Abstract
Studies of heterochronic parabiosis demonstrated that with age, the composition of the circulatory milieu changes in ways that broadly inhibit tissue regenerative capacity. In addition, local tissue niches have age-specific influences on their resident stem cells. Here we use bio-orthogonal proteome labeling for detecting in vivo proteins present only in transplanted myoblasts, but not in host tissue, and proteins exclusive to one young mouse and transferred during parabiosis to its old partner. We use a transgenic mouse strain that ubiquitously expresses a modified tRNA methionine synthase, metRS, which preferentially incorporates the methionine surrogate azido-nor-leucine (ANL) into newly generated proteins. Using click chemistry and a modified antibody array to detect ANL-labeled proteins, we identify several ‘young’ systemic factors in old regenerating muscle of the heterochronic parabiotic partners. Our approach enables the selective profiling of mammalian proteomes in mixed biological environments such as cell and tissue transplantation, apheresis or parabiosis.
Subject terms: Proteome, Transgenic organisms, Ageing
Clarifying the source of proteins in mixed biological environments, such as after transplantation or parabiosis, remains a challenge. Here, the authors address this need with a mouse strain that incorporates a methionine derivate into proteins, allowing for their detection using click chemistry and antibody arrays.
Introduction
The proteome of the cell changes with age and its associated pathologies1–4. Proteins produced by the local and systemic environments of organ stem cells broadly regulate the regeneration and maintenance of adult tissues1–4. Furthermore, the age-imposed changes in the intensity of evolutionary conserved signaling pathways interfere with productive regeneration of multiple mammalian tissues3–5. For example, the age-specific changes in the local and systemic environments of organ stem cells perturb Notch, TGF-beta/BMP, MAPK and Wnt broad-action signaling networks, which regulate the regeneration and maintenance of muscle, brain, liver, blood, etc. tissues6–8. Importantly, studies of heterochronic parabiosis (surgical joining of young and old animals) suggest both productive tissue repair and the key signal transduction pathways that control stem cell activation are restored to ‘youth’ in the old parabionts by young systemic factors9, 10.
It would be beneficial from academic and clinical stand-points to determine which proteins in tissues of parabiotically connected animals are derived from the circulation of young versus old partner. Such a database of systemic proteins that end up in specific tissues in setting of heterochronic parabiosis, would suggest potentially rejuvenating (young blood) and inhibitory (old blood) molecules with direct effects in a given tissue, also informing on the cross-tissue conservation and differences of the systemic factors from one tissue to another. While biochemical fractionation of serum and plasma can provide some characterization of the molecular differences between young and old circulatory milieu, this technique is fraught with the risk of missing proteins that act in complexes with each other and other macromolecules. In addition, serum and plasma fractionation are indirect approaches based mostly on in vitro studies. And it remains unknown whether or not the age-specific systemic proteins have direct effects in regenerating tissues with their resident stem cells. Candidate factor approaches can be tried, but they require a long time to confirm or rule out just one molecule. In the past decade, these approaches have only yielded a few pro-regenerative molecules, some of which are controversial11, 12.
Our approach of choice relies on tRNA synthase that specifically recognizes and incorporates BONCAT(Bio-Orthogonal Non-Canonical Amino acid Tagging) into proteins13–18. Specifically, the methionine surrogate azido-nor-leucine (ANL) is incorporated into newly synthesized proteins only in cells expressing this mutant methionine tRNA synthase, MetRS17. The mutant MetRS which has a single evolutionary conserved amino acid substitution: 274L→G preferentially incorporates ANL into mammalian cells and in Drosophila melanogaster tissues in vivo; and ANL-tagged proteins can be selectively conjugated to dyes or affinity probes and identified13, 19, 20.
To facilitate detection by proteomics, we have selected the BONCAT method over the CTAP (cell type-specific labeling with amino acid precursors) where proteomes are tagged with heavy isotope—labeled precursors16, 21, 22; and over the incorporation of Met analogs azidohomoalanine (AHA) and homopropargylglycine (HPG), which do not allow one to selectively profile young versus old proteomes in settings of parabiosis23.
To advance MetRSL274G ANL labeling technology to live mammals, we have developed and characterized a novel transgenic mouse strain, in which mutant MetRSL274G is broadly expressed (MetRSL274G mice). Our data demonstrate the survival and vigor of these animals as well as the effective proteome labeling with ANL of cells in vitro and all examined tissues in vivo. Importantly, ANL tagging using our dosage did not perturb the key properties of the proteins, such as the rejuvenating effects of young ANL labeled serum on the old muscle progenitor cells in vitro or the enhancement of old muscle repair in vivo while still allowing the detection by Click-western, FUNCAT and the bio-orthogonal proteomics profiling.
We have performed transplantation of MetRSL274G myoblasts into muscle of C57BL/6 mice, which demonstrated a new capability for identification of transplanted cell proteomes without their re-derivation from host tissues. And we have established parabiotic pairings between the young MetRSL274G mice and old C57BL/6 mice, which yielded data on the candidate systemic rejuvenating factors, e.g., the ‘young’ proteins that have traveled through the parabiotic circulation and were derived from the aged muscle.
Results
Characterization of the transgenic mice that broadly express MetRSL274G
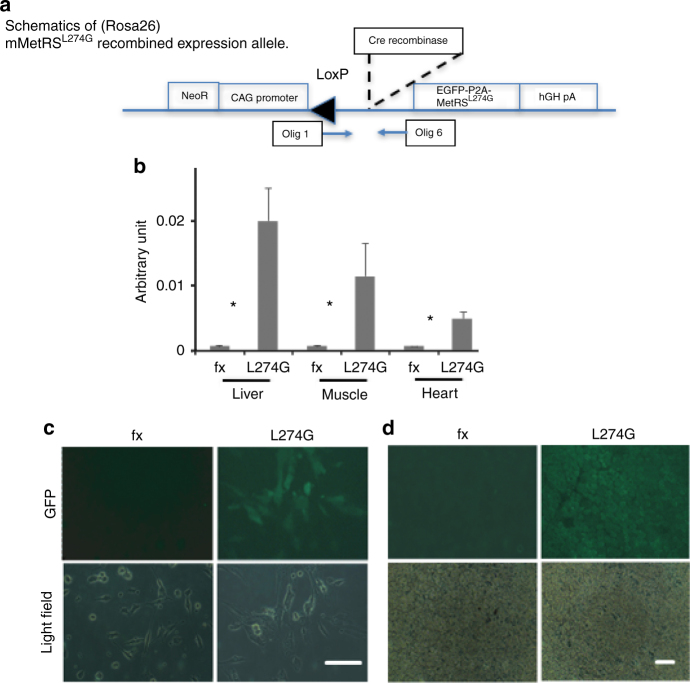
To enable the in vivo labeling of mammalian proteome via broad expression of mMetRSL274G, we crossed floxed STOP-EGFP tagged MetRSL274G mice (i.e., fx mice) with CMV-Cre mice (Fig. 1a). Fx mice were generated using conventional techniques, in which the previously published mMetRSL274G 13, 15, 24 and EGFP constructs are inserted into the Rosa26 locus with Floxed STOP sequence (Supplementary Fig. 1). Breeding pairs of these mice (generously provided by Erin Schuman, Max Planck Institute for Brain Research, Frankfurt, Germany), were genotyped and established into a colony; and animals harboring homozygous fx alleles were crossed with CMV/Cre mice (Jackson Labs, N > 10 on C57BL/6 background), thereby enabling constitutive broad expression of mMetRSL274G. F1 progeny containing both transgenes Rosawt/exCreCMV MetRSL274G (i.e., MetRSL274G mice) were identified by polymerase chain reaction (PCR) genotyping (Supplementary Fig. 1) and used for these studies, as well as to establish the founder strain. The PCR, PCR with reverse transcription and direct GFP fluorescence confirmed the excision of the STOP signal and expression of the mMetRSL274G and EGFP in the cells and tissues of these animals (Fig. 1c, d).
Fig. 1.
