Abstract
Ocean acidification is the increase in seawater pCO 2 due to the uptake of atmospheric anthropogenic CO 2, with the largest changes predicted to occur in the Arctic seas. For some marine organisms, this change in pCO 2, and associated decrease in pH, represents a climate change‐related stressor. In this study, we investigated the gene expression patterns of nauplii of the Arctic copepod Calanus glacialis cultured at low pH levels. We have previously shown that organismal‐level performance (development, growth, respiration) of C. glacialis nauplii is unaffected by low pH. Here, we investigated the molecular‐level response to lowered pH in order to elucidate the physiological processes involved in this tolerance. Nauplii from wild‐caught C. glacialis were cultured at four pH levels (8.05, 7.9, 7.7, 7.5). At stage N6, mRNA was extracted and sequenced using RNA‐seq. The physiological functionality of the proteins identified was categorized using Gene Ontology and KEGG pathways. We found that the expression of 151 contigs varied significantly with pH on a continuous scale (93% downregulated with decreasing pH). Gene set enrichment analysis revealed that, of the processes downregulated, many were components of the universal cellular stress response, including DNA repair, redox regulation, protein folding, and proteolysis. Sodium:proton antiporters were among the processes significantly upregulated, indicating that these ion pumps were involved in maintaining cellular pH homeostasis. C. glacialis significantly alters its gene expression at low pH, although they maintain normal larval development. Understanding what confers tolerance to some species will support our ability to predict the effects of future ocean acidification on marine organisms.
Keywords: ocean acidification, pH, phenotypic buffering, RNA‐seq, stress response, transcriptomics
1. INTRODUCTION
Anthropogenic CO2 emissions are increasing the pCO2 of the atmosphere and the oceans (Le Quéré, Raupach, & Canadell, 2009). Increased pCO2 in surface waters alters the carbonate chemistry of sea water, ultimately increasing hydrogen ion (H+), bicarbonate ion (HCO3 −), and dissolved inorganic carbon (DIC) concentrations and decreasing pH and carbonate ion (CO3 2−) concentrations. This process is termed ocean acidification (Caldeira & Wickett, 2003; Orr, Fabry, & Aumont, 2005). While occurring in all of the world's oceans, the largest changes in carbonate chemistry are expected to occur in the Arctic seas (Steinacher et al., 2009; Bellerby, Anderson, & Azetsu‐Scott, 2013). Low‐salinity Arctic waters have low buffering capability, resulting in lower pHs for a given amount of CO2, and projections indicate further freshening from ice melt and river input (Steinacher et al., 2009; Bellerby et al., 2013). The reduction in sea ice with climate change may also increase the atmosphere‐ocean flux of CO2 (Bates, Moran, Hansell, & Mathis, 2006; Arrigo, Pabi, Van Dijken, & Maslowski, 2010; Parmentier et al., 2013; Barber, Hop, & Mundy, 2015).
High pCO2, and associated low pH, is a stressor for some marine organisms (Kroeker, Kordas, Crim, & Singh, 2010; Wittmann & Pörtner, 2013). Many physiological processes including gas exchange, ion transport, enzyme activity, and protein function rely on a specific range of intracellular pH (Whiteley, 2011). Maintaining pH homeostasis can incur additional energetic costs when organisms must actively compensate for low extracellular pH using energy‐expensive ion pumps (Pörtner, 2008; Whiteley, 2011), leaving less energy for growth, development, and reproduction (e.g., Stumpp, Wren, Melzner, Thorndyke, & Dupont, 2011).
In calanoid copepods, exposure to pH levels projected for the next century has produced inconsistent effects on organismal‐level measures of fitness and performance. Some species show no changes in development, respiration, and feeding rates with lowered pH (Kurihara & Ishimatsu, 2008; Mayor, Everett, & Cook, 2012; McConville et al., 2013; Runge, Fields, & Thompson, 2016), whereas other calanoid species are detrimentally affected (Zhang, Li, Wang, & Guo, 2011; Fitzer et al., 2012; Pedersen, Håkedal, & Salaberria, 2014; Thor & Dupont, 2015). Effects may vary by species, but also by life stage, with eggs and early naupliar stages being most vulnerable (Cripps, Lindeque, & Flynn, 2014; Pedersen, Våge, Olsen, Hammer, & Altin, 2014).
The ability of an organism to maintain fitness‐related traits, like growth, reproduction, and development, while exposed to a stressful environment by adjusting underlying physiological processes has been referred to as phenotypic buffering (Reusch, 2014). Phenotypic buffering is crucial for tolerance of spatially and temporally variable environments, and organisms that are capable of phenotypic buffering may be able to adapt more easily to long‐term environmental change (Donelson, Munday, McCormick, & Pitcher, 2012; Miller, Watson, Donelson, McCormick, & Munday, 2012). However, depending on the mechanism, long‐term utilization of phenotypic buffering in a stressful environment may also incur an energetic cost. Phenotypic buffering may involve, among other things, altered gene expression, compensatory feeding, and changes in energy allocation (Sunday et al., 2014).
Recent progress in high‐throughput genomic technology has made possible the sequencing of entire transcriptomes (RNA‐seq), with the potential to measure expression in all transcribed genes, not limited to a targeted set of genes in a microarray. This has allowed for a universal, nontargeted exploration of gene expression (or transcriptomic) responses to environmental drivers. Investigating the transcriptomic response of species to climate change‐related environmental drivers has the potential to provide greater understanding of why and how some species are more tolerant to change than others (Franssen, Gu, & Bergmann, 2011; Reusch, 2014; Seneca & Palumbi, 2015).
The calanoid copepod Calanus glacialis is a key component of the Arctic marine ecosystem, comprising up to 90% of the zooplankton biomass in shelf regions (Conover & Huntley, 1991; Blachowiak‐Samolyk et al., 2008). It is an important prey for fish, seabirds, and baleen whales (Lowry, 1993; Karnovsky, Kwasniewski, Weslawski, Walkusz, & Beszczynska‐Möller, 2003; Hop & Gjøsæter, 2013). Calanus glacialis maintains its vital rates at normal levels when exposed to low pH. Organismal‐level measures such as respiration, ingestion, survival, gonad maturation, egg production, and naupliar growth and development are unaffected by low pH (Weydmann, Søreide, Kwasniewski, & Widdicombe, 2012; Hildebrandt, Niehoff, & Sartoris, 2014; Hildebrandt, Sartoris, Schulz, Riebesell, & Niehoff, 2015; Bailey, Thor, & Browman, 2016). In this study, we sought to characterize the molecular‐level response of C. glacialis nauplii to lowered pH to elucidate the physiological processes involved in this tolerance. Understanding the mechanisms of phenotypic buffering that allow some species to tolerate a wide range of pH is important for the field of ocean acidification research. Not only can it help detect low‐level stress potentially unnoticed in organismal‐level measurements, but it also contributes to understanding how some species, but not others, can tolerate low pH. We quantified whole transcriptome gene expression in the last naupliar stage (N6) of C. glacialis which had been exposed to lowered pH as they developed from eggs (over a month). We hypothesized that gene expression would respond to the pH treatment despite the lack of change in developmental rate, growth, and respiration observed during the same experiment (Bailey et al., 2016) and that these modulations would be related to the maintenance of growth and development. Furthermore, we expected that the physiological functions affected by pH, as indicated by altered expression of genes coding for functional groups of proteins, would be similar to those affected in other marine organisms at low pH and would, therefore, contribute to an understanding of a generalized molecular response to low pH.
2. METHODS
2.1. Collection and exposure
The collection and culturing of the copepods are detailed in Bailey et al. (2016). Briefly, C. glacialis were collected in Rijpfjorden, northeast Svalbard (80°27′54″N, 021°56′63″E) in January 2013 (−1 to 0°C) and held for 50 days prior to the experiment in 40 L flow‐through tanks in a cooling container at 2°C. Once females were mature, a pool of 1,900 females was used to inoculate 12 tanks (3 replicate tanks for each of four pCO2 treatments) of 40 L with eggs (7,039 ± 1,178 SE eggs per tank) in turn over 19 days. Females laid eggs in mesh‐bottomed buckets suspended in the tanks, which allowed eggs to settle into the tank but retained females. During inoculations, the tanks were filled with ambient sea water; the pH treatment began after the females were removed, with tanks reaching target pH in about 4 hr.
The eggs developed through six naupliar stages to the final naupliar stage, N6. Development was tracked every second day by collecting 30 individuals from each tank and identifying naupliar stages under a stereoscope. As stages N1 and N2 do not feed, the nauplii were fed with a mix of live algae mixture starting at stage N3 (Isochrysis sp. [CCAP 927/14], Rhodomonas baltica [NIVA 5/91], Chaetocerous muelleri [CCAP 1010/3], and Skeletonema costatum [NIVA BAC‐1]) at satiating concentrations (>200 μgC/L, Campbell, Wagner, Teegarden, Boudreau, & Durbin, 2001).
2.2. Treatment water
Projections of Arctic ocean acidification for 2,100 are ΔpH = −0.45 following the Intergovernmental Panel on Climate Change's (IPCC) RCP8.5/SRES A2 carbon emission scenario (Steinacher et al., 2009; Ciais, Sabine, & Bala, 2013), and ΔpH = ‐0.7 by 2,300 following the extended IPCC ERC8.5 scenario (Caldeira & Wickett, 2003; Collins, Knutti, & Arblaster, 2013). Accordingly, four pH treatments were chosen to simulate the current conditions at the collection site (high pH; pH 8.05; 320 μatm pCO2), ambient water at the research station (ambient; pH 7.90; 530 μatm), and projected conditions in Arctic seas in years 2100 (mid‐pH; pH 7.70; 800 μatm) and 2300 (low‐ pH; pH 7.50; 1,700 μatm).
For each treatment, pH was controlled in a 100 L mixing tank by a pH controller (Endress and Hauser, Liquiline CM 442) connected to a pH electrode (Endress and Hauser Orbisint CPS11D glass electrode) calibrated with NIST pH standards (pHNBS). The controller maintained the target pH by regulating the addition of either CO2‐enriched sea water (for the low‐ and mid‐pH treatments) or CO2‐stripped air (for the high‐pH treatment) to ambient sea water in the mixing tanks. The CO2‐enriched sea water (pH ~5.5) was created by bubbling pure CO2 continuously into ambient sea water. CO2‐stripped air was produced by forcing air through a CAS series CO2 adsorber (Twin Tower Engineering). Sea water from each of the four mixing tanks was pumped into header tanks, each of which fed three 40‐L experimental tanks. Inflow of treatment water to the experimental tanks was 10–20 L/min.
The pHNBS of each treatment was logged continuously by the pH electrode in each mixing tank. Additionally, water samples (100 mL) from each experimental tank were taken every few days (n = 10 during the experiment) for spectrophotometric measurement of total scale pH (pHT) with the pH‐sensitive indicator dye m‐cresol purple (MCP) (Dickson, Sabine, & Christian, 2007). Carbonate chemistry and nutrient concentrations were analyzed collected weekly intervals (n = 8) from one experimental tank of each treatment and preserved with a saturated mercuric chloride solution (Riebesell, Fabry, Hansson, & Gattuso, 2010). Temperature was measured daily in every tank. Methods of spectrophotometric pHT and carbonate chemistry measurements are described in more detail in Bailey et al. (2016). Carbonate chemistry parameters in the experiment are given in Table S9.
