Abstract
Agropyron cristatum is a wild grass of the tribe Triticeae and serves as a gene donor for wheat improvement. However, very few markers can be used to monitor A. cristatum chromatin introgressions in wheat. Here, we reported a resource of large-scale molecular markers for tracking alien introgressions in wheat based on transcriptome sequences. By aligning A. cristatum unigenes with the Chinese Spring reference genome sequences, we designed 9602 A. cristatum expressed sequence tag-sequence-tagged site (EST-STS) markers for PCR amplification and experimental screening. As a result, 6063 polymorphic EST-STS markers were specific for the A. cristatum P genome in the single-receipt wheat background. A total of 4956 randomly selected polymorphic EST-STS markers were further tested in eight wheat variety backgrounds, and 3070 markers displaying stable and polymorphic amplification were validated. These markers covered more than 98% of the A. cristatum genome, and the marker distribution density was approximately 1.28 cM. An application case of all EST-STS markers was validated on the A. cristatum 6 P chromosome. These markers were successfully applied in the tracking of alien A. cristatum chromatin. Altogether, this study provided a universal method of large-scale molecular marker development to monitor wild relative chromatin in wheat.
Introduction
Wild relatives of crops contain broad genetic diversity and serve as gene donors to crops1. In the tribe Triticeae, Agropyron cristatum is a wild relative of wheat and an elite gene donor for wheat improvement. In addition to its resistance to drought and cold and its moderate tolerance to salinity2–4, A. cristatum carries many useful traits, including resistance to barley yellow dwarf, wheat streak mosaic virus5, leaf rust6, and powdery mildew7 as well as large spike phenotype8, making it a powerful genetic reservoir for the improvement of wheat6,9.
Due to the abundance of diversified genetic resources of A. cristatum, wide crosses between common wheat and A. cristatum have been conducted to transfer desirable traits into common wheat6,8,10–16. Introducing A. cristatum alien chromosomes into common wheat depends on cytological genetic technology and molecular markers to track the alien chromosome fragment. In general, C-banding and genomic in situ hybridization (GISH) analyses are powerful cytological techniques for the detection of alien chromatin in wheat backgrounds17. However, these classic detection methods rely on complex experimental procedures and the use of fluorescence microscopes. Conversely, molecular markers can be detected rapidly and operated easily using only thermal cyclers to produce high throughput. Until now, due to the lack of genomic information, only limited molecular markers exist for A. cristatum. Only a few sequence-tagged site (STS) markers, sequence characterized amplified region (SCAR) markers from repeat sequences, random amplified polymorphic DNA (RAPD) markers and amplified fragment length polymorphism (AFLP) markers have been reported in A. cristatum 18–20. Chen et al. (1994) used a set of assigned wheat random fragment length polymorphism (RFLP) probes for detecting and establishing the homoeology of alien A. cristatum chromosomes19. Wu et al. (2010) reported three SCAR markers specific to the P genome repeat sequences of A. cristatum originating from a RAPD assay18. Luan et al. (2010) reported several 6P chromosome-specific STS markers using the suppression subtractive hybridization method16. Han et al. (2015) used the degenerate oligonucleotide-primed (DOP)-PCR products of microdissected 6PS as a fluorescence in situ hybridization (FISH) probe to identify P genome chromatin in wheat-A. cristatum introgression lines, but the method relied on cytological techniques21.
The large quantity of translocation lines carrying alien P chromatin of A. cristatum have been produced by irradiation or Aegilops gametocidal chromosome methods in our lab14–16. Hence, the heavy abundance of identification tasks requires the development of large amounts of molecular markers to efficiently track A. cristatum alien chromosome fragments in a wheat background. The goal of this study was to develop large-scale molecular markers for tracking A. cristatum alien introgressions in a wheat background based on transcriptome sequences. This study provides a comprehensive molecular marker resource and presents a universal application case of molecular marker development in wide crosses based on transcriptome sequencing technology.
Results
Sequence alignment analysis and primer design
We previously reported that more than 90 million transcriptome reads from the flag leaf and young spike tissues of the representative tetraploid A. cristatum accession No. Z559 were assembled into 73664 unigenes22. Considering a 93.8% average identity between A. cristatum unigenes and wheat transcripts, unigenes with high identity were removed to increase the polymorphic rate of primer screening. Hence, we first conducted the alignment analysis using the Basic Local Alignment Search Tool for nucleotides (BLASTn) on the 73664 unigenes with the Triticum aestivum cDNA database and 5X CS wheat genomic DNA sequences23, after which the remaining unigenes that had less than 85% identity were used for primer design according to the outline shown in Fig. 1. Finally, we selected 9602 unigenes that had low identity for primer design. All primer names and sequences are provided in Supplementary Table 1. Most primers were designed by the high-throughput web application platform BatchPrimer3, using the generic primer method and the default parameters. A few primers were designed manually that exhibited insertion-deletion (InDel) variation between A. cristatum unigenes and wheat transcripts. The workflow of the expressed sequence tag-STS (EST-STS) marker development is shown in Fig. 1.