The expression of GFP-tagged MetRSL274G upon ‘Stop signal’ excision. a Schematic representation map of the allele after CMV-Cre mediated ‘Stop signal’ excision (MetRSL274G mice). b QT PCR demonstrates that transgene expression is indeed induced upon Cre-LoxP mediated excision of the ‘STOP’ signal. *P < 0.05, Wilcoxon rank sum test. s.e.m., n = 3 for each genotype. c Cells were isolated from indicated genotypes and cultured in F10, 20% FBS for 24 h after which the cells were imaged on bright field and for GFP fluorescence. d Muscles from three separate mice were dissected, cryo-sectioned and imaged on bright field and for GFP fluorescence. 3–4 experimental replicates (sections) were assayed for each animal. Representative images are shown. Scale bar, 100 μm
In previous studies, mice fed with the methionine homolog AHA had similar body weight as the animals on a regular diet25, indicating that the mouse proteome is amenable to tagging with the non-canonical amino acids without toxicity. To confirm these results in our ANL experimental system, we demonstrate that MetRSL274G mice are viable, fertile, and do not exhibit weight loss as compared to the wild-type mice and fx littermates, both without and with ANL administration (Supplementary Fig. 2).
These results demonstrate successful in vivo expression of the MetRSL274G and establish that BONCAT does not negatively affect the health, striving and fertility of animals.
Ubiquitous MetRSL274G enables broad labeling of mammalian cells and tissues with ANL
To examine the feasibility of MetRSL274G cells for bio-orthogonal proteome labeling, we isolated neonatal fibroblasts from 1- to 2-day-old MetRSL274G pups and incubated these cells with ANL in culture. Click reaction and western blotting was used to detect ANL incorporation into proteins13. As shown in Fig. 2a, proteome labeling was observed in MetRSL274G fibroblasts in the ANL dose-dependent manner ranging from 100 micromolar to 1 millimolar. Identical low background – noise was detected in MetRSL274G cells without ANL treatment and in fx cells that were cultured with 1 mM ANL. The ANL treated MetRSL274G cells were also eagerly detected by the Clicked AlexaFluor 488 in FUNCAT (fluorescent non-canonical amino acid tagging26), as compared to the low background fluorescence of the negative controls (Supplementary Fig. 3A).
Fig. 2.
Bio-orthogonal metabolic labeling in mammalian cells in vitro and in mice in vivo. a Click-western blotting was performed as described in Methods. The in vitro BONCAT of MetRSL274G fibroblasts has resolved an ANL dose-dependent protein tagging (smears), as compared to the low background of fx cells. The nonspecific bands (arrowheads) were present even without ANL treatment in MetRSL274G samples and in fx samples. Coomassie blue image shows similar protein loading for each lane. Similar results were obtained with cells derived from three animals of each strain. b In vivo Click-western blotting, which was performed as detailed in Methods, has resolved the incorporation of ANL into all examined tissues of MetRSL274G mice (smears of proteins), as compared to the background signal of fx tissues. The same non-specific bands as described above were present in all samples (arrowheads). Coomassie blue staining is shown as loading control. Three independent experiments with each mouse strain, yielded similar results. c 10-micrometre TA muscle cryosections were prepared and assayed by FUNCAT; and the adjacent sections (~ 10 micrometre away) were stained with dystrophin antibody. Robust FUNCAT signal was detected in MetRSL274G dystrophin positive muscle, as compared with the low background fluorescence of fx muscle. Scale bar, 100 μm. Three independent experiments with each mouse strain, yielded similar results
We next studied the success of in vivo mammalian BONCAT by administering ANL via intraperitoneal (I.P.) injections to the MetRSL274G and the negative control fx mice daily for 6 days, after which brain, heart, skeletal muscle, liver, bone marrow and blood serum were isolated. Tissue lysates were prepared and Clicked as described26 and the in vivo ANL-labeling of mammalian proteome was assayed by the Click-western and FUNCAT. Click-western blotting resolved robust proteome labeling in all tissues from MetRSL274G mice as compared to the low background of fx- samples, which was similar to that of in vitro negative controls (Fig. 2b). The highest in vivo incorporation of ANL was detected in heart and skeletal muscle, which fits well with rapid protein turn over in these tissues and their rapid growth in juvenile animals27–29. In agreement with the Click-western data, the in vivo incorporation of ANL into proteins of MetRSL274G, tibialis anterior (TA) muscle was detected by FUNCAT, well above the background fluorescence of the TA from fx mice that were identically treated with ANL in vivo (Fig. 2c and Supplementary Fig. 4).
These results establish that L274G mutation enables mammalian bio-orthogonal proteome tagging and detection by Click western and fluorescence both in cell culture and in vivo.
ANL-based detection of MetRSL274G transplant without re-derivation from host muscle
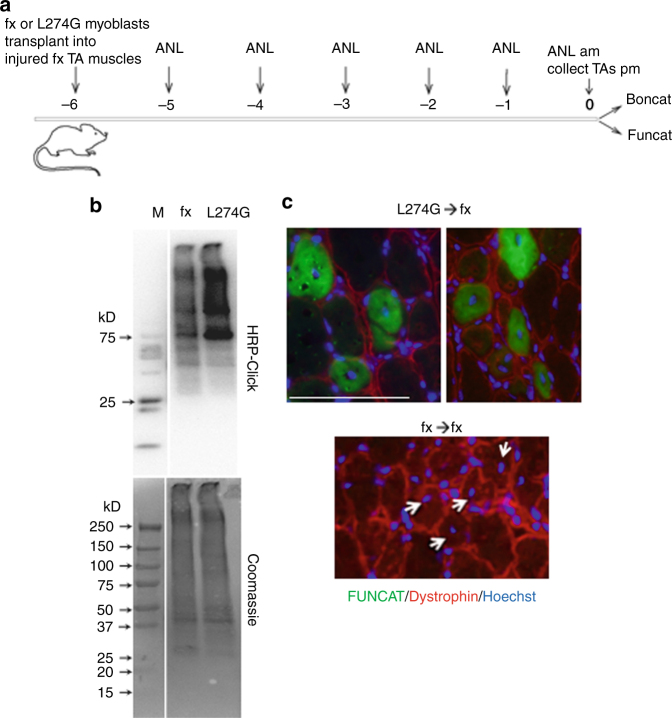
Encouraged by these findings, we decided to study whether transplanted MetRSL274G myogenic cells would be detected in host muscles without their re-derivation solely through the in vivo BONCAT. Activated by injury muscle stem cells were isolated from MetRSL274G and the negative control, fx mice and were cultured for 3–4 weeks, during which time satellite cells give rise to primary myogenic precursors30, 31. These myogenic precursors from each strain were transplanted into the fx host muscle following freeze injury1, which provides a localized permissive engraftment niche (Fig. 3a). The fx host mice were treated with ANL in vivo for 6 days, as above, after which their TA and Gastroc muscle were isolated and analyzed by Click-western (Fig. 3b) and FUNCAT (Fig. 3c). The de-novo synthesized in vivo ANL-tagged proteins were robustly detectable in fx tissues transplanted with MetRSL274G myogenic cells, as compared to the low background of fx muscle transplanted with fx cells (Fig. 3). The success of cell transplantation was confirmed by the identification of sites with clusters of newly formed centrally nucleated dystrophin positive myofibers that, as expected, were present in transplants of both genetic backgrounds and were FUNCAT + only in the case MetRSL274G transplants (Fig. 3c).
Fig. 3.
Detection of newly synthesized proteins in fx muscles transplanted with MetRSL274G primary myogenic cells. a Scheme of the experimental design. The TA of fx mice were injured with dry ice. Primary myogenic cells derived from MetRSL274G or fx mice were transplanted into the injury sites. ANL was injected for 6 consecutive days I.P. into transplanted animals after which TA muscles were collected. b Click-chemistry western blotting on lysates of fx muscles that were transplanted with MetRSL274G versus fx myogenic cells detects ANL-tagged proteins (smearing) in the MetRSL274G samples, as compared to the background signal of fx protein lysates. Coomassie blue staining served as a loading control. Similar results were obtained in three independent experiments. c FUNCAT assay in conjunction with anti-dystrophin immunofluorescence and Hoechst nuclear co-stain were performed, as above. The ANL labeling (green fluorescence) of dystrophin positive (red outlines) myofibers that have been recently regenerated (central Hoechst positive nuclei) was reliably detected in fx TAs that were transplanted with the primary MetRSL274G myoblasts. The negative controls, fx TAs that were transplanted with fx primary myobalsts were FUNCAT negative, while as expected, dystrophin positive and with central nuclei at the sites of transplantation (white arrowheads). Similar results were obtained in three independent experiments. Scale bar, 100 μm
These data demonstrate a capability to detect proteomes of transplanted cells in host tissues in vivo without re-derivation and to label de-novo translated proteins at any time after transplantation.