2.3. Nauplii sampling
Nauplii were collected from tanks when development had reached stage N6, 35–38 days post‐egg laying. The sample was gently sieved and transferred to a petri dish with sea water, where the nauplii were identified to stage by stereoscope. Stage N6 nauplii were transferred to a 10‐mL Falcon tube, and RNAlater (Qiagen) was added immediately to preserve the RNA from degradation; the time from collection to preservation was <10 min. Two pools of 10 individuals were collected from each tank. With three replicate tanks per pH, this resulted in six samples from each pH treatment (with five in the pH 7.95 treatment due to sample loss, 23 samples total). The samples were then stored at −80°C for 9 months until extraction.
2.4. RNA‐seq
Total RNA was extracted from each of the 23 samples using an RNeasy Mini kit (Qiagen). RNA concentration was measured with a high‐sensitivity QuBit fluorometric assay (Life Technologies), and its quality was tested with a denaturing MOPS gel electrophoresis assay. From total RNA, mRNA was isolated and fragmented, and the complementary DNA (cDNA) libraries were synthesized according to the TruSeq RNA sample preparation v2 low sample protocol (Illumina). Concentrations of cDNA were measured with the high‐sensitivity QuBit fluorometric assay. Barcoded index adapters were ligated, and samples were pooled equimolarly (5–6 samples per pool) and sequenced single‐end, with a 50 bp read length in four lanes in an Illumina HiSeq 2500 high‐throughput sequencing machine (High Output mode) at the Swedish National Genomics Infrastructure (SNP & SEQ platform) in Uppsala, Sweden. All raw data were submitted to National Center for Biotechnology Information's (NCBI) Short Read Archive (SRA, accession numbers SAMN05990738‐SAMN05990760; BioProject ID: PRJNA352656).
Sequenced reads were quality‐checked and used for de novo transcriptome assembly and quantification of gene expression following the pipeline in De Wit, Pespeni, and Ladner (2012) and the associated open‐access scripts (https://github.com/DeWitP/SFG; downloaded on 13 January 2016). In short, low‐quality sequenced bases (Phred score < 20) were removed using the Fastx‐toolkit (0.0.13), as were adapter sequences. Duplicate reads were identified and counted using MarkDuplicates (Picard tools 2.0.1).
A de novo transcriptome was compiled from 636,738,663 reads into contiguous sequences (contigs, putative transcripts) using CLC Genomics Workbench (Ver. 8.5.1; QIAGEN Aarhus A/S). The CLC De Novo Assembly tool was used with simultaneous mapping (minimum contig length = 200 bp, mismatch cost = 1, insertion cost = 2, deletion cost = 2, no global alignment, update contigs based on mapping) and default settings (length fraction = 0.5, similarity fraction = 0.8). Transcriptome assembly and annotation completeness were evaluated using Benchmarking Universal Single‐Copy Orthologs (BUSCO) analysis, which is based on evolutionarily informed expectations of gene content (ver 2.0; Simão, Waterhouse, Ioannidis, Kriventseva, & Zdobnov, 2015). Individual reads were mapped to this transcriptome using the Burrows‐Wheeler Aligner (0.7.12) with the following settings: maxDiff = 0.01, maxSeedDiff = 5, seedLen = 30, and nThrds = 2. As a proxy of gene expression, the counts of uniquely mapped reads mapping to each contig were compiled for each sample using a custom script from De Wit et al. (2012) with a mapping quality threshold of 20 and a read length threshold of 20 bp.
The proportion of duplicate reads was high. Removal of 110 contigs which matched known C. glacialis ribosomal sequences (1,006 sequences, downloaded from NCBI on 12 April 2016) reduced the number of duplicates, resulting in 45%–93% duplicate reads per sample. As duplicate reads may arise from highly expressed genes and may have biological significance, they were not removed (Parekh, Ziegenhain, Vieth, Enard, & Hellmann, 2015).
To associate physiological function to each of the contigs, the proteins for which the contigs coded were identified by searching the arthropod sequences in NCBI's nonredundant (nr) protein database and the entire UniProt protein database (downloaded on July 6 2016) (e‐value cutoff = 1 × 10−5; Blastx 2.3.0+). Hypothetical or predicted protein annotations in nr were excluded by discarding matches with the following words: “hypothetical,” “predicted,” “unknown,” and “putative.” The sequences corresponding to known proteins were annotated with biological process and molecular function Gene Ontologies (GO), a classification of physiological function spanning from very broad to specific descriptions for each protein, as well as Kyoto Encylopedia of Genes and Genomes (KEGG) Gene ID using their Uniprot ID and the Uniprot flatfile database (uniprot.org, downloaded on 13 July 2016, http://www.genome.jp/kegg). For each contig with KEGG Gene ID, the KEGG orthologs and their associated KEGG pathways were downloaded from the KEGG REST API using the R package KEGGREST (Tenenbaum, 2016). KEGG orthologs, and the KEGG pathways they are associated with, are not species specific and are thus designed for cross‐species comparisons, allowing their application in nonmodel species like C. glacialis.
2.5. Gene expression analyses
Differentially expressed contigs (DECs), those whose expression varied significantly with pH (treated as a continuous variable), were identified with the R package DESeq2 (version 1.10.1, Love, Huber, & Anders, 2014a). DESeq2 outperforms nearly all other HTS gene expression R packages in detecting significantly differently expressed genes (Seyednasrollah, Laiho, & Elo, 2013; Love, Huber, & Anders, 2014b; Schurch, Schofield, & Gierliński, 2016), taking into account common sources of error such as low sample size, low‐count genes and high variability using shrinkage estimators of dispersion and fold change (Love et al., 2014a). Contig expression is discussed here as gene expression.
To determine whether gene expression varied significantly along a continuous pH scale, DESeq2 fits generalized linear models (GLMs) to raw counts of each gene, with a logarithmic link (log2) and negative binomial distribution using a shrunken dispersion estimate (Love et al., 2014a). The GLM fit produces the overall expression strength of the gene and the log fold change (LFC) per unit of pH. LFCs are then shrunk to reduce heteroscedasticity caused by low‐count genes showing high LFCs, with low‐count, low sample size, or high‐dispersion genes shrunk the most. Ward tests with Benjamini‐Hochberg procedure are used to adjust for multiple testing in DESeq2. Significant differently expressed contigs were defined as those with |LFC| > 1 and adjusted p‐value (false discovery rate, FDR) <0.1.
To visualize the similarity of expression of the DECs across samples, a heatmap was created, with the raw contig counts transformed using the rlog function in DESeq2 and plotted using “aheatmap” in the R package NMF, which centers and scales expression of each contig (0.17.6, Gaujoux & Seoighe, 2010). DESeq2 and heatmap were run on R version 3.2.3 (R Core Team, 2015) and the graphics package ggplot2, version 2.0.0 (Wickham, 2009).
2.6. Gene set enrichment
FatiScan of the Babelomics5 suite was used to detect functional sets of contigs significantly affected by pH (Al‐Shahrour et al., 2007; Montaner & Dopazo, 2010; Alonso, Salavert, & Garcia‐Garcia, 2015). FatiScan is a gene set enrichment analysis (GSEA) tool sensu Khatri, Sirota, and Butte (2012). The first‐generation over‐representation tools searched for enriched pathways in a subset of genes selected by a threshold of fold change or p‐value. However, focusing only on a subset of high‐fold‐change genes can introduce bias (Dalman, Deeter, Nimishakavi, & Duan, 2012) and distracts from detecting genes that truly affect fitness, which often show mild fold changes (Evans, 2015). In contrast, second‐generation GSEA tools use a threshold‐free method that utilizes all genes and their fold changes. They can, therefore, more sensitively detect functional sets of genes (genes in the same GO or pathway) that behave in moderate, but coordinated, manner in response to treatment. Enrichment of GO and KEGG pathways in genes up‐ and downregulated with pH was analyzed by the FatiScan logistic model using the log fold changes of all genes, with our de novo transcriptome as a reference, and a significance cutoff (α) of FDR‐adjusted p‐value of .05. GOs with log‐odds ratio (LOR) < 0 were taken to be downregulated, and LOR > 0 upregulated.
To distill the most important functions affected by pH, the significant GOs were clustered based on their semantic similarity and GSEA p‐value (SimRel > 0.5; Schlicker, Domingues, Rahnenführer, & Lengauer, 2006) using ReViGO (Supek, Bošnjak, Škunca, & Šmuc, 2011). These results were represented in a network using the network‐plotting tool Cytoscape (version 3.4.0, Shannon, Markiel, & Ozier, 2003) with the Prefuse Force‐directed Layout based on SimRel weight between GO nodes.
3. RESULTS
The cDNA library sizes differed between treatments and were normalized by DESeq2's coefficient (size factors), which ranged from 0.26 to 3.33, with the size factors of the pH 7.95 treatment being consistently low (Fig. S1). There were 59,353 contigs identified in the de novo transcriptome, with a N50 of 1,019 bp (Supporting Information). BUSCO analysis indicated that the transcriptome was relatively complete: of the 1,066 arthropod orthologs queried, the transcriptome contained 75.4% complete orthologs (71.5% single and 3.9% duplicate copies), with 18.1% fragmented and only 6.5% missing. Of 59,353 contigs, 19,790 (33%) were associated with GO classifications, with an average of 3.54 classifications per contig.
Of these 59,353 contigs, 151 were identified as significantly differently expressed with pH between pH 7.5 and 8.05 (Figure 1). The majority of the DE contigs (140 of 151; 93%) were downregulated as pH decreased (hereafter referred to as downregulated), with 11 contigs upregulated at lower pH (Figure 1). Biological replicates (tanks) and, to a greater degree, technical replicates (different animals from the same tank) showed similar expression of many DECs, although certain tanks and technical replicates show different expression in parts of the profile (Figure 2).
Figure 1.
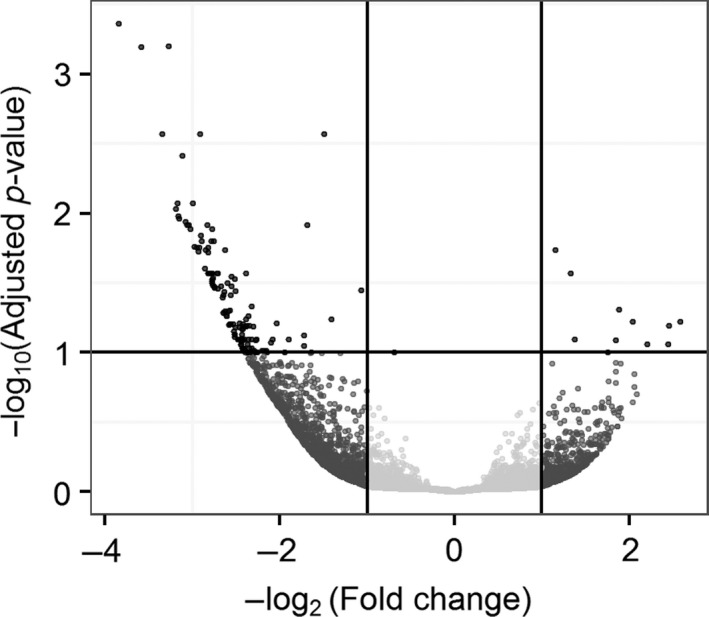
“Volcano plot” illustrating the selection of genes that were significantly differently expressed with pH in C. glacialis. Plotted are ‐log‐transformed Ward test p‐values for the slope of expression with pH, corrected for multiple tests by the Benjamini‐Hochberg method, and presented as false discovery rate (FDR), and –(log2 fold change) of expression over one unit of pH. The log2 fold change (LFC) is negative to indicate that those with a negative value are downregulated as pH is reduced. Significantly differently expressed genes (DECs, in darkest gray) were defined as those with |LFC| > 1 and FDR < 0.1
Figure 2.
