Summary
The infectious metacyclic forms of Trypanosoma brucei result from a complex development in the tsetse fly vector. When they infect mammals, they cause African sleeping sickness in humans. Due to scarcity of biological material and difficulties of the tsetse fly as an experimental system, very limited information is available concerning the gene expression profile of metacyclic forms. We used an in vitro system based on expressing the RNA binding protein 6 to obtain infectious metacyclics and determined their protein and mRNA repertoires by mass-spectrometry (MS) based proteomics and mRNA sequencing (RNA-Seq) in comparison to non-infectious procyclic trypanosomes. We showed that metacyclics are quiescent cells, and propose this influences the choice of a monocistronic variant surface glycoprotein expression site. Metacyclics have a largely bloodstream-form type transcriptome, and thus are programmed to translate a bloodstream-form type proteome upon entry into the mammalian host and resumption of cell division. Genes encoding cell surface components showed the largest changes between procyclics and metacyclics, observed at both the transcript and protein levels. Genes encoding metabolic enzymes exhibited expression in metacyclics with features of both procyclic and bloodstream forms, suggesting that this intermediate-type metabolism is dictated by the availability of nutrients in the tsetse fly vector.
Keywords: Trypanosoma brucei, metacyclic form, proteome, transcriptome, quiescence, transmission
Abbreviated summary
We used an in vitro system to obtain Trypanosoma brucei metacyclics and determined their protein and mRNA repertoires by mass-spectrometry-based proteomics and mRNA sequencing in comparison to non-infectious procyclic trypanosomes. We show that metacyclics are quiescent cells, and we propose that the transition to this quiescent state is a factor in the choice of a “minimalistic” metacyclic variant surface glycoprotein (VSG) expression site, because it requires fewer cellular resources for activation.
Introduction
Trypanosoma brucei is the causative agent of African sleeping sickness in humans. Together with T. congolense and T. vivax, it is also responsible for animal African trypanosomiasis. The life cycle of T. brucei between the mammalian host and the blood-feeding tsetse fly vector (Glossina sp.) involves complex changes in cell morphology, surface coat composition, metabolism, signaling pathways and gene expression. After infection is initiated by metacyclic trypanosomes, within the bloodstream of the mammal the parasites exist as a proliferative, slender form, which establishes parasitaemia. The variant surface glycoprotein (VSG) coat expressed in this form is the paradigm for antigenic variation (Horn, 2014, Mugnier et al., 2016). In the insect midgut, T. brucei differentiates into procyclic forms (PF) that are no longer infectious to mammals. Reacquisition of infectivity is achieved through a developmental program that culminates in the tsetse salivary glands with the generation of metacyclic trypanosomes expressing on their surface a specific subset of VSGs referred to as metacyclic VSGs (mVSGs). To progress in the life cycle and hence be a pathogen, trypanosomes must complete a developmental program in the insect vector that is both adapted to the specific nutritional environment, as well as to becoming an infectious metacyclic form (MF). The most striking and best understood changes occur at the level of the parasite surface. Whereas trypanosomes residing in the midgut possess a procyclin coat (Richardson et al., 1988, Roditi et al., 1989), epimastigote forms express brucei alanine-rich proteins (BARP) on the surface (Urwyler et al., 2007). The latter coat is then replaced by VSGs in metacyclics residing in the salivary gland (Tetley et al., 1987). Little is known about additional proteins on the surface, which in bloodstreams are referred to as invariant proteins and they include the transferrin and haptoglobin-haemoglobin receptors, families of nucleotide and sugar transporters, adenylate cyclases and type I trans-membrane domain proteins, with invariant surface glycoprotein (ISG) 65 and 75 being described more than 20 years ago (Ziegelbauer & Overath, 1992, Ziegelbauer et al., 1992). Recently, it was shown that a bloodstream stage-specific invariant surface glycoprotein family (ISG75; Tb927.5.350-400) mediates suramin uptake (Alsford et al., 2012). However, these invariant proteins remain poorly characterized and it is not known what role, if any, they play during trypanosome development in the insect vector.
A second major adjustment occurs in energy metabolism due to a change in nutrient availability. In the midgut, trypanosomes rely on oxidative phosphorylation, which occurs within a highly-branched mitochondrion. A switch to glycolysis for ATP production takes place in bloodstream form (BF) trypanosomes, which are adapted for growth in the glucose-rich environment. This transformation is reflected by mitochondrial regression (Vickerman, 1985) and the expression of high-capacity glucose transporters on the surface of the parasite (Bringaud & Baltz, 1993). The development of parasites in the insect vector also highlights a number of changes in cell morphology and positioning of organelles, including a highly unusual asymmetric division of epimastigotes (Van Den Abbeele et al., 1999, Sharma et al., 2008).
Both BF and PF T. brucei can be easily cultured in vitro and sets of data are available describing their protein (Urbaniak et al., 2012, Urbaniak et al., 2013, Gunasekera et al., 2012, Butter et al., 2013, Dejung et al., 2016) and mRNA composition (Queiroz et al., 2009, Jensen et al., 2009, Siegel et al., 2010, Nilsson et al., 2010, Kolev et al., 2010, Archer et al., 2011, Capewell et al., 2013, Fadda et al., 2014), as well as the ribosome occupancy on mRNAs (Vasquez et al., 2014, Jensen et al., 2014). In contrast, due to scarcity of biological material and difficulties of the tsetse fly as an experimental system, very limited information is available concerning the gene expression profile of the infectious MF. The most comprehensive description of metacyclic T. brucei consists of detailed morphological studies of cell ultrastructure by Keith Vickerman during the 1960s (Vickerman, 1962, Vickerman & Luckins, 1969). Expression of individual genes in T. brucei metacyclics has been assayed by immunofluorescence (Le Ray et al., 1978, Barry et al., 1979, Natesan et al., 2007, Rotureau et al., 2012), which identified examples of upregulated genes, including mVSGs (Le Ray et al., 1978, Barry et al., 1979), calflagin (Rotureau et al., 2012), Rab11 and clathrin (Natesan et al., 2007).
Recently, we developed an in vitro system for generating metacyclic T. brucei trypanosomes, based on the inducible expression of the RNA-binding protein RBP6 (Kolev et al., 2012). Metacyclics generated by this method possess a VSG coat, expressed from VSG genes positioned at metacyclic expression sites and initiate trypanosome infections in mice (Kolev et al., 2012). This system for metacyclogenesis allowed us to perform an analysis of the transcriptome and the proteome of these infectious cells causing African trypanosomiasis. We show that mature MF trypanosomes have features attributed to quiescent cells, and hypothesize that the transition to this quiescent state is a factor in the choice of a “minimalistic” VSG expression site, because it requires fewer cellular resources for activation.
Results
Metacyclics are arrested in the cell cycle and have reduced protein synthesis
The DNA content for populations of different morphological stages during the life cycle of T. brucei has been previously studied by flow cytometry, indicating that metacyclic forms derived from the tsetse fly salivary gland are non-dividing and arrested in G1/G0 (Shapiro et al., 1984). For the studies reported here, we took advantage of an established in vitro differentiation system based on the inducible expression of RBP6 in the T. brucei Lister 427 (29–13) strain (Kolev et al., 2012). This system recapitulates many of the aspects of trypanosome development in the fly, including the generation of metacyclics. Since we noted that metacyclics were not dividing in culture, we determined their position in the cell cycle by scoring the number of nuclei and kinetoplasts (mitochondrial kDNA). In trypanosomes, cells with one kinetoplast and one nucleus (1K1N) are in the G1/G0 phase of the cell cycle, 2K1N cells have segregated the kinetoplast and are at the end of S phase, and 2K2N cells are post-mitotic (Woodward & Gull, 1990). In procyclic cells, as expected for an asynchronously growing cell population, the majority of parasites were 1K1N with about 20% of cells representing dividing cells having either a 2K1N or 2K2N configuration (Fig. 1A). In contrast, in purified metacyclics (≥ 99% pure) there was no evidence of dividing cells and the population quantitatively accumulated in the 1K1N configuration (Fig. 1A), representing mainly G1/G0 cells.
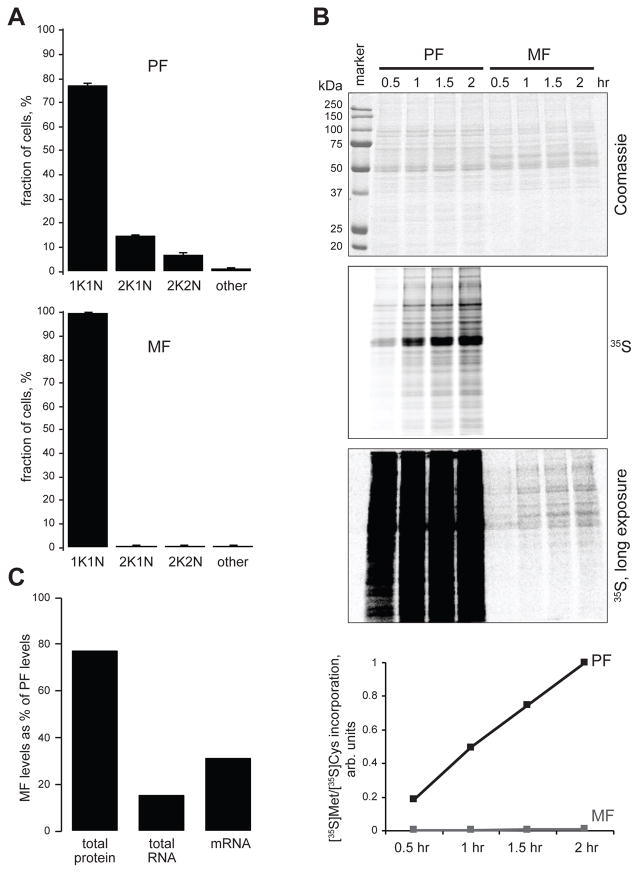
Figure 1. Nondividing metacyclic cells are quiescent.
(A) Procyclic and metacyclic cells (400 each, n=3; error bars, SEM) were scored for the presence of 1 kinetoplast and 1 nucleus (1K/1N), 2 kinetoplasts and 1 nucleus (2K/1N), 2 kinetoplasts and 2 nuclei (2K/2N) or other configurations (other). (B) Time course of [35S]Met and [35S]Cys incorporation into newly translated protein by equal number of purified metacyclic cells and procyclic cells. Coomassie staining of SDS-PAGE gel (top), short and long exposures of the dried gel to a PhosphorImager screen, and graphical representation of the quantified signals along the entire gel lanes (bottom) are shown. (C) Total protein (Bradford assay), total RNA (TRIzol extraction yield) and mRNA content (calculated with spike-in RNA-Seq control) of metacyclic cells expressed as percentage of the values in procyclic cells.