ANL selectively labels MetRSL274G proteome in settings of parabiosis with C57BL/6 mice
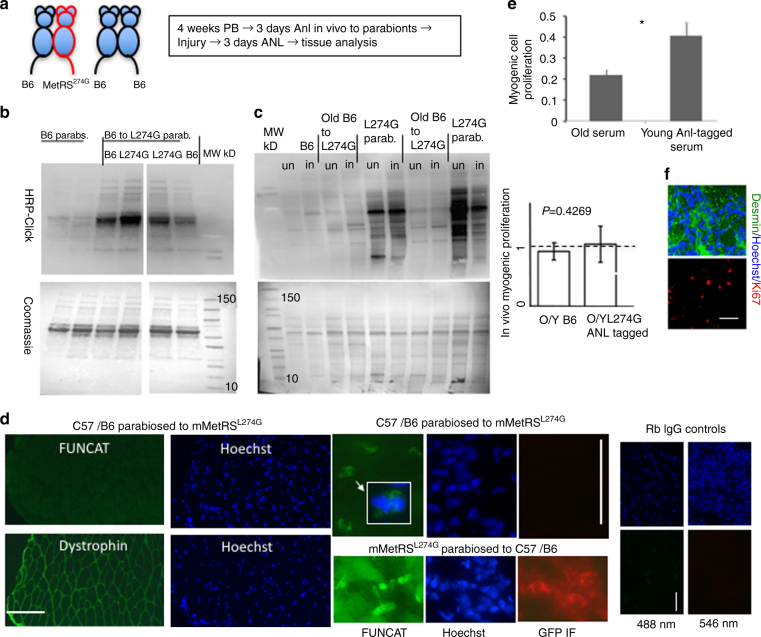
To pursue our main goal of studying BONCAT in setting of heterochronic parabiosis we have established pairs of young (3–4 month old) MetRSL274G mice joined with the old (22–24 month old) C57BL/6 mice, using six young-to-old C57BL/6 pairs as controls (Fig. 4a). The success rate for the old C57BL/6 to young MetRSL274G parabiosis was 78% (seven pairs out of nine survived), and the rate for old C57/B6 to young C57/B6 was 46% (6 of 13 pairs survived). The overall success rate was 59%, which is typical for such heterochronic parabiosis studies. Therefore, MetRSL274G transgene does not appear to increase the incidence of parabiotic disease. The pairs were maintained for 5 weeks with daily observation and weighing of the animals, and the parabionts of all genetic backgrounds were healthy, with no detectable adverse reactions, rejection or inflammation (including the MetRSL274G to old B6 pairs, with or without ANL).
Fig. 4.
Bio-orthogonal proteome labeling in the setting of heterochronic parabiosis. a Schematic representation of the study. b Click-western blotting on serum proteins from the heart-bleeds of MetRSL274G parabionts and their C57BL/6 partners and the negative control syngeneic C57BL/6 pairs. c Click-western blotting resolved the robust selective in vivo ANL-labeling of the proteomes from uninjured (un) and injured (in) Gastrocnemius muscle of MetRSL274G parabionts (Parabs) and much lower ANL-labeling of proteins that were derived from muscle of their C57BL/6 partners; however, this signal was above the background of the negative control, syngeneic C57BL/6 pairs. Coomassie blue staining demonstrates equal protein loading. Similar results were obtained in at least three independent experiments with each cohort. d FUNCAT assay in 10-micrometre muscle cryosections from the indicated parabiotic cohorts has confirmed the selectivity of ANL-labeling of MetRSL274G proteome in settings of parabiosis. A few mono-nucleated cells with FUNCAT signal above that of the nearby tissue were identified in the C57BL/6 muscle (20× image and an enlarged overlay). These FUNCAT + cells were not MetRSL274G leukocytes, as per negative GFP immunostaining (red). Scale bar, 100 μm. Similar results were obtained in three old C57BL/6 mice independently parabiosed to young MetRSL274G mice and analyzed by FUNCAT. e Proliferation rates of old C57BL/6 satellite cells were compared in culture, in presence of serum from young MetRSL274G mice that were administered with ANL in vivo versus the serum from old C57BL/6 mice. Shown are means and SD; N = 3, P < 0.05 (two-tail Student t-test). f Muscle regenerative index (the numbers of proliferating Ki67 + Desmin + myogenic cells) at 3-days post CTX injury; all parabionts were administered ANL in vivo. Representative immunostaining images are shown in right panels and quantification of the percent in vivo myogenic cell proliferation normalized to the percent myogenic cell proliferation in young muscles that is set at 1 (dashed line), is shown in the left panel bar graph. Shown are means and SD. N = 3, P = 0.426908 (two-tail Student t-test). C57BL/6 versus young MetRSL274G. Scale bar, 100 μm
After 4 weeks in parabiosis, ANL was administered to all animals for 6 days at 0.2 mmol/kg. At day 3, one TA and one Gastroc muscle per mouse were injured by cardiotoxin, as published3, 7, 32. Three days later injured and uninjured muscle were isolated, and cell-free blood serum and blood cell pellets were collected from heart bleeds.
Click-western was performed, as described above on the parabiotic serum where it resolved the in vivo ANL-tagged proteins in heart bleeds, validating the blood chimerism (Fig. 4b). And the PCR with Cre-specific primers (Supplementary Table 1) confirmed the blood chimerism in blood cells that were derived from the heart bleeds of the MetRSL274G to C57BL/6 pairs (Supplementary Fig. 5).
After confirmation of the success of parabioses, we have performed the Click-western and FUNCAT on tissues of the old C57BL/6 mice and their young MetRSL274G parabiotic partners. Click-westerns have revealed the preferential incorporation of the ANL that has been administered to parabionts in vivo into the muscle of MetRSL274G mice, confirming the selectivity of the proteome labeling by BONCAT in the mixed parabiotic environments (Fig. 4c). And interestingly, some ANL-tagged proteins were consistently detected by Click-westerns in muscle of the C57BL/6 mice sharing blood with the MetRSL274G parabionts, as compared to the background signal of the syngeneic C57BL/6 pairs (Fig. 4c).
Providing further validation of the data obtained by Click-westerns, FUNCAT demonstrated higher ANL-based protein fluorescence in muscle of the MetRSL274G parabionts as compared to the muscle of their C57BL/6 partners (Fig. 4d and Supplementary Fig. 3C—FUNCAT and dystrophin immunofluorescence). Interestingly, we detected a few rare cells in the old C57BL/6 muscle with higher FUNCAT signal than the rest of the tissue, suggesting preferential interactions with the circulatory ANL-tagged proteins from young MetRSL274G parabionts; these were not MetRSL274G- derived cells such as circulating leukocytes, as they were negative for GFP immunofluorescence (Fig. 4d). Muscle of C57BL/6 syngeneic pairs were consistently negative by FUNCAT (Supplementary Fig. 3B).
The intensity of the Click-western and FUNCAT signals in C57BL/6 muscle is limited by the dose and duration of the ANL pulse, and it is expected that with longer labeling and higher ANL dose there will be more ANL-tagged proteins in the parabiotic partners of MetRSL274G mice. The current regiment of six daily administrations allowed us to see both the selectivity of MetRSL274G BONCAT in mixed environments, e.g., muscle tissue of parabiotically joined animals, and also to detect the ANL-tagged MetRSL274G derived proteins that traveled through the shared circulation and rendered some B6 tissues labeled by Click-westerns and FUNCAT.
