates either containing or lacking MMC (Sigma, St. Louis, MO, USA) at the indicated concentration. The plates were incubated in the dark at 30°C for ∼2 days before being imaged with a flat-bed scanner. All strains were derived from the wild type YPH genetic background (MATa ura3-52 lys2-801_amber ade2-101_ochre trp1Δ63 his3Δ200 leu2Δ1) (25) by standard methods. Details of the strain constructions are available upon request.
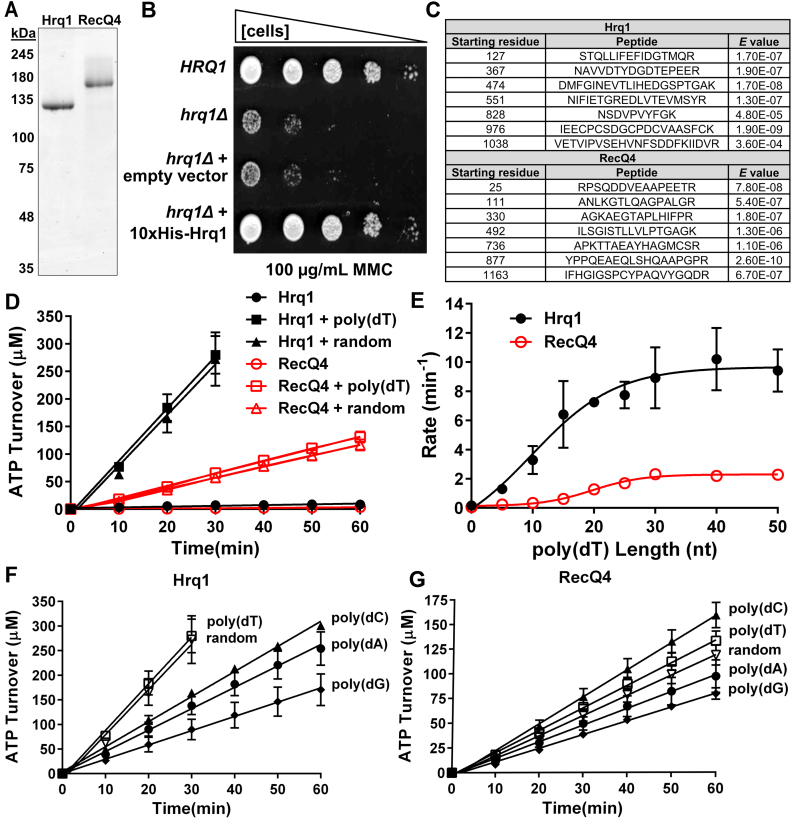
Figure 2.
Purified Hrq1 and RecQ4 are DNA-stimulated ATPases. (A) SYPRO orange-stained SDS-PAGE gel images of 1 μg of recombinant Hrq1 and RecQ4. (B) 10xHis-Hrq1 is active in vivo. Cells lacking Hrq1 (hrq1Δ) are sensitive to mitomycin C (MMC). This sensitivity is rescued to wild type (HRQ1) levels by a plasmid encoding 10xHis-Hrq1 but not by empty vector. (C) Peptide sequences demonstrating that our optimized purified protein preparations indeed contain Hrq1 or RecQ4. (D) Hrq1 (black) and RecQ4 (red) have low basal levels of ATPase activity that can be stimulated by the addition of random-sequence (random) or poly(dT) ssDNA. (E) The effect of ssDNA length on ATPase activity. The ATPase activities of Hrq1 and RecQ4 were measured in the presence of poly(dT) oligonucleotides of the indicated lengths. The slopes (rates) of the hydrolysis curves are plotted versus ssDNA length. (F and G) The effect of ssDNA sequence on Hrq1 (F) and RecQ4 (G) ATPase activity. The random-sequence, poly(dA), poly(dC), poly(dG) and poly(dT) oligonucleotides were all 50-nt long. In this and all other figures, the data points are averages from ≥3 independent experiments, and the error bars represent the standard deviation.
Construction of baculovirus transfer vectors for expression of Hqr1 and RecQ4
A DNA cassette encoding a 10× His tag followed by a human rhinovirus 3C protease (HRV 3C Protease) site (LEVLFQGP) was inserted immediately 5΄ of the BamHI site in the pKL vector (29) using the SLIC method (26), yielding the pKL-10xHis-3C vector. The DNA sequence encoding a 10x His tag followed by maltose-binding protein (MBP) and a HRV 3C site was codon optimized for expression in insect cells, synthesized, and sub-cloned into the EcoRV site of the pUC57 vector by GenScript (Piscataway, NJ), yielding the vector pUC57-10His-MBP-3C. The 10His-MBP-3C sequence was then PCR amplified using pUC57-10His-MBP-3C as the template, and the PCR product was cloned immediately 5΄ of the BamHI site of the pKL vector via the SLIC method (26), yielding the pKL-10His-MBP-3C vector.
The open reading frame (ORF) of the HRQ1 gene was PCR amplified from plasmid pMB309 (13). The PCR product was digested with RsrII and HindIII, and the DNA fragment was sub-cloned into the same sites in the pKL-10His-3C vector. DNA sequence encoding a BamHI site, the cDNA sequence for the human RecQ4 protein with a C-terminal Twin-Strep-tag (27), and a HindIII site was codon-optimized, synthesized, and sub-cloned into the EcoRV site of the pUC57 vector by GenScript (Piscataway, NJ, USA), yielding the pUC57-BamHI-RECQ4-Twin-Strep-HindIII vector. The BamHI–HindIII DNA fragment from this vector was then sub-cloned into the same sites in the pKL-10His-MBP-3C vector, yielding the pKL-10His-MBP-hRecQ4-Twin-Strep vector. Further details concerning the amplification and cloning of DNA sequences are available upon request.
Virus production and storage
The production of high-titer viruses in Sf9 cells has been described previously (28). Liquid stocks of viruses were stored at 4°C. Frozen viruses were generated as described (i.e. using the TIPS method) (29) and stored under liquid nitrogen.
Purification of recombinant S. cerevisiae Hrq1 and the catalytically inactive Hrq1-K318A mutant
Frozen cells from 500 ml insect cell culture (∼10 g) were thawed on ice and lysed at 4°C by stirring the cell pellet in 100 ml of Hrq1 lysis buffer (50 mM Na-HEPES [pH 7.6], 0.4 M NaCl, 20 mM imidazole and 10% glycerol) supplemented with fresh 5 mM β-mercaptoethanol, protease inhibitor mix (600 nM leupeptin, 2 μM pepstatin A, 2 mM benzamidine and 1 mM phenylmethanesulfonyl fluoride (PMSF)) and 20 μg/ml DNase I. The cell lysate was clarified by centrifugation at 4°C for 30 min at 14 000 rpm, and the supernatant was transferred into a 50-ml conical tube containing 2.5 ml HIS-Select Nickel Affinity Gel (Sigma) pre-equilibrated with the lysis buffer. The tube was placed onto a nutator to mix gently for 30 min at 4°C. The tube was then centrifuged at 2000 × g for 5 min at 4°C, and the bulk of the supernatant was removed. The beads were transferred to a 30-ml gravity column and washed three times (∼30 ml each) with Hrq1 lysis buffer. 10xHis-Hrq1 was eluted using five column volumes (CVs) of Hrq1 lysis buffer containing 500 mM imidazole. The purest fractions, as judged by SDS-PAGE and Coomassie staining, were pooled and dialyzed against storage buffer (25 mM Na-HEPES [pH 8], 30% glycerol, 300 mM NaOAc [pH 7.6], 25 mM NaCl, 5 mM MgOAc, 1 mM DTT and 0.1% Tween-20). The catalytically inactive 10xHis-Hrq1-K318A mutant was purified identically.
Purification of recombinant human RecQ4-Strep and the catalytically inactive RecQ4-K508A mutant
Cell lysates containing recombinant MBP-RecQ4-Strep protein or the MBP-RecQ4-K508A-Strep mutant were prepared from the frozen cell pellet from 1 l insect cell culture (∼20 g) in the same manner as described above for Hrq1 except that fresh 2 mM dithiothreitol (DTT) was used instead of β-mercaptoethanol, and no imidazole was added to the lysis buffer. Clarified lysate was mixed with 2.5 ml of amylose resin (NEB) in a 50-ml conical tube for 30 min at 4°C on a nutator. After briefly centrifuging the tube to remove the bulk of the lysate, the beads were transferred to a 30-ml gravity column as described above.
After extensive washing with RecQ4 lysis buffer, RecQ4-Strep protein was eluted from the resin by cleavage with HRV 3C protease (Novagen) overnight at 4°C. Eluate (2 CV) from the amylose resin containing RecQ4-Strep was subsequently loaded onto a Strep-Tactin Superflow column (IBA). The column was washed with 20 CVs strep wash buffer (50 mM Na-HEPES [pH 7.6], 5% glycerol, 600 mM NaOAc [pH 8.0], 5 mM MgOAc, and 0.05% Tween-20) and eluted with 6 CVs of strep wash buffer supplemented with 2.5 mM desthiobiotin. Peak elution fractions were pooled and dialyzed against storage buffer as described above.
