Abstract
A central goal of pharmacological efforts to treat central nervous system (CNS) diseases is to develop systemic therapeutics that can restore CNS homeostasis. Achieving this goal requires a fundamental understanding of CNS function within the organismal context so as to leverage the mechanistic insights on the molecular basis of cellular and tissue functions towards novel drug target identification. The immune system constitutes a key link between the periphery and CNS, and many neurological disorders and neurodegenerative diseases are characterized by immune dysfunction. We review the salient opportunities for applying computational models to CNS disease research, and summarize relevant approaches from studies of immune function and neuroinflammation. While the accurate prediction of disease-related phenomena is often considered the central goal of modeling studies, we highlight the utility of computational modeling applications beyond making predictions, particularly for drawing counterintuitive insights from model-based analysis of multi-parametric and time series data sets.
Introduction
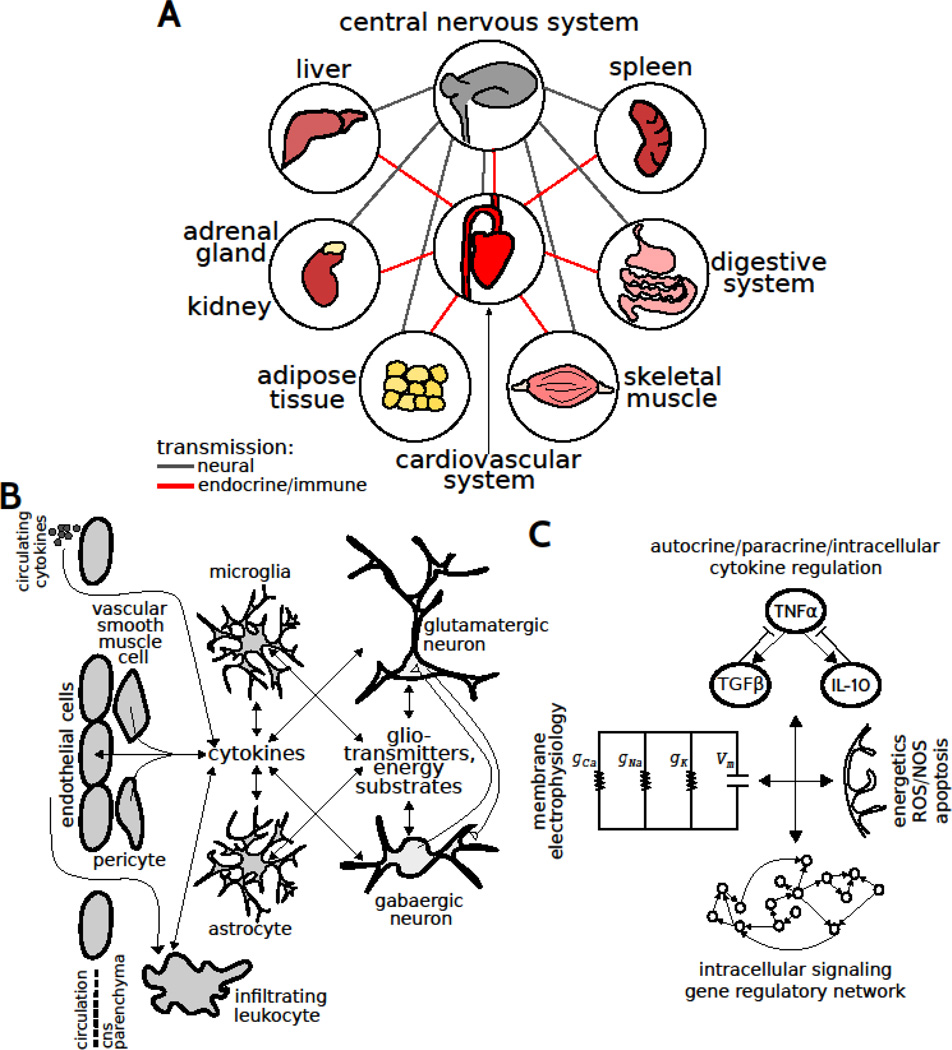
Neurological disorders and neurodegenerative diseases of the central nervous system (CNS) typically exhibit slow progression to chronic pathology mediated by a multifactorial repertoire of elements. CNS diseases involve maladaptive neural circuits (e.g., Alzheimer’s disease, AD), neuromodulation (e.g., Parkinson’s disease, PD), and/or neurodegeneration (e.g., multiple sclerosis, MS). These diseases are multigenic, non-cell autonomous, and co-morbid with a number of organismal maladaptations including heart disease, metabolic syndrome, and impaired immune function (Fig 1A). Importantly, malfunctional autonomic nervous system activity has been implicated in the pathology associated with neurodegenerative disease (Cersosimo & Benarroch, 2013). Numerous studies have established a role for neuroinflammation in CNS diseases. At the tissue level, neuroinflammation involves autocrine and paracrine cytokine signaling and interactions amongst vascular cells, infiltrating leukocytes, microglia, astrocytes, and neurons (Fig 1B). Aberrant cytokine/chemokine regulation, endoplasmic reticulum stress, and mitochondrial dysfunction driving the upregulation of reactive oxygen/nitrogen species contribute to neuroinflammation (Fig 1C). The complexity of the multiscale regulatory networks and spatiotemporally distributed factors underlying CNS diseases precludes a direct application of intuition to uncover the underlying principles and identify key control points for effective intervention.
Figure 1.
Distributed control in multiscale networks coordinating CNS function. (A) CNS function is coupled to the function of multiple organs through efferents/afferents and endocrine/immune transmission through the blood. (B) Tissue scale interactions amongst multiple cell types in the CNS. (C) Molecular scale networks regulating the integration of molecular signaling with neuronal physiology.
