Abstract
Introduction
Identification of blood-based metabolic changes might provide early and easy-to-obtain biomarkers.
Methods
We included 127 Alzheimer's disease (AD) patients and 121 control subjects with cerebrospinal fluid biomarker-confirmed diagnosis (cutoff tau/amyloid β peptide 42: 0.52). Mass spectrometry platforms determined the concentrations of 53 amine compounds, 22 organic acid compounds, 120 lipid compounds, and 40 oxidative stress compounds. Multiple signatures were assessed: differential expression (nested linear models), classification (logistic regression), and regulatory (network extraction).
Results
Twenty-six metabolites were differentially expressed. Metabolites improved the classification performance of clinical variables from 74% to 79%. Network models identified five hubs of metabolic dysregulation: tyrosine, glycylglycine, glutamine, lysophosphatic acid C18:2, and platelet-activating factor C16:0. The metabolite network for apolipoprotein E (APOE) ε4 negative AD patients was less cohesive compared with the network for APOE ε4 positive AD patients.
Discussion
Multiple signatures point to various promising peripheral markers for further validation. The network differences in AD patients according to APOE genotype may reflect different pathways to AD.
Keywords: Alzheimer's disease, Amino acids, Biomarkers, Graphical modeling, Metabolomics, Oxidative stress
Highlights
-
•
Multiple metabolic signatures point to peripheral AD markers for future validation.
-
•
AD may be described by changes in the metabolism of amines and oxidative stressors.
-
•
APOE ε4-driven AD and non- APOE ε4-driven AD represent different biochemical pathways.
-
•
Network analyses of metabolomics data enable the study of metabolic changes in AD.
1. Introduction
Accumulation of amyloid and tau proteins is considered the core pathologic hallmark for Alzheimer's disease (AD) [1], but other factors such as genetic liability, oxidative stress, inflammation, and lifestyle contribute to the complex mechanism of this disease [1], [2], [3], [4], [5]. Noninvasive measurement of disease-specific biochemical changes in living patients is difficult but may have value in terms of prognosis and identification of patients at risk for AD.
The metabolome, that is, the collection of small molecules that result from metabolic processes, is organized in biochemical pathways and is influenced by many internal and external factors, including genetics [6]. Metabolomics refers to the collective quantification of these metabolites [7]. Analytical methods have improved tremendously, with (targeted) mass spectrometry (MS) platforms now available for most compound classes. In AD, metabolomics seems of utmost importance because various alterations in metabolism, for example, higher levels of insulin and insulin resistance, are associated with an increased risk of AD [8]. Moreover, the epsilon 4 (ε4) allele of the apolipoprotein E (APOE) gene is not only an important risk factor for AD but is also related to alterations in lipid metabolism [9], [10]. Previous metabolomics studies in AD have reported alterations in lipid, antioxidant, and amino acid metabolism. However, results are not always unequivocal [11], [12], [13], [14], [15]. This is most likely due to differences in (analytical) methods, cohort selection, or context of use [16].
We aim to study AD-related metabolic change from various perspectives with the use of multiple signatures to generate hypotheses regarding dysregulated metabolic events. First, we evaluate shifts in the expression of individual metabolites using nested linear models. Afterward, we assess the classification performance of the metabolites in demarcating AD from control subjects. Finally, we use state-of-the-art graphical modeling to explore metabolic dysregulation from a network perspective. In addition, we evaluate metabolic network changes according to APOE status, to study the hypothesis that metabolic pathways are differentially dysregulated according to the genotype.
2. Methods
2.1. Patients
We selected 150 AD patients and 150 control subjects with available plasma from the Amsterdam Dementia Cohort [17]. All subjects underwent standard cognitive screening including medical history assessment; physical, neurologic, and cognitive examination; blood sampling; lumbar puncturing; and magnetic resonance imaging. Diagnoses were made in a multidisciplinary consensus meeting. Until 2012, the diagnosis “probable AD” was based on the clinical criteria formulated by the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association [18]. From 2012 onward the criteria of the National Institute on Aging-Alzheimer's Association were used [19]. Subjects with subjective cognitive decline were used as control subjects. These subjects presented with memory complaints at the VUmc memory clinic, but performed normal on cognitive testing, that is, criteria for mild cognitive impairment, dementia, or psychiatric diagnosis were not fulfilled. Clinical characteristics are provided in Table 1. All subjects gave written informed consent to use their clinical data for research purposes and to collect their blood samples for biobanking.
Table 1.
Comparison of clinical characteristics between AD and control groups
| Characteristic | AD group | Control group | P value |
|---|---|---|---|
| n (%) | 127 (51) | 121 (49) | |
| MMSE score, median (IQR) | 21 (5.5) | 29 (2) | <.001∗ |
| Anthropometric | |||
| Age, median (IQR) | 65.1 (9.1) | 62.7 (8) | .548∗ |
| Gender (female), n (%) | 63 (50) | 56 (46) | .692† |
| ≥1 APOE ε4 allele (yes), n (%) | 87 (69) | 34 (28) | <.001† |
| MAP, mean (SD) | 106.1 (11.5) | 103.9 (11.7) | .133‡ |
| BMI, mean (SD) | 24.2 (3.3) | 26.27 (3.6) | <.001‡ |
| Intoxications | |||
| Smoking | .558† | ||
| Former, n (%) | 42 (33) | 46 (38) | |
| Current, n (%) | 21 (17) | 15 (12) | |
| Alcohol (yes), n (%) | 98 (77) | 88 (73) | .509† |
| Comorbidities | |||
| Hypertension (yes), n (%) | 37 (29) | 33 (27) | .854† |
| Diabetes mellitus (yes), n (%) | 4 (3) | 14 (12) | .021† |
| Hypercholesterolemia (yes), n (%) | 14 (11) | 9 (7) | .451† |
| Medication | |||
| Cholesterol lowering (yes), n (%) | 31 (24) | 22 (18) | .298† |
| Antidepressants (yes), n (%) | 12 (9) | 15 (12) | .589† |
| Antiplatelets (yes), n (%) | 26 (20) | 19 (16) | .418† |
Abbreviations: AD, Alzheimer's disease; APOE, apolipoprotein E; BMI, body mass index; IQR, interquartile range; MAP, mean arterial pressure; MMSE, Mini–Mental State Examination; SD, standard deviation.
Mann-Whitney U test.
Pearson χ2 test.
Welch's t test.
2.2. Cerebrospinal fluid biomarkers
Amyloid β peptide 42 (Aβ42) and total tau (t-tau) were, for all subjects, measured in cerebrospinal fluid (CSF) using commercially available enzyme-linked immunosorbent assays (Innotest Aβ42 and Innotest hTAU-Ag; Innogenetics, Ghent, Belgium) [20]. The cutoff for pathologic biomarker status was defined as t-tau/Aβ42 > 0.52 [21]. Of the 300 subjects included, 263 (136 AD patients and 127 controls) had a biomarker status in concordance with their clinical diagnosis, that is, t-tau/Aβ42 > 0.52 for AD and t-tau/Aβ42 ≤ 0.52 for controls. These subjects were included for further analysis.
