Abstract
The objective of this review is to highlight the proteases required for regulated cell death mechanisms in animals and plants. Our aim is to be incisive, and not inclusive of all the animal proteases that have been implicated in various publications. We aim to focus on instances when several publications from disparate groups have demonstrated the involvement of an animal protease, and also when there is substantial biochemical, mechanistic and genetic evidence. In doing so we can cull the literature to a handful of proteases, covering most of the known regulated cell death mechanisms – apoptosis, regulated necrosis, necroptosis, pyroptosis, and NETosis in animals. In plants the literature is younger and not as extensive as for mammals, but the molecular drivers of vacuolar death, necrosis, and the hypersensitive response in plants are becoming clearer. Each of these death mechanisms has at least one proteolytic component that plays a major role in controlling the pathway, and sometimes they combine in networks to regulate cell death/survival decision nodes. Some similarities are found among animal and plant cell death proteases, but overall the pathways that they govern are kingdom-specific with very little overlap.
Keywords: apoptosis, caspase, cathepsin, metacaspase, necroptosis, necrosis, NETosis, peptidase, proteolysis, pyroptosis
Introduction
Proteases are probably one of the most ancient families of enzymes. Almost every organism, including viruses, has at least one protease. Although one could surmise that the earliest proteases were probably degrading enzymes that recycled amino acids, most of the proteases in the genomes of modern day organisms, especially complex metazoans like ourselves, don’t spend their time degrading proteins. Mostly they create very limited cuts in target proteins as essential components of signaling pathways and networks. Classic examples of such limited proteolysis would be blood coagulation, complement activation, and apoptosis.
So if we are going to review proteases and their role in cell death we should distinguish those proteases that participate in cell death by degrading components of the cell from those that precipitate cell death by either initiating, amplifying, or effecting changes in the morphology and physiology to mediate cell death. The nomenclature commission on cell death recognizes a large and variable number of physiologically distinct forms of cell death [1, 2]. However, we are going to stick to just a handful of regulated forms of cell death that have been well-characterized with respect to their engagement of proteolytic systems in the outcome of death. We will cover animal metazoan and plant cell death, comparing and contrasting the role of proteases and proteolytic systems that drive the distinct outcomes in these two divisions of the biotic universe.
Protease families and catalytic mechanisms
By way of a little biochemistry, proteins are one of the most stable biological polymers known. Their core constituent, the peptide bond, is postulated to exist in interstellar space, and therefore predates the origin of life [3]. Cleavage (hydrolysis) of a peptide bond takes years in neutral aqueous solution, but has a half-life of milliseconds in the presence of a good protein catalyst – a protease (also called a peptidase). The vast acceleration in hydrolysis (formally kcat/kuncat values in excess of 109) comes from a protease’s ability to destabilize the highly stable trigonal geometry of a peptide bond while adding water, with or without covalent participation by the protease [4].
Five mechanistic classes of proteases are recognized: cysteine, serine, and threonine proteases (forming covalent enzyme intermediates) and aspartic and metalloproteases (do not form covalent enzyme intermediates) – Fig 1. Catalysis by cysteine, serine or threonine proteases depends on the interplay of a nucleophile (the eponymous Cys, Ser or Thr residues) and a general base (usually His) - sometimes with the help of an acid (often Asp) to create the classic “catalytic triad”. In the second category general acid/base catalysis via catalytic Asp residues (in aspartic proteases) or enzyme bound Zinc (in metalloproteases) polarizes a water molecule that directly adds to a peptide bond to perform the hydrolytic reaction. In proteases of different clans (different folding types with no obvious common ancestor) within the same catalytic type, similar spatial arrangements of catalytic residues are observed, but the order of the residues in the sequence is different. This illustrates how different groups and different protein structures can achieve the same reaction [5].
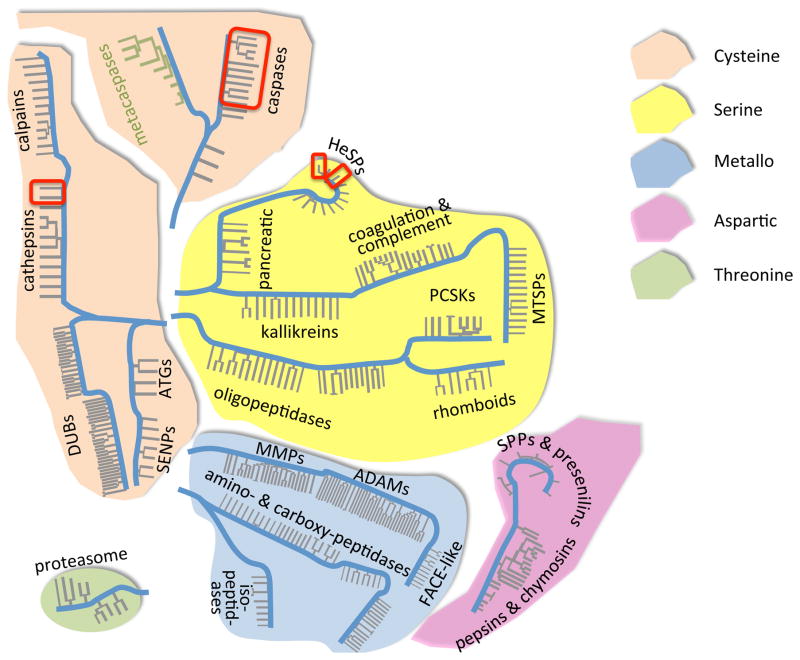
Fig. 1.
Human Protease Landscape. Protease catalytic types come from unrelated evolutionary origins. The intent of this map is to show the diversity of human proteases, based on phylogenetic relationships of most human protease catalytic domains collected from the MEROPS database. Family relationships are gathered by the grey lines, and evolutionary divergence by the blue mainlines (not to an evolutionary scale). Within a catalytic class some families and clans have no common origin – shown by disconnected blue mainlines. The thickness and spacing of the grey lines does not signify anything, and is just a device to cluster some protease families with a large number of members. A. thaliana metacaspases (green) are overlaid on this human map to display their relation to caspases. Red boxes highlight some of the human proteases considered in this review as playing roles in regulated cell death.
For biologists these distinctions in catalytic mechanism are less important than understanding the biological function of proteases, but for chemists and drug manufacturers they are essential since they dictate the type of chemistry that can be employed to devise inhibitory compounds. A fundamental property of the different catalytic types that is often overlooked is that the chemistry of the catalytic center can dictate where proteases are likely to be active. Although there are specific and important exceptions, in general the different catalytic types are environmentally restricted as follows:
Cysteine proteases – catalytic thiol readily oxidized, thus they are highly restricted to the reducing environment inside cells. Classic exception clostripain.
Serine proteases – found primarily outside cells, either as secreted or membrane associated enzymes – similar to metalloproteases. Classic exception mitochondrial rhomboids.
Threonine proteases – found exclusively inside cells. Classic exception gamma-glutamyl transferase.
Aspartic proteases – generally found in acidic or highly protected environments due to a requirement for partial protonation of the catalytic Asp dyad. Classic exception renin.
Metallo proteases – found primarily outside cells, either as secreted or membrane associated enzymes – similar to serine proteases. Classic exception Farnesylated-protein converting enzyme.
Proteases and death signaling
In reviewing which proteases could induce regulated cell death we noticed that endopepetidases (which can cleave the peptide bonds internal to proteins) are much more frequently mentioned than exopeptidases (which can only cleave peptide bonds near the end of proteins) as participants in signaling pathways – Table 1. We speculate that exopeptidases simply wouldn’t be capable of cutting in the right place to drive forward conformational changes and separate domains – typical of signaling pathways [6, 7]. There are two notable exceptions to this observation: 1) dipeptidyl peptidase 1 (DPP1 - also known as cathepsin C) and 2) the proteasome. The former is directly involved in the activation of hematopoietic serine proteases (HeSPs) - see later - and the latter is involved in the degradation of many cellular proteins that regulate cell death/survival decisions. Thus one could define the proteasome as a cell death protease, but that’s about all we will say about it and readers are directed to extensive reviews on the proteasome and its relation to cell death [8–12]. However, we note that one of the earliest described forms of programmed cell death to be characterized from a genetic viewpoint is the loss of intersegmental muscles of the moth Manduca sexta at the end of metamorphosis, and this form of cell death requires the ubiquitin/proteasome system [13, 14]. More recently it has been revealed that the death of insect cells during metamorphosis integrates this system with autophagy – see [15] for a detailed review.
Table 1.
Human proteases implicated in regulated cell death mechanisms. The intensity of the shading roughly synthesizes the involvement of each protease in a pathway reported in the literature. *Caspase-8 is involved in necroptosis in a negative context since it inactivates this pathway.
| Protease | Apoptosis | Pyroptosis | Necroptosis | Necrosis | NETosis | Unspecified |
|---|---|---|---|---|---|---|
| Caspase 3,6,7,8,9,10 | +++ | − | (Caspase-8)* | − | − | − |
| Caspase 1,4,5,11 | − | +++ | − | − | − | − |
| Granzyme A | − | − | − | − | − | ++ |
| Granzyme B | +++ | + | − | − | − | − |
| Cathepsin C (DPP1) | +++ | − | − | − | + | − |
| Cathepsin B | + | + | − | ++ | − | ++ |
| Cathepsin D | + | − | − | ++ | − | + |
| Neutrophil Elastase | − | − | − | − | + | − |
| Calpain 1,2 | + | − | − | ++ | − | + |
Proteases participate in cellular signaling, including cell death signaling, by cleaving protein substrates. The functional outcome of substrate cleavage is particular to the signaling pathway, the nature of the protein substrate itself, and the site of proteolysis. Proteases generally have more than one physiological substrate, cleaving these proteins at specific sites according to the specificity of the protease. Sometimes one can predict with some degree of confidence whether a protein is likely to be cleaved by a given protease, but this depends heavily on knowing the detailed specificity of the protease. The frequent occurrence of sequences within proteins that match individual protease consensus substrate specificities suggests a multitude of substrates in vivo – somewhere in the order of several thousand in humans alone, and the list of proteins that are reported to be cleaved by proteases in vitro proliferates rapidly. Modern focused proteomics technologies have revealed a massive database of proteins whose cleavage is observed under experimental conditions [6, 7, 16–18].
The problem is that only a few of these proteins have been rigorously established as biologically or pathologically relevant, bona fide substrates in vivo. It’s likely that most of them simply represent “innocent bystanders”, or even erroneous assignments [6]. In this review we focus on the proteases themselves, not the substrates, and we point readers to resources that discuss and report on the substrates in more detail in Table 2.
Table 2.
Protease resource websites
| General protease resources | |
| Merops | Well annotated universe of proteolytic enzymes including experimentally determined substrates – the most comprehensive of all protease databases |
| CutDB | Focuses on the annotation of individual proteolytic events, both actual and predicted |
| PoPS | A set of computational tools for investigating protease specificity |
| PROSPER | An integrated feature based server for the prediction of novel substrates and their cleavage sites |
| TopFIND | An integrated knowledgebase focused on protein termini, their formation by proteases and functional implications |
| Toppr | A store of high quality proteomics based proteolytic processed events |
| Proteasix | An open source peptide centric tool that can be used to predict in silico the proteases involved in naturally occurring peptide generation |
| Caspase-specific resources | |
| Casbah | Comprehensive list of caspase substrates and a searchable web resource |
| CaspDB | Machine learning prediction model including structural data for caspase cleavages |
| Degrabase | A description of caspases substrates found in healthy and apoptotic human cells |
Most publications on cell death focus on animals, and so a question arises as to whether cell death mechanisms are conserved across kingdoms. Is plant cell death related to animal cell death, and if so how? Plants do not contain caspases, instead they contain a series of distantly related proteases known as metacaspases, which are absent in animals (Fig 2). Are these cell death effectors? Towards the end of this review we focus on the place of proteases in plant cell death, comparing and contrasting with animal cell death mechanisms.
