Abstract
Bacteria are cosmopolitan organisms that in recent years have demonstrated many roles in maintaining host equilibrium. In this review, we discuss three roles bacteria can occupy in a host: pathogenic, symbiotic, and transient, with a specific focus on how bacterial small molecules contribute to homeostasis or dysbiosis. First, we will dissect how small molecules produced by pathogenic bacteria can be used as a source for communication during colonization and as protection against host immune responses. The ability to achieve a higher level of organization through small molecule communication gives pathogenic bacteria an opportunity for increased virulence and fitness. Conversely, in symbiotic relationships with hosts, small molecules are used in the initial acquisition, colonization, and maintenance of this beneficial population. Chemical signals can come from both the host and symbiont, and it is often observed that these interKingdom symbioses result in coevolution of both species involved. Furthermore, the transition from transient to commensal or opportunistic likely relies on molecular mechanisms. The small molecules utilized and produced by transient bacteria are desirable for both the immune and nutritional benefits they provide to the host. Finally, the advantages and disadvantages of modern analytical techniques that are available to researchers in order to study small molecules in situ is an important aspect of this review. It is our opinion that small molecules produced by bacteria are central to many biological processes and a larger focus on uncovering the function and identity of these small molecules is required to gain a deeper understanding of host-microbe associations.
Keywords: Pathogen, symbiont, transient, bacteria, specialized metabolite, analytical techniques, structure elucidation, mass spectrometry
eTOC
The relationships between bacteria and their animal hosts are key to understanding health and disease as well as environmental sustainability. Small molecules are produced by bacteria and hosts for varied purposes and these molecules are central to understanding the complex bacteria-host interplay. Study of how this chemistry translates into communication on a cellular level will move our scientific paradigms forward.
Introduction
Bacteria produce a wide variety of unique chemical entities. These molecules have been studied and exploited for the treatment of a variety of disease or human related needs and include: antimicrobials, anticancer agents, anti-aging creams, dietary supplements, and antivirals to name a few.1 Additionally, a number of labs have investigated the roles that a small subset of these molecules may exert on a specific system, such as quorum sensing2,3 or oxidation-reduction potential of electron transporting small molecules.4,5 Bacteria produce unique chemistry for specific ecological functions, and these molecules have been termed specialized metabolites.6 In the context of this review, specialized metabolites that are low in molecular weight such as nitric oxide (NO) are discussed and the largest small molecules are large peptides (~5,000 Da). The bacteria discussed in this review are implicated or contain the genetic material to produce the discussed chemical entities and we delve into the function of these small molecules in host-bacterial relationships.
Recently, a few groups have started to examine the chemical communication between epithelial cells and bacteria in vitro.7–9 Specifically, Ismail et al described an intimate relationship between mammalian epithelial cells and two different enteric microbes that have the capacity to quorum sense via the autoinducer-Lux pathway.9 In this paper, the authors described an unknown chemical entity that targets the Lux pathway of quorum sensing in both Vibrio harveyi and Salmonella typhimurium, which in turn, or as a response to the original chemical stressors, activates the bacteria to produce another chemical(s) that disrupts the epithelial tight junctions. While this research is tantalizing for generating hypotheses about interKingdom signaling pathways, it highlights the large gap in knowledge when it comes to the chemistry that bacteria utilize to adapt to host environments (Figure 1). Elucidating the structure and genetic factors responsible for the production of these specialized metabolites is not trivial but is important if we are to continue to explore the roles and expression of specific metabolites directly from the host environment. In light of the recent microbiome initiative we believe that small molecule chemistry will be imperative in studying and testing complex host-bacterial relationships.
Figure 1.
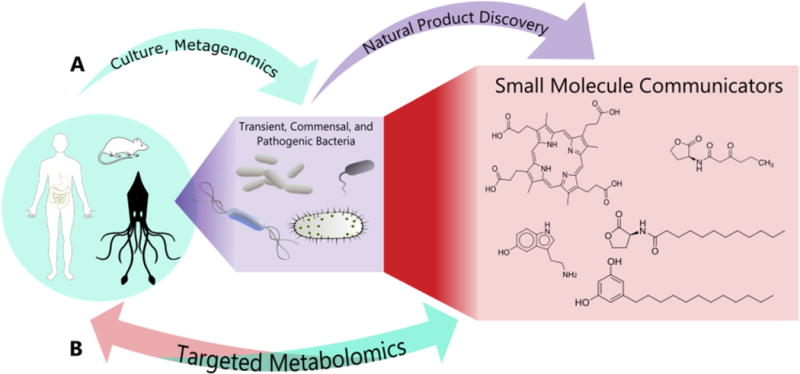
Transient, commensal, and pathogenic bacteria can reside both separately or in combination within a host. To date, (A) a majority of studies have inferred relationships based on metagenomic surveys of the microbes in or on a host, (B) but as further chemical information is learned a direct inference can be made about the host systems.
Pathogenic Relationships
In the quest for evolutionary dominance, bacteria and their hosts use the language of small molecules to cooperate and quarrel with one another. The production of specialized metabolites, commonly referred to as small molecules, secondary metabolites, or natural products, by bacteria can be used to form a higher order of organization within a bacterial community and increase resistance towards a host immune response or exogenous antibiotics. Quorum sensing is a term frequently used to describe the regulation of bacterial gene expression in response to fluctuations in cell-population density.10 The ability to communicate, and therefore regulate gene expression, allows a bacterium to rapidly adapt to survive in different environments. For example, Vibrio cholerae, the causative agent of the waterborne disease cholera, is able to survive the transition from highly aerobic freshwater sources to the anaerobic intestines of the human body. The microenvironment adaptability of V. cholerae allows for colonization of humans and continued propagation of the pathogenic bacterium to the environment.
Bacteria are experts at survival, and even in the presence of a host immune response, pathogenic bacteria have found a way to fight back (Figure 2). There is a plethora of small molecules that pathogenic bacteria have been documented to produce that allow them to communicate, infect, and grow in vivo. Sviridova et al. found that N- dodecanoyl-L-homoserine lactone (C12-HSL) (1), produced by Pseudomonas aeruginosa, was fatally toxic to human granulocytes and monocytes.11 P. aeruginosa is a tenacious bacterium that can thrive in water, soil, and animals, including humans.12 Regarding human hosts, P. aeruginosa presents the most danger to those who are immunocompromised such as burn victims and cystic fibrosis (CF) patients.12 Sviridova et al. conducted a comparison between the cytotoxic effects of 1 and an exogenous chemical control, 5- N- dodecyl resorcinol (C12-AR) (2). Despite the chemical similarities in these compounds, 2 lead to rapid lysis without apoptosis, whereas 1 lead to rapid apoptosis of human granulocytes and monocytes that were isolated from leukocyte-rich plasma.11 C12-HSL (1) and 2 are similar in molecular weight, but have different functional groups, therefore eliciting distinct host responses. Both molecules have saturated lipid chains that contain the same number of carbon atoms, but the head group of 1 is a lactone whereas 2 has a benzenediol. While both molecules cause cell death, the phenotypes would likely be linked to the chemical difference of the head groups between 1 and 5. Not surprisingly, the mechanism of action is reliant on the specificity of the molecule. Therefore, using 1, P. aeruginosa is able to defend itself from a principal opponent in the human host immune response. This example highlights a bacterium’s ability to use small molecules to achieve a specific action, such as the increase in virulence, with a high level of selectivity and efficiency.
Figure 2.
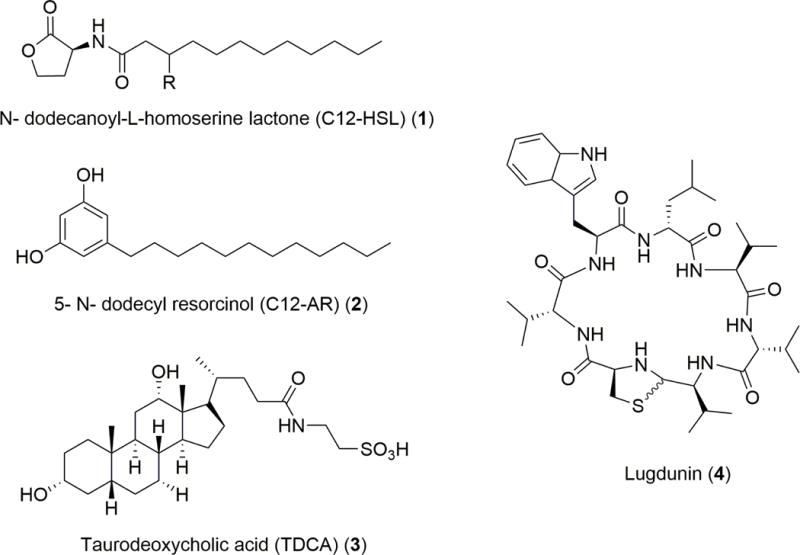
Molecular structures of compounds involved in host-pathogen interactions that elicit responses based on environmental changes.
In another example, P. aeruginosa utilizes small molecules to adapt and thrive in the human host lung of a CF patient. Once P. aeruginosa infiltrates a compromised lung, it begins to experience stress due to the highly anaerobic, low fluid shear (LS) lung microenvironment of the cystic fibrosis lung.12 LS refers to the natural flow of fluid inside the human body, in this case the lungs. Using a rotating wall vessel bioreactor, researchers can create 3D cultures with either high or low fluid flow (LS) which simulates an in vivo environment. Once P. aeruginosa senses LS, the bacterium upregulates genes involved in alginate biosynthesis, glycerol metabolism, and tryptophan biosynthesis, all of which are known to stimulate the formation of biofilms, thereby increasing virulence.12 At the same time, LS conditions downregulate genes involved in motility, phenazine biosynthesis, type VI secretion, multidrug efflux, and an extensive group of genes with an unknown functional class.12 In an RNA transcription analysis, of 240 genes with altered gene expression in CF patients, 36 and 39 unknown genes were either upregulated or downregulated, respectively.12 We believe some of these genes are likely related to specialized metabolite biosynthesis, and that further investigation into the small molecule products of these genes is necessary and alluring because of the potential to discover new chemical entities with significant ecological functions. With a vast gap of unknown chemistry responsible for the defense and survival mechanisms of pathogenic bacteria, many gene clusters have been labeled with unidentified function and are referred to as hypothetical proteins. For example, 1,169 genes out of 3,693 in the Vibrio cholerae genome are unknown regarding the structure of the protein and its corresponding biological function (Figure 3A). Of an entire bacterial genome, 32% is labeled as hypothetical with no prediction of biological purpose. Since the other 68% that have been identified include basic proteins important for viability, motility, reproduction, and virulence we believe that the remaining 32% is largely composed of specialized metabolite biosynthetic genes. We hypothesize that small molecules participate in a constantly evolving cycle in order to increase bacterial fitness and develop to fight against host defense mechanisms.
Figure 3.
