Abstract
The circadian clock directs many aspects of metabolism to separate in time opposing metabolic pathways and optimize metabolic efficiency. The master circadian clock of the suprachiasmatic nucleus synchronizes to light, while environmental cues such as temperature and feeding out of phase to the light schedule may synchronize peripheral clocks. This misalignment of central and peripheral clocks may be involved in the development of disease and the acceleration of aging, possibly in a gender specific manner. Here we discuss the interplay between the circadian clock and metabolism, the importance of the microbiome and how they relate to aging.
Keywords: Circadian clock, metabolism, microbiome, gender specificity, aging
The circadian clock
Life has adapted to energetic cycles, governed by the Earth’s rotation, by evolving molecular mechanisms that anticipate the most advantageous time of day for biological processes. As a result, the majority of biological functions exhibit daily rhythms. In mammals, these diurnal (Glossary box) oscillations are evoked by autoregulatory transcriptional and translational feedback loops known as circadian clocks [1]. The circadian clock of the suprachiasmatic nucleus (SCN) in the hypothalamus of mammals serves as the central pacemaker at the level of the organism. This has been shown directly in hamsters, where lesioning of the SCN rendered the animals arrhythmic while implantation of brain grafts containing fetal SCN restored circadian rhythms [2]. SCN neurons receive light information from specialized melanopsin expressing intrinsically photosensitive ganglion cells in the retina via the retinohypothalamic tract and synchronize the phase of their circadian clock to the phase of the light [3]. Explanted SCN is capable of maintaining robust circadian rhythmicity for many days in vitro, while peripheral tissues, although rhythmic when explanted, show much less robust rhythms that do not persist as long [4]. This suggests that the circadian clocks in peripheral tissues require continuous entrainment to remain synchronized. The SCN transmits its rhythmic information to other brain regions and peripheral organs via neuronal connections, endocrine signals, body temperature rhythms and indirect cues, provoked by oscillating behavior such as feeding rhythms (Figure 1, Key Figure) [3]. At the molecular level, the transcription factors Brain Muscle Arnt-Like Protein 1 (BMAL1) and Clock Locomotor Output Kaput (CLOCK) or Neuronal PAS domain protein 2 (NPAS2) heterodimerize during the early circadian day, bind to E-box containing elements of gene promoters and induce transcription of downstream genes. Among these genes are Period (Per) and Cryptochrome (Cry) that encode repressors of BMAL1:CLOCK/NPAS2. During the early circadian night, PER and CRY translocate to the nucleus and form large complexes [5] that repress the transcriptional activity of BMAL1:CLOCK/NPAS2, thus downregulating their own expression. Degradation of PER and CRY during the night ends this repression and allows the start of a new transcriptional cycle with a period of approximately 24 hours. In an additional feedback loop, BMAL1:CLOCK/NPAS2 activate the transcription of Rev-erb (also known as nuclear receptor subfamily 1, group D (Nr1d) and RAR-related orphan receptor (Ror) that encode the nuclear receptors REV-ERB and ROR. REV-ERBs inhibit and RORs activate the transcription of Bmal1 [6, 7]. This second feedback loop generates oscillations of Bmal1 mRNA. The rhythmic binding of BMAL1:CLOCK/NPAS2 and REV-ERB/ROR respectively on E-box and REV-ERB/ROR sequences in regulatory elements, drive the rhythmic expression of a substantial fraction of genes in any particular cell or tissue involved in many different functions [8]. The function and timing of the transcriptional feedback loops depends on post-translational modifications regulating the stability and degradation of the transcription factors [9] and epigenomic regulation of their transcriptional activity [10]. This molecular oscillator exists in almost all cells/tissues throughout mammals and the rhythmic expression of genes ultimately generates rhythms of physiological relevance [3].
Figure 1.