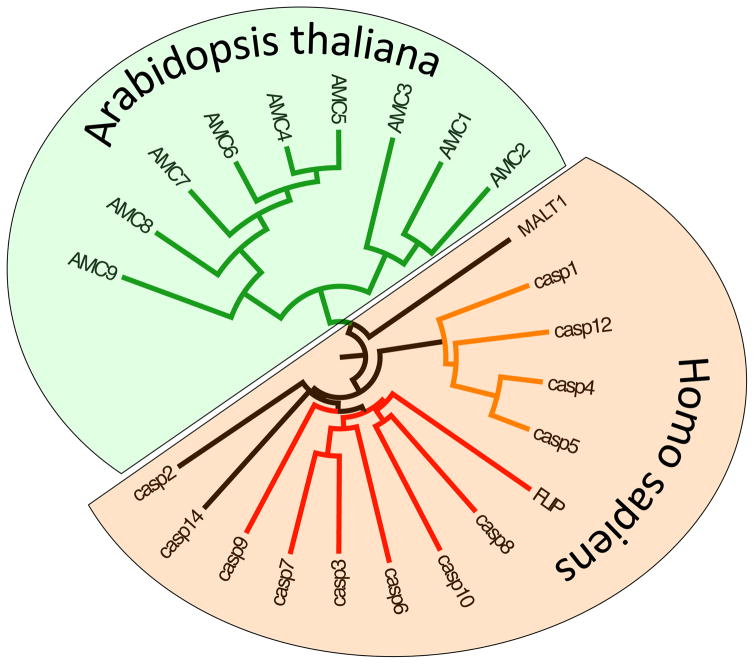
Fig. 2.
Vertebrate and Plant clan CD Proteases. In addition to the proteases identified here, plants and mammals contain two clan CD members in common: legumain and separase (not shown). Members specific for each kingdom are human caspase family, which can be divided into apoptotic caspases (red) and inflammatory caspases (orange). We have not included caspase-2 in the apoptotic caspases, as some would do, simply because the evidence for its role is so bewildering with many conflicting reports [181–185]. Caspase-14 is not an apoptotic caspase, and the closest relative to the caspases, the paracaspase MALT1 is a survival protease that operated via the NFkB pathway [186]. Two types of metacaspases can be distinguished in A. thaliana: Type I (AMC1–3) and Type II (AMC 4–9).
Apoptosis
Caspases
The biochemical events that lead to the characteristic morphology of apoptosis include membrane blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, chromosomal DNA fragmentation to oligonucleosome-sized fragments, and eventually cell death. In vivo, apoptotic cell fragments are rapidly cleared by macrophages via “find me” and “eat me” signals [19, 20], but in cell culture many of the full morphologies described as definitive can be observed. The concept is that apoptosis is an immunologically silent cell demise, indeed it may be anti-inflammatory, and therefore a complex network of events is required to dismantle and package cells for removal [20–22]. This network absolutely requires the participation of caspases.
Caspases are intracellular proteases that trigger a cascade of signaling events in programmed cell death, proliferation and inflammation. Their name derives from their defining enzymatic property as cysteine-dependent aspartate-specific protease. They can be roughly grouped into the categories of apoptotic and pro-inflammatory, wherein the apoptotic caspases can be further distinguished into initiator (caspases-8, -9 and -10) and effector (caspases-3, -6 and -7) caspases. As is common to several other protease families, caspases use a Cys side chain as nucleophile during peptide bond hydrolysis. However, they have a rare primary specificity for Asp in the P1 residue and small and uncharged residue at P1’.
Caspases are widely expressed, caspase-14 being the exception as it is limited to keratinocytes [23]. They are synthesized as single chain zymogens with an N-terminal prodomain followed by a catalytic domain. Activation or maturation of the protein leads to cleavage of the catalytic domain into a large and small subunit (Fig. 3). Both subunits interact with each other to form the active protease: The catalytic dyad residues Cys and His reside in the large subunit while the substrate binding groove is formed through residues from the small subunit.
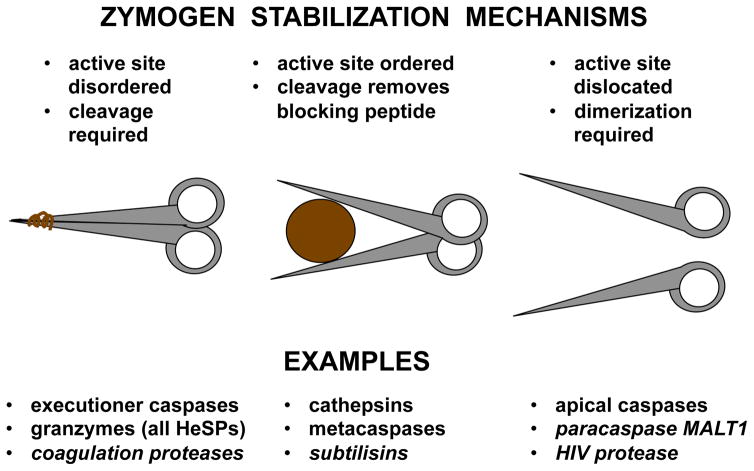
Fig. 3.
Protease Zymogen Activation Mechanisms. Schematic of most common activation mechanisms with examples covered in the review, plus other classic examples in italics. The cartoon explains the mechanism of zymogen stabilization, and the processes required to activate each example.
Initiator caspases are inert monomers in the off state and require homodimerization for activation. This is achieved through the assembly of oligomeric activation platforms following an apoptotic signal. Caspases are specifically recruited to those platforms by adaptor molecules. The local increase in concentration forces proximity-induced dimerization and therefore activation [24]. Most structural and biochemical work on activation of apical caspases has focused on caspases-8 and 9, but the same concept is thought to hold true for the activation of pro-inflammatory caspases.
Knockout of caspase-9, the apical caspase of the mitochondrial pathway (intrinsic pathway), reveals severe developmental defects and resistance to a subset of apoptotic stimuli in thymocytes, consistent with a role for this caspase in development and immune cell function [25]. Interestingly, thymocytes defects can be corrected later by an unknown mechanism, [26] leading to the realization that compensation can obscure the function of caspases and other apoptotic mediators in mouse ablation studies [27]. Tissue specific knockouts of caspase-8, the apical caspase of the death receptor pathway (extrinsic pathway), reveal an expected role for caspase-8 in T and B cell clonal expansion, and also for lymphocyte homeostasis, confirming a role in promoting apoptosis – reviewed in [28]. However, total knockout of caspase-8 discloses an unexpected role for this caspase in survival (see section on necroptosis below). It is generally accepted that the main substrates for the apoptotic initiator caspases are the zymogens of the executioner caspases, providing a two-step proteolytic activation pathway (Fig 4). Why there must be two steps is a mystery since C. elegans seems to do fine with a single apoptotic caspase, CED3 [29, 30].
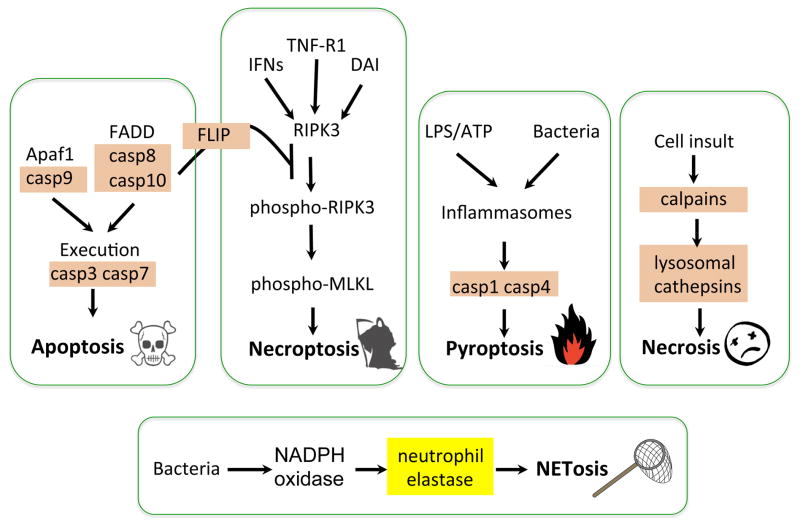
Fig. 4.
Regulated Cell Death Pathways. The schematics illustrate the involvement of cysteine proteases in most regulated cell death pathways, with the exception of NETosis, which requires the serine protease neutrophil elastase. Different sets of caspases are important for either apoptotic or pyroptotic cell death. In addition to its role in initiating the extrinsic pathway of apoptosis, caspase-8 regulates necroptosis by forming a complex with the pseudocaspase FLIP. Although the story is still quite murky in humans, regulated necrosis has been shown in C. elegans to depend on calpains and lysosomal cathepsins.
Executioner caspases are already dimeric shortly after their synthesis and are restrained by a linker separating the large and small subunit. Proteolytic processing of the linker allows formation of the catalytic site through rearrangement of characteristic mobile loops. (Auto-) proteolysis can follow activation via additional cleavage of the intrasubunit linker as well as trimming or removal of the pro-domain. This process is important for caspase stability and downstream regulatory events [24, 31].
It has proven much easier to characterize caspases, make chemical tools, and obtain detailed structural information than to find out what they actually do. Mouse knockouts of executioner caspases provide coarse level information about their role in physiology, but not much about what the key substrates that help them to orchestrate cell death actually are [6]. To date, hundreds of proteins are reported in proteomic databases to be caspase substrates, but very few have been verified by the complex logic of replacing cleavage sites in vivo [32]. Likely the complex biochemistry of apoptosis requires a number of proteins to take on gain-of-function or loss-of-function to drive the pathways needed to neatly dismantle a cell and prepare it for removal by phagocytes.
Granzymes
The term Granzyme was coined by Masson and Tschopp to define a family of serine esterases in lytic granules of cytolytic T lymphocytes [33]. Although the reagents used at the time could not distinguish between proteases and general esterases, it is now known that the granzymes represent a subset of the hematopoietic serine proteases (HeSPs) found throughout white blood cells. These proteases probably originated in tetrapods and evolved as distinct groups before the separation of tetrapods from the ray-finned fish [34]. Characteristic of the HeSPs in general, granzymes are evolving rapidly and there are substantial differences between the complement of these proteases between mice and humans. HeSPs have an unusually plastic S1 pocket (primary specificity pocket), and the ancestral protein is considered to have a despecialized specificity compared to the trypsin-like proteases that preceded it [35]. The architecture of the S1 pocket allows for rapid and radical changes in primary specificity during short evolutionary timescales. HeSPs are not found in plants, and the closest plant relatives are members of the DEG/HtrA family contained in organelles such as mitochondria and chloroplasts [36]
Granzyme B is probably the most well known of the granzyme family, and there is ample biochemical evidence that it is directly involved in apoptosis, acting primarily in a similar capacity to an apical apoptotic caspase. Indeed, granzyme B is a good example of convergent evolution where a specific biological function is subsumed by proteins with no structural similarity – the granzyme (HeSP) fold is completely unrelated to the caspase fold (Fig 1). The general concept is that granzyme B is delivered from a cytotoxic lymphocyte (CTL or NK cell) directly to the cytosol of a target cell via the pore forming perforin complex, although the exact mechanism of this delivery is still debated [37]. Upon delivery granzyme B can process executioner caspases [38] and the pro-apoptotic Bcl2-family member BID much in the same way as caspase-8 to initiate extrinsic apoptosis. This simplistic view is confounded by several studies that demonstrate cleavage of peripheral and matrix-derived proteins on the exterior of cells [39], and by the finding that the specificity of mouse and human caspases is sufficiently different in vitro [40] that clear cross-species conclusions cannot be firmly drawn. Nevertheless it is clear from mouse knockout studies that granzyme B is essential for CTL-mediated apoptosis of target cells, and from the perspective of this review we can conclude that this enzyme is a bone fide apoptotic cell death protease.
CTLs from mice deficient in granzyme B retain the ability to kill target cells, but with slower kinetics and non-apoptotic morphology compared to those from control wild type mice. Mice with a double deletion of granzymes A and B show even slower death kinetics, revealing that Granzyme A has a role to play in cell death, but not via apoptosis [41]. If we go along with the general concept that apoptosis must involve caspase-like cleavages after Asp residues it makes complete sense that granzyme A can’t induce apoptosis since it is Arg-specific, so the question then becomes how does Granzyme A kill? This is still somewhat of a controversial subject, much complicated by the classic observation that injecting trypsin into a cell will kill it [42].