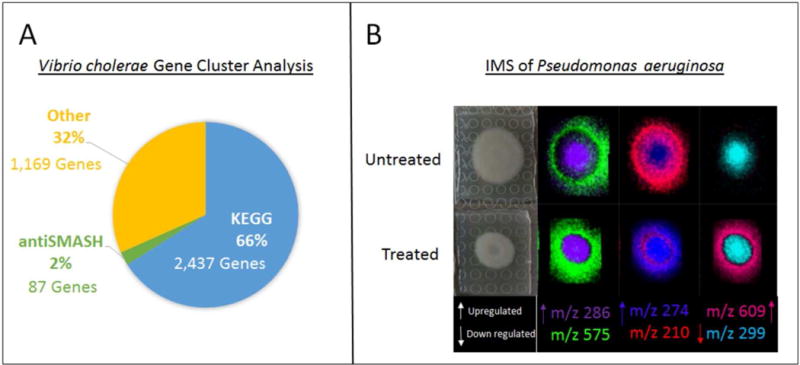
There is a vast amount of unknown chemistry involved in gene regulation of V. cholerae as depicted by the lack of identification of protein in the genome. MALDI-TOF MS is a tool which can be used to work towards identifying the unknown chemistry and aid in determining biological function of genes in these species. (A) Gene cluster analysis of V. cholerae (chromosome I: NC_00205 and chromosome II: NC_002506) using data collected annotated by antiSMASH and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (B) Imaging mass spectrometry data collected using MALDI-TOF MS. P. aeruginosa analyzed in situ and treated with a non-lethal biofilm inhibitor to visualize the spatial distribution of small molecules regulated pre and post exposure to treatment.
There has been a substantial increase in biofilm studies over the past 13 years.13 Biofilms are communities of bacteria that have encased themselves in an excreted matrix allowing for the bacteria to adhere to surfaces in unwelcoming environments such as extreme heat, acidic and basic niches, frozen glaciers, and the human body. Though there are many antibiotics to treat bacterial infections, biofilm-associated bacteria have been shown to be 100–1,000 fold more resistant to antibiotic treatment, making them difficult to eliminate.14–16 In the Sanchez laboratory we study the small molecules that are produced by Gram-negative microbes in biofilm states. In order to study the small molecules being released by bacterial species throughout the biofilm life cycle after exposure to treatment, imaging mass spectrometry (IMS) is utilized. IMS is capable of displaying the spatial distribution of molecules in agar and tissue samples. An example can be seen in Figure 3B with the bacterial species P. aeruginosa. This technique is discussed further in the Analytical Techniques section of this review. The colors are arbitrary and computationally assigned during data analysis. The focus of this image is the location of specific molecules. This technique allows for the identification of molecules which are being up- and downregulated with and without treatment in order to rapidly prioritize molecules for further analysis.
As bacteria continue to evolve new defense techniques, so do susceptible hosts. A study reported that expression of phoP, a virulence gene in Salmonella enterica, was regulated differently when S. enterica was grown in the presence or absence of physiological bile.17 Subsequently, only a subset of small molecules from physiological bile are responsible for the repression of phoP which therefore repressed S. enterica growth by 3.2 fold.17 Though the structures of bioactive compounds have not been completely elucidated the authors did run experiments showing that the mechanism of action of this subset of molecules was not due to osmolarity. The molecules had hydrophobic properties, and the most abundant compound in the bioactive fraction was taurodeoxycholic acid (TDCA) (3). Upon further analysis it was determined that 3 was not the sole cause of phoP repression, but rather a component in the subset of molecules that repress phoP. An outstanding question remains as to whether host defense is related to innate immunity of the host or an evolved protection mechanism against S. enterica. Elucidating the chemical structures of distinct host-pathogen interactions would shed light on how host chemistry influences bacteria and vice versa. Methicillin-resistant Staphylococcus aureus (MRSA) is an increasing clinical problem among CF patients and unfortunately the therapeutic options are limited.18 MRSA infections cause an accelerated decline in lung function of the already declining health of a CF patient.18 In a study of human upper airway epithelial cells and MRSA it is suggested that evolutionary pressures are the reason that upper airway epithelial cells release nitric oxide (NO), a well characterized bactericidal, after sensing the presence of MRSA.19 Variations in NO production between patients infected with S. aureus were observed and were inversely proportional to S. aureus growth. Regulating NO production, a well characterized pathway in humans, efficiently countered S. aureus colonization. In the context of S. aureus, evolution may play a large role in alternative host immune responses: 44% of the human population has never been colonized by S. aureus while the remaining 56% have experienced intermittent or persistent asymptomatic nasal cavity colonization.19 Recently, it was reported that the uncolonized human population has a type of defense mechanism against S. aureus that involves a commensal bacterium that produces lugdunin (4), a novel thiazolidine-containing cyclic peptide antibiotic, which prohibits the colonization of S. aureus.20 In this case, the human host-beneficial microbe association protects against the pathogenic species mediated through a specific small molecule.
There are also cases when competing pathogens have different adaptive abilities within one host. A study by Wiles et al. demonstrated that contractions of the human gut leads to a collapse in Aeromonas veronii population but has no effect on V. cholerae.21 Experiments showed that when V. cholerae was not present A. veronii successfully recovered after a decrease in population due to intestinal contractions.21 Thus V. cholerae plays a role in the demise of A. veronii and a logistic growth model revealed that direct bacterial competition only partially explains the observed phenomenon between these two bacterial species. To further investigate this phenomenon, using zebrafish as a host, studies were conducted to examine the population densities in situ.21 The difference in bacterial survival were due to the difference in growth and structure between the two species; in the gut, V. cholerae remains in a motile, planktonic state and therefore is completely unaffected by intestinal motility whereas A. veronii is in a rigid, highly aggregated state causing it to be forced out of the intestines upon contraction and unable to recover.21 As previously stated, this pathogenic competition is due to bacteria-bacteria communication and intestine motility. The chemistry behind both of these mechanisms is not defined and would be insightful towards discovering potential quorum sensing or antibiotic molecules. Once again reinforcing the prevalence of small molecule chemistry, interactions bacteria have with themselves, each other, and their hosts are regulated and driven using the language of small molecules. The lack of knowledge observed and demonstrated by many of the above examples is proof of how little is actually known of the pathogens that scientists so desperately try to fight. According to the World Health Organization new resistance mechanisms emerge and spread globally every day making treatment and eradication extremely difficult.22 Pathogenic bacteria are experts, having evolved and survived for millions of years prior to human civilization. Our advancement as mankind has only aided bacteria in increased colonization abilities. Decoding the language of small molecules is essential to overcoming the rapidly growing antibiotic drug resistant strains of pathogenic bacteria now found in hospitals, communities, and the environment.
Symbiosis in Nature
Interactions between animals and bacteria range from completely detrimental for one partner to thoroughly beneficial for both partners. The latter category of interaction is referred to as symbiosis or, more specifically, mutualism. While the presence and effects of pathogenic bacterial colonies are widely known, symbiotic relationships between bacterial species and animal hosts are less often recognized. Walter et al. presents an enlightened perspective on microbial interactions with eukaryotic hosts: symbioses exist on a spectrum between mutualistic and parasitic relationships.23 Most of what has been studied in animal-microbe symbiosis has been in invertebrates, namely insects24 and nematodes.25 For comprehensives overviews of insect-bacterial symbioses, we recommend reviews by Su et al.,26 Ferrari et al.,27 and Chaves et al.28
InterKingdom symbiotic relationships provide strong evidence for the coevolution of cross-Kingdom species from decades of interaction. Coevolution is an important outcome of these interKingdom relationships, signifying that they often require incredible specificity and selectivity. While novel animal-microbe symbioses continue to be discovered, a large focus has historically been on elucidating the symbiotic relationships between plants and microbes, as these interactions are both economically and agriculturally significant. See Gunatilaka for a comprehensive overview of plant-microbe symbioses.29 There is a staggering amount of evidence indicating that small molecules are involved in the establishment of plant-microbe relationships.30 Symbiotic relationships in plants have been so well studied that they have been separated into two defined categories based upon the positioning of the bacterial colony relative to the plant. Ectosymbiosis describes a bacterial colony that externally resides on the plant to provide benefits, whereas endosymbiosis signifies that the bacteria exists within the cells or tissues of the plant. Even marine algae, one step further down the evolutionary ladder from plants, have a thoroughly established relationship with symbiotic bacteria.30 However, there is a significant gap in knowledge regarding the small molecule signaling that happens in animal-microbe symbioses. Several studies have qualitatively documented bacteria as being critical to beneficial relationships in animal hosts.31–34 We hypothesize that the qualitative understanding of these symbioses must be connected to established evidence of the underlying chemical interactions to support our claim that small molecules play an important role in establishing and maintaining animal-microbe symbioses (Figure 4).
Figure 4.
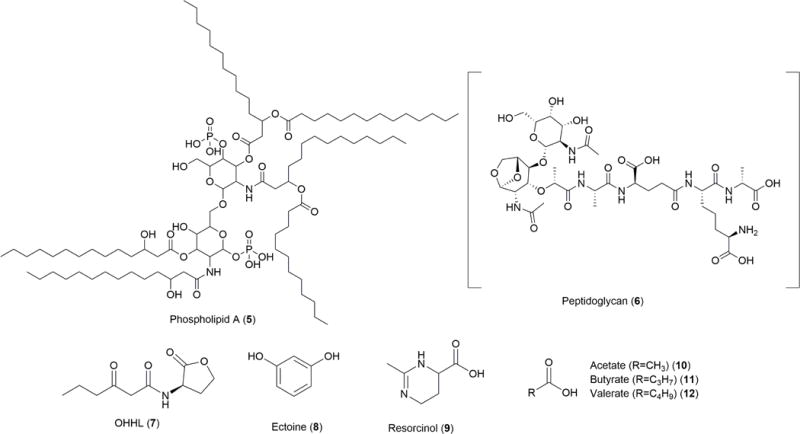
Molecular structures involved in establishment and maintenance of symbiotic relationships between microbes and animals.
Many of the described symbiotic relationships between bacteria and hosts have come from the marine environment. Just as plants benefit from microbial interactions because they have little opportunity for mobility and defense, many marine animals exist in inhospitable environments and require microbial assistance for simple survival strategies. High salinity, high pressure, high temperatures, and a lack of light and carbon create these severe habitats. Many species of mussels (Idas modiolaeformis as well as nearly all of the species in the Bathymodiolus genus) thrive in deep sea vents where the water is sulfur-rich and severely lacking carbon sources.35,36 On their own, the mussels would not be able to perform any of the faculties required for survival, but a population of psychrophilic Gram-negative Colwellia sp. have adapted to the harsh depths to aid the Bathymodiolinae populations. The Colwellia sp. establish a population in the gills of the mussels and perform sulfur oxidation as well as production of methane from the limited carbon directly from the surrounding water.36 Bacterial populations isolated from the mussel gills contained eleven times as much sulfur as any other part of the body, indicating sulfur-oxidizing activity. It is curious why these mussels would establish habitation in these incredibly volatile environments, but their existence there further serves to support the claim of coevolution. The bacterial species adapt to the limited resources and modify them to benefit the host species, while establishing a habitat of their own. The most important chemical reactions here are the production of methane and the oxidation of sulfur, both relatively simple and widespread processes, but extremely crucial to survival in these uncommon symbioses.