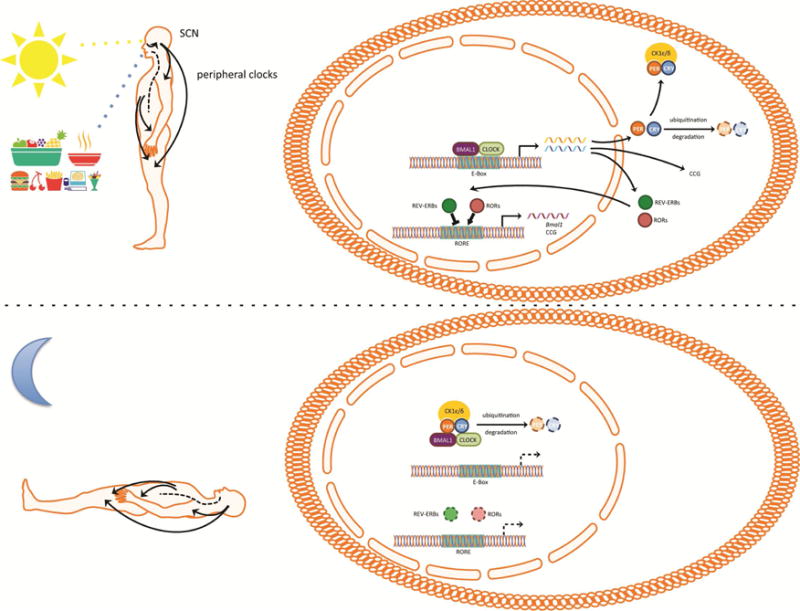
The mammalian circadian clock. Photosensitive melanopsin ganglion cells within the retina relay light information to the neurons of the suprachiasmatic nucleus (SCN). The neurons of the SCN fire with a rhythm of approximately 24 hours that is driven by autoregulatory transcriptional and translational feedback loops known as circadian clock. Circadian clocks exist in most cells and consist of transcriptional activators Brain Muscle Arnt-Like Protein 1 (BMAL1) and Clock Locomotor Output Kaput (CLOCK) that drive the expression of genes during the daytime. Among those genes are Period (Per) and Cryptochrome (Cry) that encode repressors of BMAL1 and CLOCK and Rors, Rev-erbs. PER and CRY form complexes with casein kinase 1 ε/δ (CK1ε/δ) that translocate to the nucleus during early night and repress the transcriptional activity of BMAL1 and CLOCK. Degradation of PER and CRY ends the repression on BMAL1 and CLOCK and allows the initiation of a new transcriptional cycle. Postranslational modifications of PER and CRY delay the transcriptional translational loop to a period of approximately 24 hours. The nuclear receptors RAR-related orphan receptor (ROR) (α, β, γ) and REV-ERB (α, β) translocate to the nucleus to activate and repress the expression of Bmal1. The circadian clocks of the SCN synchronize the clocks of the rest of the body to the same phase to generate 24 hour rhythms of physiology and behavior. Environmental cues such as feeding synchronize peripheral clocks but not the SCN clocks to the time of feeding.
Cellular energy is circadian
Early efforts at identifying genes with rhythmic expression under constant conditions that are under the control of the circadian clock revealed many that encoded proteins involved in metabolic processes [11, 12]. Around the same time, the idea that the circadian clock and the metabolic state of the cell are interconnected was proposed [13]. Since then, many different groups contributed to our current understanding of the crosstalk between the circadian clock and energy state within the cell.
The circadian clock regulates mitochondrial activity through temporal regulation of mitochondrial fission, mitophagy and biogenesis to maintain respiration at times of increased bioenergetic demand [14]. In addition, the circadian clock controls the NAD salvage pathway by imposing rhythms on nicotinamide phosphoribosyltransferase (NAMPT), the enzyme that catalyzes the rate-limiting step in the synthesis of NAD [15]. The circadian clock directed rhythms in NAD biosynthesis drive oscillations in the activity of the NAD+-dependent deacetylases, the sirtuins (SIRTs). As a result, the circadian clock orchestrates SIRT driven cell physiology by dictating oscillations in the energy status of the cell. The rhythmic activity of mitochondrial SIRT3 generates rhythms of mitochondrial oxidative phosphorylation [16]. SIRT3 drives the rhythmic acetylation of mitochondrial proteins to generate rhythms in mitochondrial activity [16]. In turn, the NAD+/NADH redox state of the cell influences the transcriptional activity of BMAL1:CLOCK [13]. SIRT1 binds BMAL1:CLOCK in a rhythmic manner and promotes the deacetylation of clock proteins and histones [17, 18] to create a feedback loop between the redox state and the circadian clock. SIRT6 interacts with the chromatin recruitment of BMAL1:CLOCK to regulate the expression of a set of clock controlled genes distinct from the ones influenced by SIRT1 [19]. SIRT6 also regulates chromatin recruitment of the metabolic transcription factor, sterol response element-binding protein (SREBP1) to control circadian fatty acid metabolism [19]. Aside from mitochondrial activity, ATP cellular levels are directly involved in the crosstalk between the circadian clock and energy state. ATP levels exhibit circadian rhythms in several brain regions including the SCN [20] and the ratio of ATP to AMP regulates the activity of adenosine monophosphate-activated protein kinase (AMPK) [21]. In turn, AMPK phosphorylates and destabilizes CRY1 to promote its degradation [22]. AMPK also phosphorylates casein kinase Iε (CKIε) resulting in increased CKIε activity and degradation of PER2 [23]. AMPK controls the expression of Nampt to increase cellular NAD+ levels feeding into the feedback regulation of the circadian clock through the NAD+/NADH redox state and the activity of sirtuins [24] (Figure 2). Over- and hyper-peroxidation rhythms of peroxiredoxins have been found in human red blood cells [25], which have no nucleus that are found to be driven by hemoglobin auto-oxidation rhythms and are associated with the degradation of hyperoxidized peroxiredoxins [26]. Inhibiting the pentose phosphate pathway, a critical source for NADPH, was found to influence circadian rhythms in cells across species [27], with a recent study showing an effect on the amplitude and phase of the clock [28] This work further supports a feedback from cellular energy metabolism to the transcriptional/translational circadian clock.
