Abstract
Clathrin facilitates vesicle formation during endocytosis and sorting in the trans-Golgi network (TGN)/endosomal system. Unlike in mammals, yeast clathrin function requires both the heavy (CHC) and light (CLC) chain, since Chc1 does not form stable trimers without Clc1. To further delineate clathrin subunit functions, we constructed a chimeric CHC protein (Chc-YR), which fused the N-terminus of yeast CHC (1-1312) to the rat CHC residues 1318-1675, including the CHC trimerization region. The novel CHC-YR allele encoded a stable protein that fractionated as a trimer. Chc-YR also complemented chc1Δ slow growth and clathrin TGN/endosomal sorting defects. In strains depleted for Clc1 (either clc1Δ or chc1Δ clc1Δ), CHC-YR, but not CHC1, suppressed TGN/endosomal sorting and growth phenotypes. Chc-YR-GFP localized to the TGN and cortical patches on the plasma membrane, like Chc1 and Clc1. However, Clc1-GFP was primarily cytoplasmic in chc1Δ cells harboring pCHC-YR, indicating that Chc-YR does not bind yeast CLC. Still, some partial phenotypes persisted in cells with Chc-YR, which are likely due either to loss of CLC recruitment or chimeric HC lattice instability. Ultimately, these studies have created a tool to examine non-trimerization roles for the clathrin LC.
Keywords: Clathrin, endocytosis, membrane trafficking, TGN/endosomal sorting
Introduction
Clathrin-mediated membrane trafficking events are essential for cellular metabolism, signaling and survival. Cytoplasmic clathrin is found as triskelions that consist of three clathrin heavy chains (CHC) trimerized at their C-termini, with each CHC non-covalently associated with a light chain (CLC). The polyhedral assembly of clathrin triskelions on the cytosolic face of membranes provides protein coats for vesicles forming at the plasma membrane, trans-Golgi network (TGN), and endosomes (1).
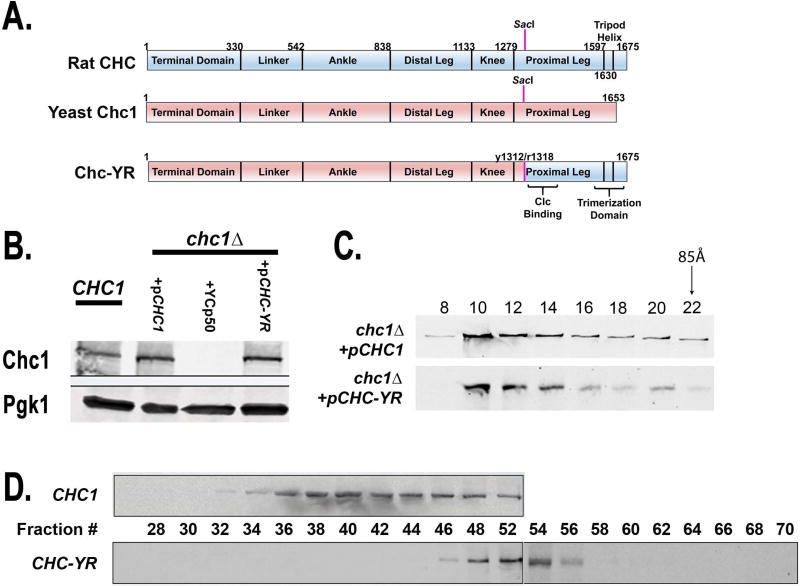
The CHC structure can be divided into several conserved domains (Figure 1A) including: an N-terminal domain (TD), an ankle, a distal leg, a knee, a proximal leg, a trimerization domain and an Hsc70 binding site (2). The final four domains make up a self-assembling fragment called the triskelion hub (3). During coat formation, interactions between the CHC-TD, which forms a seven-bladed β-propeller, and adaptor molecules hold clathrin triskelions at the membrane and these interactions are required for efficient clathrin-mediated transport (4–8). Mutations of the clathrin TD that diminish its affinity for adaptor molecules lead to ‘ephemeral’ endocytic sites that disband prior to membrane invagination (9, 10).
Figure 1.
Yeast-rat clathrin heavy chain chimera (domains and expression). A. An ‘in frame’ fusion of sequences coding for the N-terminus of yeast Chc1 (amino acids 1-1312) and the C-terminus of the rat CHC (amino acids 1318-1675) was created using digestion/ligation at a conserved SacI site. The resultant protein, Chc-YR, combines the yeast terminal domain, linker, ankle, distal leg and knee with the proximal leg and trimerization sequence from rat CHC. B. Immunoblot of cell extracts showing protein expression of Chc1 and Chc-YR. The first lane is from cells expressing CHC1 from its genomic locus (SL1463), followed by chc1Δ yeast harboring pCHC1 (SL6972), YCp50 (SL6971), or pCHC-YR (SL6973). PGK was blotted as a loading control. C. The 100,000 x g supernatant of the cell extract from chc1Δ + pCHC1 (SL7101) or + pCHC-YR (SL7102) was subjected to Superose 6 column chromatography and then fractions were immunoblotted with anti-Chc1 mouse monoclonal antibodies. D. Cell lysates from chc1Δ yeast containing either pCHC1 (SL6972) or pCHC-YR (SL6973) were subjected to the yeast clathrin coated vesicle purification protocol (see Materials and Methods). The 100,000 × g pellet was re-suspended and further fractionated on an S-1000 column. Shown are immunoblot analyses of column fractions probing with anti-Chc1 monoclonal antibodies.
The clathrin light chain is thought to hone clathrin activity by affecting CHC stability, triskelion formation, and lattice assembly. CLCs bind along the HC proximal leg, adjacent to the vertex (11). Although trimerization of mammalian CHCs occurs spontaneously, CLC binding stabilizes mammalian triskelion hub fragments (12). Structural analyses have shown that interaction of CLC with CHC promotes the bending of the CHC knee that is required for lattice assembly (13, 14), and CLC increases the rigidity of the clathrin lattice, which is favorable for membrane deformation during budding (13). CLC binding also inhibits spontaneous lattice assembly (3, 15, 16) and some recent evidence suggests it is positioned in the lattice to restrain uncoating by auxilin and Hsc70 (17). CLC’s are located on the outer surface of the clathrin lattice, such that that they are poised to interact with other cytosolic and regulatory factors (2). Studies in the slime mold Dictyostelium discoideum have shown that CHC can trimerize independently of the CLCs; although cells lacking the CLCA gene still exhibit clathrin deficient-phenotypes (18). These phenotypes could only be alleviated via expression of the C-terminal region of CLC that includes the CHC and calmodulin binding regions, supporting a regulatory role for CLCs (19). In higher eukaryotes neither depleting CLC nor over-expressing a CLC mutant lacking the N-terminus affected endocytosis, although this slowed TGN/endosomal sorting (20–22). This indicates that in animal cells, at least some clathrin functions operate independently of CLC.
The specific roles of Clc1 remain a persistently unresolved question in budding yeast, because Clc1 is needed for Chc1 trimerization and stability (23, 24). Over-expression of the yeast CLC gene, CLC1, suppresses endocytic defects in clathrin HC deficient (chc1Δ) yeast, indicating that CLC possesses CHC-independent endocytic functions (24). This ability requires a region in the CLC N-terminus that binds to Sla2 and suppresses both endocytic and temperature sensitive phenotypes of either chc1Δ or clc1Δ mutants (25, 26). Sla2 and its mammalian homologue, Hip1R, are thought to play a pivotal role during endocytosis since they bind plasma membrane phosphoinositides, the endocytic coat and the actin cytoskeleton (27–29). The N-terminus of Clc1 both binds (26) and negatively regulates Sla2’s ability to interact with F-actin (30). Consistent with findings in yeast, biochemical analysis using mammalian proteins show similar CLC interaction with HIP1/R and CLC regulation of HIP1/R interaction with F-actin (20, 31, 32). Treatments that affect the ability of CLC to interact with Hip1R in animal cells, perturb actin structures at clathrin coated membranes (9, 20, 22, 32). In vivo studies in yeast support this regulatory function of CLC, as a clc1 mutant lacking the Sla2 binding region (clc1-ΔNT) could suppress mutants causing slowed actin assembly or inefficient vesicle constriction needed for scission (30). Together these data suggest that Clc1 controls endocytic progression by regulating the timing and/or location of Sla2 anchoring of the membrane to actin at the invaginating pit.