The seminal paper from Williams and Henkart [42] showing that unrestrained proteolytic activity introduced into a recipient cell can lead to that cell’s demise complicates attempts to understand the role of granzymes. Granzymes are proteases, so does the artificial introduction into cells simply recapitulate the Williams and Henkart effect? They might also have redundant functions, and it has been difficult to translate in vitro experiments into in vivo settings. Granzyme concentrations used by different groups in vitro vary over orders of magnitude and it is unclear how these relate to in vivo settings where granzymes are delivered from a cytotoxic cell granule into the cytosol of a target. Quantitative experiments are not the only thing lacking since granzymes are different between mice and humans [37], and even within different mouse strains [43], and one would thus also expect differences in their substrates.
Dipeptidyl peptidase 1 (DPP1)
All HeSPs, like many proteases, are activated by removal of an N-terminal peptide (Fig. 3). Their main activator is DPP1 [44] – also known as cathepsin C - which is actually a cysteine protease related to the cysteine lysosomal cathepsins (Fig. 1). Activation takes place during packaging into specialized granules in hematopoietic cells where the fully active proteases await release to a cognate compartment – in the case of granzymes this is probably a target cell (see above). As its name signifies, DPP1 removes dipeptides sequentially from the N-terminus of a protein. Since the activation peptide of many (but not all) HeSPs is a dipeptide, there is a priori reason to think that DPP1 has co-evolved with the HeSPs. Actually, phylogenic analysis suggests that DPP1 predates the HeSPs, so it’s more likely that the HeSPs utilized a dipeptide activation sequence for the extant DPP1.
The structural fold of the cysteine cathepsins is adapted to confer different modes of substrate binding and cleavage [45]. This means that the family of cysteine cathepsins can act as endopeptideses, carboxypeptidases and/or aminopeptidases, and they accomplish this by evolving loops that block regions of the extended substrate-binding cleft [46]. DPP1 is an extreme example of this mechanistic plasticity since an additional domain (the exclusion domain) transforms the fundamental endopeptidase scaffold by excluding approach of a folded protein substrate and allows only an N-terminal peptide to accommodate the catalytic cleft [45]. Most endopepetidases can’t also be exopeptidases because the charged groups at protein termini preclude binding into the substrate cleft. DPP1, based on the cysteine cathepsin scaffold, overcomes this by providing the carboxylic group of Asp1 to counteract the positive charge on the N-terminal amino group of the substrate.
Mouse knockout studies implicate DPP1 as an important activator of many HeSPs and granzymes, but it is apparent that backup strategies for activation of (at least) granzyme B must exist since DPP1−/− mice retain residues active granzyme B [47]. Perhaps more tellingly, humans with mutations in the DPP1 gene suffer from Papillon-Lefevre syndrome, a rare autosomal disorder characterized by severe periodontitis and palmar plantar hyperkeratosis [48]. If DPP1 is the only granzyme B activator, one would expect these patients to have defects in the cytotoxic lymphocyte compartment. Revealingly, an analysis of cells from Papillon-Lefevre patients in vitro revealed that loss of DPP1 activity is associated with severe reduction in the activity and stability of neutrophil-derived HeSPs, but retain substantial granzyme activities in cytotoxic lymphocyte compartments [49]. Thus human DPP1 seems to be a major activator of neutrophil HeSPs, but a backup strategy must exist for granzymes.
Calpains and Cathepsins
Although it is expected that many proteases can kill a cell when introduced into the cytosol, as evidenced by the Williams and Henkart experiment [42], only a few can do this by participating in, triggering, or hijacking apoptosis. Candidates for endopeptidases that reside inside cells and therefore could have access to the apoptotic machinery are cytosolic calpains and lysosomal cathepsins. There is substantial controversy about whether these proteases can and/or do induce apoptosis under normal conditions [50], but there is more robust evidence that they participate in pathology [51]. One of the original hypothetical points at which these proteases could engage apoptosis was through activation of executioner caspases-3 and -7. Since the highly frangible intra-domain loop of these proteases needs to be cleaved for activation [52], the initial suspicion was that this would be a point of entry. However, although unstructured and highly flexible [53, 54], the intra-domain loop is surprisingly resistant to cleavage by calpains and lysosomal cathepsins [55–57]. Indeed the only cathepsin that could cleave and activate pro-caspase-7 was cathepsin G – which is not a lysosomal protease, nor a cysteine protease, but is a hematopoietic serine protease (HeSP – see Fig 1) found almost exclusively in neutrophils. It almost seems as though the executioner caspase activation loops are under negative selection for cleavage by calpains and lysosomal cathepsins in contrast to Granzyme B, which seems to have evolved for this purpose – an interesting conjecture that is evident, but difficult to test.
Calpains seem to play no clear role in developmental or physiologic forms of apoptosis, but are implicated in neurodegenerative disorders [58]. The most likely pro-apoptotic targets for the most common (ubiquitous) calpains 1 and 2 are Bcl-2 family proteins and the argument has been made that cleavage of these proteins would unleash the intrinsic apoptotic pathway upstream of mitochondria [56].
Cathepsin B has been implicated in promoting apoptosis induced by TNFα [59] but there seems to be no direct involvement of any lysosomal cathepsin in apoptosis mediated by other death receptors such as TRAIL- and CD95 [60]. Thus there may be a minor role, if any, for lysosomal cysteine cathepsins in triggering events in the extrinsic apoptosis pathway. Interestingly, the pepsin family aspartic lysosomal protease cathepsin D may participate in the extrinsic pathway through cleaving caspase-8 leading to a more robust activation on extrinsic pathway adapter platforms [61, 62]. Although caspase-8 is activated by dimerization and not by proteolysis, prior cleavage within the intradomain loop may stabilize the enzyme and therefore promote extrinsic apoptosis to some extent [63].
Naturally, and because of the highly organized topology of protein compartmentalization, any scenario involving lysosomal protease in any form of cell death, including apoptosis, requires that they are somehow released from the lysosomal compartment into the cysosol, or that they participate in an outside-in mechanism by cleaving cell surface proteins. In vitro evidence implicates lysosomal cathepsins in promoting apoptosis via a similar mechanism to calpains – cleavage of Bcl2 family proteins, but also XIAP, thereby removing controls on the progression of apoptotic pathways [57, 64]. But a more recent study using pharmacologic inhibitors has failed to find a role for lysosomal cysteine cathepsins in apoptosis in several different cell lines [60], highlighting the uncertainty in the field.
Necrosis
The term necrosis is a catch-all definition, basically of cell deaths that are not apoptotic. Necrosis is highly diverse and defined mainly at a microscopic level by cell swelling and plasma membrane rupture [2]. Consequently it is difficult, if not futile, to try to apportion all necrotic deaths into categories, but certain ones lay themselves out nicely because we recognize them as regulated by specific gene products. These currently include necroptosis and pyroptosis, and maybe NETosis. In addition to these regulated cell demises (see later) there is clear evidence for protease-regulated necrosis in humans, and animal genetic model organisms.
Genetic and siRNA screens in C. elegans have produced the best evidence to date for a pathway to necrotic cell death, and interestingly the main culprits identified are proteases. Linker cell death employs a cell-autonomous death program that is independent of apoptosis genes, and has morphologic characteristic of necrosis, and which is controlled by components of the C. elegans developmental timing pathway [65]. Gain-of-function mutations in specific ion channel genes in C. elegans produce a degenerative (necrotic-like) cell death that is independent of apoptotic genes. This necrotic degeneration requires the activity of calpain-like proteases and aspartic proteases, revealing that two catalytically-distinct classes of proteases are involved in necrotic cell death [66–68]. An orthogonal study revealed that the necrotic response of C. elegans to hypo-osmotic shock is controlled by a protease inhibitor, srp-6, that has the capacity to regulate a number of cysteine proteases of the calpain family, among other possible targets [69]. These analyses in C. elegans indicate that calpains and autophagy genes act within the same pathway (Fig. 5), but the involvement of cathepsins and cytosol acidification act synergistically with autophagy [70]
Fig. 5.
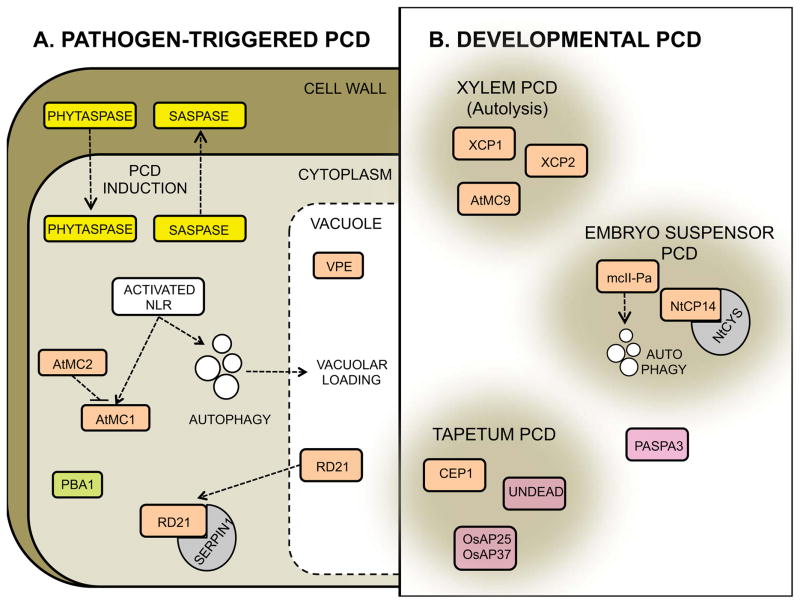
Proteases identified during different types of plant cell death. A) Pathogen-triggered cell death. The plant subtilases phytaspases and saspases, with caspase-like activity are activated during pathogen-triggered cell death. Whereas phytaspases enter the cytosol from the apoplast upon induction, saspases relocalize from the cytosol to the apoplast. The Arabidopsis metacaspase 1 (AtMC1) acts as a positive regulator of the hypersensitive response triggered upon NLR activation. AtMC2 negatively regulates this process. Autophagy acts additively to AtMC1, contributing to HR. The vacuolar VPE and the proteasome subunit PBA1 display caspase-1 and -3 activities, respectively during pathogen-triggered cell death. RD21 is a positive regulator of necrotroph-triggered cell death. The protease inhibitor SERPIN1 binds and blocks RD21 activity in the cytoplasm, which might ensure timely activation of the protease. B) A few representative types of developmental cell death are displayed with the associated proteases identified to date. The cysteine proteases XCP1, XCP2 and AtMC9 have been shown to jointly regulate post-mortem cell clearance during xylem formation. Elimination of the embryo suspensor has been shown to involve the metacaspase mcII-Pa which operates upstream of autophagy to orchestrate this type of cell death. The positive regulator of suspensor cell death NtCP14 is negatively regulated by the cystatin NtCYS, to avoid uncontrolled activation of the protease. Finally, the cysteine protease CEP1 and the aspartic proteases UNDEAD and OsAP25 and OsAP37, shown to positively regulate tapetum cell death during pollen development are highlighted. PASPA3 is included in the scheme as it is common to several types of developmental cell death. Serine proteases are displayed in yellow, cysteine proteases in orange, aspartic proteases in pink and threonine proteases in green in accordance with Fig 1.
The endosomal/lysosomal compartment may play an amplifying or supporting role in pathologic apoptosis [71] but studies in C. elegans and D. melanogaster suggest it may play a foundational role in promoting regulated necrosis [70]. It has been proposed that necrosis, unlike apoptosis, does not engage biochemical mechanisms that evolved to specifically facilitate cell death [68]. The proteins of both the apoptotic pathway and necrosis are highly conserved throughout multicellular animals [72, 73], but the necrotic effectors are likely involved in mechanisms that operate within the cell under normal conditions. Only when cells become abnormally stressed, and apoptosis somehow inactivated, do cells undergo a necrotic fate and this had once been thought to be irreversible. However, the studies in C. elegans tend to refute this argument because inhibition of presumptive necrotic effectors, proteases, rescues the cell, and in the case of srp-6, the organism. In D. melanogaster lysosomal proteases have been implicated in a non-apoptotic, non-autophagic removal of nurse cell remnants from late stage egg chambers [74]. Finally, moving to mammals, mouse mammary gland involution is dependent on Stat3-regulated cell death involving leakage of cathepsins culminating in cell death [50].