Recent technological advances have led us into a ‘omics’ era in which it is reasonably cost effective and almost routine to obtain genomic, transcriptomic, proteomic, and metabolomic scale data from even single cells. Analysis of such large-scale data has shown that mammalian tissue and cell type organization is far more complex than previously appreciated, and at the same time has begun to provide insights into the molecular and functional states of cells in vivo. For example, single cell transcriptomic data analysis uncovered 47 distinguishable neuronal phenotypes in the mouse cortex (Zeisel et al., 2015). Similarly, single cell proteomic analysis has led to new insights in the cellular hierarchy and lineage progression of immune cells (Bendall et al., 2011). A quantitative understanding of how these multifactorial and multiscale components interact in regulatory networks is still elusive. Following others, we argue that computational modeling is necessary to make the next leap towards a comprehensive understanding of mechanisms driving neurological disorders and neurodegenerative diseases, and for developing effective therapeutic strategies (Mesarovic et al., 2004; Lander, 2010).
Systems biologists have proposed to use modeling approaches to understand function in terms of principles that govern the interactions amongst biological elements, so as to facilitate a rational development of CNS disease therapeutics (Lazebnik, 2002; Fischer, 2008). A hierarchy of control mechanisms regulate complex biological systems such as the CNS, ranging from molecular/cellular events to cell-cell interactions, along with the integration of signals from the blood and lymphatic systems (Louveau et al., 2016). Hence, a comprehensive understanding of the distributed multiscale control mechanisms of homeostasis – within the context of the organism – is central to understanding and tackling the CNS pathogenesis (Iyengar, 2013; Fig 1A). Furthermore, peripheral inflammation is common in neurodegenerative disease. It is likely the case that peripheral cytokines interact with central neuroinflammation through active of the brain endothelium, and/or compromised blood-brain-barrier (BBB) (Fig 1B). Hence, understanding the mechanisms driving peripheral inflammation is highly relevant to tackle neurodegeneration.
In this review, we summarize the virtues and shortcomings of select computational modeling approaches, and highlight evidence that such approaches can be leveraged to define new biomarkers and therapeutics for CNS diseases. In particular, we review key literature related to study of neuroinflammation in CNS disorders using computational modeling, and discuss the challenges and enabled opportunities.
Cytokine networks, glial phenotypes, and immune function
Proinflammatory cytokine upregulation and morphological adaptations of glial cells are the defining features of neuroinflammation. For instance, tumor necrosis factor-α (TNFα) upregulation was observed in AD, PD, and MS (Mccoy & Tansey, 2008). TNFα has been shown to yield excitotoxicity in neurons through effects on intrinsic membrane ion channels (Park & Bowers, 2010; Fig 1C). Furthermore, TNFα is known to induce apoptosis and necrosis, thereby exacerbating neurodegeneration (Mccoy & Tansey, 2008). Importantly, cytokines have been shown to interact through complex regulatory networks (Schmitz et al., 2011). Thus, understanding the dynamics of complex cytokine interactions is critical to understanding the variegated and counterintuitive effects of glial cells on CNS phenotypes (Sriram & O'Callaghan, 2007; Lobo-Silva et al., 2016).
Microglia and astrocytes secrete and respond to an expansive repertoire of cytokines and chemokines. In vitro, cytokine networks can function through autocrine/paracrine signaling involving a single cell type. In vivo, cytokine network activity involves multiple cell types and cell-cell interactions, with prominent contributions from microglia and astrocytes (Fig 1B). The effects of cytokines such as TNFα on cellular functional states have been considered in terms of feedback loops (Schmitz, 2011). For instance, microglial activation mediated by TNFα upregulation results in the activation of multiple feedback cytokines that regulate the neuroinflammatory phenotype. The complicated crosstalk topologies of cytokine interaction networks highlight the necessity of computational approaches to disambiguate complex phenomena. It is well known that the expression level and activation timing of a cytokine determine its functional effects. However the influences of cytokine activation timing have not been thoroughly studied from an integrated network perspective in vivo in CNS disease and injury-related pathogenesis (Meyer-Hermann et al., 2009; Foteinou et al., 2010).
Cytokine network activity following the inflammatory activation is associated with functionally relevant plasticity of glial morphology. Morphological plasticity can result in phagocytosis of boutons and spines, as well as physical displacement of synaptic terminals. Inflammatory stimuli such as infection, trauma, or stroke elicit a phenotypic transition in microglia and astrocytes. Hence, both neurochemical and morphological aspects of glial activation in neuroinflammation cooperate to regulate neural network activity (Kettenmann et al., 2013), however, the mechanistic coupling between cytokine network dynamics and morphological plasticity are only beginning to be revealed (Anderson et al., Submitted-a).
Computational modeling: Conceptual motivation and implementation frameworks
Models abound in science. We all use them, though most of our models exist as implicit conceptual constructs and mental models rather than explicit formulations and mathematical representations. Understanding how CNS cellular phenotypes depend on the spatial location as well as amplitude and dynamics of molecular signals in the local microenvironment is critical for rationally developing therapeutic interventions against CNS disorders. Typical modeling studies entail modification or construction of a computational model, estimation of unknown parameters based on available data, testing the model predictions using data that were not utilized for parameter estimation, and simulation based analysis of the model. We argue that modeling is important for purposes beyond generating and testing predictions, including the following: to test/generate hypotheses, identify new questions, illuminate uncertainties, explore intervention strategies, explain complex phenomena, and achieve an integrated understanding of biological processes (Epstein, 2008). The knowledge derived from such approaches can yield important insights for drug development, such as whether a given property of a system has a physiological impact, is a consequence of another mechanism, or is an epiphenomena.
