ABSTRACT
Emerging evidence shows that the human microbiota plays a larger role in disease progression and health than previously anticipated. Helicobacter pylori, the causative agent of gastric cancer and duodenal and gastric ulcers, was early associated with gastric disease, but it has also been proposed that the accompanying microbiota in Helicobacter pylori-infected individuals might affect disease progression and gastric cancer development. In this study, the composition of the transcriptionally active microbial community and H. pylori gene expression were determined using metatranscriptomic RNA sequencing of stomach biopsy specimens from individuals with different H. pylori infection statuses and premalignant tissue changes. The results show that H. pylori completely dominates the microbiota not only in infected individuals but also in most individuals classified as H. pylori uninfected using conventional methods. Furthermore, H. pylori abundance is positively correlated with the presence of Campylobacter, Deinococcus, and Sulfurospirillum. Finally, we quantified the expression of a large number of Helicobacter pylori genes and found high expression of genes involved in pH regulation and nickel transport. Our study is the first to dissect the viable microbiota of the human stomach by metatranscriptomic analysis, and it shows that metatranscriptomic analysis of the gastric microbiota is feasible and can provide new insights into how bacteria respond in vivo to variations in the stomach microenvironment and at different stages of disease progression.
KEYWORDS: atrophic gastritis, gastric carcinogenesis, Helicobacter pylori, metatranscriptomics, stomach microbiota
INTRODUCTION
The human stomach was long thought to be sterile due to the hostile environment characterized by extremely low pH. The discovery of Helicobacter pylori in 1983 revealed that bacteria can in fact colonize the stomach (1). Since then, several studies on the stomach microbiota have been performed both in gastric juice and in mucosal biopsy specimens, using culture-dependent methods (2, 3) as well as analysis of DNA extracted from biopsy specimens (3–8). These studies show a previously unappreciated richness of the stomach bacterial flora but also that the communities are highly uneven and have large interindividual variation. Commonly detected phyla include Firmicutes, Bacteriodetes, Actinobacteria, Fusobacteria, and Proteobacteria (4, 5, 7). These phyla are represented mainly by the genera Streptococcus, Lactobacillus, Veillonella, Clostridium, Prevotella, Porphyromonas, Rothia, Neisseria, and Haemophilus, (4, 5, 9, 10). However, the Proteobacteria species H. pylori stands out as completely dominant in the microbiota of H. pylori-infected individuals, and its importance as an inhabitant of the human gastric microflora is further emphasized by the fact that it is estimated to colonize nearly 50% of the human population globally.
Helicobacter pylori is the main causative agent of gastric cancer (GC) and gastric and duodenal ulcers. Although the infection always causes an immune response and inflammation of the gastric mucosa, the majority of infected individuals remain asymptomatic. However, approximately 10 to 15% of infected individuals develop gastric or duodenal ulcers, and another 1 to 3% develop gastric cancer (11). H. pylori infection is usually acquired in childhood, and a lifelong infection is established in the absence of antibiotic treatment. In some individuals the infection leads to antrum-predominant gastritis, characterized by increased production of gastric acid (hyperchlorhydria) in the lower part of the stomach (antrum), a state associated with duodenal ulcer development. This course of disease is divergent from the development of atrophic gastritis in the upper part of the stomach (corpus), which instead leads to lower production of gastric acid, a hallmark in the progression toward the intestinal type of GC (12). The atrophy can be succeeded by intestinal metaplasia, which leads to even more drastic changes in the gastric environment, followed by dysplasia and finally gastric adenocarcinoma (13). It is well established that atrophy of the corpus mucosa, leading to decreased acid secretion, is highly associated with GC development. How the chronic infection and inflammation lead to cancer in some individuals but leave the majority without symptoms is still not outlined but is thought to depend on a combination of host susceptibility, environmental factors, and bacterial pathogenicity (14).
Recently, it has been proposed that the accompanying microbiota in H. pylori-infected individuals might affect the outcome of the infection, since the hypochlorhydric environment caused by atrophic changes in the H. pylori-infected corpus is assumed to promote gastric colonization by other microorganisms (15). Studies on the microbiota in the presence or absence of clinical H. pylori infection have generated conflicting results. While some studies could not detect any significant changes in taxonomic composition (5, 16, 17), others reported increased abundance of the genera Spirochaetae, Streptococcus, Lactobacillus, and Veillonella in response to increased stomach pH and H. pylori-induced gastric cancer (2). A recent study also found similar differences in stomach microbiota composition between individuals from well-described high-risk and low-risk areas in Colombia (18). To date, no study has been able to convincingly link the presence or absence of other bacteria to H. pylori-induced cancer development.
Recent studies have explored the gastric microbiota using DNA-based techniques, but this will not provide information about whether the detected microorganisms in human stomach biopsy specimens are viable and belong to the resident microbiota or not. We therefore performed massively parallel RNA sequencing of stomach biopsy specimens from H. pylori-uninfected controls and H. pylori-infected individuals with and without the precancerous changes atrophic gastritis and intestinal metaplasia. In this study, we chose to focus on analysis of the corpus gene expression, since histological changes in this area of the stomach are closely associated with the development of the intestinal type of gastric adenocarcinoma, for which the precancerous cascade has been described in detail (19).
The sequencing data were used for metatranscriptomic taxonomic analyses based on rRNA gene expression to determine the composition of the viable microbial community. We found that H. pylori dominated the microbiota in human stomachs, not only among infected individuals but also in individuals who were classified as H. pylori negative by conventional methods. In addition, we found a significant positive correlation between the levels of H. pylori rRNA and rRNA from Campylobacter, Deinococcus, and Sulfurospirillum. Finally, while the host response to H. pylori infection has been extensively investigated, fewer studies have focused on the bacterial transcriptional response to the changing host environment. The transcriptome of H. pylori revealed high expression levels of genes involved in pH regulation and nickel transport, indicating that H. pylori in close contact with the epithelium induces expression of virulence genes and encounters pH stress.
