Abstract
Present-day knowledge on the regulatory biology of denitrification is based on studies of selected model organisms. These show large variations in their potential contribution to NO2−, NO, and N2O accumulation, attributed to lack of genes coding for denitrification reductases, but also to variations in their transcriptional regulation, as well as to post-transcriptional phenomena. To validate the relevance of these observations, there is a need to study a wider range of denitrifiers. We designed an isolation protocol that identifies all possible combinations of truncated denitrification chains (NO3−/NO2−/NO/N2O/N2). Of 176 isolates from two soils (pH 3.7 and 7.4), 30 were denitrifiers sensu stricto, reducing NO2− to gas, and five capable of N2O reduction only. Altogether, 70 isolates performed at least one reduction step, including two DNRA isolates. Gas kinetics and electron flow calculations revealed that several features with potential impact on N2O production, reported from model organisms, also exist in these novel isolates, including denitrification bet-hedging and control of NO2−/NO/N2O accumulation. Whole genome sequencing confirmed most truncations but also showed that phenotypes cannot be predicted solely from genetic potential. Interestingly, and opposed to the commonly observed inability to reduce N2O under acidic conditions, one isolate identified as Rhodanobacter reduced N2O only at low pH.
Introduction
The past 150 years have experienced a massive anthropogenic input of reactive nitrogen to the biosphere, leading to a global acceleration of microbial nitrogen transformations (van Groenigen et al., 2015; Stokstad, 2016). One of the major processes is denitrification, by which NO3− and/or NO2− is reduced to gaseous forms of nitrogen. Complete denitrification of NO3− to N2 involves four reduction steps. These are catalyzed by four main types of functional enzymes, the nitrate reductases Nar or Nap; the NO-generating nitrite reductases NirK or NirS; the N2O generating nitric oxide reductases cNor, qNor, or qCuNor; and the two types of nitrous oxide reductases N2OR (Zumft, 1997; Shapleigh, 2013). A denitrifying organism may contain both nitrate reductases, but generally only one type of the nitrite, nitric oxide, and nitrous oxide reductases. Organisms lacking one or more of the denitrification steps occur frequently in the environment (Shapleigh, 2013; Graf et al., 2014), but physiological studies of truncated denitrifiers are scarce, yet they demonstrate prominent physiological and regulatory differences (Bergaust et al., 2008; Roco et al., 2016). Denitrification is primarily performed by facultative anaerobic bacteria, and has received substantial interest during the last decades due to its economic and environmental implications, reducing the amount of N available to the crops, and acting as the main source of the greenhouse gas N2O (Schlesinger, 2008), which is also the most important destructor of the stratospheric ozone layer (Portmann et al., 2012).
Attempts to correlate nosZ gene abundance, sequence diversity and/or transcript numbers with N2O emissions from soils show variable results (Rich and Myrold, 2004; Morales et al., 2010; Jones et al., 2014; Liu et al., 2014; Domeignoz-Horta et al., 2015), demonstrating that these parameters cannot alone predict the propensity of a soil microbial community to emit N2O. The root of the problem may be that denitrification genes in soil are not necessarily transcribed in response to anoxia, and that transcription does not necessarily lead to the production of functional enzymes. The current knowledge on the regulatory biology of denitrification is based on studies of only a few model organisms. Denitrification is widely spread among taxonomically diverse groups of bacteria and more fundamental understanding is needed from a wider range of organisms before we may, eventually, use metagenomics to predict the propensity of soils to emit N2O.
Many groups of denitrifying bacteria have as yet no cultivable representatives available in biobanks (Zumft, 1997; Janssen et al., 2002). Previous isolation efforts have been conducted under anoxic conditions in the presence of NO3− (Gamble et al., 1977; Cheneby et al., 2000; Heylen et al., 2006; Hashimoto et al., 2009). Denitrifying bacteria may, however, be unable to grow if exposed to sudden anoxia, as, if they do not have enough time to express the denitrification reductases, they may be entrapped in anoxia without energy to produce the enzymes (Højberg et al., 1997; Hassan et al., 2014). This will limit the isolates to the ones that manage such abrupt shifts. Moreover, the early isolation studies used full-strength media, which have been shown to inhibit a large fraction of viable soil bacteria (Olsen and Bakken, 1987). Even more importantly, the previous attempts to isolate denitrifying bacteria were not inclusive because: (1) they have not isolated organisms without NAR, as nitrite was generally not provided; (2) they have not isolated organisms with only N2OR; and (3) they have probably not captured organisms with only NIR since such organisms produce NO as end product and therefore will not survive in pure culture when exposed to anoxia. Moreover, NO was not measured in those studies. On the basis of this, and with the aim to isolate as broad a range of denitrifying bacteria, possessing different truncated denitrification pathways, we chose a two-pronged strategy. First, as it was also done in the study by Roco et al. (2016), we isolated the bacteria on low-nutrient agar plates under aerobic conditions, to allow the organisms to adapt to growth under these conditions before challenging them with a gradual transition to anaerobic respiration. Second, after re-streaking until pure cultures were obtained, single colonies were transferred to vials containing low-nutrient medium with NO3− and NO2−, and with micro-oxic headspace supplemented with N2O. This strategy, which was a development of the one by Roco et al. (2016), allowed us to efficiently identify bacteria performing all four denitrification steps, as well as bacteria with all possible combinations of truncated denitrification pathways (except those carrying only Nor), and those performing DNRA (dissimilatory reduction of NO3− via NO2− to NH4+).
We extracted bacteria from two agricultural peat soils of contrasting pH (pH 3.7 and pH 7.4) sampled from a long-term field experiment (Liu et al., 2010), where earlier studies have shown strong negative relationship between pH and N2O/N2 product ratio (Liu et al., 2010; Russenes et al., 2016), in accordance with findings from a wide range of other soils (Simek and Cooper, 2002; Cuhel et al., 2010; van den Heuvel et al., 2011; Bakken et al., 2012; Russenes et al., 2016). Recently, we have found that the two soils are also profoundly different regarding the transient accumulation of nitrite during anoxic incubations (an order of magnitude higher in the pH 7.4 soil), and that the low pH soil is indeed able to reduce N2O, albeit at a slower rate and much delayed compared to the high pH soils (Lim, Bakken and Frostegård, unpublished). We hypothesized that we would isolate comparable numbers of denitrifiers from both soils and that truncated denitrification chains would be common. We also expected that detailed phenotypic analyses would reveal a variety of phenotypes, not always reflecting the genomic potential of the organisms, and with examples of ‘Rapid Complete Onset (RPO)’ and ‘Progressive Onset (PO)’ of denitrification (Liu et al., 2013), as well as ‘bet hedging’, that is, that only a fraction of the cells switch to denitrification (Hassan et al., 2014; 2016). Furthermore, we hypothesized to find some organisms in the low pH soil that were capable of reducing N2O at low pH.
Materials and methods
Soil samples and isolation of bacteria
Soils were collected from a long-term research field site at Fureneset Rural Development Centre located in Fjaler on the western coast of Norway, where plots (8 × 12.5 m) had been limed to different pH levels about 40 years ago (Sognnes et al., 2006). Two soils of contrasting pH were used; one from an untreated plot (pHH2O 3.7, in the following referred to as ‘low pH soil’) and one from a plot that had received 800 m3 shell sand ha−1 (pHH2O 7.4, in the following referred to as ‘high pH soil’). The samples were sieved (4 mm mesh size) and kept at 4 °C until use (1–3 months).