Figure 1.
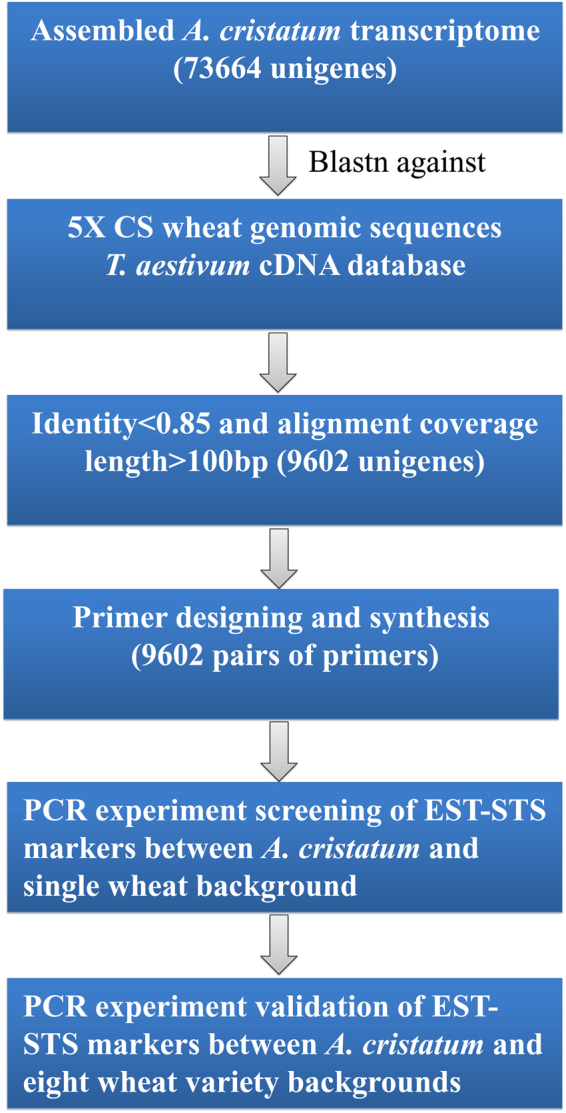
The workflow of the EST-STS marker development.
PCR experiment screening of EST-STS markers specific for the P genome in the single-receipt wheat background
We selected A. cristatum Z559 as a positive control and the common wheat parent Fukuho (from the original wide cross) as a negative control for the experimental screening of specific primers of the P genome in the wheat background. First, all 9602 primer pairs were amplified in two control DNA templates, and polymorphic PCR product bands were distinguished on non-denaturing polyacrylamide gels. Two types of markers were screened: dominant and co-dominant markers. The dominant markers amplify only within the A. cristatum template, whereas co-dominant markers amplify polymorphic products in both the A. cristatum and wheat templates. As a result, 6063 polymorphic markers were specific for the A. cristatum genome (Supplementary Table 2). The screening rate reached 64.5%. More than 30% of the markers were functionally annotated using the NCBI nonredundant (nr) protein database, but the other 70% had no homologous genes in any database. Two thousand ninety-five markers without annotation belonged to predicted long intergenic noncoding RNAs (lincRNAs) (Supplementary Table 2). We previously showed that these lincRNAs are specific to A. cristatum as opposed to other Triticeae species, including wheat, rye and barley22. Hence, these 2095 markers will be useful for identifying the A. cristatum P genome among diversified Triticeae grasses.
To evaluate the coverage rate and probable distribution density of these markers on A. cristatum chromosomes, a virtual chromosome location map was constructed by anchoring these markers onto the wheat population sequencing (POPSEQ) genetic map24. A total of 1951 EST-STS markers were successfully anchored onto the wheat POPSEQ genetic map (Supplementary Table 3). These markers are covered more than 98% of the A. cristatum genome, and the marker distribution density was 1.28 cM (Table 1). The virtual distribution map suggests these markers have high density coverage on A. cristatum chromosomes. Using the wheat POPSEQ genetic map, we isolated 584 EST-STS markers as conserved loci between the P genome and the A, B, and D genomes, which will be useful for the homologous identification of A. cristatum alien chromosomes of different addition lines.
Table 1.
Virtual chromosome location of EST-STS markers of A. cristatum in the reference wheat genome
| Homologous groups | Chromosome | Number of conserved markers | Marker coverage rate | Marker density (cM/marker) |
|---|---|---|---|---|
| 1 | 1A(106),1B(123),1D(105) | 74 | 100% | 1.43 |
| 2 | 2A(146),2B(178),2D(140) | 104 | 98% | 0.83 |
| 3 | 3A(69),3B(158),3D(99) | 53 | 98% | 1.88 |
| 4 | 4A(92),4B(111),4D(59) | 42 | 96% | 1.29 |
| 5 | 5A(77),5B(143),5D(146) | 63 | 99% | 1.48 |
| 6 | 6A(240),6B(191),6D(263) | 177 | 99% | 0.64 |
| 7 | 7A(118),7B(110),7D(195) | 71 | 97% | 1.40 |
The number within parentheses denotes the number of markers located on the corresponding wheat chromosome.