RESULTS AND DISCUSSION
H. pylori does not markedly alter the rest of the viable stomach corpus microbiota on the phylum level.
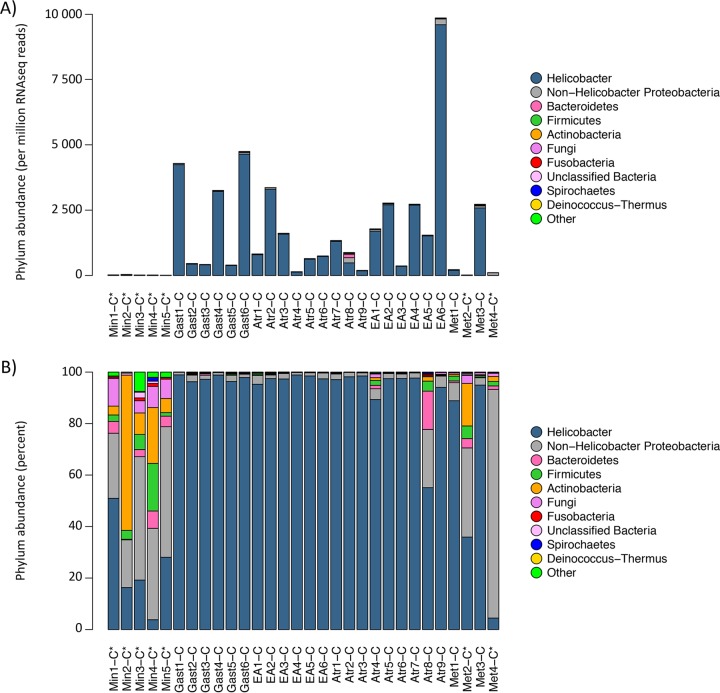
16S rRNA reads were extracted from the RNA sequencing data using Metaxa2 (20) and normalized against the total read count in each library. Applying a cutoff of at least 1 rRNA read per million reads, 9 bacterial phyla and the eukaryotic kingdom Fungi were identified in at least one of the samples (Fig. 1A). Biopsy specimens from individuals initially scored as H. pylori positive by urea breath test (UBT), serology and culture contained on average 64 times (P < 0.01) more bacterial rRNA reads than those from individuals scored as H. pylori negative (see Fig. S2 in the supplemental material), and the majority of those were assigned to the phylum Proteobacteria (Fig. 1A). Most of these reads (4.8% to 99%; average, 82.1%) were assigned to the Helicobacter genus. Although the community compositions in clinically H. pylori-positive and -negative individuals seemed to be different (Fig. 1; see Fig. S3 in the supplemental material), those effects were not significant on the phylum level. Thus, if H. pylori infection does have an effect on the composition of other bacteria on the phylum level, it seems to be minor, as differences between individuals overshadow it.
FIG 1.
Relative phylum-level abundance in the gastric corpus, based on 16S rRNA classification. (A) Phylum-level abundances (16S rRNA reads per million total RNA-seq reads). (B) Phylum-level relative abundances. For both panels, the Proteobacteria phylum has been split into the Helicobacter genus and non-Helicobacter Proteobacteria genera. Min, H. pylori-uninfected individuals; Gast, gastritis with no atrophy or metaplasia; Atr, atrophic gastritis; EA, extensive atrophy; Met, intestinal metaplasia. rRNA reads were extracted from the RNA-seq data and classified using Metaxa2. Asterisks represent individuals negative for H. pylori infection as determined by clinical tests.
Previous studies have shown that the gastric microbiota is complex, and conflicting evidence exists on whether there is a difference between the stomach microbial communities in H. pylori-positive and -negative individuals or at different disease stages (3, 5, 7, 16). Bik and colleagues found no significant differences in taxonomic composition between H. pylori-negative and -positive subjects, confirming our observations (5), while other studies have found that Helicobacter pylori-negative subjects had higher relative abundances of Firmicutes, Bacteroidetes, and Actinobacteria and that H. pylori-positive individuals had greater abundances of non-H. pylori Proteobacteria, Spirochetes, and Acidobacteria (21). Our data confirm the finding of Firmicutes, Bacteroidetes, and Actinobacteria in several of the subjects with low levels of H. pylori, with Actinobacteria being the dominating phylum in one case. These data support the findings of Maldonado-Contreras et al., who also reported higher levels of Actinobacteria in H. pylori-negative individuals (7).
Helicobacter is the most common genus found in stomach corpus biopsy specimens.
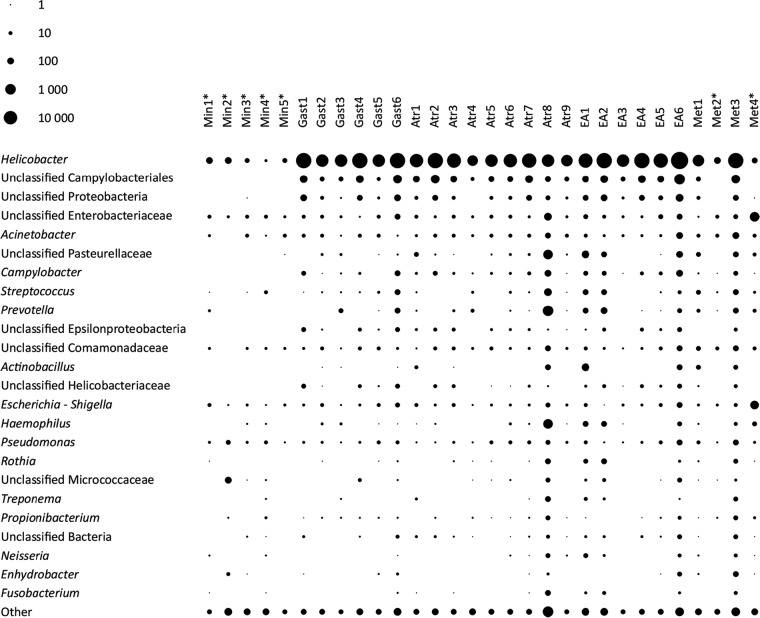
Metaxa2 analysis revealed the presence of 350 bacterial genera across all samples, of which 33 had an abundance of higher than one per million reads in at least one sample (Fig. 2). The most abundant genera found in all biopsy specimens were Helicobacter, Pseudomonas, Acinetobacter, Escherichia, and unclassified Enterobacteriaceae (Fig. 2). Interestingly, Helicobacter was present in every sample, being the dominant genus also in 5 out of the 7 samples taken from individuals classified as uninfected using conventional methods. However, the seven samples from subjects classified as H. pylori negative contained approximately 100- to 10,000-fold fewer Helicobacter-associated reads than biopsy specimens scored as H. pylori positive. These findings were further supported by one-step PCR 16S rRNA amplicon sequencing of the same samples, showing highly similar results (see Fig. S4 in the supplemental material). In general, Helicobacter rRNA reads constituted 91.0% to 99.0% of all bacterial rRNA reads in the H. pylori-positive samples, except in sample Atr8 (56.7%), while in the individuals scored as H. pylori negative, the proportion of Helicobacter reads ranged from 4.7% to as many as 63.2% of all bacterial rRNA reads. Similar observations have been reported previously (5, 7, 8, 16), but in contrast to the case for previous studies, our findings represent actual expressed sequences and imply that these bacteria are viable and transcriptionally active, although they still could constitute a transient population. However, since these samples were derived from tissue biopsy specimens, to be detected by the method used in this paper, such transient bacteria would still need to be attached to the surface, speaking in favor of an established population rather than a transient one. These findings suggest that the frequency of low-level H. pylori infection might be higher than anticipated; it is even possible that H. pylori may be present to some degree in a vast majority of individuals. However, the numbers and/or activity of these bacteria must be very low in order to go undetected by conventional methods, and such low numbers of H. pylori are probably not contributing significantly to disease risk, since only very weak signs of inflammation were seen in 2 out of 5 of the H. pylori-negative controls (see Table S1 in the supplemental material).
FIG 2.
Abundances of different genera in the gastric corpus mucosa. The diameter of each spot corresponds to the relative abundance of each genus in each sample, counted as rRNA reads belonging to that genus per million reads. Only genera having more than two reads per million in at least one sample were included in the figure. Min, H. pylori-uninfected individuals; Gast, gastritis with no atrophy or metaplasia; Atr, atrophic gastritis; EA, extensive atrophy; Met, intestinal metaplasia. The asterisks denote individuals that were H. pylori negative according to clinical methods. The rRNA transcripts were classified using Metaxa2.