In the case of the Hawaiian bobtail squid (Euprymna scolopes), a mucus secreted by the specialized winnowing structure is purported to contain signaling molecules that attract Vibrio fischeri, a bioluminescent bacterium and sole bacterial symbiont to the squid’s light organ. The unique squid is born free of any bacterial symbionts but acquires a thriving population of V. fischeri within hours after hatching. V. fischeri comprises a minute 0.02% of the surrounding bacterial population in the water column, but is the sole symbiont of E. scolopes after selection during a process called winnowing. This bioluminescent bacterium is localized to the epithelial layers of the squid’s light organ; the bacteria aggregate to a population dense enough that through quorum sensing they are able to collectively activate the luxR pathway, inducing the production of light throughout the light organ and into the external environment (Figure 4, Figure 5A). The light provides anti-predatory protection known as counterillumination as well as guidance in the squid’s own predatory efforts. This coevolved interKingdom species-specific symbiosis is crucial to the survival of both species, although little is known of the small molecules that attract the symbionts selectively to one another. Phospholipid A (PLA) (5) and peptidoglycan (PGN) (6) are excreted within hours of the squid hatching and have both been heavily implicated in the process of initial symbiotic bacterial acquisition by E. scolopes (Figure 5B). These molecules are secreted in the mucus of E. scolopes while the winnowing limb-like structures are active and attached to the mantle of the squid. Once a sufficient number of bacterial cells (often four is enough) have been guided into the organism’s light organ, the winnowing arms undergo cleavage from the rest of the body. However, even when the structure has been removed after the initial colonization event, if all of the bacteria is experimentally removed from the organism, the mucus is once again produced and excreted, indicating that the mucus and its components are essential in the acquisition process.37 Is has been proposed that signals from the squid mucus is what attracts the bacterial population to the organism before any epithelial contact is established.38 Aside from 5 and 6 that regulate the winnowing process, we hypothesize that there are many more signaling events that occur in every stage of organismal development. After acquisition of the symbiont, the detachment of the winnowing limbs occurs via an unknown mechanism.38 Although it is a ubiquitous signal and regulates many biological processes, levels of NO were observed to change during the limb detachment process. Nitric oxide synthase, the protein responsible for production of NO, is present in the squid’s internal crypts. This small molecule may be the responsible signal for the maturation step in the squid’s life cycle, but much research remains to be done.
Figure 5.
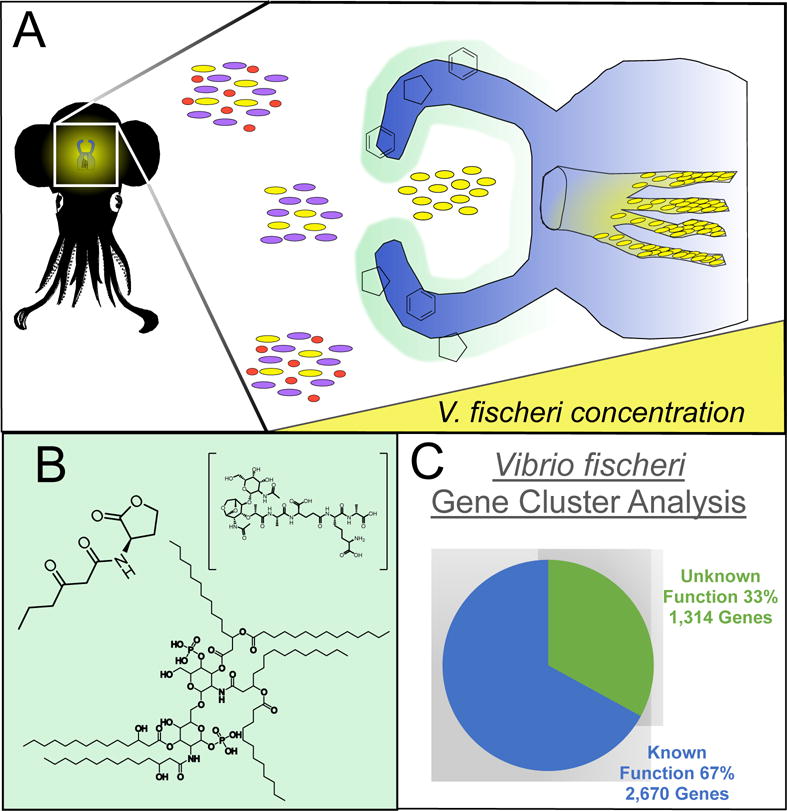
The Euprymna-Vibrio symbiosis serves as a simple system within which to relate genetics to unknown chemistry. A. An illustration of the winnowing process of Euprymna scolopes that selects for Vibrio fischeri. The limb-like winnowing structures on the squid mantle secrete a mucus that attracts the bacterial symbiont into the crypts of the internal light organ. The bacterium makes up 0.02% of the population in the surrounding sea water, but 100% of the population in the crypts of the light organ. B. Peptidoglycan (6) and PLA (5) are two mucosal molecules that attract the bacterial symbiont during acquisition. OHHL (7) is an autoinducer that activates the light-producing luxR pathway inside the light organ to activate counterillumination. C. One-third of the genes in the genome of Vibrio fischeri (Strain ES114, NCBI Reference Sequence NC_006840.2) have no observed or hypothesized function.104 Many of these many be small molecules, or specialized secondary metabolites.
The molecules discussed above are all implicated in the host-microbe acquisition process, but V. fischeri also has unique intraspecies chemical communication metabolites as well. In the introductory remarks it was mentioned that bacterial small molecules play a role in inducing the lux pathway in several host species, and in the Euprymna-Vibrio symbiosis this pathway is also mediated by at least one small molecule, an autoinducer, called N-(3-oxohexanoyl)-L-homoserine lactone (known as OHHL) (Figure 4, 7).39 It is called an autoinducer because having enough of 7 present causes V. fischeri to automatically induce the light producing pathway.40 Characteristic of quorum sensing, many bacterial cells are needed to produce enough of 7 to bind reversibly to the autoinducer receptor on the LuxR proteins. The LuxR-OHHL complex, in turn, binds to the luxICDABEG operon and transcribes the proteins required for light production. The luxAB protein, a luciferase enzyme, results in the production of light. The transcription of this illuminating protein can be regulated by the amount of 7 in the system; during the day the squid eliminates up to 95% of its bacterial symbiont population to prevent the production of high light levels and aid in camouflage as it hides in the sand. This entire orchestration is a magnificent example of quorum sensing and while the role of 7 has been well documented, there are many more small molecules crucial to this initial symbiotic establishment that have not yet been observed (Figure 5C).
Figure 7.
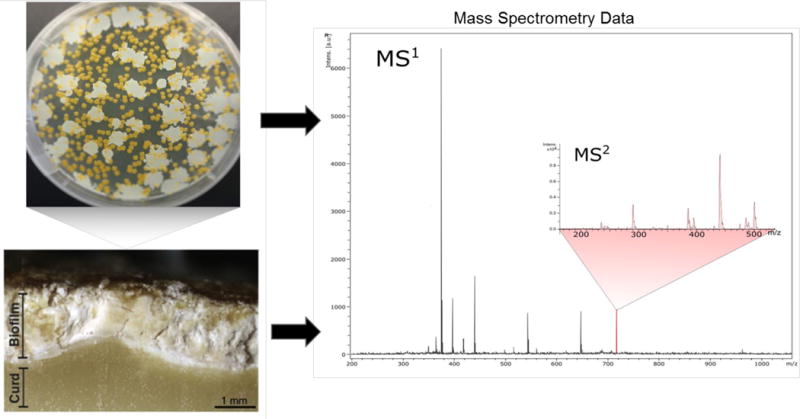
Using mass spectrometry techniques we can identify molecules produced by bacterial communities found in cheese rinds. Microbes are cultured and extracted that are normally found on naturally aged cheese rinds. Following prioritization by mass spectrometry experiments, molecules are purified and further analyzed by mass spectrometry. Tandem mass spec (MS2) of prioritized (MS1) signals reveals the molecular fragmentation for the purpose of identification of novel molecules as well as dereplication. Comparison of laboratory cultures with cheese rinds can be made by directly analyzing cheese rinds with mass spectrometry tools. Cheese rind picture reproduced with permission.69
In an assessment of the putative specialized metabolites produced by V. fischeri, we analyzed the bacterial genome using the Antibiotics and Secondary Metabolite Analysis Shell (AntiSMASH).41 AntiSMASH is an open online workspace that detects gene clusters with similarities to documented gene sequences of secondary metabolites. The program detected three noteworthy gene clusters that display strong homology to three known specialized metabolites: 7, ectoine (8), and an arylpolyene called resorcinol (9).41 We expected to see 7; its presence supports the predicted role of the molecule in the quorum sensing process. Ectoine (8) is a small molecule produced mainly by halophilic bacteria that is implicated in maintaining osmolality in a host and aiding populations attempting to survive in extreme environmental conditions.42 It prevents a host from succumbing to osmotic stress, and many bacterial species perform this balance in plants.43 The properties of 8 are so well known that they are used in many skin care products to maintain moisture in human skin.44,45 McFall-Ngai purports that V. fischeri colonizing the deep crypts of the squid are exposed to a halide peroxidase capable of producing hypohalous acids, halogen containing bactericides.38 It is possible that the bacteria are utilizing the molecules to combat the stressful environment of the mammalian epithelial crypts. Resorcinol (9), also known as 1,3-benzenediol, is recognized medically for its benefits on skin conditions. Most widely regarded is its ability to soften the skin; it is used commercially and therapeutically in products that treat acne, psoriasis, and other epithelial conditions. Although human epidermal epithelia are extremely different from the epithelium of a squid’s crypts, V. fischeri may be producing 9 in response to contact with the internal epithelia in an effort to colonize more efficiently. This is simply one possible hypothesis since the function of 9 in vivo has not yet been elucidated.
While squid epithelia differ from human epithelia, the evolutionary advantage of bacterial symbiosis is well documented in both marine life and mammals. Most topically, the microbiome of humans has been strongly linked to the gastrointestinal health of individuals, providing development of the immune system and often a source of nutrients for the host.23 Research indicates that bacterial communities that reside in the human gut aid in digestive processes.32 (Human interaction with transient or commensal bacteria is discussed in more detail in the following section.) Similar mutualistic relationships have also been observed in the guts of cows,46 giraffes,47 deer, and camels.46 All of these animals are ruminants: mammals that employ the fermentation capabilities of microbes in order to more easily digest plant matter, particularly cellulose. The class of compounds that most effectively help in the absorption of nutrients from plant matter are short-chain fatty acids (SCFA). SCFAs prioritize maintenance of the concentration gradient between the cytosol and the lumen in the ruminal epithelium.48 The concentration gradient allows the SCFAs to cross the epithelium into the gut to provide energy to the animal; without the bacterial species (mainly from the Firmicutes family) to ferment the cellulose and hemicellulose, these nutrient sources would be completely unavailable to gut absorption.47 Acetate (10), butyrate (11) and valerate (12) are all examples of SCFAs32,49 that regulate the absorption of nutrients across the ruminal epithelium.48 Although they serve mainly to facilitate digestion, when the ruminant is oversaturated with introduced carbohydrate material, the number of SCFAs produced by the microbiome is excessive and leads to inflammation.50 While SCFAs are not linked explicitly to the establishment of the symbiosis between the ruminant and the bacterial colonies, their role in the fermentation of carbohydrates results in the production of by-products that warn the ruminants of the dangers of an unhealthy diet. This example illustrates that all animal-microbe interactions lie on a spectrum between advantage and disadvantage. The small molecules utilized by the mammals in the crucial processes of digestion and metabolism are the result of established bacterial interaction within the gut, but the process of establishment still remains unknown.