Validation of EST-STS markers specific for the P genome in multiple wheat backgrounds
To validate the specificity of EST-STS markers for the P genome of A. cristatum in wheat genetic backgrounds and remove false-positive markers, we collected eight domestic and international common wheat varieties, including Fukuho, Chinese Spring (CS), Gaocheng 8901, Xiaoyan 6, Hi-Line, CMH83.605, McGuire, and Lovrin 10, for the further experimental validation. The wheat varieties that have high genetic variation served as negative controls to eliminate the false-positive disturbance from the wheat background. Accounting for 1B/1R translocations distributed widely throughout commercial varieties25,26, we also increased the representative variety Lovrin 10 that has the 1B/1R translocation for detection. A total of 4956 markers with polymorphism between A. cristatum and Fukuho were randomly selected for validation. As a result, 3070 EST-STS markers that exhibited stable amplification and clear PCR product bands were screened. The screening rate reached 61.9%. The highly stringent markers are provided in Supplementary Table 4. Based on these screening results, twenty-one markers were amplified in both A. cristatum and the Lovrin 10 1B/1R translocation line. These EST-STS markers included AgC5106, AgC12260, AgC14448, AgC23390, AgC23424, AgC24347, AgC24610, AgC24754, AgC24816, AgC25726, AgC26732, AgC30508, AgC31937, AgC36971, AgC49315, AgC50266, AgC50467, AgC52107, AgC53511, AgC54696 and AgC71190, which could be detected in both 1B/1R translocation and A. cristatum. We suggested that the 21 EST-STS markers should be ruled out in the detection of wheat backgrounds with the 1B/1R translocation.
EST-STS marker screening specific for A. cristatum chromosomes from addition lines
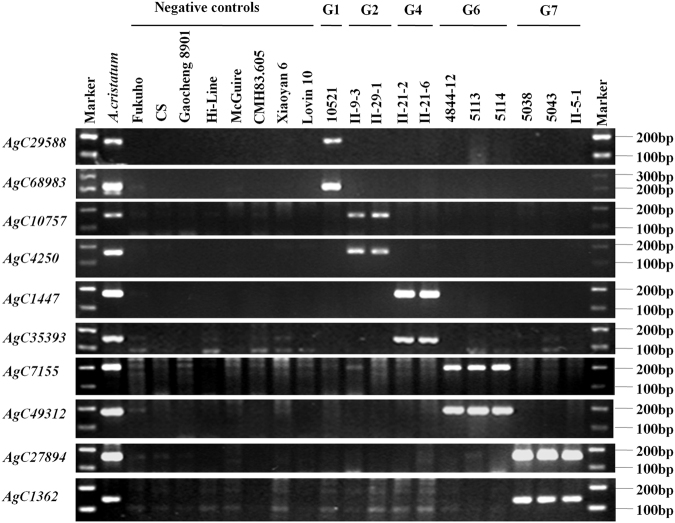
To screen the EST-STS markers specific for single chromosomes of A. cristatum in the wheat genetic background, we collected from our lab 11 addition lines, including 5 homologous groups, for screening the specific marker for P chromosomes (Table 2). Two markers were screened for each chromosome, including chromosomes 1P, 2P, 4P, 6P and 7P, from the addition lines. These markers were highly amplified in both A. cristatum and the addition lines, whereas no amplification was observed in the eight wheat varieties that have broad genetic diversity. Among these markers, AgC29588 and AgC68083 were specific for chromosome 1P, AgC10757 and AgC4250 were specific for chromosome 2P, AgC1447 and AgC35393 were specific for chromosome 4P, AgC7155 and AgC49312 were specific for chromosome 6P, and AgC27894 and AgC1362 were specific for chromosome 7P (Fig. 2). These markers will be very useful for distinguishing the specific chromosomes in wide crosses of common wheat and A. cristatum.
Table 2.