To verify these findings with deeper sequencing and complementary methods, we reverse transcribed the corpus RNA samples and amplified and sequenced the V3-V4 region of the 16S rRNA gene. This was also done for antrum samples from the same individuals. Although level were also very low using this method, we could verify the presence of Helicobacter pylori-specific amplicons in this analysis, after subtracting sequences found in no-template PCR control, in both antrum and corpus biopsy specimens.
It is possible that some of the reads classified only to the level of the Helicobacter genus derive from species other than H. pylori, some of which are not detected by traditional tests, as has been suggested previously (22). In the transcriptome sequencing (RNA-seq), the samples from some individuals contained reads classified as Helicobacter canis, although no such sequences could be detected through amplicon sequencing. However, the numbers of H. canis reads were very low, at most 0.44% of all Helicobacter reads and 0% in the clinically H. pylori-positive samples.
Finally, there is also the possibility that this is a technical artifact. Indeed, barcode leakage in Illumina sequencing is known to happen, but precautions to limit this were taken, as described previously (23). The samples were randomized in all steps of the workflow so that there would not be any batch effects from either the extraction, library preparation, or sequencing. However, this means that we cannot exclude contamination between sample groups, but we find this explanation unlikely in this case, since the transcripts detected in the clinically H. pylori-negative individuals were relatively diverse and did not represent only the most abundant transcripts overall and the 16S rRNA amplicon sequencing confirmed the finding of H. pylori rRNA in these samples. It should also be noted that Nicaragua is a country where the prevalence of H. pylori is high, around 70% (24), and it remains to be seen if these findings can be replicated in low-prevalence populations.
Presence of Helicobacter pylori is significantly associated with Campylobacter, Sulfurospirillum, and Deinococcus.
Testing for correlations between Helicobacter and the abundance of other genera, we found Campylobacter, Sulfurospirillum, and Deinococcus rRNA levels to be significantly associated with high abundance of Helicobacter (see Table S6 in the supplemental material). Deinococcus and Sulfurospirillum were also found exclusively in patients who were H. pylori positive by conventional methods. Campylobacter and Sulfurospirillum both belong to the family Campylobacteraceae and the families Helicobacteraceae and Campylobacteraceae together form the Epsilonproteobacteria subclass of Proteobacteria. It remains to be investigated whether these closely related species benefit from coexistence in the human stomach or whether the correlation is due to similar environmental preferences among Epsilonproteobacteria. Deinococcus, however, belongs to the phylum Deinococcus-Thermus and is regarded to be an environmental extremophile not expected to inhabit the human stomach, but the occurrence of Deinococcus in this study confirms one previous report (5).
Notably, we found that there seems to be a set of bacterial genera that are present in high, but similar, abundances across most of the study subjects, regardless of the levels of H. pylori, including Escherichia, Acinetobacter, Pseudomonas, and Streptococcus (Fig. 2). Contrary to previous reports (9), we did not see any significant effect of clinically determined Helicobacter status on the Shannon or Simpson diversity of other bacterial genera. However, the richness of genera was larger in Helicobacter-positive samples than in those scored negative by clinical tests (P = 0.01193). When Helicobacter was included in the analysis, genus richness was not significantly different (P = 0.4983), while the diversity and evenness estimates of the communities drastically changed (P < 10−10; specifically, for Shannon diversity P = 1.75e−15, for Simpson diversity P = 5.586e−14, and for evenness P = 1.436e−11), resulting from lower diversity indices for the H. pylori-positive individuals than for the H. pylori-negative individuals due to the very high abundance of Helicobacter rRNA. Notably, earlier studies of the stomach microbiota based on sequencing of DNA as opposed to RNA have indicated substantially higher taxonomic diversity (5, 8, 9). However, many of these taxa exist in very limited quantities and are likely to be transcriptionally inactive, suggesting that their influence on the gastric ecosystem may be insignificant. Furthermore, this study also emphasizes the very small quantities of non-Helicobacter bacterial genera in stomach biopsy specimens (Fig. S4). Such small quantities of viable bacteria increase the impact of contaminations during nucleotide extraction and PCR amplification, which must be accounted for, for example, by comparison to negative controls.
The gastric microbiota does not change in relation to atrophy level.
To determine whether the composition of the stomach microbiota was influenced by changes in the stomach environment as a result of disease progression, we first analyzed the community composition in relation to the histopathological changes as determined based on four biopsy specimens in the corpus (see Table S1 in the supplemental material). The analysis did not reveal any segregation between the groups (see Fig. S3 and S5 in the supplemental material), but all groups of H. pylori-positive individuals overlapped with each other, with only a few individuals as outliers. However, as atrophic and metaplastic changes can occur very variably across the tissue and the overall histological scores were based on other, albeit adjacent, biopsy specimens, there is a substantial likelihood that these scores may not correspond to the atrophic status of the biopsy specimen that was used for RNA extraction. Therefore, to determine the level of atrophy in the tissue independently of the pathology scores, we took advantage of the fact that we also had information about the host gene expression in the RNA-seq data.
We used the expression levels of genes associated with atrophic gastritis, i.e., ATP4A, ATP4B, GHRL, GIF, CCKRB, PGC, PGA3, and PGA4, to determine the level of “molecular atrophy” in the tissues (see Materials and Methods for details). These genes are linked mainly to the presence of parietal cells, a cell type lost in atrophic gastritis (25, 26). Also, by using the gene expression of the proton pump-encoding ATP4A and ATP4B of the human hosts, we obtained a nonsubjective measure of the acid-secreting capabilities of the microenvironment of the bacteria analyzed. Regression analysis was performed on bacterial rRNA abundance relative to degree of atrophy and mean ATP4 expression. We found that the abundance of the bacterial genera did not change significantly in relation to the level of molecular atrophy in the tissue. Furthermore, we detected no association between corpus atrophy level or disease status and differences between the communities in the antrum and corpus sample within each individual. In line with our results, Dicksved et al. reported that gastric cancer patients were dominated by the genera Streptococcus, Lactobacillus, Veillonella, and Prevotella but that there were no significant differences from healthy controls (16). Aviles-Jimenez and colleagues did find significant differences in abundances of taxa when comparing nonatrophic gastritis to gastric cancer; however, subjects having intestinal metaplasia overlapped both the gastritis and cancer groups (6). Our results were based on samples ranging from no to high-grade atrophy, and in this set we could not identify any distinct set of bacterial genera that changed with more severe atrophy.
Helicobacter pylori expresses high levels of virulence factors in vivo.