We also confirmed the phenomenon of association of systemic in vivo ANL-tagged proteins with myogenic cells by incubating cultured primary C57BL/6 myoblasts with serum from the ANL-treated MetRSL274G mice versus the negative control ANL-treated fx mice, and performing Click-western and FUNCAT (Supplementary Fig. 6).
Notably, we established that the incorporation of ANL in vivo into serum proteins (in MetRSL274G young mice) did not interfere with their rejuvenating properties. This conclusion is drawn upon the following two observations. In the first experiment, we have assayed the proliferation of old C57BL/6 satellite cells that were cultured with either their own old serum or with the young serum from the in vivo ANL-treated MetRSL274G mice; the ANL-treated young MetRSL274G serum maintained the known positive effects of young serum on the proliferation of old satellite cells4, 30, 33 (Fig. 4e, P < 0.05; two tail Student t-test). In the second study, the in vivo numbers of Ki67 + /Desmin + myogenic cells were quantified in muscle sections of old C57BL/6 parabionts. Sharing the systemic milieu with either ANL-tagged young MetRSL274G partner or with the young C57BL/6 partner produced similar enhancement of the old C57BL/6 myogenic cell proliferation2, 6, 9 (P = 0.426908, two tail Student t-test), Fig. 4f.
These results establish an approach for bio-orthogonal proteome tagging in settings of heterochronic parabiosis.
Modified for bio-orthogonality antibody arrays reveal a number of ‘young’ ANL-tagged proteins that were transferred to old muscle through heterochronic parabiosis
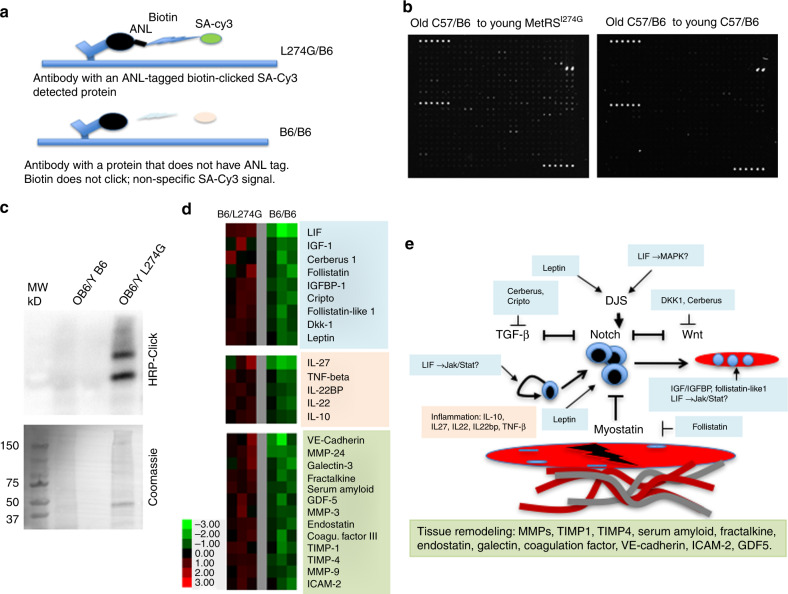
Finally, to advance the field of systemic aging and rejuvenation by direct profiling of young parabiotic proteome in old tissues, we have developed and used a modified antibody array approach on the samples of muscle from the old C57/B6 to young MetRSL274G versus young C57/B6 parabiosis. All parabionts were labeled with ANL in vivo for 6 days, including the 3 days of recovery from muscle injury induced by Cardiotoxin. In conventional proteomics array samples are labeled with biotin through primary amine and proteins are detected by the binding to their cognate antibodies and biotin-streptavidin-Cy3 fluorescence. We have deliberately omitted the amine-based biotinylation and instead Clicked the biotin-alkyne label to protein samples, postulating that without ANL incorporation the biotin-streptavidin-Cy3 background will be lower than the signal of the ANL-labeled proteins (Fig. 5a). To pursue this proteomics approach, we have used the Ray Biotech arrays, which have 301 features, with duplicates for each antibody specificity.
Fig. 5.
Proteomics of the ANL labeled proteins that are derived from muscle of old C57BL/6 parabiotic partners of young MetRSL274G mice. a Scheme of the application of antibody arrays to BONCAT focused proteomics. b Representative images of scanned antibody arrays that were performed with the muscle lysates from old C57BL/6 animals parabiosed to young MetRSL274G partners as compared to isogenic C57BL/6 pairs. The lysates were isolated from CTX injured TA and Gastroc muscles at 3 days post injury. c For these antibody arrays a Click-western blotting was performed to confirm detectable ANL-labeling in the muscle lysates from old C57BL/6 to young MetRSL274G parabionts, as compared to the background noise of the syngeneic C57BL/6 parabionts. Coomassie blue staining demonstrates equal protein loading. d Heat Map of the proteins that were detected at >2.0-fold higher levels in the muscle of old C57BL/6 mice parabiosed to young MetRSL274G, as compared to those parabiosed to young C57BL/6 animals with statistical significance of P < 0.05 (Wilcoxon rank sum test). n = 3. Color-coding: myogenic regulators (blue), inflammation regulators (pink), tissue remodeling proteins (green). e Schematic cartoon of the possible roles of the young proteins (colored boxes that correspond to protein grouping in the Heat Map) that were detected in the muscle of old parabiotic partners, grouped by canonical roles in regulating myogenic proliferation and differentiation, TGF-beta/myostatin, Notch and WNT signaling pathways, tissue remodeling and inflammation. Question marks signify putative roles
This strategy was successful, as demonstrated by the representative images where a robust signal over background is evident throughout the slide; and an equally successful hybridization and detection procedure for the experimental (Old C57BL/6 to MetRSL274G) and control (syngeneic C57BL/6) samples is confirmed by the internal positive controls, visible as the rows of bright spots (Fig. 5b). Click-westerns were performed to confirm the ANL-tagging of the proteome in the muscle of old C57BL/6 partners of young MetRSL274G parabionts, as compared to the background signal of the old C57BL/6 partners of young C57BL/6 parabionts (Fig. 5c).
Applying this strategy, we have resolved 70 ANL-tagged proteins in the regenerating after an injury muscle of old C57BL/6 mice that were parabiotically joined with the young MetRSL274G partners with statistically significant two-fold signal over the background of the identically treated B6/B6 samples (Supplementary Table 2). Three independent proteomics array experiments with the regenerating muscle from each heterochronic parabiosis cohort produced similar results, emphasizing the robustness of this BONCAT-based high throughput approach.
Further validating our findings and their significance, many of the identified proteins are known to be secreted-systemic; and interestingly, many of these soluble young proteins resolved in the old muscle have myogenic properties (Fig. 5d, e). Of these quite a few are likely to enhance myogenic proliferation34, by activating the age-specific determinant Notch signaling1 or antagonizing the TGF-beta or Wnt pathways, age-imposed elevation which inhibits old muscle repair6, 7, 35, 36, Fig. 5d, e. Follistatin-like1 positively mediates glucose uptake into skeletal muscle37, supporting homeostatic health of myofibers, and it also interacts with LIF1 in fat browning, which has multiple health-promoting effects38.
Another sub-set of young proteins that have been resolved in old muscles represent tissue remodeling factors, including MMPs and their inhibitors, TIMP proteins, VE-cadherin and VCAM2 that promote cell–cell interactions, GDF5 regulator of re-innervation as well as blood clotting and vascular remodeling factors (Serum amyloid, Fractalkine, Endostatin, Coagulation factor III).
And while some of these ‘young’ parabiotic proteins (directly identified in the old muscle) are well described for their positive and diminished with age muscle-related properties (IGF-1 and Follistatin, for example), others are less characterized in this regard (LIF, Leptin, Cerberus, GDF5, TIMP1, TIMP4).
Finally, another class of ANL-tagged proteins that have been identified in old muscle of C57/B6 mice parabiosed to the young MetRSL274G animals are the leukocyte-specific proteins: Interleukin receptors, chemokines, etc.; these are expected as blood cells travel from the young to old parabionts and are present in skeletal muscle where they contribute to adult myogenesis4, 39. Of these, IL27, IL10, IL22, IL22bp and TNF-β regulate tissue inflammation40.