Approximately 20 g of each soil were diluted 1:10 in sterile distilled water and blended in an Omni Mixer Homogenizer (Omni) for 3 × 1 min, speed 7. The homogenization was carried out on ice to prevent heating. The soil homogenates were then left to settle for 10 min, allowing the bigger aggregates to sediment. Appropriate dilutions of the supernatants were plated onto 1/10 TSA medium (Tryptic Soy Agar, Merck, Darmstadt, Germany), containing 1.5 g l−1 casein peptone and 0.5 g l−1 soy peptone, supplemented with 100 mg l−1 cycloheximide (Fluka, Steinheim, Germany) to prevent fungal growth. Bacteria from the high pH soil (series A) were plated on medium with pH 7.3. Bacteria from the low pH soil were divided in two series, in order to capture as many types of bacteria as possible. Series B was plated on 1/10 TSA medium of pH 7.3 and series C was plated on 1/10 TSA adjusted to pH 5.7 using 20 mM phosphate buffer and 1.4 mM sulfuric acid. The same buffering system was used for both pH levels, therefore the low pH was set to 5.7. The plates were incubated under oxic conditions at 22±2 °C for two weeks, and inspected regularly to detect newly appearing colonies. Colonies were picked from dilutions that gave around 50 colonies per plate, generally the 10−6 dilution. Single, well-spaced colonies were selected based on morphology (size, shape, surface structure, and color), aiming at obtaining as large a variation of bacterial isolates as possible, and streaked to purity on new plates with the same medium. Total viable counts were performed at the end of the incubation, after 14 days. Pure cultures were preserved at −80 °C in TSB medium (Tryptic Soy Broth, Merck) containing 15% glycerol, until further analysis. The total number of bacteria in the two soils was determined using microscopic direct counts after SybrGreen staining (Noble and Fuhrman, 1998).
Endpoint analyses for determination of phenotypes
The experimental setup was designed to identify complete or partial denitrifiers, as well as nitrate reducers and organisms performing DNRA (Figure 1). Three out of four electron acceptors used in nitrate reduction (NO3−) and denitrification (NO2−, N2O) were provided. Bacterial cultures were raised from frozen stocks and inoculated into serum vials containing 4 ml of 1/10 TSB medium supplemented with 1 mM NaNO3 and 1 mM NaNO2. The pH of the medium was 7.3 for the A and B series. For series C the medium was adjusted to pH 5.7 (as described above). The cultures were incubated with vigorous shaking to secure oxic conditions, thus preventing denitrification. When visible turbidity was reached, the vials were crimp-sealed with rubber septa and made anoxic by repeated evacuation and helium filling. Pure O2 (1% v/v) and pure N2O (1% v/v) was added to the headspace and the cultures were incubated at room temperature for 10 or 21 days (isolates from high and low pH soils, respectively). At the end of the incubation, headspace samples were taken to quantify NO, using a chemiluminescence NO analyser (Sievers NOA), while N2O and N2 were quantified by gas chromatography (Molstad et al., 2007). Concentrations of NO3− and NO2− were determined by measuring the amount of NO produced by the reaction with vanadium (III) chloride in HCl at 95 °C (NO2−+NO3−), and the reaction with sodium iodide in acetic acid at room temperature (NO2− only), using the purger system coupled to the Sievers Nitric oxide analyser NOA280i (Cox, 1980; Braman and Hendrix, 1989). Samples in which nitrogen imbalance was detected were investigated for ammonium concentration using a colorimetric hypochlorite method according to Bower and Holm-Hansen (1980).
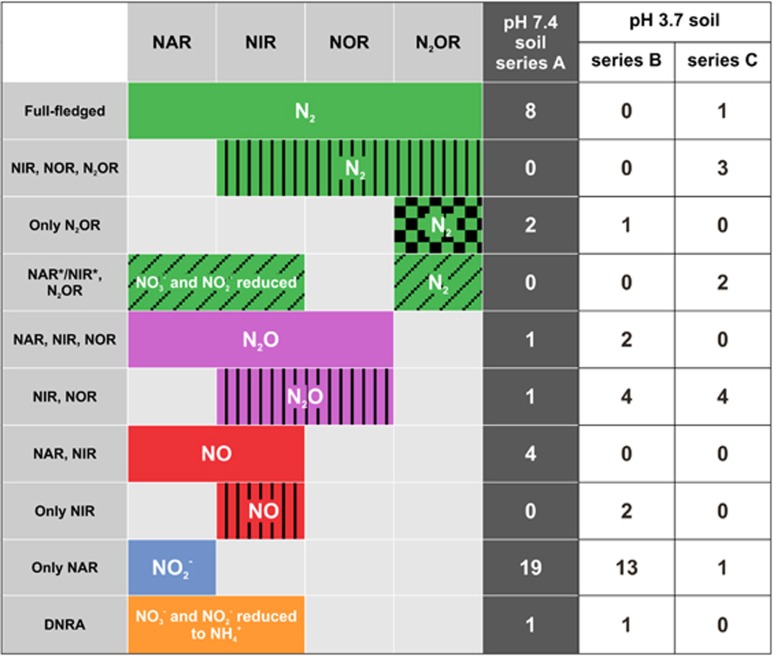
Figure 1.
Phenotypic classification of new soil isolates able to perform at least one step in the dissimilatory reduction of NO3−, NO2−, NO, and N2O. The phenotypes were designated as NAR, NIR, NOR, and N2OR. The end points (N2, N2O, NO, NO2−, or NH4+) are indicated inside the colored bars. DNRA designates dissimilatory nitrate reduction to ammonium. A total of 176 bacterial isolates were investigated; 99 from a soil of pHH2O 7.4 (dark gray) and 77 from a soil of pHH2O 3.7 (white). Bacteria from the low pH soil were isolated either on medium of pH 7.3 (Series B) or pH 5.7 (Series C). The color codes that identify the different phenotypes are used to indicate the phenotype of the isolates in the phylogenetic trees presented in Figures 2 and 3. NAR*/NIR*, N2OR indicates organisms which reduced NO3−/NO2−, but with no corresponding gases detected in the end-point analysis. End-point ammonium measurements did not classify these organisms as DNRA. They were, however, capable of reduction of externally provided N2O.
Phylogenetic analysis
Bacterial isolates that carried out at least one of the steps in the reduction of NO3− to N2 were taxonomically classified based on their 16S rRNA gene sequences. Each isolate was cultivated in 1/10 TSB medium, DNA was extracted and purified using standard procedures (QIAamp DNA Mini Kit, Qiagen, Hilden, Germany) and the 16S rRNA gene was amplified using universal eubacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Baker et al., 2003). Purified PCR products (Omega, Norcross, GA, USA) were Sanger sequenced (GATC, Germany) and a phylogenetic analysis of these and 16S rRNA genes from closely related isolates retrieved from the NCBI database, was performed based on the neighbor-joining method (Interior-branch test, number of bootstrap replications=500, site coverage cutoff=95%) using the MEGA version 6 (Tamura et al., 2013).
Whole genome sequencing
A total of 12 isolates displaying complete or partial denitrification were genome-sequenced by MiSeq (Illumina, San Diego, CA, USA). Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen), and samples were processed using the Nextera XT sample preparation kit. Sequencing was done using the MiSeq reagent kit v3 for 300 × 2 paired end sequencing. Adapters and bar codes were trimmed from the sequence reads before further preprocessing by Trimmomatic (quality threshold 20). Assembly was done using SPAdes with contig length set to 500 and minimum read coverage of two. Draft genomes were uploaded to the RAST server (http://rast.nmpdr.org/, (Aziz et al., 2008) for identification of open reading frames, and scrutinized for genes encoding the nitrogen oxide reductases. Gene annotations were confirmed through BLASTp and the presence of conserved active site motifs. The selected isolates included four full-fledged denitrifiers as determined by end-point analysis, and 8 isolates lacking the last step of the denitrification pathway (N2O to N2), thus potentially contributing to N2O emission. One isolate of special interest, which utilizes N2O as sole electron acceptor during anaerobic growth, was also investigated.