Models of inflammatory regulation include static and dynamic representations. Parameter-free simulations of network models can be implemented using Boolean logic, in which elements of the system are considered to be either ‘on’ or ‘off’ at a given time. While Boolean models provide a convenient framework for simulating the steady-state behaviors of networks inferred from high-throughput data without the necessity for estimating uncertain parameters, this approach lacks biological plausibility insofar as the on-off digital representation of the molecular variables does not capture the dynamic and graded variations that are observed in complex biological processes. Dynamic modeling formalisms can be deterministic or stochastic. Network structure identification can be accomplished through measurements of responses to perturbations or inferences of molecular interaction coefficients that encode dynamics of the respective network (Kholondenko et al., 2002; Anderson et al., Submitted-b). Deterministic models are described by systems of ordinary or partial differential equations (ODEs, PDEs). Stochastic models incorporate biological randomness inherent to systems that involve molecular fluctuations. Deterministic models are relatively simple and efficient to implement, with limited degree of analytical tractability. Whereas stochastic models incorporate biological randomness, deterministic models can be amenable to formal mathematical analyses that identify fundamental properties of the dynamic systems. Concepts from Boolean modeling and dynamic modeling can be integrated in multi-scale approaches. Agent based models (ABMs) incorporate interactions between cells that are characterized by specific states (e.g., activated, infected, or proliferating; Meier-Schellersheim 2006; Cilfone, 2015).
Overview of cell signaling and immune modeling studies
We highlight select examples with relevance to studying cytokine networks in neuroinflammation. As an example of statistical approach, Janes and colleagues (2005) examined cytokine influences on apoptosis. The key feature of this study was the use of multi-perturbation, multi-parametric, time series experimental design to yield a compendium of data. Janes et al. analyzed this compendium using statistical modeling to identify novel molecular mechanisms connecting autocrine feedback loops involving IL-1α and TGFα signaling to apoptotic responses. Subsequent studies built on these data-driven approaches to develop constrained fuzzy logic modeling of signaling (Morris et al., 2011) and gene regulatory networks (Park et al., 2015). These empirical, data-driven methods hold promise for expanding our knowledge beyond canonical cytokine signaling networks in order to fully interpret the dynamic patterns of cytokines and their molecular targets underlying neuroimmune processes.
In the context of deterministic and stochastic modeling frameworks, investigators have examined immune function and inflammation over a wide range of spatiotemporal scales. An illustrative example is the computational modeling of the receptor mediated activation of transcription factor NF-κB that transduces signaling responses to extracellular pathogens and cytokines and regulates immune functions and apoptosis. Computational modeling of NF-κB activation revealed new insights into how different feedback regulators control distinct dynamic aspects of NF-κB level, localization, and activity (Hoffman, 2002; Kearns, 2006; Paszek, 2010). An illustrative example of modeling at the tissue scale is the study of granulomas and associated immune response dynamics using a multiscale framework combining ODEs and ABMs. Cilfone (2013) modeled tuberculosis infection by simulating including macrophages and T cells functions driven by intracellular cytokine signaling mediated by TNFα and IL-10 receptor activation. An important finding of this study was that an optimal balance of these cytokines that was associated with minimized bacterial load. Simulation environments such as the Simmune tool provide systematic frameworks for representing molecular regulatory networks and cellular state transitions, and allow relating specific biochemical states to cellular scale responses such as division, death, migration, and secretion of cytokines and other factors (Meier-Schellersheim et al., 2006). Computational modeling studies such as the above illustrated cases provide key insights into the molecular regulation of emergent properties of biological systems from cells to tissues. Such model-driven insights can provide new hypotheses and predictions as well as motivate the need for reformulation of the conceptual basis of a biological phenomenon.
Computational modeling applications to study neuroinflammation and neurodegeneration
Statistical and simulation based modeling approaches have been undertaken to study CNS diseases. Zhang and colleagues (2013) applied Bayesian network inference to identify molecular network modules from gene expression analyses of human brain samples from AD patients. This approach led to the identification of TYROBP as a regulator of microglia-mediated neuroinflammation in the prefrontal cortex. A temporal logic approach, similar to boolean logic was implemented in simulations designed to examine how/why amyloid beta-stimulated microglia exhibit pro- and anti-inflammatory cytokine profiles in parallel, and how/why aged microglia fail to phagocytose elevated amyloids (Anastasio, 2015). This work provided candidate explanations for key phenomena associated with AD. ODE-based studies of microglial cell signaling revealed that heat shock proteins may protect against stroke through inhibition of NF-κB signaling (Sheppard et al., 2014). In the context of amyotrophic lateral sclerosis (ALS), modeling applications include statistical modeling of clinical data (Küffner et., 2015), tissue-scale modeling of cytokine regulatory dynamics (Shao et al., 2013), and single cell modeling of neuronal electrophysiology integrated with cellular energetics (Le Masson et al., 2014). These studies elaborated our understanding of the multiscale mechanisms underlying ALS, suggested novel treatment regimens based on perturbing cytokine regulation, and identified clinical criteria for better prediction of ALS progression.