Necroptosis
Signaling by the TNF type I receptor (TNFR-I) is quite complex. Depending on the availability of intracellular adapters (and maybe even the strength of the TNFa signal) the receptor can engage survival and inflammatory effects through gene transcription mediated by NFkB, or it can engage extrinsic apoptosis. A third outcome discovered in the mid 1990s was a necrotic outcome that depended on caspase inhibition [75] - see [76] for an extensive review. The mechanism behind this puzzling triumvirate of outcomes was deciphered a decade later when it became clear that Receptor Interacting Protein (RIP) kinases were also engaged by TNFR-I activation, and the necrotic-like outcome of this RIPK-dependent death was coined necroptosis [77].
The somewhat unexpected development that caspases could prevent cell death has been explored most intensively in the field of lymphocyte physiology where caspases, in addition to their role in mediating cell death, also play a critical and positive role in B and T cell activation and proliferation. Activation of some caspases can be detected early after T cell activation and some caspase substrates are cleaved in the absence of any signs of cell death [78–80]. Humans with mutant caspase-10 display an autoimmune lymphoproliferative syndrome caused by defective lymphocyte apoptosis [81]. Humans with mutant caspase-8, while also exhibiting defects in lymphocyte apoptosis, have pronounced defects in their ability to activate lymphocytes, with resulting immunodeficiency [82]. Importantly, the latter study revealed that caspase-8 deficiency is compatible with development in humans, although it is embryonic lethal in mice [83], and the argument made in the study is because mice lack caspase-10 [84]. These data reveal a role for mouse caspase-8 in cell survival, clearly distinct from its pro-apoptotic role.
The survival (non-death) role of caspase-8 has been most extensively explored in mice, where genetic ablation of caspase-8, its inactive homolog (FLIPL), and its adaptor molecule FADD has provided striking data that these central components of the extrinsic pathway confer a pro-life, rather than a pro-death function in certain cells. Mice ablated in any of these three genes exhibit a similar embryonic lethal defect with impaired heart muscle development [83, 85, 86]. The cause of the defect likely involves substantial underdevelopment of the cardiovascular or placental systems, implying a pro-life role for the extrinsic pathway [87].
How an apparently lethal pathway such as caspase-8 activation could paradoxically result in survival is a mystery at present, but the hypothesis has been raised that the activity of caspase-8 may be altered, or diverted, by selective cleavage of caspase substrates. Non proteolytic roles for caspase-8 in epithelial cell survival have been proposed [88, 89], but rescue of caspase-8 deficiency in the hematopoietic compartment requires the catalytically active form [90]. It was initially thought that caspase-8 is required for lymphocyte expansion through NFkB signaling [91, 92], but it now seems that caspase-8 impacts a RIPK signaling to alternative death pathway [93, 94]. It appears that caspase-8 prevents RIPK3-dependent necrosis without inducing apoptosis by functioning in a proteolytically active complex with FLIPL, and this complex is required for the protective function [95]. Indeed, the caspase-8/FLIPL heterocomplex has a distinct protease specificity compared to the caspase-8/caspase-8 homocomplex, consistent with the cleavage of pro-survival proteins [96]. Importantly, only inferential data implicates the caspase-8/FLIPL heterocomplex as the direct driver of survival, and mechanistic studies are required to define how the switch of caspase-8 for a death to a survival role occurs at the biochemical level.
Pyroptosis
Pyroptosis was first observed in response to intracellular infection of macrophages with Shigella flexneri [97] or Salmonella typhimurium [98]. Activation of this programmed cell death pathway was found to be dependent on caspase-1 [99] and was only later on recognized as morphologically and mechanistically distinct from apoptosis. Due to its highly inflammatory nature this process was termed ‘pyroptosis’ from the Greek ‘pyro’ for fire or fever and ‘ptosis’ for falling [100].
Pyroptosis is dependent on the inflammatory caspases and the pyroptotic phenotype manifests in rapid cell swelling and lysis, resulting in the release of inflammatory cellular contents, a process in clear contrast to apoptosis in which membrane integrity is observed. Neutrophils take up bacteria released during pyroptotic cell lysis and then the bacteria are destroyed through the NADPH oxidase system in the secondary mechanism of phagocytotic clearance [101]. Additionally, the release of proteolytically-processed mature members of the interleukin (IL) cytokine family induces further recruitment of innate immune cells and the innate immune response.
Pyroptosis is dependent on the activation of the inflammatory caspases-1, and -11 in mice, and 1 and 4 in humans (caspase-4 is probably orthologous with caspase-11, and mice do not contain a caspase-5 ortholog), also termed cytokine activators. The roles of the other presumptive pro-inflamatory caspases-5 and -12 are enigmatic – see below. These caspases show a similar domain structure with an N-terminal caspase-recruitment domain (CARD) followed by the large and small subunit. The CARD mediates recruitment into activation complexes or facilitates direct dimerization for activation in the same way as apical apoptotic caspases (Fig. 3).
Caspase-1 is activated in inflammasomes, which induce dimerization to an initial catalytically-competent enzyme and subsequent self-processing of capase-1 into its fully-active form [102, 103]. This process requires two distinct biochemical signals: signal 1 - a transcriptional signal, induces expression of inflammasome components, followed by signal 2 - a post-translational molecular signal that leads to assembly of inflammasomes and activation of caspase-1 [104].
The earliest studied downstream substrates of caspase-1 are the proinflammatory cytokines IL-1beta and IL-18 [102], whose expression is also upregulated during signal 1 and are processed upon caspase-1 activation after signal 2. The nature of how these mature cytokines are released from the cell had been a puzzle, as they accumulate in the cytosol for lack of an export sequence for direct secretion and many non-classical secretion mechanisms have been proposed [105]. Even though IL-1b−/−IL-18−/− mice are more susceptible to diverse pathogens, caspase-1−/− mice are even more sensitive to infection [101], highlighting other important substrates of caspase-1 during pyroptosis. Such a substrate is the recently discovered Gasdermin D, which is cleaved by caspases-1 and 11 to engage a lytic signal, providing a route for release of processed cytokines from inflammatory macrophages by cell lysis [106, 107].
Caspase-11 was initially described as an interaction partner and activator of caspase-1 [108] and further studies showed it to be activated in a non-canonical inflammasome-dependent manner [109]. However, caspase-1 is still required for clearing extracellular replicating facultative intracellular pathogens through the innate immune system [110]. Caspase-4, -5, and -11 have been proposed to bind LPS directly through their CARD domain and therefore act as cytoplasmic LPS receptors [111]. Subsequent oligomerization and cell death are a mechanism markedly different from LPS sensing and inflammasome-dependent activation of caspase-1.
Caspase-12 on the other hand seems to be expressed with a premature stop codon in the majority of the human population [112]. In mice, the expression pattern of caspase-12 is substantially different from that of caspase-1 and -11 [113]. Additionally, a residue that helps define the Asp specificity of caspases, Arg-341, is replaced by a Lys in mice. Furthermore, caspase-12 reduces the inflammatory response in mice in a mechanism similar to cFLIP [114], so it is possible that caspase-12 is not a protease per se, but rather a protease modulator.
NETosis
Neutrophils, the first cellular responder to infection, have a short half life of a few hours upon release into circulation, and die by apoptosis if they do not engage pathogens [115]. Upon recognition of bacteria neutrophils attempt to engulf a pathogen for delivery to a highly degradative phagosome that includes a number of HeSPs, initiate an oxidative burst that essentially bleaches engulfed pathogens, and ultimately extrude their nucleus to form NETs (neutrophil extracellular traps), by a process dubbed NETosis [116]. NETs are reported to use antimicrobial proteins bound to DNA to neutralize pathogens and the process of NET formation has been proposed to be dependent on the activity of the HeSP neutrophil elastase [117, 118]. The idea put forward is that elastase contributes to chromatin decondensation by cleaving histones to facilitate DNA release [117]. Indeed, NETs observed in vitro are decorated by elastase [119], but this elastase is not catalytically active [120]. Elastase appears to be inhibited on NETs, and the catalytically active protease is found only in granular compartments of neutrophils that have undergone NETosis [120]. This disconnect, and the mechanisms of NETosis, remain unexplained. Nevertheless, neutrophils contain three other HeSPs in addition to elastase, all highly restricted to neutrophils, with overlapping substrate specificities [121, 122], and the jury is still out as to whether these all, or just some of them, are implicated in driving the unique morphology of NETosis. All of these proteases requite activation by DPP1, so one could assume a two-step proteolytic cascade as part of this intriguing cell death mechanism.
Plant cell death proteases
Plant regulated cell death
Programmed cell death is fundamental for plant development and for the interactions between plants and the environment. Although plant cell death shares some similarities with animal cell death, obvious structural differences between plants and animals underscore the morphological divergence that can be observed during programmed cell death processes in both kingdoms. Plant cells are surrounded by rigid cell walls, precluding formation of apoptotic bodies. In addition, plants do not have a circulatory system with phagocytes that can dispose of cell remnants after death. This leaves plants with three main types of cell death [123]: vacuolar, necrotic and mixed, the latter combining features of both vacuolar and necrotic to different extents.
Vacuolar cell death involves massive vacuolization of the cell followed by digestion of the cytosolic content that results in complete cell clearance. Developmental cell death during morphogenesis (embryo, organs, tissues and reproductive structures) and senescence is typically vacuolar. Necrotic cell death in plants is comparable to animal necrosis, involving loss of plasma membrane integrity, mitochondrial swelling and increases in cytoplasmic calcium concentration. In contrast to animals, where early plasma membrane permeabilization results in cytosolic swelling, in plants it causes shrinkage due to the presence of the cell wall. Abiotic stress is the most common trigger of necrotic cell death in plants. Necrotic cell death with vacuolar features (mixed type of cell death) is a widespread type of programmed cell death in plants. It is typical of plant-pathogen interactions leading to cell death [124, 125] and has also been observed during pollen-pistil incompatible interactions [126].
At the molecular level, plant cell death pathways remain poorly characterized and there is a very long way to go to reach the level of detail and complexity that has been achieved in animal cell death research. Cell death receptors, “deathosomes” or executioners that lead to the dismantling of the cell have yet to be identified – and perhaps they do not exist in plants, at least as we know them in animals. In addition, although the substrates of some putative death proteases have been identified, their link to regulated cell death remains to be established [127, 128]. Thanks to a growing community of plant cell death researchers [129], characterization of this process at the molecular level has started taking off in recent years bringing to light many key regulators of developmental and pathogen-induced cell death, including proteases [130, 131].
In plants, as in animals, proteases play a key role in regulated cell death. In the next two sections we focus on the current knowledge about plant cell death proteases highlighting metacaspases, proteases with caspase-like activities in plants, and other plant proteases reported to be involved in regulated cell death.
Plant Metacaspases
Plant genomes do not encode caspases. However, a variety of caspase-like activities have been typically associated with many plant cell death types [132]. At the time of their discovery, metacaspases emerged as ideal candidates behind all these caspase-like activities. The cysteine-dependent proteinases termed metacaspases were identified in the year 2000 on a refined in silico search targeting proteins that possessed homology with caspases [133]. They share with caspases the presence of a caspase-hemoglobinase fold containing the Cys-His catalytic dyad, a defining characteristic of clan CD proteases (Fig. 2). In plants, metacaspases have been classified as Type I if they have an N-terminal prodomain or Type II if they lack the prodomain and have a long linker between the p20 and p10 chains of the catalytic domain. In the following years after their discovery, several independent reports showed that metacaspases preferentially cleave their substrates after an arginine or a lysine, instead of aspartic acid characteristic of caspases [134–137]. In addition, metacaspases do not cleave typical caspase substrates and their activity is not blocked by caspase inhibitors [138]. Crystallization of yeast and protozoan metacaspase revealed a structure that precludes dimerization – reviewed in [139]. Thus, metacaspases were finally not the long-sought “plant caspases”. However, metacaspases have been functionally linked to developmental cell death as well as death triggered by biotic or abiotic stimulus.
