Abstract
A bioassay guided investigation (cancer cell cytotoxicity) of a Moorea producens collection from Key West, Florida, led to the discovery of two new bioactive natural products [(+)-malyngamide Y and a cyclic depsipeptide, (+)-floridamide]. Their planar structures were deduced through extensive analysis of 1D and 2D NMR spectroscopic data and supported by HRFAB mass spectrometry. The new cyclic depsipeptide contains four amino acids units, including N-methyl phenylalanine (N-MePhe), proline (Pro), valine (Val) and alanine (Ala), beside the unique unit, 2,2-dimethyl-3-hydroxy-octanoic acid (Dhoaa). In addition to the discovery of these two new compounds, two previously reported metabolites were also isolated and identified from this cyanobacterial collection; (−)-C-12 lyngbic acid and the antibacterial agent (−)-malyngolide.
Keywords: Moorea producens, cytotoxicity, malyngamide, floridamide
Graphical Abstract
1. Introduction
A large number of collections of the marine cyanobacterium Moorea producens produce a class of lipopeptide metabolite known as the ‘malyngamides’(Cardellina et. al. 1978; Moore et. al. 1978, Ainslie et. al. 1985; Wright et. al. 1990; Mynderse, et. al. 1978; Gerwick et. al. 1987; Engene et. al. 2012). To date, more than thirty members belonging to this group of cyanobacterial metabolites have been discovered. There are two distinct and characteristic portions comprising the malyngamides; a methoxy fatty acid (known trivially as lyngbic acid) and a variety of functionalized amines, linked through an amide bond. These lyngbic acids have varying chain lengths, ranging from C-12 to C-20, with a methoxy group at C-7 as well as a trans double bond at C-4. The C-12 and C-14 lyngbic acids have also been detected in free form from marine cyanobacteria. The absolute stereochemistry of the lyngbic acids has been confirmed by total chemical syntheses (Praud et. al. 1993; Mesguiche et. al. 1999; Orjala et. al. 1995). The marine cyanobacterium Moorea producens is known to be a rich source of unique and bioactive peptides. Examples include the antimicrobial and actin polymerization-inhibiting lyngbyabellins A and B, the cytotoxic lyngbyastatin 2 and the pro-inflammatory lyngbyatoxins A and B (Luesch et. al. 2000, Milligan et. al. 2000; Luesch et. al. 1999; Cardellina et. al. 1979; Fujiki et. al. 1981; Aimi et. al. 1990; Youssef et. al. 2015). As part of our ongoing search for structurally and pharmacologically interesting substances from M. producens, a detailed examination of a Key West, Florida collection was undertaken. During this study two new compounds, (+)-malyngamide Y (1) and (+)-floridamide (2) have been isolated. In addition, two metabolites of known identity were isolated, namely the anti-microbial (−)-malyngolide (3) and the (−)-C-12 lyngbic acid (4).
2. Results and discussion
A collection of Moorea producens (active in H460 cancer cell cytotoxicity assay) was obtained from Key West, Florida, extracted with 2:1 CH2Cl2/MeOH and fractionated over silica gel vacuum liquid chromatography (EtOAc/hexanes gradient). Successive reversed phase SPE and HPLC fractionation resulted in the isolation of two new compounds (1, 2), in addition to two previously known compounds (3, 4). Analysis of their spectroscopic properties (UV, IR, LRMS, HRMS, 1D NMR and 2D NMR) allowed the unequivocal construction of their planar structures (Figure 1).
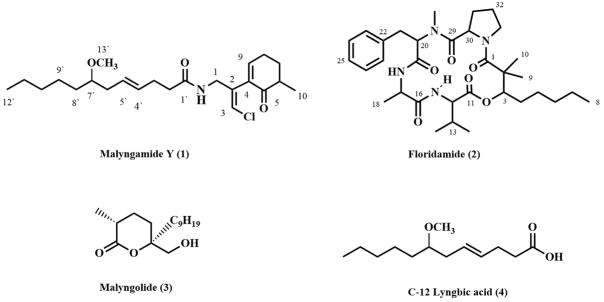
Figure 1.