In light of these endocytic-specific roles of the yeast CLC, we sought a means to further dissect new cellular roles of clathrin LC and HC. Hence, we engineered a clathrin HC chimera that could trimerize independently of yeast Clc1. This construct maintains the N-terminal adaptor interaction region of yeast CHC, but it is fused to the C-terminal segment of rat CHC, which includes the entire trimerization region (11). Our study here demonstrates that this chimeric clathrin allele, CHC-YR, efficiently complements many phenotypes of chc1Δ. CHC-YR, which does not bind to Clc1, largely suppresses endocytic and TGN/endosomal sorting defects in yeast lacking either or both clathrin subunits (chc1Δ, clc1Δ, or chc1Δ clc1Δ). However, despite remarkably complementing chc1Δ clc1Δ, Chc-YR is not able to entirely restore endocytic dynamics. While this could be due to effects on lattice stability, which is affected in vitro, it may also suggest the existence of further CLC-specific functions of clathrin.
Results
Engineering a chimeric clathrin heavy chain allele (CHC-YR) in yeast
Since yeast Chc1 trimerization and stability requires binding to Clc1 (23, 24), whereas mammalian CHC trimerizes independently of the clathrin LC (11, 12, 21, 22), we tested whether mammalian heavy chain could function in yeast and override CHC instability in yeast lacking the clathrin light chain. However, due to difficulties with expression of full length rat CHC in yeast (data not shown), we instead created a novel allele encoding a fusion protein combining the N-terminal two-thirds of yeast CHC with the C-terminus from rat CHC, taking advantage of a conserved SacI restriction site in the coding sequences (33, 34). This new engineered gene (CHC-YR) encoded the terminal domain, distal leg, knee and initial residues of the proximal leg of the Yeast CHC (amino acids 1-1312) fused in frame with the remainder of the Rat CHC (amino acids 1318-1673) including the distal leg, CLC binding region, the trimerization domain and the HSC70 binding site important for auxilin-dependent uncoating (Figure 1A)(35). The CHC-YR yeast/rat chimeric gene was cloned into a centromere-containing plasmid (YCp50) using the native yeast CHC1 promoter. This construct produced a slightly larger protein than wildtype Chc1, since the rat HC is longer (Figure 1B). Chc-YR was produced at similar levels in chc1Δ (Figure 1B) as compared to Chc1 when expressed from plasmids or as compared to chromosomally expressed Chc1 in a wild type strain.
To determine whether Chc-YR could trimerize, cellular extracts from chc1Δ strains expressing either Chc1 or Chc-YR were fractionated on a Superose 6 gel filtration column (Figure 1C). Both Chc1 and Chc-YR eluted as triskelions peaking at fractions 10–14, before the thyroglobulin size marker (fraction 22, 85Å Stokes Radius), but after the void volume (fraction 6). We also found that Chc-YR lattices sedimented in a 100,000 × g vesicle fraction, suggesting that the Chc-YR protein is competent for creating clathrin coats or lattices. But when the Chc-YR 100,000 × g pellet was subjected to S-1000 column fractionation these structures were less stable than those collected from a strain expressing CHC1 (Figure 1D). Chc-YR eluted later on the column in a position more consistent with unassembled clathrin. Hence we next tested for functional complementation.
CHC-YR rescues cell growth and TGN/endosomal sorting defects of chc1Δ
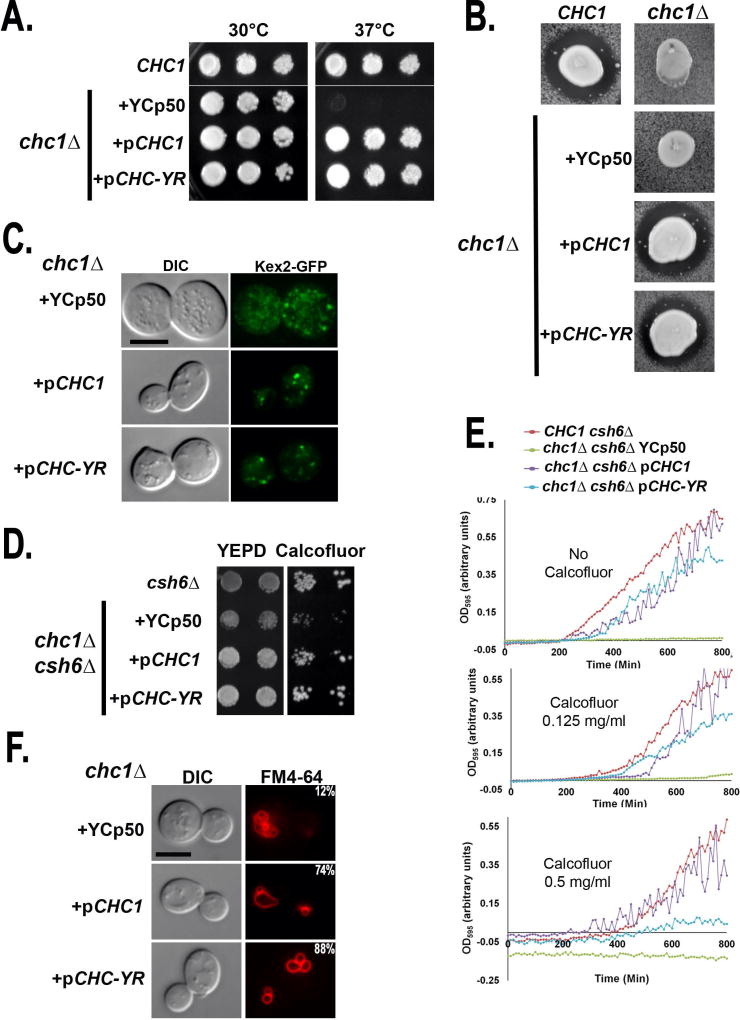
Clathrin null yeast (clc1Δ or chc1Δ) are viable, but temperature sensitive (ts) for growth. We found that chc1Δ strains, which grow poorly at 37°C, could be rescued by either YCp50-CHC1 (pCHC1) or YCp50-CHC-YR (pCHC-YR) (Figure 2A). Clathrin deficiency also causes defects in TGN/endosomal sorting. In chc1Δ MATα yeast, the mating pheromone α-factor processing enzymes, including Kex2, a subtilisin-like protease, are not retained in the TGN resulting in a secretion of an inactive precursor form of α-factor (36, 37). To determine whether Chc-YR rescued the α-factor processing defect of chc1Δ cells, halo assays were performed in which MATα yeast expressing different CHCs were tested for their ability to inhibit the growth of MATa cells supersensitive to mature α-factor. The chc1Δ cells with wildtype pCHC1 or pCHC-YR restored formation of strong halos, indicating secretion of mature α-factor and rescue of proper TGN sorting by the chimera (Figure 2B). Likewise, direct visualization of Kex2-GFP revealed that the numerous small puncta of Kex2 seen in chc1Δ cells were restored to the normal TGN appearance of Kex2 by expression of either CHC1 or CHC-YR (Figure 2C).
Figure 2.
The yeast-rat clathrin heavy chain chimera (CHC-YR) complements chc1Δ. A. Growth of wildtype (SL1463) and chc1Δ yeast harboring YCp50 (SL7100), pCHC1 (SL7101), or pCHC-YR (SL7102) on YEPD at 30°C and 37°C. B. Halo assays for mature alpha factor secretion. MATα wildtype (SL1463), chc1Δ (SL249), and chc1Δ yeast containing YCp50 (SL7100), pCHC1 (SL7101), or pCHC-YR (SL7102) were spotted over a newly seeded lawn of MATa sst1 (BJ3556) and grown at 30oC for 2 days. Shown are tiles from a single plate. Secretion of mature α-factor causes zone of growth inhibition of the tester lawn. C. Kex2-GFP localization in chc1Δ yeast harboring YCp50 (SL7100), pCHC1 (SL7101), or pCHC-YR (SL7102) all transformed with pKEX2-GFP-TRP1. Shown are medial planes from optical z-sections, following nearest neighbor deconvolution. D. Calcofluor White (CFW) sensitivity was tested by growth of chs6Δ (YRV19), and chs6Δ chc1Δ yeast containing YCp50 (SL7103), pCHC1 (SL7104) or pCHC-YR (SL7105) on YEPD ± 150 µg/ml CFW at 30°C. E. Sensitivity to CFW was assessed by dynamic growth analysis measuring optical density (OD595) over 24 hours at 30°C in liquid YEPD containing CFW at concentrations indicated. Strains shown are as in panel D. F. Vacuolar morphology: Vacuoles were stained with FM4-64 in chc1Δ with YCp50 (SL7100), pCHC1 (SL7101), or pCHC-YR (SL7102). Numbers indicate the percentage of cells with vacuoles that have fewer than three lobes (n ≥ 50).