Denitrification regulatory phenotypes
Denitrification Regulatory Phenotypes (DRP) of selected isolates were determined following Bergaust et al. (2011) and Liu et al. (2013). Pre-cultures were raised from frozen stocks that were inoculated into vials containing 1/10 TSB medium adjusted to pH 7.3 or pH 5.7 (for high and low pH isolates, respectively) and incubated with vigorous stirring until they reached the optical density (OD, 660 nm) of 0.1. This approach ensured that fully oxic conditions were maintained during pre-culturing, thus preventing expression of nitrogen oxide reductases prior to the experiment. When OD660 reached 0.1, 1 ml portions of the aerobic cultures were inoculated into new, gas tight 120 ml vials in triplicates containing 50 ml sterile medium (same pH as the respective pre-cultures) with 2 mM NO3− for the complete denitrifiers and 1 mM NO2− for those lacking nitrate reductase. The air in the headspace had been replaced with He and 1% (v/v) O2. The cultures were incubated with vigorous stirring at 23 °C in a robotized incubation system enabling the quantification of O2, CO2, NO, N2O and N2 in the headspace of respiring cultures (Molstad et al., 2007) but with a modified GC system (Qu et al., 2014). Frequent gas sampling from headspace during the transition from aerobic respiration to denitrification provided high-resolution gas kinetics which, coupled to NO2− and OD660 measurements, allowed detailed characterization of the denitrification phenotypes of the isolates under the given conditions.
Results
Total viable counts after 14 days of incubation were 4.71 × 107±8.62 × 106 (n=10) and 3.14 × 107± 3.06 × 106 (n=7) CFUs g−1 of wet soil for the high- and low pH soils, respectively. This accounted for ~0.4% and 0.3% of the total cell number, determined by epifluorescence microscopic counts (1.19 × 1010±6.15 × 109 cells g−1 soil for the high pH soil and 1.1 × 1010±4.1 × 109 cells g−1 soil for the low pH soil). Altogether, 176 isolates were obtained as pure cultures. Of these, 99 isolates were from the high pH soil (series A) and 77 isolates were from the low pH soil with 50 isolates in series B (isolated on medium with pH=7.3) and 27 isolates in series C (isolated on medium with pH=5.7). End-point analysis of NO3−, NO2−, NO, N2O, and N2 allowed the determination of a phenotype for each isolate able to perform at least one step in the reduction of NO3− to N2 (Figure 1). Isolates that could perform nitrate reduction, and/or at least one step of denitrification, were obtained in similar numbers from the low and the high pH soils, and comprised representatives of all possible truncations of the denitrification chain, except the phenotype ‘Only NOR’ which could not be detected by our isolation scheme.
Among the 99 isolates from the high pH soil, 36 could perform at least one of the reduction steps (Figure 1). For the low pH soil, this fraction was 44% (34 out of 77 isolates; 23 from the B series and 11 from the C series). There were all in all 30 isolates that qualified as denitrifiers sensu stricto, that is, with the capacity to reduce nitrite to NO, 14 from the high pH soil (A) and 16 from the low pH soil (B and C combined). Altogether, nine of these isolates were full-fledged denitrifiers (5.7% of the total 176 isolates), capable of reducing NO3− to N2. Of these, 8 were from the high-pH soil and only one was from the low-pH soil (series C). Sequencing of the 16S rRNA genes showed that all full-fledged denitrifiers were members of the Proteobacteria (Figures 2 and 3). Those that were isolated from the high pH soil represented the orders Burkholderiales, Pseudomonales and Rhizobiales, whereas the isolate from the low pH soil (isolate C04) belonged to Burkholderiales.
Figure 2.
Phylogenetic tree based on neighbor joining (MEGA 6, Tamura et al., 2013) of 16S rRNA gene sequences. The tree shows the relationship between the bacterial isolates from the high pH soil (series A), identified as having the capacity of dissimilatory reduction of at least one of the tested N oxides, and closely related isolates retrieved from the NCBI database. The color codes correspond to the ascribed phenotypes shown in Figure 1.
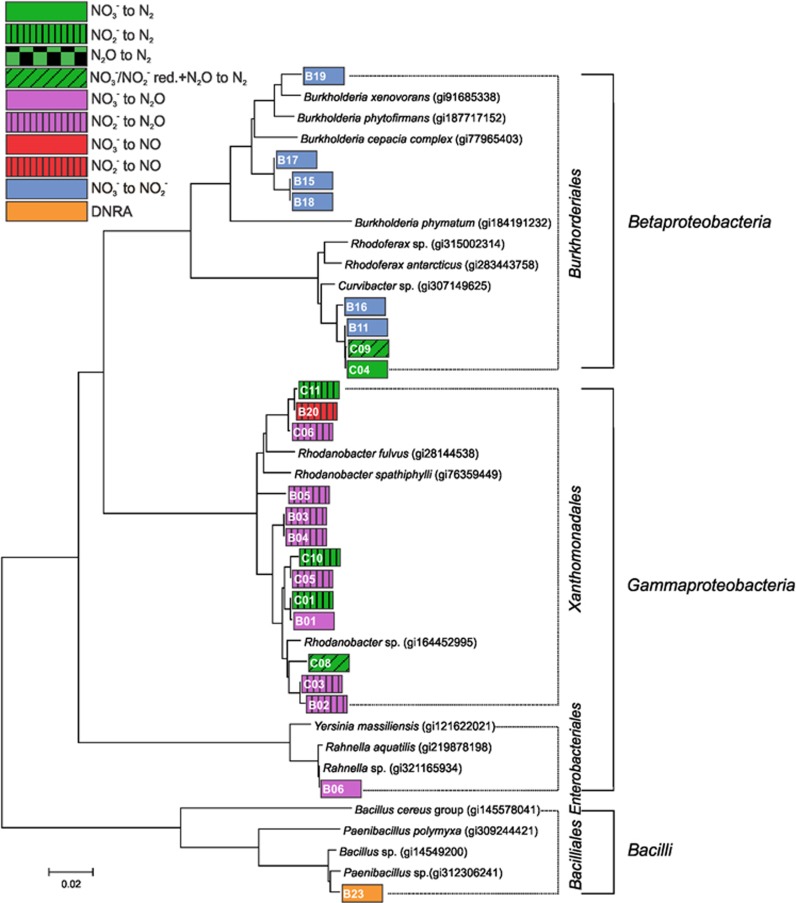
Figure 3.
Phylogenetic tree based on neighbor joining (MEGA 6, Tamura et al., 2013) of 16S rRNA gene sequences. The tree shows the relationship between the bacterial isolates from the low pH soil (series B and C), identified as having the capacity of dissimilatory reduction of at least one of the tested N oxides, and closely related isolates retrieved from the NCBI database. The color codes correspond to the ascribed phenotypes shown in Figure 1.
Twenty-one of the isolates classified as denitrifiers sensu stricto showed a truncated denitrification phenotype, meaning that they lacked the capacity to perform one or more of the reduction steps (Figure 1). Only three of these truncated denitrifiers were capable of reducing N2O. These isolates were unable to reduce NO3−, but reduced NO2− to N2 (phenotype ‘NIR, NOR, N2OR’). They were isolated from the low pH soil, series C, and classified as members of the genus Rhodanobacter (Figure 3). In addition to the twelve denitrifiers that were able to reduce N2O (9 full-fledged and the three truncated isolates), there were three isolates classified as ‘Only N2OR’ (two from series A and one from series B). One of those in series A was identified as belonging to the order Cytophagales (Figure 2). The other two with this phenotype could unfortunately not be recovered from long-term storage and were therefore not sequenced.
