Abstract
Animal development proceeds through the activity of genes and their cis-regulatory modules, working together in sets of gene regulatory networks (GRNs). Emergence of species-specific traits and novel structures results from evolutionary changes in GRNs. Recent work in a wide variety of animal models, and particularly in insects, has started to reveal the modes and mechanisms of GRN evolution. I discuss here different aspects of GRN evolution and argue that developmental system drift, in which conserved phenotype is nevertheless a result of changed genetic interactions, should regularly be viewed from the perspective of GRN evolution. Advances in methods to discover related cis-regulatory modules in diverse insect species, a critical requirement for detailed GRN characterization, are also described.
Keywords: GRN, evo-devo, developmental system drift, DSD, cis-regulatory module, enhancer evolution
GRNs and their components
King and Wilson [1], in their seminal paper demonstrating human-chimpanzee proteome similarity, popularized the notion that regulatory changes play a major role in evolution. This idea, subsequently championed by influential evolutionary developmental biologists such as Sean Carroll and the late Eric Davidson [2, 3], has been strengthened by the recent availability of hundreds of fully-sequenced animal genomes. The prevailing paradigm is that a relatively small set of common “toolkit” genes shape the animal body plan, regulated by conserved “subcircuits” (see Glossary) of regulatory interactions, themselves parts of larger gene regulatory networks (GRNs). The linkages between these modular genetic building blocks can be altered via evolutionary changes at gene regulatory sequences, leading to morphological changes and eventually, speciation.
If changes in GRNs underlie much of how evolution proceeds at the organismal scale, it follows that to understand evolution we must study how GRNs evolve at the molecular scale. GRNs (Fig. 1) consist of transcription factors (TFs) and the cis-regulatory modules (CRMs, e.g. “enhancers”) that control spatio-temporal patterns of gene expression. Signaling pathways can also be included in a GRN, where they often serve as links between GRN subcircuits. (Signaling components can also function as agents of GRN evolution—e.g. see [4]—but are not further discussed here.) For purposes of this article, I will hold to a strict definition of GRNs as requiring at a minimum both TFs and CRMs. For a GRN to be understood as a network, both edges (TFs) and nodes (CRMs) must be defined, and changes to both the TFs and the CRMs must be considered when exploring the mechanisms of GRN evolution.
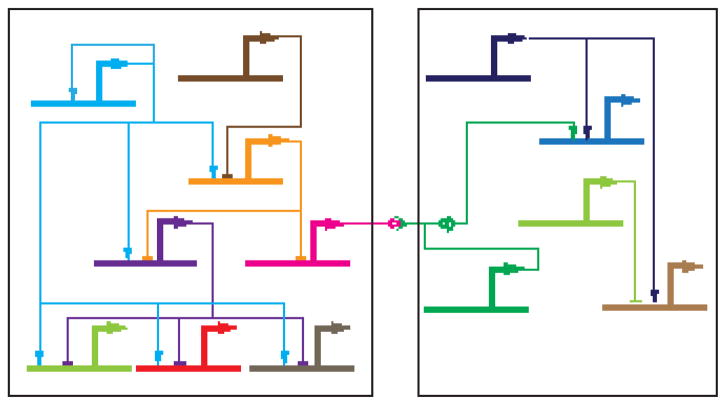
Figure 1. A gene regulatory network.
A hypothetical GRN functioning in two cells is shown. Heavy lines indicate CRMs and lighter-weight lines TF-DNA or protein-protein interactions (→, transcriptional activation; —|, transcriptional repression; —o>—, ligand-receptor binding; —o)— signal transduction process; see [62]). The TF represented by light blue coloring at the upper left is a “master regulator” whose activity impinges on the expression of many other genes in the network, including its own via autoregulation. For an introduction to GRNs, see Levine and Davidson [63].
Modes of GRN Evolution
Studying GRN evolution at a detailed molecular level is more challenging than it might initially appear. To do so, we require well-defined GRNs (i.e., with both TFs and sufficiently characterized CRMs) in not just one species, but rather in two or more related species (note that more than two species are necessary if the direction of evolutionary change is to be inferred). As there are still relatively few highly-characterized GRNs even in a single species, this represents a significant obstacle. Moreover, for efficient molecular, CRM-based analysis, the species being compared must be far enough diverged that there are clear genotypic and phenotypic differences, yet not so far diverged that homologous CRMs can no longer be identified. Despite these challenges, examples are beginning to accrue both of deep conservation of GRNs as well as of changes in GRNs leading to the evolution of new phenotypes and new structures, in organisms as diverse as sea urchins to flies to fish to mammals. A survey of these examples (many of which have recently been reviewed in more detail elsewhere [5]) suggests four basic classes of GRN evolution (Figure 2, Key Figure) [6]. These distinctions are not in all cases absolute—there can be some overlap between classes—and at times may result as much from incomplete knowledge of the GRNs under consideration as from mechanistically different modes of GRN evolution. Nevertheless, they can be instructive in considering the ways in which GRNs evolve.