Clathrin is also involved in an AP-1-dependent pathway that transports proteins including chitin synthase III (Chs3) from early endosomes back to the TGN (38). Chs3 resides in both Kex2-containing compartments and at the PM where it localizes to the mother-bud neck in order to deliver a ring of chitin at the site of the emerging buds (39). The delivery of Chs3 from the TGN to the cell surface is dependent upon the Chs5/Chs6 coat complex, hence chs5Δ or chs6Δ yeast have increased intracellular retention of Chs3 and reduced chitin deposition at the bud site (40, 41). This leads to resistance to the toxic effects of the chitin-binding compound Calcofluor White (CFW) (40). Deletion of clathrin or AP-1 subunit genes eliminates intracellular Chs3 retention and restores CFW sensitivity in the chs6Δ background (38). We performed CFW sensitivity growth assays with chc1Δ chs6Δ cells transformed with pCHC1 or pCHC-YR. First, growth assays on plates containing 150 µg/ml CFW revealed that the growth inhibition of chc1Δ chs6Δ appeared equally suppressed by either pCHC1 or pCHC-YR (Figure 2D). We tested this further using a dynamic assay, measuring growth in liquid medium containing CFW ranging as high as 1 mg/ml and found that pCHC1 and pCHC-YR showed similar suppression of calcofluor sensitivity of chc1Δ chs6Δ at or below 250 µg/ml (Figure 2E). However, chc1Δ chs6Δ yeast with pCHC-YR remained sensitive to calcofluor concentrations at or above 500 µg/ml (Figure 2E, data not shown).
Additional trafficking defects in chc1Δ mutants result in fragmentation of the yeast vacuole (42, 43). In chc1Δ vacuoles appear small and multi-lobed with only 12% of chc1Δ yeast containing vacuoles with three or fewer lobes. However, pCHC1 and pCHC-YR suppressed vacuolar fragmentation similarly increasing the percent of cells with vacuoles containing three or fewer lobes to 74% and 88%, respectively (Figure 2F). Both pCHC1 and pCHC-YR also reduced the enlarged cell size often associated with chc1Δ (Figure 2C, F).
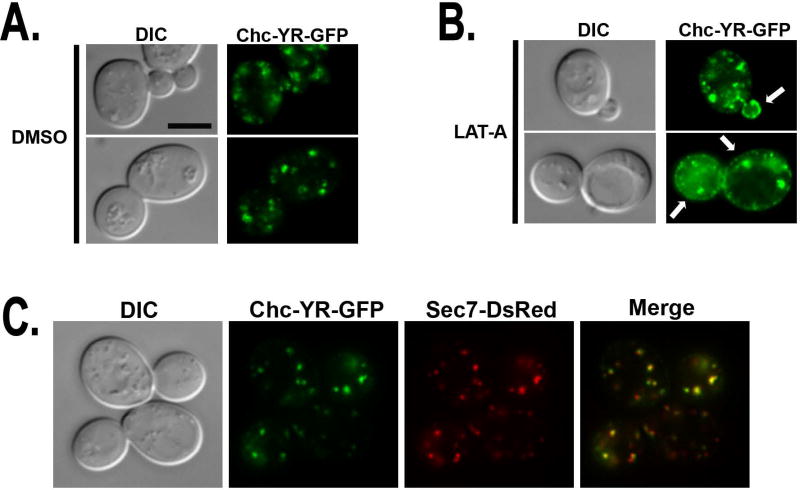
Since pCHC-YR rescued the TGN/endosomal sorting defects in chc1Δ, we expected that the Chc-YR protein would localize to the sites of clathrin function. Indeed, Chc-YR with a C-terminal GFP-tag was found primarily in large cytoplasmic puncta (Figure 3A), similar to the pattern previously seen for wild type Chc1 or Clc1 (see Figure 5A, (10, 44)). Chc-YR-GFP co-localized with the TGN marker Sec7-DsRed (Figure 3C) supporting its ability to perform TGN/endosomal functions like wildtype clathrin HC. It is typically difficult to image clathrin at the plasma membrane given the relative intensity of internal clathrin structures. To exaggerate plasma membrane/endocytic patch localization, we treated cells with Latrunculin A (LatA), a drug that sequesters actin monomers and thus causes clathrin accumulation at stalled endocytic sites. Previously we reported that following 20 minutes of LatA treatment clathrin-containing puncta could be visualized at the cell surface in ~70% of yeast expressing GFP-Clc1 (10, 44). Similarly, ~67% of chc1Δ yeast expressing Chc-YR-GFP demonstrated cortical accumulation of clathrin (Figure 3B).
Figure 3.
Chc-YR localizes to TGN/endosome and endocytic sites. A-B. Representative micrographs of chc1Δ yeast expressing pCHC-YR-GFP:TRP1 (SL7111) grown to log phase and treated for 2 hours with (A) DMSO vehicle control or (B) 200 µM Latrunculin A (LAT-A). C. Representative micrograph of chc1Δ yeast expressing Chc-YR-GFP and the TGN marker Sec7-DsRed (SL7116). Merged images show Chc-YR-GFP co-localizes with Sec7-DsRed.
Figure 5.
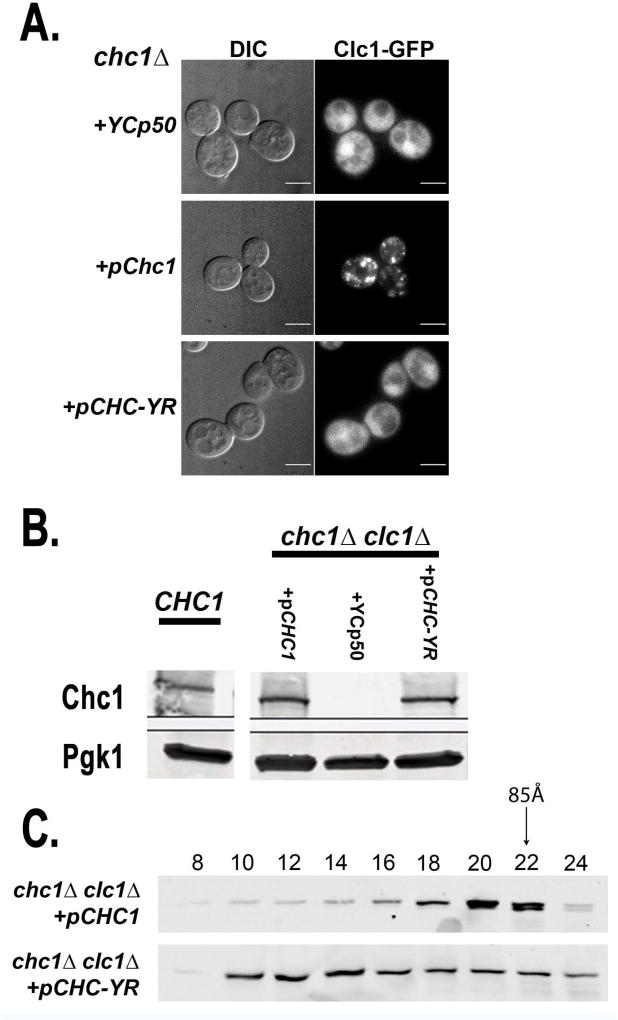
Chc-YR functions independently of clathrin light chain. A. The yeast-rat clathrin heavy chain chimera (Chc-YR) does not recruit Clc1 to membranes, but trimerizes without CLC. A. Clc1-GFP localization in chc1Δ yeast harboring YCp50 (SL6999), pCHC1 (SL7000) or pCHC-YR (SL7001). B. Immunoblot of cell extracts showing protein expression of Chc1 and Chc-YR in the absence of Clc1. From left to right are extracts from cells expressing CHC1 from its genomic locus (SL1463) and chc1Δ clc1Δ yeast harboring either pCHC1 (SL6975), YCp50 (SL6974), or pCHC-YR (SL6976). PGK was blotted as a loading control. Note the first lane (CHC1) is the same as shown in Figure 1B, as all of these samples were run on the same gel. C. The 100,000 xg supernatant of the cell extract from clc1Δ chc1Δ + pCHC1 (SL7108) or +pCHC-YR (SL7109) was subjected to Superose 6 column chromatography and then fractions were immunoblotted with anti-Chc1 mouse monoclonal antibodies.