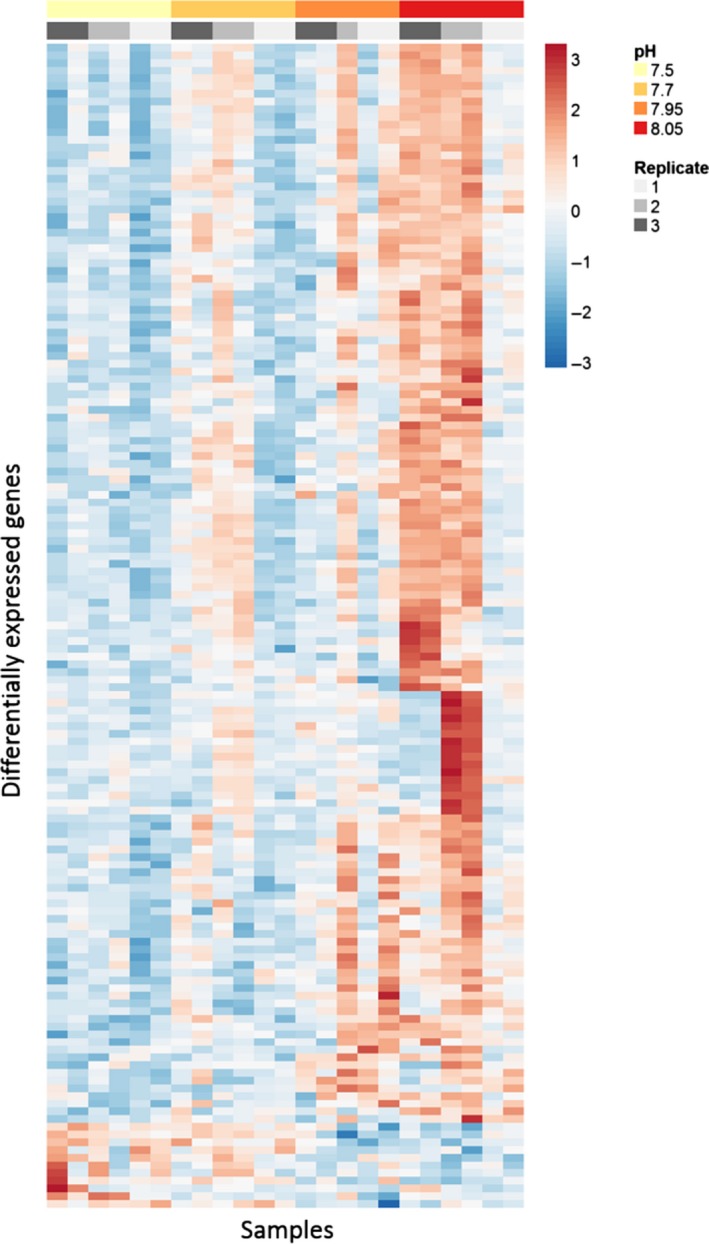
Heatmap of rlog‐transformed gene expression in C. glacialis at different pH treatments, centered, and standardized by gene for each sample (in columns). The color of the cell indicates relative gene expression, with red being higher than the gene's average, and blue lower. Only the 151 significantly differentially expressed genes are shown. The pH treatment and biological replicate (tank) are indicated in shaded red‐yellow and gray, respectively, above the heatmap. On the left vertical axis, genes are arranged according to their position in the hierarchical clustering (similarity measure was Pearson correlation coefficient; clustering by complete linkage)
Numerous cellular processes (1,001 biological process GO terms, 358 molecular function GO terms and 16 KEGG pathways) were significantly downregulated with lowered pH. Clustering these GO terms by their similarity reduced the list to 87 key biological processes, including DNA repair, cell adhesion, growth, cholesterol homeostasis, chromatin modification, development, protein glycosylation, endocytosis, mRNA splicing, regulation of transcription, regulation of protein stability, protein folding, and signaling pathways (Figure 3; Table S2). There were 184 key downregulated molecular functions, including transcription factors, serine‐type endopeptidase activity, receptor activity, oxidoreductase activity, ubiquitin‐protein transferase activity, chitinase activity, and helicase activity (Fig. S3 and Table S3). The KEGG pathways significantly downregulated at lowered pH included metabolic pathways, lysosome, cytochrome P450, amino acid metabolism, spliceosome, peroxisome, and RNA transport (Table S4).
Figure 3.

Cytoscape network of the 350 most significant downregulated biological process Gene Ontology (GO) terms in C. glacialis, linked by SimRel semantic similarity of the terms by ReViGO. The color of each GO term node (from white to dark red) indicates the adjusted p‐value, with more significant terms in darker red. Likewise, the larger the label name, the more significant the adjusted p‐value. The size of each node indicates the size of the GO term in the entire UniProt database
As fewer contigs were significantly upregulated at lowered pH, fewer GO terms (91 biological process GO terms and 39 molecular function GOs) and KEGG pathways (13) were identified as significantly upregulated. The condensed list of biological process GOs that were upregulated at lowered pH include myosin filament organization, locomotion, proteoglycan catabolic process, muscle contraction, transposition, regulation of sodium:proton antiporter, protein deamination, and keratinization (Figure 4, Table S5). Molecular function GOs upregulated at low pH included motor activity, calmodulin‐dependent protein kinase activity, and MAP kinase phosphatase activity (Fig. S4, Table S6). The upregulated KEGG pathways included tight junction, cardiac muscle contraction, pathogenic Escherichia coli infection, ribosome, AMPK signaling pathway, plant‐pathogen interaction, and regulation of actin cytoskeleton (Table S7).
Figure 4.
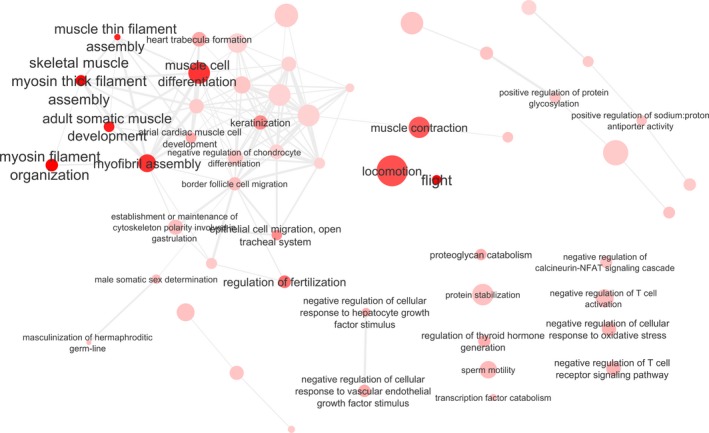
Cytoscape network of the most significant upregulated biological process Gene Ontology (GO) terms in C. glacialis, linked by SimRel semantic similarity of the terms by ReViGO. Plot components as in Figure 3
4. DISCUSSION
In a parallel study on the same experimental cultures, C. glacialis nauplii were able to develop, grow, and respire at normal rates in CO2‐acidified sea water down to pH 7.5 (Bailey et al., 2016). On the organismal level, therefore, C. glacialis nauplii were tolerant of pH levels relevant for future ocean acidification scenarios. However, in the present study, significant changes occurred at the transcriptomic level, despite the lack of an observable response on organismal‐level vital rates (Bailey et al., 2016). Altered gene expression at low pH indicated regulation of ion exchange activity, DNA transcription, molecular chaperones, and processes involved in locomotor activity in N6 nauplii. This may have allowed C. glacialis to phenotypically buffer the change in pH, thus maintaining normal nauplii development. By measuring the response to low pH at multiple levels of biological organization, both organismal and molecular, we detected physiological changes in response to low pH that were undetected in organismal measures. Additionally, there continues to be considerable inconsistency in the measured responses of marine organisms to low pH, even within taxa (Kroeker et al., 2010; Kroeker, Kordas, & Crim, 2013). Therefore, these results on the molecular response of an apparently tolerant species will improve our understanding of the physiological basis of low pH tolerance that is found in some, but not all species.
4.1. Percentage of transcriptome altered
Investigating the expression of single genes identified 151 genes as significantly differently expressed with pH between pH 7.5 and 8.05, representing ~0.25% of the genes in the transcriptome. The de novo transcriptome created in this study was of similar size to that of another C. glacialis assembly (55,562 contigs, Ramos et al., 2015) and that of Calanus sinicus (56,809, Ning, Wang, Li, & Sun, 2013) but larger than another C. glacialis transcriptome (36,880, Smolina et al., 2015). Our transcriptome had a higher N50 than the previous C. glacialis transcriptomes. Combined with a good BUSCO completeness measure, this indicates that our transcriptome is of high quality. While ~0.25% of the transcriptome significantly differentially expressed is a low percentage, it is similar to other studies using nontargeted investigations of the entire transcriptome: in a copepod in response to heat stress (0.88%; Schoville, Barreto, Moy, Wolff, & Burton, 2012), an Antarctic fish in response to pH and heat stress (0.08%–1.0%; Huth & Place, 2016), corals in response to heat stress (0.4%–0.7%; Barshis et al., 2013), and sea grass in response to heat stress (0.8%; Franssen et al., 2011). However, other studies have found greater percentages of the transcriptome affected (e.g., corals in response to low pH; 12%–19%; Moya, Huisman, & Ball, 2012; or heat shock; 27%; Seneca & Palumbi, 2015). The number of genes that have a significant impact on phenotype is very low, and the percent that modify sensitivity to abiotic stressors is even smaller (0.6%–2% in Brewer's yeast (Saccharomyces cerevisae) deletion strains) (Feder & Walser, 2005; Auesukaree et al., 2009; Evans, 2015). This indicates that even changes in a small percent of the transcriptome can have considerable physiological impact.