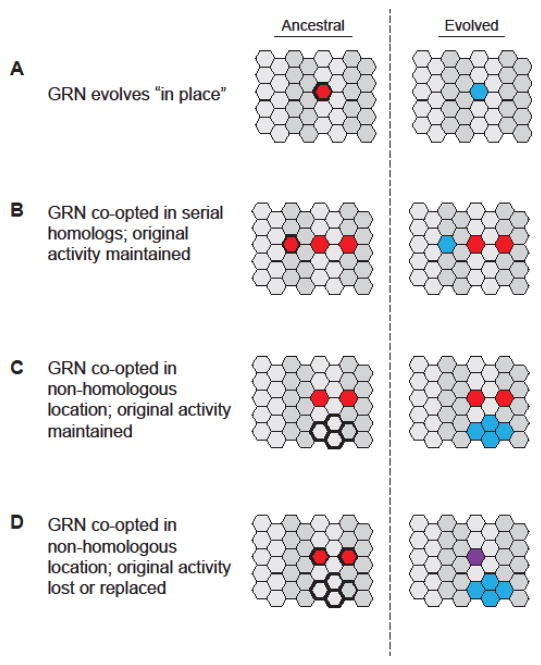
Figure 2. Modes of GRN evolution.
Examples of at least four different modes of GRN evolution have been described. Each panel depicts cells (hexagons) organized into segments or compartments (light versus dark shading). Cells in the ancestral organism (left) that will experience GRN evolution are outlined in bold. (A) GRN evolution occurs “in place” when the ancestral GRN (red) changes to create a GRN with modified activity (blue) and thus a new phenotype in directly homologous cells or tissues. Alternatively, the ancestral state can be maintained while the GRN is co-opted via one or more changes to produce a modified serial homolog of the original structure (B), or a novel structure or phenotype in a non-homologous group of cells (C). Finally, a “repeal, replace, and redeploy” scenario can occur in which the GRN (red) evolves such that the ancestral function is lost from its original location but new activity is gained in a non-homologous group of cells (blue). The original function may be lost altogether or replaced by a different GRN (purple). Adapted from [6].
Evolution “in place”
The class for which we have the greatest number of examples is GRN evolution “in place”: changes in a GRN within a common (homologous) tissue in two species (Fig. 2A). For example, development of the pelvic spine on stickleback fish requires a GRN regulated by the TF Pitx1. Inactivation of the Pitx1 CRM leads to pelvic reduction, an evolutionary event that has occurred independently in multiple stickleback lineages [7]. More dramatically, the GRN responsible for echinoderm mesoderm development consists of a conserved upstream subcircuit which is coupled to diverged downstream components in sea urchins and sea stars. This leads to significantly different mesodermal fates subsequent to similar initial specification of the mesoderm [8]. Evolution of the GRNs governing pigmentation in drosophilids, or the patterning of Drosophila larval denticles, provide further examples [9–15]. A feature of GRN evolution “in place” is that in the evolved species, the ancestral GRN is no longer active in the tissue in question. This is in contrast to the modes described below, where a GRN has evolved or been co-opted in a new tissue, but remains functional in at least some of its original locations.
Evolution in serially homologous tissues
A second mode of GRN evolution is evolution of the network in a serially homologous structure (Fig. 2B). Hox gene control of insect fore- and hind-wing development provides several examples of this. In Drosophila and other Diptera, wings develop in the second thoracic segment (T2) while in the third thoracic segment (T3), a balancing organ known as a haltere develops instead of the hindwing observed in other winged insects. The haltere is a serial homolog of the wing and can undergo homeotic transformation to wing fate upon loss of the Hox gene Ultrabithorax (Ubx), demonstrating that the wing GRN, believed to be ancestral, is intact but repressed in T3 [reviewed by 16]. Examination of several wing GRN CRMs at different levels in the GRN has shown that they are bound directly by Ubx, which can act to both activate and repress gene expression [17–20]. These data suggest that the ancestral wing GRN may have experienced evolution at numerous nodes through addition of Ubx binding sites, likely a gradual evolutionary process [21].
In beetles, such as the red flour beetle Tribolium castaneum, it is the T2 forewing that is modified, into a hardened wing cover called an elytron; the T3 hindwing maintains the familiar membranous wing form. In contrast to the case in Drosophila, Tribolium Ubx is necessary to promote, rather than repress, the wing fate. This could be accounted for by acquisition of Ubx binding sites in CRMs of multiple members of the wing GRN, which became indispensible for T3 wing gene expression [22]. Such an explanation would be consistent with the multiple gains of Ubx responsiveness observed in Drosophila, although a test of this hypothesis must await characterization of sufficient beetle wing GRN CRMs. The exoskeleton-related structure of the T2 elytron appears to be built through multiple co-options of exoskeleton genes into, or downstream of, the wing GRN [23]. Thus in T2 the wing GRN appears to remain functional at the upper levels, with heavy modification taking place toward the terminal branches. Defining the mechanisms through which the exoskeleton components have been co-opted at multiple places into the wing GRN likewise must await characterization of the relevant CRMs.
Evolution of the wing GRN has also been demonstrated in the first thoracic segment (T1) of T. castaneum [24]. Here, the carinated margin, a T1 structure, is likely a serial homolog of the wings. Importantly, elements of the wing GRN, including a CRM regulating expression of the important wing-development gene nubbin, are active in this tissue. However, nubbin itself does not appear to play a role in carinated margin development. Activity of the nubbin CRM therefore may be an evolutionary hold-over, with a disruption in the GRN someplace downstream of nubbin in the version of the network leading to carinated margin fate [24]. Note that to the extent so far determined, the GRN remains active in both serially homologous tissues, albeit with significant changes in function that still require characterization. This differs both from the situation discussed above for Drosophila, where wing GRN genes are repressed and their CRMs inactive in the non-wing forming T3 segment, and for the Tribolium T2 elytra, where the wing GRN retains its wing-building function, but also has been modified through co-option of additional elements.