Structures of the isolated compounds from Moorea producens
Malyngamide Y (1) was isolated as colorless oil from the cancer cell cytotoxic organic extract of M. producens. The isotope pattern observed for the molecular ion (FAB) indicated the presence of one chlorine atom, and HRFABMS established a molecular formula of C23H37ClNO3 (six degrees of unsaturation). The IR spectrum of 1 showed absorption bands at 3301 and 1675 cm−1, indicative of the presence of an amide functional group. Inspection of the 1H NMR spectrum indicated the presence of an olefin proton triplet signal at δ 6.87 (H-9). Another sharp singlet at δ6.18 was characteristic of the olefin proton associated with the vinyl chloride functionality found in most malyngamides (H-3). Additionally a broad triplet at δ 6.02 was observed for an amide proton coupled with a methylene group. Another two doublet of doublets at δ 4.20 and 4.06 (H1a and H1b) were seen. A sharp three proton singlet at 3.20 was observed and indicated the presence of a methoxy group (C-13′). Moreover a sharp triplet at δ 3.14 was indicative of a proton attached to a carbon carrying oxygen (H-7′), and a 3H doublet at δ 1.16, suggested a methyl group attached to methine group. Finally, a terminal methyl triplet (δ 0.89) was observed (H3-12).
The 13C NMR spectrum in CDCl3 confirmed the presence of twenty-three carbon atoms. Analysis of 13C NMR, DEPT135 and DEPT90 data revealed the presence of one ketone resonance at δ 202.40 (C-5), an amide resonance at δ 172.40 (C-1′), six olefinic carbons (δ 149.0, 138.70, 138.40, 131.00, 127.00 and 120.10), ten methylene groups (δ 63.94, 31.0, 36.9, 36.80, 33.74, 32.40, 29.10, 26.12, 25.37, 23.90), two aliphatic methine groups (81.1 and 42.5) and also three methyl groups (56.9, 15.48 and 14.5). This was in keeping with five degrees of the six degrees of unsaturation required by the molecular formula and confirmed the need for one ring to accomodate the six degrees of unsaturations.
Chemical shift arguments, 1H-1H COSY and TOCSY correlations supported by MS data and HMBC allowed the assignment of the planar structure of 1. From the 1H-1H COSY NMR spectrum of 1, it was possible to differentiate three discrete spin systems. A continuous spin system was evident in which a broad amide proton triplet (δ 6.02) was coupled to two mutually coupled midfield methylene resonances (δ 4.2 and 4.0 H-1a and H-1b), which in turn, showed couplings to an olefinic 1H resonance (δ 6.1, H-3). The proton signal at δ 6.8 (H-9) was coupled to the proton at δ 2.4 (H-8) which by itself coupled to the methylene proton at δ 2.0 (H-7). The latter proton was found to couple with the methine proton at δ 2.4 (H-6), and completed the second spin system.
The third spin system was the C-12 chain of lyngbic acid. This spin system began with the terminal methyl protons at δ 0.88 (H3-12′) which were found to couple with the methylene group at δ 1.23 (H2-11′). The latter proton signal was coupled to the protons at δ 1.22 (H2-10′), which in turn was coupled to the methylene protons at δ 1.35 (H2-9′). The H2-9′ protons were adjacent to the methylene proton at δ 1.6 (H2-8′) which coupled to a methine proton at δ 3.12; by chemical shift, this C-7′ methine carbon also carried the methoxy group. The latter proton signal was coupled to the methylene protons at δ 2.15 (H2-6′) which were adjacent to the olefinic proton at δ 5.4 (H-5′).
Coupling of H-5′ with the second olefinic proton H-4′, and of H4′ to the final methylene (H2-3′) completed the third spin system. These connections were confirmed by TOCSY correlations. HMBC correlations from δ 4.0/4.2 (Ha/b-1) to C-2, C-3, and C-4 combined with complementary HMBC correlations from H-3 to C-1, C-2, and C-4, firmly established the C-1/C-2/C-3/C-4 fragment of 1. HMBC correlations from the methyl singlet at δ 1.12 (H3-10) to C-5, C-6, C-7 and C-8 confirmed this connectivity deduced from COSY data. Securing of the cyclohexene ring structure was possible by placement of the carbonyl at C-5 due to HMBC correlations from the methyl group at δ 1.12 to C-5, C-6, C-7, and C-8. The C-2 (δ 138.7)/C-3 (δ 120.1) double bond was determined as being terminal with the chlorine at C-3 by HMBC correlations from H-4 to C-1 (δ 39.4), C-2, and C-3.