Acclimation to altered pH over the exposure period may have reduced the number of differentially expressed genes over time, as differential gene expression is often highest immediately after the introduction of the stressor. Huth and Place (2016) found that the degree of altered gene expression in fish exposed to pH and temperature stress decreased with time, being highest 7 days after the start of exposure and decreasing by 80%–90% at 28 and 56 days postexposure start (from 1.0% to 0.1% to 0.2%, respectively). Evans, Chan, Menge, and Hofmann (2013) found a strong reduction in the number of genes differentially expressed with time in purple sea urchin larvae (Strongylocentrotus purpuratus), from 153 at 44 hr of age to only five genes at 96 hr. The acclimation of gene expression can occur over even shorter timescales. Seneca and Palumbi (2015) found many coral genes that were differentially expressed 5 hr after a simulated heat wave but not after 20 hr. The copepods in this study were exposed for 35–38 days before sampling and therefore may have had larger differences in gene expression between high and low pH immediately after the pH treatments began. Investigating gene expression in N6 nauplii that have been chronically exposed to lowered pH throughout their lives, from the time of hatching through all naupliar stages, reflects an acclimation to low pH rather than the short‐term response. Short‐term gene expression changes are unlikely to have a long‐term effect on fitness, which is why we chose to focus on the long‐term, acclimation response, which more closely reflects how gene expression may respond to near‐term climate change with increased pCO2 in natural systems.
Further, mortality was not tracked in these experiments and is known to be high in broadcast spawned copepod larvae (Kiørboe & Sabatini, 1994). Our measurements, both on the organismal level, as discussed in Bailey et al. (2016), and on the transcriptomic level may therefore reflect the surviving, low pH‐tolerant portion of the population. However, this does not change the importance of these data for understanding the transcriptomic basis of tolerance to low pH.
4.2. Regulation of gene set expression
Investigating the coordinated expression of sets of genes revealed that a wide range of biological processes were affected by pH in C. glacialis N6 nauplii. Many of the biological processes downregulated by pH are key components of the evolutionarily conserved, universal cellular stress response (CSR; Kültz, 2003, 2005). Cells activate the CSR in response to damage and deformation of macromolecules (proteins, DNA, RNA, and membrane lipids) that occurs as a result of a broad range of stressors (Kültz, 2003). The universal stress response includes the following functions: redox regulation, DNA damage sensing and repair, molecular chaperones, protein degradation, fatty acid/lipid metabolism, and energy metabolism (Kültz, 2005). In a meta‐analysis of 14 studies of transcriptomic responses of oysters (genera Crassostrea, Ostrea, and Saccostrea) to various stressors, Anderson et al. (2015) defined 12 intracellular processes affected by stress as follows: cell cycle, communication/signaling, cytoskeleton, extracellular matrix, immunity, metabolism, nucleic acid regulation (DNA repair), protein regulation (protease, proteosome), membrane transport, stress (chaperones, detox, redox), transcription, and translation. The processes significantly downregulated in C. glacialis at lower pH include nearly all of the main functionalities of the CSR, including cell cycle regulation, metabolism, DNA repair, protein modification and degradation, redox regulation, and transcription regulation (Table 1).
Table 1.
Key Gene Ontology (GO) terms and Kyoto Encylopedia of Genes and Genomes (KEGG) pathways regulated significantly with pH in C. glacialis and associated with the universal cellular stress response, as outlined by Kültz (2005) and Anderson et al. (2015). Negative log‐odd ratio (LOR) indicates downregulation with lower pH. Number of contigs in the GO term or KEGG pathway (#) and adjusted p‐value (p) are also presented
| Kültz, 2005 | Anderson et al., 2015 | # | p | LOR | |
|---|---|---|---|---|---|
| Energy metabolism | Metabolism: lipid binding, lipid metabolism, electron transport, glycolysis, TCA cycle | ||||
| Fatty acid lipid metabolism | GO:0010906 | regulation of glucose metabolic process | 138 | 4.2E−03 | −0.40 |
| GO:0006099 | tricarboxylic acid cycle | 75 | 5.2E−03 | −0.53 | |
| GO:0042157 | lipoprotein metabolic process | 25 | 2.9E−03 | −0.99 | |
| GO:0005975 | carbohydrate metabolic process | 131 | 1.4E−07 | −0.77 | |
| GO:0042632 | cholesterol homeostasis | 63 | 8.3E−06 | −0.94 | |
| GO:0004467 | long‐chain fatty acid‐CoA ligase activity | 80 | 4.8E−03 | −0.52 | |
| GO:0045922 | negative regulation of fatty acid metabolic process | 2 | 5.2E−03 | −2.70 | |
| KEGG 1100 | metabolic pathways | 1,950 | 4.4E−35 | −0.47 | |
| DNA repair and damage sensing | Nucleic acid repair: DNA repair | ||||
| GO:0006281 | DNA repair | 268 | 9.9E−18 | −0.88 | |
| GO:0003684 | damaged DNA binding | 56 | 1.2E−02 | −0.56 | |
| GO:0006974 | cellular response to DNA damage stimulus | 222 | 3.4E−09 | −0.66 | |
| GO:0042769 | DNA damage response, detection of DNA damage | 5 | 1.2E−02 | −1.76 | |
| GO:0042770 | signal transduction in response to DNA damage | 13 | 3.3E−02 | −0.99 | |
| GO:0000724 | double‐strand break repair via homologous recombination | 67 | 7.2E−04 | −0.69 | |
| GO:0000731 | DNA synthesis involved in DNA repair | 17 | 1.5E−02 | −0.99 | |
| Protein degradation | Protein regulation: protease, proteosome | ||||
| GO:0004843 | ubiquitin‐specific protease activity | 59 | 1.1E−03 | −0.71 | |
| GO:0010499 | proteasomal ubiquitin‐independent protein catabolic process | 21 | 1.9E−05 | −1.51 | |
| GO:0006508 | Proteolysis | 260 | 5.9E−09 | −0.60 | |
| GO:0031647 | regulation of protein stability | 54 | 6.5E−04 | −0.77 | |
| GO:0010499 | proteasomal ubiquitin‐independent protein catabolic process | 21 | 1.9E−05 | −1.51 | |
| GO:0006511 | ubiquitin‐dependent protein catabolic process | 138 | 2.5E−05 | −0.59 | |
| Transmembrane gates, channels, pumps | |||||
| GO:0005231 | excitatory extracellular ligand‐gated ion channel activity | 2 | 3.0E−02 | −2.24 | |
| GO:0005245 | voltage‐gated calcium channel activity | 49 | 2.5E−02 | −0.53 | |
| GO:0005262 | calcium channel activity | 100 | 5.8E−03 | −0.45 | |
| Redox | Detox and intracellular stress: oxidative stress | ||||
| GO:0045454 | cell redox homeostasis | 87 | 2.2E−04 | −0.66 | |
| GO:1900408 | negative regulation of cellular response to oxidative stress | 1 | 5.1E−03 | 2.98 | |
| GO:1900034 | regulation of cellular response to heat | 25 | 6.1E−05 | −1.32 | |
| Molecular chaperones | Chaperones | ||||
| GO:0061077 | chaperone‐mediated protein folding | 29 | 1.2E−02 | −0.78 | |
| GO:0031072 | heat shock protein binding | 23 | 4.6E−02 | −0.69 | |
| GO:0006457 | protein folding | 172 | 1.2E−07 | −0.67 | |
| Transcription: nucleosomes | |||||
| GO:0006351 | transcription, DNA‐templated | 1,855 | 8.1E−20 | −0.35 | |
| GO:0006355 | regulation of transcription, DNA‐templated | 1,134 | 4.0E−15 | −0.38 | |
| GO:0016568 | chromatin modification | 164 | 4.5E−07 | −0.66 | |
| GO:0006337 | nucleosome disassembly | 10 | 4.9E−03 | −1.45 | |
| GO:0000398 | mRNA splicing, via spliceosome | 183 | 2.2E−13 | −0.91 | |
| GO:0010468 | regulation of gene expression | 118 | 4.0E−04 | −0.54 | |
| KEGG 3040 | spliceosome | 173 | 1.0E−13 | −0.95 | |
| Translation, post‐translational processing: ribosomes | |||||
| GO:0006486 | protein glycosylation | 203 | 6.8E−12 | −0.81 | |
| GO:0003743 | translation initiation factor activity | 67 | 1.1E−02 | −0.51 | |
| GO:0006446 | regulation of translational initiation | 36 | 2.6E−03 | −0.84 | |
| Cell cycle: differentiation, proliferation, apoptosis, death | |||||
| GO:0060548 | negative regulation of cell death | 47 | 1.9E−03 | −0.76 | |
| GO:0030154 | cell differentiation | 293 | 1.3E−03 | −0.31 | |
| GO:0051301 | cell division | 352 | 7.5E−10 | −0.54 | |
| GO:0007049 | cell cycle | 191 | 6.6E−07 | −0.60 | |
| GO:0007050 | cell cycle arrest | 71 | 4.7E−02 | −0.38 | |
| GO:0051302 | regulation of cell division | 6 | 4.1E−02 | −1.36 | |
| GO:0042127 | regulation of cell proliferation | 116 | 1.7E−05 | −0.66 | |
| GO:0045595 | regulation of cell differentiation | 25 | 1.2E−02 | −0.84 | |
| GO:0006915 | apoptotic process | 397 | 1.6E−03 | −0.26 | |
| GO:1902177 | pos. reg. of oxidative stress‐induced intrinsic apoptotic signaling | 2 | 2.1E−02 | −2.35 | |
| Communication: membrane receptors, intracellular signaling | |||||
| GO:0010649 | regulation of cell communication by electrical coupling | 4 | 1.5E−02 | −1.87 | |
| GO:0030518 | intracellular steroid hormone receptor signaling pathway | 11 | 2.5E−02 | −1.13 | |
| GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 4 | 2.5E−02 | −1.76 | |
| Cytoskeleton: vesicular trafficking | |||||
| GO:0030866 | cortical actin cytoskeleton organization | 32 | 5.8E−03 | −0.81 | |
| GO:0070062 | extracellular vesicular exosome | 1,506 | 4.8E−23 | −0.42 | |
| Extracellular matrix | |||||
| GO:0030198 | extracellular matrix organization | 103 | 5.8E−03 | −0.45 | |
| Immunity | |||||
| GO:0045087 | innate immune response | 156 | 5.1E−06 | −0.61 | |
| GO:0006955 | immune response | 107 | 1.7E−03 | −0.50 | |
| GO:0002682 | regulation of immune system process | 7 | 4.8E−03 | −1.69 | |
4.3. Downregulation of stress genes
Interestingly, these CSR‐related GO terms and KEGG pathways were downregulated in response to low pH, rather than upregulated. In a short‐term response to stress, an upregulation in CSR proteins (or a transcriptomic upregulation of genes coding for them) is expected (Tomanek & Somero, 2000; Kültz, 2003), as has been reported in numerous marine organisms (Lauritano, Procaccini, & Ianora, 2012; Tomanek, 2014; Huth & Place, 2016). However, several studies show both up‐ and downregulation of CSR‐related genes in response to a range of abiotic stressors (Anderson et al., 2015; Goncalves et al., 2016). Downregulation of stress‐related genes has been reported in the Sydney rock oyster (Saccostrea glomerata) in response to lowered pH (Goncalves et al., 2016). Downregulation of stress‐related genes when exposed to a stressor may also be a characteristic of tolerant populations. In a study comparing wild oysters to a selectively bred line that had developed tolerance to low pH, Goncalves et al. (2016) found that the line that had developed tolerance to low pH reacted to decreased pH by downregulating genes associated with stress response. In contrast, the sensitive wild population upregulated the same genes. Raising wild oysters in low pH water for only three generations produced oysters that responded to low pH stress in the same way as the selectively bred line (selected for fast growth and disease immunity over seven generations), namely downregulation of stress genes (Parker et al., 2012). Similarly, gene expression of an Antarctic fish, whose adaptation to cold waters involves constitutively elevated stress‐related genes, shows a downregulation of those genes when exposed to temperature stress (Huth & Place, 2016). Heat‐tolerant strains of Daphnia pulex showed a general downregulation of genes involved in transcription, translation, DNA replication, DNA repair, and core metabolic pathways in response to heat stress, a response not present in heat‐sensitive strains (Yampolsky et al., 2014). Downregulation of helicase, involved in DNA replication, was also involved in tolerance to low pH in Pseudocalanus acuspes copepods which had gained some tolerance to low pH after being exposed for three generations (De Wit, Dupont, & Thor, 2015). In this context, the downregulation of the majority of DECs in this study can reasonably be interpreted as an indication that C. glacialis is naturally tolerant of lowered pH, which is consistent with the organismal‐level tolerance measured on the same nauplii (Bailey et al., 2016).