Evolution in non-homologous tissues
A common feature of both GRN evolution in place and GRN evolution in serial homologs is the fact that in the ancestral species, the GRN would have been functional in the relevant tissue (Fig. 2A, B, bold outline), i.e., all of the necessary TFs and signaling pathways would have been present and accessible for use. In contrast, GRN evolution in a non-homologous tissue implies redeployment of a GRN in a place where it was not originally active, suggesting that one or more key regulators were absent in the new location (Fig. 2C). This appears to be the case for the evolution of the posterior lobe, a male genital outgrowth present in Drosophila melanogaster and closely related species, but not in species outside of the melanogaster subgroup. Detailed analysis has shown that posterior lobe development utilizes a GRN co-opted from development of the larval posterior spiracle [25]. Not only is the posterior lobe not a serial homolog of the spiracle, it is a novel morphological structure that develops from a non-homologous field of cells. In fact, in non-lobed species, only two of the ten genes identified as members of the shared posterior spiracle/posterior lobe GRN are expressed in the cells which in lobed species give rise to the posterior lobe. However, all seven identified posterior lobe CRMs are jointly active in both the spiracle and the posterior lobe. This suggests that in contrast to the acquisition of Ubx-dependent repression in multiple CRMs of the Drosophila wing GRN described above, trans-regulatory changes significantly outweigh cis-regulatory changes in the evolution of this network. One likely candidate for a responsible trans-acting factor is the STAT ligand encoded by unpaired (upd). upd sits near the top of the GRN and, unlike the other examined shared genes, does not appear to be regulated via a shared CRM—the sole upd posterior spiracle CRM identified to date is not active in the posterior lobe [25]. Whether introduction of upd expression alone is sufficient to activate the posterior spiracle GRN in the genitalia, or if additional TFs must also be introduced, remains to be determined.
Evolution by “repeal, replace, and redeploy”
A fourth mode of GRN evolution has been dubbed “repeal, replace, and redeploy ” (Fig. 2D) [6]. A comparison of nervous system development between Drosophila and the mosquito Aedes aegypti has revealed that a GRN involved in ventral midline development in Drosophila appears to have shifted—been “redeployed”—from medial to lateral regions in the A. aegypti late embryonic nervous system. The functional consequences of this shift are not obvious, and overall nervous system morphology appears identical. Similarly, midline development appears largely identical in the two species, suggesting that a new GRN has “replaced” the original one in the midline. Confirmation of this hypothesis and an understanding of its details must await characterization of the A. aegypti late midline GRN, currently undefined. An A. aegypti CRM (for the A. aegypti short gastrulation (sog) ortholog) putatively homologous to its Drosophila midline-active counterpart was active in the midline in transgenic Drosophila, suggesting that trans-regulatory differences further upstream in the GRN are responsible for the observed medial-to-lateral shift in GRN activity. Identification of the trans-acting factors, as well as direct tests of the model in transgenic mosquitos, are still awaiting completion.
Hidden complexity in GRN evolution
Many of the described instances of GRN evolution in place involve seemingly simple changes, such as loss of the Pitx1 enhancer in sticklebacks [7] (leading to absence of an important trans-acting regulator throughout the GRN) or modification of the bab enhancer for Drosophila dimorphic body pigmentation [10] (a cis-regulatory change affecting a downstream patterning event). Such a mechanism, GRN evolution by means of an individual cis-regulatory change, seems reasonable, as the GRN is already fully constituted and active in the ancestral species. A simple single change can therefore have immediate phenotypic consequences without serious pleiotropic effects, and we might suppose that this parsimonious mechanism would be the primary one for GRN evolution in place or in serial homologs. However, this apparent simplicity frequently masks a deeper mechanistic complexity. One study [10] identified at least five individual sequence modifications to which the inferred ancestral bab CRM would have been subjected in evolving the current forms in D. melanogaster and D. willistoni. Similarly, the wing pigmentation GRN in D. biarmipes, as compared to that in D. melanogaster, has multiple modifications in a yellow CRM as well as additional as-yet-uncharacterized changes elsewhere in the GRN [9]. Other studies indicate that different yellow CRMs appear to be involved in various species [12]. The situation in the Drosophila wing and haltere, an example of GRN evolution in serial homologs, similarly provides a cautionary counterweight to the idea of simple but far-reaching mechanisms, as the T3 wing GRN appears to have evolved through a large series of cis-regulatory changes affecting numerous nodes of the network [17, 18]. The fact that a substantial number of the known examples of GRN evolution involve multi-step and multi-locus changes suggests that there is unlikely to be a single unifying mechanism underlying any of the modes of GRN evolution, and that detailed and thorough molecular characterization will be needed for each GRN under investigation.
Putting the GRNs into DSD
Studies of evolutionary developmental biology have revealed cases in which although a developmental pathway appears to have changed, there is no corresponding change in outcome: the phenotype is maintained despite apparent genetic rewiring. This phenomenon has been referred to in the literature as “developmental system drift” (DSD), or less commonly, “phenogenetic drift” [26, 27]. Although the literature on DSD and GRN evolution has remained largely separate, it seems clear that DSD represents a form of GRN evolution, and I would argue that DSD should be discussed in that context. Using the categories described above, we can see that the situation typically described as DSD, where a GRN evolves without giving rise to phenotypic differences, constitutes the most basic form of GRN evolution “in place.” An extreme form of DSD appears to be involved in the above-referenced example of “repeal, replace, and redeploy” GRN evolution in the Drosophila and Aedes central nervous systems, where the GRN responsible for midline development in A. aegypti has not merely evolved in place but rather has undergone a wholesale substitution with either a different existing GRN, or one that has evolved de novo to take on the midline role.