Discussion
This work establishes a selective mammalian in vivo metabolic labeling of proteins in mixed environments (parabiosis and cell transplantation) and introduces a new paradigm for identifying and characterizing key determinants of health-youth versus aging-disease, at the base-line and after experimental treatments. This system does not adversely affect the animal health; and the ANL tagged proteins maintain their functionality. The C57BL/6 background of our CMV-Cre/MetRSL274G animals enables their broad immuno-compatible use in parabioses, blood exchanges and cell transplantation studies. Furthermore, these studies describe an antibody-array high throughput proteomics approach that is adapted for mammalian in vivo bio-orthogonal translation and requires less starting material than mass spectrometry: micrograms instead of milligrams.
A number of new ramifications in the biomedical arena are enabled by this study. Ubiquitous expression of MetRSL274G allows approaches where it is important to profile age-specific or disease-imposed, genetic-controlled, etc. changes, be those in global proteomes or in specific candidate protein factors. Not only circulatory, but also local tissue determinants of aging and disease can be more easily profiled: in tissue transplants, one can identify the in vivo proteome of the MetRSL274G expressing cells and tissues directly, at specific time points and without re-derivation.
The identified young circulatory proteins that were found in muscle of old mice provide the most interesting conclusion of these studies. Not just one or two, but many of these young-blood derived proteins possess rejuvenating pro-myogenic properties, and moreover, they are expected to synergize when such young factors reach the old tissue simultaneously. This suggests that not just one, but many youthful circulatory factors improve the health of old tissues in heterochronic environments, and argue against a ‘one silver bullet’ approach to systemic rejuvenation.
With respect to specific candidates, of a particular interest is LIF-1 that has been shown to enhance the repair of injured muscle, but was not well studied in the context of muscle aging and rejuvenation41. LIF-1 is well known to activate the Jak/Stat pathway, which has been shown to reduce symmetric division of myogenic precursors, which would possibly signify return to quiescence, and to promote fusion into myotubes (at the other end of myogenic lineage)42. But LIF-1 also activates the MAPK signaling, which can induce the Notch activating ligands Delta and Jagged, promoting myogenic proliferation43, 44.
Cripto and Cerberus1 are also interesting, as they act as antagonists of TGF-beta1, which increases with age and inhibits the regeneration of old muscle7, 32, 45, 46. With respect to the young derived GDF5 (a TGF-beta family member), it promotes muscle innervation that is known to decline with age47. And well-known muscle-specific TGF-beta superfamily protein, myostatin that inhibits muscle stem cell proliferation, might be antagonized by the ‘young’ follistatin48. Additionally, Cerberus1 also antagonizes the age-elevated Wnt pathway; and another young-derived Wnt inhibitor, DKK-1, is found in old muscle of old mice parabiosed to the young partners.6, 49, 50.
Of note, the ‘young’ proteins, which were transferred to old parabionts have known positive effects on other than skeletal muscle tissues51. For instance, inhibitors of TGF-beta and BMP and specifically Fractalkine and Cerberus1, were shown to play a role in brain aging and Parkinson’s Disease51, 52, as well as in rheumatoid arthritis53. LIF regulates T-cell fate54; it also has anti-oxidant properties and is implicated in health of brain, bone, etc. tissues55, 56. GDF5 promotes cardiac repair57. Follistatin-like1, which was transferred to old mice from young parabionts, is among myokines, which have been implicated in the broad positive effects of exercise on heart, brain, liver etc.58. Follistatin-like1 also protects cardiomyocites from experimentally-induced injury and promotes endothelial cell function and tissue re-vascularization59, 60.
Finally, leptin that we have identified as one ‘young’ systemic protein that is present in old muscle of parabiotically joined mice, broadly regulates hormonal networks, including those controlling reproduction and metabolism; it has numerous anti-aging effects and leptin or its’ receptor knock-out mice are diabetic, obese and short lived61, 62. Interestingly, leptin (that can be produced by many tissues, including skeletal muscle) has been shown to have direct positive effects on myogenic cell proliferation, on the expression of muscle markers, muscle-bone cross-talk and it activates Notch63, 64. Of note, leptin interacts with another endocrine hormone—oxytocin in its positive effects on the health of muscle, bone, brain and in reduction of reduces obesity64–68. Age-imposed leptin resistance69 is a known phenomenon that is linked to frailty; and since oxytocin also declines with age3, the alterations of leptin/oxytocin axis might represent a key event in mammalian aging that is rescued by heterochronic parabiosis.
Taken together, we demonstrate a useful approach for in vivo profiling of proteomes that orchestrate the young-healthy versus old-diseased tissue states. The initial proteomic data, obtained by this approach, suggest that a combination of therapeutics would be most effective to enhance resistance to age-imposed and pathological organ attrition.
Methods
Animals
All procedures were performed in accordance with the administrative panel of the Office of Laboratory Animal Care. The protocol was approved by the UC Berkeley Animal Care and Use Committee. Mice were anaesthetized with isoflurane drop and euthanized via cervical dislocation. Blood samples were collected by heart puncture. Young (2–4 mo) male C57BL/6 mice and CMV-Cre strain (Jackson labs, #00664, #006054) were purchased from the Jackson Laboratory. Twenty-two-month-old male C57BL/6 mice were purchased from the National Institute on ageing. The CAG-floxed-Stop-eGFP-mMetRSL274G (i.e., fx, Supplementary Fig. 1) breeder pairs (from Erin Shuman, Max Planck Institute for Brain Research, Frankfurt Germany) were genotyped, established as a colony and crossed with CMV-Cre strain. F1 progeny (2–4 mo males) containing both the Cre and recombined and expressed MetRSL274G were used (e.g., MetRSL274G mice).
Genotyping
Tail clips or blood cells were digested in digestion buffer (100 mM Tris-HCl (pH 8.5), 5mMEDTA, 0.2% SDS, 200 mM NaCl, 100ug/ml proteinase K). Then DNA samples were precipitated with isopropanol and then dissolved in TE buffer. To detect the Cre transgene, PCR was performed using OL2642 and OL2643 (Supplementary Table 1, primers were from Elim Biopharmaceuticals. Inc, Hayward, CA, 94545) in a reaction consisting of 2.5 units Taq polymerase (Promega) according to the manufacturer’s protocol. A 450 bp band confirmed the presence of the Cre transgene. Control samples negative for Cre did not amplify a product. PCR using Oligo 1 and 2 produced a product of ~ 441 bp while oligo 5 and 6 produced a ~ 493 bp, testing the presence of the ‘stop signal’ (Supplementary Fig. 1). Oligos 1 and 6 were used to test the excision of the ‘stop signal’. Oligo 3 and 4 were used to test the presence of eGFP tagged mMetRSL274G. Oligos 7 and 8 were used to test the presence of Rosawt allele. Ctrl1 and Ctrl2 amplified the CD79b wild-type allele producing ~ 330 and ~ 500 bp respectively. The PCR reaction was carried out using the following conditions: 95°/5 min→[95 °C/30 s→ 60 °C /30 s→ 72 °C /30 s]x 30cycles→72°/5 min → 4°/∞. Oligo 3 and 2839reverse were used to amplify a fragment, for which oligo1169reverse was used to sequence the PCR product to confirm the mouse L274G mutation.
RNA isolation and QT PCR
RNA was isolated using the QIAGEN Rneasy kit (Qiagen, Valencia, CA). RNA concentration was assessed using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE). cDNA was made with Superscript III first strand synthesis kit (Invitrogen 18080-051). QT PCR was carried out using RT2 SYBR® Green FAST Master mixes (Qiagen, cat 330602) according to manufacturer’s protocols. Oligo 3 and 162reverse (Supplementary Table 1) were used to test mMetRSL274G expression. GAPDH amplified with oligos GAPDH-F and GAPDH-R (Supplementary Table 1) was used as normalization. We use 2−∆Ct as the relative expression unit.