4.4. General downregulation
Calanus glacialis nauplii showed an overall downregulation of transcription in the contigs whose expression was significantly related to pH. Of the 151 differentially expressed contigs, 93% were downregulated at low pH. Other studies have also found a general downregulation of proteins and genes in response to low pH stress (Todgham & Hofmann, 2009; Evans & Watson‐Wynn, 2014; Dineshram et al., 2016). In a meta‐analysis of eight papers evaluating the transcriptomic response of sea urchins to low pH, Evans and Watson‐Wynn (2014) found an overwhelming downregulation, with 80% of differentially expressed genes downregulated at low pH (Evans & Watson‐Wynn, 2014). Notably, in one paper, 90%–100% of the differentially expressed genes were downregulated (Todgham & Hofmann, 2009). This included cellular stress response genes, metabolism, and apoptosis in urchins exposed to lowered pH, both at pH 7.96 and 7.88. In larvae of the Pacific oyster (Crassostrea gigas), 70% of the differentially expressed proteins were downregulated at low pH, overshadowing the influence of temperature and salinity stressors (Dineshram et al., 2016). They suggested that while the function of general downregulation of protein synthesis and transcription at low pH is unknown, protein synthesis is both energetically costly (Rolfe & Brown, 1997; Wieser & Krumschnabel, 2001; Sokolova, Frederich, Bagwe, Lannig, & Sukhotin, 2012) and produces reactive oxygen species (ROS; Tomanek, 2014). Thus, downregulating protein synthesis at low pH may be a strategy to conserve energy and minimize oxidative damage from ROS. If the downregulation of transcription of some genes observed in C. glacialis nauplii indicates a downregulation of their protein synthesis, this energy conservation strategy may at least partially explain why we did not observe an increase in metabolic rate in the larvae during this experiment (Bailey et al., 2016).
The direction of gene regulation (up‐ or downregulation) can change with time after onset of stress, with some genes being initially upregulated and then later downregulated in response to heat stress in C. gigas (Meistertzheim, Tanguy, Moraga, Thébault, & Thebault, 2007) or pH and heat stress in the staghorn coral Acropora aspera (Ogawa, Bobeszko, Ainsworth, & Leggat, 2013). In response to stress, Daphnia pulex shows an initial upregulation and then a longer‐term downregulation of stress‐related genes (Heckmann, Sibly, & Connon, 2008). This may indicate that the downregulation we measured is a result of the long‐term stress exposure prior to sampling. An upregulation of the same stress‐related genes may have occurred earlier in the exposure. Depending on the half‐life of the proteins they code for, an early upregulation of these stress genes could have produced enough protein that the transcription of more mRNA had been negatively regulated by the time we sampled, thus explaining the general downregulation of many stress‐related genes. An exciting future avenue for research would thus be investigation of protein content in combination with transcript‐level data over a temporal sampling regime.
4.5. Upregulation
Among the processes upregulated at low pH was the positive regulation of sodium:proton antiporter activity (GO:0032417). The elevated proton concentrations in low pH sea water likely increase the proton concentrations in the extracellular fluid of C. glacialis (Pörtner, 2008; Whiteley, 2011; Melzner, Thomsen, & Koeve, 2013). Aquatic crustaceans maintain pH homeostasis primarily via ion transport (Henry & Wheatly, 1992) and the sodium:proton antiporter is key to maintaining intracellular pH (Casey, Grinstein, & Orlowski, 2010). By removing a proton in exchange for a sodium ion, sodium:proton antiporters directly regulate intracellular pH. Calanus glacialis appears to increase the transcription of genes which upregulate the activity of these antiporters in response to low pH sea water. Other organisms upregulate sodium:proton antiporters in response to low pH water (fish: Catches, Burns, Edwards, & Claiborne, 2006; Huth & Place, 2016) or other ion pumps (sodium:potassium ATPase; Pan, Applebaum, & Manahan, 2015), while corals (Moya et al., 2012; Evans et al., 2013) and urchins (Todgham & Hofmann, 2009; Stumpp, Dupont, Thorndyke, & Melzner, 2011) did not upregulate them at low pH, potentially being able to regulate pH sufficiently with the existing antiporters. Our findings indicate that sodium:proton pumps were upregulated by C. glacialis nauplii in an attempt to maintain cellular pH homeostasis at low pH, although the wide range of altered gene expression at low pH indicates that they did not succeed in fully maintaining homeostasis. Unlike active ion pumps which hydrolyze ATP to move ions across a membrane, sodium:proton antiporters passively exchange ions, a process that does not require the expenditure of metabolic energy (Aharonovitz, Demaurex, Woodside, & Grinstein, 1999). The upregulation of these antiporters, therefore, does not represent an increased energetic cost of pH homeostasis for C. glacialis, indicating that it may be an energetically sustainable component of their response to low pH.
The upregulation of sodium:proton antiporters is also of interest for general copepod physiology. Little is known about the osmoregulation of copepods, and marine copepods are considered osmoconformers (Mauchline, 1998). However, the calanoid copepod Eurytemora affinis has the capability to osmoregulate via the ion‐transport enzymes V‐type H+‐ATPase and Na+/K+‐ ATPase, an ability that allowed it to colonize freshwater from marine environments (Lee, Kiergaard, Gelembiuk, Eads, & Posavi, 2011). Our findings with C. glacialis lend additional support to the osmoregulatory capacity of marine copepods, possibly via sodium:proton antiporters.
Also upregulated were molecular functions related to locomotion, actin‐dependent ATPase activity and structural constituents of cuticle and muscle, suggesting altered locomotory muscle use. While we did not measure swimming behavior or escape response speed, larvae of other crustaceans including the American lobster (Homarus americanus) show higher swimming speeds at low pH (Waller, Wahle, Mcveigh, & Fields, 2016). Actinomyosin ATPase is a considerable component of the cellular energy budget (Rolfe & Brown, 1997). If an organism is energy‐limited, upregulation of this energy‐demanding process may lead to less energy for fitness‐related processes such as growth and development, although we did not detect declines in developmental time, body weight, or C:N ratio, or an increase in respiration in the C. glacialis nauplii with low pH (Bailey et al., 2016).
4.6. Transcriptomic expression of tolerance
Understanding the transcriptomic response of an organism to an environmental variable provides information on the extent to which the variable induces a stress response and which physiological functions are induced by that stress response, indicating whether they are components of the universal stress response or specific to that stressor or species. Altered gene expression can indicate stress but also the ability to deal with stressors: Some indicate beneficial responses and some maladaptive.
In evaluating altered gene expression in response to potential stressors, it is important to consider that not all genes have a similar ability to affect fitness. For many genes, the silencing of a single gene produces no phenotypic change at all (Feder & Walser, 2005; Evans, 2015), while other genes have a disproportionate effect on fitness because they are involved in regulatory networks and therefore have the capacity to influence many more proteins than just the one protein that they code for. Additionally, genes which are differentially expressed in response to a stress are not necessarily the same as those that confer tolerance to the same stress (de Nadal, Ammerer, & Posas, 2011; Gibney, Lu, Caudy, Hess, & Botstein, 2013; Giaever & Nislow, 2014; De Wit et al., 2015; Evans, 2015). Two types of genes that usually are not differentially expressed under stress, but whose expression have far‐reaching effects that can confer tolerance to that stress, are those involved in chromatin modification and cell signaling (Gibney et al., 2013; Jarolim, Ayer, & Pillay, 2013; Evans, 2015). Interestingly, in C. glacialis, genes controlling both chromatin modification (GO:0016568) and intracellular signaling transduction (GO:0035556) were significantly affected by pH, being downregulated in lower pH. As both chromatin and signaling pathways have the potential to regulate a large number of other genes and proteins, this indicates that large physiological changes may have been altered in C. glacialis via these transcriptional changes. Chromatin modification affects DNA replication, transcription, and repair and is a critical component of an organism's response to stress (Smith & Workman, 2012) and acclimation to their environment (van Zanten, Tessadori, Peeters, & Fransz, 2013). Changes to chromatin can also be passed on to offspring via epigenetic inheritance, thus potentially affecting the response of future generations to the environment (Campos, Stafford, & Reinberg, 2014). Indeed, epigenetic acclimation to stressors may be possible in copepods. Transgenerational effects (either adaptation or epigenetic acclimation) in the copepod Pseudocalanus acuspes alleviated the negative effect of low pH after two generations in low pH (Thor & Dupont, 2015). While the temporal duration of the chromatin modifications in C. glacialis at low pH, or their ability to be epigenetically passed on to the next generation, is unknown, they may play a part in both acclimatization within a single lifetime and over several generations.
The degree of altered gene expression during stress does not necessarily correlate with a reduction in performance or fitness. Activation of the cellular stress response immediately after the onset of the stress can help protect cells against protein unfolding, DNA damage, and oxidative damage (Kültz, 2003, 2005). Differential gene expression can therefore indicate adaptive responses and contribute to tolerance. Smolina et al. (2015) suggested that a strong transcriptomic response to stress underpins tolerance at the organismal level (i.e., respiration, feeding, mortality). They showed that C. finmarchicus strongly altered their gene expression during heat stress, while C. glacialis exhibited a near lack of response, altering the expression of only one gene transcript. As a result, C. finmarchicus showed mild changes in feeding and mortality at increased temperature, while C. glacialis was strongly affected, reducing grazing and showing higher mortality rates than C. finmarchicus under heat stress. In contrast to Smolina et al.'s (2015) findings on C. glacialis’ mild gene expression response to increased temperature, we found that C. glacialis exhibited a significant change in gene expression in response to low pH. Similar to their findings, we found that significant regulation of gene expression was associated with tolerance to stress. The gene expression regulation in C. glacialis nauplii is associated with tolerance of low pH for a range of organismal‐level vital rates (Bailey et al., 2016).