Chc-YR partially suppresses the chc1Δ endocytic defects
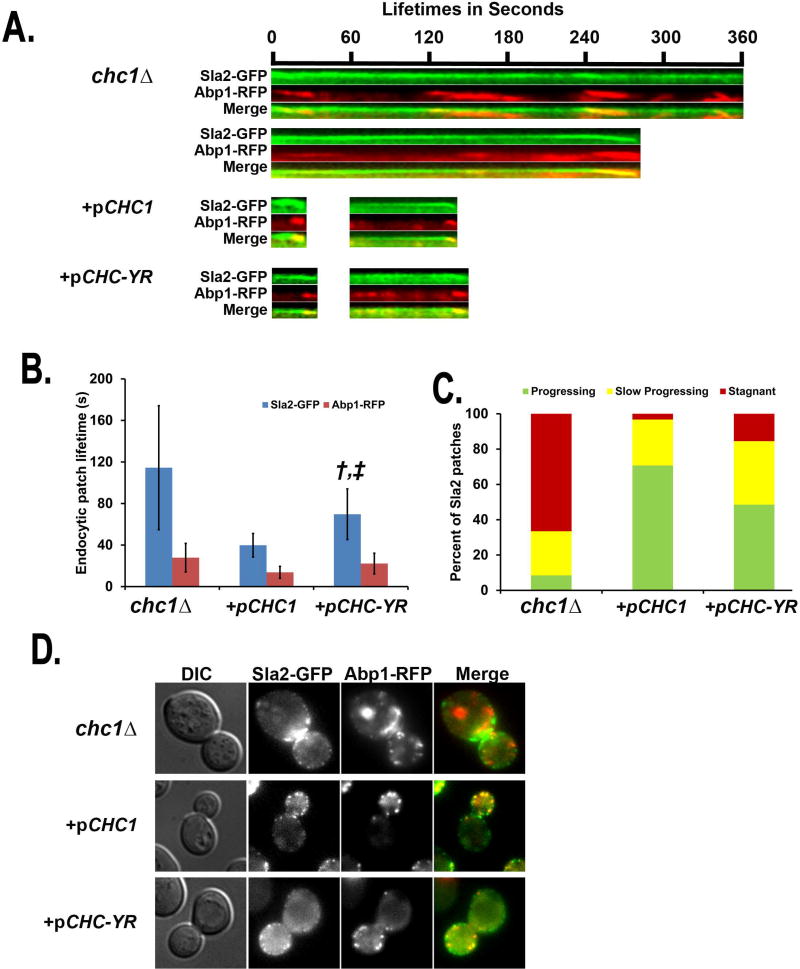
In yeast, clc1Δ or chc1Δ cause a significant impairment in internalization of plasma membrane proteins including the α-factor receptor, Ste2 (23, 24). Clathrin deficiency causes delays in endocytic progression, which can be measured by elongated lifetimes of endocytic patches (27, 45). To visualize whether CHC-YR suppresses the internalization defects of chc1Δ we examined Sla2-GFP as an endocytic coat marker and Abp1-RFP to mark the mobile/actin phase of endocytic vesicle formation by time-lapse microscopy (see Supplemental movies 1–3). The chc1Δ yeast had dramatically elongated lifetimes of Sla2-GFP of 115 ± 59 seconds (Figure 4A,B), which could be restored to wildtype rates by pCHC1 (40 ± 11 sec p ≤ 0.0001). However, this was only partially complemented by pCHC-YR (70 ± 24 seconds, p ≤ 0.0001). Also, chc1Δ leads to an elongation of Abp1-RFP lifetimes (28 ± 14 sec), which was completely suppressed by pCHC1 (to 14 ± 6 sec, ≤ 0.0001), whereas pCHC-YR only partially suppressed this delay (22 ± 10 sec) (Figure 4A,B).
Figure 4.
Dynamic endocytic defects caused by chc1Δ are ameliorated but not entirely suppressed by pCHC-YR. Strains are chc1Δ expressing SLA2-GFP and ABP1-RFP (SL5226), +pCHC1 (SL5386), and +pCHC-YR (SL5729). A. Representative kymographs of the two major categories of Sla2-GFP/Abp1-RFP patches shown in panel C. B. Fluorescence lifetimes of Sla2-GFP and Abp1-RFP. Data are reported as average ± SD (n ≥ 30). † indicates p ≤ 0.0001 compared with pCHC1; ‡ indicates p ≤ 0.0001 compared with chc1Δ. C. Three patch behaviors were seen during six minute movies. Shown are the percentage of patches that showed “normal progression” (including inward movement), “slow progression”, where patches were delayed for longer than two minutes and “stagnant” where patches persisted longer than 4 minutes without internalizing (n = 50). D. Representative micrographs of Sla2-GFP/Abp1-RFP patches.
Endocytic patches in chc1Δ yeast were categorized by three major behaviors: patches that progress normally in 2 minutes (8%); slowly progressing patches that internalize between 2 and 6 minutes (25%); and patches that remain stagnant on the cortex (67%) (Figure 4C). The percentage of endocytic patches that progress normally was dramatically increased by expressing pCHC1 (71%), but less so pCHC-YR (49%) (Figure 4C). There was also a major reduction in stagnant endocytic sites when chc1Δ was complemented with either pCHC1 (3%) or pCHC-YR (16%) (Figure 4C). In addition, expression of wildtype CHC or the yeast/rat chimera also restored actin patch polarization (Figure 4D, Abp1-RFP), although Sla2 appeared more cytoplasmic with the chimera. Overall, the rescue of chc1Δ endocytic phenotypes by Chc-YR was not as complete as for Chc1.
Yeast bearing Chc-YR show reduced requirement for the clathrin LC
Since Chc-YR contains mammalian sequences for LC binding we examined whether yeast LC was associated with Chc-YR using fluorescence microscopy. In chc1Δ cells GFP-Clc1 localization is cytosolic or found in the nucleus (Figure 5A, (44)). CHC1 expression restored GFP-Clc1 to the normal punctate localization of clathrin seen in wildtype yeast (Figure 5A). In contrast, Chc-YR could not direct GFP-Clc1 to these structures and the LC was cytosolic or nuclear, like in cells lacking clathrin heavy chain (Figure 5A).
The lack of GFP-Clc1 localization in cells expressing the chimeric clathrin, combined with the significant functional complementation of chc1Δ, strongly suggested that Chc-YR functions independently of Clc1 and possibly trimerizes in the absence of clathrin LC in yeast. To examine this, we first analyzed the stability of Chc1 and Chc-YR in extracts from cells that lacked both endogenous clathrin genes (clc1Δ chc1Δ). Chc1 and Chc-YR appeared comparably expressed and stable by immunoblot (Figure 5B). We note that in a clathrin LC mutant, CHC1 expressed from its chromosomal locus shows 5–10 fold reduction of Chc1 due to its instability in the absence of CLC (23, 24); however the protein level of Chc1 when expressed from a CEN plasmid in clc1Δ is similar to that of CLC1 CHC1 cells, likely due to compensatory plasmid amplification (24).
We also examined clathrin trimerization in cell lysates from chc1Δ clc1Δ strains expressing each CHC allele by fractionation on the Superose 6 gel filtration column (Figure 5C). In cells without clathrin LC (clc1Δ), Chc1 trimerizes poorly and elutes as a monomeric heavy chain (peak fractions 20–22), as shown previously (24). Nevertheless, the chimeric HC, Chc-YR, still eluted primarily in the triskelion factions even without any CLC.