The phenotypes ‘NAR, NIR, NOR’ and ‘NIR, NOR’ were unable to perform the last step of denitrification, and thus emitted N2O as end product. Together, these two phenotype groups comprised 12 isolates (7%) of the total number of isolates (Figure 1). Three of them belonged to the ‘NAR, NIR, NOR’ phenotype, one coming from the high pH soil (order Burkholderiales; Figure 2) and two from the low pH soil (orders Xanthomonadales and Enterobacteriales, both series B; Figure 3). The ‘NIR, NOR’ phenotype was found for 8 isolates from the low pH soil (four from the B and four from the C series), and belonged to the order Xanthomonadales with most of them clustering with the genus Rhodanobacter (Figure 3). Only one isolate with this phenotype was isolated from the high pH soil (order Flavobacteriales; Figure 2).
The phenotype ‘NAR, NIR’, which reduced NO3− to NO but showed no production of N2O or N2, was found for four isolates from the high pH soil. These isolates accumulated 0.68–2.11 μM NO (concentration in the liquid), measured at the end of the incubation. Two of the isolates were members of the orders Burkholderiales and Bacilliales, respectively, while the two others were unrecoverable from storage and thus not sequenced. None of the isolates from the low pH soil showed this phenotype. Two isolates from the low pH soil (B20 and B21) exhibited the one-step reduction of NO2− to NO (‘Only NIR’ Figure 1), as seen from their accumulation of NO reaching up to 54.8 μM at the end of the incubation (Supplementary Materials,Supplementary Table S1). This value is based on the measured NO concentration in the headspace, assuming equilibrium between gas phase and liquid phase. One isolate (B20) in the ‘Only NIR’ group clustered together with the order Xanthomonadales and was identified as belonging to the genus Rhodanobacter, the other one could not be recovered from the long-term storage and was therefore not sequenced.
Bacteria capable of only NO3− reduction to NO2− (phenotype group ‘Only NAR’ Figure 1), and thus not considered ‘denitrifiers’ sensu stricto, comprised the largest group with 19% and 18% of the total number of isolated isolate in the high- and low pH soil, respectively. Interestingly, 13 out of the 14 ‘Only NAR’ isolates from the low pH soil were isolated on pH 5.7 medium (series C). The isolates from the high pH soil were taxonomically diverse, as determined from the 16S rRNA sequences. Most of them belong to Actinomycetales, but there are also representatives from Bacilliales, Flavobacteriales and Burkholderiales. Only six isolates were sequenced from the low pH soil, all of them belonging to Actinomycetales (Figures 2 and 3). Isolates for which the end-point analysis showed nitrogen imbalance, thus suggesting DNRA (seen as NO3− reduction, but no production of relevant gases), were investigated for NH4+ accumulation. On the basis of this, only one isolate from the high pH soil and one from the low pH soil (series B) were identified as DNRA organisms (classes Actinobacteria and Bacilli, respectively).
The presence or absence of all the relevant genes encoding N-oxide reductases was determined by whole genome sequencing of the 12 isolates that were phenotypically classified as capable of reducing NO3− or NO2− to N2O or N2 (thus phenotypes ‘Full-fledged’ NIR, NOR, N2OR and ‘NAR, NIR, NOR’, Figure 1), and one isolate capable of sole reduction of N2O to N2 (Figure 6). Of the seven sequenced isolates able to reduce NO3− to NO2−, four carried narG but not napA while one isolate, belonging to the genus Bradyrhizobium, carried napA but not narG. Thus, none of the isolates carried both these genes. In addition, three of the isolates lacking narG and napA carried nasA, which is another gene for nitrate reductase involved in nitrate assimilation (Luque-Almagro et al., 2011). The sequencing results confirmed the presence in all 12 denitrifiers of genes encoding the defining reaction of denitrification (NO2− reduction to NO), catalyzed by the product of a nir gene (Zumft, 1997). Approximately 10 of these isolates carried the nirK gene coding for a Cu containing nitrite reductase (Zumft, 1997). One Rhodanobacter isolate (C06) from the low pH soil had an ~240 bp larger nirK gene than the others, containing a cytochrome C domain at the 3' end. The nirS gene, encoding the cd1 nitrite reductase (Zumft, 1997), was identified in only two isolates from the high pH soil, representing the genera Hydrogenophaga and Pseudomonas. Genes encoding respiratory NO reductases were detected in all the isolates for which NO reduction to N2O was detected (12 out of the 13 sequenced isolates). As predicted from the phenotypic analyses, no nor genes were found in isolate A40 (Algoriphagus). Generally, the isolates carrying a respiratory nor had either the qnor or the cnor type, and these were mainly divided between the low and high pH soil isolates, respectively. The only exception was isolate C04 (genus Polaromonas), which in addition to two versions of qnor, had a large norB gene which was placed downstream of a gene annotated as cytochrome c oxidase polypeptide. The atypical norB gene encoded a protein apparently carrying a ~130 aa C-terminal extension as compared to the normal length NorB found in the Pseudomonas isolate (A25). The aa sequences of the cNor in CO4 cluster with proteins of similar size in Rhodanobacter species (NCBI BLASTp, 85 and 84% identities for putative NorC and NorB, respectively), but were not found within the present collection. Some of the other isolates from the low pH soil did however also have double copies of qnor. Nine genomes harbored a nosZ gene which encodes a nitrous oxide reductase (N2OR): eight of them were classified as nosZ clade I, and one was identified as belonging to nosZ clade II (Jones et al., 2013)—the isolate A40 assigned to the genus Algoriphagus. However, only six of these isolates exhibited N2OR activity under the conditions tested (pH 5.7 or pH 7.3 for isolates from low and high pH soil, respectively). Of the low pH soil isolates able to reduce N2O, one was identified as belonging to the genus Rhodanobacter (C01) while one belonged to Polaromonas (C04) (Figure 3).
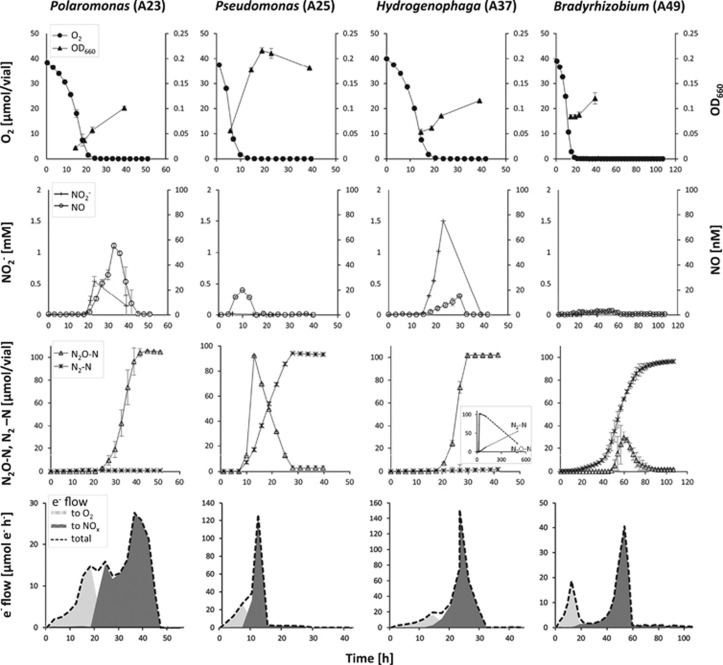
Denitrification regulatory phenotypes of selected isolates are presented in Figure 4 (high pH soil) and Figure 5 (low pH soil). Nitrate reduction was initiated as the O2 concentration in the medium reached ~5 μM for most isolates, but the Rhodanobacter isolates C01 and C05 started to denitrify at 16–17 μM O2, while the Rhodanobacter isolate C06 depleted virtually all O2 before any gas production could be detected. The isolates show clear differences in the accumulation of intermediates, revealing a large diversity in the regulation of denitrification in these organisms. Among the isolates from the high pH soil, an isolate of Hydrogenophaga (A37) accumulated NO2− as long as NO3− was present in the medium. In contrast a Bradyrhizobium isolate (A49) exhibited a rapid onset of all denitrification genes in response to decreasing O2 concentration, seen as an early increase in N2 with no detectable NO2−, and only transient and low accumulation of NO (maximum amount 7 nmol vial−1, corresponding to 5 nM concentration in the liquid) and N2O (maximum 30 μmol N vial−1). The N2O accumulation in the Bradyrhizobium culture never exceeded 40% of the total N provided, and at the end of the incubation all nitrogen was recovered as N2. Isolate A25, ascribed to the genus Pseudomonas, accumulated 100% of the provided N oxides as N2O before reducing this further to N2. Isolate A23, clustering with Polaromonas (family Comamonadaceae; order Burkholderiales) instead accumulated N2O as the end-product of denitrification with no detection of N2. The absence of a nosZ gene in this organism was confirmed by whole genome sequencing (Figure 6).