Plant materials used in this study
| Material name | Chromosome composition | Type of material | Homologous group of wild relative | Origin country |
|---|---|---|---|---|
| Z559 | 2n = 4X = 28P | A. cristatum | China | |
| 10521 | 2n = 6X = 42W + 2P | addition line | 1P | China |
| II-9-3 | 2n = 6X = 42W + 2P | addition line | 2P | China |
| II-29-1 | 2n = 6X = 42W + 2P | addition line | 2P | China |
| II-21-2 | 2n = 6X = 42W + 2P | addition line | 4P | China |
| II-21-6 | 2n = 6X = 42W + 2P | addition line | 4P | China |
| 4844-12 | 2n = 6X = 42W + 2P | addition line | 6P | China |
| 5113 | 2n = 6X = 42W + 2P | addition line | 6P | China |
| 5114 | 2n = 6X = 42W + 2P | addition line | 6P | China |
| 5038 | 2n = 6X = 42W + 2P | addition line | 7P | China |
| 5043 | 2n = 6X = 42W + 2P | addition line | 7P | China |
| II-5-1 | 2n = 6X = 42W + 2P | addition line | 7P | China |
| Fukuho | 2n = 6X = 42W | wheat landrace | Japan | |
| Chinese Spring | 2n = 6X = 42W | wheat landrace | China | |
| Gaocheng 8901 | 2n = 6X = 42W | wheat landrace | China | |
| Xiaoyan 6 | 2n = 6X = 42W | commercial wheat variety | China | |
| Hi-Line | 2n = 6X = 42W | commercial wheat variety | USA | |
| CMH83.605 | 2n = 6X = 42W | commercial wheat variety | CIMMYT | |
| McGuire | 2n = 6X = 42W | commercial wheat variety | USA | |
| Lovrin 10 | 2n = 6X = 42W | 1BL.1RS translocation | Romania |
Figure 2.
The screening of EST-STS markers specific for A. cristatum addition lines in a wheat genetic background. The amplification product of EST-STS markers were run on 2% agarose gel. G1, G2, G4, G6, and G7 indicate the homologous groups of alien chromosomes of A. cristatum. The full-length gels are included in the Supplementary Information file.
EST-STS marker validation in A. cristatum chromosome 6P
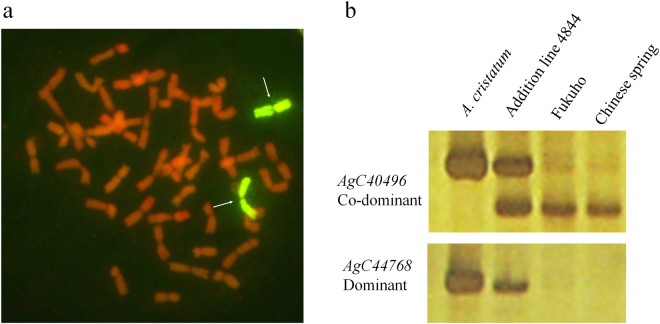
To validate the screening rate and efficiency of EST-STS markers in wheat backgrounds, we selected chromosome 6P, which carries a high number of grain-related traits, as a validation case8. We screened all EST-STS markers on chromosome 6P of addition line 4844-12. All 9602 markers were screened among the four DNA samples, including A. cristatum, 6P addition line 4844-12, Fukuho (the receiving parent of the wide cross) and Chinese Spring. GISH detection first proved addition line 4844-12 contained a pair of A. cristatum chromosome 6P (Fig. 3a). The experimental screening was then performed for all 9602 pairs of primers. PCR amplification band types of EST-STS markers were divided into co-dominant and dominant types (Fig. 3b). For example, the co-dominant marker AgC40496 amplified two different bands from A. cristatum and common wheat, whereas the dominant marker AgC44768 only amplified one band in A. cristatum and none in wheat (Fig. 3b). Finally, after high-stringency experiment screening, 680 EST-STS markers specific for chromosome 6P were screened from 9602 primers, resulting in a 7.08% screening rate (Supplementary Table 5). These markers have been widely applied for monitoring chromosome 6P segments in addition lines, deletion lines and translocation lines27–30.
Figure 3.
Agropyron cristatum chromosome 6P-specific EST-STS marker screening in a wheat background. (a) GISH detection showed 21 pairs of wheat chromosomes and one pair of 6P chromosomes in addition line 4844-12. The arrowheads indicate the addition of chromosome 6P by the green fluorescent signal. (b) The amplification product of EST-STS markers was run on 8% non-denaturing polyacrylamide gel. Agropyron cristatum and addition line 4844-12 are positive controls, and wheat varieties Fukuho and Chinese Spring are negative controls.
To evaluate the probable order of the 680 EST-STS markers on chromosome 6P, the homologous relationships of the 680 EST-STS markers on chromosome 6P were evaluated using BLASTn against wheat and barley genomes (Table 3). Three hundred fifty-nine and 259 EST-STS markers of A. cristatum chromosome 6P were anchored onto wheat and barley chromosomes, respectively. The highest percentages of anchored markers were 30.29% and 23.09% in the sixth homologous group of wheat and barley, respectively. According to the wheat POPSEQ genetic map of the A, B and D subgenomes24, three virtual genetic maps were also constructed (Supplementary Figure 1). Two hundred forty-one unique EST-STS markers were anchored onto chromosomes 6A, 6B and 6D, and 38 EST-STS markers were conserved among chromosomes 6P, 6A, 6B and 6D. The results indicate a good synteny relationship between chromosome 6P and chromosomes 6A, 6B or 6D. Hence, we confirmed the application value of all A. cristatum EST-STS markers in the detection of alien chromosomes in a wheat background.