The curious separation between GRN evolution and DSD in the literature may to some extent result from a problem of scale: while GRN-level studies by their nature look at a multitude of genes and regulatory sequences, studies of DSD have often been based on more limited data and in particular have frequently not included CRMs. This point is well-illustrated by studies carried out by Abouheif and colleagues looking at the genetics of wing polyphenism in ants [28–30]. Wing polyphenism is the ability for ants of the same genotype to be members of either winged or wingless castes, with the phenotype regulated by environmental signals. Wing polyphenism is described as “classic DSD” because expression of different wing-patterning genes is altered in different species to mediate the winged/wingless trait [30]. However, inclusion of more genes in the analysis reveals that in five studied species, one gene, brinker (brk), is always expressed in winged castes and never in wingless castes, whereas the expression of other genes, for example engrailed (en), is sometimes present and sometimes absent in the wingless case (Fig. 3).
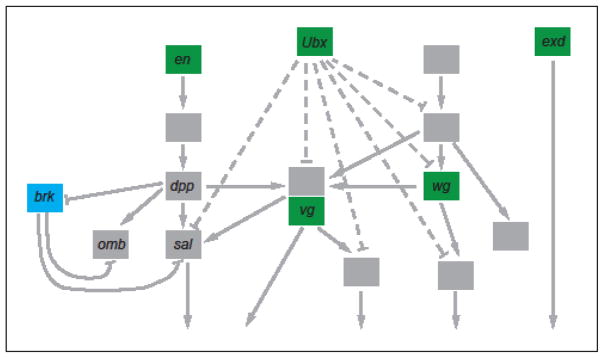
Figure 3. DSD in the ant wing GRN.
A partial schematic of the ant wing development GRN, based on data from Drosophila, is pictured, with genes represented by boxes and genetic interactions by arrows (activation) or capped lines (repression). No CRM-level data are currently available. Genes shaded in green are altered in expression among the wingless castes of at least some of the five species surveyed. Genes shown in blue are expressed identically in all five species; only brk currently meets this criterion. Genes shaded gray (some names have been omitted for simplicity) have not yet been systematically examined. Figure based on [30].
This observation has several interesting implications. One, it impacts directly on what will cause an evolutionary phenomenon to be considered DSD: if looking solely at brk expression, which tracks directly with phenotype, a determination of DSD in this “classic case” would not have been made. Two, it points to the existence of critical nodes in a GRN—in this case, regulation of brk—that are required to produce a given phenotype and thus likely under stabilizing selection. Parts of the GRN upstream of (or perhaps in parallel to) this point would have more latitude to evolve neutrally and therefore if looked at in isolation, may be confused as being causal and as exemplars of DSD. However, when seen at greater distance, using a wider GRN viewpoint, it is apparent that key regulators are in fact fixed (“non-labile” in the terminology of Shbailat and Abouheif [30]). The “labile” members of the network may play only minor functional roles and have changed simply via neutral drift [30]. In this way, the genes in a GRN can be thought of as similar to the “driver” and “passenger” genes defined for models of tumor formation [31, 32]: the driver genes are primarily responsible for the phenotype, while the passenger genes have been pulled along at some earlier point of cooption of the GRN and are slowly replaced (or not) during subsequent evolution.
Third, the studies by Abouheif and colleagues point to the value of conducting analysis in not just two but in multiple related species. Had only a single species pair been looked at, several of the labile genes would have appeared to be fixed, and the primacy of truly fixed regulators such as brk would not have come into sharp focus.
Finally, these studies underscore the importance of CRM-level data in understanding GRN evolution. Such data are not yet available in the ant wing system and preclude obtaining crucial mechanistic details of the evolutionary changes. For example, how brk expression is being downregulated in wingless castes, and whether or not this is being achieved through identical mechanisms in different species, is not yet known. In other systems, detailed CRM characterizations have yielded important insights and demonstrated the prevalence of convergent evolutionary events targeting key GRN nodes. For example, multiple independent changes in the pdm3 locus affect female-limited color dimorphism in members of the Drosophila montium subgroup [33], and multiple independent mutations in CRMs for ebony underlie the convergent evolution of male color pattern in the Drosophila ananassae subgroup [34].
Genomes and regulatory annotation
Given the importance of CRM-level data for studying GRNs and GRN evolution, to what extent is such information available? The most detailed data are for Drosophila melanogaster, for which the REDfly database has curated more than 22,000 empirically-validated regulatory sequences, over 5800 of which are CRMs identified through in vivo reporter gene analysis and which therefore have associated spatial and temporal functional annotations [35]. Similarly, the Vista Enhancer Browser [36] contains over 1400 mouse and human sequences verified through reporter gene assays in transgenic mice. Ensembl collects a number of regulatory tracks (e.g., histone modifications, open chromatin) and for the human and mouse genomes provides a “regulatory build” based primarily on chromatin segmentation analyses that predict functional regulatory regions [37]. Unlike REDfly and the Vista Enhancer Browser, these are not based on reporter gene data, and the ability of chromatin segmentation algorithms to accurately classify regulatory sequences is still an unsettled issue [38]. However, integration into the Ensembl platform, with its intuitive BioMart download capabilities, makes these data highly accessible. Chromatin-based regulatory data can also be found in many of the major model organism databases and through genomics projects such as ENCODE and modENCODE [39, 40]. Although they currently do not include regulatory data, annotated insect genomes can be accessed through the National Agricultural Library’s i5k Workspace (56 genomes)[41] and InsectBase (138 genomes)[42], providing a potentially rich source for comparative genomic analyses. Overall, however, comprehensively annotated regulatory genomes remain frustratingly lacking, and the need for CRM identification presents a major stumbling block for extensive GRN characterization and studies of GRN evolution.