To determine whether the observed arrest in the cell cycle was accompanied by repressed mRNA translation, we compared the incorporation of 35S-labeled Met and Cys in equal numbers of purified metacyclic and procyclic cells (Fig. 1B). Metacyclics exhibited only ~1.2% of the procyclic [35S]Met/[35S]Cys incorporation into newly translated proteins. Thus, taken together our results showed that metacyclics have entered a quiescent state.
Transcriptomic and proteomic data overview
We applied Illumina high-throughput cDNA sequencing (RNA-Seq) and stable isotope labeling with amino acids in cell culture (SILAC) coupled with mass spectrometry to determine changes in the transcriptome and the proteome between PF and MF trypanosomes. For RNA-Seq, RBP6 expression was induced for 10 days and metacyclics were purified on zirconia/silica beads (Ramey-Butler et al., 2015). Messenger RNA was processed for sequencing on the Illumina platform and the resulting reads were aligned to the T. brucei 11 megabase chromosomes (release 5 of the T. brucei genome strain TREU927/4 GUTat10.1). Our analysis of the transcriptomes is based on four biological replicates of each cell type with correlation coefficients of >0.98 (Fig. S1 and Table S1).
For SILAC (Ong et al., 2002), procyclic cells were labeled with heavy stable isotope-containing lysine (Lys8) and arginine (Arg10), since PF are metabolically highly active and rapidly dividing in culture. Purified light labeled metacyclics were lysed with SDS-containing buffer, combined with the same amount of lysed heavy procyclics, and prepared for protein digestion by filter-aided sample preparation FASP (Wisniewski et al., 2009). Resulting peptides were then analyzed by shotgun proteomics without further fractionation. During the time course of the experiments, we achieved 98.9% incorporation for Arg10 and 98.6% for Lys8 in procyclic cells (Fig. S2). Proteome analysis yielded the identification of 4,760 proteins (derived from 25,645 peptides) and quantified 4,524 proteins (Fig. S3 and Table S2). The quantification of H/L ratios between biological and technical replicates reached the excellent correlation of 0.90–0.97 (Fig. S4).
Comparison of protein and transcript abundance in procyclic and metacyclic trypanosomes
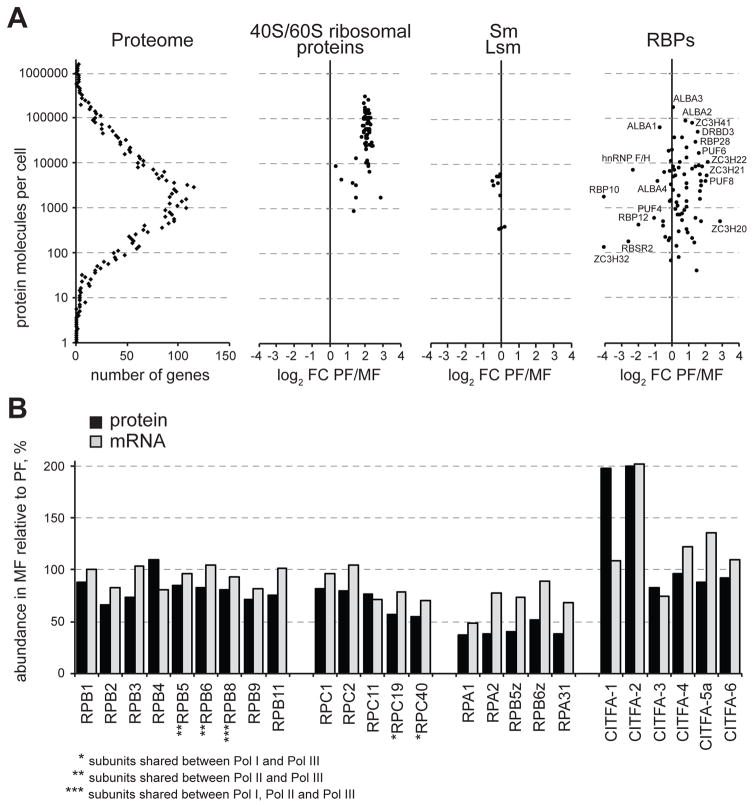
By using a spike-in control for our RNA-Seq samples, we determined that metacyclic cells possess only ~30% of the mRNA content of procyclic cells (Fig. 1C). While these cells have 77% of the protein content of procyclic cells, their total RNA amount, which is mostly rRNA, is only 15% of the value for procyclics (Fig. 1C). This low number closely matches the average abundance for 40S and 60S ribosomal proteins, determined in our SILAC-MS experiments. Metacyclics have only 16% of the cytoplasmic ribosomal proteins content of procyclics. In addition to the observations that MF are cell cycle arrested, the overall decrease of the levels of the general translational machinery in metacyclics, reflected in reduction in protein synthesis (Fig. 1B), is the hallmark of quiescent cells, shared with other single celled organisms (Marguerat et al., 2012).
We previously estimated that the median mRNA copy number per gene is three molecules per cell in procyclic trypanosomes (Kolev et al., 2010). By using the Argonaute 1 protein (Tb927.10.10850) as a reference (60,000 molecules per procyclic cell, data not shown), we estimated that the median number of expressed protein molecules per gene per cell is 1,650 in procyclics. Thus, in this stage the average protein has a 550-fold higher steady-state copy number than its corresponding mRNA. Since metacyclic cells contain only one third of the mRNA amount of procyclics, and the relative abundance distribution profile in the two cell types is very similar for both mRNAs and proteins (Fig. S5), the median abundance for mRNAs in metacyclics is around 1 molecule per cell. The median copy number for proteins in metacyclics is 1,250, thus the protein:mRNA ratio is at least twice as much as in procyclics.
Among the 200 most highly expressed mRNAs (arbitrary cut-off) in the procyclic transcriptome, the majority were coding for ribosomal proteins (106) and the abundance of each transcript is summarized in Table S3. Nevertheless, the procyclin isoforms (EP1, EP2, EP3) were by far the most abundant transcripts in the procyclic dataset. Other highly expressed mRNAs originated from genes coding for α- and β-tubulin, the procyclin isoform GPEET, five translation elongation factors, histones H1, H3 and H4, 14 transcripts encoding mitochondrial proteins and heat shock protein 70. In the metacyclic RNA-Seq sample the procyclin isoforms were replaced by the five mVSGs as highly expressed transcripts encoding surface proteins, but transcripts coding for ribosomal proteins (105/200) remained dominant.
Similar to other eukaryotes (Marguerat et al., 2012), a small proportion of the trypanosome proteome accounts for the bulk protein content of the cell in both metacyclic and procyclic T. brucei. We observed that ~15% of the different proteins detected represent ~80% of the total protein mass per cell (Fig. S6). The catalogue of the 200 most highly expressed proteins was more diverse in both procyclics and metacyclics. In procyclics 36 ribosomal proteins made the list and this number dropped to 5 in metacyclics consistent with the quiescent state of these cells. Translation factors (7) and histones (H2A, H2B, H3 and H4) were noticeable in both lists (Table S4).
Differential mRNA and protein expression between procyclic and metacyclic cells
Comparison of proteome and corresponding transcriptome changes (Fig. S7) showed a modest correlation (0.62), in agreement with previous high-throughput studies comparing bloodstream form and procyclic form trypanosomes (Urbaniak et al., 2012, Gunasekera et al., 2012, Butter et al., 2013). Protein abundance correlated better with mRNA abundance in procyclics (0.59) than in metacyclics (0.42) (Fig. S7). A comparison of metacyclic and bloodstream form (BF) proteomes and transcriptomes, using previously reported data for BF (Siegel et al., 2010, Butter et al., 2013) revealed that the transcriptomes of these two trypanosome life-cycle stages are more similar (R2 = 0.67) than their proteomes (R2 = 0.55) (Fig. S8A and S8B).
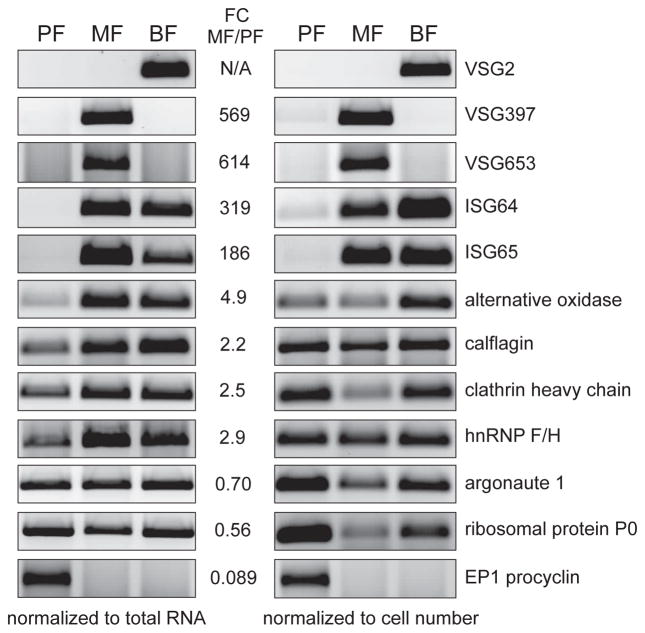
To begin to probe potential molecular differences between procyclic and metacyclic trypanosomes, we used DESeq2 (Love et al., 2014) and defined differential expression as significant when transcript abundance changed at least two-fold and with an adjusted p-value <0.05 (adjusted p-value was calculated using the Benjamini/Hochberg method). With these criteria and following filtering of pseudogenes, retrotransposon hot spot (RHS) protein genes and unlikely hypothetical proteins, 1,312 transcripts revealed significant differential expression. 703 and 609 transcripts were up- and down-regulated, respectively, in metacyclics when compared to procyclics (Tables S5 and S6). To validate a selected number of our RNA-Seq findings, we had to consider that metacyclics have only 30% of the mRNA content of procyclics. We used semi-quantitative RT-PCR and normalized the amount of cDNA template for the PCR step relative to the total RNA in the samples. Overall, the surveyed transcripts encoding VSG397, VSG653, invariant surface glycoproteins of 64 kDa and 65 kDa (ISG64, ISG65), EP1 procyclin, calflagin, hnRNP F/H, clathrin heavy chain and alternative oxidase matched our RNA-Seq data (Fig. 2).