Figure 4.
Denitrification regulatory phenotypes of selected isolates from the high pH soil. Aerobically prepared bacterial cultures were inoculated into sealed 120 ml medical vials containing 50 ml 1/10 TSB medium of pH 7.3 supplemented with 2 mM KNO3. Headspace atmosphere after repeated helium flushing was adjusted with pure O2 to 1% concentration (v/v) prior inoculation. The vials were incubated at 22 °C with vigorous stirring. Gas kinetics (O2, NO, N2O, N2), NO2− concentrations and OD660 were monitored by frequent sampling from headspace and liquid phase, respectively. Error bars represent standard deviation for three replicates. Bottom graphs depict the calculated rates of electron flow to O2, nitrogen oxides (NOx) and the total electron flow.
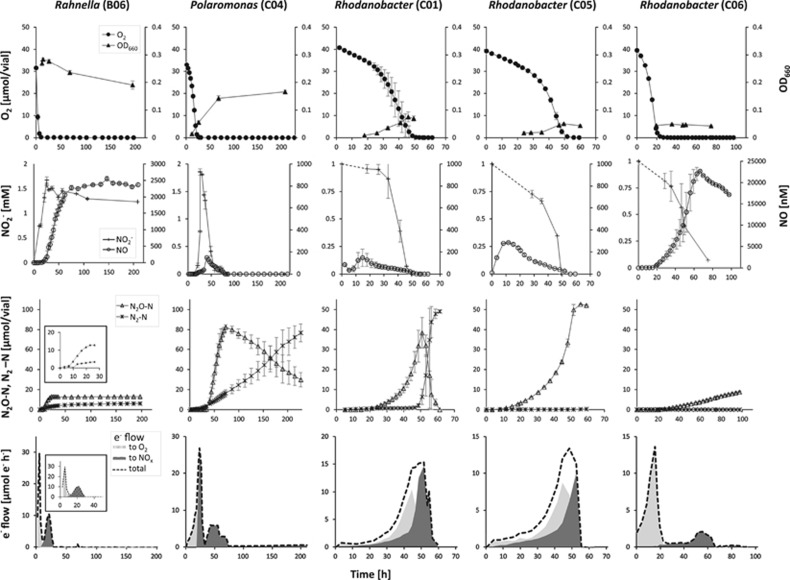
Figure 5.
Denitrification regulatory phenotypes of selected isolates from the low pH soil. Aerobically prepared bacterial cultures were inoculated into sealed 120 ml medical vials containing 50 ml 1/10 TSB medium of pH 5.7 supplemented with 2 mM KNO3 for isolates B06 and CO4, and 1 mM KNO2 for the Rhodanobacter isolates C01, C05, and C06. Headspace atmosphere after repeated helium flushing was adjusted with pure O2 to 1% initial concentration (v/v) prior inoculation. The vials were incubated at 22 °C with vigorous stirring. Gas kinetics (O2, NO, N2O, N2), NO2− concentrations and OD660 were monitored by frequent sampling from headspace and liquid phase, respectively. Error bars represent standard deviation for three replicates. Bottom graphs depict the calculated rates of electron flow to O2, nitrogen oxides (NOx) and the total electron flow.
Figure 6.
Genetic potential for denitrification. Whole genomes sequenced (MiSeq) and scrutinized for denitrification functional genes (RAST). *Large nirK containing 3’-localized cytochrome C domain. **Large norBC containing a cytochrome C domain. ***nosZ clade II. Genomes were deposited in NCBI, and are available under given accession numbers.
Five isolates from the low pH soil were whole genome sequenced (Figure 6). The denitrification regulatory phenotypes of four of these are shown in Figure 5. Three of them (isolates C01, C05, and C06), which were classified as members of the genus Rhodanobacter, were incapable of reducing NO3− but readily reduced NO2−. Genome analysis confirmed the absence of genes encoding dissimilatory nitrate reductase, and the presence of a nirK gene (Figure 6). The two other isolates, designated B06 and C04, started to respire NO3− when O2 concentrations became low (~10 μM in the liquid phase), but did not respire the NO2− produced until most, or all, of the NO3− was depleted. The isolate C04, for which the closest match was the genus Polaromonas within the order Burkholderiales, accumulated significant amounts of NO reaching a maximum of 225 nmol (vial−1), which corresponds to 0.15 μM in the liquid. Similar amounts of NO were detected in two of the Rhodanobacter cultures (C01 and C05). The Polaromonas isolate C04 accumulated up to 80% of the provided N oxyanions as N2O, before reducing it further to N2. The isolate B06, clustering with Rahnella (Figure 3) within the family Enterobacteriaceae, gradually accumulated NO until the concentration exceeded 2.5 μM in the liquid (3.7 μmol vial−1). Reduction of NO to N2O was detectable in the Rahnella culture, but came to a halt when the concentration of NO reached 0.5 μM, which apparently inhibited respiration, seen as inhibited CO2 production (Supplementary materials,Supplementary Figure S1). This was contrary to the Rhodanobacter isolate C06, which accumulated tenfold higher concentration of NO compared with Rahnella sp., but still continued respiration (seen as further reduction of NO, and CO2 evolution). All three investigated isolates of the genus Rhodanobacter were equipped with a nosZ clade I gene. Yet, all three accumulated N2O to different levels. The Rhodanobacter isolate C01, which was the only one capable of N2O reduction at pH 5.7, accumulated 80% of the provided nitrogen as N2O before reducing it further to N2. This was in contrast to the other two Rhodanobacter isolates, C05 and C06, which were unable to reduce N2O under the conditions tested. Of these, isolate C05 reduced 100% of the provided N to N2O in <60 h, whereas C06 only converted 9% of the N to N2O during 100 h incubation. The O2 consumption rate, on the other hand, was approximately three times higher in C06 than in C05 (Figure 5).
The analysis of high-resolution gas kinetics, coupled with frequent measurements of NO2−, allowed estimations of the electron flow to O2 and to the various N-oxide electron acceptors (Figures 4 and 5). Differences in electron flow patterns reveal variations in the way denitrifying organisms regulate the transition from aerobic to anaerobic respiration and reflect how large a proportion of cells in a population switches to denitrification upon depletion of oxygen (Hassan et al., 2014). The smooth patterns of total electron flow seen for isolates A25 (Pseudomonas sp.) and A37 (Hydrogenophaga sp.) from the high pH soil (Figure 4), and for isolates C01 and C05 (both Rhodanobacter sp.) from the low pH soil (Figure 5), suggest that all cells in the cultures switched to denitrification in response to anoxia. Contrary to this, a drastic drop in electron flow after the O2 depletion, as exhibited by the Bradyrhizobium isolate A49 (Figure 4), suggests that only a marginal fraction of the aerobic culture managed the transition to denitrifying respiration. The pattern for the low pH soil isolate C04 (Polaromonas sp.; Figure 5) revealed yet another phenomenon, where the electron flow dropped twice; first in response to O2 depletion, and second in response to NO3− depletion (accumulated as NO2−).