The model plant Arabidopsis encodes 9 metacaspases: AtMC1–3 are Type I and AtMC4–9 are Type II metacaspases (Figs. 2 and 5). The type II metacaspase AtMC4 was shown to act as a positive regulator of cell death mediated by oxidative stress and pathogen attack. Pathogen-triggered cell death – commonly known as the hypersensitive response (HR) - is a plant-specific, highly localized response that occurs upon pathogen recognition resulting in the death of the cells at attempted infection sites [140]. The highly homologous Type I metacaspases AtMC1 and AtMC2 display opposite functions during HR [141]. AtMC1 was shown to act as a positive regulator of HR cell death acting antagonistically to AtMC2, which negatively regulates AtMC1. These two proteins do not interact with each other and the specifics of their interplay remain to be elucidated. Interestingly, removal of the prodomain enhanced the function of both AtMC1 and AtMC2. Catalytic activity was only required for AtMC1, implying a templating role of some kind for AtMC2 – somewhat reminiscent of the pro-survival role of vertebrate FLIP and caspase-8 (see section on necroptosis). HR cell death in AtMC1 knock-out mutants was reduced only to half of the wild-type, indicating that other processes act in concert with AtMC1 to kill the cell. In this regard, it has been shown that autophagy functions additively to AtMC1 during this process [141]. Induction of autophagy has been observed during HR [142, 143], which is not surprising given the oxidative burst and massive protein synthesis that accompanies this defense response, dramatically changing the intracellular environment. Since the central vacuole is the main protease storage of the plant cell one could speculate that vacuole overloading due to autophagy induction during HR could lead to tonoplast rupture, resulting in the massive release of the stored proteases.
Regarding cell death processes triggered by abiotic stress stimulus, AtMC8 has been shown to function as a positive mediator of UV- and hydrogen peroxide-induced cell death [136]. Another Type II metacaspase, AtMC9, was involved in xylem formation, an essential developmental cell death process in plants [144]. AtMC9 contributes to clearance of the cytosolic contents from dead cells that constitute xylem vessels. However, the mechanism by which AtMC9 exerts this function has not yet been defined. Global analysis of AtMC9 substrates by N-terminal combined fractional diagonal chromatography did not point towards any cell death-related process [128]. The Type II metacaspase from Norway spruce mcII-Pa was shown to share a phylogenetically conserved substrate with caspase-3, TSN (Tudor staphylococcal nuclease) [127]. Caspase-3 and mcII-Pa processed the protein at different sites, in line with their cleavage specificities. In plants, this cleavage occurred during both developmental and stress-induced cell death, which may indicate that TSNs are common substrates of programmed cell dismissal. Autophagy was shown to act downstream of mcII-Pa during developmental cell death, contributing to cell clearance during this process [145].
Interestingly, plant metacaspases seem to be involved not only in death-related events, but also in pro-survival processes. Evidence for a pro-survival role of metacaspases emerged first from work on protozoa and yeast, where Type I metacaspases can also be found. In these organisms, metacaspases have been not only been implicated in cell death, but also in cell cycle dynamics [146–151], cell proliferation [152] and protein quality control [153, 154]. In plants, besides the negative regulator of pathogen-triggered cell death AtMC2, AtMC1 appears to also have a pro-survival role that is developmentally regulated. In senescing plants, AtMC1 has been shown to participate in clearance of the aggregates that accumulate as part of normal aging [141].
Proteases with caspase-like activities in plants
Abundant examples of caspase-like activities associated with cell death in plants can be found in the literature over the last two decades - see [132] for review. Up to now, several enzymes have been found to be associated to these activities: phytaspases, saspases, vacuolar processing enzymes (VPE) and the proteasome.
The subtilisin-like proteins (subtilases) phytaspases and saspases are serine proteases [155]. Saspases were identified from oat treated with a death-inducing pathogen, the fungus Cochliobolus victoria [156]. As a necrotroph, C. victoria feeds from dead plant tissue and to induce cell death it secretes victorin, a toxin that binds the defense-associated thioredoxin TRX-h5 and triggers an HR-like response mediated by LOV1, an atypical NLR (nucleotide-binding leucine-rich repeat) [157]. Activity of the two saspases was inhibited by a pan-caspase chloromethyl ketone inhibitor and they were able to cleave a fluorogenic pan-caspase substrate, showing no activity against caspase-2, -3, -5 and -6 substrates and only a low level for caspase-1 and -4. The natural substrates of saspases remain to be elucidated. On the other hand, tobacco phytaspase, cleaved the VirD2 protein from the phytopathogenic bacterium Agrobacterium tumefaciens upon HR induction [158]. Both tobacco and rice phytaspases were active against diverse caspase synthetic substrates, with maximum activity observed against the caspase-like substrate sequence IWLD in rice [159]. Phytaspases have been shown to be involved in cell death triggered both by biotic and abiotic stresses [158, 160]. Interestingly, both saspases and phytaspases are located both intracellularly and in the extracellular fluid (apoplast). The apoplastic localization of saspases increased upon death induction, which might be linked to their activation in that compartment [156]. In contrast, phytaspases were shown to be constitutively processed to their mature form and secreted to the apoplast, and only enter the cell after induction of cell death [158, 160]. The exact role of these two enzymes in the apoplastic fluid remains to be established. It is worth noting that most phytopathogens are extracellular, which leaves the apoplast as one of the main battlegrounds for plant-pathogen interactions wherein most proteases present await to be characterized.
Like caspases and metacaspases, VPEs (also known as legumains) are clan CD cysteine proteases. VPEs contain a signal peptide that directs them to the vacuole once synthesized. Although VPEs preferentially cleave after an asparagine residue, they have been shown to process substrates after an aspartic acid and have been linked to caspase-like activities in plants [161–163]. In particular, VPEs were active against caspase-1 substrates and were blocked by a caspase-1 inhibitor, much as seen in their mammalian counterpart [164]. In addition, VPEs were shown to act as positive regulators of virus- or bacteria-induced HR cell death [162, 163] and to participate in seed coat formation, a developmental process featuring cell death [165].
Still, caspase-3-like activities observed in plants during cell death remain to be assigned to a particular protease. A few years back, the subunit PBA1 of the proteasome was shown to display DEVDase (caspase-3-like) activity during HR triggered upon bacterial infection [166]. HR was abolished both by caspase-3 and proteasome inhibitors, indicating that the proteasome was behind the caspase-3-like activities previously observed in this particular context. Similarly, proteasome inhibition prevented xylem differentiation [167] and isolation of caspase-3-like activities detected during this process were later assigned to the proteasome [168]. Together, these findings strongly support the notion that in plants the proteasome has caspase-like activity on small substrates, similar to its mammalian counterpart [169], and this likely has led to much confusion.
The fact that most caspase-like activities in plants can be assigned to proteases which are structurally unrelated to caspases is extremely exciting and opens the door to discover other unpredicted players in cell death among the vast protease repertoire of plants with many of their members are of unknown function. Moreover, the exact role of the caspase-like enzymes identified so far in plants needs to be better defined: up to now it is not clear what their molecular triggers and in most cases, their natural substrates are, nor whether these are linked to any kind of cell death.
Other protease activities involved in plant programmed cell death
Beyond caspase-like proteins and metacaspases, several other plant proteases have been implicated in plant cell death. For instance, many vacuolar papain-like cysteine proteases of clan CA related to the mammalian lysosomal cysteine proteases (Fig. 5) have been implicated in plant developmental and pathogen-triggered HR [170]. Among them, RD21A (Responsive to Dehydration 21A) from Arabidopsis has been shown to participate in defense against necrotrophic fungi [146, 171]. To avoid uncontrolled cell death when changes in the vacuolar membrane permeability occur, RD21A activity is contained by direct interaction with the protease inhibitor Serpin1 [146]. RD21A also interacts with PDI5 (Protein Disulfide Isomerase), a positive regulator of seed endothelium cell death, which indicates a potential role in this developmentally regulated process [172]. Another bipartite module controlling cell death in plants is composed by the cathepsin H homolog from tobacco, NtCP14 and its inhibitor, the cystatin NtCYS [173]. NtCP14 is a positive regulator of suspensor cell death, a developmental process that takes place during the late stages of embryogenesis. The suspensor is a structure that connects the embryo with its feeding source, the endosperm. When no longer needed, the suspensor undergoes cell death. To prevent early degradation of the suspensor, NtCYS binds and inhibits NtCP14. Other PLCPs involved in developmental cell death include the Xylem Cysteine Proteases 1 and 2 (XCP1 and XCP2), which together with AtMC9 were shown to participate in the cellular autolysis of the xylem [144, 174]. The cysteine endopeptidase 1 (CEP1) has been recently implicated in tapetal cell death, a form of developmental cell death that takes place during pollen development [175]. CEP1 was shown to be processed into its mature, processed form in the vacuole, where the protease is transported before vacuolar bursting.
Aspartic proteases have also been commonly associated with developmental cell death processes. For example, Arabidopsis UNDEAD controls timing of cell death during tapetum development [176]. Tapetum cell death in rice has been shown to be promoted the aspartic proteases AP25 and AP37 and their transcription is tightly and dynamically regulated to prevent unfettered cell death [177]. The expression of the Plant Aspartic Protease A3 (PASPA3) is upregulated in several developmental cell death contexts [178]. Its function in cell death awaits elucidation. Finally, there is at least one plant aspartic protease that acts as a negative regulator of developmental cell death. Promotion of Cell Survival (PCS1) is an aspartic protease that acts as a negative regulator of cell death during embryogenesis and reproductive organ formation in Arabidopsis. Its absence leads to excessive cell death in these contexts [179].
Concluding Remarks
In reviewing the literature we find substantial similarities between cell death in plants and animals. Both utilize aspartic and lysosomal-type cysteine proteases to execute necrotic-type cell deaths, but whether these are actually regulated types of death, rather than pathologic “accidental” deaths in animals is still under investigation. Certainly C. elegans models speak to forms of regulated necrosis and there is early data for related pathways in vertebrates. Both kingdoms utilize proteasomal degradation via the ubiquitin/proteasome pathway to “dissolve” specific cells – in animals this is seen in the moth M. sexta via the loss of intersegmental muscles during adult development and in plants it is seen in xylem differentiation. Whether the involvement of similar proteases in necrotic deaths is an example of conservation of mechanism or convergent evolution remains unknown.
The differences between plants and animals are also clear, with apoptosis and pyroptosis confined to animals. This is entirely due to the absence of caspases in plants, required to execute these animal-specific pathways. The closest plant homologs of caspases – the metacaspases – are involved in an intricate network of death/survival signals and pathways, and it is interesting that, just as plants don’t contain caspases, metazoan animals don’t contain metacaspases (although they are found in some animal parasites).
We know that apoptosis and the specialized protein machinery of apoptosis (including caspases) is ancient, going back to some of the earliest metazoans (the cnidarians) and seems to have been a development timed with the origin of animals with specialized organs [72, 73]. But we don’t know when this apoptotic machinery was adapted to serve a role in inflammation and pyroptosis. Indeed, it’s pretty strange that apoptotic caspases could evolve to become inflammatory caspases, substituting an anti-inflammatory death with a supposedly patent inflammatory death – pyroptosis.
Clearly lots of mysteries remain, and it’s worthwhile to keep an open mind because the apparent similarities between death pathways in disparate organisms may be superficial. Popular reagents that are thought to report on one kind of cell death or one kind of biochemical pathway, but actually report entirely different phenotypes in different organisms [180], can further confound apparent similarities.
No matter what the regulated cell death mechanism, proteases frequently play a role as signaling proteins. Since most pathologies in animals and plants involve too much or too little cell death, following the protease routes should provide outstanding opportunities to modulate pathology for therapeutic outcome as well as enhance the yield and quality the crops that we and our animals eat.
Footnotes
This review is dedicated to the memory of the late Professor Christopher Froelich – a world leader in the field of granzymes, an outstanding scientist, a selfless collaborator, and a delightful human being.