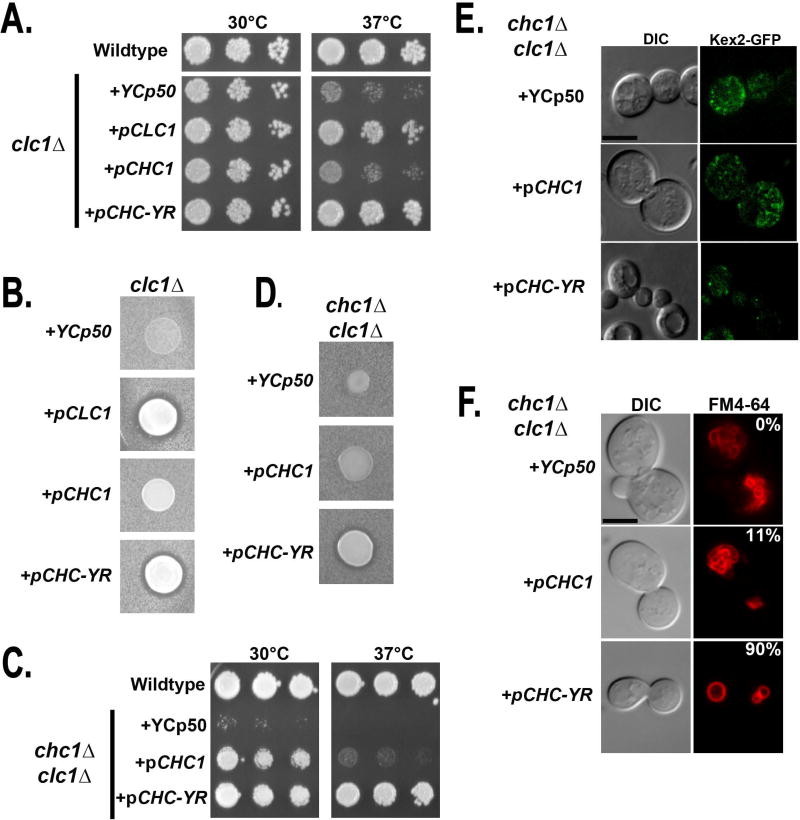
Since Chc-YR trimerizes without CLC (Figure 5C), we next sought to determine if pCHC-YR could entirely bypass the need for Clc1. Like chc1Δ, clc1Δ confers a ts growth phenotype at 37°C. The clc1 mutant was complemented by pCLC1 or the chimeric HC (pCHC-YR), but not pCHC1 (Figure 6A). Halo assays showed that pCHC-YR also suppresses the alpha factor maturation defects of clc1Δ (which also express endogenous CHC1), whereas pCHC1 cannot (Figure 6B). In order to determine if Chc-YR could function as the sole clathrin subunit in the cell, we examined the double knockout strain (clc1Δ chc1Δ) that carried either empty vector YCp50, pCHC1 or pCHC-YR. pCHC-YR dramatically restored growth of clc1Δ chc1Δ yeast at 37°C. In contrast clc1Δ chc1Δ cells with pCHC1 only suppressed the growth defects at 30°C (Figure 6C), while remaining ts at 37°C (Figure 6C). This is the expected phenotype of a clc1Δ strain, in which the trimerization defect of Chc1 is exposed (see Figure 6A). Likewise, mature alpha factor production was restored in clc1Δ chc1Δ strains expressing pCHC-YR, but not pCHC1 (Figure 6D). This was likely due to the rescue of TGN/endosomal sorting, since Kex2-GFP localization was restored in clc1Δ chc1Δ harboring pCHC-YR, but not pCHC1 (Figure 6E). Additionally, the vacuolar fragmentation and cell size defects were rescued in strains harboring pCHC-YR compared to either pCHC1 or empty vector YCp50 (Figure 6F).
Figure 6.
Chc-YR does not require Clc1 to rescue clathrin function. A. Wildtype (SL1463) and clc1Δ yeast harboring YCp50 (SL1916), pCLC1 (SL1915), pCHC1 (SL1917), or pCHC-YR (SL5936) were diluted and grown on YEPD at 30°C and 37°C. B. MATα clc1Δ yeast containing YCp50 (SL1916), pCLC1 (SL1915), pCHC1 (SL1917), or pCHC-YR (SL5936) were spotted onto a lawn of MATa sst1-2 cells (BJ3556). Plates were grown at 30°C for 2 days. Shown are tiles from a single plate. Secretion of mature α-factor causes a zone of growth inhibition of the tester lawn. C. Wildtype (SL1463) and chc1Δ clc1Δ yeast harboring YCp50 (SL7107), pCHC1 (SL7108), or pCHC-YR (SL7109) were diluted and grown on YEPD at 30°C and 37°C. D. MATα chc1Δ clc1Δ yeast containing YCp50 (SL7107), pCHC1 (SL7108), or pCHC-YR (SL7109) were each spotted on a lawn of MATa sst1-2 cells (BJ3556) for α-factor secretion halo assays as described in panel B. Shown are tiles from a single plate. E. pKEX2-GFP-TRP1 was transformed into clc1Δ chc1Δ yeast harboring YCp50 (SL7239), pCHC1 (SL7240), or pCHC-YR (SL7241) and imaged for localization of Kex2-GFP. Shown are medial planes from optical z-sections, following nearest neighbor deconvolution. F. Vacuolar morphology: chc1Δ clc1Δ with YCp50 (SL7107), pCHC1 (SL7108), or pCHC-YR (SL7109) were stained with FM4-64. Numbers indicate the percentage of yeast with vacuoles that have fewer than three lobes (n ≥ 60).
We note that the differences in the ability of pCHC1 and pCHC-YR to suppress clathrin phenotypes in the absence of Clc1 were not merely the result of differences in levels of the clathrin heavy chains, since when expressed from plasmids both were present at levels similar to that found in wild type cells (Figure 1B and 5B; (24)).
Actin phase endocytic defects are not overcome by Chc-YR expression
Previously our lab identified amino-terminal residues (amino acids 19–76) in Clc1 that bind to and negatively regulate attachment of the yeast endocytic factor Sla2 (Hip1R homologue) to F-actin (27, 30). This is thought to relieve tension at the internalizing pit and allow the progressive elongation of the endocytic tubule. Chc-YR is deficient for binding yeast CLC (Figure 5) and does not completely restore the chc1Δ endocytic defects (Figure 4). Therefore, we considered that this endocytic phenotype is associated with loss of CLC function. To examine this, we tested whether these defects are exacerbated in chc1Δ clc1Δ yeast by measuring patch lifetimes of Sla2. As expected, there was a dramatic elongation of Sla2 lifetimes in double mutant strains harboring pCHC1, since without CLC, Chc1 does not trimerize and clathrin HC function is severely impaired (Figure 7A). Importantly, the clc1Δ chc1Δ mutant harboring pCHC-YR showed similar partial rescue of Sla2 lifetime as seen in chc1Δ cells (Figure 7A). Thus, the same phenotype was observed whether endogenous CLC was present or not, consistent with the inability of Chc-YR to bind and target Clc1 to the plasma membrane for its endocytic-specific role(s).
Figure 7.
pCHC-YR only partially suppresses clc1Δ endocytic defects and cannot phenocopy clc1-Δ19-76. A. Fluorescence lifetimes of Sla2-GFP in chc1Δ (SL5226) and chc1Δ bearing pCHC1 (SL5386) or pCHC-YR (SL5729) as previously shown in figure 4B), as well as Sla2-GFP lifetimes of clc1Δ chc1Δ (SL7236) and clc1Δ chc1Δ pCHC1 (SL7237) or pCHC-YR (SL7238). Data are reported as average ± SD (n ≥ 30). B. Percent of cells that internalized Lucifer Yellow (LY) in wildtype (SL1463), clc1-Δ19-76 vrp1Δ (SL6049) and clc1Δ vrp1Δ yeast harboring YCp50 (SL7125), pCHC1 (SL7126), or pCHC-YR (SL7127) following 1 hour of uptake at 30°C, * indicates a p value ≤ 0.05 when compared to vrp1Δ clc1Δ containing the empty vector. Reported are compiled results from 3 independent experiments totaling an n ≥ 100. C. LY uptake in wildtype (SL1463), clc1-Δ19-76 rvs167Δ (SL6052), and clc1Δ rvs167Δ yeast harboring YCp50 (SL7131), pCHC1 (SL7132), or pCHC-YR (SL7133) following 1 hour at 30°C. * indicates a p value of ≤ 0.01 when compared to rvs167Δ clc1Δ containing the empty vector. # indicates a p value of ≤ 0.03 when compared to rvs167Δ clc1Δ expressing CHC1. Reported are compiled results from 3 independent experiments totaling an n ≥ 100.