In all of the described mutualistic relationships, it is clear that there is an evolutionary advantage to these symbioses. The understanding of the chemical and metabolic influence on the establishment and maintenance of these symbioses is severely lacking. A recent encouraging paper highlights the use of IMS to determine the spatial resolution of chemical mass signatures that exist at the interface between a dinoflagellate and a cnidarian species.51 The focus here is not on animal-bacterial interactions, but their approach strongly implies that at least three distinct mass signatures exist where the sea anemone and the dinoflagellates are connected. Their presence is upregulated in comparison to a strain of dinoflagellates that displays no symbiotic inclination, suggesting that the molecules are the results of the interaction between the interKingdom species. Providing further support to our review, the three most distinct chemical signals from the IMS analyses were less than 1000 Daltons and would certainly be considered small molecules. Most of the studies that focus on symbiotic relationships characterize the interaction on a genetic level, with many of the experimental procedures relying on bacterial characterization by 16s RNA sequencing. While understanding the species that are involved in the interaction is a crucial foundation to symbiosis research, the next step must be elucidating the molecules that are involved in developing the specific relationships. With striking evidence that both partners in symbiotic relationships benefit from their mutualism, symbioses have long received attention, albeit mostly from a biological standpoint, from scientists. Only recently have transient animal-microbe relationships garnered their own respective spotlight, as discussed next.
Dysbiosis and Homeostasis
Transient exposure to bacterial species can result in a number of outcomes for a host organism: colonization, infection, or clearance. The outcome between a transient bacterium and a host depends on a number of factors. When transient bacteria colonize the body, they cease to be considered transient and become a normal part of the microbiota, whereas commensals and symbionts are usually a permanent component of the microbiome that contribute to homeostasis. Commensals and symbionts have, however, also been documented to undergo transitional periods. When discussing what happens when transients are encountered, it is helpful to first understand how transitions occur between the microbial categories discussed above. In this section, focus will be on mammalian microbiota and how the host-bacteria interplay can change temporally.
The transition from commensal microbe to pathogen is complex and the underlying biochemical mechanisms can be elusive. It typically only occurs under particular circumstances and those events have been well studied from a genetic approach. One aspect of the commensal-pathogen transition that is lacking data however, is that of the chemical information exchanged. Quorum sensing is one way in which microbes can make the transition from commensal bacteria to pathogen. This mechanism allows microbes to sense adequate growth through production of autoinducers needed to initiate biofilm formation and production of virulence factors. It is a finely tuned mechanism for bacteria to communicate and conserve energy under circumstances where numbers are not high enough to fend off a host immune response. While sufficient growth of bacteria is characteristic of this shift and may seem to be the driving force behind virulence, the process by which this happens is regulated by small molecules. A weakened immune system, an altered microbiota, or breached integumentary barriers are all factors that can allow overgrowth of bacteria and thus quorum sensing initiation of virulent gene expression. For example, the common skin bacteria P. aeruginosa is normally harmless but becomes a serious health threat when the skin barrier is breached, the most common case being burn victims. Perhaps understanding the mechanisms underlying a transition from commensal to pathogenic bacteria requires an understanding of how the small molecule landscape changes under these compromised situations.
Quorum sensing studies demonstrate that the gene expression of virulence factors occurs upon reaching a certain titer of bacterial cells. For bacterial growth to reach that titer they must pass the lag phase of growth and sometimes the sequence of exposure of the host to different strains matters greatly. In other words, the early bird gets the worm; those strains that can colonize and utilize nutrients first often establish dominance. Even when environmental factors are identical, the order of species colonization of a particular niche can result in divergence of communities.52 Sometimes physical barriers to surface adhesion can explain this phenomenon. Leathem et al. demonstrated that in a murine gut model, commensal E. coli spp. prevent subsequently encountered E. coli spp. (in some cases pathogenic E. coli) from colonizing the gut.53 This is partially explained by limited nutrients preventing a second species from passing the lag phase in growth and partially by adhesion of the initial colony to the intestinal wall. This study used subspecies of E. coli that utilize different sugars for metabolism and showed that mice colonized by an E. coli species were resistant to infection from that same E. coli spp. but not from different subspecies. This study provides evidence supporting two different mechanisms: physical attachment to the intestinal wall provides advantage as does nutritional utilization. Medellin-Pena and Griffiths additionally demonstrated in vivo that probiotic strains can release molecules that prevent enterohaemorrhagic E. coli (EHEC) adherence to epithelial cells.54 Presumably by producing small bioactive molecules, these probiotics can interfere with pathogenic quorum sensing and adherence.
There are other possibilities for molecular modulation of symbiont/pathogen interactions (a point that is also acknowledged by the authors), some of which are evidenced in other situations (Figure 6). For example, Mazmanian et al. investigated the factors that lead to Helicobacter hepaticus-induced development of human gut disorders such as colitis and inflammatory bowel disease (IBD).55 When this opportunistic pathogen and the symbiont Bacteroides fragilis are co-colonized, hosts are protected significantly from disease. Host protection occurs because B. fragilis produces polysaccharide A (PSA, 13), a molecule consisting of hundreds of repeating tetrasaccharide units, that possesses various immunomodulatory effects. PSA (13) is a special bacterial polysaccharide because the repeating units contain both a positively charged amino group and a negatively charged carboxyl group. The unique electrostatic nature of 13 allows direct interaction with both CD4+T cells and B cells, eliciting a multifaceted immune response to 13 exposure.56 Further support of this is provided in this study in that B. fragilis strains that did not produce 13 failed to protect mammalian hosts against the inflammation caused by H. hepaticus and subsequent inflammatory bowel disease and colitis. Interestingly, it was not by clearance of H. hepaticus that B. fragilis protected its host but simply by producing a molecule to counteract the negative effects of H. hepaticus. This study successfully traced the beneficial effects of a symbiotic bacterium back to a bioactive molecule (13).
Figure 6.
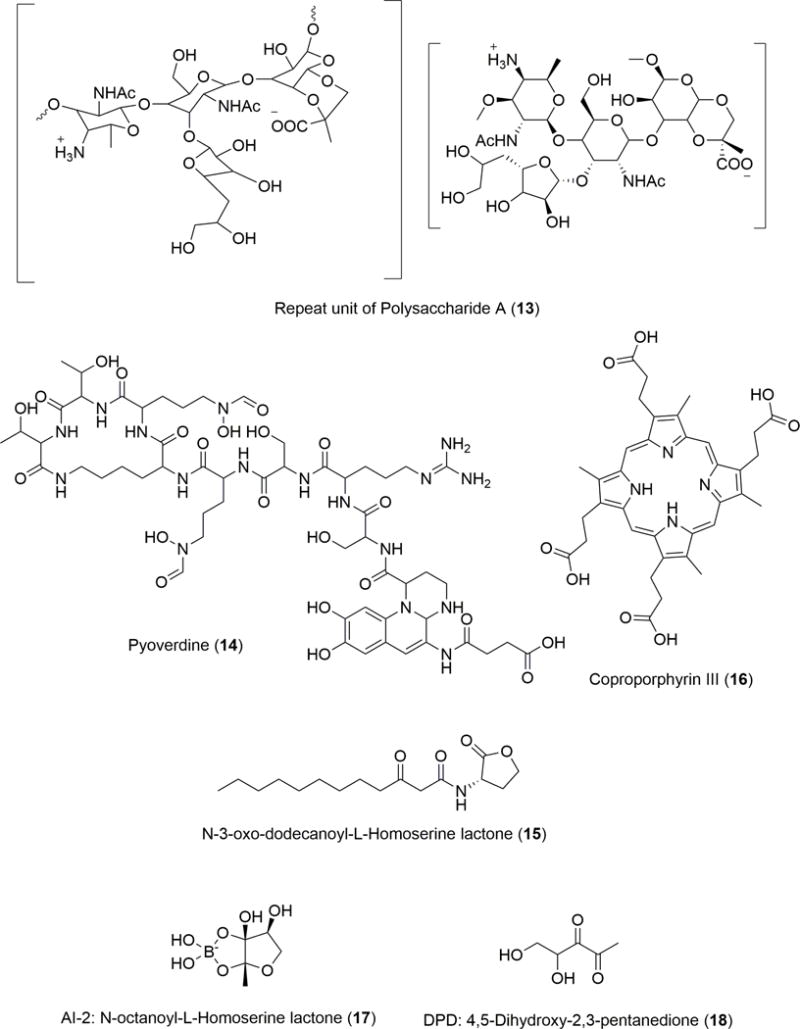
Molecules transiently involved in human and bacterial interactions
Competition for iron, production of antimicrobial compounds, and environmental modification are all factors that influence interactions between commensal and transient bacterial strains. One of the limiting factors for bacterial growth is the amount of iron available in the environment. Particularly in a host system where molecules such as lactoferrin and transferrin regulate the availability of free iron, the concentration of iron is well below optimal levels for bacterial growth. In order to cope with this, bacteria produce siderophores that chelate iron and transport it into the cells. Upon a host encountering a pathogen, a competition for iron ensues. Jimenez et al. studied the small molecules produced by Pseudomonas sp. under iron limiting conditions and identified a gene (pvdQ) that is located within the pyoverdine (Figure 6, 14) locus and encodes an acylase PvdQ. Its location in the pvdQ locus suggests that it is also involved in production of the siderophore 14 which is known to have a high affinity for iron.57 The exact role of pvdQ in the biosynthesis of 14 is unclear, but it is a necessary component as evidenced by the fact that deletion strains do not produce 14. Pyoverdine (14) itself consists of a fluorescent chromophore linked to a peptide moiety. The peptide moiety exists in different forms, creating more than 40 different structural variants of 14.58 The peptidic portion of 14 imparts specificity for a particular Pseudomonas spp., usually the producing strain, thus preventing other bacterial species from being able to uptake iron-bound 14 that were originally produced by a Pseudomonas spp. Perhaps most interesting about the activation of this gene is the accompanied activation of virulence factors. PvdQ also increased motility, controlled 14 production and enhanced biofilm formation. Another known function of PvdQ is as a quorum quencher. As a member of the N-terminal nucleophile hydrolases and a homolog to a β-lactam acylase, PvdQ possesses the ability to degrade Pseudomonas’ own quorum sensing molecule, 3-oxo-C12-HSL (15). Although 15 bears great similarity to 1, presence of the additional ketone on the acyl tail distinguishes them. The small chemical difference between 1 and 15 highlights the importance of the specificity of these molecules. Although they are both homoserine lactones, 1 activates the lux pathway and 15 activates the las pathway. However, this study showed that quorum quenching by PvdQ is only noticeable when the pvdQ gene is expressed under iron-limiting conditions and 15 production increases.57 The ability of an organism to switch on and off virulence factors is key in describing the switch from commensal to pathogen in P. aeruginosa. The signal for these transitions comes from nutrient limitation and results in production and degradation of various small molecules.