References
- 1.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi, Bravo-San Pedro L, Vitale JM, Aaronson I, Abrams SA, Adam JM, Alnemri D, Altucci ES, Andrews L, Annicchiarico-Petruzzelli D, Baehrecke M, Bazan EH, Bertrand NG, Bianchi MJ, Blagosklonny K, Blomgren MV, Borner K, Bredesen C, Brenner DE, Campanella C, Candi M, Cecconi E, Chan F, Chandel FK, Cheng NS, Chipuk EH, Cidlowski JE, Ciechanover JA, Dawson A, Dawson TM, De Laurenzi VL, De Maria V, Debatin R, Di Daniele KM, Dixit N, Dynlacht VM, El-Deiry BD, Fimia WS, Flavell GM, Fulda RA, Garrido S, Gougeon C, Green ML, Gronemeyer DR, Hajnoczky H, Hardwick G, Hengartner JM, Ichijo MO, Joseph H, Jost B, Kaufmann PJ, Kepp T, Klionsky O, Knight DJ, Kumar RA, Lemasters S, Levine JJ, Linkermann B, Lipton A, Lockshin SA, Lopez-Otin RA, Lugli C, Madeo E, Malorni F, Marine W, Martin JC, Martinou SJ, Medema JC, Meier JP, Melino P, Mizushima S, Moll N, Munoz-Pinedo U, Nunez C, Oberst G, Panaretakis A, Penninger T, Peter JM, Piacentini ME, Pinton M, Prehn P, Puthalakath JH, Rabinovich H, Ravichandran GA, Rizzuto KS, Rodrigues R, Rubinsztein CM, Rudel DC, Shi T, Simon Y, Stockwell HU, Szabadkai BR, Tait G, Tang SW, Tavernarakis HL, Tsujimoto N, Vanden Berghe Y, Vandenabeele T, Villunger P, Wagner AEF, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22:58–73. doi: 10.1038/cdd.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buhl D, Snyder LE, Schwartz PR, Edrich J. HNCO in the Galactic Centre. Nature. 1973;243:513–514. [Google Scholar]
- 4.Drag M, Salvesen GS. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polgar L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005;62:2161–72. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- 7.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–87. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 8.Bernassola F, Ciechanover A, Melino G. The ubiquitin proteasome system and its involvement in cell death pathways. Cell Death Differ. 2010;17:1–3. doi: 10.1038/cdd.2009.189. [DOI] [PubMed] [Google Scholar]
- 9.Groll M, Huber R, Moroder L. The persisting challenge of selective and specific proteasome inhibition. Journal of peptide science : an official publication of the European Peptide Society. 2009;15:58–66. doi: 10.1002/psc.1107. [DOI] [PubMed] [Google Scholar]
- 10.Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–62. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Almond JB, Cohen GM. The proteasome: a novel target for cancer chemotherapy. Leukemia. 2002;16:433–43. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- 12.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz LM. The role of cell death genes during development. Bioessays. 1991;13:389–95. doi: 10.1002/bies.950130805. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz LM, Smith SW, Jones ME, Osborne BA. Do all programmed cell deaths occur via apoptosis? Proc Natl Acad Sci U S A. 1993;90:980–4. doi: 10.1073/pnas.90.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Baehrecke EH. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol. 2015;25:376–87. doi: 10.1016/j.tcb.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overall CM, Dean RA. Degradomics: systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 2006;25:69–75. doi: 10.1007/s10555-006-7890-0. [DOI] [PubMed] [Google Scholar]
- 17.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–91. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Impens F, Colaert N, Helsens K, Plasman K, Van Damme P, Vandekerckhove J, Gevaert K. Mass spectrometry-driven protease substrate degradomics. Proteomics. 2010 doi: 10.1002/pmic.200900418. [DOI] [PubMed] [Google Scholar]
- 19.Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, Armstrong AJ, Penuela S, Laird DW, Salvesen GS, Isakson BE, Bayliss DA, Ravichandran KS. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature. 2010;467:863–7. doi: 10.1038/nature09413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harbor perspectives in biology. 2013;5:a008748. doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henson PM, Bratton DL. Antiinflammatory effects of apoptotic cells. J Clin Invest. 2013;123:2773–4. doi: 10.1172/JCI69344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin SJ, Henry CM, Cullen SP. A perspective on mammalian caspases as positive and negative regulators of inflammation. Mol Cell. 2012;46:387–97. doi: 10.1016/j.molcel.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Denecker G, Hoste E, Gilbert B, Hochepied T, Ovaere P, Lippens S, Van den Broecke C, Van Damme P, D’Herde K, Hachem JP, Borgonie G, Presland RB, Schoonjans L, Libert C, Vandekerckhove J, Gevaert K, Vandenabeele P, Declercq W. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat Cell Biol. 2007;9:666–74. doi: 10.1038/ncb1597. [DOI] [PubMed] [Google Scholar]
- 24.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–13. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 25.Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–37. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 26.Marsden VS, O’Connor L, O’Reilly LA, Silke J, Metcalf D, Ekert PG, Huang DC, Cecconi F, Kuida K, Tomaselli KJ, Roy S, Nicholson DW, Vaux DL, Bouillet P, Adams JM, Strasser A. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature. 2002;419:634–7. doi: 10.1038/nature01101. [DOI] [PubMed] [Google Scholar]
- 27.Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, Lazebnik Y, Flavell RA. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat Med. 2000;6:1241–7. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- 28.Salvesen GS, Walsh CM. Functions of caspase 8: the identified and the mysterious. Semin Immunol. 2014;26:246–52. doi: 10.1016/j.smim.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HM. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 30.Taylor RC, Brumatti G, Ito S, Hengartner MO, Derry WB, Martin SJ. Establishing a blueprint for CED-3-dependent killing through identification of multiple substrates for this protease. J Biol Chem. 2007;282:15011–21. doi: 10.1074/jbc.M611051200. [DOI] [PubMed] [Google Scholar]
- 31.Fuentes-Prior P, Noeske-Jungblut C, Donner P, Schleuning WD, Huber R, Bode W. Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc Natl Acad Sci U S A. 1997;94:11845–50. doi: 10.1073/pnas.94.22.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chau BN, Borges HL, Chen TT, Masselli A, Hunton IC, Wang JY. Signal-dependent protection from apoptosis in mice expressing caspase-resistant Rb. Nat Cell Biol. 2002;4:757–65. doi: 10.1038/ncb853. [DOI] [PubMed] [Google Scholar]
- 33.Masson D, Tschopp J. A family of serine esterases in lytic granules of cytolytic T lymphocytes. Cell. 1987;49:679–685. doi: 10.1016/0092-8674(87)90544-7. [DOI] [PubMed] [Google Scholar]
- 34.Wernersson S, Reimer JM, Poorafshar M, Karlson U, Wermenstam N, Bengtén E, Wilson M, Pilström L, Hellman L. Granzyme-like sequences in bony fish shed light on the emergence of hematopoietic serine proteases during vertebrate evolution. Developmental & Comparative Immunology. 2006;30:901–918. doi: 10.1016/j.dci.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Wouters MA, Liu K, Riek P, Husain A. A despecialization step underlying evolution of a family of serine proteases. Mol Cell. 2003;12:343–54. doi: 10.1016/s1097-2765(03)00308-3. [DOI] [PubMed] [Google Scholar]
- 36.Schuhmann H, Huesgen PF, Adamska I. The family of Deg/HtrA proteases in plants. BMC plant biology. 2012;12:52. doi: 10.1186/1471-2229-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol. 2015;15:388–400. doi: 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Stennicke HR, Wang B, Green DR, Janicke RU, Srinivasan A, Seth P, Salvesen GS, Froelich CJ. Granzyme B mimics apical caspases. Description of a unified pathway for trans-activation of executioner caspase-3 and -7. J Biol Chem. 1998;273:34278–83. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 39.Froelich CJ, Zhang X, Turbov J, Hudig D, Winkler U, Hanna WL. Human granzyme B degrades aggrecan proteoglycan in matrix synthesized by chondrocytes. J Immunol. 1993;151:7161–7171. [PubMed] [Google Scholar]
- 40.Cullen SP, Adrain C, Luthi AU, Duriez PJ, Martin SJ. Human and murine granzyme B exhibit divergent substrate preferences. J Cell Biol. 2007;176:435–44. doi: 10.1083/jcb.200612025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Susanto O, Stewart SE, Voskoboinik I, Brasacchio D, Hagn M, Ellis S, Asquith S, Sedelies KA, Bird PI, Waterhouse NJ, Trapani JA. Mouse granzyme A induces a novel death with writhing morphology that is mechanistically distinct from granzyme B-induced apoptosis. Cell Death Differ. 2013;20:1183–93. doi: 10.1038/cdd.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams MS, Henkart PA. Apoptotic cell death induced by intracellular proteolysis. J Immunol. 1995;153:4247–4255. [PubMed] [Google Scholar]
- 43.Andoniou CE, Sutton VR, Wikstrom ME, Fleming P, Thia KY, Matthews AY, Kaiserman D, Schuster IS, Coudert JD, Eldi P, Chaudhri G, Karupiah G, Bird PI, Trapani JA, Degli-Esposti MA. A natural genetic variant of granzyme B confers lethality to a common viral infection. PLoS Pathog. 2014;10:e1004526. doi: 10.1371/journal.ppat.1004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGuire MJ, Lipsky PE, Thiele DL. Generation of active myeloid and lymphoid granule serine proteases requires processing by the granule thiol protease dipeptidyl peptidase I. J Biol Chem. 1993;268:2458–2467. [PubMed] [Google Scholar]
- 45.Turk D, Janjic V, Stern I, Podobnik M, Lamba D, Dahl SW, Lauritzen C, Pedersen J, Turk V, Turk B. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20:6570–82. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton VR, Waterhouse NJ, Browne KA, Sedelies K, Ciccone A, Anthony D, Koskinen A, Mullbacher A, Trapani JA. Residual active granzyme B in cathepsin C-null lymphocytes is sufficient for perforin-dependent target cell apoptosis. J Cell Biol. 2007;176:425–33. doi: 10.1083/jcb.200609077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hart TC, Hart PS, Bowden DW, Michalec MD, Callison SA, Walker SJ, Zhang Y, Firatli E. Mutations of the cathepsin C gene are responsible for Papillon-Lefevre syndrome. Journal of medical genetics. 1999;36:881–7. [PMC free article] [PubMed] [Google Scholar]
- 49.Pham CT, Ivanovich JL, Raptis SZ, Zehnbauer B, Ley TJ. Papillon-Lefevre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J Immunol. 2004;173:7277–81. doi: 10.4049/jimmunol.173.12.7277. [DOI] [PubMed] [Google Scholar]
- 50.Sargeant TJ, Lloyd-Lewis B, Resemann HK, Ramos-Montoya A, Skepper J, Watson CJ. Stat3 controls cell death during mammary gland involution by regulating uptake of milk fat globules and lysosomal membrane permeabilization. Nat Cell Biol. 2014;16:1057–68. doi: 10.1038/ncb3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aits S, Jaattela M. Lysosomal cell death at a glance. J Cell Sci. 2013;126:1905–12. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Q, Salvesen GS. Activation of pro-caspase-7 by serine proteases includes a non-canonical specificity. Biochem J. 1997;324:361–364. doi: 10.1042/bj3240361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riedl SJ, Fuentes-Prior P, Renatus M, Kairies N, Krapp S, Huber R, Salvesen GS, Bode W. Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci U S A. 2001;98:14790–5. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chai J, Wu Q, Shiozaki E, Srinivasula SM, Alnemri ES, Shi Y. Crystal structure of a procaspase-7 zymogen. Mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 55.Wolf BB, Goldstein JC, Stennicke HR, Beere H, Amarante-Mendes GP, Salvesen GS, Green DR. Calpain functions in a caspase-independent manner to promote apoptosis- like events during platelet activation. Blood. 1999;94:1683–92. [PubMed] [Google Scholar]
- 56.Gil-Parrado S, Fernandez-Montalvan A, Assfalg-Machleidt I, Popp O, Bestvater F, Holloschi A, Knoch TA, Auerswald EA, Welsh K, Reed JC, Fritz H, Fuentes-Prior P, Spiess E, Salvesen GS, Machleidt W. Ionomycin-activated calpain triggers apoptosis. A probable role for Bcl-2 family members. J Biol Chem. 2002;277:27217–26. doi: 10.1074/jbc.M202945200. [DOI] [PubMed] [Google Scholar]
- 57.Stoka V, Turk B, Schendel SL, Kim TH, Cirman T, Snipas SJ, Ellerby LM, Bredesen D, Freeze H, Abrahamson M, Bromme D, Krajewski S, Reed JC, Yin XM, Turk V, Salvesen GS. Lysosomal protease pathways to apoptosis: cleavage of bid, not pro-caspases, is the most likely route. J Biol Chem. 2001;276:3149–3157. doi: 10.1074/jbc.M008944200. [DOI] [PubMed] [Google Scholar]
- 58.Momeni HR. Role of calpain in apoptosis. Cell journal. 2011;13:65–72. [PMC free article] [PubMed] [Google Scholar]
- 59.Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, Kaufmann SH, Gores GJ. Cathepsin B contributes to TNFa–mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J Clin Invest. 2000;106:1127–1137. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spes A, Sobotic B, Turk V, Turk B. Cysteine cathepsins are not critical for TRAIL- and CD95-induced apoptosis in several human cancer cell lines. Biol Chem. 2012;393:1417–31. doi: 10.1515/hsz-2012-0213. [DOI] [PubMed] [Google Scholar]