Figure 2.
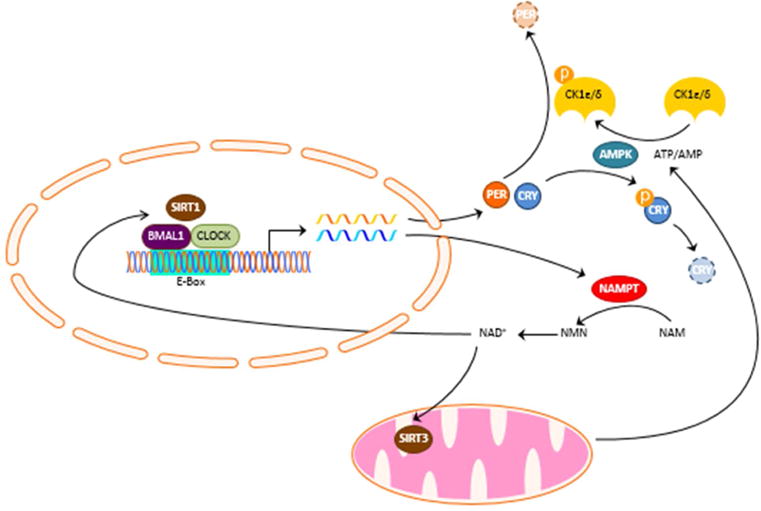
Interplay between the circadian clock and cellular energy state. The circadian clock imposes rhythms of nicotinamide phosphoribosyltransferase (NAMPT) that catalyzes the synthesis of NAD+ from the conversion of nicotinamide (NAM) to nicotinamide mononucleotide (NMN). The rhythmic synthesis of NAD+ drives rhythms in the activity of the NAD+-dependent deacetylases sirtuins (SIRTs). SIRT3 generates rhythms of mitochondrial oxidative phosphorylation by rhythmic acetylation of mitochondrial proteins. Rhythmic ATP production imposes a rhythm in the phosphorylation of casein kinase 1 ε/δ (CK1ε/δ) and Cryptochrome (CRY) by adenosine monophosphate-activated protein kinase (AMPK). SIRT1 binds BMAL1:CLOCK in a rhythmic manner and promotes the deacetylation of clock proteins.
Cellular metabolism is under the control of the clock
The circadian clock regulates cellular metabolism through the control of transcription, by imposing rhythmic expression on genes that encode metabolic enzymes. BMAL1 directly targets genes that encode rate-limiting enzymes such as phosphoenolpyruvate carboxykinase (PEPCK) for gluconeogenesis [29], long-chain fatty acid elongase 6 (Elovl6) for fatty acid biosynthesis and triacylglycerol hydrolase for triglyceride breakdown [30]. Clock proteins control the rhythmic expression or rhythmic activity of other transcription factors, which then impose a rhythm in the expression of metabolically relevant genes. For example, the transcription factors DBP, TEF and HLF are under the control of BMAL1:CLOCK and regulate rhythms in xenobiotic metabolism through the control of the nuclear constitutive androstane receptor [31]. REV-ERBα controls transcription both by direct binding to DNA and indirectly through interaction with other transcription factors [32]. Repression of circadian clock gene transcription requires the DNA binding domain of REV-ERBα, while transcription of lipid metabolic genes is independent of the DNA binding domain and involves tethering of REV-ERBα to DNA by other transcription factors [32]. In addition, REV-ERBα controls the circadian transcription of many factors involved in metabolism. For example, REV-ERBα influences the circadian transcription of Insig2, encoding a trans-membrane protein that sequesters SREBP proteins to the endoplasmic reticulum and thereby controlling their activity to regulate cholesterol and lipid metabolism [33]. PER2 interacts directly with Peroxisome proliferator-activated receptor alpha (PPARα) to rhythmically modulate its target gene G6pc encoding glucose-6-phosphatase [34]. PER2 also blocks Peroxisome proliferator-activated receptor gamma (PPARγ) recruitment to target promoters to promote lipogenesis [35]. CRYs compete with glucocorticoids for the glucocorticoid response element in the phosphoenolpyruvate carboxykinase 1 promoter to regulate the conversion of energy substrates to glucose [36]. Clock proteins also interact with chromatin-modifying factors to achieve cyclical activation and repression of expression. REV-ERBα dictates the genomic recruitment of Histone deacetylase 3 (HDAC3) to direct a circadian rhythm of histone acetylation and expression of genes involved in lipid metabolism [37]. Aside from transcription, the circadian clock regulates rhythmic mRNA translation into protein. Rhythmic phosphorylation of BMAL1 by ribosomal S6 protein kinase 1 mediates the association of BMAL1 with the translational machinery and results in rhythmic protein synthesis [38]. In addition to the transcriptional and translational control of metabolism, the circadian clock controls protein phosphorylation to regulate the timing of metabolic pathways. The activity of kinases downstream of the insulin/insulin-like growth factor 1 (IGF1) receptor, measured as the phosphorylation of their substrates, is high during the active phase [39]. Phosphorylation of proteins by the kinases, glycogen synthase kinase 3 (GSK3) and AMPK, including the rate-limiting enzymes of glycolysis (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3)), glycogen synthesis (glycogen synthase 2 (GYS2)), and fatty acid biosynthesis (acetyl-CoA carboxylase 1 (ACACA)) peaked during the inactive phase [39]. The modulation of key rate-limiting enzymes through phosphorylation permits the activation of glucose anabolic metabolism, protein synthesis during the active phase and the stimulation of glycolysis, repression of glycogen synthesis and inhibition of lipid biosynthesis during the inactive phase. Rhythmic oscillations also exist in the phosphorylation of transcriptional regulators of metabolism, such as the glucocorticoid receptor and oxysterol receptor LXRa [39].
Circadian organization of metabolism at the level of the organism
The profound regulation of cellular metabolism by the circadian clock is reflected at the systemic level of organisms as rhythmic cycles of metabolic processes that can be measured as oscillations of circulating and tissue metabolites. Recent advances in metabolomic technologies allowed the profiling of metabolites across the daily cycle and revealed large scale oscillations of metabolic pathways [40–42]. Lipids oscilate in adipose tissue under the control of the circadian clock [43]. The importance of circadian organization of metabolism has been illustrated by the use of genetic animal models of circadian clock disruption. Early studies showed that universal whole body inactivation of BMAL1 and CLOCK induces hypoglycemia in response to insulin during the inactive phase [29]. Subsequent studies confirmed these findings and showed that hypoglycemia can be at least partially attributed to the inactivation of the liver clock [44, 45]. The role of the circadian clock in maintenance of glucose homeostasis extends to the secretion of insulin by the pancreatic β cells. Inactivation of pancreatic clocks causes glucose intolerance due to defects in the synthesis and secretion of insulin [46, 47]. Circadian organization is critical for energy homeostasis. Both whole body Clock mutants [48] and Bmal1 knockouts [45] develop obesity characterized by increased adiposity. Inactivation of the circadian clock of adipocytes is at least partially responsible for both the increased adiposity and increased body weight [30]. The inactivation of the adipocyte clock compromised a time-dependent feedback from adipose to the brain that appears to be necessary for the timing of feeding behavior [30]. The importance of the circadian clock in metabolic fitness has been demonstrated in liver and heart, where loss of circadian rhythmicity in mitochondrial function leads in reduced respiratory output, increased oxidative damage, and abnormal mitochondrial morphology [14, 49]. Similarly, deletion of the transcriptional activators of the circadian clock in neurons of the central nervous system results in increased oxidative damage in the brain [50].
The importance of the circadian clocks in metabolic fitness raises the question of how the circadian clock may be implicated in the development of the metabolic syndrome. As discussed above, peripheral clocks require regular entrainment to stay in synchrony and are subject to entrainment by both the SCN and cues such as metabolites and temperature that are influenced by changes in activity and behavior. Indeed, both the sleep-wake cycle [51] and feeding patterns [52] have strong effects on circadian metabolic oscillations. It has been suggested that many aspects of the modern lifestyle, such as shift work and jet lag, can throw circadian clocks out of synchrony to accelerate metabolic dysfunction and poise towards the development of the metabolic syndrome [53]. Indeed, time restricted feeding is protective against diet induced obesity and diabetes [54]. Studies of induced circadian misalignment in humans show impaired glucose tolerance, reduced plasma leptin and increased blood pressure [55, 56].