Discussion
The proposed isolation strategy allowed us to capture organisms capable of nitrate reduction only, denitrification sensu stricto (reduction of nitrite to gaseous N), and/or DNRA (reduction of nitrite to ammonium) from both soils, representing not only a wide phenotypic but also a large taxonomic diversity. Most of them belonged to four phyla: Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria. The protocol includes some significant improvements compared to most previously used strategies for isolation of denitrifiers. Most importantly, the truncated phenotypes listed in Figure 1 would not be captured by traditional isolation schemes as: (1) Organisms incapable of reducing nitrate (14 of the 30 isolates) would not be isolated using schemes that only provide nitrate (Gamble et al., 1977; Cheneby et al., 2000; Hashimoto et al., 2009). Heylen et al. (2006) did compare media containing NO3− and NO2− for isolation of denitrifiers from activated sludge, but the effect was not clear. (2) Organisms without NOR would not be detected by any of the old approaches, none of which measured NO. Moreover, NO accumulation can result in denitrification coming to a halt long before all NO3−/NO2− is reduced (Bergaust et al., 2008). (3) Organisms unable to grow on high strength media would not be captured. The nutrient concentration is an important factor, as demonstrated by Hashimoto et al. (2009). This is relevant for Gamble et al. (1977), but not for Heylen et al. (2006) and Cheneby et al. (2000) who used medium strength media, comparable to ours.
To compare the frequency of denitrifying organism with that found by others, we must consider the techniques and criteria used. Cheneby et al. (2000) enumerated and isolated denitrifying bacteria by most probable number technique in the presence of NO3−, using N2O production in the presence of acetylene as the criterion for denitrification. Thus, they would capture only organisms possessing NAR, NIR, and NOR, with or without N2OR, but would overlook the phenotypes ‘NIR, NOR, N2OR’ ‘Only N2OR’ ‘NIR, NOR’ ‘Only NIR’ or ‘DNRA’ listed in Figure 1. They found that the frequency of denitrifiers in different soils ranged from 0.9 to 4.7% of the total number of aerobes (which were also enumerated by most probable number). In comparison, our culture collection contained altogether 12 isolates with NAR, NIR, NOR±N2OR, which was 6.8% of the total number of isolates. Thus, our frequency is higher than the highest value recorded by Cheneby et al. (2000), suggesting that our isolation strategy is at least as inclusive as their most probable number scheme. Higher frequencies of denitrifying bacteria (9.7%) were reported by Gamble et al. (1977). This is, however, the percentage of cells growing anaerobically, and thus not comparable with our results since the number of viable anaerobes is lower than the number of viable aerobes in most soils (Cheneby et al., 2000). In addition to the isolates in the studies by Gamble et al. (1977) and Cheneby et al. (2000), our isolation scheme also captured isolates lacking NAR but with the ability to reduce nitrite to NO, thus performing the defining step of denitrification. If all the phenotypes that had ‘NIR’ are included, they comprised 16% of all the isolates in our study.
Taken together, this suggests that isolation on low nutrient plates under fully oxic conditions, followed by a gentle transition to anaerobic respiration in liquid medium, as done in the present study, was a successful strategy that allowed us to capture a large number of diverse truncated and complete denitrifiers. The implementation of a gentle transition is in line with the observations by Højberg et al. (1997), showing that an abrupt decrease in oxygen may leave denitrifiers entrapped in anoxia without energy, and thus unable to produce colonies. Another important factor which apparently increased the number of different isolates was the adjustment of pH of the medium for the isolates from the low pH soil. This may reflect that many bacteria have a relatively narrow pH range for growth (Bååth, 1996). It is somewhat surprising that many organisms from the low pH soil grew well on neutral pH medium, but were not found among the isolates on the low pH plates. One explanation could be that bacteria live in microsites with locally increased pH.
The frequent occurrence of truncated denitrification pathways among the isolates is in line with the vast information found in databases about the genetic potential of denitrifying bacteria (Shapleigh, 2013; Graf et al., 2014). Full-fledged denitrifiers were almost exclusively found in the high pH soil, with only one out of nine isolates being from the low pH soil, series C (Figure 1) and none from the B series (bacteria from low pH soil spread on high pH medium). A truncated denitrification phenotype can be due to the absence of, or mutations in, the functional genes encoding the reductases, or genes involved in their regulation. In addition, transcriptional regulation as well as post-transcriptional phenomena may come into play. The end-point analysis in which all possible electron acceptors for denitrification were provided, except NO, allowed us to identify all different combinations of truncated denitrification phenotypes, apart from the tentative phenotype ‘Only NOR’ (Figure 1).
Organisms that carry a N2O reductase, but lack one or more of the other denitrification genes, are of particular interest since they are potential sinks for the greenhouse gas N2O. The ‘NIR, NOR, N2OR’ phenotype was found in only three organisms isolated from the low pH soil, all belonging to the genus Rhodanobacter. If living in a complex system, these bacteria would use NO2−, as well as N2O, produced by other members of the microbial community. They could thus act both as a source and a sink for N2O. Three isolates were sole N2O reducers (phenotype ‘Only N2OR’), leaving nitrate and nitrite untouched in the end-point analysis. As such, they will not produce any N2O, but can reduce N2O produced by other organisms. Unfortunately, only one of them was viable after storage in −80 °C. Genome sequencing confirmed that this isolate, belonging to the genus Algoriphagus in the phylum Bacteroidetes, carried a nosZ gene (clade II) but lacked the other denitrification genes. Such organisms should be searched for since they are interesting as components in plant growth promoting bacterial inocula, where their role would be to reduce N2O emissions from cultured fields (Gao et al., 2016).
Altogether twelve isolates were found which had N2O as end-point (Figure 1). This phenotype was explained by lack of the nosZ gene for the two isolates from the high pH soil (A23 and A02), and also for one of the Rhodanobacter isolates (B04) from the low pH soil, although this latter contained the nosL gene which may be a remnant from a more complete operon. Three of the whole genome sequenced isolates from the low pH soil were unable to reduce N2O at neither low nor high pH, yet they did carry the nosZ gene. They all belonged to the genus Rhodanobacter, reported to be dominant members of various denitrifying communities in low pH environments (van den Heuvel et al., 2010; Green et al., 2012). The inability of nosZ-carrying bacteria to reduce N2O at acidic pH, despite producing nosZ transcripts at similar levels as nirS transcripts, was reported for Paracoccus denitrificans (Bergaust et al., 2010). Similar results were obtained from intact soil and extracted soil bacterial communities (Liu et al., 2010; 2014). This suggests a general, post-transcriptional effect, most likely caused by unsuccessful assembly of the N2O reductase at low pH (Bakken et al., 2012). One of the Rhodanobacter isolates (C01) did, however, reduce N2O at low pH, an interesting observation that calls for detailed biochemical investigations that may shed new light on the regulation of denitrification and N2O emissions in low pH soils.
All the Rhodanobacter isolates lacked genes coding for the dissimilatory NO3− reductases (Nar or Nap). Such organisms have probably been overlooked in isolation studies where NO3−, but not NO2−, was provided. In the present study, most of the isolates that lacked NAR were from the low pH soil. It can be speculated that this is a way for organisms living in acidic environments to avoid being exposed to high concentrations of toxic N oxides. Nitrite produced from the reduction of NO3− would be released into the environment and, if not being taken up again for reduction, chemical decomposition of NO2− could result in high concentrations of toxic NO in the vicinity of the cells (Bancroft et al., 1979; van Cleemput and Samater, 1996). The fact that 14 isolates from the low pH soil were classified in the phenotype group ‘Only NAR’ contradicts, however, this explanation and raises the question if these organisms have other ways to protect themselves from toxic nitrogenous compounds. One possibility could be that they are equipped with two or several NO reductases as seen in some of the sequenced isolates from the low pH soil, which carried two copies of qnor (Figure 6). Alternatively, they exist in close assemblages with other cells in their natural environment, which carry NO reductases and thus keep NO concentrations low. The latter may explain the presence of several isolates with the phenotype ‘NAR, NIR’ or ‘Only NIR’, with NO as end point product of denitrification. This phenotype, which was also found in a study by Falk et al. (2010), is most likely dependent on other organisms and will not survive when grown in anaerobic batches as pure cultures.