Figure 2. Selected examples of mRNAs changing in abundance between MF and PF.
Semi-quantitative RT-PCR confirmation of RNA-Seq data using equal amount of total RNA per sample (left) or equal number of trypanosomes per sample (right). Inverted images of EtBr-stained agarose gels are shown. Listed are the RNA-Seq fold-change values obtained by the DESeq2 analysis (see Experimental Procedures).
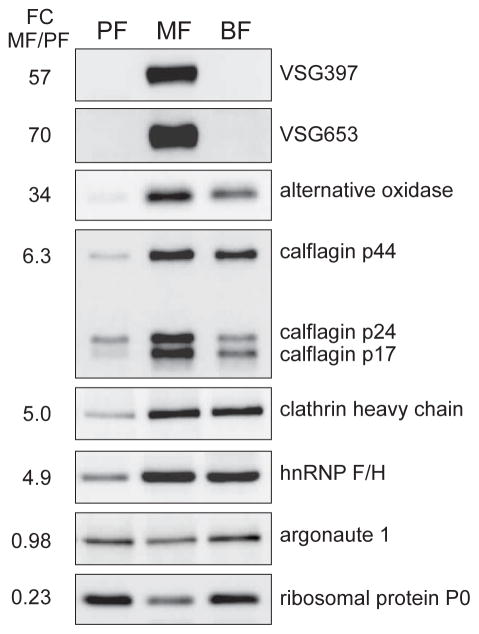
The abundance of 59 proteins (1.3%) changed dramatically (10-fold or higher, arbitrary cut-off) between metacyclics and procyclics, and the abundance of 1,281 proteins (28%) varied 2-fold or more between the two life cycle stages (Table S7). To validate our proteomic results, we performed western blots against proteins for which antibodies are available and also included in vitro cultured BF cells (strain Tb427) in our analysis (Fig. 3). Since the protein content of metacyclics is close to the one of procyclics, we used the same number of cells in our samples. Argonaute 1, which according to our proteomic data is represented almost equally in procyclics and metacyclics, did not show much variation in abundance across the tested life-cycle stages. As expected, we observed expression of two of the mVSGs (VSG397 and VSG653) only in metacyclics, but not in bloodstream trypanosomes, which express VSG2 (previous nomenclature VSG221). We detected a lower abundance of ribosomal protein P0 in metacyclics, when compared to bloodstream forms, which are dividing cells and possess amounts of P0 similar to growing procyclics. As measured by proteomics, clathrin heavy chain and hnRNP F/H were more abundant in metacyclics compared to procyclics, with levels for these proteins similar to BF abundance. Metacyclic calflagin p44 protein levels were comparable to BF and higher than procyclics. Calflagin p24 and p17 were more abundant in metacyclics relative to both procyclics and BF. The mitochondrial alternative oxidase was confirmed to be highly up-regulated in metacyclics, even in comparison to cultured bloodstream trypanosomes.
Figure 3. Examples of proteins with changing abundance in MF and PF.
Western blotting confirmation of proteomics data with equal number of cells per sample. Listed are the fold-change values from the high-throughput analysis.
Functional categorization by Gene Ontology (GO) analysis identified classes of transcripts and proteins significantly enriched in MF. Notably, both up-regulated transcripts and proteins highlighted categories related to integral component of membrane, endosome and vesicle-mediated transport (see Tables S8 and S9 for full list of GO terms). GO analysis of the differentially down-regulated transcripts and proteins revealed statistically significant biological processes, including translation, and mitochondrion as a cellular component.
In the following sections, we highlight a few of our findings and relate them to known T. brucei biology.
Our proteome and transcriptome data revealed that differences in cytoskeleton/flagellum components are in agreement with the very early observation that MF cells have a motility closely resembling that of BSF trypanosomes (Vickerman, 1985). In addition to cytoskeleton/flagellum, MF also have BF-type membrane trafficking system, required for the rapid recycling of the cell surface. One of the two highly-homologous dynamins (Tb927.3.4720) was 14-fold more abundant in MF than in PF. This protein is involved not only in endocytosis (Chanez et al., 2006), but also in mitochondrial fission (Morgan et al., 2004, Chanez et al., 2006), hinting at a potential role in the process of mitochondrial regression which is associated with differentiation to MF.
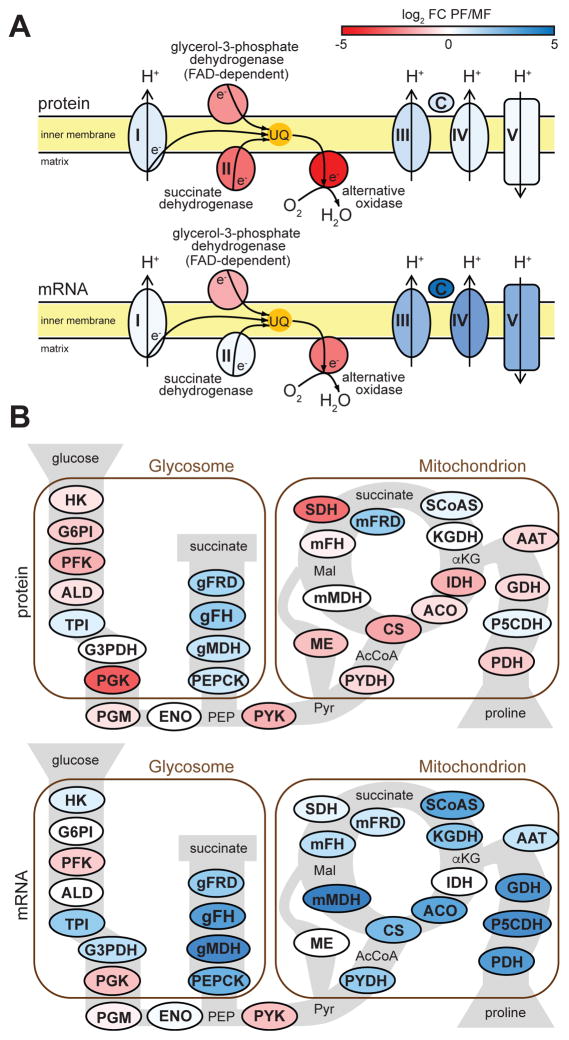
Trypanosomes in the mammalian bloodstream rely exclusively on glycolysis for meeting their energy needs and PF use the TCA cycle and oxidative phosphorylation for ATP generation (Bringaud et al., 2006). Unexpectedly, MF had similar protein levels for components of complexes I, III, IV and V of the electron transport chain to PF. However, they differed in the amounts of glycerol-3-phosphate dehydrogenase, succinate dehydrogenase and especially alternative oxidase, 5-, 7- and 34-fold higher, respectively (Fig. 4A). This indicated that electrons from both the cytoplasm and the mitochondrial matrix of MF are preferentially channeled directly to oxygen via the alternative pathway. At the mRNA level, complexes III, IV and V were strongly down-regulated in MF (Fig. 4A) and the entire electron transport chain exhibited a BF transcript pattern. Similarly, mRNAs for almost all components of the TCA cycle were down-regulated in MF (Fig. 4B), with no down-regulation of their protein counterparts. Surprisingly, the protein levels of citrate synthase, isocitrate dehydrogenase and succinate dehydrogenase were higher in MF in comparison to PF (Fig. 4B). The glycolytic pathway enzymes had elevated protein abundance, but in general, presented unaltered mRNA levels. Phosphoglycerate kinase C and pyruvate kinase, both generating ATP, were among the most up-regulated (9- and 2.5-fold, respectively) glycolytic proteins in MF, and exhibited higher abundance of their mRNAs in MF (Fig. 4B).
Figure 4. Changes in energy metabolism in metacyclics.
(A) Components of the mitochondrial respiratory chain (multisubunit complexes are represented by a single protein component in the figure): complex I, NADH-ubiquinone oxidoreductase (Tb927.5.450); glycerol-3-phosphate dehydrogenase (Tb927.11.7380); succinate dehydrogenase (Tb927.9.5960); alternative oxidase (Tb927.10.7090), complex III, rieske iron-sulfur protein (Tb927.9.14160); cytochrome c (Tb927.8.5120); complex IV, cytochrome oxidase subunit IV (Tb927.1.4100); complex V, ATP synthase alpha chain (Tb927.7.7420). (B) Glycolysis, TCA cycle and proline metabolism (multisubunit complexes are represented by a single protein component in the figure): HK, hexokinase (Tb927.10.2010); G6PI, glucose-6-phosphate isomerase (Tb927.1.3830); PFK, phosphofructokinase (Tb927.3.3270); ALD, aldolase (Tb927.10.5620); TPI, triosephosphate isomerase (Tb927.11.5520); G3PDH, glyceraldehyde 3-phosphate dehydrogenase (Tb927.6.4280); PGK, phosphoglycerate kinase (Tb927.1.700); PGM, phosphoglycerate mutase (Tb927.10.7930); ENO, enolase (Tb927.10.2890); PEPCK, phosphoenolpyruvate carboxykinase (Tb927.2.4210); gMDH, glycosomal malate dehydrogenase (Tb927.10.15410); gFH, glycosomal fumarate hydratase (Tb927.3.4500); gFRD, glycosomal fumarate reductase (Tb927.5.930); PYK, pyruvate kinase (Tb927.10.14140); PYDH, pyruvate dehydrogenase (Tb927.3.1790); CS, citrate synthase (Tb927.10.13430); ACO, aconitase (Tb927.10.14000); IDH, isocitrate dehydrogenase (Tb927.8.3690); KGDH, α-ketoglutarate dehydrogenase (Tb927.11.1450); SCoAS, succinyl-CoA synthetase (Tb927.3.2230); SDH, succinate dehydrogenase (Tb927.9.5960); mFRD, mitochondrial fumarate reductase (Tb927.5.940); mFH, mitochondrial fumarate hydratase (Tb927.11.5050); mMDH, mitochondrial malate dehydrogenase (Tb927.10.2560); ME, malic enzyme (Tb927.11.5450); PDH, proline dehydrogenase (Tb927.7.210); P5CDH, pyrroline-5 carboxylate dehydrogenase (Tb927.10.3210); GDH, glutamate dehydrogenase (Tb927.9.5900); AAT, alanine aminotransferase (Tb927.1.3950). The RNA-Seq fold-change values used here were obtained by the DNASTAR analysis (see Experimental Procedures).