We have integrated computational modeling with experimentation in cellular, tissue-level, and organismal-scale studies of cytokine networks. At the cellular level, we modeled a microglia-specific autocrine/paracrine cytokine interactions (Anderson et al., 2015). Surprisingly, we found that negative feedback inhibitors of TNFα showed divergent influences dependent on their relative dynamics. While TGFβ exhibited slower kinetics and facilitated the adaptation of TNFα to a sustained inflammatory stimulus, relatively fast IL-10 mediated feedback was associated with a counterintuitive decrease in adaptation. This finding was observed in experiments involving LPS-induced cytokine response of bone marrow derived macrophages in vitro (Anderson et al., 2015). We developed a tissue-scale model of neuroinflammation including microglial and astrocytic contributions, and our simulations and experiments showed that IL-10 reduced TNFα adaptation in vivo (Anderson et al., Submitted-a). Furthermore, we analyzed single cell multivariate microglial morphology data and found that morphological properties related to the shapes of somata and processes showed IL-10-dependent adaptation patterns. These model-driven studies demonstrated a novel link between cytokine network dynamics and morphological features of microglia. We designed organismal scale models to simulate the development of dysregulated homeostasis in the context of autonomic nervous system dysfunction (Anderson et al., Submitted-b). We applied systems identification techniques to infer dynamic multi-tissue gene regulatory networks involving cytokine transcripts in health and disease. Our analyses revealed that autonomic disease development was associated with a rewired network structure and divergent activity patterns. We identified key regulatory elements with disease-specific molecular interactions and dynamic profiles, thereby providing candidates for further evaluation of compensatory responses to disease conditions, biomarker potential, and therapeutic interventions.
General principles of system function obtained from computational modeling and analysis
Design and control principles of CNS function elude simple intuition, necessitating an integrative and quantitative perspective. Computational modeling has highlighted the mutual influences of molecular kinetics and network structures in the context of process control mechanisms (e.g., adaptation and tolerance) and information processing (e.g., encoding and decoding). Multiple topologically similar but kinetically distinct feedback loops can exhibit differentiated functions to allow fine-tuning of system responses (Bachmann et al., 2011; Yang et al., 2011; Longo et al., 2013; Anderson, et al., 2015). Through expansive searches of network structures, specific topological motifs have been implicated in emergent properties including adaptation to a sustained stimulus and priming/tolerance to repeated stimuli (Ma et al., 2009; Fu et al., 2012), both of which are critical to immune function and have been studied in cytokine networks (Day et al., 2006; Anderson, et al., 2015). However, as a note of caution, some of these findings may be dependent on the mathematical framework utilized (e.g., Hill equations to describe interaction rates) and do not necessarily generalize for application on identical topological motifs with different mathematical formulations of the dynamics (Barzel & Barabási, 2013). The encoding and decoding properties of a given signal are governed by dynamic characteristics including delay, onset duration, amplitude, signal duration, deactivation, and frequency (Purvis & Lahav, 2013; Makadia et al., 2015). Stimulus-specific feedback interactions have been shown to impart cytokine stimulus-specific coding properties as reflected by distinct dynamic and transcriptional responses (Werner et al., 2005; Braun et al., 2013). Individual features of signaling dynamics can be insufficient for determining system response profiles (Makadia et al., 2015), thereby providing a quantitative explanation of why individual therapeutics, as opposed to combination therapies, are often insufficient to revert diseases. Furthermore, these findings support the notion that dynamic properties of signal can be considered as effective targets for therapeutic intervention (Behar et al., 2013). These examples highlight the complex mapping between stimuli and network responses that can be unraveled using computational modeling.
Opportunities for applications in CNS drug discovery
CNS disease research could greatly benefit from integrated computational and experimental approaches for identifying novel diagnostic/prognostic biomarkers, drug targets, dosing regimens, adverse drug responses, and patient-specificity. Here we outline a few opportunities to facilitate the use of computational approaches based on acquiring necessary data, taking advantage of approaches utilized in other fields, and reformulating the conceptual paradigms of disease initiation, progression and response to intervention. Overall, the future directions suggested to advance drug discovery for neuroinflammation and neurodegeneration include addressing a set of questions aimed at identifying key elements, interactions, and dynamics that serve as control points for effective intervention: What are the critical molecular mechanisms that govern cytokine regulation within the local neural tissue microenvironment? How to integrate contributions of multiple cell types within the neural tissue across relevant timescales? How to couple functional responses of distinct neural and immune cell subpopulations to changes in neuronal physiology? What are the relative contributions of genotype and physiological phenotype in shaping response to neuroinflammation? How to account for the intrinsic and inter-individual variation in inflammatory responses and progression of neurodegenerative disease?
