Abstract
Melioidosis, caused by Burkholderia pseudomallei, is a potentially lethal infection with no licensed vaccine. There is little understanding of why some exposed individuals have no symptoms, while others rapidly progress to sepsis and death, or why diabetes confers increased susceptibility. We prospectively recruited a cohort of 183 acute melioidosis patients and 21 control subjects from Northeast Thailand and studied immune parameters in the context of survival status and the presence or absence of diabetes. HLA-B*46 (one of the commonest HLA class I alleles in SE Asia) and HLA-C*01 were associated with an increased risk of death (odds ratio 2.8 and 3.1 respectively). Transcriptomic analysis during acute infection in diabetics indicated the importance of interplay between immune pathways including those involved in antigen presentation, chemotaxis, innate and adaptive immunity and their regulation. Survival was associated with enhanced T cell immunity to nine of fifteen immunodominant antigens analysed including AhpC (BPSL2096), BopE (BPSS1525), PilO (BPSS1599), ATP binding protein (BPSS1385) and an uncharacterised protein (BPSL2520). T cell immunity to GroEL (BPSL2697) was specifically impaired in diabetic individuals. This characterization of immunity associated with survival during acute infection offers insights into correlates of protection and a foundation for design of an effective multivalent vaccine.
Introduction
Melioidosis is a disease caused by the Gram-negative soil-dwelling bacterium Burkholderia pseudomallei (Bp). A major cause of mortality in Southeast Asia and Northern Australia, it is now recognised as a significant cause of disease in tropical regions worldwide1,2. In-hospital mortality in Northeast Thailand is 40%3. Using a model that mapped cases and environmental Bp, the estimated global burden of melioidosis is 165,000 human melioidosis cases/pa, causing 89,000 deaths2. Furthermore, the model predicts melioidosis under-reporting. Key risk factors are diabetes mellitus, alcohol consumption, chronic renal and lung disease, and increasing age4. A vaccine targeting at risk groups is an urgent unmet need.
Studies in patients and mouse models have sought to identify immune correlates of protection from Bp, but there is a lack of consensus about the qualities required of protective vaccine candidates. Bp is an intracellular pathogen5, so an effective vaccine is likely to require induction of humoral and cellular immunity. T cell immunity to several Bp antigens has been described in exposed individuals6–8 and survivors of acute melioidosis9.
Mapping the immunomics of Bp exposure poses significant challenges: the Bp genome is over seven megabase pairs in size and contains more than 5,000 coding sequences10. Felgner and colleagues used a protein array platform to screen 1,205 proteins11 against 747 individual human sera and identified forty-nine immunoreactive Bp proteins. Based on data indicating that these forty-nine proteins contained both B cell and T cell epitopes, work within the NIH/NIAID-funded Immune Epitope Database (IEDB) consortium then sought to develop this into a systematic approach for Bp T cell antigen discovery, predominantly in HLA transgenic mice and in asymptomatic, seropositive human donors12–14. Of the 49 candidate, T cells antigens, a short-list of 15 was here selected for study, based on ease of synthesis and solubility, and prior experience in terms of their ability to elicit T cell responses. Thus, in the present study, the short-listed, 15 Bp proteins were chosen for screening of T cell immunogenicity by interferon-gamma (IFNγ) ELISpots in patient cohorts from northeast Thailand with culture-confirmed acute melioidosis, and again twelve weeks after recovery, alongside endemic controls seronegative for Bp by indirect haemagglutination assay (IHA). Results were analysed by survival and diabetic status. HLA-B*46, and HLA-C*01, were associated with increased risk of death. T cell immunity to GroEL (BPSL2697) was specifically impaired in diabetic individuals and transcriptomic analysis during acute infection in diabetics demonstrated the importance of immune pathways involved in antigen presentation, chemotaxis, innate and adaptive immunity and their regulation. We identified five protein antigens that are immunodominant in survivors: BPSS1525 (BopE) – a Type III secreted protein, BPSL2096 (AhpC) – the alkyl hydroperoxide reductase, BPSL2520 – an uncharacterised secreted protein, BPSS1385 (ATP binding protein) – a Type III effector protein, and BPSS1599 (PilO) – a Type IV pilus biosynthesis protein. T cell responses to these antigens offer insights into correlates of protection making them attractive candidates for development of a vaccine platform.
Results
Survival following acute melioidosis is associated with enhanced T cell responses to immunodominant Bp antigens
Bp-specific T cell immunity was assessed in 44 melioidosis patients from Ubon Ratchathani, a melioidosis endemic region of Thailand, compared to 21 local, seronegative healthy controls identified from blood donors at Sunpasitthiprasong Hospital. Of the 44 melioidosis patients, 13 died within 28 days of admission to hospital. Peripheral blood was collected during the acute illness and again from survivors at 12 weeks. Diabetic status at the time of diagnosis of melioidosis was recorded.