RNA-binding proteins (RBPs) and RNA polymerase I transcription
The generally accepted view is that T. brucei does not appear to employ transcriptional control to regulate the abundance level of Pol II-transcribed mRNAs and adaptation to different environments. Although various post-transcriptional mechanisms could be invoked, there is increasing evidence that RNA-binding proteins (RBPs) play a central role in the regulation of gene expression (Clayton, 2013, Kolev et al., 2014). Consistent with this notion, we discovered that ten RBPs and seventeen zinc finger proteins, most with unknown functions were up-regulated in MF at the transcript level (Table S5). Similarly, at the protein level, five RBPs and five zinc finger proteins were up-regulated in metacyclics (Table S6). Most increased in protein abundance was a member of the RNA-recognition motif (RRM) family, RBP10 (Tb927.8.2780), and a member of the C3H zinc finger family, ZC3H32 (Tb927.10.5250), both up-regulated 16-fold (Fig. 5A). RBP10 mRNA was increased 5-fold in metacyclics. HnRNP F/H (Tb927.2.3880), the first pre-mRNA trans-splicing regulator shown to be differentially expressed in African trypanosomes, also has a role in the cytoplasm by affecting mRNA stability (Gupta et al., 2013) and is up-regulated in metacyclics 5-fold at the protein level (Fig. 5A). Both RBP10 and hnRNP F/H have been shown to be highly up-regulated in bloodstream form trypanosomes. Interestingly, RBP10 was shown to be able to remodel the T. brucei transcriptome from a procyclic to a more “bloodstream form pattern” (Wurst et al., 2012, De Pablos et al., 2017). Thus, its elevated expression in metacyclics could suggest a role for RBP10 in shaping the transcriptome in these infectious trypanosomes in preparation for mammalian host invasion.
Figure 5. Abundance differences for RNA-binding proteins and RNA polymerase subunits between MF and PF trypanosomes.
(A) Abundance profile and changes in expression for 40S and 60S ribosomal proteins, Sm and Lsm spliceosomal snRNP proteins, and RBPs with RRM, ALBA, Pumilio and C3H1 zinc finger domains. Shown for comparison is the abundance distribution for the entire proteome (left). (B) Abundance of RNA polymerases subunits and subunits of the class I transcription factor A (CITFA). Shown are only the components detected by both proteomics and RNA-Seq. The RNA-Seq fold-change values used here were obtained by the DNASTAR analysis (see Experimental Procedures).
Five RBPs (Tb927.2.4710, Tb927.6.4440, Tb927.8.6650, Tb927.8.710,, Tb927.11.350), which includes RBP42 previously shown to target mRNAs involved in energy metabolism (Das et al., 2012), and two putative Zn-finger proteins (Tb927.7.2670, Tb927.7.2680) were down-regulated at the transcript level. Tb927.2.4710 may have an influence on transcription as it is a chromatin modulator (Naguleswaran et al., 2015). However, at the protein level, and perhaps consistent with the contracted total RNA and mRNA content in metacyclics, fifteen RBPs and twelve Zn-finger proteins were down-regulated (Fig. 5A), including RBP42 and five members of the pumilio family of RBPs.
All of the RBPs that we detected up-regulated in metacyclics are proteins of moderate abundance (Fig. 5A), represented with few hundred to few thousand molecules per cell. These copy numbers are similar to the representation of Sm and Lsm proteins, which are components of the spliceosomal snRNPs. In contrast, we found that the most abundant RBPs in trypanosome cells, rivaling the representation of 60S and 40S ribosomal proteins, are members of the ALBA family, which showed little variation in abundance between metacyclics and procyclics. Importantly, T. brucei ALBA proteins were previously shown to localize to cytoplasmic mRNP granules upon starvation (Mani et al., 2011, Subota et al., 2011) and in Plasmodium ALBA proteins were shown to be enriched in cytoplasmic mRNP bodies, termed maternal P granules, and to stabilize translationally silent mRNAs (Mair et al., 2010).
The dramatic decrease in rRNA content in MF is coincident with nucleolar dispersal that we reported previously in these cells (Ramey-Butler et al., 2015) and is likely the result, at least partially, of decreased protein levels of RNA Pol I subunits (Fig. 5B). While Pol II and Pol III subunits showed modest change in abundance in MF compared to PF, Pol I specific subunits (RPA1, RPA2, RPA31, RPB5z, RPB6z) and subunits shared between Pol I and Pol III (RPC19, RPC40) were reduced to at least ~50% of their PF levels. This observation is relevant not only for the expression of rRNA and ribosomes, but also for the production of the VSG coat in metacyclics, since the mVSG genes are transcribed by Pol I (Alarcon et al., 1994, Graham & Barry, 1995, Ginger et al., 2002) and in contrast to rRNA, they are highly upregulated in MF. Subunits of the general class I transcription factor A (CITFA), previously shown to recognize the promoters for both rRNA and protein coding Pol I genes in T. brucei (Brandenburg et al., 2007), also exhibited changes in abundance between PF and MF. Six CITFA subunits were identified in our proteomic data and two of them, CITFA-2 and CITFA-1, were up-regulated ~2-fold in MF (Fig. 5B). Both CITFA-1 and CITFA-2 have been shown to directly contact the VSG promoter in bloodstream VSG expression sites (BESs) (Kirkham et al., 2016) and CITFA-2 associates preferentially with the active BES VSG promoter (Nguyen et al., 2014).
Remodeling of the trypanosome cell surface
One of the requirements for re-acquisition of infectivity by African trypanosomes in the salivary glands of tsetse is the biosynthesis of a VSG coat. We previously showed that the five predominantly expressed VSG genes in in vitro generated metacyclics have the features associated with metacyclic VSG expression sites, i.e. they are monocistronic and have a metacyclic type RNA Pol I VSG promoter (Kolev et al., 2012). We determined that the abundance of the five individual mVSG proteins in the metacyclic population corresponds well to their respective transcript abundances (Fig. 6). Although our methods were not designed to specifically detect protein phosphorylation, we found modification sites in VSG653, phospho-Ser at positions 394 or 393 (Fig. S9), and phospho-Thr at positions 389 and 390 (Table S2). The significance of mVSG phosphorylation is at present unclear, but the same type of modification was also detected in VSG2, expressed in cultured slender BF trypanosomes (Urbaniak et al., 2012). For reasons previously described (Urbaniak et al., 2012, Gunasekera et al., 2012, Butter et al., 2013), we could not detect EP and GPEET procyclins in our proteomics data. However, at the transcript level procyclin mRNAs were dramatically reduced in metacyclics when compared to procyclics (Fig. 2 and Table S5). It is worth noting that whereas the level of procyclic transcripts decreased, transcripts encoding procyclin-associated genes (PAGs) substantially increased in metacyclics. These genes are located not only in the same transcription units as the procyclin genes, but also elsewhere in the genome in putative Pol II transcription units. PAG genes are not essential for trypanosome transmission by the tsetse fly (Haenni et al., 2006) and their function remains an enigma.
Figure 6. Metacyclic VSG expression.
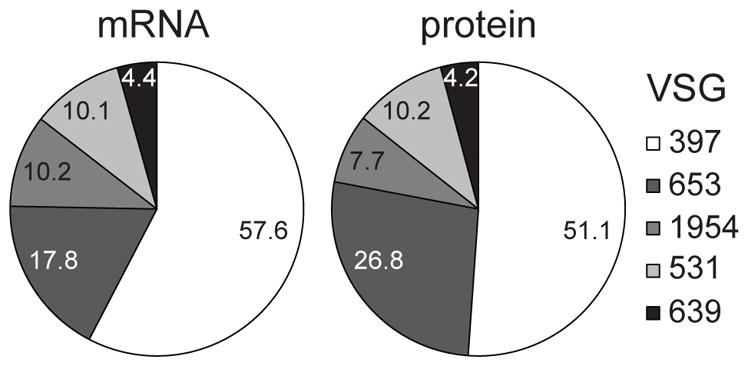
Shown are the percentages for mRNA and protein for the five T. brucei Lister427 VSG genes with a metacyclic-type Pol I promoter (set to 100%), expressed in metacyclic cells.
In addition to VSGs, metacyclics had higher levels of ISGs and almost all (see below) expression site associated genes (ESAGs), which must be expressed from locations different from the active mVSG expression sites (Graham & Barry, 1991), because these transcription units are monocistronic (Kolev et al., 2012). Metacyclics did not appear to have increased levels of the transferrin receptor ESAG6/ESAG7, which is a major difference in comparison with BF (Bitter et al., 1998), but they had highly elevated (17-fold) protein levels of the trypanosome homolog (Tb927.8.6010) of the Leishmania heme receptor LHR1 (Huynh et al., 2012). In stark contrast, the mRNA levels for Tb927.8.6010 were diminished 5-fold in metacyclics. The mRNA for haptoglobin/hemoglobin receptor (Tb927.6.440) was increased 6-fold. The bloodstream-form-specific glucose transporter (THT1, Tb927.10.8440-8480; (Bringaud & Baltz, 1992) was up-regulated at both the protein (8-fold) and mRNA level (2.5-fold) in metacyclics. Of the proteins associated with differentiation (PAD, carboxylate-transporter family), implicated as sensors of environmental stimuli and triggers of differentiation (Dean et al., 2009), PAD1, PAD6 and PAD8 (Tb927.7.5930, 5980 and 6000) were elevated in metacyclics at the protein (23-, 10- and 4-fold, respectively), whereas PAD1, PAD2, PAD3, PAD5 and PAD7 (Tb927.7.5930, 5940, 5950. 5970 and 5990) were down-regulated at the mRNA level (2-, 4-, 3.5-, 4.5- and 4-fold, respectively). PAD2 and PAD4 proteins (Tb927.7.5940 and 5960) were down-regulated in metacyclics 3- and 2-fold, respectively.