Protein concentrations were quantified based on the intensities of protein bands on SDS-PAGE gels stained with SYPRO orange (Sigma) and using a dilution series of known concentrations of BSA to plot a standard curve. Gel imaging and analysis were performed using a Typhoon 9210 Variable Mode Imager (Amersham Biosciences).
Mass spectrometry (MS) analysis or recombinant proteins
MS analyses using an LTQ Orbitrap XL (Thermo Fisher) were performed on in-solution samples of recombinant Hrq1 (2 μg) and RecQ4 (1 μg) by the Laboratory for Biological Mass Spectrometry at Indiana University. Peptides were searched against S. cerevisiae and human databases for Hrq1 and RecQ4, respectively, using Protein Prospector version 5.16.0 (University of California San Francisco; http://prospector.ucsf.edu/prospector/mshome.htm). Peptides were also searched against the Drosophila melanogaster database to identify contaminating peptides from insect cell culture.
NADH-coupled ATPase assay
ATPase reactions were performed in ATPase buffer (25 mM Na-HEPES [pH 8.0], 5% glycerol, 50 mM NaOAc [pH 7.5], 150 μM NaCl and 0.01% NP-40 substitute) including the following reagents: 5 mM ATP (pH 7.0) (DOT Scientific), 5 mM MgCl2, 0.5 mM phosphor(enol)pyruvic acid (Sigma), 0.4 mM NADH (MP Biomedicals, LLC), 5 U/ml rabbit pyruvate kinase (Roche), and 8 U/ml lactate dehydrogenase from oyster (Sigma). Unless otherwise stated, the helicase concentration was 10 nM in all reactions, and DNA was used at 1 μM. To minimize possible confounding effects caused by secondary structures that the oligonucleotides may form, the substrates were diluted to their working concentrations, boiled, and snap-cooled prior to addition to the ATPase assays. Absorbance at 340 nm was read at 37°C in 96-well plates using a BioTek Synergy H1 microplate reader. Absorbance readings were converted to ATP turnover based on NADH concentration. It was assumed that 1 μM NADH oxidized is proportional to 1 μM ATP hydrolysed.
DNA substrates
Substrates were made by 5΄-end labeling the oligonucleotides indicated in Supplemental Table S1 with T4 polynucleotide kinase (T4 PNK; NEB) and γ[32P]-ATP. Labeled oligonucleotides were separated from free label using illustra ProbeQuant G-50 micro columns (GE Healthcare) following the manufacturer's instructions. Oligonucleotides were annealed by incubating complementary or partially complementary oligonucleotides overnight at 37°C in Annealing Buffer (20 mM Tris–HCl [pH 8], 4% glycerol, 0.1 mM EDTA, 40 μg/ml BSA, 10 mM DTT and 10 mM MgOAc) (30). The G4 DNA substrate was folded by incubating oligonucleotide MB819 (Supplemental Table S1) in 1 M NaCl at 60°C for 48 h. The folded product was 5΄-end labeled as above and gel-purified on an 8% 19:1 acrylamide:bis-acrylamide gel run in 1× TBE buffer (90 mM Tris–HCl [pH 8.0], 90 mM boric acid, and 2 mM EDTA [pH 8.0]) at 10 V/cm. The gel slice containing the folded G4 substrate was placed into a microcentrifuge tube containing TBE buffer, and the DNA was allowed to diffuse into the buffer overnight at room temperature. The G4 DNA was then ethanol-precipitated and resuspended in 1× TE buffer (90 mM Tris–HCl [pH 8.0] and 2 mM EDTA [pH 8.0]).
DNA binding
DNA binding was measured using electrophoretic mobility shift assays (EMSAs). The helicases were incubated at the indicated concentrations with 0.1 nM radiolabeled DNA for 30 min at 30°C in binding buffer (25 mM Na-HEPES [pH 8.0], 5% glycerol, 50 mM NaOAc [pH 7.6], 150 μM NaCl, 7.5 mM MgOAc and 0.01% Tween-20). Protein–DNA complexes were separated from unbound DNA on 8% 19:1 acrylamide:bis-acrylamide gels in TBE buffer at 10 V/cm. Gels were dried under vacuum and imaged using a Typhoon 9210 Variable Mode Imager. DNA binding was quantified using ImageQuant 5.2 software.
Helicase assay
DNA unwinding was assessed by incubating the indicated concentrations of helicase with 5 mM ATP, 0.1 nM radiolabeled DNA and 1× binding buffer. RecQ4 helicase assays were performed in the presence of 15 nM cold ssDNA trap to observe unwinding, which is otherwise masked by annealing (31). ATP was required to observe unwinding, suggesting that strand exchange was not the activity being measured (data not shown) (32). The presence or absence of cold trap did not affect Hrq1 unwinding efficiency and was therefore not used in Hrq1 helicase assays (data not shown) (13). Reactions were incubated at 37°C for 30 min and stopped with the addition of 1× Stop-Load dye (5% glycerol, 20 mM EDTA, 0.05% SDS and 0.25% bromophenol blue) supplemented with 400 μg/ml SDS-Proteinase K followed by a 1-min incubation at 37°C. Unwound DNA was separated on 8% 19:1 acrylamide:bis-acrylamide gels in TBE buffer at 10 V/cm and imaged as for the DNA binding assay. Unwinding time courses were performed under the same conditions as above but with a single helicase concentration of 100 nM. Reactions were incubated at 37°C for times ranging from 0 to 30 min and stopped with the addition of 1× Stop-Load dye and SDS-Proteinase K as above.
Transmission electron microscopy (TEM)
To prepare negatively stained grids, 4 μl of protein solution (∼20 ng/μl) was applied to glow-discharged 300-mesh carbon-coated copper grids (Electron Microscopy Sciences). Excess solution was removed by blotting with filter paper, and subsequently, the grid was stained with 0.75% uranyl formate (Electron Microscopy Sciences) and allowed to air dry. To minimize potential beam-induced damage to the protein samples, the grids were imaged under low-dose conditions (≤20e−/Å2) using a JEOL 300-kV 3200FS transmission electron microscope with the energy filter operated at an energy slit of 20 eV. The digital images were collected using a Gatan 4k × 4k CCD camera at a nominal magnification of 80 000×, yielding a 1.5 Å/pixel resolution in the final images. For Hrq1, a total of 139 digital micrographs were collected, and 7623 particles were selected in 128 (pixels) × 128 (pixels) boxes using the e2boxer.py program (33). For RecQ4, a total of 5025 particles were boxed. Defocus estimation and CTF (contrast transfer function) correction were performed with EMAN2 software. The phase contrast-corrected particle sets were imported into Relion for reference-free 2D classification (34).
To generate the human RecQ1 projection images for comparison, only one chain of the crystal structure of RecQ1 (PDB 2V1X, A chain) was used. Briefly, the crystal structure was low-pass filtered to 30 Å and a pixel size of 1.5 Å in a 128 × 128 × 128 (pixels) volume using e2pdb2mrc.py (33). The projection images were than generated using e2project3d.py with C1 symmetry. As the RecQ1 structure contains 591 residues, it was used as a size standard to compare Hrq1 (1077 residues) and RecQ4 (1208 residues).
Statistical analyses
All data were analysed and plotted using GraphPad Prism 6 (GraphPad Software, Inc). The plotted values are averages, and the error bars were calculated as the standard deviation from three or more independent experiments. P-values were determined by analysis of variance (ANOVA). We defined statistical significance as P < 0.01.
RESULTS
Hrq1 functions in the Pso2 ICL repair pathway
In metazoans, DNA ICLs are preferentially repaired by the Fanconi anaemia (FA) pathway, which includes many proteins that, when mutated, cause the disease FA (reviewed in (35)). However, mutations in non-FA proteins such as RecQ4 (11) also sensitize cells to ICL-inducing agents. This suggests that an alternate mechanism exists for ICL repair apart from the FA pathway. S. cerevisiae lacks homologs of many FA proteins (36), so this situation is reversed: a pathway defined by the nuclease Pso2 is preferentially used to repair ICLs rather than the minimalist FA-like pathway (Figure 1B). We previously demonstrated that like pso2Δ cells, yeast lacking HRQ1 are also sensitive to the ICL-inducing drug mitomycin C (MMC) and that hrq1Δ is epistatic to pso2Δ (see (13) and Figure 1C). However, it is unclear where Hrq1 functions in this repair pathway.