Further HMBC correlations from Ha/b-1 to C-1′ established the C-1/N-H/C-1′ connection. 13C NMR and 2D-NMR established the fatty acid as a 12-carbon chain with unsaturation at the 4′-position. Placement of the methoxy group (δH 3.4 and δC 56.9) was possible through an HMBC correlation from the methoxy proton singlet to C-7′ (δ 81.1). Strong HMBC correlations from δ 6.1 (H-3) to C-1 confirmed the stereochemistry of the vinyl chloride group to be E, and completed the planar structure of malyngamide Y (1) (McPhail et. al. 2003).
Floridamide (2) was isolated as a colorless oil with a molecular formula of C33H50N4O6 as determined by HRFABMS (observed [M+Na]+ at m/z 621.9). The IR spectrum of floridamide (2) gave characteristic absorption bands at 3293, 1734, 1650, 1625 cm−1, indicative of ester/amide carbonyl functionalities. Of the 11 degrees of unsaturation inherent to the molecular formula, four could be accounted for by a phenyl group as suggested in the 1H NMR spectrum. In addition, the peptidic nature of 2 was indicated by exchangeable NH protons resonating at δ 8.6 and δ 6.7. One distinct N-CH3 proton singlet was also observed in the 1H NMR data at δ 2.95. Two other high field CH3 proton singlets were also observed at δ 0.92 and δ 1.4. Thirty-three carbon signals were observed in the 13C NMR data of floridamide (2), which included signals for a mono-substituted phenyl ring as well as five signals belonging to amide/ester carbonyls in the 169–173 ppm range. One oxygenated sp3 carbon resonating at δ 77.9 was also detected in the HSQC spectrum.
From 1D and 2D NMR data, including HMBC and TOCSY, the presence of two conformers of compound 2 were observed in a ratio of 2:1. Five substructures were assembled for the major conformer of floridamide (2), including four amino acids (N-MePhe, Pro, Val and Ala) and one hydroxy acid, 2,2-dimethyl-3-hydroxy-octanoic acid (Dhoaa). The latter hydroxy acid, Dhoaa, is a unique unit previously reported from cyanobacterial depsipeptides and was deduced in floridamide from HMBC and TOCSY data.14 The sequence of these five residues in floridamide (2) was established mainly from CIMS and HMBC correlations. Sequential HMBC correlations in CD2Cl2 were observed between H-3, H-28, H-1/C-4; H-17, H-3, NH (δ 6.7)/C-19; H-12, H-3/C-11; H-3, H-9, H-10, H-1/C-1 and H-17, H-12, NH (δ 8.6)/C-16 which gave rise to the Pro/N-MePhe/Ala/Val/Dhoaa sequence. The overall cyclic structure of floridamide (2) was deduced by consideration of the downfield chemical shifts of alpha protons for each residue and consideration of the overall molecular formula. The absolute stereochemistry of floridamide (2) was not determined due to lack of available material.
Malyngamide Y (1) was found to have cytotoxic activity (EC50 = 1.45 x10−5 μM/ml) to a human lung cancer cell line (NCI-H460) as well as to the mouse neuro-2a neuroblastoma cell line. However, floridamide (2) had weaker cytotoxic activity (EC50 = 1.89 x10−5 μM/ml) in these cell lines. Neither of these compounds were found to have blocking or activating activity in a sodium channel modulation assay (Manger et. al. 1995).
3. Experimental
3.1. General Experimental Procedures
Optical rotations were measured on a Perkin-Elmer 141 polarimeter. IR and UV spectra were recorded on Nicolet 510 and Beckman DU640B spectrophotometers, respectively. NMR spectra were recorded on a Bruker DPX400 spectrometer, with the solvent (CDCl3 at δC 77.2, δH 7.26) used as an internal standard. Mass spectra were recorded on a Kratos MS50TC mass spectrometer, and HPLC isolations were performed using Waters Millipore model 515 pumps and a Waters 969 diode array detector.