The glycosylphosphatidylinositol-specific phospholipase C (Mensa-Wilmot et al., 1990) (GPI-PLC, Tb927.2.6000) was in higher abundance (6- and 8-fold at protein and mRNA levels, respectively) in metacyclics, while the major surface protease MSP-B (Tb927.8.1640), the protease primarily responsible for the release of VSG molecules from the plasma membrane (LaCount et al., 2003), was decreased in abundance (protein 7-fold and mRNA 3-fold). We also found increased protein levels in metacyclics for other members of the MSP family, MSP-A (Tb927.11.7640, 7750, 7710; 17-, 12- and 6-fold, respectively) and MSP-C (Tb927.10.2410; 5-fold), a member of the Fam73 family of the African trypanosome cell surface phylome (Jackson et al., 2013) (Tb927.5.4580; 23-fold), a member of Fam5 (Tb927.3.600; 20-fold), a member of Fam63 (Tb927.3.3870, containing a predicted lipase domain; 17-fold) and a member of Fam10 (Tb927.7.6580; 17-fold), all examples of genes with elevated expression in BF cells relative to procyclic cells (Urbaniak et al., 2012, Gunasekera et al., 2012, Butter et al., 2013). Thus, many genes encoding predicted cell surface components had highly elevated levels of both mRNA and protein in metacyclics, making these cells similar in regard to their surface to slender BF in the mammalian host.
Does genome location contribute to gene expression levels?
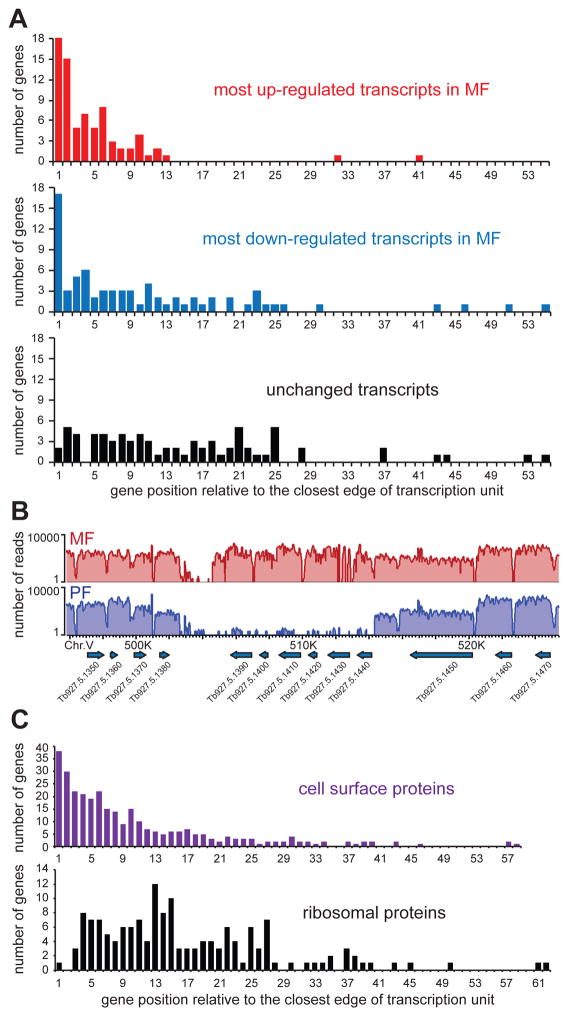
Since a) cell surface genes are overrepresented in the groups of most elevated or decreased transcripts in metacyclics (Table S5), and b) it was previously noted that some of the gene families belonging to the African trypanosome cell surface phylome are located at transcription strand switch regions of the genome (Jackson et al., 2013), we decided to analyze the position of the most differentially expressed genes relative to the edges of putative Pol II transcription units. For this analysis, we excluded potential Pol I-transcribed genes present in the cell surface phylome of T. brucei, i.e. VSGs, VSG-related, ESAGs and genes related to ESAGs (GRESAGs). We then analyzed the genes representing the top 75 (arbitrary cut-off) up-regulated (42 of them members of the cell surface phylome) and the 75 most down-regulated (19 members of the cell surface phylome) transcripts in metacyclics. Interestingly, these putative Pol II-transcribed genes were clearly enriched in locations close to the edges of transcription units (beginning or end, irrespectively; Figs. 7A, B and Fig. S10), whereas a control group of genes (40S and 60S subunits ribosomal proteins) showed no specific enrichment (Fig. 7C). The trend was also not observed for any genes whose transcripts do not change in abundance between metacyclics and procyclics (Fig. 7A), but was clearly present for genes of the African trypanosome cell surface phylome (Fig. 7C). In addition to location proximal to the edges of transcription units, there was also a modest tendency for the most differentially expressed genes to be in shorter (in absolute length) transcription units. Although we cannot speculate about the underlying biological mechanism (transcription or co-transcriptional pre-mRNA processing) or the reason necessitating the enrichment of cell surface related genes at the beginnings and ends of transcription units, this indicates that control of gene expression is easier achieved at such chromatin locations. In agreement with this notion is the number of transcripts changing their levels dramatically between life-cycle stages and positioned at such loci.
Figure 7. The beginning and end of Pol II transcription units are preferred location for genes with the largest difference in abundance between MF and PF.
(A) Histograms of the number of genes for the top 75 most upregulated (top), down-regulated (middle), or unchanged (bottom) transcripts at a specific position in a transcription unit relative to its closest edge (beginning or end, irrespectively). (B) Genome browser view of an example at the end of a transcription unit with several genes showing great differences in mRNA levels. The plots show the number of RNA-Seq reads (log2 scale) aligning to a specific genomic location. (C) Histograms of the number of genes from the African trypanosome cell surface phylome (Jackson et al., 2013) (top) and 40S and 60S ribosomal proteins (bottom) at a specific position relative to the closest edge of a transcription unit.
Discussion
The organization of genes in trypanosomatids into polycistronic transcription units necessitates control of gene expression primarily at post-transcriptional steps (pre-mRNA processing, mRNA stability, mRNA localization, translation, post-translational modifications and protein stability). Our data provide further evidence supporting the important role of post-transcriptional gene-expression regulators in controlling T. brucei development. Among factors with previously demonstrated regulatory activity are the RNA-binding proteins RBP10 and hnRNP F/H, two proteins with clear functions in establishing a bloodstream-form type transcriptome (Wurst et al., 2012, Gupta et al., 2013, De Pablos et al., 2017), which are highly up-regulated in metacyclic trypanosomes (Fig. 5A). Additionally, a recently identified repressor of differentiation kinase 2 (RDK2, Tb924.4.5310) was shown to be required for maintenance of the bloodstream form, since it’s downregulation led to differentiation of bloodstream form trypanosomes to procyclics (Jones et al., 2014). RDK2 is the protein kinase most up-regulated in metacyclic cells (18-fold higher protein levels than in procyclics), suggesting that it also has a function during the differentiation to quiescent metacyclic cells.
Focusing on the most differentially abundant mRNAs, we detected that the genes they are transcribed from are preferentially positioned at the edges (both beginning and end) of transcription units (Fig. 7A, B). This finding potentially adds another layer of complexity to gene-expression regulation. Similar observations, however not distinguishing the putative identity of the transcribing polymerase, were previously made for specific transcription units when comparing BF and procyclic mRNA levels (Veitch et al., 2010, Daniels et al., 2010). Only a subset of polycistronic units showed this trend, and at present we cannot determine what distinguishes them from the rest of the transcription units. Nevertheless, metacyclic cells have higher levels (2.3-fold) of a histone variant, H4v (Tb927.2.2670), previously shown to be enriched at the ends of transcription units and suspected to contribute to the chromatin signature marking the regions for transcription termination (Siegel et al., 2009). In addition, in vitro generated metacyclic T. brucei showed up-regulated J-binding proteins 1 and 2 (JBP1 and JBP2, Tb927.11.13640 and Tb927.7.4650), to levels similar to bloodstream form trypanosomes. J is a hypermodified nucleobase in genomic trypanosomatid DNA, which has been found only in bloodstream form, but not in procyclic T. brucei, and is enriched at subtelomeric regions of the genome that are transcriptionally silenced and at regions of divergent or convergent transcription (Cliffe et al., 2010). The interplay between base J, H4v and other variant and modified histones distribution likely holds the key to deciphering the mechanism of selective up-regulation or down-regulation of genes at the edges of transcription units. Transcripts whose levels change upon heat shock have been previously reported to be derived from genes that are preferentially located close to transcription initiation sites (for down-regulated mRNAs) or distant from such sites (for up-regulated mRNAs) (Kelly et al., 2012). It was suggested that the modulation of the expression of these genes is the direct result of temperature change on the ability of the Pol II machinery to initiate transcription. Since there is no change in ambient temperature during the transformation to metacyclics in our in vitro system for differentiation, we cannot exclude the possibility that not only transcription, but also other steps in gene expression, e.g. pre-mRNA trans-splicing and polyadenylation, are responsible for controlling genes at the edges of transcription units. However, we were unable to find specific underlying sequence signatures for pre-mRNA processing in the differentially expressed genes. Although the biological mechanism and physiological significance of these findings is yet unclear, the observations that the most-differentially expressed T. brucei mRNAs (developmentally or under stress conditions) are produced at or close to transcription unit edges suggest that tight transcriptional or co-transcriptional control of gene expression might be easier achieved at such chromatin locations in trypanosomatids.