Others have suggested that Hrq1 participates in nucleotide excision repair (NER), which is the initial step in both of the yeast ICL repair pathways (Figure 1B) (37). If that is the case, then hrq1Δ should also be epistatic to deletion of the genes encoding the FA-like proteins Mph1 and Chl1 (35). To test this hypothesis, we deleted CHL1, HRQ1 and MPH1 individually and in combination and assayed for MMC sensitivity. For comparison, we also repeated the hrq1Δ and pso2Δ epistasis analysis. As reported (13), hrq1Δ and pso2Δ cells are both sensitive to chronic exposure to 50 μg/ml MMC, but there was no additive effect in the hrq1Δ pso2Δ double mutant (Figure 1C, top). In contrast, chl1Δ and mph1Δ cells displayed relatively little sensitivity to MMC compared to hrq1Δ, even at concentrations of 100 μg/ml (Figure 1C, bottom). There was no additive sensitivity in the chl1Δ mph1Δ double mutant, in agreement with the model in which they function in the same ICL repair pathway (Figure 1B and (36)). However, the chl1Δ hrq1Δ mph1Δ triple mutant grew well in rich medium but was exquisitely sensitive to MMC.
These results indicate that hrq1Δ is not epistatic to perturbations in the FA-like pathway and suggest that Hrq1 functions downstream of NER in the Pso2 ICL repair pathway. This mirrors the ICL sensitivity of recq4 mutant cell lines (11) and the lack of RecQ4 involvement in the FA pathway. However, it is still unclear how Hrq1 and RecQ4 contribute to ICL repair. Do they function immediately upstream of nuclease incision, in concert with the nuclease, or perhaps downstream during recombination? In the absence of detailed biochemical analyses, it is difficult to speculate how these enzymes participate in various DNA transactions. Thus, to gain mechanistic insight into their potential functions, we next sought to characterize and compare the biochemical activities of Hrq1 and RecQ4 in vitro.
Purification of full-length Hrq1 and RecQ4
One difficulty in performing biochemical experiments with RecQ4 sub-family helicases is obtaining sufficient amounts of recombinant protein. Both Hrq1 and RecQ4 are large (123 and 133 kDa, respectively), and their N-terminal domains are predicted to be natively disordered (Supplemental Figure S1A and B), making over-expression troublesome. To overcome this, we optimized expression and purification schemes for both helicases (see the Online Methods for details). Briefly, over-expressing N-terminally 10× His-tagged Hrq1 in insect cell culture ultimately yielded 20–40 mg protein with a final concentration of 35–40 μM per liter of Hi5 cells. When expressed in S. cerevisiae, this tagged construct rescued the sensitivity of hrq1Δ cells to MMC (Figure 2B), and recombinant 10× His-Hrq1 displayed indistinguishable biochemical activities compared to C-terminally 6xHis-tagged Hrq1 overexpressed in Escherichia coli (data not shown; (13)).
RecQ4 expression and purification was more demanding. The RECQ4 cDNA was codon optimized for expression in insect cells, and purification was facilitated by N-terminal MBP (ultimately removed) and C-terminal Twin-Strep-tags (27). A typical purification from 1 l of Hi5 cell culture yielded 50–100 μg protein with a final concentration of ∼1.2 μM.
Fluorescent staining of our protein preparations revealed that they are highly (>99%) pure (Figure 2A). To verify their identity and purity, samples of Hrq1 and RecQ4 were analysed by mass spectrometry (MS). The results demonstrated good coverage of both Hrq1 (61% coverage by 86 unique peptides) and RecQ4 (54% coverage by 78 unique peptides) with high confidence peptides (Figure 2C and Supplemental Table S2). Furthermore, both protein preparations were highly pure and absent of enzymes that could confound the results of the enzymatic assays presented below (Supplemental Table S2). Overall, these data indicated that our novel Hrq1 and RecQ4 over-expression and purification schemes yielded copious amounts of recombinant protein suitable for rigorous biochemistry.
Hrq1 and RecQ4 ATPase activity is stimulated by ssDNA
We first assayed for the ATPase activity of our recombinant enzymes. As previously reported (12,20,38), the wild-type proteins each displayed weak ATPase activity that was stimulated by the addition of random-sequence or poly(dT) ssDNA 50mer oligonucleotides (Figure 2D). Because there was no significant difference in the stimulation of ATPase activity by the poly(dT) versus random-sequence ssDNA (P > 0.01), subsequent experiments were performed with poly(dT) oligonucleotides (unless stated otherwise) to avoid unwanted DNA secondary structure formation. We also verified that the ATPase activity of each enzyme was not due to a spurious contaminant from our overexpression and purification procedure because Walker A box lysine-to-alanine substitution mutants (Hrq1-K318A and RecQ4-K508A) purified identically to the wild-type proteins did not display ATP hydrolysis above background levels (Supplemental Figure S2).
ATPase stimulation was apparent with substrates as short as 5 nt and increased with increasing ssDNA length up to a maximum of ∼50-fold over reactions lacking ssDNA (Figure 2E). For both helicases, maximum stimulation was achieved with 30–40 nt ssDNA. Previously, it was reported that maximal RecQ4 ATPase stimulation requires ∼60 nt of poly(dT), whereas the reported DNA binding site size is 20–40 nt (12). It is unclear why the ssDNA stimulation of RecQ4 ATPase activity plateaued at ∼30 nt here, but additionally increasing the substrate length to up to 100 nt had no further effect on stimulation (data not shown).
DNA sequence affects Hrq1 activity in vitro (13), so we also quantitated the effect of ssDNA sequence on Hrq1 and RecQ4 ATPase activity using equimolar concentrations of poly(dA), (dC), (dG), (dT), or random sequence 50mer oligonucleotides (Figure 2F and G). For both helicases, polypyrimidine ssDNA preferentially stimulated ATPase activity compared to polypurine ssDNA. However, for Hrq1, the poly(dT) and random sequence substrates both maximally stimulated ATPase activity (Figure 2F), whereas for RecQ4, addition of the poly(dC) substrate yielded maximum ATP hydrolysis (Figure 2G).
Hrq1 and RecQ4 bind a similar suite of DNA substrates in vitro
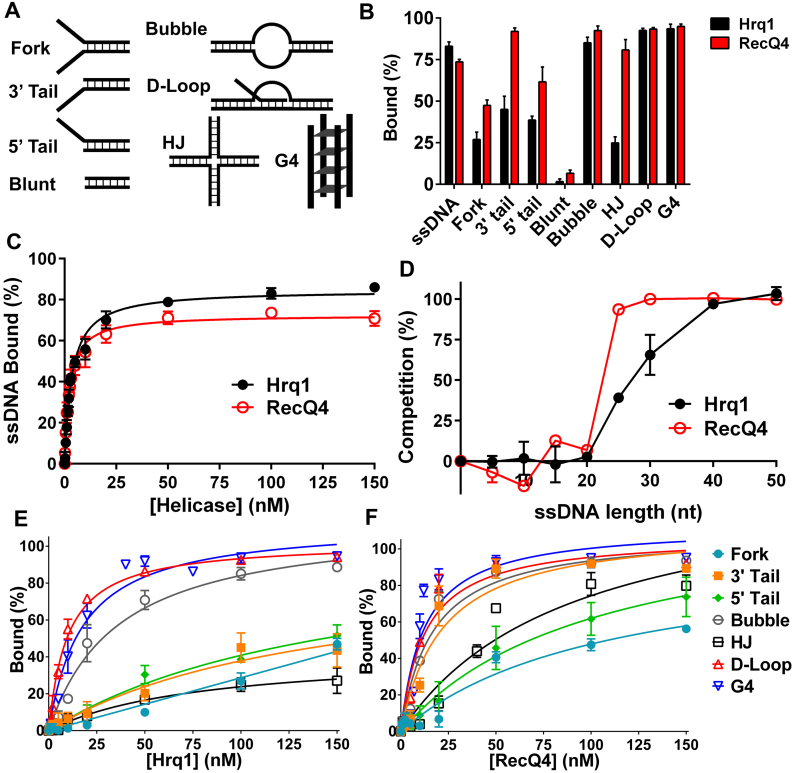
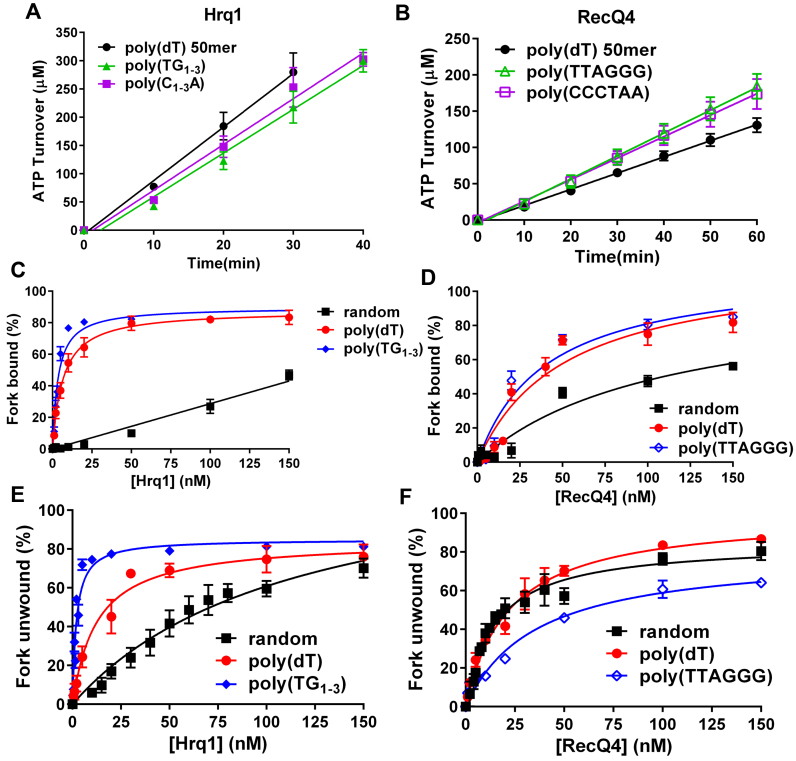
Using electrophoretic mobility shifts assays (EMSAs), we previously demonstrated that recombinant Hrq1 binds ssDNA substrates with high affinity (13). Here, the Hrq1 and RecQ4 overexpressed and purified using our optimized scheme also displayed high affinity for 40mer poly(dT) ssDNA (Hrq1 Kd = 3.45 ± 0.19 nM, RecQ4 Kd = 2.38 ± 0.15 nM; Figure 3B and C) but bound poorly to blunt-ended dsDNA (Figure 3A and B). Performing competition assays with radiolabeled 40mer poly(dT) ssDNA and a 100-fold molar excess of unlabeled poly(dT) substrates of various lengths, we found that only substrates ≥ 25 nt could compete for binding (Figure 3D), suggesting that these helicases have large binding site sizes. This also corresponds to the maximal stimulation of ATPase activity exerted by ssDNA ≥ 30 nt in length (Figure 2E) and the previously reported footprint of RecQ4 (12).