2.3. APOE genotyping
DNA was isolated from 7 to 10 mL ethylenediaminetetraacetic acid (EDTA) blood. Subsequently, samples were subjected to polymerase chain reaction. A QIAxcel DNA Fast Analysis kit (Qiagen, Venlo, The Netherlands) was used to check for size. Sequencing was performed using Sanger sequencing on an ABI130XL.
2.4. Metabolic profiling
Nonfasting EDTA plasma samples were, within 2 hours of collection, centrifuged at 1800g for 10 minutes at room temperature and stored at −80°C in polypropylene tubes (Sarstedt, Nurmberg, Germany). Metabolic profiling of the samples was performed on four MS platforms, that is, amines, lipids, and oxidative stress compounds were identified using ultraperformance liquid chromatography-tandem MS, and organic acids were analyzed with gas chromatography-MS [7], [22], [23], [24]. Reproducibility of individual metabolites was assessed in terms of the relative standard deviation of quality control (RSDQC) samples. Metabolites with RSDQC >30% were deemed to fail acceptance criteria. After QC correction, 53 amine compounds, 22 organic acid compounds, 120 lipid compounds, and 40 oxidative stress compounds were considered detected. See Supplementary Text 1 and its accompanying tables for details on the profiling methods and detected compounds.
2.5. Data processing
Metabolites with more than 10% missing observations were removed, leading to the removal of four lipid compounds and one oxidative stress compound. Three data samples (i.e., observed metabolite abundance profiles stemming from corresponding plasma samples) were removed as their (plasma) quality was deemed unsure. These samples had many (30 or more) concentrations below the limit of detection that could not be attributed to instrumental errors. Twelve additional data samples were removed because of instrumental errors in one or more platforms. Hence, we only retained data samples that were free of instrumental errors across all four different MS platforms. The remaining missing values are attributable to concentrations failing the limit of detection. These (feature-specific) missing values were imputed by half of the lowest observed value (for the corresponding metabolic feature). The final metabolic data set thus contained n = 263−3−12 = 248 data samples (127 AD patients and 121 controls) and P = 235−5 = 230 metabolites.
The possible confounding effects of the clinical characteristics regarding anthropometrics, intoxications, comorbidities, and medication were evaluated in the expression and classification signatures demarcating the AD and control groups (see Section 2.6). Table 1 contains the full list of characteristics, and Table S2.1 of Supplementary Text 2 (SMT2) contains additional information on measurement. The missing observations on these variables (<6%) were imputed. Continuous variables were imputed on the basis of Bayesian linear regression, polytomous variables were imputed on the basis of polytomous regression, and binary variables were imputed on the basis of logistic regression [25]. See Section 1 of SMT2 for additional information on data processing.
2.6. Statistical analysis
Differences in clinical characteristics between AD patients and control subjects were evaluated through chi-square, Mann-Whitney U, and t tests. Differential metabolic expression between AD patients and control subjects was assessed by using nested linear models. We tested, for each individual metabolite, whether its addition to a model containing clinical characteristics significantly contributed to model fit. One then assesses if, conditional on the effects of the clinical characteristics, metabolic expression does indeed differ between the AD and control groups. This entails an F test for nested models (see Section 2.1 of SMT2 for details). The conditioning sets were (1) sex and age, and (2) all clinical characteristics. We adjusted for multiple testing by controlling the false discovery rate (FDR) [26] at 0.05.
Subsequently, metabolic classification signatures for the prediction of group membership (AD or control) were constructed by way of penalized logistic regression with a Lasso penalty [27]. The Lasso penalty enables estimation in our setting where the metabolite to sample ratio (230/248) is too high for standard logistic regression. It also achieves automatic feature selection. Two settings were considered: (1) the Lasso selects among the metabolites without considering the clinical characteristics; and (2) the Lasso selects among the metabolites while the clinical characteristics go unpenalized. The resulting models were compared with an unpenalized logistic regression model that considered only the clinical characteristics. The optimal penalty parameter in the penalized models was determined on the basis of leave-one-out cross-validation of the model likelihood. Predictive performance of all models was assessed by way of (the comparison of) receiver operating characteristic (ROC) curves and the area under the ROC curves (AUCs). ROC curves and AUCs for all models were produced by 10-fold cross-validation. See Section 2.2 of SMT2 for additional information.
A metabolic pathway can be thought of as a collection of metabolites originating from all over the metabolome that work interdependently to regulate biochemical (disease) processes. Hence, a pathway is a network. We additionally used network extraction techniques to examine regulatory signatures, that is, dysregulation in metabolic biochemical pathways pertaining to the AD disease process. From a network perspective, molecular pathway dysregulation is likely characterized by the loss of normal (wanted) molecular interactions and the gain of abnormal (unwanted) molecular interactions. From this perspective, the network topologies of the AD and control groups are expected to primarily share the same structure, while potentially differing in a number of (topological) locations of interest. Network extraction was based on graphical modeling, more specifically, on targeted fused ridge estimation of inverse covariance (i.e., scaled partial correlation) matrices [28]. This method (1) can deal with our metabolite to sample ratio (230/248, which is too high for standard graphical modeling), and (2) explicitly takes into account that there are multiple groups of interest for which the shared network structures should be fused whereas the unique network structures should be distinguished. The resulting networks are to be interpreted as conditional independence graphs, that is, the nodes represent metabolic compounds and the edges connecting the nodes represent substantive partial correlations. Extracted networks were subjected to subsequent analyses aimed at detecting hub compounds, group structures, and differential metabolic connections between groupings of interest. Our efforts first juxtaposed metabolic networks for AD patients and control subjects. Subsequently, we compared networks according to APOE genotype. See Sections 2.3 and 2.4 of SMT2 for additional detail.
3. Results
3.1. Clinical characteristics
Table 1 contains an overview of the clinical characteristics per diagnostic group. The Mini–Mental State Examination score [29] of AD patients was lower compared with control subjects. AD patients were more often carrier of at least one APOE ε4 allele. Moreover, AD patients had a lower body mass index and were less likely to have diabetes.