To study H. pylori gene transcription at the site of infection, we extracted the H. pylori transcriptome from the RNA-seq data. Due to the low proportion of bacterial RNA in the samples, we used only the expression values for the genes passing certain threshold criteria (see Materials and Methods), since expression estimates of genes with lower expression were at risk of be highly influenced by noise. The thresholds applied allowed differential expression analysis of 285 genes (see Table S5 in the supplemental material). In line with the rRNA classification, we detected H. pylori transcripts in all of the individuals, but the total amount varied substantially between the samples (Fig. S2), and for the clinically H. pylori-negative individuals, fewer than 100 reads per million reads mapped to the H. pylori genes. The number of reads mapping to Helicobacter pylori mRNA per million total reads was strongly correlated with the number of bacterial 16S rRNA sequences per million reads (r2 = 0.76, P < 0.0001).
Similar to the results of previous studies (27), we found high expression of several of the classical toxins and adhesion factors of H. pylori (Table 1). The napA and flaA genes were among the most highly expressed genes, while, for example, vacA, babA, and sabA had lower expression levels. Interestingly, HP1192, a secreted protein involved in flagellar motility, was the most highly expressed gene after the rRNA genes. Among the H. pylori genes with highest overall expression were genes involved in pH regulation and ammonia production, including those encoding urease subunits A and B (ureA and ureB), a pH-regulated permease (ureI), and an aliphatic amidase responsible for ammonia production, acylamide amidohydrolase (amiE). Also, two genes encoding the recently characterized histidine-rich, metal-binding polypeptides Hpn (HP1427) and Hpn2 (HP1432) (28), involved in sequestration and intracellular trafficking of nickel, were among the most highly expressed genes.
TABLE 1.
The 20 most highly expressed Helicobacter pylori genes, based on median value over all samples
| Gene | Symbol(s) | Description | Median tpma |
|---|---|---|---|
| 23S rRNA | 23S rRNA | 1,496.94 | |
| 16S rRNA | 16S rRNA | 1,204.63 | |
| HP1192 | Secreted protein involved in flagellar motility HP1192 | 199.80 | |
| Transfer-mRNA | ssrA | Transfer-mRNA SsrA | 93.69 |
| HP0073 | ureA | Urease subunit alpha | 33.27 |
| HP1432 | hpn2 | Histidine and glutamine-rich protein Hpn2 | 33.02 |
| HP1204 | rpL33 | 50S ribosomal protein L33 | 31.09 |
| Nic01_A_00859 | Hypothetical protein | 30.31 | |
| HP1563 | tsaA | Alkyl hydroperoxide reductase TsaA | 29.02 |
| HP1427 | hpn | Histidine-rich, metal-binding polypeptide Hpn | 27.82 |
| tRNA-Ser(gct) | tRNA-Ser(gct) | 20.50 | |
| HP0072 | ureB | Urease subunit beta | 17.03 |
| HP0875 | katA | Catalase KatA | 14.90 |
| HP0294 | amiE | Acylamide amidohydrolase AmiE | 10.73 |
| HP1161 | fldA | Flavodoxin FldA | 9.89 |
| HP0243 | napA | Neutrophil-activating protein NapA (bacterioferritin) | 8.72 |
| HP0824 | trx1, trxA | Thioredoxin TrxA | 8.48 |
| HP0835 | hup | DNA-binding protein HU | 8.37 |
| HP0601 | flaA | Flagellin A, FlaA | 8.32 |
| HP0129 | Hypothetical protein HP0129 | 8.16 |
tpm, reads mapping to the transcripts per million reads.
Urease hydrolyzes urea to ammonia and carbon dioxide, which neutralizes the pH in the acidic stomach. Although our samples showed a progression toward corpus-predominant gastritis and intestinal metaplasia, conditions that usually are associated with an increase of pH, we could not observe a significant change in ureA and ureB expression at later stages of disease. This implies that H. pylori need to maintain high expression levels of urease even during progression toward gastric cancer. The high expression of hpn and hpn2 is also very interesting in this context, since nickel is essential for urease activity and these two proteins regulate the availability of nickel in the cell.
Ammonia is of great importance as a nitrogen source and as a cytotoxic molecule. In addition to urease, H. pylori possess two aliphatic amidases responsible for ammonia production: AmiE, a classical amidase, and AmiF, a new type of formamidase. The amiE and amiF genes were highly expressed in the biopsy specimens, confirming the importance of ammonia and pH regulation for H. pylori infection and suggesting that H. pylori, when associated with the corpus epithelium, faces extensive pH stress (29). In addition, genes involved in protection against oxidative damage, encoding catalase (katA), thioredoxin (trxA), alkyl hydroperoxide reductase (tsaA), flavodoxin (fldA), and superoxide dismutase (sodB), were highly expressed.
Helicobacter pylori virulence gene expression varies with atrophic changes in the tissue.
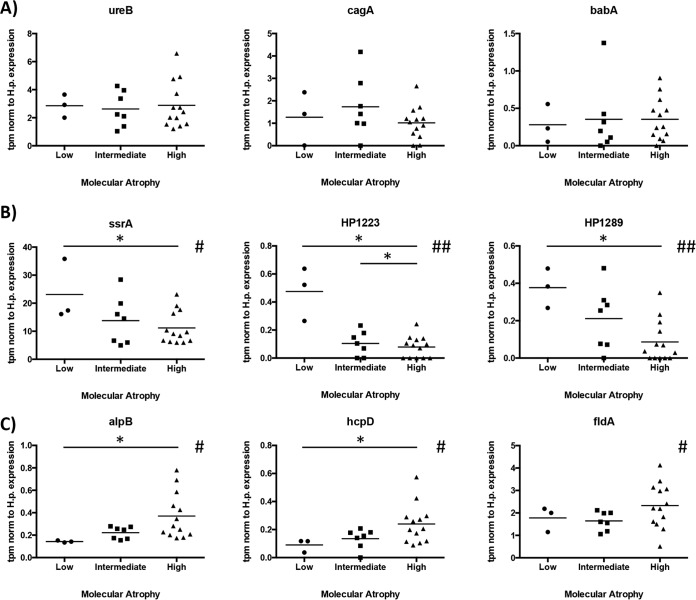
While the absolute levels of expression are important, they do not explain how H. pylori regulates its gene expression in response to the host environment and progressing disease stages, which change the microenvironment at the site of infection. To study the bacterial gene expression at sites of premalignant tissue changes, we compared the expression levels of the 285 identified H. pylori genes to the tissue atrophy levels determined as described above. Albeit not significant after correction for multiple testing, several H. pylori genes showed clear trends of expression related to the degree of atrophy in the tissue (Fig. 3; see Table S7 in the supplemental material). Several of these genes also showed differential expression between the disease stages gastritis and extensive atrophy, but apart from this, few changes were seen associated with the pathology groups. Furthermore, the genes encoding outer membrane protein AlpB, flavodoxin FldA, and beta-lactamase HcpD were more highly expressed at high atrophy than at low atrophy levels (Fig. 3). Several genes were also negatively correlated with the level of atrophy, such as the genes encoding transfer-mRNA SsrA and hypothetical proteins HP1289 and HP1223.
FIG 3.