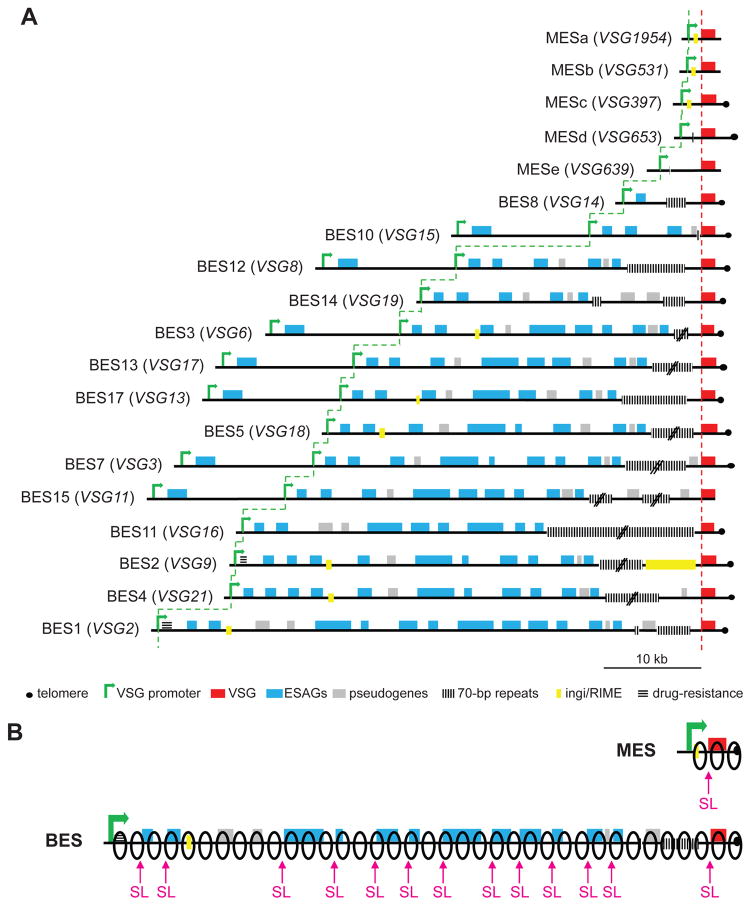
The existence of two types of VSG expression sites in T. brucei, metacyclic (MES) and bloodstream (BES), has been perplexing since their discovery (Alarcon et al., 1994, Graham & Barry, 1995, Barry et al., 1998). While the organization and composition of BESs and MESs (Fig. 8A) has been thoroughly addressed experimentally, the question about the need for the monocistronic MESs in the parasite genome has remained unanswered. Why don’t cells developing to infectious metacyclics synthesize their VSG coat de novo from the available repertoire of BESs? There are documented cases when BF trypanosomes use MESs (Alarcon et al., 1994), but the expression of BES in a developing metacyclic cell has not been observed. Our finding that metacyclic trypanosomes are quiescent provides a possible explanation. Previous experiments in BF cells have demonstrated that VSGs cannot be expressed from two different expression sites at the same high level without a penalty on cellular fitness (Chaves et al., 1999, Batram et al., 2014). Additionally, BF cells developing to quiescent stumpy forms (Vassella et al., 1997, Capewell et al., 2013, Rico et al., 2013) have the VSG coat present on their surface, in contrast to MF cells that synthesize the VSG coat de novo during their transition to quiescence. Since a) both MESs and BESs are Pol I gene expression units that produce about 10% of the mRNA and protein in the cell, b) compete for binding to the same general transcription initiation factor CITFA (Kolev et al., 2017) and c) contain very similar VSG pre-mRNA trans-splicing and polyadenylation signals, the choice of MESs over BESs in these cells transitioning to quiescence indicates that not only they cannot support the expression of more than one VSG expression site, but they also cannot support the expression of a single 40–60 kb long BES. A typical MES will require only ~1/10 of ribonucleoside triphosphates and Pol I molecules for transcription compared to a typical BES (Fig. 8B), and it contains on average 10-times fewer trans-splice sites that consume SL RNA for processing. In the light of a proposed “winner-takes-all” model for monoallelic VSG expression (Glover et al., 2016), in cells transitioning to quiescence, a “minimalistic” MES wins the competition with only one tenth (relative to a BES) of the diminished resources in these cells with limited biosynthetic capacity. Our hypothesis correlates well with data for monoallelic expression in other organisms. Notably, the var genes in Plasmodium falciparum and the olfactory receptor genes in mammals are other examples of “high-demand” monoallelic expression units that produce highly abundant mRNAs and proteins. Competition for transcriptional and splicing machineries appears to influence monoallelic var gene expression is revealed by the critical roles of the var promoter and intron (Deitsch et al., 2001, Voss et al., 2006). It is increasingly evident that epigenetic marks have a critical role in the maintenance of active and silent states in monoallelic expression, but the accumulated evidence over a large number of years suggests that competition for general gene expression factors (i.e. transcriptional apparatus, splicing machinery, ribosomes and translation factors) might be a contributing force in limiting the expression of these highly abundant gene products to a single allele/gene. While there are numerous examples of partial de-repression of “silent” members of monoallelically expressed multigene families, most notably by alteration of the levels of chromatin modifiers, there are no examples that more than one gene containing the same features can be expressed simultaneously and at the same level (absolute mRNA and protein amounts) as a singular expression. Thus, it should be considered that the “winner-takes-all” model likely involves an additional level of competition for gene expression factors not only between the many members of a single gene family, but also between the “active” member of the family and all remaining genes in the genome.
Figure 8.
(A) Diagram of the known expression sites in T. brucei Lister 427 (according to (Hertz-Fowler et al., 2008, Cross et al., 2014, Kolev et al., 2012). Note that the distance from the closest promoter to the VSG start codon can range from 0.84 kb (top) to 56.6 kb (bottom). (B) Depiction of the difference in the number of Pol I complexes (black outline ovals) and spliced leader (SL) RNA molecules required to transcribe and process a primary transcript from a ‘minimalistic’ MES and a ‘full-length’ BES, assuming similar rates of VSG mRNA production.
Experimental Procedures
T. brucei cell culture, production and purification of MF cells, SILAC
T. brucei Lister 427(29-13) strain, carrying an inducibly-expressed RBP6 (Tb927.3.2930) transgene (Kolev et al., 2012) was cultured at 28°C and 5% CO2 in Cunningham’s medium supplemented with 10% Tet-system-approved heat-inactivated FBS and 2 mM L-Gln. RBP6 expression was induced for 10 days with 10 μg ml−1 of doxycycline and cultures were diluted daily with medium containing doxycycline to 5 × 106 cells ml−1. Metacyclics were purified at room temperature on 0.1 mm diameter zirconia/silica beads columns prepared in Pasteur pipettes in BBSG buffer (50 mM bicine, 50 mM NaCl, 5 mM KCl, 70 mM glucose, pH 8.0) (Ramey-Butler et al., 2015). Purity of the metacyclic cells was determined by the morphology of live cells, combined with DNA staining and assessing the position of the kinetoplast DNA. Metacyclics were allowed to recover for 2 hours at 28°C and 5% CO2 in filtered medium from the original culture, prior to harvesting.
For SILAC, Cunningham’s medium was prepared without Arg and Lys, and supplemented with 2 mM L-Gln and 10% dialyzed FBS. MF cells were purified from cultures grown in medium supplemented with 40 mg l−1 light L-Arg (Sigma) and 40 mg l−1 light L-Lys (Sigma). PF cells were grown in parallel for 10 days in medium without doxyxycline and supplemented with 40 mg l−1 heavy (13C6/15N4) L-Arg (Cambridge Isotope Labs) and 40 mg l−1 heavy (13C6/15N2) L-Lys (Cambridge Isotope Labs).
35S metabolic labeling of proteins
PF and purified MF cells (4 × 107 each) were collected and resuspended in 4 ml Cunningham’s medium prepared without Met and Cys and supplemented with 2 mM L-Gln and 10% dialyzed FBS. Each cell sample was aliquoted into four 1.5 ml Eppendorf tubes and incubated at room temperature for 30 min. Cells were spun down and resuspended in 1 ml of the minus Met/Cys, dialyzed serum medium. To each tube, 5 μl (~50 μCi) of protein labeling mix containing both [35S]Met and [35S]Cys (PerkinElmer) were added and samples were incubated at 28°C for the indicated time. Cells were washed twice with PBSG (PBS containing 0.7 g l−1 glucose), lysed with 200 μl of SDS-PAGE loading buffer, and 20 μl were separated on 10% SDS-PAGE. The gel was stained with Coomassie, destained, dried, and exposed for analysis with Typhoon FLA 7000 PhosphorImager (GE Healthcare Life Sciences).
RNA preparation, RNA-Seq, read processing and data analysis
Total RNA was prepared from 2 × 107 PF, or purified MF cells. Biological replicates represent experiments performed at least two weeks apart. As a spike-in RNA control, the equivalent of 1/1000th of the analyzed PF or MF cells, i.e. 2 × 104 BF Lister 427 (single marker) cells in 10 μl of TRIzol reagent, were added to the PF and MF samples lysed with 1 ml of TRIzol reagent. The VSG2 mRNA is not expressed in our procyclic or metacyclic cells, and is present in sufficient amount in the small number of added BF cells to serve as the spike-in control. The spike-in control was used only to estimate the relative mRNA content of PF and MF cells by comparing the number of VSG2 mRNA reads to the total number of all mRNA reads in the corresponding PF and MF samples. The RNA was prepared with the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Four independent samples, i.e. biological replicas, were prepared from PF and MF cells and isolation of poly(A)+ mRNA, library preparation and sequencing on an Illumina HiSeq4000 platform were performed at the Yale Center for Genome Analysis. The resulting reads of 75 nt in length were mapped to the T. brucei 11 megabase chromosomes (GeneDB version 5) using the Lasergene 13.1 software package from DNASTAR as described (Savage et al., 2016). Briefly, the SeqMan NGen layout algorithm by DNASTAR is based on a local match percentage and the match percentage threshold has to be met in each overlapping window of 50 bases. We used a minimum aligned length of 35 nt and allowed a maximum of two mismatches. Reads with multiple matches in the genome were distributed randomly between the identical sequences, and the genes were counted as separate. For normalization, we used RPKM (reads assigned per kilobase of target per million mapped reads), where the signal values for each experiment are divided by the total bases of target sequence divided by one thousand; and the resulting number divided by the total number of mapped reads divided by one million. The RPKM expression data were further tested by Pearson correlation analysis, and all correlation co-efficiencies between biological pairs were >0.98 (Figure S1). Only predicted open reading frames were used to calculate RPKM values. The RPKM values and the total raw counts are listed in Table S1.
Two different statistical methods were used for differential gene expression analysis: DESeq2 package version 1.16.1 (Love et al., 2014) and the Lasergene 13.1 software package from DNASTAR (Savage et al., 2016) with an excellent correlation of 0.97. For DESeq2 the quantified transcript read files were imported into Bioconductor (version 3.5), an open-source software project based on the R programming language (Gentleman et al., 2004). Adjusted p-value (false discovery rate, FDR) was calculated using the Benjamini/Hochberg method for multi-comparison (Love et al., 2014). DEGs were filtered for fold-change >2 and adjusted p-value < 0.05. Results of the DESeq2 analysis are shown in Table S5 (DESeq2 DEG All data), Table S6 (DESeq2 DEG 2-fold changes), and Table S8 (DESeq2 GO Analysis).
The Lasergene software package from DNASTAR uses the moderated t-Test to compare the means of gene expression values for two individual replicates or two groups of replicates for a given gene, and p-values were adjusted with the FDR (Benjamini/Hochberg) method (Love et al., 2014). Results of the DNASTAR analysis are shown in Table S10 (DNASTAR RNA-Seq All data) and Table S11 (DNASTAR RNA-Seq DEG 2-fold changes).
Differentially expressed genes and proteins were analyzed for functional annotation using the Gene Ontology (GO) enrichment tool on the TriTrypDB webserver (http://tritrypdb.org/). GO terms were further submitted to REVIGO, a web server that summarizes and condenses long lists of GO terms by removing redundant entries (Supek et al., 2011).