An N-terminal deletion of the clathrin LC (clc1-Δ19–76) was previously shown to suppress endocytic defects in mutants that: (1) caused slowed actin assembly during the mobile phase of endocytosis (e.g. verprolin mutant, vrp1Δ), or (2) prevented narrowing at the neck of endocytic tubules (e.g. amphiphysin mutant, rvs167Δ) (30). It was hypothesized that this rescue resulted from an inability of the mutant CLC to bind and negatively regulate Sla2, thus prolonging/stabilizing Sla2-mediated attachment between the membrane and actin. The inability of Chc-YR to target LC to sites of CME suggested that the CHC-YR allele may suppress the endocytic defects in vrp1Δ and rvs167Δ to similar degrees as clc1-Δ19–76, by also prolonging attachment between Sla2 and actin. Hence, we examined whether bulk fluid phase endocytosis in vrp1Δ and rvs167Δ could be restored by pCHC-YR using a Lucifer Yellow (LY) uptake assay (Figure 7B,C). Partial suppression was seen in both clc1Δ vrp1Δ and clc1Δ rvs167Δ yeast when transformed with pCHC-YR. With CHC-YR more clc1Δ vrp1Δ cells were LY positive in the vacuole (15%), compared to those expressing an empty vector (8% p ≤ 0.05) or pCHC1 (7%, not significant) (Figure 7B). LY uptake was similarly rescued in clc1Δ rvs167Δ cells expressing the CHC-YR allele (25%) compared to YCp50 (5%, p ≤ 0.01) or pCHC1 (14%, p ≤ 0.03) (Figure 7C). Although Chc-YR provided some rescue, it did not reach the levels seen in cells expressing the clc1-Δ19–76 allele, which yielded 43% and 50% LY positive cells for vrp1Δ and rvs167Δ, respectively. The remaining defect could be due to clathrin lattice assembly/disassembly defects or failure to recruit clathrin LC (see discussion).
Discussion
Although the clathrin LC exists throughout eukaryotes, its proposed functions are diverse. Biochemical studies suggest that mammalian CLC prevents coat assembly (15), stabilizes clathrin hubs (12), and affects uncoating (17, 46). Crystallography and microscopy studies show that LC alters the pitch of clathrin in lattices, providing stability to a range of clathrin coat architectures (13, 14). Mammalian studies suggest that depletion of CLCs using RNAi knockdowns causes defects in cation-independent mannose-6 phosphate receptor (CI-MPR) recycling, delays in Cathepsin-D maturation and actin rearrangements (21, 22), but do not affect internalization of CI-MPR or EGFR or total numbers of CCVs (21, 22). In yeast, our studies have identified endocytic specific roles of CLC, which rely on the N-terminal residues that interact with the Hip1R homologue, Sla2 (24, 26, 30). This work agrees with mammalian clathrin studies demonstrating that binding of CLC to Hip1R alters Hip1R affinity for F-actin (32). However, in contrast to mammalian cells, mutation of the Sla2 binding site on yeast CLC is not associated with disruption of the actin cytoskeleton, nor endosomal sorting (22, 30). It remains possible that these discrepancies are more due to differential uses of actin assembly in membrane traffic between yeast and mammals. TGN/endosomal sorting appears more dependent upon actin in animal cells than in yeast, perhaps do to tighter regulation required by more complex cells. In yeast, actin is essential for internalization, likely due to the attachment needed to overcome the turgor pressure that is maintained by the cell (47). Actin plays a lesser role in mammalian CME, except at sites of cell attachment to the substratum.
A full understanding the yeast CLC function has been complicated by the reliance of yeast clathrin heavy chain on CLC for trimerization and stability (23, 24). Again, this is in stark contrast to CHCs of mammals or Dictyostelium, which assemble trimers independently of CLCs (12, 18). This incongruous role of yeast CLC has impeded our ability to more rigorously test for CLC-specific functions in yeast.
We initially attempted to express the full length rat CHC in yeast, in order to test whether it could complement Chc1 function, thus avoiding the complication of CHC instability in clc1Δ yeast. However, for unknown reasons, this construct was not well expressed and instead we engineered a chimeric allele encoding the N-terminus of yeast Chc1 and the C-terminal rat CHC sequence. This novel yeast/rat chimeric clathrin heavy chain allele (CHC-YR) allowed us to further delineate the roles of clathrin heavy chain and light chain in yeast. This allele produced a stable protein, which was capable of localizing to the same membrane surfaces at the TGN and plasma membrane as wildtype clathrin (10, 44). However, in contrast to wildtype Chc1, it was stable and trimerized in the absence of CLCs. Likewise, we were able to sediment clathrin lattices or clathrin-coated vesicle (CCV’s)/small membranes from cells bearing only the chimeric CHC-YR allele suggesting formation of CCV’s in cells; although, these structures seem less stable in vitro than wildtype CCVs.
Isolation of fragile or incomplete clathrin lattices or CCV’s from Chc-YR cells may be explained by the misalignment of the four key histidine residues in the distal and proximal legs of the clathrin triskelions needed for lattice stability (2, 48). Since the junction of the Chc-YR fusion lies at yeast amino acid 1312, it contains the one conserved histidine residue in the yeast distal leg (H1285, conserved with rat H1279) and two histidine residues (H1458, H1432) in the rat CHC proximal leg. It is possible that these are misaligned or simply not sufficient to maintain stable hydrogen bonding and lattice structure under the conditions of isolation. We surmise that Chc-YR does not directly bind yeast LC, because Chc-YR lacks the yeast CLC binding regions and fails to direct the yeast clathrin light chain to the sites of clathrin transport (TGN/plasma membrane). Thus instability of assembled Chc-YR structures in vitro may also be explained by the need for CLC to maintain the proper clathrin lattice pitch (13, 14).
Remarkably, despite Chc-YR appearing to be less competent to maintain stable lattices in vitro, it suppressed nearly all phenotypes of chc1Δ yeast. Replacing endogenous CHC1 with the CHC-YR allele alleviated defects in TGN/endosomal sorting, including the fragmented vacuolar morphology, mis-localization of Kex2, and failure to produce mature α-factor. Also, expression of this chimeric clathrin restored growth of chc1Δ chs6Δ on Calcofluor White (at concentrations at or below 250 µg/ml), indicating relatively robust retention of chitin synthase III in the cell.
Since we found that Chc-YR does not direct yeast CLC to membrane sites, we examined more closely the requirement for CLC1. In chc1Δ clc1Δ yeast Kex2 localization and α-factor secretion were restored by expression of CHC-YR, but not CHC1. Likewise, CHC-YR expression reversed the vacuolar fragmentation associated with clathrin deficiency, whereas CHC1 expression did not. Taken together, these data suggest that the chimeric CHC functions efficiently at the TGN/endosome independently of the clathrin LC.
Still we found that the CHC-YR allele was less able to restore the endocytic defects in chc1Δ yeast, as compared to CHC1. The Sla2 lifetimes in chc1Δ harboring CHC-YR mirrored those in clc1Δ chc1Δ yeast with CHC-YR, perhaps reflecting the inability of Chc-YR to recruit Clc1 to the endocytic site. We tested if CHC-YR could suppress defects in vesicle scission (rvs167Δ) or actin assembly (vrp1Δ) in the absence of CLC, since it no longer directs the Sla2 regulatory region of Clc1 to the endocytic site (30). As such, we hypothesized that CHC-YR expression might phenocopy the suppression by clc-Δ19–76, which prolongs Sla2 anchoring of actin to the endocytic coat (30). However, the suppression was modest as assessed by LY uptake. Thus, Clc1 is not needed to promote trimerization of Chc-YR, but lasting phenotypes, particularly endocytic defects, persist in cells expressing the chimera without or without Clc1. One explanation could be that CLC has other endocytic specific roles, which are lost when the chimeric HC is unable to deliver CLC to sites of CME. Whether these are directly due to Clc1 is still yet to be proven, and other explanations exist. These residual defects could be due to lattice instability seen with the Chc-YR or even interference of the endogenous CHC. However, we believe the latter is unlikely because chromosomally expressed Chc1 is highly unstable without Clc1. Future studies will be needed to explore these alternative possibilities.
In sum, these studies demonstrate that in yeast, the C-termini of yeast and rat clathrin heavy chains are roughly interchangeable for function despite the latter not binding yeast clathrin LC. When trimerization of CHC is restored artificially, trafficking is largely reestablished in yeast, even in the absence of Clc1. With this chimeric heavy chain and other tools in hand, we can now tease out the other ways that Clc1 is specifically regulating and altering clathrin mediated trafficking in yeast.
Methods
Yeast strains and growth assays
Standard methods and media were employed for genetic manipulations, growth, and transformation of yeast (49). A list of strains and plasmids that were used or generated for these studies are included in Supplemental Table S1.