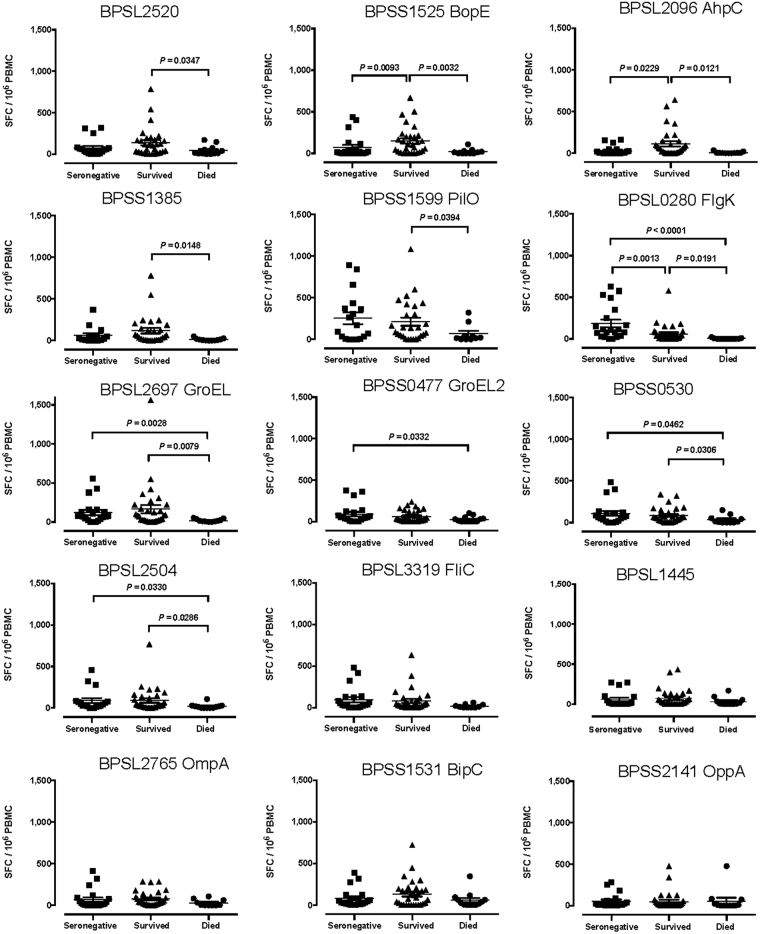
We studied T cell correlates of disease outcome by comparing T cell immunity in patients who died with that of survivors. Bp protein antigen-specific T cell responses at the time of acute infection were lower in patients who died. In particular, T cell responses to 9 of the 15 antigens studied (BPSL2520, BPSS1525, BPSL2096, BPSS1385, BPSS1599, BPSL0280, BPSL2697, BPSS0530, BPSL2504) were significantly lower (Figs 1 and 2). One of these, BPSL2096 (AhpC) has previously been reported by us as a strong antigen across diverse HLA types14.
Figure 1.
Enhanced adaptive immunity to 9 Burkholderia pseudomallei antigens during acute melioidosis in survivors compared to fatal cases. IFNγ ELISpot assays for HIA-Bp or one of 15 Bp antigens were performed on cryopreserved PBMC samples from 18 seronegative controls (black squares), 30 acute melioidosis survivors (black triangles) and 12 fatal cases (black circles). Data shown for each antigen is the number of spot forming cells (SFC) per 106 PBMC following subtraction of the number of SFC from culture with media alone. Error bars represent mean + SEM and statistical significance between groups was determined using the Mann Whitney test.
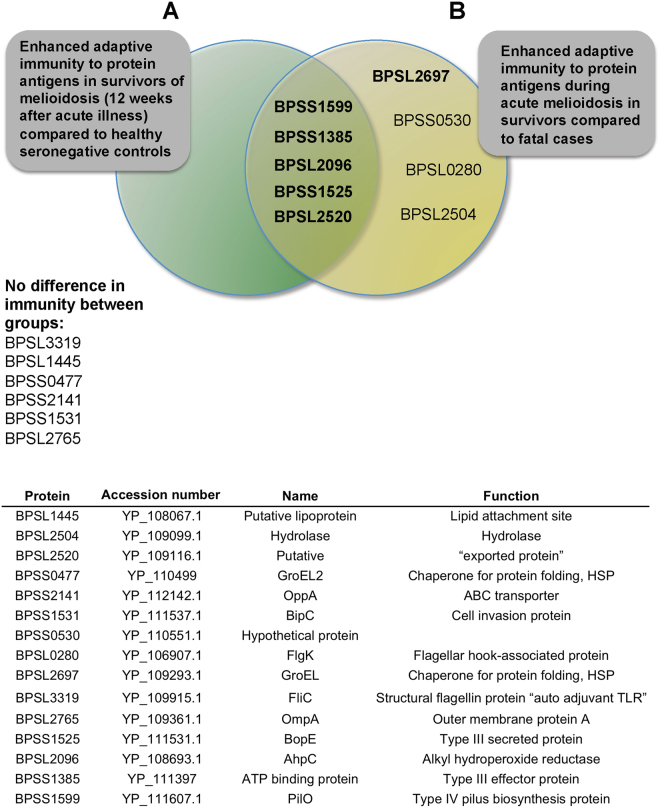
Figure 2.