Table 3.
Comparative analysis of homologous relationships between A. cristatum and wheat and barley chromosomes based on the alignment of A. cristatum 6P-specific EST-STS markersa.
| Species | Homologous groups | Number of anchored unigenes | Average identity (%) | Average coverage length (bp) | Percent of anchored markers |
|---|---|---|---|---|---|
| Wheat | 1A,1B,1D | 20 | 89.6 | 251 | 2.94% |
| 2A,2B,2D | 18 | 89.6 | 211 | 2.65% | |
| 3A,3B,3D | 30 | 88.4 | 243 | 4.41% | |
| 4A,4B,4D | 19 | 87.1 | 260 | 2.79% | |
| 5A,5B,5D | 30 | 90.1 | 258 | 4.41% | |
| 6A,6B,6D | 206 | 91.4 | 335 | 30.29% | |
| 7A,7B,7D | 36 | 88.5 | 256 | 5.29% | |
| No hits | 321 | 47.21% | |||
| Barley | 1H | 17 | 89.4 | 184 | 2.50% |
| 2H | 15 | 88.6 | 165 | 2.21% | |
| 3H | 24 | 90.6 | 207 | 3.53% | |
| 4H | 11 | 89.0 | 216 | 1.62% | |
| 5H | 17 | 88.4 | 170 | 2.50% | |
| 6H | 157 | 91.3 | 286 | 23.09% | |
| 7H | 18 | 89.1 | 189 | 2.65% | |
| No hits | 421 | 61.91% |
aBLASTn alignments were conducted with an e-value threshold less than 1e-10 and an alignment coverage length of more than 100 bp.
Discussion
Traditionally, the detection of alien chromosomes has mainly depended on chromosome banding and GISH techniques17. However, the two methods require complex instruments and expensive reagents. Molecular markers provide a rapid, reliable, easy, high-throughput method for detecting the alien chromosome segments. We previously showed that repeat sequence markers can detect A. cristatum alien chromosomes in a common wheat background18,21. However, repeat sequence markers are distributed throughout a whole chromosome and cannot distinguished specific chromosome fragment loci. EST-STS markers have been effective in similar studies involving Haynaldia villosa, Secale cereale, and Psathyrostachys huashanica 31–33. These markers successfully tracked specific chromosomes or homologous groups. In the present study, we demonstrated the successful development of large-scale EST-STS markers based on transcriptome sequences for tracking A. cristatum chromosome fragments in a wheat background. These markers originated from native transcriptome sequences of A. cristatum and showed three main characteristics: (i) They are monomorphic and can be tracked to specific chromosome segments; (ii) They have an estimated average marker coverage density of 98% of the A. cristatum genome; and (iii) They consist of native markers from the A. cristatum transcriptome. Among these EST-STS markers, 6063 can be used in the identification of wide crosses in the single-receipt Fukuho parent background, and 3070 A. cristatum-specific markers in eight wheat variety backgrounds can be applied to molecular marker-assisted selection of A. cristatum introgressions into commercial varieties. In the present study, these markers have been applied successfully for the physical mapping of 2P, 6P and 7P alien chromatin fragments in A. cristatum–wheat introgression lines27,30,34,35.
We previously showed that 4831 A. cristatum unigenes were predicted as lincRNAs and were A. cristatum specific compared with other Triticeae species22. In the present study, 2095 markers were predicted to be lincRNAs and accounted for 43% of all A. cristatum-specific lincRNAs. The results imply that lincRNAs should be the first choice for marker development and have the advantage of species specificity for monitoring wild relative chromatin in wheat backgrounds. Taken together, this study reported a universal method of large-scale molecular marker development for monitoring wild relative chromatin in the common wheat background based on transcriptome sequences.
In the study of wide crosses, identifying the homologous group of alien chromosomes from the Triticeae tribe is a major challenge. Successful identification leads to the effective selection and utilization of homoeologous recombination translocation lines between wheat and wild relatives36. In the present study, we used 6P addition line 4844-12 as a case for large-scale marker development of chromosome 6P specifically in a common wheat background and for homologous group identification. A total of 680 high-quality 6P-specific markers in the wheat genetic background provided a rapid detection platform for chromosome engineering applications, and these markers were used to track the alien 6P fragment of A. cristatum in the common wheat background. The positive marker screening rate in addition line 4844-12 was close to 1/14 (680/9602≈1/14) rather than 1/7, suggesting that the A. cristatum tetraploid in our study is not an autotetraploid but rather a segmental allotetraploid, which was in accordance with the results of a previous study37. After comparing 680 sequences with wheat draft genomes, a good synteny relationship was observed between A. cristatum chromosome 6P and wheat chromosomes 6A, 6B and 6D. The present study also supports the previous cytogenetic results regarding the replacement of chromosome 6D of wheat with chromosome 6P of A. cristatum in wheat–A. cristatum chromosome substitution lines8. The study provides good prospective applications for the homologous group identification of alien chromosomes in addition lines of other wild relatives based on transcriptome sequences.