Figure 3.
Hrq1 and RecQ4 bind a broad range of DNA substrates. (A) Cartoon of the various DNA substrates used in the EMSA assays (not shown: ssDNA). (B) DNA binding by 100 nM Hrq1 (black) and RecQ4 (red). (C) ssDNA binding as a function of helicase concentration. Both helicases bind tightly to poly(dT) ssDNA. (D) Hrq1 and RecQ4 have a binding site size of 20–40 nt. Competition assays were performed using radiolabeled poly(dT) 40mer and a 100-fold molar excess of unlabeled poly(dT) substrates of the indicated lengths. (E and F) DNA binding curves for Hrq1 (E) and RecQ4 (F) for the indicated substrates.
Next, we assayed for binding to a variety of DNA substrates with different structures. Both helicases bound to dsDNA substrates containing either a 5΄ or 3΄ ssDNA tail, a substrate containing both tails (fork), a substrate containing a ssDNA bubble, a model D-loop, a Holliday junction (HJ), and a G4 structure (Figure 3A and B, Supplemental Figures S3 and S4). The dissociation constant (Kd) for each substrate is listed in Table 1, and a comparison of binding at 100 nM helicase is presented in Figure 3B. Overall, Hrq1 and RecQ4 bound to the same structures, though their relative affinities varied by up to eight-fold.
Table 1. Dissociation constants (Kd) and apparent Michaelis constants (KM) for Hrq1 and RecQ4.
| Hrq1 | RecQ4 | |||
|---|---|---|---|---|
| Substrate | Binding Kd (nM) | Unwinding KM (nM) | Binding Kd (nM) | Unwinding KM (nM) |
| Poly(dT) ssDNA1 | 3.45 ± 0.19 | N/A | 2.38 ± 0.15 | N/A |
| Random Fork | N/D | 113.9 ± 19.5 | 107.7 ± 32.5 | 14.7 ± 1.1 |
| 3΄ tail | 140.7 ± 59.4 | 111.4 ± 43.4 | 21.8 ± 4.2 | 98.1 ± 17.8 |
| 5΄ tail | 154.5 ± 44.6 | - | 101.6 ± 29.4 | - |
| Blunt dsDNA | - | - | - | - |
| Bubble | 36.1 ± 4.9 | 23.6 ± 2.2 | 16.8 ± 2.3 | 143 ± 56.1 |
| HJ | 79.2 ± 22.7 | - | 90.1 ± 23.4 | - |
| D-loop | 9.9 ± 0.7 | 1.6 ± 0.1 | 12.6 ± 1.8 | 5.7 ± 0.8 |
| G4 | 17.6 ± 2.2 | 14.1 ± 0.9 | 12.3 ± 2.5 | 38.4 ± 8.7 |
| Poly(dT) Fork | 6.5 ± 0.5 | 12.6 ± 1.7 | 50.9 ± 10.4 | 21.7 ± 1.1 |
| Yeast Tel Fork | 3.5 ± 0.4 | 2.0 ± 0.2 | 18.5 ± 3.1 | 9.2 ± 1.6 |
| Human Tel Fork | 15.7 ± 1.7 | 23.0 ± 1.5 | 39.7 ± 8.3 | 35.4 ± 5.4 |
1Abbreviations used: ssDNA, single-stranded DNA; N/D, not determined; N/A, not applicable; dsDNA, double-stranded DNA; HJ, Holliday junction; G4, G-quadruplex; and Tel, telomere repeat sequence.
Hrq1 and RecQ4 display similar helicase substrate specificity
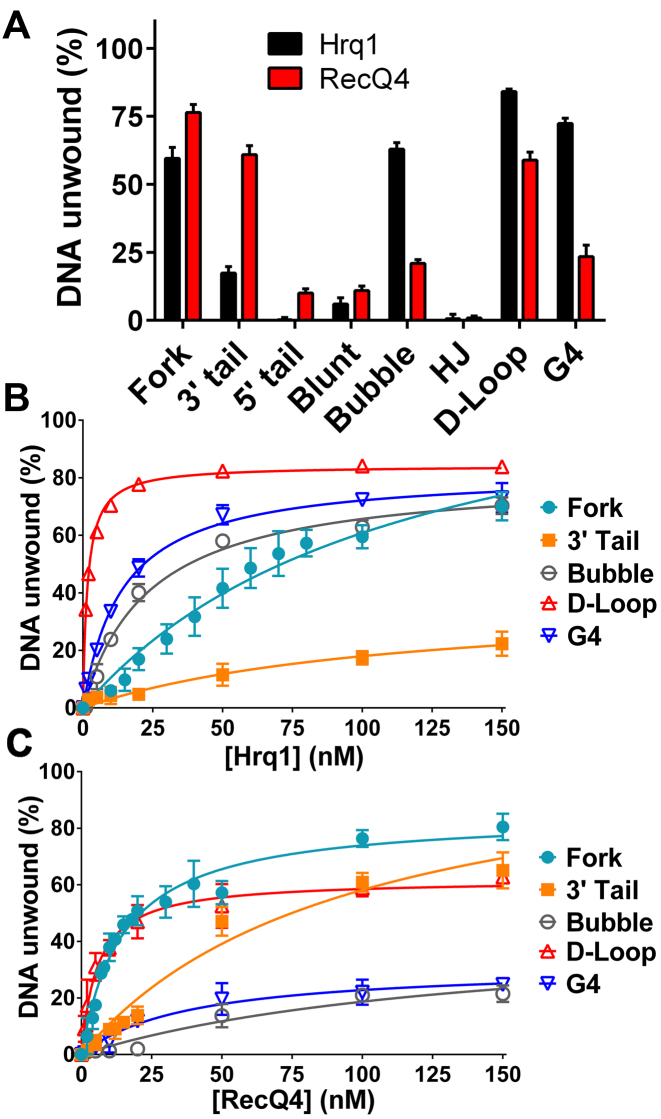
We next investigated the unwinding of our DNA substrates (Supplemental Figures S5 and S6). As shown in Figure 4A, at 100 nM protein, both helicases displayed appreciable (>17%) unwinding activity on all substrates except for the 5΄-tail, blunt dsDNA, and HJ structures. This was neither surprising for the blunt dsDNA due to the poor binding of this substrate by the helicases (Figure 3B) nor for the 5΄ tail substrate given the 3΄-5΄ directionality of both Hrq1 and RecQ4 (13). Similarly, RecQ4 was previously shown not to unwind HJs (12,39). Some differences between Hrq1 and RecQ4 were also noted. For instance, Hrq1 was highly active on the bubble substrate, but RecQ4 was less able to unwind this structure. This may be due to the strong annealing activity of RecQ4 (32) and the two separate regions of complementary DNA comprising the bubble.
Figure 4.