In evaluating the implication of transcriptomic regulation on tolerance, it is important to consider the time course of changes in gene expression. With the emergence of transcriptomics studies in investigating organismal responses to environmental stressors, the concept of “transcriptomic resilience” has recently been introduced (Franssen et al., 2011). They propose that the quick return to normal gene expression after exposure to a stressor is a sign that an organism can tolerate environmental changes. Seneca and Palumbi (2015) support this theory, showing that gene expression in corals from a tolerant population returned to control levels to a greater extent than in corals from a sensitive population, in response to heat stress. Those individuals whose gene expression returned to control levels exhibited less bleaching than those whose gene expression remained altered. Tracking the temporal signal of differential gene expression is therefore of utmost importance when comparing the stress tolerance of different species or the effect of different stressors. To achieve this, we suggest that future studies on gene expression responses to stressors collect samples over time, starting immediately after exposure initiation, in order to track the temporal change in gene expression.
The concentration of specific proteins is not only the result of transcription, but also the balance between mRNA degradation, translation, and protein degradation (Schwanhäusser, Busse, & Li, 2011). While stress can alter translation (Picard, Loubière, Girbal, & Cocaign‐Bousquet, 2013), mRNA degradation (Huch & Nissan, 2014), protein degradation (Flick & Kaiser, 2012), and protein activity (Krebs & Holbrook, 2001; Pan et al., 2015), transcriptomic regulation nonetheless can show a clear pattern of response to environmental stressors and, thus, indicate the involvement of genes and physiological processes in the organism's response (de Nadal et al., 2011; Evans & Hofmann, 2012; Evans & Watson‐Wynn, 2014; Anderson et al., 2015). With six measurements at each pH (five in the pH 7.95 treatment) and samples consisting of pooled individuals, we were able to conservatively select only the consistent signals in gene expression (Schurch et al., 2016). Furthermore, few papers examine differential gene expression over >2 pH treatments or use pH as a continuous variable (i.e., Barshis et al., 2013; Seneca & Palumbi, 2015; Huth & Place, 2016). By treating pH as a continuous variable (as opposed to a factor) and collecting data from four pHs, we narrowed the focus of this study to genes differentially expressed consistently over the investigated pH range, rather than those reacting to specific treatments, and reduced the chance of false positives.
Altered gene expression allowed C. glacialis to phenotypically buffer the change in pH, thereby maintaining normal nauplii development. The observed downregulation of stress‐related genes in response to stress appears to be a characteristic of stress‐tolerant populations and species. Additionally, the upregulation of energy‐neutral ion exchangers may contribute to C. glacialis’ response to low pH without adding additional cost. In predicting the effects of ocean acidification on marine organisms, understanding the molecular mechanisms that confer tolerance to some species will support our ability to predict the effects of future ocean acidification on marine organisms.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
HIB, PT, HH, AB, DMF, JR: Concept and idea. AB, PDW, PT: Study design and methods. AB, PDW, SS, CT, RB: Data gathering. AB, PDW: Data analyses and interpretation. AB: Manuscript writing. PDW, HIB, PT, HH, DMF, JR: Manuscript interpretation/comments/revisions.
Supporting information
ACKNOWLEDGMENTS
The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure. We thank NOTUR (UNINETT Sigma2) for computing time (project NN9428K) and support on the high performance computing cluster Stallo (UiT The Arctic University of Norway) and the Institute of Marine Research's marine laboratory at Austevoll, Norway, for housing and supporting the copepod cultures. We also thank Mats Töpel at University of Gothenburg for providing computing infrastructure (Albiorix). This research was supported by the Research Council of Norway (HAVKYST Project #225279 to PT, HH, and HIB), the Institute of Marine Research (Project #83192‐01 to HIB), and Fram Centre (FRAM—High North Research Centre for Climate and the Environment) funding (Project #14591‐02 to AB, HIB, HH, and PT). JR and CT were supported by NSF award OCE‐1459087. The publication charges for this article have been funded by a grant from the publication fund of UiT The Arctic University of Norway.
DATA ACCESSIBILITY
All short Illumina reads were submitted to NCBI SRA (accession numbers SAMN05990738‐SAMN05990760; BioProject ID: PRJNA352656). The transcriptome assembly is provided in the Supporting Information.
Bailey A, De Wit P, Thor P, et al. Regulation of gene expression is associated with tolerance of the Arctic copepod Calanus glacialis to CO2‐acidified sea water. Ecol Evol. 2017;7:7145–7160. https://doi.org/10.1002/ece3.3063
REFERENCES
- Aharonovitz, O. , Demaurex, N. , Woodside, M. , & Grinstein, S. (1999). ATP dependence is not an intrinsic property of Na+/H+ exchanger NHE1: requirement for an ancillary factor. American Physiological Society‐ Cell Physiology, 276, 1303–1311. [DOI] [PubMed] [Google Scholar]
- Alonso, R. , Salavert, F. , Garcia‐Garcia, F. , Carbonell‐Caballero, J. , Bleda, M. , Garcia‐Alonso, L. , ... Dopazo, J. (2015). Babelomics 5.0: Functional interpretation for new generations of genomic data. Nucleic Acids Research, 43, W117–W121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Shahrour, F. , Arbiza, L. , Dopazo, H. , Huerta‐Cepas, J. , Mínguez, P. , Montaner, D. , & Dopazo, J. (2007). From genes to functional classes in the study of biological systems. BMC Bioinformatics, 8, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, K. , Taylor, D. A. , Thompson, E. L. , Melwani, A. R. , Nair, S. V. , & Raftos, D. A. (2015). Meta‐analysis of studies using suppression subtractive hybridization and microarrays to investigate the effects of environmental stress on gene transcription in oysters. PLoS ONE, 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo, K. R. , Pabi, S. , Van Dijken, G. L. , & Maslowski, W. (2010). Air‐sea flux of CO2 in the Arctic Ocean, 1998–2003. Journal of Geophysical Research: Biogeosciences, 115, 1998–2003. [Google Scholar]
- Auesukaree, C. , Damnernsawad, A. , Kruatrachue, M. , Pokethitiyook, P. , Boonchird, C. , Kaneko, Y. , & Harashima, S. (2009). Genome‐wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae . Journal of Applied Genetics, 50, 301–310. [DOI] [PubMed] [Google Scholar]
- Bailey, A. , Thor, P. , Browman, H. I. , Fields, D. M. , Runge, J. A. , Vermont, A. , ... Hop, H. (2016). Early life stages of the Arctic copepod Calanus glacialis are unaffected by increased seawater pCO2 . ICES Journal of Marine Science. https://doi.org/10.1093/icesjms/fsw066 [Google Scholar]
- Barber, D. G. , Hop, H. , Mundy, C. J. , Else, B. , Dmitrenko, I. A. , Tremblay, J. E. , ... Rysgaard, S. (2015). Selected physical, biological and biogeochemical implications of a rapidly changing Arctic Marginal Ice Zone. Progress in Oceanography, 139, 122–150. [Google Scholar]
- Barshis, D. J. , Ladner, J. T. , Oliver, T. A. , Seneca, F. O. , Traylor‐Knowles, N. , & Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proceedings of the National Academy of Sciences of the United States of America, 110, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, N. R. , Moran, S. B. , Hansell, D. A. , & Mathis, J. T. (2006). An increasing CO2 sink in the Arctic Ocean due to sea‐ice loss. Geophysical Research Letters, 33, 1–7. [Google Scholar]
- Bellerby, R. , Anderson, L. , Azetsu‐Scott, K. , Croot, P. , Macdonald, R. , Miller, L. A. , ... Steiner, N. (2013) Acidification in the Arctic Ocean In: AMAP assessment 2013: Arctic ocean acidification, pp. 9–33. [Google Scholar]
- Blachowiak‐Samolyk, K. , Søreide, J. E. , Kwasniewski, S. , Sundfjord, A. , Hop, H. , Falk‐Petersen, S. , & Hegseth, E. N. (2008). Hydrodynamic control of mesozooplankton abundance and biomass in northern Svalbard waters (79–81°N). Deep Sea Research Part II: Topical Studies in Oceanography, 55, 2210–2224. [Google Scholar]
- Caldeira, K. , & Wickett, M. E. (2003). Oceanography: Anthropogenic carbon and ocean pH. Nature, 425, 365. [DOI] [PubMed] [Google Scholar]
- Campbell, R. G. , Wagner, M. M. , Teegarden, G. J. , Boudreau, C. A. , & Durbin, E. G. (2001). Growth and development rates of the copepod Calanus finmarchicus reared in the laboratory. Marine Ecology Progress Series, 221, 161–183. [Google Scholar]
- Campos, E. I. , Stafford, J. M. , & Reinberg, D. (2014). Epigenetic inheritance: Histone bookmarks across generations. Trends in Cell Biology, 24, 664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, J. R. , Grinstein, S. , & Orlowski, J. (2010). Sensors and regulators of intracellular pH. Nature Reviews Molecular Cell Biology, 11, 50–61. [DOI] [PubMed] [Google Scholar]
- Catches, J. S. , Burns, J. M. , Edwards, S. L. , & Claiborne, J. B. (2006). Na+/H+ antiporter, V‐H+‐ATPase and Na+/K+‐ATPase immunolocalization in a marine teleost (Myoxocephalus octodecemspinosus). The Journal of Experimental Biology, 209, 3440–3447. [DOI] [PubMed] [Google Scholar]
- Ciais, P. , Sabine, C. , Bala, G. , Bopp, L. , Brovkin, V. , Canadell, J. , ... Thornton, P. (2013). Carbon and other biogeochemical cycles In Stocker T. F., Qin D., Plattner G.‐K., Tignor M., Allen S. K., Boschung J., Nauels A., Xia Y., Bex V., & Midgley P. M. (Eds.), Climate change 2013: The physical science basis. Contribution of working group I to the fifth assessment report of the intergovernmental panel on climate change (pp. 465–570). Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press. [Google Scholar]
- Collins, M. , Knutti, R. , Arblaster, J. , Dufresne, J.‐L. , Fichefet, T. , Friedlingstein, P. , ... Wehner, M. (2013). Long‐term climate change: Projections, commitments and irreversibility In Intergovernmental Panel on Climate Change (Eds.), Climate change 2013: The physical science basis. Contribution of working group I to the fifth assessment report of the intergovernmental panel on climate change (pp. 1029–1136). Cambridge, UK: Cambridge University Press. [Google Scholar]
- Conover, R. J. , & Huntley, M. (1991). Copepods in ice‐covered seas–Distribution; adaptations to seasonally limited food; metabolism; growth patterns and life cycle strategies in polar seas. Journal of Marine Systems, 2, 1–41. [Google Scholar]
- Cripps, G. , Lindeque, P. , & Flynn, K. J. (2014). Have we been underestimating the effects of ocean acidification in zooplankton? Global Change Biology, 20, 3377–3385. [DOI] [PubMed] [Google Scholar]