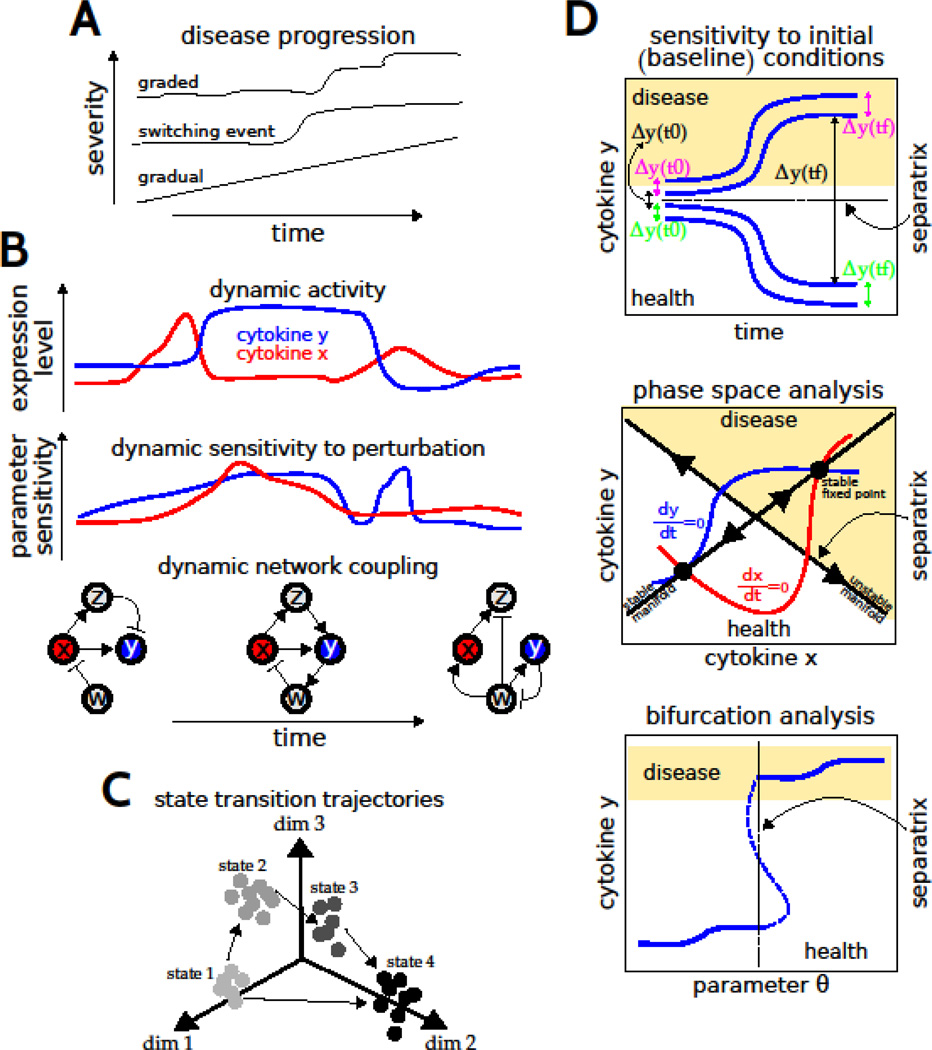
The time courses of disease progression, and variability thereof across human populations, are not thoroughly understood. Multiple temporal profiles could exist, with distinct implications for therapeutic approaches (Fig 2A). In this regard, understanding the dynamics of the molecular and cellular networks may provide a more tractable way to differentiate disease risk and treatment viability than considering genetics alone (Civelek & Lusis, 2014). In understanding the intrinsic variation, a critical unresolved issue is the question of cell type contributions to the inflammatory milieu in the CNS. In addition to microglia and astrocytes, other cells including neurons, endothelial cells, and pericytes are known to secrete cytokines. Similarly, vascular smooth muscle cells have been demonstrated to undergo phenotypic transitions into macrophage-like cells under inflammatory conditions (Bennett et al., 2016). However, the extent of cell phenotype plasticity and variability, and relative contributions of diverse cell types to tissue levels of cytokines, are unknown. These questions can be addressed through gene expression profiling of single cells in tissue sections using laser capture microdissection (Park et al., 2014, 2016). In addition, proteomic analysis optimized for single-cell scale samples would be very useful for determining the actual levels of the cytokines within the localized microenvironment. In general, this issue of cell type contributions to overall cytokine levels must be resolved to achieve a comprehensive understanding of cytokine network function and the mechanisms of neuroinflammation. This information is also critical to appropriately specify the structure and parameterize a computational model of the multicellular network underlying neuroinflammation (Fig 1B). Similarly, understanding the mechanisms underlying multi-organ communication could be informative to formulate and parameterize corresponding computational models to study how neuroinflammation is regulated in an organismal context (Fig 1A).
Figure 2.
Modeling disease dynamics. (A) Potential dynamics of disease progression. (B) Illustrative examples of dynamics in molecular levels, parameter sensitivity, and molecular regulatory network structure. (C) Illustrative trajectories of state transitions derived from statistical analysis of high-dimensional data and visualized in a reduced dimensional space. (D) Graphical representations of mathematical analysis to identify thresholds of disease - or separatrices - based on changes in initial conditions, variations in levels of system elements (e.g., cytokines x and y), and variations in parameters (e.g., network interaction coefficients).
Understanding the dynamic mechanisms of disease is critical for identification of new biomarkers and drug targets. Given an ODE model of a CNS disease, computational studies can help to elucidate the relationship between the temporal evolution of a system response and the sensitivities of the underlying elements to a therapeutic perturbation (Fig 2B). Sensitivity analyses have shown that specific molecular elements are predominantly important for system behavior at particular times, whereas interventions at other times were ineffectual (Miller et al., 2010). Importantly, these results suggest that successful precision medicine requires an understanding of when a specific molecular intervention should be applied for optimal effect. It is becoming increasingly clear that varying stages of a disease progression are associated with distinct molecular interactions or ‘differential networks’ (Fig 2B; Zickenrott et al., 2016). Hence, models of CNS disease progression need to consider cellular and molecular networks that are defined by dynamic rather than static connectivity structures.
From a statistical perspective, global molecular configurations can be defined by projecting high dimensional multivariate data onto a lower dimensional subspace that can be analyzed for trajectories of state transitions associated with disease processes, as well as for mapping alternative trajectories that could account for inter-individual variability and therapeutic responses (Fig 2C). Such approaches have provided important insights into both developmental processes and drug responses (Bendall et al., 2011; Marco et al., 2014). Network modeling based on protein-protein interactions has also facilitated understanding of potential molecular underpinnings of seemingly unrelated diseases. Furthermore, the module of disease genes identified through network analysis was enriched for targets of drugs with adverse event profiles (Berger et al., 2010). These data-driven methods could be integrated with dynamic modeling by distilling high-throughput data sets into sets of response profiles that are used to constrain network structures and fine tune model parameters, as well as experimentally characterize the landscape of state transitions (Fig 2D).