Semi-quantitative RT-PCR was performed to confirm RNA-Seq results for selected mRNAs with SuperScript II reverse transcriptase (Invitrogen) and Platinum Pfx DNA polymerase (Invitrogen) following manufacturer’s instructions. First strand cDNA synthesis was performed with random hexamers and the sequences of the PCR primers for the individual transcripts analyzed are available upon request.
MS sample preparation, MS measurement and data analysis
Heavy PF and light MF, from two independent biological replicates (2 × 107 and 6 × 107 cells, respectively) were washed three times with PBSG and lysed in lysis buffer (50 mM Tris-HCl, pH 9.0, 5% SDS, and 100 mM DTT) for 30 min at 55°C. Lysates were cleared via centrifugation at 17,000 × g for 10 min. Protein concentrations were measured using Bradford assay and heavy and light cell lysate were mixed to a 1:1 (H:L) ratios. Combined supernatants were diluted with buffer UA (8 M urea, 0.1 M Tris-HCl, pH 8.5) to a final concentration of 0.5% SDS. Proteins were digested with LysC and trypsin following the protocol for filter aided sample preparation (Wisniewski et al., 2009). Recovered peptides were acidified by trifluoroacetic acid and spun to remove precipitates. 1.5 μg of peptides were injected into the high-performance liquid chromatograph. The first biological replicate was analyzed with 3 technical replicates and the second biological replicate was analyzed with 8 technical replicates.
Reversed phase chromatography was performed on a Thermo Easy nLC 1000 system connected to a Q Exactive mass spectrometer (Thermo) through a nanoelectrospray ion source. Peptides were separated on 50-cm columns (New Objective) with an inner diameter of 75 μm packed in house with 1.9 μm C18 resin (Dr. Maisch GmbH). Peptides were eluted from 50-cm columns with a linear gradient of acetonitrile from 5–30% in 0.1% formic acid for 240 min at a constant flow rate of 250 nl min−1. The column temperature was kept at 40°C by an oven (Sonation GmbH, Germany) with a Peltier element. Eluted peptides from the column were directly electrosprayed into the mass spectrometer. Mass spectra were acquired on the Q Exactive in a data dependent-mode to automatically switch between full scan MS and up to 10 data dependent MS/MS scans. The maximum injection time for full scans was 20 ms with a target value of 3,000,000 at a resolution of 70,000 at m/z=200. The 10 most intense multiple charged ions (z≥2) from the survey scan were selected with an isolation width of 3Th and fragment with higher energy collision dissociation (HCD) with normalized collision energies of 25. Target values for MS/MS were set to 100,000 with a maximum injection time of 120 ms at a resolution of 17,500 at m/z=200. To avoid repetitive sequencing, the dynamic exclusion of sequenced peptides was set to 45 sec.
The resulting MS and MS/MS spectra from all biological and technical replicates were analyzed together using MaxQuant (version 1.3.0.5), utilizing its integrated ANDROMEDA search algorithms. Peak lists were searched against the UNIPROT databases for Trypanosoma brucei TREU927 (version 3.3, 9826 entries) downloaded from tritrypDB (http://tritrypdb.org) with common contaminants added. The search included carbamidomethylation of cysteines as fixed modification, and methionine oxidation, phosphorylation and N-terminal acetylation as variable modifications. Maximum allowed mass deviation for MS peaks was set to 6ppm and 20ppm for MS/MS peaks. Maximum missed cleavages were 3. The false discovery rate was determined by searching a reverse database. Maximum false discovery rates were 0.01 both on peptide and protein levels. Minimum required peptide length was 6 residues. Proteins with at least two peptides (one of them unique) were considered identified. The “match between runs” option was enabled with a time window of 1 min to match identification between replicates.
Western blotting validation of the results for selected proteins used the following dilutions of primary antibodies: VSG397 – 1:5,000; VSG653 – 1:1,000; alternative oxidase – 1:200; calflagin – 1:1,000; clathrin heavy chain – 1:1,000; hnRNP F/H – 1:5,000; argonaute 1 – 1:1,000; ribosomal protein P0 – 1:10,000. Horseradish peroxidase (HRP)-conjugated secondary antibodies were used for chemiluminescent detection.
RNA-Seq data from this study have been submitted to the NCBI Sequence Read Archive - SRA at http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi - under accession number SRP103532. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2013) partner repository with the dataset identifier PXD006197.
Supplementary Material
Acknowledgments
We thank Mingzhou Fu for performing the DESeq2 analysis and M. Chaudhuri, D. Engman, S. Michaeli and M. Field for antibodies. This work was supported by National Institutes of Health (http://www.nih.gov) grants AI028798 and AI110325 to C.T. T.C.W is an investigator of the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no financial conflict of interest.
Footnotes
Author contributions
Conceptualization and design of the study: EU, TCW, CT, RC, NGK; data acquisition: RC, NGK, HS; data analysis: RC, NGK, CT; writing – original draft: NGK, CT, RC; writing – review and editing: NGK, RC, TCW, CT.
References
- Alarcon CM, Son HJ, Hall T, Donelson JE. A monocistronic transcript for a trypanosome variant surface glycoprotein. Mol Cell Biol. 1994;14:5579–5591. doi: 10.1128/mcb.14.8.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, Eckert S, Baker N, Glover L, Sanchez-Flores A, Leung KF, et al. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Inchaustegui D, Queiroz R, Clayton C. The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One. 2011;6:e18425. doi: 10.1371/journal.pone.0018425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JD, Graham SV, Fotheringham M, Graham VS, Kobryn K, Wymer B. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol Biochem Parasitol. 1998;91:93–105. doi: 10.1016/s0166-6851(97)00193-x. [DOI] [PubMed] [Google Scholar]
- Barry JD, Hajduk SL, Vickerman K, Le Ray D. Detection of multiple variable antigen types in metacyclic populations of Trypanosoma brucei. Trans R Soc Trop Med Hyg. 1979;73:205–208. doi: 10.1016/0035-9203(79)90213-x. [DOI] [PubMed] [Google Scholar]
- Batram C, Jones NG, Janzen CJ, Markert SM, Engstler M. Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife. 2014;3:e02324. doi: 10.7554/eLife.02324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W, Gerrits H, Kieft R, Borst P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- Brandenburg J, Schimanski B, Nogoceke E, Nguyen TN, Padovan JC, Chait BT, et al. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 2007;26:4856–4866. doi: 10.1038/sj.emboj.7601905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Baltz T. A potential hexose transporter gene expressed predominantly in the bloodstream form of Trypanosoma brucei. Mol Biochem Parasitol. 1992;52:111–121. doi: 10.1016/0166-6851(92)90040-q. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Baltz T. Differential regulation of two distinct families of glucose transporter genes in Trypanosoma brucei. Mol Cell Biol. 1993;13:1146–1154. doi: 10.1128/mcb.13.2.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Riviere L, Coustou V. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149:1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Butter F, Bucerius F, Michel M, Cicova Z, Mann M, Janzen CJ. Comparative proteomics of two life cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite’s host adaptation machinery. Mol Cell Proteomics. 2013;12:172–179. doi: 10.1074/mcp.M112.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capewell P, Monk S, Ivens A, Macgregor P, Fenn K, Walrad P, et al. Regulation of Trypanosoma brucei Total and Polysomal mRNA during Development within Its Mammalian Host. PLoS One. 2013;8:e67069. doi: 10.1371/journal.pone.0067069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanez AL, Hehl AB, Engstler M, Schneider A. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J Cell Sci. 2006;119:2968–2974. doi: 10.1242/jcs.03023. [DOI] [PubMed] [Google Scholar]
- Chaves I, Rudenko G, Dirks-Mulder A, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C. The regulation of trypanosome gene expression by RNA-binding proteins. PLoS Pathog. 2013;9:e1003680. doi: 10.1371/journal.ppat.1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe LJ, Siegel TN, Marshall M, Cross GA, Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GA, Kim HS, Wickstead B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol Biochem Parasitol. 2014;195:59–73. doi: 10.1016/j.molbiopara.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Daniels JP, Gull K, Wickstead B. Cell biology of the trypanosome genome. Microbiol Mol Biol Rev. 2010;74:552–569. doi: 10.1128/MMBR.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Morales R, Banday M, Garcia S, Hao L, Cross GA, et al. The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. RNA. 2012;18:1968–1983. doi: 10.1261/rna.033829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pablos LM, Kelly S, de Freitas Nascimento J, Sunter J, Carrington M. Characterization of RBP9 and RBP10, two developmentally regulated RNA-binding proteins in Trypanosoma brucei. Open Biol. 2017:7. doi: 10.1098/rsob.160159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S, Marchetti R, Kirk K, Matthews KR. A surface transporter family conveys the trypanosome differentiation signal. Nature. 2009;459:213–217. doi: 10.1038/nature07997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Dejung M, Subota I, Bucerius F, Dindar G, Freiwald A, Engstler M, et al. Quantitative Proteomics Uncovers Novel Factors Involved in Developmental Differentiation of Trypanosoma brucei. PLoS Pathog. 2016;12:e1005439. doi: 10.1371/journal.ppat.1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda A, Ryten M, Droll D, Rojas F, Farber V, Haanstra JR, et al. Transcriptome-wide analysis of trypanosome mRNA decay reveals complex degradation kinetics and suggests a role for co-transcriptional degradation in determining mRNA levels. Mol Microbiol. 2014;94:307–326. doi: 10.1111/mmi.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger ML, Blundell PA, Lewis AM, Browitt A, Gunzl A, Barry JD. Ex vivo and in vitro identification of a consensus promoter for VSG genes expressed by metacyclic-stage trypanosomes in the tsetse fly. Eukaryot Cell. 2002;1:1000–1009. doi: 10.1128/EC.1.6.1000-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L, Hutchinson S, Alsford S, Horn D. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc Natl Acad Sci U S A. 2016;113:7225–7230. doi: 10.1073/pnas.1600344113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SV, Barry JD. Expression site-associated genes transcribed independently of variant surface glycoprotein genes in Trypanosoma brucei. Mol Biochem Parasitol. 1991;47:31–41. doi: 10.1016/0166-6851(91)90145-v. [DOI] [PubMed] [Google Scholar]