Hrq1 and ReQ4 preferentially unwind D-loops. (A) DNA unwinding by 100 nM Hrq1 (black) and RecQ4 (red). (B and C) Helicase activity as a function of Hrq1 (B) or RecQ4 (C) concentration. Both helicases display the highest unwinding activity on the D-loop substrate.
Helicase activity was next measured at a variety of enzyme concentrations to determine the apparent Michaelis constant (KM) for each substrate that was appreciably unwound in Figure 4A and, thus, the relative specificities of the helicases for these substrates. The data are plotted for Hrq1 and RecQ4 in Figure 4B and C, respectively, and the KM values (i.e. the concentration of enzyme needed to unwind 50% of the substrate) are listed in Table 1. Hrq1 preferentially unwound the D-loop substrate (KM = 1.6 ± 0.1 nM), with the most to least preferred substrates being D-loop > G4 > bubble > fork ≈ 3΄ tail. RecQ4 was also most active on the D-loop (KM = 5.7 ± 0.8 nM) but displayed a somewhat different substrate preference (D-loop > fork > G4 > 3΄ tail > bubble) compared to Hrq1. Again, we hypothesize that this discrepancy is due to the strong annealing activity of RecQ4.
DNA helicase substrate preference can also be revealed by the kinetics of unwinding, with more rapid kinetics indicating a more preferred substrate. Therefore, we measured DNA unwinding as a function of time for Hrq1 and RecQ4. We focused on the D-loop and fork substrates because both helicases displayed the lowest KM for the D-loop (Table 1), and the extent of unwinding for the fork was high by both enzymes (Figure 4A–C). Hrq1 and RecQ4 unwound the fork with similar kinetics, with the time necessary to unwind 50% of the substrate (t1/2) equal to 2.9 ± 0.3 and 5.6 ± 0.4 min, respectively (Supplemental Figure S7A). Consistent with their preference for the D-loop substrate based on KM values, both helicases displayed faster unwinding of the D-loop compared to the fork (t1/2 = 1.0 ± 0.1 min for both; Supplemental Figure S7B), suggesting that it is it truly a preferred substrate. Although the duplex portions of the fork and D-Loop are the same length (20 bp) with nearly identical predicted melting temperatures (Tm = 53.4°C and 53.3°C, respectively) and similar G+C content (50% versus 45%), we did find that DNA sequence affects Hrq1 and RecQ4 activity (Figure 2F and G). Thus, the more rapid unwinding kinetics of the D-loop could be due to the sequence of the dsDNA portion of this substrate relative to the fork.
Stimulation by telomeric repeat sequence
Aside from ICL repair, Hrq1 (13) and RecQ4 (19) also function in telomere maintenance. Thus, we sought to determine if yeast telomeric repeat (TG1-3/C1-3A) and human telomeric repeat (TTAGGG/CCCTAA) sequences would also stimulate Hrq1 and RecQ4 biochemical activities. Inclusion of ∼50-nt ssDNA comprised of either the G-strand or C-strand telomeric repeat sequences in the ATPase assay revealed that Hrq1 ATP hydrolysis was significantly less stimulated by poly(TG1-3) (P = 0.0005128) and poly(C1-3A) (P = 0.007746) than poly(dT) DNA (Figure 5A). In contrast, RecQ4 ATPase activity was significantly more stimulated by poly(TTAGGG) (P < 0.0001) and poly(CCCTAA) (P < 0.0001) ssDNA than poly(dT) ssDNA (Figure 5B).
Figure 5.
Hrq1 helicase activity is stimulated by the S. cerevisiae telomeric DNA repeat sequence. (A and B) Stimulation of ATP hydrolysis by telomeric repeat sequence ssDNA. (C) DNA binding as a function of Hrq1 concentration for random-sequence, poly(dT), and yeast telomeric repeat sequence ssDNA. Hrq1 binds yeast telomeric repeat sequence ssDNA with the highest affinity. (D) DNA binding as a function of RecQ4 concentration for random-sequence, poly(dT), and human telomeric repeat sequence ssDNA. The binding affinities for poly(dT) and human telomeric repeat sequence forks do not significantly differ. (E) Helicase as a function of Hrq1 concentration for DNA forks with random-sequence, poly(dT), and yeast telomeric repeat sequence ssDNA tails. Hrq1 displays preferential unwinding of the yeast telomeric repeat sequence fork. (F) Helicase activity as a function of RecQ4 concentration for DNA forks with random-sequence, poly(dT), and yeast telomeric repeat sequence ssDNA tails. The presence of the human telomeric repeat sequence as the ssDNA tails of the fork inhibited RecQ4 unwinding.
Using fork substrates with both ssDNA tails comprised of G-strand telomeric repeats, we next performed EMSAs to assess the effects of the telomeric sequence on binding affinity. As shown in Figure 5C and Table 1, the yeast telomeric sequence greatly increased Hrq1 binding affinity compared to a fork containing 25-nt random sequence tails (yeast telomeric fork Kd = 3.5 ± 0.4 nM versus an apparent Kd > 150 nM for the 25-nt random sequence fork) and nearly two-fold compared to a fork containing 25-nt poly(dT) tails (Figure 5C and Table 1). Similarly, RecQ4's binding affinity for the human telomeric sequence fork increased 2.7- and 1.3-fold compared to the 25-nt random sequence and poly(dT) forks, respectively (Figure 5D and Table 1).
Increases in binding affinity do not always equate to increases in helicase activity. A substrate can be bound with such high affinity that it decreases or eliminates helicase activity (data not shown), ostensibly because translocation along the ssDNA toward the dsDNA to be unwound is inhibited. Therefore, we also assessed Hrq1 and RecQ4 helicase activity on the tightly bound telomeric fork substrates. The yeast telomeric fork was more readily unwound by Hrq1, decreasing the KM 57- and 6.3-fold compared to the 25-nt random sequence and poly(dT) forks, respectively (Figure 5E and Table 1). In contrast, RecQ4's KM for the human telomeric fork was increased 2.4- and 1.6-fold relative to the 25-nt random sequence and poly(dT) forks, respectively (Figure 5F and Table 1), suggesting that human telomeric repeat sequence is not a preferred RecQ4 helicase substrate.
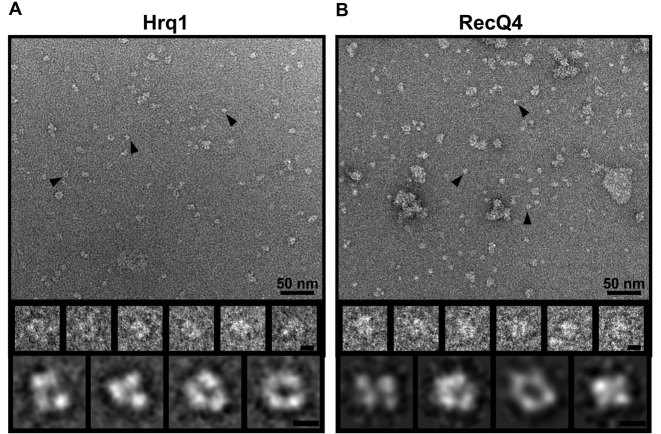
Hrq1 and RecQ4 display similar tertiary conformations
Hrq1 and RecQ4 are predicted to have similar domain architectures (Figure 1A). Combined with their in vivo functions and in vitro biochemical activities, we hypothesized that they also fold into similar tertiary structures. To investigate this, we performed TEM analysis followed by single-particle 2D structural class averaging. In both cases, the negatively stained protein preparations appeared to be uniform and homogeneous on grids, again verifying their purity (Figure 6A and B). However, class averaging indicated that the particles encompassed a narrow range of sizes, with the minority of smaller particles likely either representing low-profile views of the protein structures or degradation products. By modeling human RecQ1 (Supplemental Figure S8), which contains ∼50% the residues of Hrq1 and RecQ4 and is thus predicted to have a smaller 3D structure, we were able to eliminate class averages that were too small to be full-length Hrq1 or RecQ4. The remaining 2D class averages of Hrq1 and RecQ4 show that they have remarkably similar structures. The three lobes of electron density in Figure 6A suggest that Hrq1 is comprised of three globular domains arranged in a horseshoe shape around an empty central cavity. The larger RecQ4 appears to consist of four globular domains forming another U-shape (or possibly a ring) with an opening in the center (Figure 6B). Furthermore, the majority of observed particles for both helicases are monomeric, suggesting this is the predominant oligomeric state for each protein and consistent with the monomeric state of Drosophila RecQ4 predicted by sedimentation velocity (40).
Figure 6.