The denitrification regulatory phenotypes of selected isolates demonstrate a large variation in the way different denitrifying bacteria handle the transition from aerobic to anaerobic respiration (Figures 4 and 5). Yet, assembling the denitrification machinery comes with an energetic cost. Moreover, production and consumption of the toxic intermediates NO2− and NO should ideally be synchronized to avoid accumulation of these compounds, which requires strict regulatory control. The regulatory network of denitrification has only been studied in detail for a few model organisms, revealing a large number of transcriptional regulator enzymes and ancillary factors (Zumft, 1997; Kunak et al., 2004; Wunsch et al., 2005; Zumft and Kroneck, 2007; Bergaust et al., 2012). Although denitrifying organisms may harbor similar regulatory proteins, their regulatory phenotypes may be profoundly different. Some organisms such as isolate C04 (and most likely A37) in this study, reduce all the available NO3− to NO2− before further reduction to gaseous end-products. Such a progressive onset of the denitrification enzymes was earlier found among some isolates of Thauera (Liu et al., 2013), and appeared to be caused by a strict, NO3− dependent transcriptional repression of nirS encoding NO2− reductase. The details for this mechanism remain, however, to be elucidated. This strategy may be dangerous in a low pH environment, since it may lead to high NO concentrations due to chemical decomposition of NO2−. On the other hand, it may be an adequate ‘bet hedging’ strategy: if oxygen returns before the cells have produced the entire denitrification proteome, they would save energy. Control of the gaseous intermediates NO and N2O also varied grossly between the isolates; while some had strict control of NO, reaching maxima of 10–20 nmol vial−1, others produced 0.2–3.5 μmol NO (B06, C04, C05 and C06). Despite such high levels, most isolates managed to reduce the NO, albeit slowly, except B06 which was apparently inactivated (or killed) by NO. The N2O levels varied from organisms showing a transient accumulation of 30 μmol vial−1 (A49), to those that reduced all N oxides to N2O before further reduction to N2 (A25 and A37) thus being potentially strong N2O sources in an environmental setting.
The large variation in the regulatory biology of the isolates is also reflected in the electron flow kinetics (bottom graphs in Figures 4 and 5). Several organisms performed a smooth transition from aerobic to anaerobic respiration, while some (A23 and A49) showed a dip in electron flow when transferring to using N oxide as electron acceptor. The latter is consistent with findings for P. denitrificans, where apparently only a fraction of the cells in the population switch to denitrification. This can be regarded as a bet-hedging strategy, which is expected to increase the chances of survival of the population; if the anoxic period is long-lasting it is advantageous to produce a complete denitrification proteome, but if the anoxic spell is short it may be more energy saving to only produce one or two of the reductases (Hassan et al., 2014). From an environmental point of view such bet-hedging populations, in which the major fraction of cells produce only NAR, or NAR+N2OR, as is the case of P. denitrificans, will be strong sinks for N2O produced by other organisms (Hassan et al., 2016).
The contribution of molecular techniques to the field of microbial ecology has greatly improved the insight into the structure and genetic potential of complex microbial consortia. Yet, this cannot replace physiological and biochemical studies of organisms grown in pure cultures under controlled conditions. As pointed out by (Rocca et al., 2015), the correlation between gene abundance and the corresponding process is often weak. This was also demonstrated in the present study, where we found a mismatch between the genetic potential and observed N2O reduction in 23% of the isolates. The discrepancies between genetic potential and measured function are most plausibly the results of transcriptional and metabolic regulation of the denitrification genes and their products, demonstrating that the presence of a gene cannot alone be used to predict a function. The large variation in the regulation of denitrification found among the isolates pinpoints the need for more detailed knowledge about a wider range of denitrifiers than that obtained from model bacteria, and the present study contributes to this by finding new representative organisms to include in ecophysiological studies of denitrification. The long-term, practical goal of these efforts should be to find ways to mitigate the gas emissions from these organisms by stimulating phenotypes that act as sinks for N2O, and counteracting those that act as sources.
Acknowledgments
Pawel Lycus was financed by NORA MCS ITN from the EU 7th Framework Programme under the Grant agreement no 316472. Linda Bergaust was financed by Research Council of Norway, within FRIMEDBIO project EnDReg, no 231282/F20.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008). The RAST server: rapid annotations using subsystems technology. BMC Genom 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. (2003). Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55: 541–555. [DOI] [PubMed] [Google Scholar]
- Bakken LR, Bergaust L, Liu B, Frostegard A. (2012). Regulation of denitrification at the cellular level: a clue to the understanding of N2O emissions from soils. Philos Trans R Soc B 367: 1226–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft K, Grant IF, Alexander M. (1979). Toxicity of NO2: effect of nitrite on microbial activity in an acid soil. Appl Environ Microbiol 38: 940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergaust L, Shapleigh J, Frostegard A, Bakken L. (2008). Transcription and activities of NOx reductases in Agrobacterium tumefaciens: the influence of nitrate, nitrite and oxygen availability. Environ Microbiol 10: 3070–3081. [DOI] [PubMed] [Google Scholar]
- Bergaust L, Mao YJ, Bakken LR, Frostegard A. (2010). Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrous oxide reductase in paracoccus denitrificans. Appl Environ Microbiol 76: 8285–8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergaust L, Bakken LR, Frostegard A. (2011). Denitrification regulatory phenotype, a new term for the characterization of denitrifying bacteria. Biochem Soc Trans 39: 207–212. [DOI] [PubMed] [Google Scholar]
- Bergaust L, van Spanning RJM, Frostegard A, Bakken LR. (2012). Expression of nitrous oxide reductase in Paracoccus denitrificans is regulated by oxygen and nitric oxide through FnrP and NNR. Microbiology 158: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower EC, Holm-Hansen T. (1980). A salicylate–hypochlorite method for determining ammonia in seawater. Can J Fish Aquat Sci 37: 794–798. [Google Scholar]
- Braman SR, Hendrix SA. (1989). Nanogram nitrate and nitrite determination in environmental and biological materials by vanadium (III) reduction with chemiluminescence detection. Anal Chem 61: 2715–2718. [DOI] [PubMed] [Google Scholar]
- Bååth E. (1996). Adaptation of soil bacterial communities to prevailing pH in different soils. Fems Microbiol Ecol 19: 227–237. [Google Scholar]
- Cheneby D, Philippot L, Hartmann A, Henault C, Germon J. (2000). 16S rDNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microbiol Ecol 34: 121–128. [DOI] [PubMed] [Google Scholar]
- Cox RD. (1980). Determination of nitrate and nitrite at the parts per billion level by chemiluminescence. Anal Chem 52: 332–335. [Google Scholar]
- Cuhel J, Simek M, Laughlin RJ, Bru D, Cheneby D, Watson CJ et al (2010). Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl Environ Microbiol 76: 1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeignoz-Horta LA, Spor A, Bru D, Breuil MC, Bizouard F, Leonard J et al (2015). The diversity of the N2O reducers matters for the N2O:N2 denitrification end-product ratio across an annual and a perennial cropping system. Front Microbiol 6: 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Liu B, Braker G. (2010). Isolation, genetic and functional characterization of novel soil nirK-type denitrifiers. Syst Appl Microbiol 33: 337–347. [DOI] [PubMed] [Google Scholar]