3.2. Differential expression signature
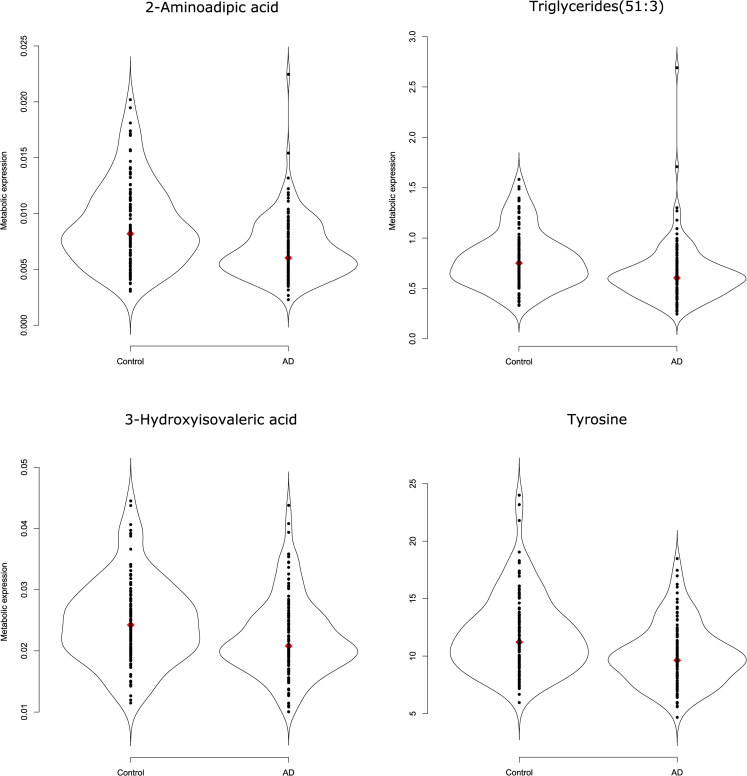
A global test [30] indicates that, given sex and age, the overall metabolic expression profile differs between AD patients and control subjects (P = 2.12 × 10−6). This difference in overall metabolic expression profile upholds when correcting for all clinical characteristics (P = 4.69 × 10−5). The metabolites listed in Table 2 pass multiple testing correction on the F test for nested models (Section 2.6) with an FDR <0.05. The third column gives the ranking (in terms of raw P value) of 51 metabolites that survive FDR correction when adjusting for sex and age only. The fourth column analogously ranks the 26 metabolites that survive FDR correction when additionally adjusting for all clinical characteristics. Triglycerides (TGs) and amines dominate the latter compounds' list. Among its top compounds, in terms of (adjusted) P value, are the amines 2-aminoadipic acid (2-AAA) and tyrosine, the TG (51:3), and the organic acid 3-hydroxyisovaleric acid. Their distributions in the AD and control groups are depicted in Fig. 1. We see that these compounds are underexpressed in the AD group relative to the control group. This relative underexpression in the AD group also holds for the remaining compounds in column 4 of Table 2, except for the sphingomyelin (d18:1/20:1), which is overexpressed in the AD group relative to the control group (see Figs. S2.1–S2.3 in SMT2).
Table 2.
Differentially expressed metabolites that survive FDR adjustment
| Metabolite | Compound class | Ranking | |
|---|---|---|---|
| 2-Aminoadipic acid | Amines | 1 | 1 |
| Valine | Amines | 2 | 16 |
| Tyrosine | Amines | 3 | 4 |
| Methyldopa | Amines | 4 | 9 |
| Lysine | Amines | 5 | |
| Methylmalonic acid | Organic acids | 6 | 14 |
| S-3-Hydroxyisobutyric acid | Organic acids | 7 | 7 |
| TG (48:0) | Lipids: TGs | 8 | 21 |
| TG (50:4) | Lipids: TGs | 9 | 6 |
| TG (48:2) | Lipids: TGs | 10 | 13 |
| TG (51:3) | Lipids: TGs | 11 | 2 |
| TG (54:6) | Lipids: TGs | 12 | 5 |
| TG (50:3) | Lipids: TGs | 13 | 17 |
| TG (50:2) | Lipids: TGs | 14 | |
| TG (50:1) | Lipids: TGs | 15 | |
| TG (48:1) | Lipids: TGs | 16 | 25 |
| TG (52:4) | Lipids: TGs | 17 | 18 |
| TG (48:3) | Lipids: TGs | 18 | 11 |
| Leucine | Amines | 19 | |
| LPC (18:1) | Lipids: lysophosphatidylcholine | 20 | |
| TG (46:2) | Lipids: TGs | 21 | 15 |
| TG (50:0) | Lipids: TGs | 22 | |
| TG (52:5) | Lipids: TGs | 23 | 19 |
| TG (52:3) | Lipids: TGs | 24 | |
| TG (51:2) | Lipids: TGs | 25 | |
| TG (56:8) | Lipids: TGs | 26 | 8 |
| Isoleucine | Amines | 27 | |
| 2-Hydroxybutyric acid | Organic acids | 28 | |
| 3-Hydroxyisovaleric acid | Organic acids | 29 | 3 |
| TG (51:1) | Lipids: TGs | 30 | |
| SM (d18:1/20:1) | Lipids: SMs | 31 | 24 |
| TG (52:1) | Lipids: TGs | 32 | |
| 8-iso-PGF2a (15-F2t-IsoP) | Oxidative stress: isoprostane | 33 | 10 |
| Proline | Amines | 34 | |
| TG (54:5) | Lipids: TGs | 35 | |
| TG (56:7) | Lipids: TGs | 36 | 20 |
| PGD2 | Lipids: prostaglandins | 37 | |
| TG (46:1) | Lipids: TGs | 38 | |
| PC (O-44:5) | Lipids: plasmalogen phosphatidylcholine | 39 | |
| LPA C14:0 | LPA | 40 | |
| PC (O-34:1) | Lipids: plasmalogen phosphatidylcholine | 41 | |
| LPC(20:4) | Lipids: lysophosphatidylcholine | 42 | |
| SM (d18:1/24:2) | Lipids: SMs | 43 | |
| 8,12-iPF2a IV | Oxidative stress: isoprostane | 44 | |
| TG (46:0) | Lipids: TGs | 45 | |
| 5-iPF2a VI | Oxidative stress: isoprostane | 46 | |
| TG (52:2) | Lipids: TGs | 47 | |
| SM (d18:1/16:0) | Lipids: SMs | 48 | |
| TG (58:10) | Lipids: TGs | 49 | 26 |
| Ornithine | Amines | 50 | 22 |
| Histidine | Amines | 51 | |
| O-Acetylserine | Amines | 12 | |
| SM (d18:1/23:0) | Lipids: SMs | 23 | |
Abbreviations: FDR, false discovery rate; LPA, lysophosphatidic acid; SM, sphingomyelin; TG, triglyceride.
The third column ranks (in terms of raw P value) the metabolites that survive FDR correction in the model that adjusts for sex and age only. The fourth column ranks (in terms of raw P value) the metabolites that survive FDR correction in the model that adjusts for all clinical characteristics. See Tables 2.2 and 2.3 of SMT2 for additional information.
Fig. 1.
Violin plots of the top four metabolites in terms of P value. Violin plots [31] combine the familiar box plot with a kernel density to better represent the distribution of the data. We see relative underexpression in the Alzheimer's disease group for all depicted metabolites. The associated adjusted P values can be found in Table 2. The violin plots of the remaining differentially expressed metabolites can be found in Figs. S2.1–S2.3 of SMT2.