Hrq1 and RecQ4 display conserved tertiary structural features. Image analysis of Hrq1 (A) and RecQ4 (B). (Upper panels) Representative TEM images of negatively stained recombinant protein. Examples of boxed particles for 2D image analysis are marked with black arrowheads. A variable number of aggregates were observed and were likely due to the dehydration process that was used to prepare the negatively stained specimens. Scale bar, 50 nm. (Middle panels) Selected particles are shown in enlarged views. Scale bar, 5 nm. (Lower panels) Selected reference-free 2D class averages of Hrq1 (A) and RecQ4 (B) from different views. From the class averages, Hrq1 appears to be comprised of three domains that form a U-shape with an opening in the centre. Likewise, RecQ4 class averages contain four globular densities indicative of four domains that also form a U- or ring-shape with an opening in the middle. Scale bar, 5 nm.
DISCUSSION
Hrq1 is a functional homolog of RecQ4
There is a need for a model system to determine the mechanism(s) by which human RecQ4 helps to maintain genome integrity and why RECQ4 mutations result in three different diseases. Barea et al. suggest that the S. cerevisiae Hrq1 may serve as this model, but due to poor sequence conservation and a lack of experimental insight into both RecQ4 and Hrq1, it is important to determine if Hrq1 is truly a functional homolog of RecQ4. Our previous work (13) and that presented in Figure 1 indicate that in vivo, Hrq1 displays similar functions to RecQ4. Here, by performing a detailed side-by-side biochemical comparison of the two helicase and observing their structures by TEM, we found that Hrq1 and RecQ4 also share conserved biochemical activities and structural features. Thus, S. cerevisiae Hrq1 is a true functional homolog of the human RecQ4 helicase.
In 2014, we showed that Hrq1 functions in vivo in Pso2 ICL repair pathway (13), but it was unclear at which step in the repair process (Figure 1B) Hrq1 activity is needed. It has been postulated that Hrq1 functions early in ICL repair during NER (37). This hypothesis is based on the sensitivity of hrq1Δ cells to cisplatin and 4-nitroquinoline-1-oxide (4NQO) and hrq1Δ epistasis with deletions of genes that encode products involved in NER (RAD4 and RAD10), though the effects are rather weak. To address this, we tested the effects of HRQ1 deletion on both ICL repair pathways in yeast and found that hrq1Δ is epistatic to pso2Δ (Pso2 pathway) but not to chl1Δ and mph1Δ (FA-like pathway). This places Hrq1 downstream of NER in ICL repair and demonstrates that, like RecQ4, Hrq1 does not participate in the FA/FA-like pathway (Figure 1B). However, it is still unclear what role(s) these helicases play in the non-FA ICL repair pathway.
To establish a foundation upon which more mechanistic details of repair processes could be elucidated, we characterized the biochemical activities of Hrq1 and RecQ4 on various types of DNA substrates, including ones mimicking repair intermediates. After developing optimized protein over-expression and purification protocols, we found that Hrq1 and RecQ4 are DNA-stimulated ATPases and that both DNA length and sequence similarly affect ATP hydrolysis by these enzymes (Figure 2). Further, Hrq1 and RecQ4 bound a diverse and comparable set of DNA structures (Figure 3), including non-B form DNA such as the TP G4 structure from the mouse immunoglobulin locus (41). This is characteristic of RecQ family helicases, many of which bind a variety of DNA structures in vitro (reviewed in (22)). Binding appears to be structure specific rather than an effect of Hrq1 and RecQ4 binding to the simple ssDNA or B-form dsDNA portion(s) of more complicated structures. For instance, neither helicase appreciably bound blunt dsDNA, but both bound to the HJ substrate (Figure 3B), which is largely composed of four blunt dsDNA arms (Figure 3A).
All substrates containing dsDNA that were bound by Hrq1 and RecQ4 were also unwound by the helicases, with the exception of the 5΄ tail and HJ substrates. HJ processing is a hallmark of RecQ family helicases (22,42–44), but in S. cerevisiae and humans, the Sgs1 and BLM RecQ helicases perform this function, respectively. Thus, while RecQ4 sub-family helicases may not be HJ resolvases, their high affinity for HJ structures (previously noted for RecQ4 (45)) suggests that they may still have a role in HJ processing in vivo. Perhaps in the presence of additional protein cofactors they are capable of unwinding branched dsDNA molecules, or they may serve a non-catalytic role by binding to HJs and recruiting other recombination factors. This catalytic vs. structural dichotomy has been previously demonstrated for Hrq1's roles in DNA ICL repair and telomere maintenance, respectively (13).
Despite these advances in our knowledge of Hrq1 and RecQ4 biochemistry, it is still unclear what function(s) they serve in ICL repair. Our findings that Hrq1 and RecQ4 bind HJs and D-loops (Figures 3 and 4) and that D-loops appear to be preferred helicase substrates (Supplemental Figure S7) indicate that Hrq1 and RecQ4 may function at the level of DNA recombination during ICL repair. This would be consistent with the known roles of nearly all RecQ helicases in recombination (1). Alternatively, they may unwind a bubble of DNA surrounding an ICL to stimulate the nuclease activity of enzymes such as Pso2 or simply provide them with better access to the lesion to be excised. This would be consistent with the binding affinity and unwinding activity of Hrq1 and RecQ4 on bubble-like substrates (Figures 3 and 4). Helicases and nucleases can stimulate the activity of their counterpart by creating a preferred substrate. For instance, Escherichia coli DNA end resection functions in this manner, with the RecQ helicase creating 5΄ ssDNA tails for RecJ exonuclease activity, which in turn creates 3΄ overhangs, a preferred substrate of RecQ (46).
Hrq1 and RecQ4 adopt similar structures
Aside from the NMR structure of a portion of the RecQ4 N-terminus (47), no high resolution structural data exist for either RecQ4 or Hrq1. Here, we report the first 2D EM class averages of the full-length human RecQ4 (Figure 6B) and the parallel comparison with Hrq1 (Figure 6A). There are a number of details that can be inferred from this analysis. Importantly, both proteins appear to fold into similar overall structures, with three-to-four globular domains surrounding an empty cavity. This horseshoe shape is distinct from that of our RecQ1 model, which also displays three-to-four lobes of electron density but in a more elongated ‘wide V’ shape (Supplemental Figure S8). Thus, the observed Hrq1/RecQ4 horseshoe structure is not universally adopted by RecQ helicases, suggesting that it is unique to RecQ4-subfamily helicases and again supports the notion that Hrq1 and RecQ4 are homologous enzymes. Notably, RecQ4 appears to have an extra domain relative to Hrq1, which could correspond to the Sld2-like portion of the RecQ4 N-terminus (Figure 1A).
Most of the Hrq1 and RecQ4 particles appeared to be monomeric in size. This agrees with the predicted oligomeric state of RecQ4 in solution (40), but we previously reported that recombinant Hrq1 predominantly forms heptameric rings (13). Although some larger ring-like particles were observed on Hrq1 grids, they were in the extreme minority relative to the monomeric particles. The addition of ATP, ATPγS, and/or ssDNA did not shift the monomers to higher order oligomers (data not shown). This discrepancy may be explained by the different Hrq1 expression systems used. The largely heptameric Hrq1 was produced by over-expression in E. coli (13), but the recombinant Hrq1 analysed here was over-expressed in insect cells. If the Hrq1 ring-like structure is an artifact of over-expression, a prokaryotic expression system may be more prone to producing these structures compared to a eukaryotic system, which would be predicted to more easily fold eukaryotic proteins into native conformations. Future work using TEM and cryo-EM will address these and other issues concerning the Hrq1 and RecQ4 structures.
Hrq1 is preferentially stimulated by telomeric repeat sequence DNA
All five human RecQ family helicases are implicated in telomere maintenance (19,48–54), though the links to RecQ5 are tenuous (reviewed in (55)). The two S. cerevisiae RecQs, Sgs1 and Hrq1, are no different. Sgs1 promotes telomere replication (56) and functions in recombination-mediated telomere maintenance in cells lacking telomerase (57,58) Similarly, Hrq1 inhibits telomerase activity and promotes type I survivor formation in cells lacking telomerase (13). Such in vivo results are also supported by biochemical evidence. For instance, in the case of the human WRN helicase, it preferentially unwinds strand invasion intermediate structures, and this activity is enhanced by the presence of telomeric repeat sequence on the invading strand (53). Likewise, we found that yeast telomeric repeat sequence DNA stimulated the DNA binding and DNA unwinding activities of Hrq1 (Figure 5). Due to the G-rich nature of the yeast telomeric repeat sequence (TG1-3), these results were surprising given the poor stimulation of Hrq1 ATPase activity by poly(dG) ssDNA (Figure 2F). However, poly(TG1-3) oligonucleotides can fold into G4 structures (59), and Hrq1 displayed robust DNA binding (Figure 3E) and unwinding (Figure 4B) activity on G4 DNA. Because many RecQ helicases unwind G4 DNA in vitro (reviewed in (60) and (41)) and are associated with multiple nuclear processes impacted by DNA sequence motifs capable of forming such structures in vivo (51,61–64), the connections between Hrq1 and G4 motifs in S. cerevisiae should be further explored. It should also be noted that we observed weak unwinding of the G4 substrate by RecQ4. Previous reports have failed to demonstrate the resolution of G4 structures by RecQ4 (32,39), but these assays used more DNA and included KCl. Both higher DNA concentration and the presence of K+ ions favour the formation of G4 DNA (60), which could have stabilized the substrate against unwinding by RecQ4 or led to the spontaneous refolding of the G4 structure in vitro after RecQ4 unwinding. Either scenario would mask G4 DNA unwinding in gel-based assays.