Mathematical analyses of dynamic models facilitate a comprehensive analysis of the landscape of transitions between health and disease, to both identify and elucidate the mechanistic basis of transition control points, in time or molecular space, that demarcate qualitative shifts from health to disease (Fig 2D; Gross & Blasius, 2008). For instance, the initial levels of system components have been shown to regulate cytokine network dynamics and apoptotic responses to cytokine treatment (Aldridge et al., 2006; Anderson et al., 2015; Fig 2D, top). Similarly, related approaches involving phase space analysis (Fig 2D, middle) and bifurcation analysis (Fig 2D, bottom) can be pursued to understand the conditions underlying critical transitions to pathological states (Liu et al., 2015; Hat et al., 2016). Such analyses further aid in identifying biomarkers that distinguish disease trajectories (as opposed to instantaneously observed states), for identifying early-warning signals of critical transitions driven by stochasticity, and for predicting effective targets that can prevent transitions to disease or reverse course towards a healthy state. Such approaches can be utilized to enhance our understanding of the molecular trajectories through which cytokine dynamics promote robust neuroinflammation and CNS disease states.
A recent application of computational models for exploring disease dynamics is the simulation of a population of “virtual patients”. The objective is to account for inter-individual variability by considering a large set of simulations based on population-relevant and physiologically meaningful ranges of parameters (Kassab et al., 2016). These approaches typically utilize statistical and clustering analyses to identify patterns in network dynamics and attempt to relate the patterns of disease dynamics to distinct underlying parameters, providing new biomarker and drug target candidates. These models can differentiate disease subtypes and the patient-specific drug responses (Liu et al., 2016). Considering the intrinsic stochasticity and extent of uncertainty in the neuroinflammation network structures and parameters, such model-based large-scale exploration is crucial to evaluate the very many possibilities in which disease dynamics may unfold. The computational model-based simulation of virtual patient populations holds promise in advancing neuroinflammation and neurodegeneration research by taking an unbiased perspective.
Conclusions
While methodological advances, increased software availability, and enhanced computational speed facilitate the integration of high throughput experimental data acquisition, analysis, and computational modeling, paradigm shifts that transform understanding are driven by conceptual advances. Understanding pathophysiology is typically formulated as a problem of deconvolving cause from consequence of CNS disease. As stated, this task is particularly difficult in human diseases. We argue that the ‘cause versus consequence’ dichotomy is a flawed notion when applied to progressive disorders that involve multiple levels of a complex hierarchical network. Conventional experimental designs may be inadequate for generating understanding of CNS disease. For instance, targeted knockouts (KO) or overexpression experiments may not permit an unambiguous understanding of the functional role of the targeted molecule in disease due to adaptation and compensation. This difficulty is due to a dynamic sensitivity of the system response to perturbations. The advancement of experimental studies tracking the molecular, cellular, and physiological dynamics of disease progression through fine-grained time-series and interpreting these data using multiscale modeling can lead to new insights into neuroinflammatory process dynamics and control principles. Combining such an approach with typical manipulation experiments will help to elucidate the contributions of molecular and cellular elements to disease dynamics, significantly advancing the quest for novel intervention strategies and therapeutic targets in CNS diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The author(s) have no conflict of interest to declare.
References
- Aldridge BB, Haller G, Sorger PK, Lauffenburger DA. Direct Lyapunov exponent analysis enables parametric study of transient signalling governing cell behaviour. Syst Biol. 2006;153:425–432. doi: 10.1049/ip-syb:20050065. [DOI] [PubMed] [Google Scholar]
- Anastasio TJ. Temporal-logic analysis of microglial phenotypic conversion with exposure to amyloid-β. Mol Biosyst. 2015;11:434–453. doi: 10.1039/c4mb00457d. [DOI] [PubMed] [Google Scholar]
- Anderson WD, Greenhalgh A, Takwale A, David S, Vadigepalli R. Mathematical modelling of cytokine networks and multivariate analysis of morphology reveals Interleukin-10 is a key regulator of microglia in vivo. (Submitted-a) [Google Scholar]
- Anderson WD, DeCicco D, Schwaber J, Vadigepalli R. Dissecting the pathogenesis of physiological homeostasis: A multi-organ time-series analysis of complex disease. (Submitted-b) [Google Scholar]