Differential expression of H. pylori genes relative to the molecular atrophy state of the tissue. (A) Unchanged genes. cagA, cytotoxin-associated gene A; ureB, urease subunit beta; babA, blood group-binding adhesin. (B) Genes with trends of negative correlation to atrophy level. ssrA, transfer-mRNA; HP1223, hypothetical protein HP1223. (C) Genes with trends of positive correlation to atrophy level. alpB, outer membrane protein AlpB; hcpD, beta-lactamase HcpD; fldA, flavodoxin. All values are shown as transcripts per million reads, normalized to total reads mapping to Helicobacter pylori genes. The atrophy score was determined by the combined relative expression of six genes (ATP4A, ATP4B, GHRL, GIF, CCKRB, PGC, PGA3, and PGA4) associated with atrophic gastritis. Asterisks denote differential expression between atrophy stages significant in the limma analysis before correction for multiple testing (*, P < 0.05). Number signs denote a significant association between mean atrophy score and expression value (#, unadjusted P < 0.05; ##, unadjusted P < 0.01).
Few studies on Helicobacter pylori gene expression in vivo exist to date and only a few genes have been studied in relationship to pathological changes in the mucosa (30, 31). When the gastric corpus mucosa becomes atrophic, several factors change in the local environment for stomach bacteria due to changes in gland cell type composition, e.g., decreased acid secretion due to loss of parietal cells. Several studies have investigated the transcriptional response to acid and other environmental stressors in vitro, showing that katA as well as cagA, vacA, and alpB expression levels are affected by acidic stress. HP1289, which encode an H. pylori-specific protein of unknown function, also has been shown to be regulated by pH (32).
AlpB is a colonization factor and binds to laminin (33). Upregulation of alpB, but not other adhesion factors, at high levels of atrophy might indicate a specific role for this adhesion factor. As laminins are located in the extracellular matrix of the gastric basement membrane (34), the Alp adhesins might play more prominent roles in areas of tissue damage or in bacterial invasion of the tissue. Laminins have also been shown to be differentially distributed in different parts of the healthy stomach and differentially expressed in premalignant and malignant lesions (35), suggesting that it might be beneficial for the bacteria to adapt to these changes.
In the differential expression analysis, several genes showed a tendency of being influenced by tissue atrophy levels, albeit not significantly after correction for multiple testing. Our sample groups were small, particularly the group with low levels of atrophy, and thus the statistical power may not be sufficient for detection of more subtle changes. Additional studies using larger sample groups and sequencing depths will probably uncover additional biomarkers for severe atrophy and H. pylori-associated disease progression. It would also be valuable to study this in subjects from several geographical locations, since the microbiota has been shown to vary in different areas of the world (36). Furthermore, the low levels of bacterial RNA in the samples in relation to human RNA resulted in exclusion of many genes from the analysis due to the fact that their read counts were below the detection limit in many of the samples, which could be alleviated by increasing sequencing depth or by depletion of human RNA in further studies.
This study shows that metatranscriptomic analysis of the gastric microbiota is feasible and can provide new insights into how bacteria behave in vivo in different stages of disease progression and under different environmental conditions. Coupling metatranscriptomic data with host gene expression levels also gives the possibility to study host-bacterial interaction in an unprecedented manner. Here, we describe the viable stomach mucosa-associated microbiota in an unbiased fashion, and at the same time we in detail study a specific species, Helicobacter pylori, in its niche in individuals with different forms of disease-associated histological changes. We found that although H. pylori infection does not change the bacterial diversity, there are correlations between the presence of Helicobacter and the Campylobacter, Sulfurospirillum, and Deinococcus genera. Analysis of viable H. pylori at the site of infection showed that expression of several genes showed a tendency to vary with atrophy levels in the infected stomach mucosa. However, the low proportion of bacterial RNA in the biopsy specimens calls for further development of enrichment techniques for microbial RNA in complex samples. Better enrichment strategies would further contribute to the use of in vivo large-scale transcriptomics analyses and would shed essential light on the host-microbial interaction at different stages of disease.
MATERIALS AND METHODS
Patients.
Patients were selected from a cohort of 149 patients undergoing endoscopy during 2010 and 2011 at the Hospital Escuela Lenin Fonseca in Managua, Nicaragua, an area of high H. pylori and gastric cancer prevalence. The sample collection was approved by the Human Research Ethics Committee at the University of Gothenburg (approval number 176-10) as well as by Universidad National Autonóma de Nicaragua, Managua (UNAN). All individuals included in the cohort agreed to participate in the study, and informed oral and written consent was obtained from each patient before participation.
All patients were tested for Helicobacter pylori infection by the urea breath test (UBT) using an IR300 spectrophotometer (Otsuka, MD, USA), by H. pylori serology according to procedures described previously (37), and by H. pylori culturing. From all patients, several endoscopic biopsy specimens were collected from different anatomic locations of the stomach, together with serum samples and whole blood to enable a variety of analyses on each patient. From this cohort, 25 patients were selected based on pathology grading of 4 antrum and 4 corpus biopsy specimens from each individual. The biological groups selected were (i) patients with gastritis, i.e., no signs of atrophy in any of the corpus biopsy specimens (Gast), (ii) patients with low-grade atrophic gastritis in the corpus (Atr), (iii) patients with extensive corpus atrophy but no signs of metaplasia (EA), and (iv) patients with intestinal metaplasia in the corpus (Met). In addition to this, we also selected five subjects who had no Helicobacter pylori infection as determined by UBT, serology, and culturing and no or very low signs of inflammation in the corpus mucosa as a control group (Min). A brief description of the subjects is given in Table 2 and more detailed clinical data in Table S1 in the supplemental material.
TABLE 2.
Subjects included in the study
| HEALFa ID | RNA-seq ID | Age (yr) | Sexb | H. pylori isolatec | Atrophy score by RNA-seqd |
|---|---|---|---|---|---|
| HEALF19432 | Min1 | 47 | M | NA | Low |
| HEALF24260 | Min2 | 34 | F | NA | Low |
| HEALF23836 | Min3 | 54 | F | NA | High |
| HEALF04977 | Min4 | 23 | M | NA | Low |
| HEALF05581 | Min5 | 51 | F | NA | Low |
| HEALF25546 | Gast1 | 27 | F | + | Intermediate |
| HEALF06010 | Gast2 | 35 | F | + | Intermediate |
| HEALF23466 | Gast3 | 30 | F | + | Intermediate |
| HEALF16065 | Gast4 | 36 | F | + | Low |
| HEALF12846 | Gast5 | 44 | F | + | Low |
| HEALF00138 | Gast6 | 24 | F | + | High |
| HEALF19582 | Atr1 | 47 | F | + | High |
| HEALF09107 | Atr2 | 41 | M | NA | Intermediate |
| HEALF03953 | Atr3 | 42 | F | NA | Intermediate |
| HEALF27875 | Atr4 | 27 | M | + | Intermediate |
| HEALF19868 | Atr5 | 53 | F | + | High |
| HEALF19162 | Atr6 | 58 | M | + | Low |
| HEALF08173 | Atr7 | 32 | F | + | High |
| HEALF01245 | Atr8 | 35 | F | + | High |
| HEALF08149 | Atr9 | 27 | F | + | High |
| HEALF24875 | EA1 | 48 | M | + | High |
| HEALF27688 | EA2 | 66 | F | + | High |
| HEALF02414 | EA3 | 29 | F | + | High |
| HEALF24293 | EA4 | 27 | M | + | High |
| HEALF14993 | EA5 | 47 | F | + | High |
| HEALF00568 | EA6 | 44 | F | NA | High |
| HEALF23215 | Met1 | 47 | M | + | High |
| HEALF03191 | Met2 | 38 | F | NA | Low |
| HEALF02475 | Met3 | 53 | F | + | Intermediate |
| HEALF03442 | Met4 | 57 | F | NA | High |
HEALF, Hospital Escuela Antonio Lenin Fonseca.