- Gamble TN, Betlach MR, Tiedje JM. (1977). Numerically dominant denitrifying bacteria from world soils. Appl Environ Microbiol 33: 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Shen W, Kakuta H, Tanaka N, Fujiwara T, Nishizawa T et al (2016). Inoculation with nitrous oxide (N2O)-reducing denitrifier strains simultaneously mitigates N2O emission from pasture soil and promotes growth of pasture plants. Soil Biol Biochem 97: 83–91. [Google Scholar]
- Graf DR, Jones CM, Hallin S. (2014). Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS ONE 9: e114118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SJ, Prakash O, Jasrotia P, Overholt WA, Cardenas E, Hubbard D et al (2012). Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl Environ Microbiol 78: 1039–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Koga M, Masaoka Y. (2009). Advantages of a diluted nutrient broth medium for isolating N-2-producing denitrifying bacteria of alpha-Proteobacteria in surface and subsurface upland soils. Soil Sci Plant Nutr 55: 647–659. [Google Scholar]
- Hassan J, Bergaust LL, Wheat ID, Bakken LR. (2014). Low probability of initiating nirS transcription explains observed gas kinetics and growth of bacteria switching from aerobic respiration to denitrification. PLoS Comput Biol 10: e1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan J, Qu Z, Bergaust LL, Bakken LR. (2016). Transient accumulation of NO2− and N2O during denitrification explained by assuming cell diversification by stochastic transcription of denitrification genes. PLoS Comput Biol 12: e1004621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heylen K, Vanparys B, Wittebolle L, Verstraete W, Boon N, De Vos P. (2006). Cultivation of denitrifying bacteria: optimization of isolation conditions and diversity study. Appl Environ Microbiol 72: 2637–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højberg O, Jorgen Binnerup S, Sørensen J. (1997). Growth of silicone-immobilized bacteria on polycarbonate membrane filters, a technique to study microcolony formation under anaerobic conditions. Appl Environ Microbiol 63: 2920–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. (2002). Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions acidobacteria, actinobacteria, proteobacteria, and verrucomicrobia. Appl Environ Microbiol 68: 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Graf DR, Bru D, Philippot L, Hallin S. (2013). The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J 7: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Spor A, Brennan FP, Breuil M-C, Bru D, Lemanceau P et al (2014). Recently identified microbial guild mediates soil N2O sink capacity. Nat Clim Change 4: 801–805. [Google Scholar]
- Kunak M, Kucera I, van Spanning RJ. (2004). Nitric oxide oscillations in Paracoccus denitrificans: the effects of environmental factors and of segregating nitrite reductase and nitric oxide reductase into separate cells. Arch Biochem Biophys 429: 237–243. [DOI] [PubMed] [Google Scholar]
- Liu B, Morkved PT, Frostegard A, Bakken LR. (2010). Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol Ecol 72: 407–417. [DOI] [PubMed] [Google Scholar]
- Liu B, Mao YJ, Bergaust L, Bakken LR, Frostegard A. (2013). Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ Microbiol 15: 2816–2828. [DOI] [PubMed] [Google Scholar]
- Liu B, Frostegard A, Bakken LR. (2014). Impaired reduction of N2O to N-2 in acid soils is due to a posttranscriptional interference with the expression of nosZ. Mbio 5: e01383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Almagro VM, Gates AJ, Moreno-Vivian C, Ferguson SJ, Richardson DJ, Roldan MD. (2011). Bacterial nitrate assimilation: gene distribution and regulation. Biochem Soc Trans 39: 1838–1843. [DOI] [PubMed] [Google Scholar]
- Molstad L, Dorsch P, Bakken LR. (2007). Robotized incubation system for monitoring gases (O2, NO, N2O N2 in denitrifying cultures. J Microbiol Methods 71: 202–211. [DOI] [PubMed] [Google Scholar]
- Morales SE, Cosart T, Holben WE. (2010). Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J 4: 799–808. [DOI] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. (1998). Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microbial Ecol 14: 113–118. [Google Scholar]
- Olsen RA, Bakken LR. (1987). Viability of soil bacteria: Optimization of plate-counting technique and comparison between total counts and plate counts within different size groups. Microb Ecol 13: 59–74. [DOI] [PubMed] [Google Scholar]
- Portmann RW, Daniel JS, Ravishankara AR. (2012). Stratospheric ozone depletion due to nitrous oxide: influences of other gases. Philos Trans R Soc Lond B Biol Sci 367: 1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Wang J, Almoy T, Bakken LR. (2014). Excessive use of nitrogen in Chinese agriculture results in high N(2) O/(N(2) O+N(2)) product ratio of denitrification, primarily due to acidification of the soils. Glob Chang Biol 20: 1685–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JJ, Myrold DD. (2004). Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biol Biochem 36: 1431–1441. [Google Scholar]
- Rocca JD, Hall EK, Lennon JT, Evans SE, Waldrop MP, Cotner JB et al (2015). Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J 9: 1693–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roco CA, Bergaust LL, Bakken LR, Yavitt JB, Shapleigh JP. (2016). Modularity of nitrogen-oxide reducing soil bacteria: linking phenotype to genotype. Environ Microbiol; e-pub ahead of print 23 February 2016, doi: doi:10.1111/1462-2920.13250. [DOI] [PubMed]
- Russenes AL, Korsaeth A, Bakken LR, Dörsch P. (2016). Spatial variation in soil pH controls off-season N2O emission in an agricultural soil. Soil Biol Biochem 99: 36–46. [Google Scholar]
- Schlesinger WH. (2008). On the fate of anthropogenic nitrogen. Proc Natl Acad Sci 106: 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapleigh J. (2013)Denitrifying Prokaryotes. In: Rosenbers E (ed). The Prokaryotes, Fourth edn. Springer-Verlag, Berlin, Heidelberg, pp 405–425. [Google Scholar]
- Simek M, Cooper JE. (2002). The influence of soil pH on denitrification: progress towards the understanding of this interaction over the last 50 years. Eur J Soil Sci Total Environ 53: 345–354. [Google Scholar]
- Sognnes LS, Fystro G, Opstad SL, Arstein A, Borresen T. (2006). Effects of adding moraine soil or shell sand into peat soil on physical properties and grass yield in western Norway. Acta Agr Scand 56: 161–170. [Google Scholar]
- Stokstad E. (2016). The nitrogen fix. Science 353: 1225–1227. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Cleemput O, Samater AH. (1996). Nitrite in soils: accumulation and role in the formation of gaseous N compounds. Fertilizer Res Microbiol 1996: 81–89. [Google Scholar]
- van den Heuvel RN, van der Biezen E, Jetten MS, Hefting MM, Kartal B. (2010). Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ Microbiol 12: 3264–3271. [DOI] [PubMed] [Google Scholar]
- van den Heuvel RN, Bakker SE, Jetten MS, Hefting MM. (2011). Decreased N2O reduction by low soil pH causes high N2O emissions in a riparian ecosystem. Geobiology 9: 294–300. [DOI] [PubMed] [Google Scholar]
- van Groenigen JW, Huygens D, Boeckx P, Kuyper TW, Lubbers IM, Rütting T et al (2015). The soil N cycle: new insights and key challenges. Soil 1: 235–256. [Google Scholar]
- Wunsch P, Korner H, Neese F, van Spanning RJ, Kroneck PM, Zumft WG. (2005). NosX function connects to nitrous oxide (N2O) reduction by affecting the Cu(Z) center of NosZ and its activity in vivo. FEBS Lett 579: 4605–4609. [DOI] [PubMed] [Google Scholar]
- Zumft WG. (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG, Kroneck PM. (2007). Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv Microb Physiol 52: 107–227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.