In some other cases, the signal can come from commensal microbes as well.59,60 For example, the commensal Propionibacterium spp. produce porphyrins that can sometimes act as metal chelators and can also serve as inflammatory markers.61 Increased porphyrin production in some strains over others are often associated with increased risk of acne or Staphylococcus aureus growth. Typically health-associated Propionibacterium strains produce less porphyrins than those associated with acne, suggesting that the ultimate difference between a “healthy” vs. “disease-inducing” bacteria lies in the metabolites produced.61 Likewise, Wollenberg et al. showed that coproporphyrin III (16) produced by Propionibacterium acnes in the nasal cavity induce S. aureus virulence by enhancing S. aureus aggregation and biofilm formation.59 Together with the knowledge that certain strains of Propionibacterium spp. produce higher levels of porphyrins, perhaps future studies could examine strain-specific interactions to see if all Propionibacterium strains induce the same level of S. aureus virulence. This draws into question the efficacy of using broad spectrum antibiotics to treat such disease states. If the disruption of normal microbiota is significant enough upon treatment with antibiotics, this could allow overgrowth of opportunistic pathogens. A study done by Thompson et al. investigated the effects of introducing an interspecies autoinducer (AI-2, 17)-producing E. coli into streptomycin-treated gut microbiota.62 The study initially found that streptomycin treatment greatly decreased normal host community diversity, favoring Bacteroidetes phyla over Firmicutes. This decreased diversity is usually associated with lower pathogenic resistance and thus seen as an undesirable outcome of antibiotic treatment. To address this concern, the authors also wanted to investigate how 17 might be used by commensal bacteria to maintain homeostasis. AI-2 refers to a class of furanosyl borate diesters (17) derived from cyclization of 4,5-dihydroxy-2,3-pentanedione (DPD, 18).63 The DPD precursor (18) can result in different structures or stereoisomers of 17 depending on the species producing it, giving the AI-2 class some level of specificity despite being widely produced and recognized by many different bacterial phyla. Overall, however, 17 is considered to mediate interspecies microbial chemical communication because of the ubiquity of the 18 synthase. By constructing E. coli mutants that either accumulated or degraded extracellular 17 and introducing them into streptomycin-treated mice they were able to reveal that increasing the levels of 17 in vivo resulted in partial restoration of diversity due to selective increase of Firmicutes. The density of various mutant strains of E. coli that were used to alter availability of 17 interestingly did not respond to 17 level variations, indicating that this model could be used to study the effects of 17 on community composition without interference from the effects of E. coli overgrowth. In light of such studies, perhaps future treatment for bacterial diseases should be focused on exploiting the difference between nutrient requirements and chemical communication pathways of disease-associated microbes and health-associated strains in order to promote or restore a beneficial community balance.
It is clear that some strains of bacteria confer health benefits to their host. If it is desirable to allow these strains to colonize, it is important to understand how transient exposure to these strains results in permanent colonization. One of the first experiences a human has with transient exposure to bacteria is during gestation. Many studies have addressed the ways that microbial composition are inherited from the mother, and it is well established that immune factors are influenced by neonatal microbiota.64 However, few studies have looked at the molecular mechanisms governing how inheritance affects the fetal immune system. In a recent paper, Gomez de Agüero et al. describe how transient infection with E. coli in pregnant mothers can produce the fetal innate immune composition normally associated with inherited gut microbiota in germ-free mouse pups.65 This information transfer occurs through production of bacterial molecules, possibly short-chain fatty acids, that can act as ligands for the aryl hydrocarbon receptor (AhR) and are passed from mother to fetus through the placenta prenatally and postnatally through milk. This was one of the identified pathways but the AhR receptor is only one aspect of immune development and it is possible that there are more microbial molecules involved in shaping immunity. It is interesting to note that a temporary bacterial colonization produced lasting effects upon innate immunity. This is suggestive that the metabolites produced by these bacteria rather than the bacteria themselves are responsible for immune regulation.
During an organism’s lifespan, many different microbes are presented to it and can be introduced in a variety of ways. One of the main methods of exposure is through ingestion. A number of studies show that diet can alter the composition of the microbiome, indicating the importance of understanding how this process occurs. In some cases this has been shown to have lasting effects upon the microbial composition.66 There are instances in which certain strains of bacteria are desirable for colonization due to the immune and nutritional benefits they provide to the host.54,67 A recent study done by David et al. highlighted the different bacterial flora patterns seen in subjects with divergent diets.66 The study showed that dietary exposure can transiently alter the composition of the gut microbiota. This in turn will alter the chemical landscape created by the microbiota. If diet changes the microbiome perhaps studying the chemistry of food microbes could help determine what a “healthy” diet is and what role the small molecules play in this relationship. Studies done on the effect of probiotic strains could help shed light on this. For example, antibiotic treatment eliminating Clostridia spp. (normal microbiota) promotes Salmonella spp. growth.68 The reason for this is the change in metabolites available when Clostridia spp. are depleted. Clostridia spp. (a normal part of the human gut flora) normally produce butyrate (11) that is oxidized by colonocytes to produce carbon dioxide (CO2), creating a hypoxic luminal environment. When 11 is not available to colonocytes, they obtain energy by fermenting glucose to lactose which allows oxygen to accumulate where it would otherwise be used in the conversion to CO2. This shift in metabolism creates an aerobic environment in which Salmonella can flourish. Other studies also support the idea that commensal gut microbes confer host protection by production of small molecules.61 This points to the importance of restoration of normal microbiota after disruptions such as antibiotic treatment.
Our lab focuses on communication between microbiota with a focus on how bacteria use chemistry to communicate. Part of the studies involve investigation of the molecules produced by different types of food microbiota (specifically those found on cheese).69 In order to deduce the effect that these microbes have, we must first pull apart the underlying molecular interactions. By using different mass spectrometry techniques along with NMR we can identify molecules produced in response to the presence of other species and attempt to determine the effect that those molecules have upon the environment (Figure 7).
First, the use of MALDI-TOF IMS helps to visualize the spatial distribution of molecular signals for prioritization. Then, extraction of biomolecules and separation via HPLC with subsequent LC-MS/MS and NMR analysis can definitively identify the molecules responsible for events such as bacterial inhibition and interKingdom interactions. By putting together a picture of the molecular interactions, we hope to understand the mechanisms behind how bacterial species and hosts affect each other.
Analytical Techniques
In order to access the molecules discussed in this review and those that have yet to be characterized, modern analytical chemistry techniques are required. We believe it is important to study the chemistry using in situ or in vivo techniques whenever possible, but we acknowledge that this is still a current technological limitation. Since not all studies can be completed in vivo, many have turned to chemical engineering to create an authentic in vitro environment which emulates the characteristics of the GI tract or allows for natural exposure of bacteria to occur. Reviews covering and literature describing in vitro techniques such as “bioproduction breadboard” for studying cell-cell communication and microfluidic flow cell devices are cited here for those looking for in vitro techniques.70,71 Since our review focuses on in vivo chemical communication we provide a brief review of the analytical techniques that have been used to study the molecules discussed in this review and other analytical techniques such as NMR, MS, atomic resolution microscopy, spatial light interference microscopy, colorimetric and fluorometric methods, and UV-Vis spectra. The advantages, disadvantages, and sample preparation of each method is discussed in each section.
NMR
One method for studying compounds in complex backgrounds is the use of 1D and 2D NMR experiments. NMR has remained a gold standard for identifying and elucidating chemical structures from biological backgrounds because the rules for interpreting chemical shifts have been well established and certain molecule classes give characteristic NMR profiles. Additionally the use of microprobes, microcoils, and cryoprobes has dramatically increased the sensitivity of the instruments allowing for nanomolar scale structure elucidations.72 The range of 2D experiments have also expanded in recent years. Of note, Schroeder and coworkers frequently employ differential analyses by 2D NMR spectroscopy (DANS) in order to interrogate complex samples without the need to pursue complex chemical isolations from media or organisms.73,74 However, some of the drawbacks inherent to NMR are sample quantity requirements, limited spin-active nuclei, and a lack of public database repositories.75 In general, NMR has not been used to profile chemistry directly in or on a host, but rather requires chemical extractions in order to introduce the sample into the instrument. However, NMR for biofluids has proven successful especially when 2D experiments are employed such as the dqfCOSY. In an elegant example from Meisel et al., the authors were able to identify specific specialized metabolites produced by Pseudomonas aeruginosa that serve to promote avoidance by the animal hosts.76 In other complex biofluids we recommend the recent review by van Duynhoven and Jacobs.77
Mass Spectrometry
Mass spectrometry is an ideal method when trying to identify specialized metabolites from complex biological sources. The advances in sensitivity, ease of use, and ionization methods has greatly increased the utility for in situ analyses. It’s important to note that all mass spectrometry methods are limited by a specific molecule’s ability to ionize. Typically, in dealing with microbial communication, volatile compounds are excluded from the analyses discussed below. A number of LC-MS/MS methods have been recently described, but these workflows tend to lose spatial correlations with a host or are very low resolution (millimeters via cotton swabbing a surface).78,79 However a handful of methods have shown great promise for detecting chemistry in a host that maintain the integrity of the host tissue. These methods can be categorized by ionization being either ambient or in vacuo.
Recently, Takats and coworkers have worked to adapt rapid evaporative ionization mass spectrometry (REIMS) to be compatible with surgical tools to develop iKnife. This intelligent knife (iKnife) utilizes the smoke released by electrosurgical tools to capture chemical profiles from tissues as the surgeon is operating.80 The release of smoke is essentially rapid ionization, similar to but distinct from electrospray ionization. A variety of small molecule structural classes have been observed with this technique. In cancerous tissue applications, the lipid profiles have been shown to differentiate the tissue margins, and in bacterial colonies on agar, molecules such as surfactin, pseudomonas quorum signal (PQS), and phosphatidyl-glycerol were all observed.81 While this method has yet to be applied to the identification of bacterial communication signals in a host, it has great promise because it is easily incorporated in surgical tools. In another example of ambient ionization tools, Mascuch et al. utilized a fluorescence microscope interfaced with a nano desorption electrospray ionization (nanoDESI) in order to directly observe the causative agent of white-nosed syndrome, Pseudogymnoascus destructans, on infected Myotis lucifugus (little brown bat) wings. In this paper, the authors were able to directly sample areas with fluorescent lesions (indicative of fungal infection) to demonstrate that the WNS bats present high-affinity siderophore production (ferrichrome and triacetylfusarinine C) at the lesion sites.82 Lastly, Cooks and coworkers have developed a variety of ambient ionization techniques coupled with field-ready mini mass spectrometers.83 In pursuing the detection of microbial signals from a host, pure cultures of Streptococcus pyogenes, the causative agent of strep throat, were compared to patient throat swabs, and introduced via touch spray ionization to reveal overlapping lipid signatures.84 While these ambient ionization techniques hold great promise, so far they have been limited to detecting microbes where they are known to exist. The methods have yet to be extended to study the chemistry bacteria utilize to colonize surfaces, and we believe that ambient techniques fail to capture chemistry at the spatial level bacteria occupy (submicron levels).