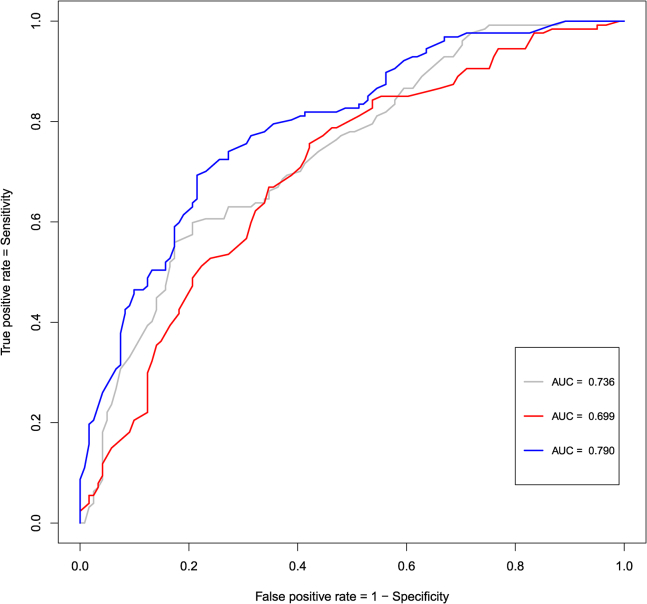
3.3. Classification signature
Subsequently, penalized logistic regression models were used to evaluate the ability of metabolites to distinguish AD patients from control subjects. Classification performances can be found in Fig. 2. The prediction model carrying the clinical variables only resulted in an AUC of approximately 0.74 (95% bootstrap confidence interval [CI]: 0.67–0.79). The model that used the Lasso for selection among the metabolites sorts a comparable classification performance, yielding an AUC of approximately 0.70 (95% bootstrap CI: 0.63–0.76). The added value of the metabolites is reflected in the prediction model that adds a (Lasso-based) selection of metabolites to the clinical variables as it improves predictive performance, sorting an AUC of 0.79 (95% bootstrap CI: 0.73–0.84). A one-tailed bootstrap test for correlated ROC curves [32] indicates that the AUC for this latter model is indeed higher than the AUC for the metabolites-only model (P = .002) and the AUC for the clinical-variables-only model (P = .005). This test also indicates that the AUCs for the metabolites-only and clinical-variables-only models do not differ significantly (P = .203). Metabolites consistently selected as top predictors (in terms of their absolute regression coefficient) in both penalized models that also occur in the differential expression signature are the amines O-acetylserine and methyldopa, the TG (51:5), and the organic methylmalonic acid. Furthermore, oxidative stress compounds were selected by the Lasso on the basis of their predictive power, especially the prostaglandin PGD2, the isoprostane 8,12-iPF2a IV, and the nitro-fatty acid NO2-aLA (C18:3). See Tables S2.4 and S2.5 of SMT2 for additional detail.
Fig. 2.
ROC curves for the classification models. The gray line represents the ROC curve for the unpenalized logistic regression model that entertains the clinical characteristics only. The red line represents the ROC curve for the logistic model in which the Lasso performed variable selection among the metabolites (and that does not consider the clinical characteristics). The blue line represents the ROC curve of the logistic model in which the clinical characteristics are present while the Lasso may select among the metabolites. The clinical variables are listed in Table 1. Abbreviation: ROC, receiver operating characteristic.
3.4. Regulatory signature
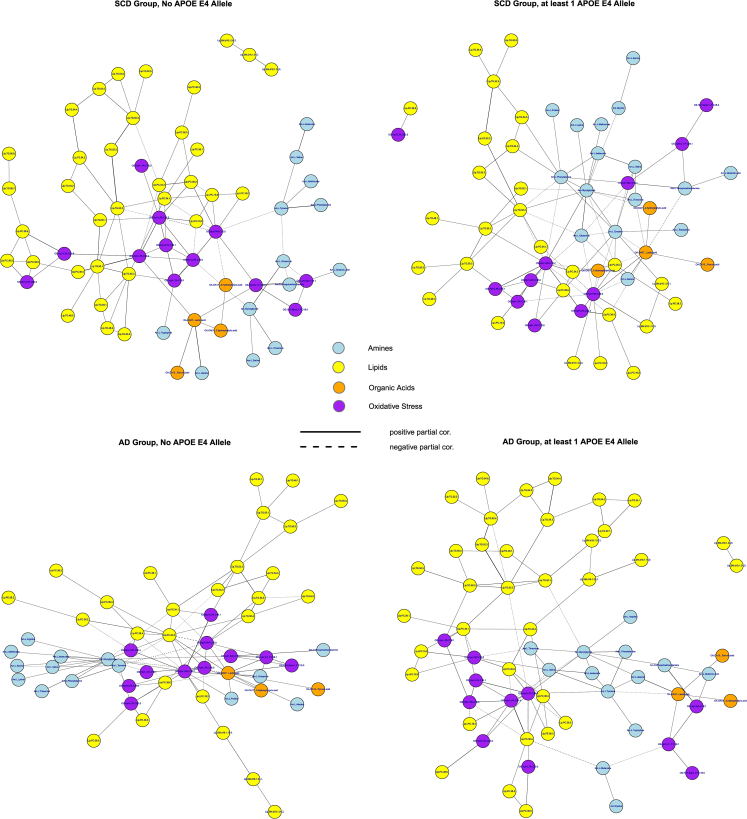
Next, graphical modeling was used to explore metabolite networks. Section 2.3.3 of SMT2 contains visualizations of the extracted networks for AD patients and control subjects. These networks convey that the strongest connections implicate metabolites from all four considered compound classes. The metabolite-network for the control patients seems stronger locally connected (Section 2.3.4 of SMT2), but both the AD and control networks are cohesive in the sense that they can be decomposed into clear communities (groups) of metabolites (Section 2.3.6 of SMT2). Hub compounds (i.e., metabolites of high regulatory importance as indicated by their centrality in a network) concur to some degree between the AD and control networks, with both having the lysophosphatidic acid (LPA) C18:2 (an oxidative stress compound) as the strongest hub. In the AD network however, as opposed to the control network, the amines glycylglycine and tyrosine are additionally indicated as central metabolites (Section 2.3.5 of SMT2). LPA C18:2, glycylglycine and tyrosine are among the metabolites whose regulatory functioning (in terms of differential connections) seems to change the most between the AD and control networks (Section 2.3.7 of SMT2).