Identification of ANL-labeled proteins through Click-western blotting
Azidonorleucine (ANL) was purchased from Jena Biosciences (catalog number CLK-AA009, CAS#159610-92-1). ANL was added into the media at indicated concentration or I.P. injected into mice at indicated dose. Protein extraction was carried out by homogenizing samples or cells in RIPA buffer (50mMTris pH7.8, 150 mM NaCl, 0.1%SDS, 0.5% sodium deoxycholate, 1% triton-100, 1 mM PMSF, appropriate proteinase inhibitors) without EDTA + Benzonase (Sigma, ≥ 250 units per ml). Homogenates were incubated for 20 min at room temperature. After incubation for 1 h at 4 °C under constant agitation, lysates were spun down at 3000 g, 5 min. at 4 °C. The resulting supernatants (‘lysates’) were precipitated using 5 volumes of cold acetone, washed with 80% acetone and pellets were air-dried and dissolved in 1%SDS in 50 mMTris˙HCL PH8.0 with 1 mM PMSF freshly added. Protein quantification was performed using Nanodrop and RED 660 protein assay (Cat# 786-676, E-bioscience, St. Louis, MO). Similar amounts of protein samples (~ 20 ug) were then clicked with biotin labeled alkyne according to manufacturer’s protocol (ClickIT biotin-alkyne labeling kit, Life Technologies, Molecular Probes C33372) followed by Western blotting. The Western blots were then incubated with 1:200 dilution of horseradish peroxidase (HRP) conjugated streptavidin and imaged with HRP substrates (Advansta, Western Bright ECL k-12045-D20) following manufacturer’s protocols.
Visualization of ANL labeled proteins by FUNCAT
Cryosections or cells cultured in chamber slide were fixed with methanol and subsequently 4% paraformaldehyde (in phosphate buffered saline, PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH7.8). Following fixation, slides were washed three times with PBS. Slides were incubated in 0.1% Triton X-100and washed three times with PBS at room temperature under gentle agitation. Slides were blocked with PBS and 1% serum at 4 °C for 1 h. ANL-labeled proteins were clicked by mixing triazole ligand (THPTA, Sigma-Aldrich, CAS Number 760952-88-3, 3 mM), Alexa Flour 488 Alkyne (Life Technologies, Cat. A-10267, 2uM in dimethylsulfoxide), sodium ascorbate (Sigma-Aldrich, CAS134-03-2, 5 mM), and CuSO4 solution (Sigma-Aldrich, 600 uM) in PBS pH 7.8. After each addition, the solution was vortexed thoroughly. Slides were incubated with the click mixture for 3.5 h at room temperature under gentle agitation. Subsequently, slides were washed twice with PBS-Tween (PBS + 0.05% v/v Tween-20) plus 0.5 mM EDTA solution for ~ 15 min each at room temperature under gentle agitation before incubation with Hoechst DNA staining solution or antibodies following regular immunostaining protocol.
Immunofluorescence
Muscle sections attached to positively charged frosted glass microscope slides were fixed with 70% ethanol at 4 °C overnight and rinsed with 1× phosphate-buffered saline (PBS) the next day. The sections were then blocked for 30 min in 1% staining buffer (1% calf serum in 1× PBS). Samples were permeabilized with 0.1% Triton X-100 in 1X PBS for 5 min at room temperature and subsequently rinsed with 1% staining buffer 3 times, 5 min per rinse. The sections were incubated with primary antibodies diluted to 0.5–1 μg/ml at 4 °C overnight. The slides were rinsed as stated and then coated with secondary antibodies and Hoechst nuclear DNA stain for 2 h at room temperature in the dark. Samples were rinsed with PBS and mounted with Fluoromount. A Zeiss Axioscope fluorescent microscope was used for imaging. antibodies: rabbit anti-GFP antibody (abcam290,1:5000), rabbit anti-Ki67(ab16667, 1:200), and mouse anti-dystrophin antibody (Santa Cruz Biotechnology 58754,1:200) were used at dilutions according to manufacturer’s instructions. Secondary fluorochrome conjugated antibodies were from Life Technologies: goat anti-rabbit 546 (A11010, 1:2000) and goat anti-mouse 488 (A11029, 1:2000). DNA was stained by Hoechst 33342 from Sigma Aldrich (B2261) 1:500 at 1 μg/mL.
Preparation and culture of primary cells
Primary myogenic cells were prepared and cultured from muscle samples as previously described30, 32. Briefly, harvested muscles underwent enzymatic digestion in DMEM (Mediatech) containing collagenase type II (250U/ml; Sigma-Aldrich), 10 mM HEPES and penicillin/streptomycin antibiotics (500 IU/ml, 0.1 mg/ml; MP Biomedicals) at 37 °C for 20 min under agitation. Fat pads and tendons were removed after a quick wash with PBS and repeated rounds of muscle trituration and sedimentation were performed to purify myofibres. Myofiber suspensions were vortexed for 1 min to release satellite cells from digested fibers, passed through a 40mm cell strainer (BD Biosciences) and plated on Matrigel coated (0.04 ug/ml in PBS) dishes and cultured at 37 °C, 5% CO2 in growth medium (Ham’s F-10 (Mediatech), penicillin/streptomycin antibiotics (500 IU/ml, 0.1 mg/ml; MP Biomedicals), and 20% Bovine Growth Serum (Life Technologies/Hyclone), supplemented with FGF-2 (6 ng/ml)). Neonatal fibroblasts were derived as follows: The pups were euthanized and washed sequentially with sterile PBS, 70% ethanol, and then sterile PBS. The pups were then minced and digested with trypsin-EDTA (Sigma, T4049) for 30 min at 37 °C. The tissue was filtered through a sterile mesh and washed with DMEM (Fisher Scientific, 11330032). The cell pellets were resuspended, cultured and expanded in DMEM with 10%FBS (GE healthcare, SH30396) for two weeks. 70% confluent cells were incubated with ANL at indicated concentrations for 24 h. The primary cells were used soon after their derivation from animals that are routinely checked (found negative) for mycoplasma and other pathogens.
Parabiosis
Animals were connected surgically in parabiosis as described9. Briefly, the pair was prepared for surgery with matching incisions made along the proximal flanks, inner limbs ligated at the elbows and knees, and skin ligated between partners to join the two together and establish anastomosis. Post-operative medication included Baytril antibiotic administered twice daily for the first 3 days, and Meloxicam NSAID administered daily for the first week, as directed by veterinarians; anecdotally, we suggest these may help reduce the incidence of parabiotic disease. The parabioses were not set up all at the same time and the study was done as separate experiments: each pair was individually treated with ANL in vivo and then tissues from individual mice were individually isolated and processed for the BONCAT, FUNCAT, muscle regeneration assays, etc. (with at least three experiments with individual animals and 4–5 experimental replicates within each assay).
Proteomics
Antibody arrays (Mouse L308 Array; RayBiotech AAM-BLG-1-2, GA, 30092) was used to profile the proteins circulating from young L274G parabionts to old C57/B6 parabionts. The samples were run on three different arrays (each of which was done with the two comparative cohorts), but the samples were different overlapping pools of proteins from individual mice of that cohort (for example, A, B, C for one pool, A, B, D for the second pool, etc.). Each protein was examined in duplicates and the means for each of the 301 proteins were compared between the mutant and the WT results for all proteins; and those found to be elevated by 2 fold in the mutant with P > 0.05 were considered to be significant and are reported in Fig. 5 and Supplementary Table 2. Proteins were clicked with biotin labeled alkyne as indicated above and dissolved in 1% SDS 50mMTris-HCl(pH8). Nanodrop was used to quantify the clicked product. The arrays were blocked with blocking solution, incubated with clicked protein (33 micrograms) in PBS(pH8), washed and detected with Cy3 labeled streptavidin, as recommended by the manufacturer. Arrays were scanned using genePix 4000Bscanner (Molecular Devices, Sunnyvale, CA, 94089) at 532 nm. Feature extraction was done using genePixpro6.1 software (Molecular Devices, Sunnyvale, CA, 94089). For each protein, duplicate spots on each array were averaged, local background fluorescence was subtracted and resulting fluorescence signal was normalized by the internal positive controls on the arrays. Average signals from all 6 arrays (3 experiments for positive and negative samples) were set to 1. Log2 transformation was done so that signal value > 0 is shown as red and signal value < 0 is shown as green. Wilcoxon rank sum test was done between Old C57BL/6/Young MetRSL274G group and Old C57BL/6/Young C57BL/6 group. P-value = 0.05 and 2-fold Old C57BL/6/Young MetRSL274G signal over Old C57BL/6/Young C57BL/6 background were selected as the cutoffs for the identified ‘young’ proteins that have been resolved in the old muscle in heterochronic parabioses.