Matrix-assisted laser desorption/ionization (MALDI) methods offer finer spatial resolution at the expense of ease of sample preparation. Ambient ionization techniques are ideal because the chemistry can be directly detected with minimal sample preparation. With MALDI techniques, as the name infers, matrix must be applied to the sample in order to achieve ionization. The matrices for MALDI are typically small, aromatic acids such as 2,5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA). The choice of matrix can greatly influence the type of molecule that is ionized as well as the matrix application method and was recently reviewed and discussed by Palmer and Alexandrov.85 Currently the spatial resolution achieved in a MALDI experiment is reliant upon the ability to focus the laser, the stage movement mechanics, and size of the co-crystallized matrix and analyte. Newer commercially available instruments have the capacity to focus the laser to five micron resolution and in an in-house modified instrument, sub-micron spatial resolution has been achieved.86,87 Aside from these technical advances, MALDI allows for high spatial resolution imaging mass spectrometry (IMS) compared to ambient techniques such as nanoDESI which are spatially limited by the size of the constant solvent droplet between the fused capillaries. This allows for users to interface a number of images to create a comprehensive view of a system. For instance, one could imagine co-registering an optical image, confocal image, and a molecular image to view a GFP tagged microbe in a cryosectioned tissue or host. This allows for a more discreet and thorough analysis of the microbe and its chemistry in relation to the host. Given that microscopy is still superior from a spatial consideration to mass spectrometry, MALDI allows for the two to be interfaced most efficiently, but microscopy lacks in its ability to identify unknown molecules.
Atomic Resolution Microscopy
One of the newer ways to investigate bacterial molecules is through atomic-resolution microscopy techniques. Techniques such as atomic force microscopy (AFM) and energy-dispersive X-ray spectroscopy (EDS, EDX, or XEDS) allow molecular characterization through imaging rather than through analytical chemistry. EDX along with either transmission electron microscopy (TEM) or scanning electron microscopy (SEM) provides information about the chemical composition of surfaces. Since this method provides analysis of the elements present in a sample, it could be used to detect nutrient utilization and spatial distribution of metabolic byproducts and waste. Biological samples for this technique are typically prepared by freeze-drying and sputter coating with gold, allowing users to obtain a snapshot of elemental localization within a cell or microenvironment. EDX with SEM has been used in some instances to detect the presence of sulfur reducing bacteria (SRB)88 and EDS with TEM can specifically determine the effect that metals can have upon SRB by visualizing the spatial distribution of metal precipitates.88 The ability to detect accumulation and detoxification of nutrients (iron availability, sulfur oxidation) could also provide insight into how various bacterial pathogens are able to colonize their hosts.89 However, this method provides little information about specialized metabolites, and would only be appropriate for studies focused on the distribution of elements and micronutrients.
Another atomic resolution technique, AFM, has been used to evaluate the mechanistic properties of bacteria and the environmental and antibacterial factors that influence those properties.90 While many of the models used in interpretation of AFM data need to be improved and adapted to use for biological cells, the technique provides a baseline for cellular mechanics. The scale at which AFM is performed allows single cell analysis as well as biofilm analysis at different stages of development. For example, the ability to detect changes in cell wall elasticity or adhesion to biological substrates could shed light on the mechanisms behind various antibiotics and biofilm formation.91 AFM can also quantify hydrophobic and glycopolymer interactions, as well as detect fluctuations in metabolism.92 For example, Longo et al. used AFM technology to evaluate the response of E. coli and S. aureus when treated with antibiotics. Their study showed that treatment with different concentrations of antibiotic resulted in varying mechanical fluctuations, and hypothesized that these fluctuations were due to metabolic changes. This hypothesis was supported by the fact that metabolic fluctuations could be detected by cantilever deflection. This demonstrates the ability of AFM to provide metabolic information about a living system. Another advantage that this method provides over other analytical techniques is the ability to evaluate liquid cultures with little to no preparation, thereby more closely mimicking natural environments. However, as previously stated atomic resolution techniques should be used to evaluate molecular interactions as opposed to molecular identifications.
Spatial light interference microscopy (SLIM)
Microscopy is a very well known and practiced technique which can be used for visualizing cell interactions in vitro and in vivo. SLIM is a fairly new optical microscopy technique that is capable of measuring nanoscale structures and dynamics in live cells over time.93 Therefore cells, and their corresponding interactions, can be monitored in real time through several cell cycles.93 Combining two types of light imaging, SLIM is able to give topographic accuracy that is comparable to that of atomic force microscopy and with an acquisition speed that is 1,000 times faster.93 One study has used this valuable technique to determine the growth patterns of a single E. coli cell in order to increase knowledge of cell cycle-dependent growth.94 SLIM allows for the visualization of cell-cell interactions in real time which can be very beneficial when trying to analyze the molecular interactions of bacteria cells and visualize the movement of small molecules. Although like all microscopy techniques, it does not aid in the identification of unknown molecules.
Colorimetric and fluorometric techniques
Light and color based tests are the main methods of nitric oxide (NO) detection in biological samples. The Griess Reaction indirectly detects the products NO3− and NO2−, two stable molecules that are produced when NO decomposes. The product of this reaction is blue when there is NO present.95 The second most common means of identifying NO in a biological sample is a fluorometric approach that exploits the production of fluorescent compounds for facile detection. While the Griess reaction can be carried out in biologically relevant conditions, diaminonaphthalene (DAN) and diaminofluoroscein-2 (DAF-2) assays may be performed in vitro and in situ. The interaction of NO with DAF-2 results in a triazole, a fluorescent compound that is the product of an internal rearrangement of the original nitrosamine product. Fluorometric detection is the most commonly used method of NO detection because the instrumentation used for fluorescent imaging can be easily adapted to work in conjugation with tissue imaging systems. Sample preparation can prove difficult if the molecules being detected are not inherently fluorescent. In these cases a fluorescent tag must be bound.
Although the two aforementioned methods have been established as dependable and accurate detection techniques, researchers have developed tools that are more easily adapted to assess biological pathways. For example, for detection of NO in E. scolopes, Ruby et al. measured the binding affinity of the historically named heme NO/oxygen-binding protein (H-NOX) to NO (theoretical evaluations have ruled out the protein’s possible affinity of O2 binding). Ruby measured the binding affinity of H-NOX in E. scolopes, which provided a direct correlation to the amount of NO in the system.96
UV-Vis Spectrum
Fluorometric and colorimetric analyses produce an output that can be directly visualized, whereas the detection of ultraviolet (UV) rays requires specialized equipment. The UV wavelength range is much lower than visible light, and is not detectable by the unaided human eye. As a comparison, the theory behind fluorescence claims that the visible fluorescing cue is produced by electrons in a sample moving from a higher energy state to a ground state, emitting fluorescence as it is lowered. An ultraviolet-visible (UV-vis) spectrometer uses a similar but opposite calculation: when electrons in a sample are exposed to UV rays they jump from their resting ground state to an excited state as they absorb the energy from varying UV wavelengths, therefore absorbance is measured. The instruments are also capable of producing wavelengths in the visible light range, as the structure of compounds vary so much that they absorb wavelengths in both ranges. Sample preparation for this technique is extremely easy and simply requires the dissolution of a sample in a compatible solution, often only at 1 mg/mL. Once the compound has been exposed to the preselected discrete UV wavelengths, their electrons will absorb the wavelengths that correspond to their electrons’ excitability, and a spectrum of peaks will be produced, mapping absorbance versus time.97 While this technique provides extensive information for structural determination of molecules, only ever can this application be used in vitro. A UV-vis spectrum can aid in structure elucidation of an unknown compound because the reactions of the electrons in the structure can be correlated to known structural features. Certain functional groups such as and double and triple bonds, respond to particular wavelengths of light because all of the electrons in these characteristic structures have different levels of ground state energy, which leaves a unique fingerprint in the output spectrum. For example, UV-vis spectroscopy was used to determine a new class of compounds from a Streptomyces sp. These compounds, the iromycins, were found to inhibit nitric oxide synthases (NOS), a critical protein implicated in NO signaling in bacteria.98
Raman Spectroscopy
Rayleigh and Raman scattering are the two types of light scattering that allow perception of vibrant and bold colors. Rayleigh scattering describes how light scatters when it encounters molecules in a medium. The interaction that occurs between a photon and the molecules in the medium can lead to a gain or loss in energy. This causes a shift in frequency of the photons and leads to another type of scattering referred to as Raman scattering. Light scattering depends on the polarizability and stretching of the molecules in the medium. Thus vibrational energy plays a large role in Raman light scattering. Raman spectroscopy applies these theories to collect data from gas samples and usually requires no sample preparation making it compatible for in vivo gas phase studies. Raman spectroscopy is used in clinical settings to detect atherosclerosis, cancer, and inflammatory diseases.99 Another popular use of Raman spectroscopy is for bacterial identification.100–102 One study used surface-enhanced Raman spectra (SERS) to work on metabolomics profiling of ten different bacterial species. SERS spectra of bacteria are created from a mix of vibrational modes of cell wall components.99 The components being detected include peptidoglycans, lipids, lipopolysaccharides, membrane proteins, and nucleic acids. If molecules with low molecular weights such as nucleic acids can be detected through vibrational modes, then it is possible this technique could be applied to identify other small molecules in bacteria. For a summary of studies using Raman spectroscopy for bacterial identification we recommend a review by Pahlow et al.103 Advantage of Raman spectroscopy lies in the ability to identify molecules due to their specific light scattering fingerprint in a non-destructive manner. Unlike mass spectrometry the sample is not destroyed in the process of collecting data therefore this represents an ideal alternative. This also allows for multiple, consecutive analyses without any damage that could alter the chemistry. Raman is not an ideal technique for structure elucidation since Raman is similar to IR spectroscopy, and if the sample being analyzed is a mixture of molecules the spectra output would be too complex to deconvolute without computational tools such as the 2D experiments discussed in the NMR section.
Future Directions
Now that we have truly begun to catalog bacteria and their presence or absence in different microenvironments we must begin to test function in order to continue mapping the complex relationships bacteria have with their hosts. We believe that chemistry represents the functional endpoint for both genes and proteins. Therefore, by investing discovery efforts to understand these small organic communication molecules we will be able to observe how ubiquitous or specific these chemistries are to different systems. Connecting disparate systems through small molecules (chemistry) will lead to fundamental biological processes and reinvigorate studies in the molecular mechanisms that produce these signalling molecules. In the age of the microbiome, the chemistry should be considered as important as the metagenomics and genomics, intimately connected to host health.
While not all bacteria fall under only one category, we define here what is meant by the use of each term in this review:
Pathogen - a microorganism that has historically been associated with causing disease.
Symbiont - one of two organisms that live in symbiosis with each other, a mutually beneficial relationship.
Commensal - relationship in which one organism benefits and the other is not affected in any way. Human flora are often thought of as commensals but may be more accurately seen as symbionts as evidence points out positive influences of the microbiota.
Transient - microorganisms that are encountered but are not a normal part of the resident microbiota. May have difficulty colonizing a host or only be capable of temporary colonization.
Probiotic - microorganism that is considered to confer health benefits to a host. Usually used to denote an ingested microorganism.
Opportunistic - a microorganism that is capable of causing disease only under unusual circumstances, such as an impaired immune system.
Chemical Communication - information transfer among microbes via molecules
Quorum Sensing - a specific subtype of chemical communication that uses autoinducers to activate LuxR superfamily gene pathway
The Bigger Picture.