Curiously, we did not observe significant stimulation of RecQ4's helicase activity by human telomeric forked DNA above that of random-sequence or poly(dT) forks (Figure 5). This discrepancy between the two helicases may simply be the result of not uncovering the proper telomeric substrate for RecQ4 and/or the lack of additional protein partners. It has been shown that RecQ4 helicase activity is stimulated by D-loops containing telomeric sequence, as well as by components of the telomeric shelterin complex (19,65). Further, RecQ4 can act cooperatively with the WRN helicase to unwind telomeric DNA substrates containing an 8-oxoguanine lesion (19), and RecQ4 interacts with BLM (66). Because RecQ4 displays a preference for DNA structures found in recombination, we plan to survey the ability of recombination intermediates, other humans RecQs, and telomeric proteins for their ability to stimulate RecQ4 activity in the future.
Differences in Hrq1 and RecQ4 biochemistry are likely due to the RecQ4 Sld2-like domain
Despite the many biochemical similarities between Hrq1 and RecQ4 that we observed, distinct differences in activity were also found. These differences are most likely due to the Sld2-like domain present in the RecQ4 N-terminus, which enables RecQ4 to anneal complementary ssDNA sequences into dsDNA. This is the opposite of helicase activity and occurs in an ATP-independent fashion, resulting in an equilibrium between DNA unwinding and re-annealing in our in vitro helicase assays. This phenomenon complicates interpretation of the biochemical results of RecQ4 unwinding for any multi-duplex region substrate, such as bubbles or D-loops, because intra-molecular annealing always competes with the unwinding of intermediates. Indeed, a molar excess of unlabeled ssDNA trap must be used in RecQ4 helicase reactions to maintain the unwound radiolabeled product in a single-stranded form for detection in our gel-based assays.
In contrast to RecQ4, Hrq1 is devoid of annealing activity (Supplemental Figure S9A). This is not surprising because Hrq1 lacks a Sld2-like domain (Figure 1A), which is required for RecQ4 annealing activity (32,45). Indeed, one can separate the annealing and unwinding activity of RecQ4 by truncating the protein at the beginning of its helicase domain (32). Soluble N-terminal fragments (e.g. aa 1–388) display strong annealing activity but lack helicase activity. Conversely, the C-terminal helicase portion of the enzyme (aa 464–1208) is nearly devoid of annealing activity but still able to unwind DNA. Thus, RecQ4464-1208 functions more like Hrq1 in vitro.
Hypothetically then, fusing S. cerevisiae Sld2 (which also displays DNA annealing activity (15)) to the N-terminus of Hrq1 should render the chimera more RecQ4-like than Hrq1 alone. We attempted this in yeast cells, but the fusion construct did not support viability in the absence of the essential wild type SLD2 gene (data not shown). This chimera was a blunt fusion of the SLD2 sequence directly onto the 5΄ end of the HRQ1 gene, which may be translated into a protein that is not folded or processed correctly, conformationally constrains Sld2 and disrupts its biochemical functions, or introduces steric hindrance that disrupts critical interactions of Sld2 with the yeast replication initiation machinery. Similarly, we sought to generate a recombinant Sld2-Hrq1 fusion protein for in vitro experiments, but expression levels were poor, even in our optimized baculovirus expression scheme (Supplemental Figure S9B). The Sld2-Hrq1 chimera is larger (∼180 kDa) than RecQ4, making it more difficult to over-express at high levels. Despite scaling up our expression cultures, we were not able to obtain sufficiently pure protein for biochemical experiments. Thus, technical challenges remain to address this hypothesis.
In conclusion, we report optimized over-expression and purification methods for the full-length Hrq1 and RecQ4 helicases and establish Hrq1 as a functional homolog of RecQ4 for biochemical investigations. We also report the first extensive biochemical analysis of Hrq1 and show that it binds and unwinds a diverse array of DNA substrates. Although there are some discrepancies in the enzymatic activities of Hrq1 and RecQ4, we hypothesize that many of the differences are due to RecQ4's strong annealing activity. Regardless, because Hrq1 can be over-expressed to levels many times that of RecQ4, we feel that it is the preferred biochemical system for investigation of the functions of RecQ4 sub-family helicases. Because Hrq1 is not involved in DNA replication, the yeast system is also a favourable model for mutational analysis in vivo. Thus, Hrq1 will be an important tool for future investigations of the roles of RecQ4 dysfunctions in human disease.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Laboratory for Biological Mass Spectrometry at Indiana University for assistance with identifying Hrq1 and RecQ4 in our recombinant protein preparations. We thank K. Yamada for his help cloning the human RecQ4 gene into the transfer vector. We also thank members of the Bochman and van Kessel labs, as well as Anthony Schwacha and Soni Lacefield, for comments and suggestions regarding this manuscript.
Footnotes
Present address: Tsuyoshi Imasaki, Riken Center for Life Science Technologies, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045 Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
College of Arts and Sciences, Indiana University [to M.L.B.]; Indiana University Collaborative Research Grant fund of the Office of the Vice President for Research [to M.L.B. and Y.T.]; American Cancer Society [RSG-16-180-01-DMC to M.L.B.]; National Institutes of Health [R01 GM111695 to Y.T.]; National Science Foundation [MCB-1157688 to Y.T.]. Funding for open access charge: American Cancer Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Croteau D.L., Popuri V., Opresko P.L., Bohr V.A.. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014; 83:519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Larizza L., Roversi G., Volpi L.. Rothmund-Thomson syndrome. Orphanet. J. Rare Dis. 2010; 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y. Rothmund-Thomson syndrome helicase, RECQ4: On the crossroad between DNA replication and repair. DNA Repair (Amst.). 2010; 9:325–330. [DOI] [PubMed] [Google Scholar]
- 4. Temtamy S.A., Aglan M.S., Nemat A., Eid M.. Expanding the phenotypic spectrum of the Baller-Gerold syndrome. Genet Couns. 2003; 14:299–312. [PubMed] [Google Scholar]
- 5. Suhasini A.N., Brosh R.M. Jr. Disease-causing missense mutations in human DNA helicase disorders. Mutat. Res. 2013; 752:138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dietschy T., Shevelev I., Stagljar I.. The molecular role of the Rothmund-Thomson-, RAPADILINO- and Baller-Gerold-gene product, RECQL4: recent progress. Cell. Mol. Life Sci. 2007; 64:796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Croteau D.L., Rossi M.L., Canugovi C., Tian J., Sykora P., Ramamoorthy M., Wang Z.M., Singh D.K., Akbari M., Kasiviswanathan R. et al. . RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell. 2012; 11:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamimura Y., Masumoto H., Sugino A., Araki H.. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 1998; 18:6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Capp C., Wu J., Hsieh T.S.. RecQ4: the second replicative helicase. Crit. Rev. Biochem. Mol. Biol. 2010; 45:233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cabral R.E., Queille S., Bodemer C., de Prost Y., Neto J.B., Sarasin A., Daya-Grosjean L.. Identification of new RECQL4 mutations in Caucasian Rothmund-Thomson patients and analysis of sensitivity to a wide range of genotoxic agents. Mutat. Res. 2008; 643:41–47. [DOI] [PubMed] [Google Scholar]
- 11. Jin W., Liu H., Zhang Y., Otta S.K., Plon S.E., Wang L.L.. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum. Genet. 2008; 123:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macris M.A., Krejci L., Bussen W., Shimamoto A., Sung P.. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst.). 2006; 5:172–180. [DOI] [PubMed] [Google Scholar]
- 13. Bochman M.L., Paeschke K., Chan A., Zakian V.A.. Hrq1, a homolog of the human RecQ4 helicase, acts catalytically and structurally to promote genome integrity. Cell Rep. 2014; 6:346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu X., Liu Y.. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009; 28:568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanter D.M., Kaplan D.L.. Sld2 binds to origin single-stranded DNA and stimulates DNA annealing. Nucleic Acids Res. 2011; 39:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barea F., Tessaro S., Bonatto D.. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput. Biol. Chem. 2008; 32:349–358. [DOI] [PubMed] [Google Scholar]
- 17. Larsen N.B., Hickson I.D.. RecQ helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 2013; 767:161–184. [DOI] [PubMed] [Google Scholar]
- 18. Marino F., Vindigni A., Onesti S.. Bioinformatic analysis of RecQ4 helicases reveals the presence of a RQC domain and a Zn knuckle. Biophys. Chem. 2013; 177-178:34–39. [DOI] [PubMed] [Google Scholar]
- 19. Ghosh A.K., Rossi M.L., Singh D.K., Dunn C., Ramamoorthy M., Croteau D.L., Liu Y., Bohr V.A.. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012; 287:196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kwon S.H., Choi D.H., Lee R., Bae S.H.. Saccharomyces cerevisiae Hrq1 requires a long 3΄-tailed DNA substrate for helicase activity. Biochem. Biophys. Res. Commun. 2012; 427:623–628. [DOI] [PubMed] [Google Scholar]