CRM discovery in multiple species
Fortunately, methods for CRM discovery, both empirical and computational, have improved dramatically in the past several years [reviewed by 43], and examples of GRN evolution have now been observed in all of the common metazoan model species. Particularly strong examples are available from the echinoderms, due to the extensive cis-regulatory analysis that has been performed on sea urchin development [44], but several well-described vertebrate instances exist as well. Many of the best examples have come from studies of flies within the genus Drosophila. This is perhaps not surprising given the large number (over 20) of fully-sequenced drosophilid genomes, the extensive knowledge of Drosophila developmental genetics gained from over 100 years of Drosophila research, and the many tools available for experimental studies in flies. However, there are inherent limitations as to how far studying only drosophilids will take us. In particular, the need to use sequence alignment as a guide to homologous regulatory element discovery has kept these studies confined to exploring relatively small phenotypic shifts among organisms with highly similar body plans and moderately diverged genomes.
Even when non-coding sequences can effectively be aligned between species, functionally homologous CRMs can be difficult to find. Chromatin profiling methods have revolutionized the field of CRM discovery over the past several years, allowing for high-quality predictions of CRM location in an unbiased fashion [reviewed by 43]. Applying these methods to profile CRMs active in liver tissue across 20 mammalian species found that the majority of CRM sequences are not conserved between species [45]. This suggests that once genomes have evolved past the distances that have allowed for successful Drosophila studies using closely related species, merely selecting an aligned portion of the genome will not ensure identification of a homologous CRM.
How then to study GRN evolution across greater distances? SCRMshaw, a machine-learning computational approach to CRM discovery developed by the Sinha and Halfon labs [46–48], has been used to root out deep CRM homologies in distantly-related insect species (Fig. 4A) [47]. SCRMshaw takes advantage of the enormous wealth of Drosophila melanogaster CRM data [35] to develop a computational model for CRMs of a particular function—e.g., wing development, heart development, etc.—that can then be applied to CRM discovery in either Drosophila or a more diverged, sequenced insect. Computational and empirical testing indicate that the method works for CRM discovery over large evolutionary distances, effective at least throughout the ~345 million years of holometabolous insect evolution. SCRMshaw was able to predict, using conservative parameters, 12 out of 16 already-known enhancers from the mosquito Anopheles gambiae, the honeybee Apis mellifera, and the beetle T. castaneum [47]. Empirical testing using transgenic Drosophila validated over twenty additional predictions from those species as well as the wasp Nasonia vitripennis and the mosquito Aedes aegypti [6, 47]. Overall, testing of the SCRMshaw method in a cross-species setting yielded close to an 80% true-positive success rate. In a further test of the method, ~1200 high-stringency predictions were made for T. castaneum based on training data from Drosophila wing imaginal discs, and compared to T. castaneum CRMs predicted by FAIRE-seq, an empirical, chromatin-based assay for CRM discovery. A comparison of these two data sets, which were obtained using very different methods, found that 76% of SCRMshaw predictions overlapped FAIRE-seq regions (Fisher’s Exact P ≈ 0; MSH and Y. Tomoyasu, unpublished results). Not only does this confirm the efficacy of SCRMshaw as a cross-species CRM discovery tool, it suggests that merging SCRMshaw with open-chromatin profiling or other generic CRM-discovery methods will be a powerful combination for increasing the specificity and sensitivity of function-specific CRM prediction (Fig. 4C). Trained, targeted approaches other than SCRMshaw (e.g., Imogene [49], CLARE [50]) are also likely to be effective, although have not yet been demonstrated to work in a cross-species manner.
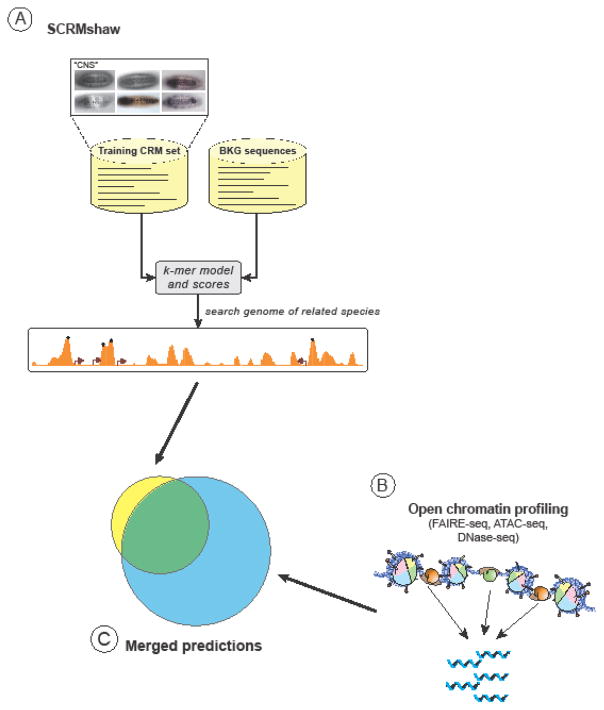
Figure 4. CRM discovery across distantly-related species.