The explosion of microbiome studies surveying members of various microbes across environments and hosts has led to an increase in unanswered questions from both the scientific and public communities. What leads to dysbiosis in a host? How stable are microbial communities across or within individuals? Does a host’s innate microbiome contribute to or confer immunity to the host when challenged by bacterial pathogens? Microbiome studies have laid a foundation of support for bacteria as major players in host health, but the exact mechanisms by which the bacteria influence or protect hosts represent a major gap in knowledge. It has been postulated that bacteria produce unique chemical entities for exquisitely specific purposes and we have only touched the tip of the iceberg as to uncovering the specific ecological function of these specialized metabolites. We believe the small molecule chemistry will be imperative in laying foundation to studying host health and disease.
Summary.
In the age of the microbiome, it is clear that bacteria play essential dual roles in maintaining host health and contributing to host illness. Our fundamental understanding of how a host and bacterium communicate to cause vastly different system-wide outcomes is lacking. This review seeks to document what work has been done in vivo to characterize chemical communication in three very different types of interactions: pathogenic, symbiotic, and transient. Largely, we find that many systems point to a chemical interplay, yet these molecules have remained uncharacterized. In instances when the chemistry is known or elucidated, important host health implications can be hypothesized and tested. We are hopeful that this review will prompt others to catalog the chemistry that microbes produce to communicate with their hosts.
Acknowledgments
ARC is supported by an NSF Illinois Louis Stokes Alliance for Minority Participation Bridges to the Doctorate Fellowship. This publication was made possible by UIC Startup funds and Grant Number K12HD055892 from the National Institute of Child Health and Human Development (NICHD) and the National Institutes of Health Office of Research on Women’s Health (ORWH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. 2016;79(3):629–61. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Hawver LA, Jung SA, Ng WL. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev. 2016;40(5):738–752. doi: 10.1093/femsre/fuw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195(7):1371–80. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez ME, Newman DK. Extracellular electron transfer. Cell Mol Life Sci. 2001;58(11):1562–71. doi: 10.1007/PL00000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies J. Specialized microbial metabolites: functions and origins. J Antibiot. 2013;66(7):361–4. doi: 10.1038/ja.2013.61. [DOI] [PubMed] [Google Scholar]
- 7.Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun. 2007;75(9):4597–607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zargar A, Quan DN, Carter KK, Guo M, Sintim HO, Payne GF, et al. Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. MBio. 2015;6(2):e00025. doi: 10.1128/mBio.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ismail AS, Valastyan JS, Bassler BL. A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe. 2016;19(4):470–80. doi: 10.1016/j.chom.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176(2):269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sviridova T, Deryabin D, Cyganok O, Chereshnev V. Cytotoxicity of N-dodecanoyl-L-homoserine lactone and 5-N-dodecyl resorcinol to human granulocytes and monocytes: a comparative study. Centr Eur J Immunol. 2013;38(3):310–316. [Google Scholar]
- 12.Dingemans J, Monsieurs P, Yu SH, Crabbé A, Förstner KU, Malfroot A, et al. Effect of Shear Stress on Pseudomonas aeruginosa Isolated from the Cystic Fibrosis Lung. MBio. 2016;7(4):e00813–16. doi: 10.1128/mBio.00813-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allewell NM. Introduction to Biofilms Thematic Minireview Series. J Biol Chem. 2016;291(24):12527–8. doi: 10.1074/jbc.R116.734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anwar H, Costerton JW. Enhanced activity of combination of tobramycin and piperacillin for eradication of sessile biofilm cells of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34(9):1666–71. doi: 10.1128/aac.34.9.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moskowitz SM, Foster JM, Emerson J, Burns JL. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol. 2004;42(5):1915–22. doi: 10.1128/JCM.42.5.1915-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjarnsholt T, Kirketerp-Møller K, Kristiansen S, Phipps R, Nielsen AK, Jensen PØ, et al. Silver against Pseudomonas aeruginosa biofilms. APMIS. 2007;115(8):921–8. doi: 10.1111/j.1600-0463.2007.apm_646.x. [DOI] [PubMed] [Google Scholar]
- 17.Antunes LCM, Wang M, Andersen SK, Ferreira RBR, Kappelhoff R, Han J, et al. Repression of Salmonella enterica phoP expression by small molecules from physiological bile. J Bacteriol. 2012;194(9):2286–96. doi: 10.1128/JB.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderhelst E, De Wachter E, Willekens J, Piérard D, Vincken W, Malfroot A. Eradication of chronic methicillin-resistant Staphylococcus aureus infection in cystic fibrosis patients. An observational prospective cohort study of 11 patients. J Cyst Fibros. 2013;12(6):662–6. doi: 10.1016/j.jcf.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Carey RM, Workman AD, Chen B, Adappa ND, Palmer JN, Kennedy DW, et al. Staphylococcus aureus triggers nitric oxide production in human upper airway epithelium. Int Forum Allergy Rhinol. 2015;5(9):808–13. doi: 10.1002/alr.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535(7613):511–6. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 21.Wiles TJ, Jemielita M, Baker RP, Schlomann BH, Logan SL, Ganz J, et al. Host Gut Motility Promotes Competitive Exclusion within a Model Intestinal Microbiota. PLoS Biol. 2016;14(7):e1002517. doi: 10.1371/journal.pbio.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. Antibiotic resistance. 2016 Sep 2; [cited 2016 Oct 21]; Available from: http://www.who.int/mediacentre/factsheets/antibiotic-resistance/en/
- 23.Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci USA. 2011;108(1):4645–52. doi: 10.1073/pnas.1000099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goettler W, Kaltenpoth M, Herzner G, Strohm E. Morphology and ultrastructure of a bacteria cultivation organ: the antennal glands of female European beewolves, Philanthus triangulum (Hymenoptera, Crabronidae) Arthropod Struct Dev. 2007;36(1):1–9. doi: 10.1016/j.asd.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Rae RG, Tourna M, Wilson MJ. The slug parasitic nematode Phasmarhabditis hermaphrodita associates with complex and variable bacterial assemblages that do not affect its virulence. J Invertebr Pathol. 2010;104(3):222–6. doi: 10.1016/j.jip.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Su Q, Zhou X, Zhang Y. Symbiont-mediated functions in insect hosts. Commun Integr Biol. 2013;6(3):e23804. doi: 10.4161/cib.23804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari J, Vavre F. Bacterial symbionts in insects or the story of communities affecting communities. Philos Trans R Soc Lond B Biol Sci. 2011;366(1569):1389–400. doi: 10.1098/rstb.2010.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaves S, Neto M, Tenreiro R. Insect-symbiont systems: from complex relationships to biotechnological applications. Biotechnol J. 2009;4(12):1753–65. doi: 10.1002/biot.200800237. [DOI] [PubMed] [Google Scholar]
- 29.Gunatilaka AAL. Natural products from plant-associated microorganisms: distribution, structural diversity, bioactivity, and implications of their occurrence. J Nat Prod. 2006;69(3):509–26. doi: 10.1021/np058128n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seyedsayamdost MR, Carr G, Kolter R, Clardy J. Roseobacticides: small molecule modulators of an algal-bacterial symbiosis. J Am Chem Soc. 2011;133(45):18343–9. doi: 10.1021/ja207172s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frese SA, Mackenzie DA, Peterson DA, Schmaltz R, Fangman T, Zhou Y, et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013;9(12):e1004057. doi: 10.1371/journal.pgen.1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinken A, Sahoo S, Fleming RMT, Thiele I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes. 2013;4(1):28–40. doi: 10.4161/gmic.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011;7(2):e1001314. doi: 10.1371/journal.pgen.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–76. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duperron S, Lorion J, Samadi S, Gros O, Gaill F. Symbioses between deep-sea mussels (Mytilidae: Bathymodiolinae) and chemosynthetic bacteria: diversity, function and evolution. C R Biol. 2009;332(2–3):298–310. doi: 10.1016/j.crvi.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Duperron S, Gros O. Colwellia and sulfur-oxidizing bacteria: An unusual dual symbiosis in a Terua mussel (Mytilidae: Bathymodiolinae) from whale falls in the Antilles arc. Deep Sea Res Part I. 2016;115:112–22. [Google Scholar]
- 37.Heath-Heckman EAC, Foster J, Apicella MA, Goldman WE, McFall-Ngai M. Environmental cues and symbiont MAMPs function in concert to drive the daily remodeling of the crypt-cell brush border of the Euprymna scolopes light organ. Cell Microbiol. 2016;18(11):1642–1652. doi: 10.1111/cmi.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2(8):632–42. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 39.Kambam PKR, Sayut DJ, Niu Y, Eriksen DT, Sun L. Directed evolution of LuxI for enhanced OHHL production. Biotechnol Bioeng. 2008;101(2):263–72. doi: 10.1002/bit.21901. [DOI] [PubMed] [Google Scholar]
- 40.Miyashiro T, Ruby EG. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol. 2012;84(5):795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43(W1):W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jebbar M, Sohn-Bösser L, Bremer E, Bernard T, Blanco C. Ectoine-induced proteins in Sinorhizobium meliloti include an Ectoine ABC-type transporter involved in osmoprotection and ectoine catabolism. J Bacteriol. 2005;187(4):1293–304. doi: 10.1128/JB.187.4.1293-1304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanekop N, Höing M, Sohn-Bösser L, Jebbar M, Schmitt L, Bremer E. Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine. J Mol Biol. 2007;374(5):1237–50. doi: 10.1016/j.jmb.2007.09.071. [DOI] [PubMed] [Google Scholar]
- 44.Marini A, Reinelt K, Krutmann J, Bilstein A. Ectoine-containing cream in the treatment of mild to moderate atopic dermatitis: a randomised, comparator-controlled, intra-individual double-blind, multi-center trial. Skin Pharmacol Physiol. 2014;27(2):57–65. doi: 10.1159/000351381. [DOI] [PubMed] [Google Scholar]
- 45.Graf R, Anzali S, Buenger J, Pfluecker F, Driller H. The multifunctional role of ectoine as a natural cell protectant. Clin Dermatol. 2008;26(4):326–33. doi: 10.1016/j.clindermatol.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Godoy-Vitorino F, Goldfarb KC, Karaoz U, Leal S, Garcia-Amado MA, Hugenholtz P, et al. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J. 2012;6(3):531–41. doi: 10.1038/ismej.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roggenbuck M, Sauer C, Poulsen M, Bertelsen MF, Sørensen SJ. The giraffe (Giraffa camelopardalis) rumen microbiome. FEMS Microbiol Ecol. 2014;90(1):237–46. doi: 10.1111/1574-6941.12402. [DOI] [PubMed] [Google Scholar]