- 21. Bochman M.L. Roles of DNA helicases in the maintenance of genome integrity. Mol. Cell. Oncol. 2014; 1:e963429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bernstein K.A., Gangloff S., Rothstein R.. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010; 44:393–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thompson J.D., Higgins D.G., Gibson T.J.. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garnier J., Gibrat J.F., Robson B.. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996; 266:540–553. [DOI] [PubMed] [Google Scholar]
- 25. Sikorski R.S., Hieter P.. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989; 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M.Z., Elledge S.J.. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007; 4:251–256. [DOI] [PubMed] [Google Scholar]
- 27. Schmidt T.G., Batz L., Bonet L., Carl U., Holzapfel G., Kiem K., Matulewicz K., Niermeier D., Schuchardt I., Stanar K.. Development of the Twin-Strep-tag(R) and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr. Purif. 2013; 92:54–61. [DOI] [PubMed] [Google Scholar]
- 28. Fitzgerald D.J., Berger P., Schaffitzel C., Yamada K., Richmond T.J., Berger I.. Protein complex expression by using multigene baculoviral vectors. Nat. Methods. 2006; 3:1021–1032. [DOI] [PubMed] [Google Scholar]
- 29. Wasilko D.J., Lee S.E., Stutzman-Engwall K.J., Reitz B.a., Emmons T.L., Mathis K.J., Bienkowski M.J., Tomasselli A.G., Fischer H.D.. The titerless infected-cells preservation and scale-up (TIPS) method for large-scale production of NO-sensitive human soluble guanylate cyclase (sGC) from insect cells infected with recombinant baculovirus. Protein Expression Purif. 2009; 65:122–132. [DOI] [PubMed] [Google Scholar]
- 30. Kanter D.M., Kaplan D.L.. Sld2 binds to origin single-stranded DNA and stimulates DNA annealing. Nucleic Acids Res. 2011; 39:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu X., Liu Y.. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009; 28:568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keller H., Kiosze K., Sachsenweger J., Haumann S., Ohlenschlager O., Nuutinen T., Syvaoja J.E., Gorlach M., Grosse F., Pospiech H.. The intrinsically disordered amino-terminal region of human RecQL4: multiple DNA-binding domains confer annealing, strand exchange and G4 DNA binding. Nucleic Acids Res. 2014; 42:12614–12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J.. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007; 157:38–46. [DOI] [PubMed] [Google Scholar]
- 34. Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mamrak N.E., Shimamura A., Howlett N.G.. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McHugh P.J., Ward T.A., Chovanec M.. A prototypical Fanconi anemia pathway in lower eukaryotes?. Cell Cycle. 2012; 11:3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi D.H., Min M.H., Kim M.J., Lee R., Kwon S.H., Bae S.H.. Hrq1 facilitates nucleotide excision repair of DNA damage induced by 4-nitroquinoline-1-oxide and cisplatin in Saccharomyces cerevisiae. J. Microbiol. 2014; 52:292–298. [DOI] [PubMed] [Google Scholar]
- 38. Yin J., Kwon Y.T., Varshavsky A., Wang W.. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 2004; 13:2421–2430. [DOI] [PubMed] [Google Scholar]
- 39. Rossi M.L., Ghosh A.K., Kulikowicz T., Croteau D.L., Bohr V.A.. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst). 2010; 9:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Capp C., Wu J., Hsieh T.S.. Drosophila RecQ4 has a 3΄-5΄ DNA helicase activity that is essential for viability. J. Biol. Chem. 2009; 284:30845–30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mendoza O., Bourdoncle A., Boule J.B., Brosh R.M. Jr., Mergny J.L.. G-quadruplexes and helicases. Nucleic Acids Res. 2016; 44:1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cejka P., Kowalczykowski S.C.. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J. Biol. Chem. 2010; 285:8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cejka P., Plank J.L., Bachrati C.Z., Hickson I.D., Kowalczykowski S.C.. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010; 17:1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raynard S., Bussen W., Sung P.. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J. Biol. Chem. 2006; 281:13861–13864. [DOI] [PubMed] [Google Scholar]
- 45. Sedlackova H., Cechova B., Mlcouskova J., Krejci L.. RECQ4 selectively recognizes Holliday junctions. DNA Repair. 2015; 30:80–89. [DOI] [PubMed] [Google Scholar]
- 46. Morimatsu K., Kowalczykowski S.C.. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5133–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohlenschlager O., Kuhnert A., Schneider A., Haumann S., Bellstedt P., Keller H., Saluz H.P., Hortschansky P., Hanel F., Grosse F. et al. . The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif. Nucleic Acids Res. 2012; 40:8309–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh D.K., Ghosh A.K., Croteau D.L., Bohr V.A.. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res. 2012; 736:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Popuri V., Hsu J., Khadka P., Horvath K., Liu Y., Croteau D.L., Bohr V.A.. Human RECQL1 participates in telomere maintenance. Nucleic Acids Res. 2014; 42:5671–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barefield C., Karlseder J.. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012; 40:7358–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Drosopoulos W.C., Kosiyatrakul S.T., Schildkraut C.L.. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J. Cell Biol. 2015; 210:191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mendez-Bermudez A., Hidalgo-Bravo A., Cotton V.E., Gravani A., Jeyapalan J.N., Royle N.J.. The roles of WRN and BLM RecQ helicases in the Alternative Lengthening of Telomeres. Nucleic Acids Res. 2012; 40:10809–10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Edwards D.N., Machwe A., Chen L., Bohr V.A., Orren D.K.. The DNA structure and sequence preferences of WRN underlie its function in telomeric recombination events. Nat. Commun. 2015; 6:8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li B., Jog S.P., Reddy S., Comai L.. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol. Cell. Biol. 2008; 28:1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Popuri V., Tadokoro T., Croteau D.L., Bohr V.A.. Human RECQL5: guarding the crossroads of DNA replication and transcription and providing backup capability. Crit. Rev. Biochem. Mol. Biol. 2013; 48:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hardy J., Churikov D., Geli V., Simon M.N.. Sgs1 and Sae2 promote telomere replication by limiting accumulation of ssDNA. Nat. Commun. 2014; 5:5004. [DOI] [PubMed] [Google Scholar]
- 57. Huang P., Pryde F., Lester D., Maddison R., Borts R., Hickson I., Louis E.. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 2001; 11:125–129. [DOI] [PubMed] [Google Scholar]
- 58. Johnson F., Marciniak R., McVey M., Stewart S., Hahn W., Guarente L.. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001; 20:905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A.. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013; 497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang W., Robles A.I., Beyer R.P., Gray L.T., Nguyen G.H., Oshima J., Maizels N., Harris C.C., Monnat R.J. Jr. The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Hum. Mol. Genet. 2016; 25:2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zimmermann M., Kibe T., Kabir S., de Lange T.. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014; 28:2477–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nguyen G.H., Tang W., Robles A.I., Beyer R.P., Gray L.T., Welsh J.A., Schetter A.J., Kumamoto K., Wang X.W., Hickson I.D. et al. . Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9905–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Damerla R.R., Knickelbein K.E., Strutt S., Liu F.J., Wang H., Opresko P.L.. Werner syndrome protein suppresses the formation of large deletions during the replication of human telomeric sequences. Cell Cycle. 2012; 11:3036–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ferrarelli L.K., Popuri V., Ghosh A.K., Tadokoro T., Canugovi C., Hsu J.K., Croteau D.L., Bohr V.A.. The RECQL4 protein, deficient in Rothmund-Thomson syndrome is active on telomeric D-loops containing DNA metabolism blocking lesions. DNA Repair (Amst.). 2013; 12:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Singh D.K., Popuri V., Kulikowicz T., Shevelev I., Ghosh A.K., Ramamoorthy M., Rossi M.L., Janscak P., Croteau D.L., Bohr V.A.. The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res. 2012; 40:6632–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bruck I., Kanter D.M., Kaplan D.L.. Enabling association of the GINS protein tetramer with the mini chromosome maintenance (Mcm)2-7 protein complex by phosphorylated Sld2 protein and single-stranded origin DNA. J. Biol. Chem. 2011; 286:36414–36426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ward T.A., Dudasova Z., Sarkar S., Bhide M.R., Vlasakova D., Chovanec M., McHugh P.J.. Components of a Fanconi-like pathway control Pso2-independent DNA interstrand crosslink repair in yeast. PLoS Genet. 2012; 8:e1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.