Methods
Plant materials
The tetraploid A. cristatum accession No. Z559 was used as the donor parent for a wide cross between wheat and A. cristatum in our previous study10. The tetraploid A. cristatum (2n = 4X = 28 P) Z559 population was also selected for de novo transcriptome sequencing in our previous study22. Common wheat varieties including Fukuho, Chinese Spring, Gaocheng 8901, Xiaoyan 6, Hi-Line, CMH83.605, McGuire, and Lovrin 10 served as negative controls and wheat genetic backgrounds for screening P genome-specific markers. Among these varieties, Fukuho was the original maternal parent of the wide cross between common wheat and A. cristatum, Chinese Spring and Gaocheng 8901 were Chinese landraces, and Xiaoyan 6 was the commercial variety of China. Hi-Line, CMH83.605, McGuire and Lovrin 10 were introduced varieties from other parts of the world. Disomic addition lines of wheat and A. cristatum were selected for A. cristatum chromosome-specific marker screening in a wheat background. These addition lines include 10521 (homologous group 1), II-9-3 and II-29-1 (homologous group 2), II-21-2 and II-21-6 (homologous group 4), 4844-12, 5113 and 5114 (homologous group 6), and 5038, 5043 and II-5-1 (homologous group 7). Plant material information is listed in Table 2.
Sequence analysis and primer design
The assembled 73664 unigenes of the A. cristatum transcriptome22 were used for alignment analysis by BLASTn against 5X CS wheat genome raw data sequences23 and the T. aestivum cDNA database (ftp://ftp.ensemblgenomes.org/pub/release-23/plants/fasta/). After removing high-identity sequences, we selected 9602 unigenes that had a nucleotide identity less than 85% for primer design to increase the polymorphism of nucleotide sequences. The large-scale resource of primer pairs was designed using the BatchPrimer3 website (http://probes.pw.usda.gov/cgi-bin/batchprimer3/batchprimer3.cgi) with the following parameters: product length of 100–300 bp, primer size of 18–24 bp (20 bp optimum) and melting temperature between 57 and 63 °C (60 °C optimum)38. A small amount of primers were designed manually that exhibited InDel variation between A. cristatum unigenes and wheat transcripts. Detailed primer lists are shown in Supplementary Table 1.
DNA extraction, PCR experiments and STS marker screening
DNA was extracted from young leaf tissue at the seedling stage using cetyltrimethylammonium bromide (CTAB) extraction buffer according to published methods39. PCR experiments were performed in a 10-μL reaction volume containing 10X PCR buffer, 0.2 mmol/L dNTP, 2.5 mmol/L Mg2+, 0.25 μmol/L primer, 0.6 U of Taq DNA polymerase, and 50 ng of template DNA. The PCR cycling parameters consisted of one cycle at 94 °C for 3 min, 35 cycles at 94 °C for 1 min, the appropriate annealing temperature for 1 min (depending on the individual primer pair), and 72 °C for 1 min; and a final extension for 7 min at 72 °C. PCR products were electrophoresed on 8% non-denaturing polyacrylamide gels (Acr/Bis = 19:1) and detected by the silver staining method40 or on 2% agarose gels. The DNA template from A. cristatum Z559 and wheat–A. cristatum disomic addition lines served as positive controls, and the DNA template from the other eight common wheat varieties served as negative controls. The primers were screened for polymorphisms according to the size of PCR amplification products.
Anchoring EST-STS markers on wheat chromosome map
Anchoring A. cristatum EST-STS markers onto the wheat chromosome map was performed by BLASTn alignment of A. cristatum unigene sequences against the Chinese Spring reference genome in accordance with the wheat POPSEQ genetic map of the A, B and D subgenomes24 and the barley H genome41. High-stringency alignment was considered when the statistics of an e-value threshold less than 1e-10 and an alignment coverage length greater than 100 bp occurred.
Functional annotation of EST-STS markers
The functional annotation of A. cristatum EST-STS markers was performed in our previous study using the NCBI nr protein sequences, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database, the Pfam domain database and lincRNAs22.
GISH analysis
GISH detection in addition line 4844-12 was performed in root tip cells using A. cristatum genomic DNA as a probe. Wheat genomic DNA from the receiving parent Fukuho was used for blocking. Root tip preparation and hybridization procedures were performed as described by Han et al. (2004)42.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China 2016YFD0100102 and 2016YFD0102000.