- 48.Penner GB, Steele MA, Aschenbach JR, McBride BW. Ruminant Nutrition Symposium: Molecular adaptation of ruminal epithelia to highly fermentable diets. J Anim Sci. 2011;89(4):1108–19. doi: 10.2527/jas.2010-3378. [DOI] [PubMed] [Google Scholar]
- 49.Mao SY, Huo WJ, Zhu WY. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ Microbiol. 2016;18(2):525–41. doi: 10.1111/1462-2920.12724. [DOI] [PubMed] [Google Scholar]
- 50.Khafipour E, Li S, Plaizier JC, Krause DO. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microbiol. 2009;75(22):7115–24. doi: 10.1128/AEM.00739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kopp C, Wisztorski M, Revel J, Mehiri M, Dani V, Capron L, et al. MALDI-MS and NanoSIMS imaging techniques to study cnidarian-dinoflagellate symbioses. Zoology. 2015;118(2):125–31. doi: 10.1016/j.zool.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77(3):342–56. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. Precolonized Human Commensal Escherichia coli Strains Serve as a Barrier to E. coli O157:H7 Growth in the Streptomycin-Treated Mouse Intestine. Infect Immun. 2009;77(7):2876–86. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medellin-Peña MJ, Griffiths MW. Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157:H7 colonization. Appl Environ Microbiol. 2009;75(4):1165–72. doi: 10.1128/AEM.01651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 56.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat, Rev, Immunol. 2006;6(11):849–58. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 57.Nadal Jimenez P, Koch G, Papaioannou E, Wahjudi M, Krzeslak J, Coenye T, et al. Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology. 2010;156(Pt 1):49–59. doi: 10.1099/mic.0.030973-0. [DOI] [PubMed] [Google Scholar]
- 58.Meyer JM. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol. 2000;174(3):135–42. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- 59.Wollenberg MS, Claesen J, Escapa IF, Aldridge KL, Fischbach MA, Lemon KP. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. MBio. 2014;5(4):e01286–14. doi: 10.1128/mBio.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antunes LCM, McDonald JAK, Schroeter K, Carlucci C, Ferreira RBR, Wang M, et al. Antivirulence activity of the human gut metabolome. MBio. 2014;5(4):e01183–14. doi: 10.1128/mBio.01183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson T, Kang D, Barnard E, Li H. Strain-Level Differences in Porphyrin Production and Regulation in Propionibacterium acnes Elucidate Disease Associations. mSphere. 2016;1(1):e00023–15. doi: 10.1128/mSphere.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10(11):1861–71. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 63.Lowery CA, Salzameda NT, Sawada D, Kaufmann GF, Janda KD. Medicinal chemistry as a conduit for the modulation of quorum sensing. J Med Chem. 2010;53(21):7467–89. doi: 10.1021/jm901742e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fulde M, Hornef MW. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev. 2014;260(1):21–34. doi: 10.1111/imr.12190. [DOI] [PubMed] [Google Scholar]
- 65.Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351(6279):1296–302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 66.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mullaney JA, Kelly WJ, McGhie TK, Ansell J, Heyes JA. Lactic acid bacteria convert glucosinolates to nitriles efficiently yet differently from enterobacteriaceae. J Agric Food Chem. 2013;61(12):3039–46. doi: 10.1021/jf305442j. [DOI] [PubMed] [Google Scholar]
- 68.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe. 2016;19(4):443–54. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolfe BE, Button JE, Santarelli M, Dutton RJ. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell. 2014;158(2):422–33. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J, Liu Y, Kim E, March JC, Bentley WE, Payne GF. Electrochemical reverse engineering: A systems-level tool to probe the redox-based molecular communication of biology. Free Radic Biol Med. 2016 doi: 10.1016/j.freeradbiomed.2016.12.029. [Internet]. Available from: http://dx.doi.org/10.1016/j.freeradbiomed.2016.12.029. [DOI] [PubMed]
- 71.Zargar A, Payne GF, Bentley WE. A “bioproduction breadboard”: programming, assembling, and actuating cellular networks. Curr Opin Biotechnol. 2015;36:154–60. doi: 10.1016/j.copbio.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 72.Molinski TF. Nanomole-scale natural products discovery. Curr Opin Drug Discov Devel. 2009;12(2):197–206. [PubMed] [Google Scholar]
- 73.Forseth RR, Schroeder FC. NMR-spectroscopic analysis of mixtures: from structure to function. Curr Opin Chem Biol. 2011;15(1):38–47. doi: 10.1016/j.cbpa.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mahanti P, Bose N, Bethke A, Judkins JC, Wollam J, Dumas KJ, et al. Comparative metabolomics reveals endogenous ligands of DAF-12, a nuclear hormone receptor, regulating C. elegans development and lifespan. Cell Metab. 2014;19(1):73–83. doi: 10.1016/j.cmet.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wist J. Complex mixtures by NMR and complex NMR for mixtures: experimental and publication challenges. Magn Reson Chem. 2017;55(1):22–28. doi: 10.1002/mrc.4533. [DOI] [PubMed] [Google Scholar]
- 76.Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell. 2014;159(2):267–80. doi: 10.1016/j.cell.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Duynhoven JPM, Jacobs DM. Assessment of dietary exposure and effect in humans: The role of NMR. Prog Nucl Magn Reson Spectrosc. 2016;96:58–72. doi: 10.1016/j.pnmrs.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34(8):828–37. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bouslimani A, Porto C, Rath CM, Wang M, Guo Y, Gonzalez A, et al. Molecular cartography of the human skin surface in 3D. Proc Natl Acad Sci USA. 2015;112(17):E2120–9. doi: 10.1073/pnas.1424409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balog J, Sasi-Szabó L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med. 2013;5(194):194ra93. doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
- 81.Golf O, Strittmatter N, Karancsi T, Pringle SD, Speller AVM, Mroz A, et al. Rapid evaporative ionization mass spectrometry imaging platform for direct mapping from bulk tissue and bacterial growth media. Anal Chem. 2015;87(5):2527–34. doi: 10.1021/ac5046752. [DOI] [PubMed] [Google Scholar]
- 82.Mascuch SJ, Moree WJ, Hsu CC, Turner GG, Cheng TL, Blehert DS, et al. Direct detection of fungal siderophores on bats with white-nose syndrome via fluorescence microscopy-guided ambient ionization mass spectrometry. PLoS One. 2015;10(3):e0119668. doi: 10.1371/journal.pone.0119668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pulliam CJ, Wei P, Snyder DT, Wang X, Ouyang Z, Pielak RM, et al. Rapid discrimination of bacteria using a miniature mass spectrometer. Analyst. 2016;141(5):1633–6. doi: 10.1039/c5an02575c. [DOI] [PubMed] [Google Scholar]
- 84.Jarmusch AK, Pirro V, Kerian KS, Cooks RG. Detection of strep throat causing bacterium directly from medical swabs by touch spray-mass spectrometry. Analyst. 2014;139(19):4785–9. doi: 10.1039/c4an00959b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palmer AD, Alexandrov T. Serial 3D imaging mass spectrometry at its tipping point. Anal Chem. 2015;87(8):4055–62. doi: 10.1021/ac504604g. [DOI] [PubMed] [Google Scholar]
- 86.Ogrinc Potočnik N, Porta T, Becker M, Heeren RMA, Ellis SR. Use of advantageous, volatile matrices enabled by next-generation high-speed matrix-assisted laser desorption/ionization time-of-flight imaging employing a scanning laser beam. Rapid Commun Mass Spectrom. 2015;29(23):2195–203. doi: 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- 87.Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. J Mass Spectrom. 2012;47(11):1473–81. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 88.Sun C, Jiang F, Gao W, Li X, Yu Y, Yin X, et al. Scanning electron microscopy coupled with energy-dispersive X-ray spectrometry for quick detection of sulfur-oxidizing bacteria in environmental water samples. Chin J Oceanol Limnol. 2016;35(1):185–191. [Google Scholar]
- 89.Ward SK, Heintz JA, Albrecht RM, Talaat AM. Single-cell elemental analysis of bacteria: quantitative analysis of polyphosphates in Mycobacterium tuberculosis. Front Cell Infect Microbiol. 2012;2:63. doi: 10.3389/fcimb.2012.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aguayo S, Bozec L. Mechanics of Bacterial Cells and Initial Surface Colonisation. Adv Exp Med Biol. 2016;915:245–60. doi: 10.1007/978-3-319-32189-9_15. [DOI] [PubMed] [Google Scholar]
- 91.Beaussart A, El-Kirat-Chatel S, Herman P Alsteens D, Mahillon J Hols P, et al. Single-cell force spectroscopy of probiotic bacteria. Biophys J. 2013;104(9):1886–92. doi: 10.1016/j.bpj.2013.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Longo G, Alonso-Sarduy L, Rio LM, Bizzini A, Trampuz A, Notz J, Dietler G, Kasas S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nature Nanotech. 2013;8(7):522–526. doi: 10.1038/nnano.2013.120. [DOI] [PubMed] [Google Scholar]
- 93.Wang Z, Millet L, Mir M, Ding H, Unarunotai S, Rogers J, et al. Spatial light interference microscopy (SLIM) Opt Express. 2011;19(2):1016–26. doi: 10.1364/OE.19.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mir M, Wang Z, Shen Z, Bednarz M, Bashir R, Golding I, et al. Optical measurement of cycle-dependent cell growth. Proc Natl Acad Sci USA. 2011;108(32):13124–9. doi: 10.1073/pnas.1100506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bryan NS, Grisham MB. Methods to detect nitric oxide and its metabolites in biological samples. Free Radic Biol Med. 2007;43(5):645–57. doi: 10.1016/j.freeradbiomed.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci USA. 2010;107(18):8375–80. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shi YM, Cai SL, Li XN, Liu M, Shang SZ, Du X, et al. LC-UV-Guided Isolation and Structure Determination of Lancolide E: A Nortriterpenoid with a Tetracyclo[5.4.0.0(2,4).0(3,7)]undecane-Bridged System from a “Talented” Schisandra. Plant Org Lett. 2016;18(1):100–3. doi: 10.1021/acs.orglett.5b03334. [DOI] [PubMed] [Google Scholar]
- 98.Surup F, Wagner O, von Frieling J, Schleicher M, Oess S, Müller P, et al. The iromycins, a new family of pyridone metabolites from Streptomyces sp I Structure, NOS inhibitory activity, and biosynthesis. J Org Chem. 2007;72(14):5085–90. doi: 10.1021/jo0703303. [DOI] [PubMed] [Google Scholar]
- 99.Eberhardt K, Stiebing C, Matthäus C, Schmitt M, Popp J. Advantages and limitations of Raman spectroscopy for molecular diagnostics: an update. Expert Rev Mol Diagn. 2015;15(6):773–87. doi: 10.1586/14737159.2015.1036744. [DOI] [PubMed] [Google Scholar]
- 100.Popp J. Identification of micro-organisms by Raman spectroscopy. SPIE Newsroom [Internet] 2007 Available from: http://www.spie.org/x16083.xml.
- 101.Stöckel S, Kirchhoff J, Neugebauer U, Rösch P, Popp J. The application of Raman spectroscopy for the detection and identification of microorganisms. J Raman Spectrosc. 2016;47(1):89–109. [Google Scholar]
- 102.Premasiri WR, Lee JC, Sauer-Budge A, Théberge R, Costello CE, Ziegler LD. The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS. Anal Bioanal Chem. 2016;408(17):4631–47. doi: 10.1007/s00216-016-9540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pahlow S, Meisel S, Cialla-May D, Weber K, Rösch P, Popp J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv Drug Deliv Rev. 2015;89:105–20. doi: 10.1016/j.addr.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 104.Brooks JF, 2nd, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, et al. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci USA. 2014;111(48):17284–9. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]