(A) The SCRMshaw method [46–48] can be used to identify CRMs in diverged insect species. Training data composed of the sequences of known Drosophila melanogaster CRMs with a common function (e.g., expression in the central nervous system (“CNS”), top of figure) are compared to randomly selected non-coding sequences (“BKG” or “background” sequences). A scoring model (“k-mer model and scores”) is then generated that can be used to search the genome of another insect species to predict CRMs. The method has been shown to be effective at least through 345 million years of holometabolous insect evolution. (C) To help reduce the roughly 25% false-positive prediction rate, SCRMshaw predictions can be merged with putative CRM regions predicted from open chromatin profiling methods (B) such as FAIRE-seq [58] or ATAC-seq [64].
Chromatin profiling has the advantage of not requiring training and therefore not being dependent on already-known CRMs. This can be important when only very few CRMs belonging to a GRN of interest are known, or in cases where CRM composition has diverged beyond the point where SCRMshaw is useful (e.g., the sim CRM discussed in [6]). These methods may also have superior sensitivity relative to SCRMshaw, although this remains to be explored rigorously. On the other hand, chromatin profiling alone has limited ability to predict spatio-temporal specificity of a given CRM, even when performed in a tissue-specific manner [e.g. 51], and moreover, cannot be focused to a particular GRN. Given that genes, and developmental genes in particular, frequently have multiple CRMs, determining the right set of CRMs necessary to study a GRN of interest—especially in several species—can be a challenge. SCRMshaw incorporates GRN specificity, but has the drawback of being a predictive method with a false-positive rate that, although low for a computational CRM discovery method, will still provide incorrect predictions. Intersecting results from both SCRMshaw (or a similar method) and chromatin profiling should allow for higher-confidence predictions while maintaining SCRMshaw’s increased function-specificity. Together, this powerful combination of methods has the potential to rapidly accelerate the pace of CRM discovery in insects, providing the crucial but largely missing cis-regulatory component necessary for in-depth GRN studies.
Concluding Remarks: An exciting time for studies of GRN evolution
Deep regulatory homologies, at distances where genomes have diverged too far to make use of sequence alignment—even trans-phyletically—have been reported previously [47, 52], but the challenges inherent in identifying homologous CRMs have kept examples to a minimum (see Outstanding Questions). Furthermore, conservation of function does not always mean conservation of mechanism. Several studies have reported instances of CRMs with related function that contain similar TF binding sites, but mutagenesis of these sites has not affected activity equivalently in each CRM [53, 54]. SCRMshaw’s high rate of success as a multi-species CRM discovery tool provides compelling evidence that deep regulatory homologies are extensive throughout the Insecta, and because the method relies on multiple shared DNA features among the identified CRMs, suggests that regulatory mechanisms will at least in part be conserved. This is a welcome finding, as the insects are a group which bears great promise for unlocking the mysteries of GRN evolution. The insects include a wealth of species covering over 440 million years of evolution, with countless adaptations and morphological novelties that provide a rich ground for evolutionary studies. Over 200 species have been fully sequenced, with more on the way [55]. The sequenced species include several groups with multiple close relatives, including the drosophilids, anopheline mosquitoes, bees, and ants. This will allow for studies of GRN evolution at close, moderate, and large divergence times, which, along with phylogenetic relationships, have recently been clarified in detail for the insects [56]. A broad exploration of GRN evolution across considerable evolutionary distances is therefore now within reach, and the next several years promise exciting progress toward unlocking the mysteries of how GRNs have evolved.
Outstanding Questions Box.
Identification of homologous CRMs across species is a critical limiting step for studies of GRN evolution. Will SCRMshaw or similar approaches continue to prove effective for a broader range of developmental networks and at greater evolutionary distances? Can other methods for cross-species CRM discovery be developed that will be more effective?
The number of GRNs characterized in detail in more than one species is small but growing. Will general principles of GRN evolution emerge as more examples are defined? Many current examples compare GRNs over relatively short evolutionary distances. Will different principles or mechanisms predominate at larger degrees of divergence?
To what extent are similar regulatory mechanisms found over long evolutionary time spans? How much of this is due to evolutionary convergence versus direct descent?
Is DSD a major or a minor evolutionary phenomenon? How often is DSD “extreme” in that the GRN leading to a phenotype is completely substituted, compared to partial whereby “driver” genes and their regulation are conserved but “passenger” genes have diverged?
Trends Box.
To better understand the evolution of phenotype, we should investigate the evolution of underlying gene regulatory networks (GRNs)
GRNs evolve at both cis- and trans- levels, but discerning between the two is sometimes difficult and a matter of perspective. Identification of relevant cis-regulatory modules is essential for making this determination, and for understanding other aspects of GRN evolution as well.
GRN evolution can be classified into multiple modes, but there are no apparent defining mechanisms for any mode
Developmental system drift (DSD) is a form of GRN evolution and should be considered accordingly
Recent advances in cis-regulatory module discovery, especially for insects, makes this group of species ideal for exploring the mechanisms of GRN evolution
Acknowledgments
I thank Yoshinori Tomoyasu and Courtney Clark-Hachtel for helpful comments on the manuscript. Support for this work comes from USDA grant 2012-67013-19361 and NIH grant R21 AI125918.