- Graham SV, Barry JD. Transcriptional regulation of metacyclic variant surface glycoprotein gene expression during the life cycle of Trypanosoma brucei. Mol Cell Biol. 1995;15:5945–5956. doi: 10.1128/mcb.15.11.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera K, Wuthrich D, Braga-Lagache S, Heller M, Ochsenreiter T. Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics. 2012;13:556. doi: 10.1186/1471-2164-13-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Kosti I, Plaut G, Pivko A, Tkacz ID, Cohen-Chalamish S, et al. The hnRNP F/H homologue of Trypanosoma brucei is differentially expressed in the two life cycle stages of the parasite and regulates splicing and mRNA stability. Nucleic Acids Res. 2013;41:6577–6594. doi: 10.1093/nar/gkt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni S, Renggli CK, Fragoso CM, Oberle M, Roditi I. The procyclin-associated genes of Trypanosoma brucei are not essential for cyclical transmission by tsetse. Mol Biochem Parasitol. 2006;150:144–156. doi: 10.1016/j.molbiopara.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. Antigenic variation in African trypanosomes. Mol Biochem Parasitol. 2014;195:123–129. doi: 10.1016/j.molbiopara.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh C, Yuan X, Miguel DC, Renberg RL, Protchenko O, Philpott CC, et al. Heme uptake by Leishmania amazonensis is mediated by the transmembrane protein LHR1. PLoS Pathog. 2012;8:e1002795. doi: 10.1371/journal.ppat.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Allison HC, Barry JD, Field MC, Hertz-Fowler C, Berriman M. A cell-surface phylome for African trypanosomes. PLoS Negl Trop Dis. 2013;7:e2121. doi: 10.1371/journal.pntd.0002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BC, Ramasamy G, Vasconcelos EJ, Ingolia NT, Myler PJ, Parsons M. Extensive stage-regulation of translation revealed by ribosome profiling of Trypanosoma brucei. BMC Genomics. 2014;15:911. doi: 10.1186/1471-2164-15-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics. 2009;10:482. doi: 10.1186/1471-2164-10-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NG, Thomas EB, Brown E, Dickens NJ, Hammarton TC, Mottram JC. Regulators of Trypanosoma brucei cell cycle progression and differentiation identified using a kinome-wide RNAi screen. PLoS Pathog. 2014;10:e1003886. doi: 10.1371/journal.ppat.1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Kramer S, Schwede A, Maini PK, Gull K, Carrington M. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012;2:120033. doi: 10.1098/rsob.120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham JK, Park SH, Nguyen TN, Lee JH, Gunzl A. Dynein Light Chain LC8 Is Required for RNA Polymerase I-Mediated Transcription in Trypanosoma brucei, Facilitating Assembly and Promoter Binding of Class I Transcription Factor A. Mol Cell Biol. 2016;36:95–107. doi: 10.1128/MCB.00705-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Gunzl A, Tschudi C. Metacyclic VSG expression site promoters are recognized by the same general transcription factor that is required for RNA polymerase I transcription of bloodstream expression sites. Mol and Biochem Parasitol. 2017 doi: 10.1016/j.molbiopara.2017.07.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ullu E, Tschudi C. The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell Microbiol. 2014;16:482–489. doi: 10.1111/cmi.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount DJ, Gruszynski AE, Grandgenett PM, Bangs JD, Donelson JE. Expression and function of the Trypanosoma brucei major surface protease (GP63) genes. J Biol Chem. 2003;278:24658–24664. doi: 10.1074/jbc.M301451200. [DOI] [PubMed] [Google Scholar]
- Le Ray D, Barry JD, Vickerman K. Antigenic heterogeneity of metacyclic forms of Trypanosoma brucei. Nature. 1978;273:300–302. doi: 10.1038/273300a0. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair GR, Lasonder E, Garver LS, Franke-Fayard BM, Carret CK, Wiegant JC, et al. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010;6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J, Guttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, et al. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensa-Wilmot K, Hereld D, Englund PT. Genomic organization, chromosomal localization, and developmentally regulated expression of the glycosyl-phosphatidylinositol-specific phospholipase C of Trypanosoma brucei. Mol Cell Biol. 1990;10:720–726. doi: 10.1128/mcb.10.2.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan GW, Goulding D, Field MC. The single dynamin-like protein of Trypanosoma brucei regulates mitochondrial division and is not required for endocytosis. J Biol Chem. 2004;279:10692–10701. doi: 10.1074/jbc.M312178200. [DOI] [PubMed] [Google Scholar]
- Mugnier MR, Stebbins CE, Papavasiliou FN. Masters of Disguise: Antigenic Variation and the VSG Coat in Trypanosoma brucei. PLoS Pathog. 2016;12:e1005784. doi: 10.1371/journal.ppat.1005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguleswaran A, Gunasekera K, Schimanski B, Heller M, Hemphill A, Ochsenreiter T, Roditi I. Trypanosoma brucei RRM1 is a nuclear RNA-binding protein and modulator of chromatin structure. MBio. 2015;6:e00114. doi: 10.1128/mBio.00114-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan SK, Peacock L, Matthews K, Gibson W, Field MC. Activation of endocytosis as an adaptation to the mammalian host by trypanosomes. Eukaryot Cell. 2007;6:2029–2037. doi: 10.1128/EC.00213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, Muller LS, Park SH, Siegel TN, Gunzl A. Promoter occupancy of the basal class I transcription factor A differs strongly between active and silent VSG expression sites in Trypanosoma brucei. Nucleic Acids Res. 2014;42:3164–3176. doi: 10.1093/nar/gkt1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, et al. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics. 2009;10:495. doi: 10.1186/1471-2164-10-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey-Butler K, Ullu E, Kolev NG, Tschudi C. Synchronous expression of individual metacyclic variant surface glycoprotein genes in Trypanosoma brucei. Mol Biochem Parasitol. 2015;200:1–4. doi: 10.1016/j.molbiopara.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP, Beecroft RP, Tolson DL, Liu MK, Pearson TW. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol Biochem Parasitol. 1988;31:203–216. doi: 10.1016/0166-6851(88)90150-8. [DOI] [PubMed] [Google Scholar]
- Rico E, Rojas F, Mony BM, Szoor B, Macgregor P, Matthews KR. Bloodstream form pre-adaptation to the tsetse fly in Trypanosoma brucei. Front Cell Infect Microbiol. 2013;3:78. doi: 10.3389/fcimb.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roditi I, Schwarz H, Pearson TW, Beecroft RP, Liu MK, Richardson JP, et al. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol. 1989;108:737–746. doi: 10.1083/jcb.108.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotureau B, Subota I, Buisson J, Bastin P. A new asymmetric division contributes to the continuous production of infective trypanosomes in the tsetse fly. Development. 2012;139:1842–1850. doi: 10.1242/dev.072611. [DOI] [PubMed] [Google Scholar]
- Savage AF, Kolev NG, Franklin JB, Vigneron A, Aksoy S, Tschudi C. Transcriptome profiling of Trypanosoma brucei development in the tsetse fly vector Glossina morsitans. PLoS One. 2016;11:e0168877. doi: 10.1371/journal.pone.0168877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SZ, Naessens J, Liesegang B, Moloo SK, Magondu J. Analysis by flow cytometry of DNA synthesis during the life cycle of African trypanosomes. Acta Trop. 1984;41:313–323. [PubMed] [Google Scholar]
- Sharma R, Peacock L, Gluenz E, Gull K, Gibson W, Carrington M. Asymmetric cell division as a route to reduction in cell length and change in cell morphology in trypanosomes. Protist. 2008;159:137–151. doi: 10.1016/j.protis.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, et al. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subota I, Rotureau B, Blisnick T, Ngwabyt S, Durand-Dubief M, Engstler M, Bastin P. ALBA proteins are stage regulated during trypanosome development in the tsetse fly and participate in differentiation. Mol Biol Cell. 2011;22:4205–4219. doi: 10.1091/mbc.E11-06-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetley L, Turner CM, Barry JD, Crowe JS, Vickerman K. Onset of expression of the variant surface glycoproteins of Trypanosoma brucei in the tsetse fly studied using immunoelectron microscopy. J Cell Sci. 1987;87:363–372. doi: 10.1242/jcs.87.2.363. [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Guther ML, Ferguson MA. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One. 2012;7:e36619. doi: 10.1371/journal.pone.0036619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak MD, Martin DM, Ferguson MA. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res. 2013;12:2233–2244. doi: 10.1021/pr400086y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S, Studer E, Renggli CK, Roditi I. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol Microbiol. 2007;63:218–228. doi: 10.1111/j.1365-2958.2006.05492.x. [DOI] [PubMed] [Google Scholar]
- Van Den Abbeele J, Claes Y, van Bockstaele D, Le Ray D, Coosemans M. Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology. 1999;118(Pt 5):469–478. doi: 10.1017/s0031182099004217. [DOI] [PubMed] [Google Scholar]
- Vasquez JJ, Hon CC, Vanselow JT, Schlosser A, Siegel TN. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res. 2014;42:3623–3637. doi: 10.1093/nar/gkt1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci. 1997;110(Pt 21):2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- Veitch NJ, Johnson PC, Trivedi U, Terry S, Wildridge D, MacLeod A. Digital gene expression analysis of two life cycle stages of the human-infective parasite, Trypanosoma brucei gambiense reveals differentially expressed clusters of co-regulated genes. BMC Genomics. 2010;11:124. doi: 10.1186/1471-2164-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman K. The mechanism of cyclical development in trypanosomes of the Trypanosoma brucei sub-group: an hypothesis based on ultrastructural observations. Trans R Soc Trop Med Hyg. 1962;56:487–495. doi: 10.1016/0035-9203(62)90072-x. [DOI] [PubMed] [Google Scholar]
- Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- Vickerman K, Luckins AG. Localization of variable antigens in the surface coat of Trypanosoma brucei using ferritin conjugated antibody. Nature. 1969;224:1125–1126. doi: 10.1038/2241125a0. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, et al. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- Wurst M, Seliger B, Jha BA, Klein C, Queiroz R, Clayton C. Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol Microbiol. 2012;83:1048–1063. doi: 10.1111/j.1365-2958.2012.07988.x. [DOI] [PubMed] [Google Scholar]
- Ziegelbauer K, Multhaup G, Overath P. Molecular characterization of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J Biol Chem. 1992;267:10797–10803. [PubMed] [Google Scholar]
- Ziegelbauer K, Overath P. Identification of invariant surface glycoproteins in the bloodstream stage of Trypanosoma brucei. J Biol Chem. 1992;267:10791–10796. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.