- de Nadal, E. , Ammerer, G. , & Posas, F. (2011). Controlling gene expression in response to stress. Nature Reviews Genetics, 12, 833–845. [DOI] [PubMed] [Google Scholar]
- Dalman, M. R. , Deeter, A. , Nimishakavi, G. , & Duan, Z.‐H. (2012). Fold change and p‐value cutoffs significantly alter microarray interpretations. BMC Bioinformatics, 13, S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, P. , Dupont, S. , & Thor, P. (2015). Selection on oxidative phosphorylation and ribosomal structure as a multigenerational response to ocean acidification in the common copepod Pseudocalanus acuspes . Evolutionary Applications, 9, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, P. , Pespeni, M. H. , Ladner, J. T. , Barshis, D. J. , Seneca, F. , Jaris, H. , ... Palumbi, S. R. (2012). The simple fool's guide to population genomics via RNA‐Seq: An introduction to high‐throughput sequencing data analysis. Molecular Ecology Resources, 12, 1058–1067. [DOI] [PubMed] [Google Scholar]
- Dickson A. G., Sabine C. L., & Christian J. R.(eds.) (2007) Guide to best practices for ocean CO2 measurements, Vol 3. PICES Special Publication 3, 191 pp. [Google Scholar]
- Dineshram, R. , Chandramouli, K. , Ko, G. W. K. , Zhang, H. , Qian, P. Y. , Ravasi, T. , & Thiyagarajan, V. (2016). Quantitative analysis of oyster larval proteome provides new insights into the effects of multiple climate change stressors. Global Change Biology, 2, 2054–2068. [DOI] [PubMed] [Google Scholar]
- Donelson, J. M. , Munday, P. L. , McCormick, M. I. , & Pitcher, C. R. (2012). Rapid transgenerational acclimation of a tropical reef fish to climate change. Nature Climate Change, 2, 30–32. [Google Scholar]
- Evans, T. G. (2015). Considerations for the use of transcriptomics in identifying the “genes that matter” for environmental adaptation. Journal of Experimental Biology, 218, 1925–1935. [DOI] [PubMed] [Google Scholar]
- Evans, T. G. , Chan, F. , Menge, B. A. , & Hofmann, G. E. (2013). Transcriptomic responses to ocean acidification in larval sea urchins from a naturally variable pH environment. Molecular Ecology, 22, 1609–1625. [DOI] [PubMed] [Google Scholar]
- Evans, T. G. , & Hofmann, G. E. (2012). Defining the limits of physiological plasticity: How gene expression can assess and predict the consequences of ocean change. Philosophical Transactions of the Royal Society B: Biological Sciences, 367, 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, T. G. , & Watson‐Wynn, P. (2014). Effects of seawater acidification on gene expression: Resolving broader‐scale trends in sea urchins. The Biological Bulletin, 226, 237–254. [DOI] [PubMed] [Google Scholar]
- Feder, M. E. , & Walser, J. (2005). The biological limitations of transcriptomics in elucidating stress and stress responses. Journal of Evolutionary Biology, 18, 901–910. [DOI] [PubMed] [Google Scholar]
- Fitzer, S. C. , Caldwell, G. S. , Close, A. J. , Clare, A. S. , Upstill‐Goddard, R. C. , & Bentley, M. G. (2012). Ocean acidification induces multi‐generational decline in copepod naupliar production with possible conflict for reproductive resource allocation. Journal of Experimental Marine Biology and Ecology, 418–419, 30–36. [Google Scholar]
- Flick, K. , & Kaiser, P. (2012). Protein degradation and the stress response. Seminars in Cell & Developmental Biology, 23, 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen, S. U. , Gu, J. , Bergmann, N. , Winters, G. , Klostermeier, U. C. , Rosenstiel, P. , ... Reusch, T. B. H. (2011). Transcriptomic resilience to global warming in the seagrass Zostera marina, a marine foundation species. Proceedings of the National Academy of Sciences, 108, 19276–19281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux, R. , & Seoighe, C. (2010). A flexible R package for nonnegative matrix factorization. BMC Bioinformatics, 11, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G. , & Nislow, C. (2014). The yeast deletion collection: A decade of functional genomics. Genetics, 197, 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney, P. A. , Lu, C. , Caudy, A. A. , Hess, D. C. , & Botstein, D. (2013). Yeast metabolic and signaling genes are required for heat‐shock survival and have little overlap with the heat‐induced genes. Proceedings of the National Academy of Sciences of the United States of America, 110, E4393–E4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves, P. , Anderson, K. , Thompson, E. L. , Melwani, A. , Parker, L. , Ross, P. M. , & Raftos, D. A. (2016). Rapid transcriptional acclimation following transgenerational exposure of oysters to ocean acidification. Molecular Ecology, 25, 4836–4849. [DOI] [PubMed] [Google Scholar]
- Heckmann, L.‐H. , Sibly, R. M. , Connon, R. , Hooper, H. L. , Hutchinson, T. H. , Maund, S. J. , ... Callaghan, A. (2008). Systems biology meets stress ecology: Linking molecular and organismal stress responses in Daphnia magna . Genome Biology, 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, R. P. , & Wheatly, M. G. (1992). Interaction of respiration, ion regulation, and acid‐base balance in the everyday life of aquatic crustaceans. Integrative and Comparative Biology, 32, 407–416. [Google Scholar]
- Hildebrandt, N. , Niehoff, B. , & Sartoris, F. J. (2014). Long‐term effects of elevated CO2 and temperature on the Arctic calanoid copepods Calanus glacialis and C. hyperboreus . Marine Pollution Bulletin, 80, 59–70. [DOI] [PubMed] [Google Scholar]
- Hildebrandt, N. , Sartoris, F. J. , Schulz, K. G. , Riebesell, U. , & Niehoff, B. (2015). Ocean acidification does not alter grazing in the calanoid copepods Calanus finmarchicus and C. glacialis . ICES Journal of Marine Science, 73, 927–936. [Google Scholar]
- Hop, H. , & Gjøsæter, H. (2013). Polar cod (Boreogadus saida) and capelin (Mallotus villosus) as key species in marine food webs of the Arctic and the Barents Sea. Marine Biology Research, 9, 878–894. [Google Scholar]
- Huch, S. , & Nissan, T. (2014). Interrelations between translation and general mRNA degradation in yeast. Wiley Interdisciplinary Reviews: RNA, 5, 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth, T. J. , & Place, S. P. (2016). Transcriptome wide analyses reveal a sustained cellular stress response in the gill tissue of Trematomus bernacchii after acclimation to multiple stressors. BMC Genomics, 17, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarolim, S. , Ayer, A. , Pillay, B. , Gee, A. C. , Phrakaysone, A. , Perrone, G. G. , ... Dawes, I. W. (2013). Saccharomyces cerevisiae genes involved in survival of heat shock., G3: Genes|Genomes|Genetics(3), 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky, N. J. , Kwasniewski, S. , Weslawski, J.‐M. , Walkusz, W. , & Beszczynska‐Möller, A. (2003). Foraging behavior of little auks in a heterogeneous environment. Marine Ecology Progress Series, 253, 289–303. [Google Scholar]
- Khatri, P. , Sirota, M. , & Butte, A. J. (2012). Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Computational Biology, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiørboe, T. , & Sabatini, M. (1994). Reproductive and life cycle strategies in egg‐carrying cyclopoid and free‐spawning calanoid copepods. Journal of Plankton Research, 16, 1353–1366. [Google Scholar]
- Krebs, R. A. , & Holbrook, S. H. (2001). Reduced enzyme activity following Hsp70 overexpression in Drosophila melanogaster . Biochemical Genetics, 39, 73–82. [DOI] [PubMed] [Google Scholar]
- Kroeker, K. J. , Kordas, R. L. , Crim, R. , Hendriks, I. E. , Ramajo, L. , Singh, G. S. , ... Gattuso, J.‐P. (2013). Impacts of ocean acidification on marine organisms: Quantifying sensitivities and interaction with warming. Global Change Biology, 19, 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker, K. J. , Kordas, R. L. , Crim, R. N. , & Singh, G. G. (2010). Meta‐analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecology Letters, 13, 1419–1434. [DOI] [PubMed] [Google Scholar]
- Kültz, D. (2003). Evolution of the cellular stress proteome: From monophyletic origin to ubiquitous function. The Journal of Experimental Biology, 206, 3119–3124. [DOI] [PubMed] [Google Scholar]
- Kültz, D. (2005). Molecular and evolutionary basis of the cellular stress response. Annual Review of Physiology, 67, 225–257. [DOI] [PubMed] [Google Scholar]
- Kurihara, H. , & Ishimatsu, A. (2008). Effects of high CO2 seawater on the copepod (Acartia tsuensis) through all life stages and subsequent generations. Marine Pollution Bulletin, 56, 1086–1090. [DOI] [PubMed] [Google Scholar]
- Lauritano, C. , Procaccini, G. , & Ianora, A. (2012). Gene expression patterns and stress response in marine copepods. Marine Environmental Research, 76, 22–31. [DOI] [PubMed] [Google Scholar]
- Le Quéré, C. , Raupach, M. R. , Canadell, J. G. , Marland, G. , Bopp, L. , Ciais, P. , ... Woodward, F. I. (2009). Trends in the sources and sinks of carbon dioxide. Nature Geoscience, 2, 831–836. [Google Scholar]
- Lee, C. E. , Kiergaard, M. , Gelembiuk, G. W. , Eads, B. D. , & Posavi, M. (2011). Pumping ions: Rapid parallel evolution of ionic regulation following habitat invasions. Evolution, 65, 2229–2244. [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014a). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. , Huber, W. , & Anders, S. (2014b). Assessment of DESeq2 performance through simulation. DESeq2 vignette. [Google Scholar]
- Lowry, L. (1993). Foods and feeding ecology In Montague J. J., & Cowles C. J. (Eds.), The Bowhead whale, 2nd edn. (pp. 201–238). Lawrence, KS: Society of Marine Mammalogy, Allen Press Inc.. [Google Scholar]
- Mauchline, J. (1998). The biology of calanoid copepods. San Diego, USA: Academic Press. [Google Scholar]
- Mayor, D. J. , Everett, N. R. , & Cook, K. B. (2012). End of century ocean warming and acidification effects on reproductive success in a temperate marine copepod. Journal of Plankton Research, 34, 258–262. [Google Scholar]
- McConville, K. , Halsband, C. , Fileman, E. S. , Somerfield, P. J. , Findlay, H. S. , & Spicer, J. I. (2013). Effects of elevated CO2 on the reproduction of two calanoid copepods. Marine Pollution Bulletin, 73, 428–434. [DOI] [PubMed] [Google Scholar]
- Meistertzheim, A.‐L. , Tanguy, A. , Moraga, D. , Thébault, M.‐T. , & Thebault, M. T. (2007). Identification of differentially expressed genes of the Pacific oyster Crassostrea gigas exposed to prolonged thermal stress. FEBS Journal, 274, 6392–6402. [DOI] [PubMed] [Google Scholar]
- Melzner, F. , Thomsen, J. , Koeve, W. , Oschlies, A. , Gutowska, M. A. , Bange, H. W. , ... Arne, H. (2013). Future ocean acidification will be amplified by hypoxia in coastal habitats. Marine Biology, 160, 1875–1888. [Google Scholar]
- Miller, G. M. , Watson, S.‐A. , Donelson, J. M. , McCormick, M. I. , & Munday, P. L. (2012). Parental environment mediates impacts of increased carbon dioxide on a coral reef fish. Nature Climate Change. Change, 2, 858–861. [Google Scholar]