- 61.Conus S, Perozzo R, Reinheckel T, Peters C, Scapozza L, Yousefi S, Simon HU. Caspase-8 is activated by cathepsin D initiating neutrophil apoptosis during the resolution of inflammation. J Exp Med. 2008;205:685–98. doi: 10.1084/jem.20072152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conus S, Pop C, Snipas SJ, Salvesen GS, Simon HU. Cathepsin D primes caspase-8 activation by multiple intra-chain proteolysis. J Biol Chem. 2012;287:21142–51. doi: 10.1074/jbc.M111.306399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pop C, Fitzgerald P, Green DR, Salvesen GS. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry. 2007;46:4398–407. doi: 10.1021/bi602623b. [DOI] [PubMed] [Google Scholar]
- 64.Cirman T, Oresic K, Mazovec GD, Turk V, Reed JC, Myers RM, Salvesen GS, Turk B. Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J Biol Chem. 2004;279:3578–87. doi: 10.1074/jbc.M308347200. [DOI] [PubMed] [Google Scholar]
- 65.Abraham MC, Lu Y, Shaham S. A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev Cell. 2007;12:73–86. doi: 10.1016/j.devcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Syntichaki P, Xu K, Driscoll M, Tavernarakis N. Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature. 2002;419:939–44. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 67.Artal-Sanz M, Samara C, Syntichaki P, Tavernarakis N. Lysosomal biogenesis and function is critical for necrotic cell death in Caenorhabditis elegans. J Cell Biol. 2006;173:231–9. doi: 10.1083/jcb.200511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Syntichaki P, Tavernarakis N. The biochemistry of neuronal necrosis: rogue biology? Nat Rev Neurosci. 2003;4:672–84. doi: 10.1038/nrn1174. [DOI] [PubMed] [Google Scholar]
- 69.Luke CJ, Pak SC, Askew YS, Naviglia TL, Askew DJ, Nobar SM, Vetica AC, Long OS, Watkins SC, Stolz DB, Barstead RJ, Moulder GL, Bromme D, Silverman GA. An intracellular serpin regulates necrosis by inhibiting the induction and sequelae of lysosomal injury. Cell. 2007;130:1108–19. doi: 10.1016/j.cell.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCall K. Genetic control of necrosis - another type of programmed cell death. Curr Opin Cell Biol. 2010;22:882–8. doi: 10.1016/j.ceb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Repnik U, Cesen MH, Turk B. The endolysosomal system in cell death and survival. Cold Spring Harbor perspectives in biology. 2013;5:a008755. doi: 10.1101/cshperspect.a008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zmasek CM, Zhang Q, Ye Y, Godzik A. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8:R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bender CE, Fitzgerald P, Tait SW, Llambi F, McStay GP, Tupper DO, Pellettieri J, Sanchez Alvarado A, Salvesen GS, Green DR. Mitochondrial pathway of apoptosis is ancestral in metazoans. Proc Natl Acad Sci U S A. 2012;109:4904–9. doi: 10.1073/pnas.1120680109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson JS, McCall K. Combined inhibition of autophagy and caspases fails to prevent developmental nurse cell death in the Drosophila melanogaster ovary. PLoS One. 2013;8:e76046. doi: 10.1371/journal.pone.0076046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W, Grooten J, Fiers W, Vandenabeele P. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med. 1998;187:1477–85. doi: 10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15:135–47. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 77.Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–9. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 78.Alam A, Cohen LY, Aouad S, Sekaly RP. Early activation of caspases during T lymphocyte stimulation results in selective substrate cleavage in nonapoptotic cells. J Exp Med. 1999;190:1879–90. doi: 10.1084/jem.190.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maelfait J, Beyaert R. Non-apoptotic functions of caspase-8. Biochem Pharmacol. 2008;76:1365–73. doi: 10.1016/j.bcp.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 80.Kang TB, Oh GS, Scandella E, Bolinger B, Ludewig B, Kovalenko A, Wallach D. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J Immunol. 2008;181:2522–32. doi: 10.4049/jimmunol.181.4.2522. [DOI] [PubMed] [Google Scholar]
- 81.Wang J, Zheng L, Lobito A, Chan FK, Dale J, Sneller M, Yao X, Puck JM, Straus SE, Lenardo MJ. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 82.Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, Skoda-Smith S, Atkinson TP, Straus SE, Lenardo MJ. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–9. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 83.Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P, Wallach D. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–76. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 84.Reed JC, Doctor K, Rojas A, Zapata JM, Stehlik C, Fiorentino L, Damiano J, Roth W, Matsuzawa S, Newman R, Takayama S, Marusawa H, Xu F, Salvesen G, Godzik A. Comparative analysis of apoptosis and inflammation genes of mice and humans. Genome Res. 2003;13:1376–88. doi: 10.1101/gr.1053803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A, Mirtsos C, Suzuki N, Bonnard M, Goeddel DV, Mak TW. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–42. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 86.Yeh WC, Pompa JL, McCurrach ME, Shu HB, Elia AJ, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, El-Deiry WS, Lowe SW, Goeddel DV, Mak TW. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science. 1998;279:1954–8. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 87.Weinlich R, Oberst A, Dillon CP, Janke LJ, Milasta S, Lukens JR, Rodriguez DA, Gurung P, Savage C, Kanneganti TD, Green DR. Protective roles for caspase-8 and cFLIP in adult homeostasis. Cell reports. 2013;5:340–8. doi: 10.1016/j.celrep.2013.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Finlay D, Howes A, Vuori K. Critical role for caspase-8 in epidermal growth factor signaling. Cancer Res. 2009;69:5023–9. doi: 10.1158/0008-5472.CAN-08-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mielgo A, Torres VA, Schmid MC, Graf R, Zeitlin SG, Lee P, Shields DJ, Barbero S, Jamora C, Stupack DG. The death effector domains of caspase-8 induce terminal differentiation. PLoS One. 2009;4:e7879. doi: 10.1371/journal.pone.0007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leverrier S, Salvesen GS, Walsh CM. Enzymatically active single chain caspase-8 maintains T-cell survival during clonal expansion. Cell Death Differ. 2011;18:90–8. doi: 10.1038/cdd.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Su H, Bidere N, Zheng L, Cubre A, Sakai K, Dale J, Salmena L, Hakem R, Straus S, Lenardo M. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 2005;307:1465–8. doi: 10.1126/science.1104765. [DOI] [PubMed] [Google Scholar]
- 92.Lemmers B, Salmena L, Bidere N, Su H, Matysiak-Zablocki E, Murakami K, Ohashi PS, Jurisicova A, Lenardo M, Hakem R, Hakem A. Essential role for caspase-8 in Toll-like receptors and NFkappaB signaling. J Biol Chem. 2007;282:7416–23. doi: 10.1074/jbc.M606721200. [DOI] [PubMed] [Google Scholar]
- 93.Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs KA, Morrissette NS, Walsh CM. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci U S A. 2008;105:16677–82. doi: 10.1073/pnas.0808597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ch’en IL, Beisner DR, Degterev A, Lynch C, Yuan J, Hoffmann A, Hedrick SM. Antigen-mediated T cell expansion regulated by parallel pathways of death. Proc Natl Acad Sci U S A. 2008;105:17463–8. doi: 10.1073/pnas.0808043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–7. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pop C, Oberst A, Drag M, Van Raam BJ, Riedl SJ, Green DR, Salvesen GS. FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem J. 2011;433:447–57. doi: 10.1042/BJ20101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zychlinsky A, Prevost MC, Sansonetti PJ. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–9. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]
- 98.Monack DM, Raupach B, Hromockyj AE, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci U S A. 1996;93:9833–8. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen Y, Smith MR, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 1996;15:3853–60. [PMC free article] [PubMed] [Google Scholar]
- 100.Cookson BT, Brennan MA. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9:113–4. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- 101.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–42. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, Elliston KO, Ayala JM, Casano FJ, Chin J, Ding GJF, Egger LA, Gaffney EP, Limjuco G, Palyha OC, Raju SM, Rolando AM, Salley JP, Yamin TT, Tocci MJ. A novel heterodimeric cysteine protease is required for interleukin-1beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 103.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–26. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 104.Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–7. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eder C. Mechanisms of interleukin-1beta release. Immunobiology. 2009;214:543–53. doi: 10.1016/j.imbio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 106.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signaling. Nature. 2015 doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 107.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015 doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 108.Wang S, Miura M, Jung Y-K, Zhu H, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 109.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–21. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 110.Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–91. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–92. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 112.Fischer H, Koenig U, Eckhart L, Tschachler E. Human caspase 12 has acquired deleterious mutations. Biochem Biophys Res Commun. 2002;293:722–6. doi: 10.1016/S0006-291X(02)00289-9. [DOI] [PubMed] [Google Scholar]
- 113.Kalai M, Lamkanfi M, Denecker G, Boogmans M, Lippens S, Meeus A, Declercq W, Vandenabeele P. Regulation of the expression and processing of caspase-12. J Cell Biol. 2003;162:457–67. doi: 10.1083/jcb.200303157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saleh M, Mathison JC, Wolinski MK, Bensinger SJ, Fitzgerald P, Droin N, Ulevitch RJ, Green DR, Nicholson DW. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature. 2006;440:1064–8. doi: 10.1038/nature04656. [DOI] [PubMed] [Google Scholar]
- 115.Remijsen Q, Kuijpers TW, Wirawan E, Lippens S, Vandenabeele P, Vanden Berghe T. Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ. 2011;18:581–8. doi: 10.1038/cdd.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–5. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 117.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–91. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Parker H, Dragunow M, Hampton MB, Kettle AJ, Winterbourn CC. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J Leukoc Biol. 2012;92:841–9. doi: 10.1189/jlb.1211601. [DOI] [PubMed] [Google Scholar]
- 119.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–41. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kasperkiewicz P, Poreba M, Snipas SJ, Parker H, Winterbourn CC, Salvesen GS, Drag M. Design of ultrasensitive probes for human neutrophil elastase through hybrid combinatorial substrate library profiling. Proc Natl Acad Sci U S A. 2014;111:2518–23. doi: 10.1073/pnas.1318548111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Korkmaz B, Attucci S, Juliano MA, Kalupov T, Jourdan ML, Juliano L, Gauthier F. Measuring elastase, proteinase 3 and cathepsin G activities at the surface of human neutrophils with fluorescence resonance energy transfer substrates. Nat Protoc. 2008;3:991–1000. doi: 10.1038/nprot.2008.63. [DOI] [PubMed] [Google Scholar]
- 122.Kasperkiewicz P, Poreba M, Snipas SJ, Lin SJ, Kirchhofer D, Salvesen GS, Drag M. Design of a Selective Substrate and Activity Based Probe for Human Neutrophil Serine Protease 4. PLoS One. 2015;10:e0132818. doi: 10.1371/journal.pone.0132818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van Doorn WG. Classes of programmed cell death in plants, compared to those in animals. Journal of experimental botany. 2011;62:4749–61. doi: 10.1093/jxb/err196. [DOI] [PubMed] [Google Scholar]
- 124.Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot. 2008;59:501–20. doi: 10.1093/jxb/erm239. [DOI] [PubMed] [Google Scholar]
- 125.Curtis MJ, Wolpert TJ. The victorin-induced mitochondrial permeability transition precedes cell shrinkage and biochemical markers of cell death, and shrinkage occurs without loss of membrane integrity. The Plant journal : for cell and molecular biology. 2004;38:244–59. doi: 10.1111/j.1365-313X.2004.02040.x. [DOI] [PubMed] [Google Scholar]
- 126.Geitmann A, Franklin-Tong VE, Emons AC. The self-incompatibility response in Papaver rhoeas pollen causes early and striking alterations to organelles. Cell death and differentiation. 2004;11:812–22. doi: 10.1038/sj.cdd.4401424. [DOI] [PubMed] [Google Scholar]