M, male; F, female.
+, individual with a whole-genome-sequenced H. pylori isolate; NA, individual from whom no H. pylori isolate could be obtained.
The atrophy score was determined by the combined relative expression of six genes (ATP4A, ATP4B, GHRL, GIF, CCKRB, PGC, PGA3, and PGA4) associated with atrophic gastritis.
Of the 30 subjects, the majority (22/30) were women, and the median age was 41.5 years, ranging from 23 to 66 years (Table 2). All the subjects with gastritis and corpus atrophy were H. pylori positive based on UBT and serology, but from one of the corpus atrophy patients (Atr2) it was not possible to render any viable H. pylori colonies. Two of the individuals with intestinal metaplasia (Met2 and Met4) were H. pylori negative in both serology and culture even though one of them displayed a positive UBT test (Met4).
RNA extraction.
Biopsy specimens for RNA extraction were obtained during endoscopy and were snap-frozen in RNAlater (Ambion) and shipped frozen on dry ice to Sweden. For this study, we used one corpus biopsy specimen per individual described in Table 2. The content of the vials was spun down to collect all cells and mucus, and the RNAlater was removed carefully. RNA was extracted using the RNeasy minikit (Qiagen) with the addition of lysozyme and proteinase K to the lysis buffer prior to disruption of the tissues using the TissueLyser (Qiagen). The RNA was subjected to double DNase treatment to ensure that the analysis would not be confounded by DNA contamination, as described previously (38). The integrity of the resulting RNA was assessed using a BioAnalyser (Agilent) with an RNA integrity number (RIN) of >6 as a cutoff, and the RNA concentration was determined by Qubit (Invitrogen) measurement just prior to the library preparation procedure.
Library preparation of cDNA was performed using the strand-specific ScriptSeq v2 RNA sample preparation kit (Epicentre Biotechnologies), including RiboZero eukaryotic rRNA depletion treatment (Epicentre). After library preparation, libraries were quantified, pooled for clustering, and sequenced using the Illumina HiScanSQ instrument, rendering for each sample a total of 20 to 30 million paired-end reads of 100 bp each. The RNA sequencing was performed at the Genomics Core Facility, Sahlgrenska Academy, University of Gothenburg.
16S rRNA amplicon analysis from cDNA.
To verify the presence of low-abundance Helicobacter rRNA in the clinically H. pylori-negative samples, RNA samples were reverse transcribed using the QuantiTect kit with accompanying genomic DNA (gDNA) depletion (Qiagen). In addition to the corpus samples, RNA was extracted from antral biopsy specimens from the same individuals using the same procedures as described above. For three individuals, Min1, Min3, and Min4, no antrum biopsy specimens were available for RNA extraction.
16S rRNA amplicon libraries were constructed from the cDNA samples using the universal primers 341F and 805R to amplify the V3-V4 region. The libraries were generated in a single PCR step with 25 cycles using a high-fidelity polymerase (Phusion Hot Start II high-fidelity PCR master mix; Thermo Scientific). Subsequent purification was carried out using a magnetic bead capture kit (AMPure; Agencourt) and quantified utilizing a fluorometric kit (QuantIT PicoGreen; Invitrogen). The purified amplicons were normalized to 4 nM and pooled equimolarly or, if the library was of lower concentration, using the same volume as if the library had been 4 nM. Sequencing was performed on the MiSeq platform using v3 chemistry, generating 2 × 300-bp pair-end reads. Negative PCR controls without template were also sequenced to monitor the background from the 16S rRNA PCR amplification.
Bioinformatics analysis.
Raw RNA-seq reads were trimmed using TrimGalore! version 0.3.5 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with a quality cutoff of Q30, Illumina adapter trimming, and removal of reads shorter than 30 bp and reads left unpaired. In addition to this, low-complexity reads were filtered out using PrinSeq version 0.20.4 (39), applying the DUST algorithm (40) to avoid unspecific matches in the alignment analyses. After quality filtering, 17 to 36 Mreads (average 25) remained for each sample.
Taxonomic analysis using rRNA transcripts.
To analyze the microbial composition of the biopsy specimen tissues, we used the Metaxa2 software version 2.1.1 (20) to extract and classify small-subunit (SSU) rRNA transcripts from the RNA-seq data. Metaxa2 was applied to the quality trimmed and filtered reads using default settings, with the exception of using blast+ rather than the default blastall program and setting the minimum read length to be included in the analysis to 75 bp to ensure a high specificity. Genus annotation in Metaxa2 is done by an internal database, based on the high-quality SILVA database, release 111 (July 2012), which consists of SSU rRNA sequences from the Bacteria, Archaea, and Eukaryota domains (41). The resulting abundance files on the different taxonomic levels were filtered so that only taxa with more than one read per million total reads were used for downstream analysis. The raw genus counts of reads classified as 16S rRNA for each sample are given in Table S2 in the supplemental material. Principal-component analysis (PCA) and other statistical analyses were performed in the R software v3.3.0. Relationships between Helicobacter abundance and the abundance of other genera were assessed using Spearman's rank correlation, and P values were corrected for multiple testing using the Benjamini and Hochberg method (significance cutoff, 0.05). Significant effects of Helicobacter presence (as assessed by traditional tests) on overall community structure on the phylum and genus levels were assessed using metaxa2_uc, part of the Metaxa2 Diversity Tools (42) (default options). In this analysis, the within-group and between-group Bray-Curtis dissimilarities were tested using subsampling for 10,000 iterations (1,000 sampled reads per iteration). Simpson and Shannon diversities as well as richness of genera in the samples were assessed using the R package Vegan version 2.0-10 (43). Significant diversity differences between clinically classified H. pylori-positive and -negative individuals were detected using Student's t test. Regression to human ATP4 gene expression and atrophy scores (see below) was performed using fitting of linear models in R. P values were corrected for multiple testing using the Benjamini and Hochberg method, and corrected P values were considered significant when they were below 0.05.
16S rRNA data analysis.
After trimming, 16S rRNA amplicon reads were trimmed using Cutadapt (44), merged using vsearch –fastq_mergepairs (45), and thereafter processed using Metaxa2 as described above. Raw 16S rRNA read counts classified on the genus level are given in Table S2. PCA plots were constructed in the R Studio software using log-transformed raw counts from Metaxa2. To assess differences between corpus and antrum samples, metaxa2_uc was used to calculate Bray-Curtis dissimilarity between samples and within each corpus/antrum sample pair (additional options, “-g none -r 100 –table T –matrix T”). The within- and between-sample dissimilarities were related to disease stage as well as human ATP4 gene expression.
Helicobacter pylori gene expression analysis.