Glossary
- Carinated margin
An outgrowth of the body wall in the first thoracic segment of beetles such as Tribolium castaneum (red flour beetle). There is evidence to suggest that the carinated margin is a wing serial homolog [24]
- Developmental system drift (DSD)
The phenomenon by which evolutionary changes in genetic pathways do not affect the resultant phenotype
- cis-Regulatory module (CRM)
Sequences on the DNA that bind transcription factors to regulate gene expression. “Enhancers” comprise one common type of CRMs
- Driver and Passenger Genes
A concept borrowed from the cancer literature and applied here to developmental system drift. “Driver” genes (or driver mutations) refer to the genes which when mutated (or to the specific mutations) promote cancer, e.g. by providing a selective growth advantage. “Passenger” genes (or mutations) are those that are present in the genetic background but which do not themselves contribute meaningfully to the cancer phenotype
- Elytron (pl. elytra)
The hardened forewing or “wing cover” found in beetles
- FAIRE-seq
FAIRE-seq, or Formaldehyde-Assisted Identification of Regulatory Elements followed by sequencing, is a method for isolating regions of “open,” or nucleosome-depleted, chromatin and determining their genomic locations via next-generation DNA sequencing [57, 58]. The isolated genomic regions have been shown to be enriched for regulatory sequences such as CRMs
- Gene Regulatory Network (GRN)
The linkages between a set of genes, and in particular between transcription factors and their interactions with the regulatory DNA of other genes, that describe the molecular genetic control of a specific cell function
- Haltere
A balancing organ that takes the place of the hindwing in the third thoracic segment of Dipteran flies
- Pleiotropic effects
Multiple effects produced by the activity of an individual gene, often in diverse organs or tissues
- Polyphenism
The phenomenon by which two or more different phenotypes are produced from the same genotype
- Subcircuit
A modular component of a gene regulatory network (GRN; see above). A GRN can contain multiple subcircuits, each of which carries out a particular developmental task that is a sub-function of the process regulated by the GRN. For more thorough discussion, see [59–61]
Footnotes
Resources
REDfly: http://redfly.ccr.buffalo.edu
Vista Enhancer Browser: https://enhancer.lbl.gov
Ensembl: http://www.ensembl.org/index.html
ENCODE: https://www.encodeproject.org
modENCODE: http://www.modencode.org
i5K: http://i5k.github.io
InsectBase: http://www.insect-genome.com
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Carroll SB, et al. From DNA to Diversity. Molecular Genetics and the Evolution of Animal Design. Blackwell Publishing; 2005. [Google Scholar]
- 3.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Academic Press; 2006. [Google Scholar]
- 4.Wang X, Sommer RJ. Antagonism of LIN-17/Frizzled and LIN-18/Ryk in nematode vulva induction reveals evolutionary alterations in core developmental pathways. PLoS Biol. 2011;9:e1001110. doi: 10.1371/journal.pbio.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rebeiz M, et al. Unraveling the Tangled Skein: The Evolution of Transcriptional Regulatory Networks in Development. Annu Rev Genomics Hum Genet. 2015;16:103–131. doi: 10.1146/annurev-genom-091212-153423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suryamohan K, et al. Redeployment of a conserved gene regulatory network during Aedes aegypti development. Dev Biol. 2016;416:402–413. doi: 10.1016/j.ydbio.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCauley BS, et al. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev Biol. 2010;340:200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Gompel N, et al. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 10.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong S, et al. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Prud’homme B, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 13.Jeong S, et al. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 14.Rebeiz M, Williams TM. Using Drosophila pigmentation traits to study the mechanisms of cis-regulatory evolution. Current Opinion in Insect Science. 2017;19:1–7. doi: 10.1016/j.cois.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel N, et al. Conserved regulatory architecture underlies parallel genetic changes and convergent phenotypic evolution. Proc Natl Acad Sci U S A. 2012;109:20975–20979. doi: 10.1073/pnas.1207715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomoyasu Y. Ultrabithorax and the evolution of insect forewing/hindwing differentiation. Current Opinion in Insect Science. 2017;19:8–15. doi: 10.1016/j.cois.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Hersh BM, et al. The UBX-regulated network in the haltere imaginal disc of D. melanogaster. Dev Biol. 2007;302:717–727. doi: 10.1016/j.ydbio.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weatherbee SD, et al. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlopoulos A, Akam M. Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis. Proc Natl Acad Sci U S A. 2011;108:2855–2860. doi: 10.1073/pnas.1015077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohit P, et al. Modulation of AP and DV signaling pathways by the homeotic gene Ultrabithorax during haltere development in Drosophila. Dev Biol. 2006;291:356–367. doi: 10.1016/j.ydbio.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Angelini DR, Kaufman TC. Comparative developmental genetics and the evolution of arthropod body plans. Annual review of genetics. 2005;39:95–119. doi: 10.1146/annurev.genet.39.073003.112310. [DOI] [PubMed] [Google Scholar]
- 22.Tomoyasu Y, et al. Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature. 2005;433:643–647. doi: 10.1038/nature03272. [DOI] [PubMed] [Google Scholar]