Quantification and statistical analysis
On the basis of the published body of work that generated statistically significant results in similar experimental set-ups and minimizes the unnecessary use of vertebrate animals, a minimum of 10 mice of each genetic background, MetRSL274G and fx, were analyzed for the recombination of the genetic locus, 3 mice for expression and function of MetRSL274G, and 10 mice for the viability and vigor of the animals (without and with ANL). A minimum of 3 independent Click-westerns and FUNCAT assays were performed with MetRSL274G and fx cells and tissues with replicates within each assay. 9 and 13 heterochronic parabioses were established for the MetRS274G to C57BL/6 and C57BL/6 to C57BL/6, respectively; with subsequent derivation and analysis of serum, cells and tissues in minimum of 3 independent Click-Chemistry Westerns, FUNCAT assays and Ray Biotech proteomic antibody arrays for each cohort with replicates within each assay. 3 independent myogenic cell transplantation experiments for each genetic background were performed with subsequent multiple analyses by FUNCAT in tissue cryosecitons and by Click-western blotting with replicates within each assay. No data points were excluded from quantification. The data was processed as means and standard deviations. P values of <0.05 that were obtained in Student T Test and Wilcoxon rank sum test were considered statistically significant.
Data availability
The fx and MetRSL274G mouse strains will be shared with all interested researchers after publication of these studies. The authors declare that all other data supporting the findings of this study are available within the paper and its Supplementary Information files, or available from the authors upon reasonable request.
Electronic supplementary material
Acknowledgements
We thank Erin Schuman (Max Planck) for providing breeding pairs of mice with Floxed mMetRSL274G 13, 15, 24; and David Tirrell and Shannon Stone (Caltech) for helpful discussion of our data. The NIHR56, Rogers’ family and Calico Awards to I.M.C. and CAPES - Ciências sem Fronteiras Brazilian government program to T.C.S supported this work.
Author contributions
Y.L. performed the experiments shown in all figures, established the MetRSL274G strain and the well-genotyped colony of the fx mice, analyzed the data and co-wrote the manuscript. M.J.C. performed the experiments shown in Figs 3–5, analyzed the data, contributed to planning of these studies and co-wrote the manuscript. M.M. performed the experiments shown in Fig. 4, Yu.L. contributed to the experiments shown in Figs 1, 2 and Supplementary Fig. 1. T.P.T. performed the experiments shown in Supplementary Figs 4, 5. A.B. contributed to the experiments shown in Fig. 5. P.R. performed the experiments shown in Fig. 4c. T.C.S. performed background studies with in vitro BONCAT western blotting. I.M.C. designed, planned and integrated the study, interpreted the data and wrote the manuscript.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/10/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Electronic supplementary material
Supplementary Information accompanies this paper at doi:10.1038/s41467-017-00698-y.
References
- 1.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- 2.Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 3.Elabd C, et al. Oxytocin is an age-specific circulating hormone that is necessary for muscle maintenance and regeneration. Nat. Commun. 2014;5:4082. doi: 10.1038/ncomms5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paliwal P, Pishesha N, Wijaya D, Conboy IM. Age dependent increase in the levels of osteopontin inhibits skeletal muscle regeneration. Aging. 2012;4:553–566. doi: 10.18632/aging.100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Han S, Cousin W, Conboy IM. Age-specific functional epigenetic changes in p21 and p16 in injury-activated satellite cells. Stem Cells. 2015;33:951–961. doi: 10.1002/stem.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 7.Carlson ME, et al. Relative roles of TGF-beta1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell. 2009;8:676–689. doi: 10.1111/j.1474-9726.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson ME, et al. Molecular aging and rejuvenation of human muscle stem cells. EMBO Mol. Med. 2009;1:381–391. doi: 10.1002/emmm.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conboy IM, Rando TA. Heterochronic parabiosis for the study of the effects of aging on stem cells and their niches. Cell Cycle. 2012;11:2260–2267. doi: 10.4161/cc.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conboy IM, Conboy MJ, Rebo J. Systemic problems: a perspective on stem cell aging and rejuvenation. Aging. 2015;7:754–765. doi: 10.18632/aging.100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egerman MA, et al. GDF11 Increases with age and inhibits skeletal muscle regeneration. Cell. Metab. 2015;22:164–174. doi: 10.1016/j.cmet.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith SC, et al. GDF11 does not rescue aging-related pathological hypertrophy. Circ. Res. 2015;117:926–932. doi: 10.1161/CIRCRESAHA.115.307527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erdmann I, et al. Cell-selective labelling of proteomes in Drosophila melanogaster. Nat. Commun. 2015;6:7521. doi: 10.1038/ncomms8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngo JT, et al. Cell-selective metabolic labeling of proteins. Nat. Chem. Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahdavi A, et al. Engineered aminoacyl-tRNA synthetase for cell-selective analysis of mammalian protein synthesis. J. Am. Chem. Soc. 2016;138:4278–4281. doi: 10.1021/jacs.5b08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone SE, Glenn WS, Hamblin GD, Tirrell DA. Cell-selective proteomics for biological discovery. Curr. Opin. Chem. Biol. 2017;36:50–57. doi: 10.1016/j.cbpa.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link AJ, et al. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc. Natl Acad. Sci. USA. 2006;103:10180–10185. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuet KP, et al. Cell-specific proteomic analysis in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2015;112:2705–2710. doi: 10.1073/pnas.1421567112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in Mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc. Natl Acad. Sci. USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller A, Stellmacher A, Freitag CE, Landgraf P, Dieterich DC. Monitoring astrocytic proteome dynamics by cell type-specific protein labeling. PLoS ONE. 2015;10:e0145451. doi: 10.1371/journal.pone.0145451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauthier NP, et al. Cell-selective labeling using amino acid precursors for proteomic studies of multicellular environments. Nat. Methods. 2013;10:768–773. doi: 10.1038/nmeth.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiger T, et al. Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Mol. Cell Proteomics. 2013;12:1709–1722. doi: 10.1074/mcp.M112.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calve S, Witten AJ, Ocken AR, Kinzer-Ursem TL. Incorporation of non-canonical amino acids into the developing murine proteome. Sci. Rep. 2016;6:32377. doi: 10.1038/srep32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngo JT, Schuman EM, Tirrell DA. Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc. Natl Acad. Sci. USA. 2013;110:4992–4997. doi: 10.1073/pnas.1216375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClatchy DB, et al. Pulsed azidohomoalanine labeling in mammals (PALM) detects changes in liver-specific LKB1 knockout mice. J. Proteome Res. 2015;14:4815–4822. doi: 10.1021/acs.jproteome.5b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat. Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brockmann GA, et al. QTLs for pre- and postweaning body weight and body composition in selected mice. Mamm. Genome. 2004;15:593–609. doi: 10.1007/s00335-004-3026-4. [DOI] [PubMed] [Google Scholar]
- 29.Wiesmann F, et al. Developmental changes of cardiac function and mass assessed with MRI in neonatal, juvenile, and adult mice. Am. J. Physiol. Heart Circ. Physiol. 2000;278:H652–H657. doi: 10.1152/ajpheart.2000.278.2.H652. [DOI] [PubMed] [Google Scholar]
- 30.Conboy MJ, Conboy IM. Preparation of adult muscle fiber-associated stem/precursor cells. Methods Mol. Biol. 2010;621:149–163. doi: 10.1007/978-1-60761-063-2_10. [DOI] [PubMed] [Google Scholar]