Circadian clock regulation of the microbiota and metabolism
Although the mechanisms by which circadian misalignment causes metabolic dysfunction are largely unknown, one of the changes in response to misalignment that has been implicated in metabolic dysfunction is the change in the enteral microbiota [57, 58]. Several studies of enteral microbiota composition reveal circadian oscillations in the relative abundance of approximately 15% of various bacterial taxa representing 60% of the total gut bacteria [57–62]. At least one species of enteral microbiota, enterobacter aerogenes, expresses diurnal rhythms when isolated outside of the human gut [63]. Enterobacter aerogenes is sensitive to the melatonin secreted into the gastrointestinal lumen and expresses circadian patterns of swarming and motility [63]. Disruption of the circadian clock of the host, either by mutations of clock genes or by jet-lag, abolishes the diurnal oscillation in composition of the intestinal microbiota [58, 59, 62]. Evidently, the control by the host’s circadian clock of enteral microbiota oscillations is mostly dictated by clock imposed diurnal rhythmicity in feeding [57, 58] (Figure 3). Animal models of genetic disruption of the circadian clock display attenuated feeding rhythms and restoring the feeding rhythms rescues the oscillation of enteral microbiota abundance in these animals [58].
Figure 3.
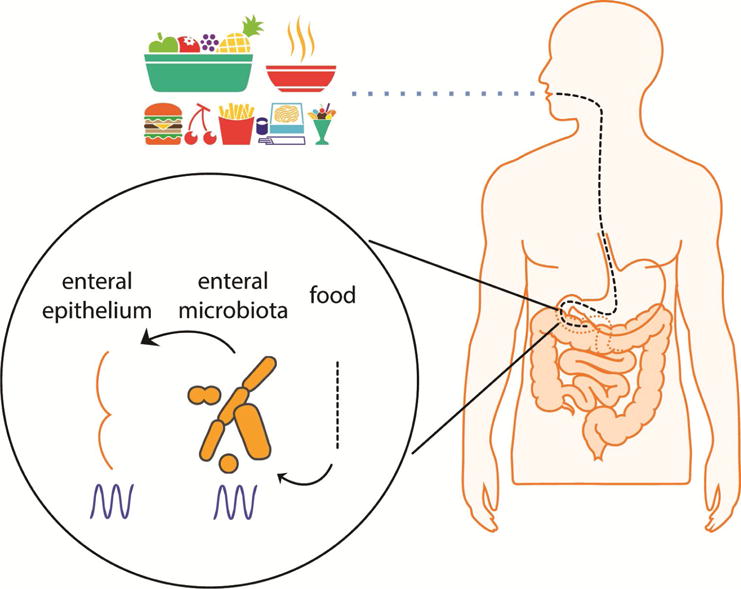
Circadian clock and the enteral microbiome. The circadian clock imposes a rhythm in the abundance of enteral microbiota mainly through feeding rhythms. In turn rhythms in the composition and localization of the enteral microbiota generate rhythmic exposure of the intestinal epithelium to different bacterial species and their metabolites. This rhythmic exposure modulates circadian gene expression in enteral epithelium and remote tissues such as the liver.
Interestingly, there is feedback from the microbiota to the circadian clocks of the host as well. Integrity of the microbiota is essential for normal circadian rhythmicity of gene expression in gut epithelium [64]. Rhythmic expression of toll-like receptors (TLRs) in intestinal epithelial cells generates rhythmic detection of microbial metabolites, which in turn mediate rhythmic TLR signaling and maintains circadian clock function in the cells [64]. In addition, the microbiota exhibits rhythmic localization, adherence to the intestinal epithelium and rhythmic metabolite secretion that determine the rhythmic exposure of the intestinal epithelium to different bacterial species and their metabolites [60, 61]. This rhythmic exposure reprograms chromatin and transcriptional oscillations, not only in the host intestinal cells but also in liver, which then modulates hepatic circadian gene expression and detoxification reactions [60, 61]. Specific microbial metabolites, in particular, short-chain fatty acids such as butyrate, can modulate circadian gene expression in hepatocytes [61]. Experiments in germ-free mice, confirm the requirement of the microbiome for hepatic circadian clock oscillators and identify changes in downstream nuclear receptors that regulate metabolic gene expression [65]. A direct association between circadian oscillations of gut microbiota and metabolic disease has been provided in studies of chronic phase shifting [58]. This maneuver ablates microbiota compositional oscillations and causes the metabolic syndrome in mice, while chronically shifted mice whose gut flora had been eliminated with antibiotics are protected [58]. Similarly, jet-lag-induced dysbiosis between host and microbiota in both mice and humans promotes glucose intolerance and obesity that are transferrable to germ-free mice upon fecal transplantation [58]. It will be interesting to determine whether gender may influence these effects, given the differences in the microbiome between males and females [62]. In addition, the changes in enteral microbiota in response to host circadian clock disruption were different between genders [62]. Male and female mice show significant differences in gut microbiota composition and sex hormones can mediate change in the microbiota [66]. Administration of testosterone after gonadectomy prevented the significant gonadectomy-associated changes in gut microbiota composition and reversed gender specific differences in bile acid composition [66]. Production of testosterone and other steroids requires the circadian clock protein BMAL1 [67]. Bile acids are regulators of intestinal microbiota [68] and their production and secretion is under the control of the circadian clock [69, 70]. Hormones such as testosterone and bile acids may relate to some of the mechanisms that explain gender differences in the microbiome and suggest an interaction between the circadian clock and gender that shapes the circadian rhythmicity and composition of the enteral microbiota. In humans, the gut microbiota differs in men and women at the bacterial phyla level, at the genus level, and at the species level and there is an association between intestinal microbiota composition and obesity that is dependent on gender [71]. Autoimmune diseases have pronounced gender specificity with the majority appearing with higher frequency in women compared to men [72]. Although the direct effect of the microbiome on the development of autoimmune diseases has not been tested in humans, the importance of the microbiota in immune system function is well established (reviewed in [73]). Given that the circadian clock controls several aspects of immunity (reviewed in [74]), the interaction between the circadian clock and the microbiome may be one of the mechanisms responsible for the gender specificity in the development of autoimmune diseases.