3.2. Cyanobacterial Collection
The marine cyanobacterium Moorea producens (voucher specimen available as collection number KWN-18/NOV/05-01) was collected by hand using SCUBA in Key West Florida, USA. The material was stored in 2-propanol at −3 °C until extraction.
3.3. Extraction and Isolation
Approximately 40 g dry weight of the cyanobacterium was extracted repeatedly with 2:1 CH2Cl2/MeOH to produce 0.99 g of crude organic extract. A portion of the extract (0.97 g) was then fractionated by silica gel vacuum liquid chromatography. The fractions eluting with 60% EtOAc in hexanes was further purified with a C18 solid phase extraction (SPE) cartridge (8:2 MeOH/H2O) and reversed-phase HPLC (9:1 CH3OH/H2O, Phenomenex Spheroclone 5 μ ODS) to yield 2.4 mg of C-12 lyngbic acid (3), 1.3 mg of malyngamide Y (1) and 1.0 mg of malyngolide (4). A second fraction eluting with 80% EtOAc in hexanes was further purified with a C18 solid phase extraction (SPE) cartridge (8:2 MeOH/H2O) and reversed-phase HPLC (9:1 CH3OH/H2O, Phenomenex Sphereclone 5 μ ODS) to yield 2.0 mg of floridamide (2).
3.4. Spectral data
Malyngamide Y (1): colorless oil; [α]25D +14° (c 0.09, CHCl3); UV (MeOH) λmax 250 (ε = 174); IR νmax(film) 3301, 2928, 2859, 1675, 1537, 1455, 1370, 1095cm−1; 1H and 13C NMR data in CDCl3, see Table S1; HRFABMS (3-NBA) obsd [M + H]+m/z 410.2467 (calc. for C23H37ClNO3 410.2462).
Floridamide (2): colorless oil; [α]25D +56° (c 0.1, CHCl3); UV (MeOH) λmax 220 (log ε = 5.467), 278 (log ε = 4.453); IR νmax (film) 3293, 2924, 2854, 2360, 2337, 1734,1650, 1625, 1510, 1458, 1175 cm−1; 1H and 13C NMR data in CDCl3, see Table S2; HRFABMS [M + H]+m/z 599.3815 (calc. for C33H51N4O6 599.3808).
(−)-Malyngolide (3) colorless oil, [α]25D-10° (c 0.10, CHCl3), literature value −12.0°;1 UV, IR, 1H, 13C NMR, and MS data were similar to literature values (Cardllina et. al. 1979).
(−)-C-12 lyngbic acid (4): colorless oil; [α]25D –8° (c 0.1, CHCl3); with remaining physical and spectroscopic properties identical to those previously reported (Kwan et. al. 2010).
3.5. Cytotoxicity against NCI-H460 human lung cancer and neuro-2a neuroblastoma cell line (Alley et. al. 1988)
The method of Alley et. al. was used to determine cell viability in NCI-H460 human lung tumor cells and mouse neuro-2a blastoma cells by MTT reduction. Cells were seeded in 96-well plates at 5000 and 8000 cells/well in 180 μl for H460 and neuro-2a cells, respectively. Twenty-four hours later, the test chemical dissolved in DMSO and diluted into medium without fetal bovine serum was added at 3 μg/well. DMSO was less than 1% final concentration. After 48 h, the medium was removed and cell viability determined.