- 31.Paliwal P, Conboy IM. Inhibitors of tyrosine phosphatases and apoptosis reprogram lineage-marked differentiated muscle to myogenic progenitor cells. Chem. Biol. 2011;18:1153–1166. doi: 10.1016/j.chembiol.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousef H, et al. Systemic attenuation of the TGF-beta pathway by a single drug simultaneously rejuvenates hippocampal neurogenesis and myogenesis in the same old mammal. Oncotarget. 2015;6:11959–11978. doi: 10.18632/oncotarget.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conboy MJ, Cerletti M, Wagers AJ, Conboy IM. Immuno-analysis and FACS sorting of adult muscle fiber-associated stem/precursor cells. Methods Mol. Biol. 2010;621:165–173. doi: 10.1007/978-1-60761-063-2_11. [DOI] [PubMed] [Google Scholar]
- 34.Charville GW, et al. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Rep. 2015;5:621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brack AS, Conboy IM, Conboy MJ, Shen J, Rando TA. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Naito AT, et al. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell. 2012;149:1298–1313. doi: 10.1016/j.cell.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HJ, et al. Kalirin, a GEF for Rac1, plays an important role in FSTL-1-mediated glucose uptake in skeletal muscle cells. Cell Signal. 2017;29:150–157. doi: 10.1016/j.cellsig.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez A, Becerril S, Ezquerro S, Mendez-Gimenez L, Fruhbeck G. Crosstalk between adipokines and myokines in fat browning. Acta Physiol. 2017;219:362–381. doi: 10.1111/apha.12686. [DOI] [PubMed] [Google Scholar]
- 39.Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- 40.Stumhofer JS, Hunter CA. Advances in understanding the anti-inflammatory properties of IL-27. Immunol. Lett. 2008;117:123–130. doi: 10.1016/j.imlet.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnard W, Bower J, Brown MA, Murphy M, Austin L. Leukemia inhibitory factor (LIF) infusion stimulates skeletal muscle regeneration after injury: injured muscle expresses lif mRNA. J. Neurol. Sci. 1994;123:108–113. doi: 10.1016/0022-510x(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 42.Price FD, et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat. Med. 2014;20:1174–1181. doi: 10.1038/nm.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicola NA, Babon JJ. Leukemia inhibitory factor (LIF) Cytokine Growth Factor Rev. 2015;26:533–544. doi: 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felling RJ, Covey MV, Wolujewicz P, Batish M, Levison SW. Astrocyte-produced leukemia inhibitory factor expands the neural stem/progenitor pool following perinatal hypoxia-ischemia. J. Neurosci. Res. 2016;94:1531–1545. doi: 10.1002/jnr.23929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guardiola O, et al. Cripto regulates skeletal muscle regeneration and modulates satellite cell determination by antagonizing myostatin. Proc. Natl Acad. Sci. USA. 2012;109:E3231–E3240. doi: 10.1073/pnas.1204017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macpherson PC, Farshi P, Goldman D. Dach2-Hdac9 signaling regulates reinnervation of muscle endplates. Development. 2015;142:4038–4048. doi: 10.1242/dev.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilson H, et al. Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. Am. J. Physiol. Endocrinol. Metab. 2009;297:E157–E164. doi: 10.1152/ajpendo.00193.2009. [DOI] [PubMed] [Google Scholar]
- 49.Cruciat CM, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seib DR, et al. Loss of Dickkopf-1 restores neurogenesis in old age and counteracts cognitive decline. Cell Stem Cell. 2013;12:204–214. doi: 10.1016/j.stem.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Yousef H, et al. Age-associated increase in BMP signaling inhibits hippocampal neurogenesis. Stem Cells. 2015;33:1577–1588. doi: 10.1002/stem.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimmig B, Morganti J, Nash K, Bickford PC. Immunomodulators as therapeutic agents in mitigating the progression of Parkinson’s disease. Brain Sci. 2016;6:E41. doi: 10.3390/brainsci6040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nanki T, Imai T, Kawai S. Fractalkine/CX3CL1 in rheumatoid arthritis. Modern Rheumatol. 2017;27:392–397. doi: 10.1080/14397595.2016.1213481. [DOI] [PubMed] [Google Scholar]
- 54.Metcalfe SM. LIF in the regulation of T-cell fate and as a potential therapeutic. Genes Immun. 2011;12:157–168. doi: 10.1038/gene.2011.9. [DOI] [PubMed] [Google Scholar]
- 55.Davis SM, Pennypacker KR. Targeting antioxidant enzyme expression as a therapeutic strategy for ischemic stroke. Neurochem. Int. 2016;107:23–32. doi: 10.1016/j.neuint.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaji H. Interaction between muscle and bone. J. Bone Metab. 2014;21:29–40. doi: 10.11005/jbm.2014.21.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaidi SH, Huang Q, Momen A, Riazi A, Husain M. Growth differentiation factor 5 regulates cardiac repair after myocardial infarction. J. Am. Coll. Cardiol. 2010;55:135–143. doi: 10.1016/j.jacc.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 58.Di Raimondo D, et al. Are the myokines the mediators of physical activity-induced health benefits? Curr. Pharm. Des. 2016;22:3622–3647. doi: 10.2174/1381612822666160429121934. [DOI] [PubMed] [Google Scholar]
- 59.Chen W, et al. Follistatin-like 1 protects cardiomyoblasts from injury induced by sodium nitroprusside through modulating Akt and Smad1/5/9 signaling. Biochem. Biophys. Res. Commun. 2016;469:418–423. doi: 10.1016/j.bbrc.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 60.Ouchi N, et al. Follistatin-like 1, a secreted muscle protein, promotes endothelial cell function and revascularization in ischemic tissue through a nitric-oxide synthase-dependent mechanism. J. Biol. Chem. 2008;283:32802–32811. doi: 10.1074/jbc.M803440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang B, Chandrasekera PC, Pippin JJ. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Curr. Diabetes Rev. 2014;10:131–145. doi: 10.2174/1573399810666140508121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarmento-Cabral A, et al. Adipokines (leptin, adiponectin, resistin) differentially regulate all hormonal cell types in primary anterior pituitary cell cultures from two primate species. Sci. Rep. 2017;7:43537. doi: 10.1038/srep43537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez A, et al. Leptin administration activates irisin-induced myogenesis via nitric oxide-dependent mechanisms, but reduces its effect on subcutaneous fat browning in mice. Int. J. Obes. 2015;39:397–407. doi: 10.1038/ijo.2014.166. [DOI] [PubMed] [Google Scholar]
- 64.Hamrick MW. Role of the cytokine-like hormone leptin in muscle-bone crosstalk with aging. J. Bone Metab. 2017;24:1–8. doi: 10.11005/jbm.2017.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balazova L, et al. Metabolic effects of subchronic peripheral oxytocin administration in lean and obese zucker rats. J. Physiol. Pharmacol. 2016;67:531–541. [PubMed] [Google Scholar]
- 66.Altirriba J, Poher AL, Rohner-Jeanrenaud F. Chronic oxytocin administration as a treatment against impaired leptin signaling or leptin resistance in obesity. Front. Endocrinol. 2015;6:119. doi: 10.3389/fendo.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perello M, Raingo J. Leptin activates oxytocin neurons of the hypothalamic paraventricular nucleus in both control and diet-induced obese rodents. PLoS ONE. 2013;8:e59625. doi: 10.1371/journal.pone.0059625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morton GJ, et al. Peripheral oxytocin suppresses food intake and causes weight loss in diet-induced obese rats. Am. J. Physiol. Endocrinol. Metab. 2012;302:E134–E144. doi: 10.1152/ajpendo.00296.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lana A, et al. Leptin concentration and risk of impaired physical function in older adults: the Seniors-ENRICA cohort. Age Ageing. 2016;45:819–826. doi: 10.1093/ageing/afw142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The fx and MetRSL274G mouse strains will be shared with all interested researchers after publication of these studies. The authors declare that all other data supporting the findings of this study are available within the paper and its Supplementary Information files, or available from the authors upon reasonable request.