- 127.Sundstrom JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA, Jr, Valineva T, Saarikettu J, Frilander MJ, Suarez MF, Zavialov A, Stahl U, Hussey PJ, Silvennoinen O, Sundberg E, Zhivotovsky B, Bozhkov PV. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol. 2009;11:1347–54. doi: 10.1038/ncb1979. [DOI] [PubMed] [Google Scholar]
- 128.Tsiatsiani L, Timmerman E, De Bock PJ, Vercammen D, Stael S, van de Cotte B, Staes A, Goethals M, Beunens T, Van Damme P, Gevaert K, Van Breusegem F. The Arabidopsis metacaspase9 degradome. The Plant cell. 2013;25:2831–47. doi: 10.1105/tpc.113.115287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stael S, Nowack MK, Van Breusegem F, Valls M, Coll NS. The death of plant cells: from proteases to field applications. Cell death and differentiation. 2014;21:1178–9. doi: 10.1038/cdd.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Van Hautegem T, Waters AJ, Goodrich J, Nowack MK. Only in dying, life: programmed cell death during plant development. Trends in plant science. 2015;20:102–13. doi: 10.1016/j.tplants.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 131.Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell death and differentiation. 2011;18:1247–56. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bonneau L, Ge Y, Drury GE, Gallois P. What happened to plant caspases? Journal of experimental botany. 2008;59:491–9. doi: 10.1093/jxb/erm352. [DOI] [PubMed] [Google Scholar]
- 133.Uren AG, O’Rourke K, Aravind LA, Pisbarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient fmilies of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 134.Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inze D, Van Breusegem F. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. The Journal of biological chemistry. 2004;279:45329–36. doi: 10.1074/jbc.M406329200. [DOI] [PubMed] [Google Scholar]
- 135.Watanabe N, Lam E. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. The Journal of biological chemistry. 2005;280:14691–9. doi: 10.1074/jbc.M413527200. [DOI] [PubMed] [Google Scholar]
- 136.He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. The Journal of biological chemistry. 2008;283:774–83. doi: 10.1074/jbc.M704185200. [DOI] [PubMed] [Google Scholar]
- 137.Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA, Jr, Rodriguez-Nieto S, Zhivotovsky B, Smertenko A. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14463–8. doi: 10.1073/pnas.0506948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan JA, De Rycke R, Brackenier A, Inze D, Harris JL, Van Breusegem F. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. Journal of molecular biology. 2006;364:625–36. doi: 10.1016/j.jmb.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 139.McLuskey K, Mottram JC. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem J. 2015;466:219–32. doi: 10.1042/BJ20141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P. Arabidopsis type I metacaspases control cell death. Science. 2010;330:1393–7. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- 141.Coll NS, Smidler A, Puigvert M, Popa C, Valls M, Dangl JL. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell death and differentiation. 2014;21:1399–408. doi: 10.1038/cdd.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–77. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 143.Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jorgensen LB, Jones JD, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–83. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 144.Bollhoner B, Zhang B, Stael S, Denance N, Overmyer K, Goffner D, Van Breusegem F, Tuominen H. Post mortem function of AtMC9 in xylem vessel elements. The New phytologist. 2013;200:498–510. doi: 10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- 145.Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, Smertenko A, Bozhkov PV. Autophagy and metacaspase determine the mode of cell death in plants. The Journal of cell biology. 2013;203:917–27. doi: 10.1083/jcb.201307082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lampl N, Alkan N, Davydov O, Fluhr R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. The Plant journal : for cell and molecular biology. 2013;74:498–510. doi: 10.1111/tpj.12141. [DOI] [PubMed] [Google Scholar]
- 147.Laverriere M, Cazzulo JJ, Alvarez VE. Antagonic activities of Trypanosoma cruzi metacaspases affect the balance between cell proliferation, death and differentiation. Cell death and differentiation. 2012;19:1358–69. doi: 10.1038/cdd.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ambit A, Fasel N, Coombs GH, Mottram JC. An essential role for the Leishmania major metacaspase in cell cycle progression. Cell death and differentiation. 2008;15:113–22. doi: 10.1038/sj.cdd.4402232. [DOI] [PubMed] [Google Scholar]
- 149.Helms MJ, Ambit A, Appleton P, Tetley L, Coombs GH, Mottram JC. Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. Journal of cell science. 2006;119:1105–17. doi: 10.1242/jcs.02809. [DOI] [PubMed] [Google Scholar]
- 150.Proto WR, Castanys-Munoz E, Black A, Tetley L, Moss CX, Juliano L, Coombs GH, Mottram JC. Trypanosoma brucei metacaspase 4 is a pseudopeptidase and a virulence factor. The Journal of biological chemistry. 2011;286:39914–25. doi: 10.1074/jbc.M111.292334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee RE, Puente LG, Kaern M, Megeney LA. A non-death role of the yeast metacaspase: Yca1p alters cell cycle dynamics. PloS one. 2008;3:e2956. doi: 10.1371/journal.pone.0002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Szallies A, Kubata BK, Duszenko M. A metacaspase of Trypanosoma brucei causes loss of respiration competence and clonal death in the yeast Saccharomyces cerevisiae. FEBS letters. 2002;517:144–50. doi: 10.1016/s0014-5793(02)02608-x. [DOI] [PubMed] [Google Scholar]
- 153.Lee RE, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13348–53. doi: 10.1073/pnas.1006610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hill SM, Hao X, Liu B, Nystrom T. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science. 2014;344:1389–92. doi: 10.1126/science.1252634. [DOI] [PubMed] [Google Scholar]
- 155.Vartapetian AB, Tuzhikov AI, Chichkova NV, Taliansky M, Wolpert TJ. A plant alternative to animal caspases: subtilisin-like proteases. Cell death and differentiation. 2011;18:1289–97. doi: 10.1038/cdd.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Coffeen WC, Wolpert TJ. Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. The Plant cell. 2004;16:857–73. doi: 10.1105/tpc.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng SC, Maier CS, Wolpert TJ. Tricking the guard: exploiting plant defense for disease susceptibility. Science. 2012;338:659–62. doi: 10.1126/science.1226743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chichkova NV, Kim SH, Titova ES, Kalkum M, Morozov VS, Rubtsov YP, Kalinina NO, Taliansky ME, Vartapetian AB. A plant caspase-like protease activated during the hypersensitive response. The Plant cell. 2004;16:157–71. doi: 10.1105/tpc.017889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Galiullina RA, Kasperkiewicz P, Chichkova NV, Szalek A, Serebryakova MV, Poreba M, Drag M, Vartapetian AB. Substrate Specificity and Possible Heterologous Targets of Phytaspase, a Plant Cell Death Protease. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.M115.675819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chichkova NV, Shaw J, Galiullina RA, Drury GE, Tuzhikov AI, Kim SH, Kalkum M, Hong TB, Gorshkova EN, Torrance L, Vartapetian AB, Taliansky M. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. The EMBO journal. 2010;29:1149–61. doi: 10.1038/emboj.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hiraiwa N, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS letters. 1999;447:213–6. doi: 10.1016/s0014-5793(99)00286-0. [DOI] [PubMed] [Google Scholar]
- 162.Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;305:855–8. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- 163.Rojo E, Martin R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sanchez-Serrano JJ, Baker B, Ausubel FM, Raikhel NV. VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Current biology : CB. 2004;14:1897–906. doi: 10.1016/j.cub.2004.09.056. [DOI] [PubMed] [Google Scholar]
- 164.Sexton KB, Witte MD, Blum G, Bogyo M. Design of cell-permeable, fluorescent activity-based probes for the lysosomal cysteine protease asparaginyl endopeptidase (AEP)/legumain. Bioorg Med Chem Lett. 2007;17:649–53. doi: 10.1016/j.bmcl.2006.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I. A vacuolar processing enzyme, deltaVPE, is involved in seed coat formation at the early stage of seed development. The Plant cell. 2005;17:876–87. doi: 10.1105/tpc.104.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes & development. 2009;23:2496–506. doi: 10.1101/gad.1825209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Woffenden BJ, Freeman TB, Beers EP. Proteasome inhibitors prevent tracheary element differentiation in zinnia mesophyll cell cultures. Plant physiology. 1998;118:419–30. doi: 10.1104/pp.118.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Han JJ, Lin W, Oda Y, Cui KM, Fukuda H, He XQ. The proteasome is responsible for caspase-3-like activity during xylem development. The Plant journal : for cell and molecular biology. 2012;72:129–41. doi: 10.1111/j.1365-313X.2012.05070.x. [DOI] [PubMed] [Google Scholar]
- 169.Kisselev AF, Garcia-Calvo M, Overkleeft HS, Peterson E, Pennington MW, Ploegh HL, Thornberry NA, Goldberg AL. The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J Biol Chem. 2003;278:35869–77. doi: 10.1074/jbc.M303725200. [DOI] [PubMed] [Google Scholar]
- 170.Shindo T, Van der Hoorn RA. Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Molecular plant pathology. 2008;9:119–25. doi: 10.1111/j.1364-3703.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shindo T, Misas-Villamil JC, Horger AC, Song J, van der Hoorn RA. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PloS one. 2012;7:e29317. doi: 10.1371/journal.pone.0029317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Andeme Ondzighi C, Christopher DA, Cho EJ, Chang SC, Staehelin LA. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. The Plant cell. 2008;20:2205–20. doi: 10.1105/tpc.108.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhao P, Zhou XM, Zhang LY, Wang W, Ma LG, Yang LB, Peng XB, Bozhkov PV, Sun MX. A bipartite molecular module controls cell death activation in the Basal cell lineage of plant embryos. PLoS biology. 2013;11:e1001655. doi: 10.1371/journal.pbio.1001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Avci U, Petzold HE, Ismail IO, Beers EP, Haigler CH. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. The Plant journal : for cell and molecular biology. 2008;56:303–15. doi: 10.1111/j.1365-313X.2008.03592.x. [DOI] [PubMed] [Google Scholar]
- 175.Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. The Plant cell. 2014;26:2939–61. doi: 10.1105/tpc.114.127282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Phan HA, Li SF, Parish RW. MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant molecular biology. 2012;78:171–83. doi: 10.1007/s11103-011-9855-0. [DOI] [PubMed] [Google Scholar]
- 177.Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nature communications. 2013;4:1445. doi: 10.1038/ncomms2396. [DOI] [PubMed] [Google Scholar]
- 178.Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guerin CJ, Krebs M, Schumacher K, Nowack MK. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Current biology : CB. 2014;24:931–40. doi: 10.1016/j.cub.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 179.Ge X, Dietrich C, Matsuno M, Li G, Berg H, Xia Y. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO reports. 2005;6:282–8. doi: 10.1038/sj.embor.7400357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hardwick JM, Cheng WC. Mitochondrial programmed cell death pathways in yeast. Dev Cell. 2004;7:630–2. doi: 10.1016/j.devcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 181.Bouchier-Hayes L, Green DR. Caspase-2: the orphan caspase. Cell Death Differ. 2012;19:51–7. doi: 10.1038/cdd.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fava LL, Bock FJ, Geley S, Villunger A. Caspase-2 at a glance. J Cell Sci. 2012;125:5911–5. doi: 10.1242/jcs.115105. [DOI] [PubMed] [Google Scholar]
- 183.Olsson M, Forsberg J, Zhivotovsky B. Caspase-2: the reinvented enzyme. Oncogene. 2015;34:1877–82. doi: 10.1038/onc.2014.139. [DOI] [PubMed] [Google Scholar]
- 184.Puccini J, Dorstyn L, Kumar S. Caspase-2 as a tumour suppressor. Cell Death Differ. 2013;20:1133–9. doi: 10.1038/cdd.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Troy CM, Shelanski ML. Caspase-2 redux. Cell Death Differ. 2003;10:101–7. doi: 10.1038/sj.cdd.4401175. [DOI] [PubMed] [Google Scholar]
- 186.Hachmann J, Salvesen GS. The Paracaspase MALT1. Biochimie. 2015 doi: 10.1016/j.biochi.2015.09.018. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]