Aging
Disrupted metabolism as a result of circadian misalignment may lead to accelerated aging. Rhythmic behaviors begin to fragment with age, suggesting that aging has an adverse effect on the circadian clock. The best example of such rhythmic behavior is sleep that often becomes less consolidated as age progresses. Animals show an impairment of rest-activity rhythms with age, similar to humans. Experiments in flies show that the core clock in the central pacemaker continues to oscillate with robust amplitude in older age [75] despite age-related changes in expression of individual clock proteins [76–78]. Although the SCN is relatively resistant to age at the level of the molecular clock, it undergoes significant age-related degradation at the network level. The total number of SCN neurons is unchanged with aging but elderly rats have significantly fewer vasopressinergic cells [79], a change that is likely to have consequences for downstream signaling. A greater proportion of SCN cells are silent in older animals ex vivo [80] and SCN neurons display decreased circadian phase coherence with age [81]. This results in desynchronization [81] and decreased amplitude of electrical activity rhythms [82] at the network level. Together these are predicted to reduce the strength of output rhythms and could account for the attenuation of behavioral rhythms in older animals.
Although the deteriorating effect of aging on circadian clock output is evident, whether circadian dysfunction promotes aging is less clear. The reduced amplitude in peripheral oscillators with age, whether from loss of intrinsic clock function or decline in entraining signals from the SCN, could contribute to the aging process [83]. Although changes in clock gene expression are still not causally linked to pathology associated with aging, we note that several clock targets are relevant to much of this pathology. The C. elegans BMAL1 homolog, Aryl Hydrocarbon receptor Associated protein (AHA-1) regulates oxidative metabolism and extends lifespan [14]. Disruption of the circadian clock in Drosophila shortens life expectancy after shortterm oxidative stress with higher accumulation of oxidative damage, lower climbing ability and increased neuronal degeneration [84]. Double-mutant flies containing both the period null mutant and a mutation in sniffer, which leads to oxidative stress and a neurodegenerative phenotype, displayed accelerated neuronal degeneration and a reduced lifespan [85]. The Drosophila clock mutants, period and timeless, (homologs of mammalian Per and Cry) are less sensitive to the lifespan-extending effects of dietary restriction (DR) [86]. Together with the observation that DR strengthens circadian oscillations in peripheral tissues, these findings suggest that clocks, at least those in peripheral tissues, are relevant to the determination of lifespan under some conditions.
In mice, disruption of the clock gene Bmal1 predisposes to accelerated aging characterized by premature bone and muscle mass and hair loss, development of cataracts and reduced lifespan [87]. Augmented accumulation of reactive oxygen species occurs in Bmal1 knockout mice and correlates with age-dependent pathology in specific tissues [87]. In support of an increased stress response that leads to accelerated aging, supplementation of these Bmal1 knockout mice with the antioxidant N-acetyl-L-cysteine partially rescues their decreased lifespan and age-dependent pathology [88]. Deletion of the transcriptional activators of the circadian clock Bmal1, or Npas2 together with Clock, predisposes to neurodegeneration, gliosis, loss of inter-regional connectivity and increased oxidative stress in the brain of mice [50]. More recently, postnatal deletion of Bmal1 was found to attenuate some but not all features of the accelerated aging phenotype observed in the conventional knockouts [89]. Most importantly, the short lifespan characterizing conventional Bmal1 knockout mice is normal when deletion of Bmal1 is delayed until the postnatal period. This finding implicates Bmal1 during development in conditioning health for the rest of the lifespan. Further investigation of the role of the circadian clock in aging and the metabolic syndrome is required.
Concluding Remarks
Further studies to improve our understanding of how circadian misalignment predisposes to the metabolic syndrome are needed so that we can get closer to using the circadian clock as a therapeutic tool (see Outstanding Questions). The possibility of improving metabolic health by manipulating the timing of behavior, or the timing of administration of already existing drugs is a very exciting and cost effective therapeutic approach. Small organic molecules that act as ‘clock-reinforcing’ compounds may also prove to be new tools with which to interrogate the complex interplay between obesity and metabolism [90].