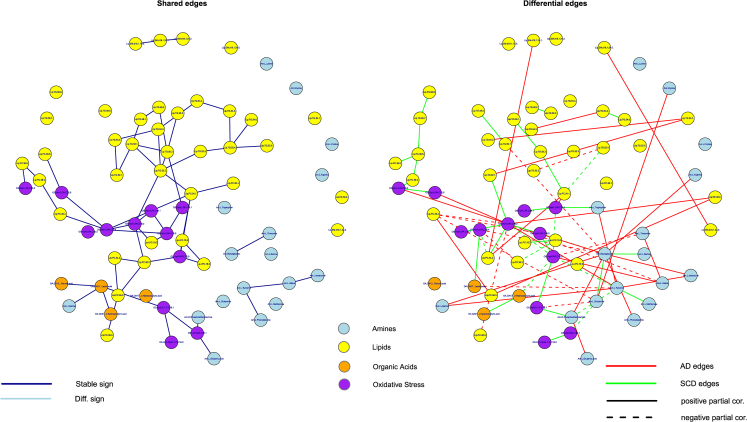
Overall, the AD and control networks seem to imply a shifting importance toward amine and oxidative stress compounds and their connections in the former. This picture becomes more pronounced when the networks are stratified according to APOE genotype (Section 2.4 of SMT2). Fig. 3 contains visualizations of the extracted networks for APOE ε4 negative control subjects and AD patients, as well as APOE ε4 positive control subjects and AD patients. The networks for APOE ε4 positive control subjects and APOE ε4 negative AD patients seem more random and less cohesive than the networks for APOE ε4 negative control subjects and APOE ε4 positive AD patients. Comparing the cohesive networks for APOE ε4 negative control subjects and APOE ε4 positive AD patients (Section 2.4 of SMT2), we see that all amines belong to the peripheral structure in the former whereas many amines belong to the core structure in the latter. This might imply that biochemical functioning in the APOE ε4 positive AD group is more reliant on amines. Hub compounds concur to some degree between these networks, with again (o.a.) LPA C18:2 as a strong hub. In the network for APOE ε4 positive AD patients the amines glycylglycine and tyrosine are consistently indicated as central metabolites. Fig. 4 presents the networks of shared and differential connections between the APOE ε4 negative control and APOE ε4 positive AD groups. The oxidative stress compounds LPA C18:2 and platelet-activating factor C16:0, and the amines glycylglycine, tyrosine, and glutamine seem to change their regulatory function the most between the APOE ε4 negative control and APOE ε4 positive AD groups (also see Table S2.12 in SMT2). From the network perspective the APOE ε4–driven AD state can be characterized (vis-à-vis the control state without APOE ε4 alleles) by a loss of connections involving platelet-activating factor C16:0, a gain of connections involving glycylglycine, and the differential wiring (both loss of normal and gain of alternative connections) of tyrosine, glutamine, and LPA C18:2.
Fig. 3.
Class-specific networks visualized with the Fruchterman-Reingold [33] algorithm. The upper left panel contains the network for the control group with no APOE ε4 allele. The upper right panel contains the network for the control group with at least one APOE ε4 allele. The lower left panel represents the network for the AD group with no APOE ε4 allele. The lower right panel represents the network for the AD group with at least one APOE ε4 allele. The metabolite compounds are colored according to the metabolite family: blue for amines, yellow for lipids, orange for organic acids, and purple for oxidative stress. Solid edges represent positive partial correlations, whereas dashed edges represent negative partial correlations. Abbreviations: AD, Alzheimer's disease; APOE, apolipoprotein E.
Fig. 4.
Common and differential networks for the control group with no APOE ε4 allele versus the AD group with at least one APOE ε4 allele. The left-hand panel contains the network consisting of the edges (solid and colored blue) that are shared between these groups. The right-hand panel contains the network consisting of the edges that are unique for either of the groups. Red edges represent connections that are present in the APOE ε4 positive AD group only. Green edges represent connections that are present in the APOE ε4 negative control group only. Solid edges represent positive partial correlations, whereas dashed edges represent negative partial correlations. The metabolite compounds are colored according to metabolite family: blue for amines, yellow for lipids, orange for organic acids, and purple for oxidative stress. Abbreviations: AD, Alzheimer's disease; APOE, apolipoprotein E.
3.5. CSF discordant subjects
Subjects whose clinical diagnosis was discordant from their CSF biomarker status have an insecure disease status and were therefore excluded from the analyses previously. A total of 37 subjects had both a complete metabolite profile and a discordant CSF biomarker status, that is, these subjects were either clinically diagnosed with AD although their CSF markers were normal (t-tau/Aβ42 ≤ 0.52) or clinically diagnosed as normal although their CSF markers indicated AD (t-tau/Aβ42 > 0.52). For purposes of comparison, we also obtained the expression and classification signatures when considering data from all n = 285 (263 + 37) subjects with a complete metabolite profile. The results—that accede to some degree with the results given in 3.2, 3.3—can be found (with discussion) in Supplementary Text 3.
4. Discussion
In this study, with CSF biomarker–confirmed AD and control cases, we show that profiling metabolic alterations in AD can highlight disease-specific biochemical changes. We assessed three metabolic signatures to highlight different aspects of metabolic change. The expression signature shows the metabolites with relative underexpression or overexpression in AD versus control subjects. This signature involved 26 metabolites, dominated by decreased levels of TGs and amines in AD. We then evaluated classification signatures: collections of clinical and metabolite markers that can successfully demarcate AD cases from control subjects. The top predictors concur (also in their sign) with metabolites found in the differential expression signature. In addition, markers of oxidative stress were identified as strong predictors. Finally, graphical modeling was used to evaluate regulatory signatures: exploratory networks of complex differential metabolite dependencies between the AD and control groups. Possible regulatory markers were again found in the amine and oxidative stress compound classes. Stratifying for APOE ε4 status, the network for APOE ε4 negative AD subjects was less cohesive compared with the network for APOE ε4 positive AD subjects. This suggests alternative biochemical dysregulation involved in these patient groups. Each signature gives a different but complementary perspective on AD-related metabolic events. We propose the combination of these three signatures as a new approach to (1) studying the complex mechanism of metabolic change, (2) defining characteristics involved in subtypes of AD, and (3) selecting robust markers of interest for further research. Subsequently, we discuss and embed the findings related to each signature.
4.1. Differential expression signature
We show in Table 2 that additional adjustments for clinical characteristics shorten the list and changes the ranking of metabolites that survive FDR correction. This underlines the effects of clinical variables, such as medication, on the metabolome. It also suggests that substantive corrections harness against overoptimistic expression signatures. Subsequently, we will focus on the 26 metabolites listed in the expression signature adjusted for all clinical variables. We found, in concordance with previous findings in both CSF and plasma, that AD is associated with decreased levels of amino acids and lipids [11], [34].
Sixteen lipids, of which 14 TGs, were underexpressed, whereas only one lipid—sphingomyelin (d18:1/20:1)—was overexpressed in AD. This is in agreement with a large and recent lipidomics study that reported a decrease in most plasma lipids in AD and in particular an association of long-chain TGs with AD [34]. Moreover, supplementation of medium-chain TGs has been tested in AD to correct neuronal hypometabolism and might show some benefit for APOE ε4 negative AD patients [35].