Author Contributions
L.L.L. and Z.J.P. conceived the experiment(s); Z.J.P., L.W.H., L.Y.Q., L.Q.X., Y.X.M. and L.X.Q. conducted the experiment(s); and Z.J.P. analyzed the results and wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12219-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ford-Lloyd BV, et al. Crop Wild Relatives-Undervalued, Underutilized and under Threat? BioScience. 2011;61:559–565. doi: 10.1525/bio.2011.61.7.10. [DOI] [Google Scholar]
- 2.Dewey, D. R. The genomic system of classification as a guide to intergeneric hybridization with the perennial TriticeaeinGene Manipulation in Plant Improvement. 16th Stadler Genetics Symposium. (ed. Gustafson, J. P.), 209–279 (Plenum 1984).
- 3.Asay KH, Johnson DA. Genetic Variances for Forage Yield in Crested Wheatgrass at Six Levels of Irrigation. Crop Sci. 1990;30:79–82. doi: 10.2135/cropsci1990.0011183X003000010018x. [DOI] [Google Scholar]
- 4.Limin AE, Fowler DB. Cold hardiness of forage grasses grown on the Canadian prairies. Can. J. Plant Sci. 1987;67:1111–1115. doi: 10.4141/cjps87-150. [DOI] [Google Scholar]
- 5.Sharma HC, Gill BS, Uyemoto JK. High levels of resistance in Agropyron species to barley yellow dwarf and wheat streak mosaic viruses. Journal of Phytopathology. 1984;110:143–147. doi: 10.1111/j.1439-0434.1984.tb03402.x. [DOI] [Google Scholar]
- 6.Ochoa V, Madrid E, Said M, Rubiales D, Cabrera A. Molecular and cytogenetic characterization of a common wheat-Agropyron cristatum chromosome translocation conferring resistance to leaf rust. Euphytica. 2014;201:89–95. doi: 10.1007/s10681-014-1190-5. [DOI] [Google Scholar]
- 7.Li Q, et al. Characterization of a Novel Wheat-Agropyron cristatum 2P Disomic Addition Line with Powdery Mildew Resistance. Crop Sci. 2016;56:2390–2400. doi: 10.2135/cropsci2015.10.0638. [DOI] [Google Scholar]
- 8.Wu J, et al. The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. TheorAppl Genet. 2006;114:13–20. doi: 10.1007/s00122-006-0405-0. [DOI] [PubMed] [Google Scholar]
- 9.Dong YS, et al. Desirable characteristics in perennial Triticeae collected in China for wheat improvement. Hereditas. 1992;116:175–178. doi: 10.1111/j.1601-5223.1992.tb00819.x. [DOI] [Google Scholar]
- 10.Li LH, Yang XM, Li XQ, Dong YC, Chen XM. Introduction of desirable genes from Agropyron cristatum into common wheat by intergeneric hybridization. SciAgric Sin. 1998;31:1–6. [Google Scholar]
- 11.Jubault M, et al. Attempts to induce homoeologous pairing between wheat and Agropyron cristatum genomes. Genome. 2006;49:190–193. doi: 10.1139/g05-074. [DOI] [PubMed] [Google Scholar]
- 12.Martín A, et al. A fertile amphiploid between diploid wheat (Triticum tauschii) and crested wheat grass (Agropyron cristatum) Genome. 1999;42:519–524. doi: 10.1139/gen-42-3-519. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Jahier J, Cauderon Y. Intergeneric hybrids between Triticum aestivum and three crested wheatgrasses: Agropyron mongolicum, A. michnoi, and A. desertorum. Genome. 1990;33:663–667. doi: 10.1139/g90-098. [DOI] [Google Scholar]
- 14.Liu WH, et al. Production and identification of wheat - Agropyron cristatum(1.4P) alien translocation lines. Genome. 2010;53:472–481. doi: 10.1139/G10-023. [DOI] [PubMed] [Google Scholar]
- 15.Song L, et al. Efficient induction of Wheat-Agropyron cristatum 6P translocation lines and GISH detection. PLoS One. 2013;8:e69501. doi: 10.1371/journal.pone.0069501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan Y, et al. Production and identification of wheat-Agropyron cristatum 6P translocation lines. Planta. 2010;232:501–510. doi: 10.1007/s00425-010-1187-9. [DOI] [PubMed] [Google Scholar]
- 17.Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS. Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica. 1996;91:59–87. doi: 10.1007/BF00035277. [DOI] [Google Scholar]
- 18.Wu M, et al. Cloning and characterization of repetitive sequences and development of SCAR markers specific for the P genome of Agropyron cristatum. Euphytica. 2010;172:363–372. doi: 10.1007/s10681-009-0033-2. [DOI] [Google Scholar]
- 19.Chen Q, Lu YL, Jahier J, Bernard M. Identification of wheat-Agropyron cristatum monosomic addition lines by RFLP analysis using a set of assigned wheat DNA probes. TheorAppl Genet. 1994;89:70–75. doi: 10.1007/BF00226985. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Li X, Ma Y, Yu Z, Li Z. A genetic linkage map of crested wheatgrass based on AFLP and RAPD markers. Genome. 2012;55:327–335. doi: 10.1139/g2012-014. [DOI] [PubMed] [Google Scholar]