Trends Box.
The circadian clock directs rhythms of cellular energy metabolism, while cellular energy status regulates circadian clock driven transcription
Advances in metabolomics and metagenomics allow the quantification of enteral microbiota and permit the characterization of diurnal rhythms in microbiota abundance.
Changes in cellular energy metabolism associated with aging may be driven by reduced amplitude of circadian rhythms in older age.
Glossary Box
- Diurnal
A rhythm recurring every 24 hours under rhythmic environmental conditions. If it characterizes an organism it refers to organisms that are active during the day (in contrast to nocturnal organisms that are active during the night)
- Suprachiasmatic nucleus
Located in the hypothalamus of the brain and positioned directly above the optic chiasm. In mammals, the suprachiasmatic nucleus contains the master circadian clock that maintains synchrony of circadian clocks throughout the organism with the phase of environmental light
- Entrainment
is the process of synchronization of the circadian clock to a specific phase
- Lipogenesis
is the biosynthesis of lipids. Lipogenesis is an anabolic metabolic process that requires energy to be completed
- Enteral microbiota
is the population of microorganisms that reside in the enteral lumen of the digestive system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehman MN, et al. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci. 1987;7(6):1626–38. doi: 10.1523/JNEUROSCI.07-06-01626.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dibner C, et al. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 5.Kim JY, et al. Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol Cell. 2014;56(6):738–48. doi: 10.1016/j.molcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 6.McNamara P, et al. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105(7):877–89. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 7.Preitner N, et al. The orphan nuclear receptor REV-ERB alpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhang R, et al. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehra A, et al. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34(10):483–90. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahar S, Sassone-Corsi P. The epigenetic language of circadian clocks. Handb Exp Pharmacol. 2013;(217):29–44. doi: 10.1007/978-3-642-25950-0_2. [DOI] [PubMed] [Google Scholar]
- 11.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 12.Rudic RD, et al. Bioinformatic analysis of circadian gene oscillation in mouse aorta. Circulation. 2005;112(17):2716–2724. doi: 10.1161/CIRCULATIONAHA.105.568626. [DOI] [PubMed] [Google Scholar]
- 13.Rutter J, et al. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293(5529):510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 14.Jacobi D, et al. Hepatic Bmal1 Regulates Rhythmic Mitochondrial Dynamics and Promotes Metabolic Fitness. Cell Metab. 2015;22(4):709–20. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakahata Y, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peek CB, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342(6158):1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 18.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masri S, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158(3):659–72. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki S, et al. Circadian rhythms of adenosine triphosphate contents in the suprachiasmatic nucleus, anterior hypothalamic area and caudate putamen of the rat—negative correlation with electrical activity. Brain Res. 1994;664(1–2):237–40. doi: 10.1016/0006-8993(94)91978-x. [DOI] [PubMed] [Google Scholar]
- 21.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 22.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–40. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Um JH, et al. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282(29):20794–8. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- 24.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho CS, et al. Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci U S A. 2014;111(33):12043–8. doi: 10.1073/pnas.1401100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rey G, et al. The Pentose Phosphate Pathway Regulates the Circadian Clock. Cell Metab. 2016;24(3):462–73. doi: 10.1016/j.cmet.2016.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putker M, et al. Mammalian Circadian Period, But Not Phase and Amplitude, Is Robust Against Redox and Metabolic Perturbations. Antioxid Redox Signal. 2017 doi: 10.1089/ars.2016.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2(11):e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paschos GK, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18(12):1768–77. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gachon F, et al. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4(1):25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science. 2015;348(6242):1488–92. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Martelot G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmutz I, et al. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24(4):345–57. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimaldi B, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12(5):509–20. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamia KA, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480(7378):552–6. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–9. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipton JO, et al. The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell. 2015;161(5):1138–51. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robles MS, et al. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017;25(1):118–127. doi: 10.1016/j.cmet.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Dallmann R, et al. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109(7):2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnaiah SY, et al. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab. 2017;25(4):961–974 e4. doi: 10.1016/j.cmet.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castro C, et al. A metabolomic study of adipose tissue in mice with a disruption of the circadian system. Mol Biosyst. 2015;11(7):1897–906. doi: 10.1039/c5mb00032g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamia KA, et al. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105(39):15172–7. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi SQ, et al. Circadian disruption leads to insulin resistance and obesity. Curr Biol. 2013;23(5):372–81. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perelis M, et al. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350(6261):aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadacca LA, et al. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54(1):120–4. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohsaka A, et al. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS One. 2014;9(11):e112811. doi: 10.1371/journal.pone.0112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Musiek ES, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013;123(12):5389–400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weljie AM, et al. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci U S A. 2015;112(8):2569–74. doi: 10.1073/pnas.1417432112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106(50):21453–8. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paschos GK. Circadian clocks, feeding time, and metabolic homeostasis. Front Pharmacol. 2015;6:112. doi: 10.3389/fphar.2015.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaix A, et al. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20(6):991–1005. doi: 10.1016/j.cmet.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris CJ, et al. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci U S A. 2015;112(17):E2225–34. doi: 10.1073/pnas.1418955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheer FA, et al. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106(11):4453–8. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarrinpar A, et al. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20(6):1006–17. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–29. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 59.Voigt RM, et al. Circadian disorganization alters intestinal microbiota. PLoS One. 2014;9(5):e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thaiss CA, et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell. 2016;167(6):1495–1510 e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 61.Leone V, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17(5):681–9. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang X, et al. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci U S A. 2015;112(33):10479–84. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paulose JK, et al. Human Gut Bacteria Are Sensitive to Melatonin and Express Endogenous Circadian Rhythmicity. PLoS One. 2016;11(1):e0146643. doi: 10.1371/journal.pone.0146643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mukherji A, et al. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153(4):812–27. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 65.Montagner A, et al. Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci Rep. 2016;6:20127. doi: 10.1038/srep20127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Org E, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7(4):313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvarez JD, et al. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J Biol Rhythms. 2008;23(1):26–36. doi: 10.1177/0748730407311254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Islam KB, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–81. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 69.Ma K, et al. Circadian dysregulation disrupts bile acid homeostasis. PLoS One. 2009;4(8):e6843. doi: 10.1371/journal.pone.0006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferrell JM, Chiang JY. Short-term circadian disruption impairs bile acid and lipid homeostasis in mice. Cell Mol Gastroenterol Hepatol. 2015;1(6):664–677. doi: 10.1016/j.jcmgh.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haro C, et al. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS One. 2016;11(5):e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eaton WW, et al. Epidemiology of autoimmune diseases in Denmark. J Autoimmun. 2007;29(1):1–9. doi: 10.1016/j.jaut.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thaiss CA, et al. The microbiome and innate immunity. Nature. 2016;535(7610):65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 74.Man K, et al. Immunity around the clock. Science. 2016;354(6315):999–1003. doi: 10.1126/science.aah4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo W, et al. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell. 2012;11(3):428–38. doi: 10.1111/j.1474-9726.2012.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolker DE, et al. Aging alters circadian and light-induced expression of clock genes in golden hamsters. J Biol Rhythms. 2003;18(2):159–69. doi: 10.1177/0748730403251802. [DOI] [PubMed] [Google Scholar]
- 77.Wyse CA, Coogan AN. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res. 2010;1337:21–31. doi: 10.1016/j.brainres.2010.03.113. [DOI] [PubMed] [Google Scholar]
- 78.Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153(7):1448–60. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roozendaal B, et al. Changes in vasopressin cells of the rat suprachiasmatic nucleus with aging. Brain Res. 1987;409(2):259–64. doi: 10.1016/0006-8993(87)90710-4. [DOI] [PubMed] [Google Scholar]
- 80.Nygard M, et al. Age-related changes in electrophysiological properties of the mouse suprachiasmatic nucleus in vitro. Brain Res Bull. 2005;65(2):149–54. doi: 10.1016/j.brainresbull.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 81.Farajnia S, et al. Evidence for neuronal desynchrony in the aged suprachiasmatic nucleus clock. J Neurosci. 2012;32(17):5891–9. doi: 10.1523/JNEUROSCI.0469-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakamura TJ, et al. Age-related decline in circadian output. J Neurosci. 2011;31(28):10201–5. doi: 10.1523/JNEUROSCI.0451-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamazaki S, et al. Effects of aging on central and peripheral mammalian clocks. Proc Natl Acad Sci U S A. 2002;99(16):10801–6. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krishnan N, et al. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging (Albany NY) 2009;1(11):937–48. doi: 10.18632/aging.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krishnan N, et al. Loss of circadian clock accelerates aging in neurodegeneration-prone mutants. Neurobiol Dis. 2012;45(3):1129–35. doi: 10.1016/j.nbd.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katewa SD, et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab. 2016;23(1):143–54. doi: 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kondratov RV, et al. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20(14):1868–73. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kondratov RV, et al. Antioxidant N-acetyl-L-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging (Albany NY) 2009;1(12):979–87. doi: 10.18632/aging.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang G, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med. 2016;8(324):324ra16. doi: 10.1126/scitranslmed.aad3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Z, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A. 2012;109(1):101–6. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]