For identification of Helicobacter pylori transcripts, we created a custom mapping reference. For 20 of the individuals, we had previously whole-genome sequenced one Helicobacter pylori isolate from the antrum and one from the corpus (Table 2). In addition, from one individual we had sequenced two antrum isolates, from one subject two corpus isolates, and from one other subject an antrum isolate only (see Table S3 in the supplemental material). From one patient who was Helicobacter pylori positive based on UBT and serology, we could not obtain any viable colonies in culture (46). From these isolates, draft genomes were prepared by de novo assembly using SPAdes v3.7.0 (47) and annotated as described previously using the prokka annotation pipeline (48) with a recent annotation of strain 26695 (49) as the primary annotation source, further curated with respect to outer membrane proteins (50) and factors involved in DNA transfer and competence (51). A complete list of the annotation is given in Table S4 in the supplemental material. From the annotated draft genomes, FASTA files of nucleotide coding sequences (CDSs) with in total 82,661 CDSs were clustered on 100% identity using USearch v8.0.1623 (52) in cluster_fast mode, rendering a FASTA file of 46,515 centroid sequences. This file was concatenated with a file of all transcripts of the human genome annotation GRCh38, release 79. The combined FASTA file was used to generate a reference for kallisto (53) with a kmer length of 31 bp. Kallisto mapping was performed to the combined reference and generated transcript per million (tpm) counts for each transcript in the reference. To aggregate the CDSs of the different H. pylori strains, counts were first aggregated on gene name, i.e., that they had been annotated to the same source (e.g., a certain CDS in the 26695 file or a specific UniProt number). To further group the different H. pylori CDSs into orthologous groups of genes, we again performed USearch clustering, this time on the protein FASTA output files from prokka, with an identity cutoff of 80%. The H. pylori counts were then aggregated for each such orthologous group to collect all counts.
Determining the level of atrophic changes based on gene expression.
To determine the extent of atrophic changes in the tissue, we used eight tissue-specific human genes for areas of atrophic changes: ATP4A, ATP4B, GHRL, GIF, CCKRB, PGC, PGA4, and PGA3 (25, 26). Based on the RNA-seq counts per million reads for these genes, every sample received a score on a three-level scale for each gene, where score 1 corresponded to expression lower than the 95% confidence interval (CI) of the expression of that gene across all 30 individuals, 2 corresponded to expression within the 95% CI, and 3 corresponded to expression greater than the 95% CI. The overall molecular atrophy score was determined using the median value of the scores, where a median of 1 was considered high-grade atrophy, a median of between 1.5 and 2.5 intermediate atrophy, and a median of 3 low-grade atrophy. Atrophy scores, mean ATP4 (ATP4A and ATP4B) expression for each sample, and the correlation between the two are shown in Fig. S1 in the supplemental material.
Differential expression analysis of H. pylori during atrophic changes.
On the kallisto counts, we applied a cutoff a median tpm higher than 1 for a gene to be included in further analysis, resulting in a list of 285 H. pylori genes (see Table S5 in the supplemental material). Since the reads mapping to H. pylori accounted for only a small fraction of the total reads, the kallisto tpm for each gene was normalized to the total tpm mapping to H. pylori genes in that sample. Samples from individuals clinically H. pylori negative were excluded from differential expression analysis due to radically lower total H. pylori counts which otherwise would confound the analysis. Differential expression analysis was performed using limma v3.28.20 (54, 55), comparing the normalized expression values (i) between pathology groups, (ii) between groups of molecular atrophy, and relative to (iii) mean ATP4 expression (ATP4A and ATP4B) and (iv) mean atrophy score.
Availability of data.
The RNA-seq data set has been deposited to the ArrayExpress public repository (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3689.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by Swedish Research Council grant no. K2012-56X-22029-01-3, VINNOVA grant no. 2011-03491, and Swedish Foundation for Strategic Research (SSF) grant no. SB12-0072 to Å.S., Swedish Research Council grant no. 2012-3329 to S.B.L., grants from the Knut and Alice Wallenberg Foundation and the Bioinformatics Infrastructure for Life Sciences (BILS) to I.N., and Assar Gabrielsson foundation grant no. FB11-68 and FB12-84 to K.T.
We thank the gastroenterologists and pathologists at Hospital Escuela Antonio Lenin Fonseca, Managua, Nicaragua, who made the sample collection possible. Sequencing was performed at the Genomics Core Facility, Sahlgrenska Academy, University of Gothenburg, and the Center of Translational Microbiome Research (CTMR), Karolinska Institutet, Stockholm, Sweden. All bioinformatics analyses were performed on resources provided by SNIC through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00031-17.
REFERENCES
- 1.Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311–1315. [DOI] [PubMed] [Google Scholar]
- 2.Sjostedt S, Kager L, Heimdahl A, Nord CE. 1988. Microbial colonization of tumors in relation to the upper gastrointestinal tract in patients with gastric carcinoma. Ann Surg 207:341–346. doi: 10.1097/00000658-198803000-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delgado S, Cabrera-Rubio R, Mira A, Suarez A, Mayo B. 2013. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol 65:763–772. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- 4.Zilberstein B, Quintanilha AG, Santos MA, Pajecki D, Moura EG, Alves PR, Maluf Filho F, de Souza JA, Gama-Rodrigues J. 2007. Digestive tract microbiota in healthy volunteers. Clinics (Sao Paulo) 62:47–54. doi: 10.1590/S1807-59322007000100008. [DOI] [PubMed] [Google Scholar]
- 5.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. 2014. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep 4:4202. doi: 10.1038/srep04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. 2011. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J 5:574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK. 2017. Alterations in gastric microbiota after H. pylori eradication and in different histological stages of gastric carcinogenesis. Sci Rep 7:44935. doi: 10.1038/srep44935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, Lau JY, Sung JJ, Ding C. 2009. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One 4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yakirevich E, Resnick MB. 2013. Pathology of gastric cancer and its precursor lesions. Gastroenterol Clin North Am 42:261–284. doi: 10.1016/j.gtc.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. 2001. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 13.Piazuelo MB, Correa P. 2013. Gastric cancer: overview. Colomb Med (Cali) 44:192–201. [PMC free article] [PubMed] [Google Scholar]
- 14.Amieva M, Peek RM Jr. 2016. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 150:64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wroblewski LE, Peek RM Jr. 2016. Helicobacter pylori, cancer, and the gastric microbiota. Adv Exp Med Biol 908:393–408. doi: 10.1007/978-3-319-41388-4_19. [DOI] [PubMed] [Google Scholar]