Multiple amino acids were also decreased in AD, among which were 2-AAA (an intermediate of the lysine pathway) and tyrosine. Plasma disturbances of the lysine pathway have been suggested to differentiate control subjects from mild cognitive impairment and AD patients [11]. Decreased tyrosine (a precursor for the neurotransmitters dopamine and norepinephrine) levels were also reported in an earlier study comparing metabolite levels in serum samples of AD patients and healthy control subjects [36]. Moreover, vanylmandellic acid—an end-product of the tyrosine pathway—was found to be increased in the CSF of AD patients [37], suggesting disturbances of the tyrosine pathway. Dopamine has been associated with cognitive control [38], and oral supplementation of tyrosine has been shown to improve working memory and information processing during demanding situations in healthy human adults [39]. Experimental studies are needed to establish if the alterations in peripheral tyrosine metabolism we found in our study also affect the function of dopamine and tyrosine in the central nervous system of AD patients.
4.2. Classification signature
The metabolites have added value in demarcating AD cases from control subjects. This is reflected by the significant improvement in predictive performance when adding a selection of metabolites to the clinical characteristics and APOE status. Metabolite panels to monitor disease are of great interest for the clinic. Especially when easy-to-obtain as with blood samples. We here hint that a metabolite panel could be of added value to the yet available clinical variables and therefore might hold promise for use in, for example, clinical effect monitoring.
Oxidative stress has been widely established to play a role in the pathogenesis of AD [2]. Defining the right markers to measure oxidative stress in vivo is, however, still an ongoing process, especially for peripheral markers in AD. We found three markers of oxidative stress to have strong predictive power in demarcating AD patients from control subjects: the isoprostane-pathway derivatives [40] 8,12-iPF-2a IV, and PGD2, and the nitro-fatty acid NO2-aLA (C18:3). This result highlights again that oxidative stress is of strong influence in AD [2], [41].
4.3. Regulatory signature
The network models revealed another oxidative stress marker, LPA C18:2, to be one of the central players in both the AD and control networks. It was prominently differentially related to other metabolites in AD versus control networks, perhaps representing a central player of metabolic change. Previously, oxidized lipoproteins have been identified as a possible oxidative stressor in the brain leading to neuronal cell death in AD [42]. LPA is the most bioactive fraction of oxidized low-density lipoprotein [43]. It has an important signal function and has been linked to the pathogenesis of AD as in vitro results suggest they support tau phosphorylation and raise levels of β-secretase, leading to increased Aβ production [43], [44], [45]. Moreover, LPAs have been identified as important factors in vascular development, atherosclerosis, and atherotrombogenesis [46], [47], [48]. As LPAs are a modulating factor in both AD and vascular changes, it could be of special interest to further study the role of vascular factors in AD.
Network models for APOE ε4 positive AD subjects were more cohesive and less random in comparison to APOE ε4 negative AD subjects. This suggests the possibility of structured, APOE ε4–driven changes in metabolism. The lack of cohesiveness for the APOE ε4 negative AD group may be natural as this group is likely heterogeneous in disease etiology. Hence, profiling metabolic subtypes is of interest for personalized clinical research.
4.4. Strengths and limitations
One strength of our study is that we used CSF biomarkers (Aβ and tau) to support the clinical diagnosis of AD and control subjects. This makes the metabolic alterations we describe more likely to be AD-specific. Moreover, with the semitargeted MS techniques referred here, we were able to integrate data of four different compound classes and to replicate many findings from other recent metabolite studies in AD.
We note that the different signatures are not completely concordant. This is explained by the different properties studied in each signature. A differential expression signature explores, for individual metabolites, shifts in distribution. A classification signature explores which conjunction of metabolites achieves an appreciable predictive performance. A regulatory signature, then, assesses which metabolites are central in the complex network of metabolite interactions. We pose the examination of multiple signatures as a strength as it uncovers metabolites of interest at the expression, prediction, and regulatory levels. Assessing only the differential expression signature, for example, would imply that many metabolites of interest would go unnoticed.
Among the potential limitations of the study is the relatively small sample size (n = 248) in comparison to the large number of metabolites studied (P = 230). However, we used novel statistical methods designed to account for high numbers of variables with limited case numbers. Moreover, we used nonfasting plasma samples, although nutritional intake and medications are known to influence metabolite levels [6], [49]. However, we corrected our results for multiple medication classes.
4.5. Future directions
Peripheral changes in metabolism in AD are of interest because it could highlight factors that are influential in the disease process on a systemic level. The regulatory signature might be of added value to explore metabolic dysregulation. However, these results are explorative and further work should focus on providing (dis)confirmation of hypotheses regarding (the effect of) network changes. Experimental studies are needed to establish if the found alterations in peripheral metabolism are related to the function of metabolites in the central nervous system of AD patients. In addition, effort should be directed to disentangle if these metabolic alterations are associated with AD-related risk factors and secondary changes (e.g., malnutrition, aging, diabetes) or with AD pathology. Moreover, integrating genomics and metabolomics could be of interest, as well as an in-depth study of the effect of patient-related and preanalytical variation in the metabolome. When its dynamics in terms of patient and preanalytical influences are fully understood, it can be a powerful tool for monitoring ongoing biology. Metabolites as identified in this study, such as for example tyrosine and 2-AAA, could then serve as biological effect-monitoring tools in clinical trials.
5. Conclusions
We show that peripheral metabolism is altered in AD patients compared with control subjects and between carriers and noncarriers of the APOE ε4 allele. Moreover, we show the added value of not only studying metabolic expression signatures, but to paint the full picture of metabolic change by also exploring classification and regulatory signatures. These additional signatures can highlight possible prediction and regulatory markers that may be overlooked when studying expression signatures alone. The consistent elements over all signatures are the changes in the metabolism of amino acids and markers of oxidative stress. In particular, the amino acid tyrosine and the oxidative stress compound LPA C18:2 were identified as possible key players of metabolic change. This is in concordance with the previous literature describing disturbances of the tyrosine pathway in AD and oxidized lipoproteins as oxidative stressors in the AD brain [36], [37], [42]. Further research is needed to validate these results and to further specify their role in AD-specific metabolic alteration.
Research in Context.
-
1.
Systematic review: Molecular aberrations tend to be amplified along the omics cascade. Hence, there is increasing interest in finding biomarkers for Alzheimer's disease (AD) in peripheral fluids such as plasma. Present study adds to a small body of the literature on potential metabolite markers stemming from plasma.
-
2.
Interpretation: Our data are used in a systematic effort to find differential expression, classification, and network deregulation signatures that demarcate AD from control cases. These signatures point to certain amines and oxidative stress markers as drivers behind AD-related metabolic deregulation.
-
3.
Future directions: The results hold promise for the development of a biomarker panel. Further studies are warranted for replication and panel development.