- Anderson WD, Makadia HK, Greenhalgh AD, Schwaber JS, David S, Vadigepalli R. Computational modeling of cytokine signaling in microglia. Mol Biosyst. 2015;11:3332–3346. doi: 10.1039/c5mb00488h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann J, Raue A, Schilling M, Böhm ME, Kreutz C, Kaschek D, Busch H, Gretz N, Lehmann WD, Timmer J, Klingmüller U. Division of labor by dual feedback regulators controls JAK2/STAT5 signaling over broad ligand range. Mol Syst Biol. 2011;7:516. doi: 10.1038/msb.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzel B, Barabási AL. Universality in network dynamics. Nat Phys. 2013;9 doi: 10.1038/nphys2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar M, Barken D, Werner SL, Hoffmann A. The dynamics of signaling as a pharmacological target. Cell. 2013;155:448–461. doi: 10.1016/j.cell.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir el-AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe'er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SI, Ma'ayan A, Iyengar R. Systems pharmacology of arrhythmias. Sci Signal. 2010;3:ra30. doi: 10.1126/scisignal.2000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Fribourg M, Sealfon SC. Cytokine response is determined by duration of receptor and signal transducers and activators of transcription 3 (STAT3) activation. J Biol Chem. 2013;288:2986–2993. doi: 10.1074/jbc.M112.386573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cersosimo MG, Benarroch EE. Central control of autonomic function and involvement in neurodegenerative disorders. Handb Clin Neurol. 2013;117:45–57. doi: 10.1016/B978-0-444-53491-0.00005-5. [DOI] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilfone NA, Perry CR, Kirschner DE, Linderman JJ. Multi-scale modeling predicts a balance of tumor necrosis factor-α and interleukin-10 controls the granuloma environment during Mycobacterium tuberculosis infection. PLoS One. 2013;8:e68680. doi: 10.1371/journal.pone.0068680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J, Rubin J, Vodovotz Y, Chow CC, Reynolds A, Clermont G. A reduced mathematical model of the acute inflammatory response II. Capturing scenarios of repeated endotoxin administration. J Theor Biol. 2006;242:237–256. doi: 10.1016/j.jtbi.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Epstein JM. Why Model? Journal of Artificial Societies and Social Simulation. 2008;11:12. doi: 10.18564/jasss.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer HP. Mathematical modeling of complex biological systems: From parts lists to understanding systems behavior. Alcohol Res Health. 2008;31:49–59. [PMC free article] [PubMed] [Google Scholar]
- Foteinou PT, Calvano SE, Lowry SF, Androulakis IP. Multiscale model for the assessment of autonomic dysfunction in human endotoxemia. Physiol Genomics. 2010;42:5–19. doi: 10.1152/physiolgenomics.00184.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Glaros T, Zhu M, Wang P, Wu Z, Tyson JJ, Li L, Xing J. Network topologies and dynamics leading to endotoxin tolerance and priming in innate immune cells. PLoS Comput Biol. 2012;8:e1002526. doi: 10.1371/journal.pcbi.1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross T, Blasius B. Adaptive coevolutionary networks: a review. J R Soc Interface. 2008;5:259–271. doi: 10.1098/rsif.2007.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hat B, Kochańczyk M, Bogdał MN, Lipniacki T. Feedbacks, Bifurcations, and Cell Fate Decision-Making in the p53 System. PLoS Comput Biol. 2016;12:e1004787. doi: 10.1371/journal.pcbi.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Iyengar R. Complex diseases require complex therapies. EMBO Rep. 2013;14:1039–1042. doi: 10.1038/embor.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- Kassab GS, An G, Sander EA, Miga MI, Guccione JM, Ji S, Vodovotz Y. Augmenting Surgery via Multi-scale Modeling and Translational Systems Biology in the Era of Precision Medicine: A Multidisciplinary Perspective. Ann Biomed Eng. 2016;44:2611–2625. doi: 10.1007/s10439-016-1596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Kirchhoff F, Verkhratsky A. Microglia: new roles for the synaptic stripper. Neuron. 2013;77:10–18. doi: 10.1016/j.neuron.2012.12.023. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Kiyatkin A, Bruggeman FJ, Sontag E, Westerhoff HV, Hoek JB. Untangling the wires: a strategy to trace functional interactions in signaling and gene networks. Proc Natl Acad Sci U S A. 2002;99:12841–12846. doi: 10.1073/pnas.192442699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küffner R, Zach N, Norel R, Hawe J, Schoenfeld D, Wang L, Li G, Fang L, Mackey L, Hardiman O, Cudkowicz M, Sherman A, Ertaylan G, Grosse-Wentrup M, Hothorn T, van Ligtenberg J, Macke JH, Meyer T, Schölkopf B, Tran L, Vaughan R, Stolovitzky G, Leitner ML. Crowdsourced analysis of clinical trial data to predict amyotrophic lateral sclerosis progression. Nat Biotechnol. 2015;33:51–57. doi: 10.1038/nbt.3051. [DOI] [PubMed] [Google Scholar]
- Lander AD. The edges of understanding. BMC Biol. 2010;8:40. doi: 10.1186/1741-7007-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik Y. Can a biologist fix a radio?--Or, what I learned while studying apoptosis. Cancer Cell. 2002;2:179–182. doi: 10.1016/s1535-6108(02)00133-2. [DOI] [PubMed] [Google Scholar]
- Le Masson G, Przedborski S, Abbott LF. A computational model of motor neuron degeneration. Neuron. 2014;83:975–988. doi: 10.1016/j.neuron.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Chen P, Aihara K, Chen L. Identifying early-warning signals of critical transitions with strong noise by dynamical network markers. Sci Rep. 2015;5:17501. doi: 10.1038/srep17501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Y, Ji H, Aihara K, Chen L. Personalized characterization of diseases using sample-specific networks. Nucleic Acids Res. 2016;44:e164. doi: 10.1093/nar/gkw772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Silva D, Carriche GM, Castro AG, Roque S, Saraiva M. Balancing the immune response in the brain: IL-10 and its regulation. J Neuroinflammation. 2016;13:297. doi: 10.1186/s12974-016-0763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo DM, Selimkhanov J, Kearns JD, Hasty J, Hoffmann A, Tsimring LS. Dual delayed feedback provides sensitivity and robustness to the NF-κB signaling module. PLoS Comput Biol. 2013;9:e1003112. doi: 10.1371/journal.pcbi.