- 23.Tomoyasu Y, et al. Repeated co-options of exoskeleton formation during wing-to-elytron evolution in beetles. Curr Biol. 2009;19:2057–2065. doi: 10.1016/j.cub.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Clark-Hachtel CM, et al. Insights into insect wing origin provided by functional analysis of vestigial in the red flour beetle, Tribolium castaneum. Proc Natl Acad Sci U S A. 2013;110:16951–16956. doi: 10.1073/pnas.1304332110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glassford WJ, et al. Co-option of an Ancestral Hox-Regulated Network Underlies a Recently Evolved Morphological Novelty. Dev Cell. 2015;34:520–531. doi: 10.1016/j.devcel.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 27.Weiss KM, Fullerton SM. Phenogenetic drift and the evolution of genotype-phenotype relationships. Theoretical population biology. 2000;57:187–195. doi: 10.1006/tpbi.2000.1460. [DOI] [PubMed] [Google Scholar]
- 28.Abouheif E, Wray GA. Evolution of the gene network underlying wing polyphenism in ants. Science. 2002;297:249–252. doi: 10.1126/science.1071468. [DOI] [PubMed] [Google Scholar]
- 29.Nahmad M, et al. The dynamics of developmental system drift in the gene network underlying wing polyphenism in ants: a mathematical model. Evol Dev. 2008;10:360–374. doi: 10.1111/j.1525-142X.2008.00244.x. [DOI] [PubMed] [Google Scholar]
- 30.Shbailat SJ, Abouheif E. The wing-patterning network in the wingless castes of Myrmicine and Formicine ant species is a mix of evolutionarily labile and non-labile genes. Journal of experimental zoology. Part B, Molecular and developmental evolution. 2013;320:74–83. doi: 10.1002/jez.b.22482. [DOI] [PubMed] [Google Scholar]
- 31.Pon JR, Marra MA. Driver and passenger mutations in cancer. Annual review of pathology. 2015;10:25–50. doi: 10.1146/annurev-pathol-012414-040312. [DOI] [PubMed] [Google Scholar]
- 32.Stratton MR, et al. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yassin A, et al. The pdm3 Locus Is a Hotspot for Recurrent Evolution of Female-Limited Color Dimorphism in Drosophila. Curr Biol. 2016;26:2412–2422. doi: 10.1016/j.cub.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Signor SA, et al. Genetic Convergence in the Evolution of Male-Specific Color Patterns in Drosophila. Curr Biol. 2016;26:2423–2433. doi: 10.1016/j.cub.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 35.Gallo SM, et al. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 2011;39:D118–123. doi: 10.1093/nar/gkq999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visel A, et al. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerbino DR, et al. Ensembl regulation resources. Database: the journal of biological databases and curation. 2016;2016 doi: 10.1093/database/bav119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwasnieski JC, et al. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014 doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.modENCODE Consortium, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poelchau M, et al. The i5k Workspace@NAL--enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 2015;43:D714–719. doi: 10.1093/nar/gku983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin C, et al. InsectBase: a resource for insect genomes and transcriptomes. Nucleic Acids Res. 2016;44:D801–807. doi: 10.1093/nar/gkv1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suryamohan K, Halfon MS. Identifying transcriptional cis-regulatory modules in animal genomes. Wiley Interdisciplinary Reviews: Developmental Biology. 2015;4:59–84. doi: 10.1002/wdev.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinman VF, Cheatle Jarvela AM. Developmental gene regulatory network evolution: insights from comparative studies in echinoderms. Genesis. 2014;52:193–207. doi: 10.1002/dvg.22757. [DOI] [PubMed] [Google Scholar]
- 45.Villar D, et al. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kantorovitz MR, et al. Motif-blind, genome-wide discovery of cis-regulatory modules in Drosophila and mouse. Dev Cell. 2009;17:568–579. doi: 10.1016/j.devcel.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kazemian M, et al. Evidence for deep regulatory similarities in early developmental programs across highly diverged insects. Genome biology and evolution. 2014;6:2301–2320. doi: 10.1093/gbe/evu184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kazemian M, et al. Improved accuracy of supervised CRM discovery with interpolated Markov models and cross-species comparison. Nucleic Acids Res. 2011;39:9463–9472. doi: 10.1093/nar/gkr621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouault H, et al. Imogene: identification of motifs and cis-regulatory modules underlying gene co-regulation. Nucleic Acids Res. 2014;42:6128–6145. doi: 10.1093/nar/gku209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taher L, et al. CLARE: Cracking the LAnguage of Regulatory Elements. Bioinformatics. 2012;28:581–583. doi: 10.1093/bioinformatics/btr704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearson JC, et al. Chromatin profiling of Drosophila CNS subpopulations identifies active transcriptional enhancers. Development. 2016;143:3723–3732. doi: 10.1242/dev.136895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maeso I, et al. Deep conservation of cis-regulatory elements in metazoans. Philos Trans R Soc Lond B Biol Sci. 2013;368:20130020. doi: 10.1098/rstb.2013.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen WC, et al. Dissection of a Ciona regulatory element reveals complexity of cross-species enhancer activity. Dev Biol. 2014;390:261–272. doi: 10.1016/j.ydbio.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halfon MS, et al. Erroneous attribution of relevant transcription factor binding sites despite successful prediction of cis-regulatory modules. BMC Genomics. 2011;12:578. doi: 10.1186/1471-2164-12-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.i5k Consortium. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. The Journal of heredity. 2013;104:595–600. doi: 10.1093/jhered/est050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 57.Giresi PG, et al. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erwin DH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- 60.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davidson EH. Network design principles from the sea urchin embryo. Curr Opin Genet Dev. 2009;19:535–540. doi: 10.1016/j.gde.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li E, Davidson EH. Building developmental gene regulatory networks. Birth defects research. Part C, Embryo today: reviews. 2009;87:123–130. doi: 10.1002/bdrc.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buenrostro JD, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]