Acknowledgments
This research was supported by Janssen Pharmaceuticals Stellar Initiative: Stellar Neurodegeneration Collaboration Project, Call 2, No. 3 (An Integrated MetaboloMic, Epidemiologic and genetic approach to DIscover clinically relevant biomarkers for Alzheimers Disease: IMMEDIAD). Research of the VUmc Alzheimer center is part of the neurodegeneration research program of Amsterdam Neuroscience. The VUmc Alzheimer center is supported by Alzheimer Nederland and Stichting VUmc fonds. The clinical database structure was developed with funding from Stichting Dioraphte. F.A.d.L is appointed at the NWO-FCB project NUDAD (project number 057-14-004).
Footnotes
Disclosures: F.A.d.L, C.F.W.P., M.I.K., A.C.H., E.A.S., H.W.T.v.V., S.J.v.d.L., M.A.v.d.W. report no relevant conflicts of interest. T.H. and C.M.v.D. work on the CoSTREAM project (http://www.costream.eu/), funded by the European Union's Horizon 2020 research and innovation programme (grant agreement 667375). A.D. and C.M.v.D. are members of PRECEDI Marie Curie exchange programme. A.D. is supported by a Veni grant (2015) from ZonMw. P.S. has received grant support (for the institution) from GE Healthcare, Danone Research, Piramal and MERCK. In the past 2 years he has received consultancy/speaker fees (paid to the institution) from Lilly, GE Healthcare, Novartis, Forum, Sanofi, Nutricia. W.M.v.d.F. has been an invited speaker at Boehringer Ingelheim. Research programs of W.M.v.d.F. have been funded by ZonMW, NWO, EU-FP7, Alzheimer Nederland, CardioVascular Onderzoek Nederland, Stichting Dioraphte, Gieskes-Strijbis fonds, Boehringer Ingelheim, Piramal Neuroimaging, Roche BV, Janssen Stellar, Combinostics. All funding is paid to her institution. C.E.T. serves on the advisory board of Fujirebio and Roche, performed contract research for IBL, Shire, Boehringer, Roche and Probiodrug; and received lecture fees from Roche and Axon Neurosciences.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2017.07.006
Contributor Information
Francisca A. de Leeuw, Email: f.deleeuw@vumc.nl.
Carel F.W. Peeters, Email: cf.peeters@vumc.nl.
Supplementary data
References
- 1.Scheltens P., Blennow K., Breteler M.M.B., de Strooper B., Frisoni G.B., Salloway S. Alzheimer's disease. Lancet. 2016;388:505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 2.Cervellati C., Wood P.L., Romani A., Valacchi G., Squerzanti M., Maria Sanz J.M. Oxidative challenge in Alzheimer's disease: state of knowledge and future needs. J Investig Med. 2016;64:21–32. doi: 10.1136/jim-2015-000017. [DOI] [PubMed] [Google Scholar]
- 3.Garciía-Mesa Y., Colie S., Corpas R., Cristòfol R., Comellas F., Nebrada A.R. Oxidative stress is a central target for physical exercise neuroprotection against pathological brain aging. J Gerontol Biol Sci Med Sci. 2015;71:40–49. doi: 10.1093/gerona/glv005. [DOI] [PubMed] [Google Scholar]
- 4.Alcolea D., Martínez-Lage P., Sánchez-Juan P., Olazarán J., Antúnez C., Izagirre A. Amyloid precursor protein metabolism and inflammation markers in preclinical Alzheimer disease. Neurology. 2015;85:626–633. doi: 10.1212/WNL.0000000000001859. [DOI] [PubMed] [Google Scholar]
- 5.Guerreiro R., Hardy J. Genetics of Alzheimer's disease. Neurotherapeutics. 2014;11:732–737. doi: 10.1007/s13311-014-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes E., Wilson I., Nicholson J. Metabolic phenotyping in health and disease. Cell. 2014;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Koek M.M., Jellema R.H., van der Greef J., Tas A.C., Hankemeier T. Quantitative metabolomics based on gas chromatography mass spectrometry: status and perspectives. Metabolomics. 2011;7:307–328. doi: 10.1007/s11306-010-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrijvers E., Witteman J., Sijbrands E., Hofman A., Koudstaal P., Breteler M. Insulin metabolism and the risk of Alzheimer's disease: the Rotterdam Study. Neurology. 2010;75:1982–1987. doi: 10.1212/WNL.0b013e3181ffe4f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahley R., Rall S. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 10.Horsburgh K., McCarron M., White F., Nicoll J. The role of apolipoprotein E in Alzheimer's disease, acute brain injury and cerebrovascular disease: evidence on common mechanisms and utility in animal models. Neurobiol Aging. 2000;21:245–255. doi: 10.1016/s0197-4580(00)00097-x. [DOI] [PubMed] [Google Scholar]
- 11.Trushina E., Dutta T., Persson X.M.T., Mielke M.M., Peterson R.C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimers disease using metabolomics. PLoS One. 2013;8:e63644. doi: 10.1371/journal.pone.0063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong Y., Liu Y., Zhou L., Di X., Li W., Li Q. A UHPLC-TOF/MS method based metabonomic study of total ginsenosides effects on Alzheimer disease mouse model. J Pharm Biomed Anal. 2015;115:174–182. doi: 10.1016/j.jpba.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 13.González-Domínguez R., García-Barrera T., Vitorica J., Gómez-Ariza J.L. Metabolomic investigation of systemic manifestations associated with Alzheimer's disease in the APP/PS1 transgenic mouse model. Mol Biosyst. 2015;11:2429–2440. doi: 10.1039/c4mb00747f. [DOI] [PubMed] [Google Scholar]
- 14.Graham S.F., Chevallier O.P., Elliott C.T., Hölscher C., Johnston J., McGuinness B. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L-arginine metabolism in mild cognitive impairment subjects converting to Alzheimers disease. PLoS One. 2015;10:e0119452. doi: 10.1371/journal.pone.0119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis B., Hye A., Snowden S.G. Metabolic modifications in human biofluids suggest the involvement of sphingolipid, antioxidant, and glutamate metabolism in Alzheimer's disease pathogenesis. J Alzheimers Dis. 2015;46:313–327. doi: 10.3233/JAD-141899. [DOI] [PubMed] [Google Scholar]
- 16.O'Bryant S., Mielke M., Rissman R., Lista S., Vanderstichele H., Zetterberg H., the Biofluid Based Biomarker Professional Interest Area Blood-based biomarkers in Alzheimer disease: current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimers Dement. 2017;13:45–58. doi: 10.1016/j.jalz.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Flier W.M., Pijnenburg Y.A., Prins N., Lemstra A.W., Bouwman F.H., Teunissen C.E. Optimizing patient care and research: the Amsterdam Dementia Cohort. J Alzheimers Dis. 2014;41:313–327. doi: 10.3233/JAD-132306. [DOI] [PubMed] [Google Scholar]
- 18.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimers disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimers Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 19.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H. The diagnosis of dementia due to Alzheimers disease: recommendations from the National Institute on Aging-Alzheimers Association workgroups on diagnostic guidelines for Alzheimers disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jongbloed W., Kester M.I., van der Flier W.M., Veerhuis R., Scheltens P., Blankenstein M.A. Discriminatory and predictive capabilities of enzyme-linked immunosorbent assay and multiplex platforms in a longitudinal Alzheimer's disease study. Alzheimers Dement. 2013;9:276–283. doi: 10.1016/j.jalz.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Duits F.H., Teunissen C.E., Bouwman F.H., Visser P.J., Mattsson N., Zetterberg H. The cerebrospinal fluid “Alzheimer profile”: easily said, but what does it mean? Alzheimers Dement. 2014;10:713–723. doi: 10.1016/j.jalz.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Hu C., van Dommelen J., van der Heijden R., Spijksma G., Reijmers T.H., Wang M. RPLC-ion-trap-FTMS method for lipid profiling of plasma: method validation and application to p53 mutant mouse model. J Proteome Res. 2008;7:4982–4991. doi: 10.1021/pr800373m. [DOI] [PubMed] [Google Scholar]
- 23.Noga M.J., Dane A., Shi S., Attali A., van Aken H., Suidgeest E. Metabolomics of cerebrospinal fluid reveals changes in the central nervous system metabolism in a rat model of multiple sclerosis. Metabolomics. 2012;8:253–263. doi: 10.1007/s11306-011-0306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Kloet F.M., Bobeldijk I., Verheij E.R., Jellema R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J Proteome Res. 2009;8:5132–5141. doi: 10.1021/pr900499r. [DOI] [PubMed] [Google Scholar]
- 25.van Buuren S., Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J Stat Softw. 2011;45:67. [Google Scholar]
- 26.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 27.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Series B Stat Methodol. 1996;58:267–288. [Google Scholar]
- 28.Bilgrau AE, Peeters CFW, Eriksen PS, Bøgsted M, van Wieringen WN. Targeted fused ridge estimation of inverse covariance matrices from multiple high-dimensional data classes. 2015;arXiv:1509.07982v1 [stat.ME].