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau A, Da Mesquita S, Kipnis J. Lymphatics in Neurological Disorders: A Neuro-Lympho-Vascular Component of Multiple Sclerosis and Alzheimer's Disease? Neuron. 2016;91:957–973. doi: 10.1016/j.neuron.2016.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makadia HK, Schwaber JS, Vadigepalli R. Intracellular Information Processing through Encoding and Decoding of Dynamic Signaling Features. PLoS Comput Biol. 2015;11:e1004563. doi: 10.1371/journal.pcbi.1004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan GC. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci USA. 2014;111:E5643–E5650. doi: 10.1073/pnas.1408993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MK, Tansey MG. TNF signaling inhibition in the CNS: implications for normal brain function and neurodegenerative disease. J Neuroinflammation. 2008;5:45. doi: 10.1186/1742-2094-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesarovic MD, Sreenath SN, Keene JD. Search for organising principles: understanding in systems biology. Syst Biol. 2004;1:19–27. doi: 10.1049/sb:20045010. [DOI] [PubMed] [Google Scholar]
- Meyer-Hermann M, Figge MT, Straub RH. Mathematical modeling of the circadian rhythm of key neuroendocrine-immune system players in rheumatoid arthritis: a systems biology approach. Arthritis Rheum. 2009;60:2585–2594. doi: 10.1002/art.24797. [DOI] [PubMed] [Google Scholar]
- Meier-Schellersheim M, Xu X, Angermann B, Kunkel EJ, Jin T, Germain RN. Key role of local regulation in chemosensing revealed by a new molecular interaction-based modeling method. PLoS Comput Biol. 2006;2:e82. doi: 10.1371/journal.pcbi.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GM, Ogunnaike BA, Schwaber JS, Vadigepalli R. Robust dynamic balance of AP-1 transcription factors in a neuronal gene regulatory network. BMC Syst Biol. 2010;4:171. doi: 10.1186/1752-0509-4-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman JR, Delos JB, Flower AA, Cao H, Kovatchev BP, Richman JS, Lake DE. Cardiovascular oscillations at the bedside: early diagnosis of neonatal sepsis using heart rate characteristics monitoring. Physiol Meas. 2011;32:1821–1832. doi: 10.1088/0967-3334/32/11/S08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MK, Saez-Rodriguez J, Clarke DC, Sorger PK, Lauffenburger DA. Training signaling pathway maps to biochemical data with constrained fuzzy logic: quantitative analysis of liver cell responses to inflammatory stimuli. PLoS Comput Biol. 2011;7:e1001099. doi: 10.1371/journal.pcbi.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namas RA, Vodovotz Y, Almahmoud K, Abdul-Malak O, Zaaqoq A, Namas R, Mi Q, Barclay D, Zuckerbraun B, Peitzman AB, Sperry J, Billiar TR. Temporal Patterns of Circulating Inflammation Biomarker Networks Differentiate Susceptibility to Nosocomial Infection Following Blunt Trauma in Humans. Ann Surg. 2016;263:191–198. doi: 10.1097/SLA.0000000000001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KM, Bowers WJ. Tumor necrosis factor-alpha mediated signaling in neuronal homeostasis and dysfunction. Cell Signal. 2010;22:977–983. doi: 10.1016/j.cellsig.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Brureau A, Kernan K, Starks A, Gulati S, Ogunnaike B, Schwaber J, Vadigepalli R. Inputs drive cell phenotype variability. Genome Res. 2014;24:930–941. doi: 10.1101/gr.161802.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Ogunnaike B, Schwaber J, Vadigepalli R. Identifying functional gene regulatory network phenotypes underlying single cell transcriptional variability. Prog Biophys Mol Biol. 2015;117:87–98. doi: 10.1016/j.pbiomolbio.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Zhu H, O'Sullivan S, Ogunnaike BA, Weaver DR, Schwaber JS, Vadigepalli R. Single-Cell Transcriptional Analysis Reveals Novel Neuronal Phenotypes and Interaction Networks Involved in the Central Circadian Clock. Front Neurosci. 2016;10:481. doi: 10.3389/fnins.2016.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek P, Ryan S, Ashall L, Sillitoe K, Harper CV, Spiller DG, Rand DA, White MR. Population robustness arising from cellular heterogeneity. Proc Natl Acad Sci U S A. 2010;107:11644–11649. doi: 10.1073/pnas.0913798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, He Y, Li KC, Zhou X. A system mathematical model of a cell-cell communication network in amyotrophic lateral sclerosis. Mol Biosyst. 2013;9:398–406. doi: 10.1039/c2mb25370d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz ML, Weber A, Roxlau T, Gaestel M, Kracht M. Signal integration, crosstalk mechanisms and networks in the function of inflammatory cytokines. Biochim Biophys Acta. 2011;1813:2165–2175. doi: 10.1016/j.bbamcr.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Sheppard PW, Sun X, Khammash M, Giffard RG. Overexpression of heat shock protein 72 attenuates NF-κB activation using a combination of regulatory mechanisms in microglia. PLoS Comput Biol. 2014;10:e1003471. doi: 10.1371/journal.pcbi.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram K, O'Callaghan JP. Divergent roles for tumor necrosis factor-alpha in the brain. J Neuroimmune Pharmacol. 2007;2:140–153. doi: 10.1007/s11481-007-9070-6. [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- Yang Q, Calvano SE, Lowry SF, Androulakis IP. A dual negative regulation model of Toll-like receptor 4 signaling for endotoxin preconditioning in human endotoxemia. Math Biosci. 2011;232:151–163. doi: 10.1016/j.mbs.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, Linnarsson S. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zickenrott S, Angarica VE, Upadhyaya BB, del Sol A. Prediction of disease-gene-drug relationships following a differential network analysis. Cell Death Dis. 2016;7:e2040. doi: 10.1038/cddis.2015.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]