3.6. Sodium channel modulation (Alley et. al. 1988)
Isolated compounds, were evaluated for their capacity to either activate or block sodium channels using the following modifications to the cell-based bioassay of Manger et. al. Twenty-four hours prior to chemical testing, mouse neuro-2a blastoma cells were seeded in 96-well plates at 8 X 104 cells/well in a volume of 30 μl. Test chemicals dissolved in DMSO were serially diluted in medium without fetal bovine serum and added at 10 μl/well. DMSO was less than 1% final concentration. Plates to evaluate sodium channel activating activity received 3 μl/well of either a mixture of 3 mM quabain and 0.3 mM veratridine (Sigma Chemical Co.) in 5 mM HCl in addition to the test chemical. Plates were incubated for 18 hr and results compared to similarly treated solvent controls with 10 μl medium added in lieu of the test chemical. The sodium channel activator brevetoxin PbTx-1 (Calbiochem) was used as the positive control and added at 10 ng/well in 10 μl medium. Sodium channel blocking activity was assessed in a similar manner except that ouabain and veratridine were 5.0 and 0.5 mM, respectively, and the sodium channel blocker saxitoxin (Calbiochem) was used as the positive control. Plates were incubated for approximately 22 hour.
4. Conclusions
Bioassay guided investigation (cancer cell cytotoxicity) of a Moorea producens, led to the discovery of (+)-malyngamide Y and (+)-floridamide. The new cyclic depsipeptide contains four amino acids units, including N-methyl phenylalanine (N-MePhe), proline (Pro), valine (Val) and alanine (Ala), beside the unique unit, 2,2-dimethyl-3-hydroxy-octanoic acid (Dhoaa).
Supplementary Material
Acknowledgments
We gratefully acknowledge Jeff. More (Chemistry, OSU) for mass spectral data, the Government of Florida State for permission to make these collections. Financial support for this work came from the National Institute of Health (GM 63554 and CA52955).
Footnotes
Supplementary material relating to this article is available online
References
- Aimi N, Odaka H, Sakai S, Fujiki H, Suganuma M, Moore RE, Patterson GML. Lyngbyatoxins B and C, two new irritants from Lyngbyamajuscula. J Nat Prod. 1990;53:1593–1596. doi: 10.1021/np50072a035. [DOI] [PubMed] [Google Scholar]
- Ainslie RD, Barchi JJ, Kuniyoshi M, Moore RE, Mynderse JS. Structure of malyngamide C. J Org Chem. 1985;50:2859–2862. [Google Scholar]
- Alley MC, Scudiero DA. Feasibility of Drug Screening with Panels of Human Tumor Cell Lines Using a Microculture Tetrazolium Assay. Cancer Research. 1988;48:589–601. [PubMed] [Google Scholar]
- Cardellina JH, II, Dalietos D, Marner F, Mynderse JS, Moore RE. (−)-trans-7(S)-Methoxytetradec-4-enoic acid and related amides from the marine cyanophyte Lyngbyamajuscula. Phytochemistry. 1978;17:391–395. [Google Scholar]
- Cardellina JH, Marner FJ, Moore RE. Seaweed Dermatitis: Structure of Lyngbyatoxin A. Science. 1979;34:193–195. doi: 10.1126/science.107586. [DOI] [PubMed] [Google Scholar]
- Fujiki H, Mori M, Nakayasu M, Terada M, Sugimura T, Moore RE. Indole alkaloids: dihydroteleocidin B, teleocidin, and lyngbyatoxin A as members of a new class of tumor promoters. Proc Natl Acad Sci USA. 1981;78:3872–3876. doi: 10.1073/pnas.78.6.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwick WH, Reyes S, Alvarado B. Two malyngamides from the Caribbean Lyngbya majuscula. Phytochemistry. 1987;26:1701–1704. [Google Scholar]
- Cardllina John H, Moore Richard E. Structure and Absolute Configuration of Malyngolide, an Antibiotic from the Marine Blue-Green Alga Lyngbya majuscula Gomont. J Org Chem. 1979;44:4039–4042. [Google Scholar]