- 16.Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. 2009. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol 58:509–516. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- 17.Jo HJ, Kim J, Kim N, Park JH, Nam RH, Seok YJ, Kim YR, Kim JS, Kim JM, Kim JM, Lee DH, Jung HC. 2016. Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter 21:364–374. doi: 10.1111/hel.12293. [DOI] [PubMed] [Google Scholar]
- 18.Yang I, Woltemate S, Piazuelo MB, Bravo LE, Yepez MC, Romero-Gallo J, Delgado AG, Wilson KT, Peek RM, Correa P, Josenhans C, Fox JG, Suerbaum S. 2016. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep 6:18594. doi: 10.1038/srep18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Correa P, Piazuelo MB. 2012. The gastric precancerous cascade. J Dig Dis 13:2–9. doi: 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH. 2015. Metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour 15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 21.Wu WM, Yang YS, Peng LH. 2014. Microbiota in the stomach: new insights. J Dig Dis 15:54–61. doi: 10.1111/1751-2980.12116. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Amado MA, Al-Soud WA, Borges-Landaez P, Contreras M, Cedeno S, Baez-Ramirez E, Dominguez-Bello MG, Wadstrom T, Gueneau P. 2007. Non-pylori Helicobacteraceae in the upper digestive tract of asymptomatic Venezuelan subjects: detection of Helicobacter cetorum-like and Candidatus Wolinella africanus-like DNA. Helicobacter 12:553–558. doi: 10.1111/j.1523-5378.2007.00526.x. [DOI] [PubMed] [Google Scholar]
- 23.Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porras C, Nodora J, Sexton R, Ferreccio C, Jimenez S, Dominguez RL, Cook P, Anderson G, Morgan DR, Baker LH, Greenberg ER, Herrero R. 2013. Epidemiology of Helicobacter pylori infection in six Latin American countries (SWOG trial S0701). Cancer Causes Control 24:209–215. doi: 10.1007/s10552-012-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HJ, Nam KT, Park HS, Kim MA, Lafleur BJ, Aburatani H, Yang HK, Kim WH, Goldenring JR. 2010. Gene expression profiling of metaplastic lineages identifies CDH17 as a prognostic marker in early stage gastric cancer. Gastroenterology 139:213–225 e213. doi: 10.1053/j.gastro.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nookaew I, Thorell K, Worah K, Wang S, Hibberd ML, Sjovall H, Pettersson S, Nielsen J, Lundin SB. 2013. Transcriptome signatures in Helicobacter pylori-infected mucosa identifies acidic mammalian chitinase loss as a corpus atrophy marker. BMC Med Genomics 6:41. doi: 10.1186/1755-8794-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janzon A, Bhuiyan T, Lundgren A, Qadri F, Svennerholm AM, Sjoling A. 2009. Presence of high numbers of transcriptionally active Helicobacter pylori in vomitus from Bangladeshi patients suffering from acute gastroenteritis. Helicobacter 14:237–247. doi: 10.1111/j.1523-5378.2009.00692.x. [DOI] [PubMed] [Google Scholar]
- 28.Vinella D, Fischer F, Vorontsov E, Gallaud J, Malosse C, Michel V, Cavazza C, Robbe-Saule M, Richaud P, Chamot-Rooke J, Brochier-Armanet C, De Reuse H. 2015. Evolution of Helicobacter: acquisition by gastric species of two histidine-rich proteins essential for colonization. PLoS Pathog 11:e1005312. doi: 10.1371/journal.ppat.1005312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 30.Peek RM, Jr van Doorn LJ, Donahue JP, Tham KT, Figueiredo C, Blaser MJ, Miller GG. 2000. Quantitative detection of Helicobacter pylori gene expression in vivo and relationship to gastric pathology. Infect Immun 68:5488–5495. doi: 10.1128/IAI.68.10.5488-5495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semino-Mora C, Doi SQ, Marty A, Simko V, Carlstedt I, Dubois A. 2003. Intracellular and interstitial expression of Helicobacter pylori virulence genes in gastric precancerous intestinal metaplasia and adenocarcinoma. J Infect Dis 187:1165–1177. doi: 10.1086/368133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol 188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senkovich OA, Yin J, Ekshyyan V, Conant C, Traylor J, Adegboyega P, McGee DJ, Rhoads RE, Slepenkov S, Testerman TL. 2011. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect Immun 79:3106–3116. doi: 10.1128/IAI.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Virtanen I, Tani T, Back N, Happola O, Laitinen L, Kiviluoto T, Salo J, Burgeson RE, Lehto VP, Kivilaakso E. 1995. Differential expression of laminin chains and their integrin receptors in human gastric mucosa. Am J Pathol 147:1123–1132. [PMC free article] [PubMed] [Google Scholar]
- 35.Tani T, Karttunen T, Kiviluoto T, Kivilaakso E, Burgeson RE, Sipponen P, Virtanen I. 1996. Alpha 6 beta 4 integrin and newly deposited laminin-1 and laminin-5 form the adhesion mechanism of gastric carcinoma. Continuous expression of laminins but not that of collagen VII is preserved in invasive parts of the carcinomas: implications for acquisition of the invading phenotype. Am J Pathol 149:781–793. [PMC free article] [PubMed] [Google Scholar]
- 36.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcon O, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattsson A, Tinnert A, Hamlet A, Lonroth H, Bolin I, Svennerholm AM. 1998. Specific antibodies in sera and gastric aspirates of symptomatic and asymptomatic Helicobacter pylori-infected subjects. Clin Diagn Lab Immunol 5:288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicklasson M, Sjoling A, von Mentzer A, Qadri F, Svennerholm AM. 2012. Expression of colonization factor CS5 of enterotoxigenic Escherichia coli (ETEC) is enhanced in vivo and by the bile component Na glycocholate hydrate. PLoS One 7:e35827. doi: 10.1371/journal.pone.0035827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgulis A, Gertz EM, Schaffer AA, Agarwala R. 2006. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol 13:1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 41.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bengtsson-Palme J, Thorell K, Wurzbacher C, Sjoling A, Nilsson RH. 2016. Metaxa2 diversity tools: easing microbial community analysis with Metaxa2. Ecological Informatics 33:45–50. doi: 10.1016/j.ecoinf.2016.04.004. [DOI] [Google Scholar]
- 43.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos PM, Stevens HH, Szoecs E, Wagner H. 2017. vegan: community ecology package. https://cran.r-project.org/web/packages/vegan/.
- 44.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 45.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. 2016. VSEARCH: a versatile open source tool for metagenomics. Peer J 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorell K, Hosseini S, Palacios Gonzales RV, Chaotham C, Graham DY, Paszat L, Rabeneck L, Lundin SB, Nookaew I, Sjoling A. 2016. Identification of a Latin American-specific BabA adhesin variant through whole genome sequencing of Helicobacter pylori patient isolates from Nicaragua. BMC Evol Biol 16:53. doi: 10.1186/s12862-016-0619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Resende T, Correia DM, Rocha M, Rocha I. 2013. Re-annotation of the genome sequence of Helicobacter pylori 26695. J Integr Bioinform 10:233. doi: 10.2390/biecoll-jib-2013-233. [DOI] [PubMed] [Google Scholar]
- 50.Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun 68:4155–4168. doi: 10.1128/IAI.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez-Gonzalez E, Backert S. 2014. DNA transfer in the gastric pathogen Helicobacter pylori. J Gastroenterol 49:594–604. doi: 10.1007/s00535-014-0938-y. [DOI] [PubMed] [Google Scholar]
- 52.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 53.Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 54.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonsson V, Osterlund T, Nerman O, Kristiansson E. 2016. Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genomics 17:78. doi: 10.1186/s12864-016-2386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.