To perform growth plating assays overnight log-phase liquid cultures were diluted to a starting concentration of 5 × 107 cells per ml and then five-fold serially diluted in 96 well plates. Diluted cells were pinned with a multi-prong frog onto YEPD plates and grown at indicated temperatures for 48 to 60 hours.
To assess Calcofluor White (CFW) sensitivity, log phase cultures were diluted to 107 cells/ml, and serial 4-fold dilutions were spotted onto YEPD plates with or without 150 µg/ml CFW and grown for 48 hours at 30°C. Dynamic CFW sensitivity assays were performed in 96-well format using TECAN Multimode micro-plate reader. Cells were maintained at 30°C in YEPD liquid in CFW concentrations ranging from 0 – 1 mg/ml. Optical density was measured every 10 minutes for 24 hours. Graphs were generated of the resultant OD readings following background correction (subtracted from medium alone).
For the α-factor halo assay, a YEPD plate was first seeded with 5 × 105 BJ3556 cells in an agar overlay to generate a MATa sst1 lawn. Liquid cultures of MATα test cells and controls were diluted to 107 cells/ml and spotted onto the previously seeded plate and then incubated at 30ºC for 48 hours.
Plasmids
Plasmids are listed in Supplemental Table S1B. The yeast-rat clathrin heavy chain chimera clone YCp50-CHC-YR (pCHC-YR) was generated taking advantage of a conserved SacI restriction site in both CHC1 and the rat CHC cDNA resulting in a novel allele encoding amino acids 1-1312 of yeast Chc1 and 1318-1675 of the rat CHC (Figure 1A). To generate YCp50-CHC-YR (pCHC-YR) a 2.49 KB BamH1-Dra1 fragment encoding the C-terminus of the rat CHC was obtained from a rat CHC cDNA clone (gift of T. Kirchhausen) and inserted into YCp50 (URA3) cut with BamH1 and Nru1 to generate pAP8. A BamH1-Sac1 fragment from CHC1 encoding the N-terminal region of the yeast CHC was inserted into pAP8 cut with BamH1 and Sac1 to yield the chimera clone, pCHC-YR. The plasmid encoding the GFP tagged version of CHC-YR was made by homologous recombination between a PCR fragment amplified from pFA6a-GFP-TRP1 and YCp50-CHC-YR, using previously described methods (50, 51). A marker swap was performed to generate pKEX2-GFP-TRP1 using the parent vector pRS426-KEX2-GFP (URA3) (gift of Todd Graham).
Biochemical methods
For immunoblots of Chc1 and Chc-YR, cultures were grown to log phase (5 × 106 cells/ml) at 30°C and 2.0 × 108 cells were lysed by glass bead homogenization in 1ml 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris (pH 8.0) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and a protease inhibitor cocktail (52). Lysates were centrifuged at 4°C for 10 min at 10,000 × g, and total protein concentration of the supernatant was measured via Bradford Assay (Pierce). Equivalent protein amounts were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on 7% polyacrylamide gels (Invitrogen), and immunoblotted using anti-Chc1 mouse monoclonal antibodies (53) or anti-PGK1 mouse monoclonal antibodies (Molecular Probes, Eugene OR) as a loading control. Antibody decoration was detected by an Odyssey Infared Imaging System (LiCor, Lincoln, NE) utilizing IRDye700 or IRDye800 conjugated secondary antiserum (LiCor, Lincoln, NE).
Triskelion analysis was performed essentially as described previously using Superose 6 column chromatography (24). Briefly, cells (~3×109) were grown to log phase in C-Ura medium, pelleted, washed and lysed with glass beads in a Braun homogenizer for 3 minutes in 1 ml Tris buffer A, which contains a 1:1 volume ratio of 1.0 M Tris HCl, pH 7.0:buffer A (0.1 M MES, pH 6.5, 0.5 mM MgCl2, 1.0 mM EGTA, 0.2 mM DTT, 0.02% NaN3) in the presence of protease inhibitors (100 mM TPCK, 500 mM E64, 1 mM benzamidine HCl, 25 mM pepstatin A, 4 mM leupeptin). Extracts were centrifuged for 30 minutes at 29,000 × g and for 1 hour at 100,000 × g. Then 200 µl of the supernatants were analyzed on a 1 cm × 24 cm Superose 6 column (Pharmacia, Sweden) at a flow rate of 0.3 ml/minute, collecting 0.3 ml fractions starting 10 minutes after sample injection. Every other fraction was run on a 7.5% SDS PAGE gel and blotted with anti-Chc1 mouse monoclonal antibodies as described above.
Microscopy and image analysis
All microscopy was carried out on an Olympus fluorescence BX61 upright microscope equipped with Nomarski differential interference contrast (DIC) optics, a Uplan S Apo 100x objective (NA 1.4), a Roper CoolSnap HQ camera, and Sutter Lambda 10-2 excitation and emission filter wheels, and a 175 watt Xenon remote source with liquid light guide. Image capture was automated using Intelligent Imaging Innovations Slidebook 4.01 for the Mac.
To image Chc-YR-GFP, yeast were grown to log phase at 25°C, the treated in synthetic medium supplemented with 200 µM latrunculin A (LAT-A) (Enzo, BML-T119) or similar concentrations of carrier (dimethyl sulfoxide) for 2 hours at 30°C. Cells were mounted in synthetic medium containing 1.6% agarose, still images were captured, and photobleach corrected. Representative micrographs are shown. Kex2-GFP localization was performed on yeast grown to log phase in synthetic medium, then mounted in 1.6% agarose. Captured serial sections of cells were photobleach corrected, and subjected to nearest neighbor deconvolution. Shown are representative medial-planes images. To image vacuolar morphology log-phase yeast were concentrated to 1 × 107 cells per µl and incubated in YEPD with 40 µM FM4-64 stain (Life Technologies) for 20 minutes at 25°C. Yeast were then washed and re-suspended in synthetic medium for 1 hour to concentrate dye at the vacuole. Cells were mounted on coverslips in 1.6% agarose for imaging. Greater than 60 cells per genotype were scored for number of vacuolar lobes.
Live cell imaging of endocytosis was carried out as described previously (54). Cells were grown to log phase at 25°C in synthetic medium, concentrated, immobilized on poly-lysine coated coverslips, mounted on slides in 1.6% agarose in synthetic dextrose medium, and then imaged at 25°C. Following capture, all movies were photo-bleach corrected in Slidebook using the exponential correction function. Average patch lifetimes and standard deviations were determined from 30–40 patches for each strain. The student’s t-test was used to calculate p-values. All kymographs, projection images and example micrographs were generated in Slidebook and then exported to Adobe Photoshop for figure assembly. Lucifer Yellow (LY) (Molecular Probes, Carlsbad, CA) uptake was performed at 30°C for one hour as described previously (55) and 100 cells were counted per genotype.
Supplementary Material
Acknowledgments
We would like to thank Tom Kirchhausen, Todd Graham, Maria Isabel Geli and Greg Odorizzi for plasmids. We thank Alexandra Pellicena-Palle for technical assistance. This work was supported primarily by the National Institute of Health grants to SKL (R01-GM055796), DRB (F32-GM084677) and an institutional training grant supporting VAS (T32-HL07188). Further support was provided by the American Cancer Society fellowship to JC (FL Summer Research Fellowship), American Heart Association Postdoctoral fellowship to JRC (6-6253Y), and a European Molecular Biology Organization Short Term Fellowship to ND (EMBO145-2012).
ABBREVIATIONS
- ts
Temperature Sensitivity
- GFP
Green Fluorescent Protein
- RFP
Red Fluorescent Protein
- CLC
Clathrin Light Chain
- CHC
Clathrin Heavy Chain
- Chc-YR
Clathrin heavy chain Yeast/Rat chimera
- F-Actin
filamentous actin
- CME
Clathrin Mediated Endocytosis
- NT-
N-terminus
- TGN
Trans Golgi Network
Footnotes
The authors have no conflicts of interest to report.