- Kwan JC, Teplitski M, Gunasekera SP, Paul VJ, Luesch H. Isolation and Biological Evaluation of 8-epi-Malyngamide C from the Floridian Marine Cyanobacterium Lyngbya majuscula. J Nat Prod. 2010;73:463–466. doi: 10.1021/np900614n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H, Yoshida WY, Moore RE, Paul VJ. Isolation and Structure of the Cytotoxin Lyngbyabellin B and Absolute Configuration of Lyngbyapeptin A from the Marine Cyanobacterium Lyngbyamajuscula. J Nat Prod. 63:1437–1439. doi: 10.1021/np000104n. 200. [DOI] [PubMed] [Google Scholar]
- Luesch H, Yoshida WY, Moore RE, Paul VJ. Lyngbyastatin 2 and Norlyngbyastatin2, Analogues of Dolastatin G and Nordolastatin G from the Marine Cyanobacterium Lyngbya majuscula. J Nat Prod. 1999;62:1702–1706. doi: 10.1021/np990310z. [DOI] [PubMed] [Google Scholar]
- Luesch H, Yoshida WY, Moore RE, Paul VJ, Mooberry SL. Isolation, Structure Determination, and Biological Activity of Lyngbyabellin A from the Marine Cyanobacterium Lyngbya majuscule. J Nat Prod. 2000;63:611–615. doi: 10.1021/np990543q. [DOI] [PubMed] [Google Scholar]
- Manger RL, Leja LSJ. Detection of sodium channel toxins: directed cytotoxicity assays of purified ciguatoxins, brevetoxins, saxitoxins, and seafood extracts. AOAC Int. 1995;78:521–527. [PubMed] [Google Scholar]
- McPhail KL, Gerwick WH. Three New Malyngamides from a Papua New Guinea Collection of the Marine Cyanobacterium Lyngbya majuscula. J Nat Prod. 2003;66:132–135. doi: 10.1021/np0204186. [DOI] [PubMed] [Google Scholar]
- Mesguiche V, Valls R, Piovetti L, Peiffer G. Characterization and synthesis of (−)-7-methoxydodec-4(E)-enoic acid, a novel fatty acid isolated from Lyngbya majuscule. Tetrahedron Lett. 1999;74:73–7476. [Google Scholar]
- Milligan KE, Marquez BL, Williamson RT, Gerwick WH. Lyngbyabellin B, a Toxic and Antifungal Secondary Metabolite from the Marine Cyanobacterium Lyngbya majuscula. J Nat Prod. 2000;63:1440–1443. doi: 10.1021/np000133y. [DOI] [PubMed] [Google Scholar]
- Moore RE, Cardellina JH, II, Marner F. Malyngamide A, a novel chlorinated metabolite of the marine cyanophyte Lyngbya majuscula. J Am Chem Soc. 1978;101:240–242. [Google Scholar]
- Mynderse JS, Moore RE. (−)-Trans-7(s)-methoxytetradec-4-enoic acid and related amides from the marine cyanophyte Lyngbya majuscule. J Org Chem. 1978;43:4359–4363. [Google Scholar]
- Nunnery JK, Suyama TL, Linington RG, Gerwick WH. Expedient synthesis of α,α-dimethyl-β-hydroxy carbonyl scaffolds via Evans’ aldol reaction with a tertiary enolate. Tetrahedron Lett. 2011;52:2929–2932. doi: 10.1016/j.tetlet.2011.03.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjala J, Nagle D, Gerwick WH. Malyngamide H, an ichthyotoxic amide possessing a new carbon skeleton from the Caribbean cyanobacterium Lyngbya majuscule. J Nat Prod. 1995;58:764–768. doi: 10.1021/np50119a019. [DOI] [PubMed] [Google Scholar]
- Praud A, Valls R, Piovetti L, Banaigs B. MalyngamideG : Proposition de Structure pour un Nouvel Amide chlorc d’une Algueblewverte Epiphyte de Cystoseiru crinite. Tetrahedron Lett. 1993;34:5437–5440. [Google Scholar]
- Wright AD, Coll JC, Price IR. Tropical marine algae, vii. The chemical composition of marine algae from north Queensland waters. J Nat Prod. 1990;53:845–861. [Google Scholar]
- Youssef DT, Shaala LA, Mohamed GA, Ibrahim SR, Banjar ZM, Badr JM, McPhail KL, Risinger AL, Mooberry SL. 2, 3-Seco-2, 3-dioxo-lyngbyatoxin A from a Red Sea strain of the marine cyanobacterium Moorea producens. Natural product research. 2015;29:703–709. doi: 10.1080/14786419.2014.982647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.