- 21.Han H, Zhang Y, Liu W, Hu Z, Li L. Degenerate Oligonucleotide Primed-Polymerase Chain Reaction-Based Chromosome Painting of P Genome Chromosomes in Agropyron cristatum and Wheat-A. cristatum Addition Lines. Crop Science. 2015;55:2798–2805. doi: 10.2135/cropsci2015.03.0183. [DOI] [Google Scholar]
- 22.Zhang J, et al. De novo transcriptome sequencing of Agropyron cristatum to identify available gene resources for the enhancement of wheat. Genomics. 2015;106:129–136. doi: 10.1016/j.ygeno.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson PA, et al. CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinformatics. 2012;13:219. doi: 10.1186/1471-2105-13-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The International Wheat Genome Sequencing Consortium. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788 (2014). [DOI] [PubMed]
- 25.Lukaszewski AJ. Frequency of 1RS.1AL and 1RS.1BL Translocations in United States Wheats. Crop Science. 1990;30:1151–1153. doi: 10.2135/cropsci1990.0011183X003000050041x. [DOI] [Google Scholar]
- 26.Xu X, et al. Inheritance of 1BL/1RS of the core parent Lovrin 10 in its derivative varieties. Journal of Triticeae Crops. 2010;30:221–226. [Google Scholar]
- 27.Song L, et al. Physical mapping of Agropyron cristatum chromosome 6P using deletion lines in common wheat background. TheorAppl Genet. 2016;129:1023–1034. doi: 10.1007/s00122-016-2680-8. [DOI] [PubMed] [Google Scholar]
- 28.Dai C, et al. Development of EST markers specific to Agropyron cristatum chromosome 6P in common wheat background. Acta Agronomica Sinica. 2012;38:1791–1801. doi: 10.3724/SP.J.1006.2012.01791. [DOI] [Google Scholar]
- 29.Han H, et al. Genetic rearrangements of six wheat-Agropyron cristatum 6P addition lines revealed by molecular markers. PLoS One. 2014;9:e91066. doi: 10.1371/journal.pone.0091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, et al. Introgression of Agropyron cristatum 6P chromosome segment into common wheat for enhanced thousand-grain weight and spike length. TheorAppl Genet. 2015;128:1827–1837. doi: 10.1007/s00122-015-2550-9. [DOI] [PubMed] [Google Scholar]
- 31.Zhao RH, et al. Development of EST-PCR Markers for the Chromosome 4V of Haynaldia villosa and Their Application in Identification of 4V Chromosome Structural Aberrants. Journal of Integrative Agriculture. 2014;13:282–289. doi: 10.1016/S2095-3119(13)60359-7. [DOI] [Google Scholar]
- 32.Wang CM, et al. Development and Application of EST-STS Markers Specific to Chromosome 1RS of Secale cereale. Cereal Research Communications. 2009;37:13–21. doi: 10.1556/CRC.37.2009.1.2. [DOI] [Google Scholar]
- 33.Du W, et al. Molecular cytogenetic identification of a wheat–Psathyrostachys huashanica Keng 5Ns disomic addition line with stripe rust resistance. Molecular Breeding. 2013;31:879–888. doi: 10.1007/s11032-013-9841-0. [DOI] [Google Scholar]
- 34.Li H, et al. Production and Identification of Wheat-Agropyron cristatum 2P Translocation Lines. PLoS One. 2016;11:e0145928. doi: 10.1371/journal.pone.0145928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu M, et al. Transferring Desirable Genes from Agropyron cristatum 7P Chromosome into Common Wheat. PLoS ONE. 2016;11:e0159577. doi: 10.1371/journal.pone.0159577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi L, Friebe B, Zhang P, Gill BS. Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res. 2007;15:3–19. doi: 10.1007/s10577-006-1108-8. [DOI] [PubMed] [Google Scholar]
- 37.Schulz-Schaeffer J, Allderdice PW, Creel GC. Segmental Allopolyploidy in Tetraploid and Hexaploid Agropyron Species of the Crested Wheatgrass Complex (Section Agropyron) Crop Science. 1963;3:525–530. doi: 10.2135/cropsci1963.0011183X000300060021x. [DOI] [Google Scholar]
- 38.You FM, et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protocols. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 40.Bassam BJ, Caetano-Anolles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-I. [DOI] [PubMed] [Google Scholar]
- 41.The International Barley Genome Sequencing Consortium. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716 (2012). [DOI] [PubMed]
- 42.Han F, Liu B, Fedak G, Liu Z. Genomic constitution and variation in five partial amphiploids of wheat-Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. TheorAppl Genet. 2004;109:1070–6. doi: 10.1007/s00122-004-1720-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.