- 29.Folstein M.F., Folstein S.E., McHugh P.R. Mini-mental state: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 30.Goeman J.J., van de Geer S.A., de Kort F., van Houwelingen J.C. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 31.Hintze J.L., Nelson R.D. Violin plots: a box plot-density trace synergism. Am Stat. 1998;52:181–184. [Google Scholar]
- 32.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fruchterman T.M.J., Reingold E.M. Graph drawing by force-directed placement. Softw Pract Exp. 1991;21:1129–1164. [Google Scholar]
- 34.Proitsi P., Kim M., Luke W., Simmons A., Sattlecker M., Velayudhan L. Association of blood lipids with Alzheimer's disease: a comprehensive lipidomics analysis. Alzheimers Dement. 2017;13:140–151. doi: 10.1016/j.jalz.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Sharma A., Bemis M., Desilets A. Role of medium chain triglycerides (Axona©) in the treatment of mild to moderate Alzheimer's disease. Am J Alzheimers Dis Other Demen. 2014;5:409–414. doi: 10.1177/1533317513518650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.González-Domínquez R., García-Barrera T., Gómez-Ariza J.L. Metabolite profiling for the identification of altered metabolic pathways in Alzheimer's disease. J Pharm Biomed Anal. 2015;107:75–81. doi: 10.1016/j.jpba.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Kadurrah-Daouk R., Zhu H., Sharma S., Bogdanov M., Rozen S.G., Matson W., Pharmacometabolomics Research Network Alterations in metabolic pathways and networks in Alzheimer's disease. Transl Psychiatry. 2013;3:e244. doi: 10.1038/tp.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cools R., D'Esposito M. Inverted-U-shaped dopamine actions on human working memory and cognitive control. Biol Psychiatry. 2011;69:e113–e125. doi: 10.1016/j.biopsych.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hase A., Jung S.E., aan het Rot M. Behavioral and cognitive effects of tyrosine intake in healthy human adults. Pharmacol Biochem Behav. 2015;133:1–6. doi: 10.1016/j.pbb.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Gao L., Zackert W., Hasford J., Danekis M., Milne G., Remmert C. Formation of prostaglandisn E2 and D2 via the isoprostane pathway: a mechanism for the generation of bioactive prostaglandins independent of cyclooxygenase. J Biol Chem. 2003;1:28479–28489. doi: 10.1074/jbc.M303984200. [DOI] [PubMed] [Google Scholar]
- 41.Teunissen C., de Vente J., Steinbusch H., de Bruijn C. Biochemical markers related to Alzheimer's dementia in serum and cerebrospinal fluid. Neurobiol Aging. 2002;23:485–508. doi: 10.1016/s0197-4580(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 42.Draczynska-Lusiak B., Doung A., Sun A.Y. Oxidized lipoproteins may play a role in neuronal cell death in Alzheimer disease. Mol Chem Neuropathol. 1998;33:139–148. doi: 10.1007/BF02870187. [DOI] [PubMed] [Google Scholar]
- 43.Shi J., Dong Y., Cui M., Xu X. Lysophosphatidic acid induces increased BACE1 expression and Aβ formation. Biochim Biophys Acta. 2015;1832:29–38. doi: 10.1016/j.bbadis.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yung Y., Stoddard N., Mirendil H., Chun J. Lysophosphatidic acid signaling in the nervous system. Neuron. 2015;85:669–682. doi: 10.1016/j.neuron.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayas C.L., Moreno-Flores M.T., Avila J., Wandosell F. The neurite retraction induced by lysophosphatidic acid increases Alzheimer's disease-like Tau phosphorylation. J Biol Chem. 1999;247:37046–37052. doi: 10.1074/jbc.274.52.37046. [DOI] [PubMed] [Google Scholar]
- 46.Teo S.T., Yung Y.C., Herr D.R., Chun J. Lysophosphatidic acid in vascular development and disease. IUBMB Life. 2009;61:791–799. doi: 10.1002/iub.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siess W., Zangl K.J., Essler M., Bauer M., Brandl R., Corrinth C. Lysophosphatidic acid mediates the rapid activation of platelets and endothelial cells by mildly oxidized low density lipoprotein and accumulates in human atherosclerotic lesions. Proc Natl Acad Sci U S A. 1999;96:6931–6936. doi: 10.1073/pnas.96.12.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi K., Takahashi M., Nishida W., Yoshida K., Ohkawa Y., Kitabatake A. Phenotypic modulation of vascular smooth muscle cells induced by unsaturated lysophosphatidic acids. Circ Res. 2001;89:251–258. doi: 10.1161/hh1501.094265. [DOI] [PubMed] [Google Scholar]
- 49.Carayol M., Licaj I., Achaintre D., Sacerdote C., Vineis P., Key T.J. Reliability of serum metabolites over a two-year period: a targeted metabolomic approach in fasting and non-fasting samples from EPIC. PLoS One. 2015;10:e0135437. doi: 10.1371/journal.pone.0135437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.