- Montaner, D. , & Dopazo, J. (2010). Multidimensional gene set analysis of genomic data. PLoS ONE, 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya, A. , Huisman, L. , Ball, E. E. , Hayward, D. C. , Grasso, L. C. , Chua, C. M. , … Miller, D. J. (2012). Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2‐driven acidification during the initiation of calcification. Molecular Ecology, 21, 2440–2454. [DOI] [PubMed] [Google Scholar]
- Ning, J. , Wang, M. , Li, C. , & Sun, S. (2013). Transcriptome sequencing and de novo analysis of the copepod Calanus sinicus using 454 GS FLX. PLoS ONE, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, D. , Bobeszko, T. , Ainsworth, T. , & Leggat, W. (2013). The combined effects of temperature and CO2 lead to altered gene expression in Acropora aspera . Coral Reefs, 32, 895–907. [Google Scholar]
- Orr, J. C. , Fabry, V. J. , Aumont, O. , Bopp, L. , Doney, S. C. , Feely, R. a. , ... Yool, A. (2005). Anthropogenic ocean acidification over the twenty‐first century and its impact on calcifying organisms. Nature, 437, 681–686. [DOI] [PubMed] [Google Scholar]
- Pan, T.‐C. F. , Applebaum, S. L. , & Manahan, D. T. (2015). Experimental ocean acidification alters the allocation of metabolic energy. Proceedings of the National Academy of Sciences, 112, 4696–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh, S. , Ziegenhain, C. , Vieth, B. , Enard, W. , & Hellmann, I. (2015). The impact of amplification on differential expression analyses by RNA‐seq. Scientific Reports, 6, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, L. M. , Ross, P. M. , O'Connor, W. A. , Borysko, L. , Raftos, D. A. , & Pörtner, H.‐O. (2012). Adult exposure influences offspring response to ocean acidification in oysters. Global Change Biology, 18, 82–92. [Google Scholar]
- Parmentier, F.‐J. W. , Christensen, T. R. , Sørensen, L. L. , Rysgaard, S. , McGuire, A. D. , Miller, P. A. , & Walker, D. A. (2013). The impact of lower sea‐ice extent on Arctic greenhouse‐gas exchange. Nature Climate Change, 3, 195–202. [Google Scholar]
- Pedersen, S. A. , Håkedal, O. J. , Salaberria, I. , Tagliati, A. , Gustavson, L. M. , Jenssen, B. M. , ... Altin, D. (2014). Multigenerational exposure to ocean acidification during food limitation reveals consequences for copepod scope for growth and vital rates. Environmental Science & Technology, 48, 12275–12284. [DOI] [PubMed] [Google Scholar]
- Pedersen, S. A. , Våge, V. T. , Olsen, A. J. , Hammer, K. M. , & Altin, D. (2014). Effects of elevated carbon dioxide (CO2) concentrations on early developmental stages of the marine copepod Calanus finmarchicus Gunnerus (Copepoda: Calanoidae). Journal of Toxicology and Environmental Health Part A, 77, 535–549. [DOI] [PubMed] [Google Scholar]
- Picard, F. , Loubière, P. , Girbal, L. , & Cocaign‐Bousquet, M. (2013). The significance of translation regulation in the stress response. BMC genomics, 14, 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pörtner, H.‐O. (2008). Ecosystem effects of ocean acidification in times of ocean warming: A physiologist's view. Marine Ecology Progress Series, 373, 203–217. [Google Scholar]
- R Core Team . (2015). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org/. [Google Scholar]
- Ramos, A. A. , Weydmann, A. , Cox, C. J. , Canário, A. V. M. , Serrão, E. A. , & Pearson, G. A. (2015). A transcriptome resource for the copepod Calanus glacialis across a range of culture temperatures. Marine Genomics, 23, 27–29. [DOI] [PubMed] [Google Scholar]
- Reusch, T. B. H. (2014). Climate change in the oceans: Evolutionary versus phenotypically plastic responses of marine animals and plants. Evolutionary Applications, 7, 104–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebesell U., Fabry V. J., Hansson L., & Gattuso J.‐P. (Eds.) (2010). Guide to best practices for ocean acidification research and data reporting. Luxembourg: Publications Office of the European Union. 260 pp. [Google Scholar]
- Rolfe, D. , & Brown, G. (1997). Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiological Reviews, 77, 731–758. [DOI] [PubMed] [Google Scholar]
- Runge, J. A. , Fields, D. M. , Thompson, C. R. S. , Shema, S. D. , Bjelland, R. M. , Durif, C. M. F. , ... Browman, H. I. (2016). End of the century CO2 concentrations do not have a negative effect on vital rates of Calanus finmarchicus, an ecologically critical planktonic species in North Atlantic ecosystems. ICES Journal of Marine Science, 73, 937–950. [Google Scholar]
- Schlicker, A. , Domingues, F. S. , Rahnenführer, J. , & Lengauer, T. (2006). A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics, 7, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoville, S. D. , Barreto, F. S. , Moy, G. W. , Wolff, A. , & Burton, R. S. (2012). Investigating the molecular basis of local adaptation to thermal stress: Population differences in gene expression across the transcriptome of the copepod Tigriopus californicus . BMC Evolutionary Biology, 12, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch, N. J. , Schofield, P. , Gierliński, M. , Cole, C. , Sherstnev, A. , Singh, V. , ... Barton, G. J. (2016). How many biological replicates are needed in an RNA‐seq experiment and which differential expression tool should you use? RNA, 22, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser, B. , Busse, D. , Li, N. , Dittmar, G. , Schuchhardt, J. , Wolf, J. , ... Selbach, M. (2011). Global quantification of mammalian gene expression control. Nature, 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Seneca, F. O. , & Palumbi, S. R. (2015). The role of transcriptome resilience in resistance of corals to bleaching. Molecular Ecology, 24, 1467–1484. [DOI] [PubMed] [Google Scholar]
- Seyednasrollah, F. , Laiho, A. , & Elo, L. L. (2013). Comparison of software packages for detecting differential expression in RNA‐seq studies. Briefings in Bioinformatics, 16, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N. S. , Wang, J. T. , Ramage, D. , ... Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão, F. A. , Waterhouse, R. M. , Ioannidis, P. , Kriventseva, E. V. , & Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single‐copy orthologs. Bioinformatics, 31, 3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith, K. T. , & Workman, J. L. (2012). Chromatin proteins: Key responders to stress. PLoS Biology, 10, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolina, I. , Kollias, S. , Møller, E. F. , Lindeque, P. , Sundaram, A. Y. M. , Fernandes, J. M. O. , & Hoarau, G. (2015). Contrasting transcriptome response to thermal stress in two key zooplankton species, Calanus finmarchicus and C. glacialis . Marine Ecology Progress Series, 534, 79–93. [Google Scholar]
- Sokolova, I. M. , Frederich, M. , Bagwe, R. , Lannig, G. , & Sukhotin, A. A. (2012). Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Marine Environmental Research, 79, 1–15. [DOI] [PubMed] [Google Scholar]
- Steinacher, M. , Joos, F. , Fr, T. L. , Frölicher, T. L. , Plattner, G.‐K. , & Doney, S. C. (2009). Imminent ocean acidification projected with the NCAR global coupled carbon cycle‐climate model. Biogeosciences Discussions, 6, 515–533. [Google Scholar]
- Stumpp, M. , Dupont, S. , Thorndyke, M. C. , & Melzner, F. (2011). CO2 induced seawater acidification impacts sea urchin larval development II: Gene expression patterns in pluteus larvae. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 160, 320–330. [DOI] [PubMed] [Google Scholar]
- Stumpp, M. , Wren, J. , Melzner, F. , Thorndyke, M. C. , & Dupont, S. (2011). CO2 induced seawater acidification impacts sea urchin larval development I: Elevated metabolic rates decrease scope for growth and induce developmental delay. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 160, 331–340. [DOI] [PubMed] [Google Scholar]
- Sunday, J. M. , Calosi, P. , Dupont, S. , Munday, P. L. , Stillman, J. H. , & Reusch, T. B. H. (2014). Evolution in an acidifying ocean. Trends in Ecology & Evolution, 29, 117–125. [DOI] [PubMed] [Google Scholar]
- Supek, F. , Bošnjak, M. , Škunca, N. , & Šmuc, T. (2011). Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum, D. (2016). KEGGREST: Client‐side REST access to KEGG. R package version, 1(12), 2. [Google Scholar]
- Thor, P. , & Dupont, S. (2015). Transgenerational effects alleviate severe fecundity loss during ocean acidification in a ubiquitous planktonic copepod. Global Change Biology, 21, 2261–2271. [DOI] [PubMed] [Google Scholar]
- Todgham, A. E. , & Hofmann, G. E. (2009). Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2‐driven seawater acidification. Journal of Experimental Biology, 212, 2579–2594. [DOI] [PubMed] [Google Scholar]
- Tomanek, L. (2014). Proteomics to study adaptations in marine organisms to environmental stress. Journal of Proteomics, 105, 92–106. [DOI] [PubMed] [Google Scholar]
- Tomanek, L. , & Somero, G. N. (2000). Time course and magnitude of synthesis of heat‐shock proteins in congeneric marine snails (Genus Tegula) from different tidal heights. Physiological and Biochemical Zoology, 73, 249–256. [DOI] [PubMed] [Google Scholar]
- van Zanten, M. , Tessadori, F. , Peeters, A. J. M. , & Fransz, P. (2013). Environment‐induced chromatin reorganisation and plant acclimation In Grafi G., & Ohad N. (Eds.), Epigenetic memory and control in plants (pp. 21–40). Berlin Heidelberg: Springer. [Google Scholar]
- Waller, J. D. , Wahle, R. A. , Mcveigh, H. , & Fields, D. M. (2016). Linking rising pCO2 and temperature to the larval development and physiology of the American lobster (Homarus americanus). ICES Journal of Marine Science, fsw154 https://doi.org/10.1093/icesjms/fsw154 [Google Scholar]
- Weydmann, A. , Søreide, J. E. , Kwasniewski, S. , & Widdicombe, S. (2012). Influence of CO2‐induced acidification on the reproduction of a key Arctic copepod Calanus glacialis . Journal of Experimental Marine Biology and Ecology, 428, 39–42. [Google Scholar]
- Whiteley, N. (2011). Physiological and ecological responses of crustaceans to ocean acidification. Marine Ecology Progress Series, 430, 257–271. [Google Scholar]
- Wickham, H. (2009). ggplot2: Elegant graphics for data analysis. New York: Springer. [Google Scholar]
- Wieser, W. , & Krumschnabel, G. (2001). Hierarchies of ATP‐consuming processes: Direct compared with indirect measurements, and comparative aspects. The Biochemical Journal, 355, 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, A. C. , & Pörtner, H.‐O. (2013). Sensitivities of extant animal taxa to ocean acidification. Nature Climate Change, 3, 995–1001. [Google Scholar]
- Yampolsky, L. Y. , Zeng, E. , Lopez, J. , Williams, P. J. , Dick, K. B. , Colbourne, J. K. , & Pfrender, M. E. (2014). Functional genomics of acclimation and adaptation in response to thermal stress in Daphnia . BMC Genomics, 15, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Li, S. , Wang, G. , & Guo, D. (2011). Impacts of CO2‐driven seawater acidification on survival, egg production rate and hatching success of four marine copepods. Acta Oceanologica Sinica, 30, 86–94. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