References
- 1.Traub LM. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim Biophys Acta. 2005;1744(3):415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432(7017):573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 3.Liu SH, Wong ML, Craik CS, Brodsky FM. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell. 1995;83(2):257–267. doi: 10.1016/0092-8674(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 4.Dell’Angelica EC. Clathrin-binding proteins: got a motif? Join the network! Trends Cell Biol. 2001;11(8):315–318. doi: 10.1016/s0962-8924(01)02043-8. [DOI] [PubMed] [Google Scholar]
- 5.Drake MT, Traub LM. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J Biol Chem. 2001;276(31):28700–28709. doi: 10.1074/jbc.M104226200. [DOI] [PubMed] [Google Scholar]
- 6.Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat Struct Mol Biol. 2004;11(3):242–248. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- 7.Lundmark R, Carlsson SR. Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J Biol Chem. 2003;278(47):46772–46781. doi: 10.1074/jbc.M307334200. [DOI] [PubMed] [Google Scholar]
- 8.Ramjaun AR, McPherson PS. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J Neurochem. 1998;70(6):2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- 9.Saffarian S, Cocucci E, Kirchhausen T. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 2009;7(9):e1000191. doi: 10.1371/journal.pbio.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collette JR, Chi RJ, Boettner DR, Fernandez-Golbano IM, Plemel R, Merz AJ, Geli MI, Traub LM, Lemmon SK. Clathrin functions in the absence of the terminal domain binding site for adaptor-associated clathrin-box motifs. Mol Biol Cell. 2009;20(14):3401–3413. doi: 10.1091/mbc.E08-10-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ybe JA, Ruppel N, Mishra S, VanHaaften E. Contribution of cysteines to clathrin trimerization domain stability and mapping of light chain binding. Traffic. 2003;4(12):850–856. doi: 10.1046/j.1600-0854.2003.0139.x. [DOI] [PubMed] [Google Scholar]
- 12.Ybe JA, Perez-Miller S, Niu Q, Coates DA, Drazer MW, Clegg ME. Light Chain C-Terminal Region Reinforces the Stability of Clathrin Heavy Chain Trimers. Traffic. 2007 doi: 10.1111/j.1600-0854.2007.00597.x. [DOI] [PubMed] [Google Scholar]
- 13.Dannhauser PN, Platen M, Boning H, Ungewickell H, Schaap IA, Ungewickell EJ. Effect of clathrin light chains on the stiffness of clathrin lattices and membrane budding. Traffic. 2015;16(5):519–533. doi: 10.1111/tra.12263. [DOI] [PubMed] [Google Scholar]
- 14.Wilbur JD, Hwang PK, Ybe JA, Lane M, Sellers BD, Jacobson MP, Fletterick RJ, Brodsky FM. Conformation switching of clathrin light chain regulates clathrin lattice assembly. Dev Cell. 2010;18(5):841–848. doi: 10.1016/j.devcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ungewickell E, Ungewickell H. Bovine brain clathrin light chains impede heavy chain assembly in vitro. J Biol Chem. 1991;266(19):12710–12714. [PubMed] [Google Scholar]
- 16.Ybe JA, Greene B, Liu SH, Pley U, Parham P, Brodsky FM. Clathrin self-assembly is regulated by three light-chain residues controlling the formation of critical salt bridges. EMBO J. 1998;17(5):1297–1303. doi: 10.1093/emboj/17.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young A, Stoilova-McPhie S, Rothnie A, Vallis Y, Harvey-Smith P, Ranson N, Kent H, Brodsky FM, Pearse BM, Roseman A, Smith CJ. Hsc70-induced changes in clathrin-auxilin cage structure suggest a role for clathrin light chains in cage disassembly. Traffic. 2013;14(9):987–996. doi: 10.1111/tra.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Virta VC, Riddelle-Spencer K, O’Halloran TJ. Compromise of clathrin function and membrane association by clathrin light chain deletion. Traffic. 2003;4(12):891–901. doi: 10.1046/j.1600-0854.2003.00144.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Wang Y, O'Halloran TJ. Clathrin light chain: importance of the conserved carboxy terminal domain to function in living cells. Traffic. 2006;7(7):824–832. doi: 10.1111/j.1600-0854.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen CY, Brodsky FM. Huntingtin-interacting protein 1 (Hip1) and Hip1-related protein (Hip1R) bind the conserved sequence of clathrin light chains and thereby influence clathrin assembly in vitro and actin distribution in vivo. J Biol Chem. 2005;280(7):6109–6117. doi: 10.1074/jbc.M408454200. [DOI] [PubMed] [Google Scholar]
- 21.Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279(16):16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 22.Poupon V, Girard M, Legendre-Guillemin V, Thomas S, Bourbonniere L, Philie J, Bright NA, McPherson PS. Clathrin light chains function in mannose phosphate receptor trafficking via regulation of actin assembly. Proc Natl Acad Sci U S A. 2008;105(1):168–173. doi: 10.1073/pnas.0707269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu DS, Pishvaee B, Payne GS. The light chain subunit is required for clathrin function in Saccharomyces cerevisiae. J Biol Chem. 1996;271(51):33123–33130. doi: 10.1074/jbc.271.51.33123. [DOI] [PubMed] [Google Scholar]
- 24.Huang KM, Gullberg L, Nelson KK, Stefan CJ, Blumer K, Lemmon SK. Novel functions of clathrin light chains: clathrin heavy chain trimerization is defective in light chain-deficient yeast. J Cell Sci. 1997;110(Pt 7):899–910. doi: 10.1242/jcs.110.7.899. [DOI] [PubMed] [Google Scholar]
- 25.Henry KR, D'Hondt K, Chang J, Newpher T, Huang K, Hudson RT, Riezman H, Lemmon SK. Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of sla2p in yeast. Mol Biol Cell. 2002;13(8):2607–2625. doi: 10.1091/mbc.E02-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newpher TM, Idrissi FZ, Geli MI, Lemmon SK. Novel function of clathrin light chain in promoting endocytic vesicle formation. Mol Biol Cell. 2006;17(10):4343–4352. doi: 10.1091/mbc.E06-07-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newpher TM, Lemmon SK. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7(5):574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Kaksonen M, Madden DT, Schekman R, Drubin DG. Interaction of Sla2p’s ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol Biol Cell. 2005;16(2):717–730. doi: 10.1091/mbc.E04-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S, Cope MJ, Drubin DG. Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol Biol Cell. 1999;10(7):2265–2283. doi: 10.1091/mbc.10.7.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol Biol Cell. 2011;22(19):3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, McPherson PS. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280(7):6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- 32.Wilbur JD, Chen CY, Manalo V, Hwang PK, Fletterick RJ, Brodsky FM. Actin binding by Hip1 (huntingtin-interacting protein 1) and Hip1R (Hip1-related protein) is regulated by clathrin light chain. J Biol Chem. 2008;283(47):32870–32879. doi: 10.1074/jbc.M802863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchhausen T, Harrison SC, Chow EP, Mattaliano RJ, Ramachandran KL, Smart J, Brosius J. Clathrin heavy chain: molecular cloning and complete primary structure. Proc Natl Acad Sci U S A. 1987;84(24):8805–8809. doi: 10.1073/pnas.84.24.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemmon SK, Pellicena-Palle A, Conley K, Freund CL. Sequence of the clathrin heavy chain from Saccharomyces cerevisiae and requirement of the COOH terminus for clathrin function. J Cell Biol. 1991;112(1):65–80. doi: 10.1083/jcb.112.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchhausen T, Owen D, Harrison SC. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harbor perspectives in biology. 2014;6(5):a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne GS, Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245(4924):1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- 37.Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118(3):531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2(3):283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 39.Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135(3):597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9(6):1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136(1):95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Black MW, Pelham HR. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J Cell Biol. 2000;151(3):587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemmon SK, Freund C, Conley K, Jones EW. Genetic instability of clathrin-deficient strains of Saccharomyces cerevisiae. Genetics. 1990;124(1):27–38. doi: 10.1093/genetics/124.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9(1):87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 45.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123(2):305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 46.Schmid SL, Braell WA, Schlossman DM, Rothman JE. A role for clathrin light chains in the recognition of clathrin cages by ‘uncoating ATPase’. Nature. 1984;311(5983):228–231. doi: 10.1038/311228a0. [DOI] [PubMed] [Google Scholar]
- 47.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11(8):1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bocking T, Aguet F, Rapoport I, Banzhaf M, Yu A, Zeeh JC, Kirchhausen T. Key interactions for clathrin coat stability. Structure (London, England : 1993) 2014;22(6):819–829. doi: 10.1016/j.str.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- 50.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 51.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13(11):1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 52.Stepp JD, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon SK. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol Biol Cell. 1995;6(1):41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemmon S, Lemmon VP, Jones EW. Characterization of yeast clathrin and anticlathrin heavy-chain monoclonal antibodies. J Cell Biochem. 1988;36(4):329–340. doi: 10.1002/jcb.240360403. [DOI] [PubMed] [Google Scholar]
- 54.Boettner DR, D’Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19(23):1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Methods enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.