Enhanced adaptive immunity to Bp antigens are seen in melioidosis survivors, both at the time of acute infection and 12 weeks after recovery from acute infection. The Venn diagram shows the overlap between enhanced T cell responses at the time of acute melioidosis in survivors compared to fatal cases (9 Bp antigens) and at 12 weeks after recovery in survivors compared to seronegative controls (5 Bp antigens). T cell responses to 6 Bp proteins showed no difference between the patient groups tested (A). The table shows the B. pseudomallei strain K96243 protein antigens selected for investigation (B).
Some seronegative controls showed T cell responses to Bp antigens (Fig. 1), possibly reflecting cross-reactivity with environmental species such as B. thailandensis. For example, BPSS1599 (PilO) responses were raised in controls; there is 91% homology between PilO of pseudomallei and thailandensis (http://www.mgc.ac.cn/cgi-bin/VFs/compvfs.cgi?Genus=Burkholderia). Comparisons were made with healthy control subjects from the region with antibody titers by IHA assay below 1:40. However, not everyone exposed to Bp has a positive IHA titre. An Australian longitudinal study of culture-confirmed cases of melioidosis, reported 13% of subjects seronegative (IHA < 1:40)15.
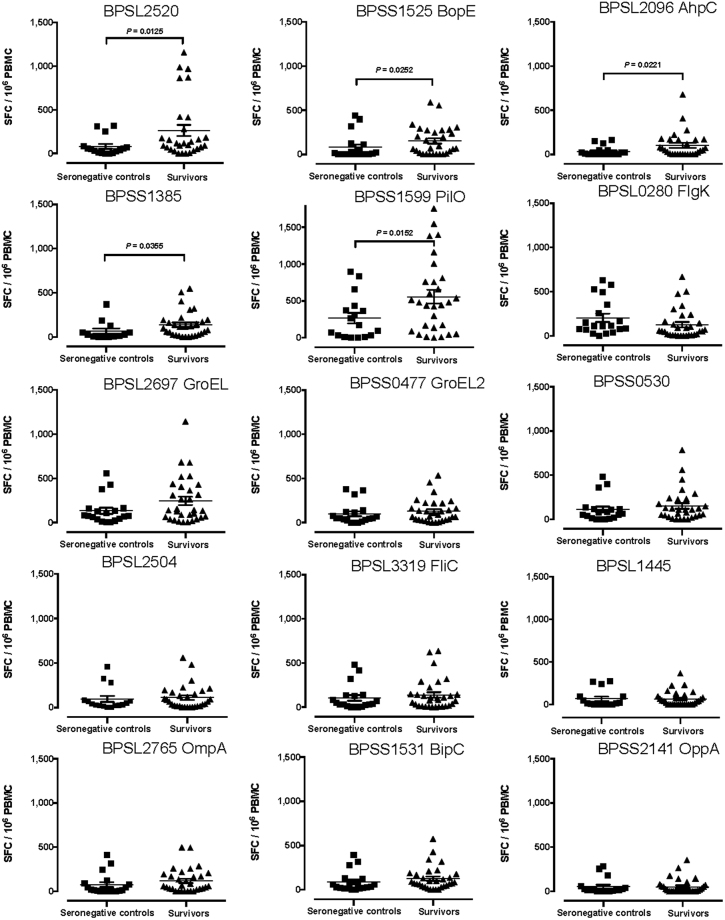
We then analysed T cell responses to Bp protein antigens in patients who survived infection, 12 weeks after admission with acute melioidosis. Five of fifteen Bp antigens tested showed significantly greater responses in convalescent melioidosis patients compared to seronegative controls (Figs 2 and 3). The five protein antigens identified as immunodominant in survivors were: BPSS1525 (Type III secreted protein, BopE), BPSL2096 (alkyl hydroperoxide reductase, AhpC), BPSL2520 (uncharacterised secreted protein), BPSS1385 (type III effector protein, ATP binding protein), and BPSS1599 (Type IV pilus biosynthesis protein, PilO). This is in line with the notion that a large and diverse T cell repertoire controls infection.
Figure 3.
Enhanced adaptive immunity to 5 Burkholderia pseudomallei antigens in survivors of melioidosis, 12 weeks after acute illness, compared to healthy seronegative controls. IFNγ ELISpot assays for HIA-Bp or one of 15 Bp proteins were performed on cryopreserved PBMC from 16 seronegative controls (black squares) and 30 acute melioidosis survivors (black triangles). PBMC samples from melioidosis survivors were taken 12 weeks after acute illness. Data shown for each protein antigen is the number of spot forming cells (SFC) per 106 PBMC following subtraction of the number of SFC from culture with media alone. Error bars represent mean + SEM and statistical significance between groups was determined using the Mann Whitney test.
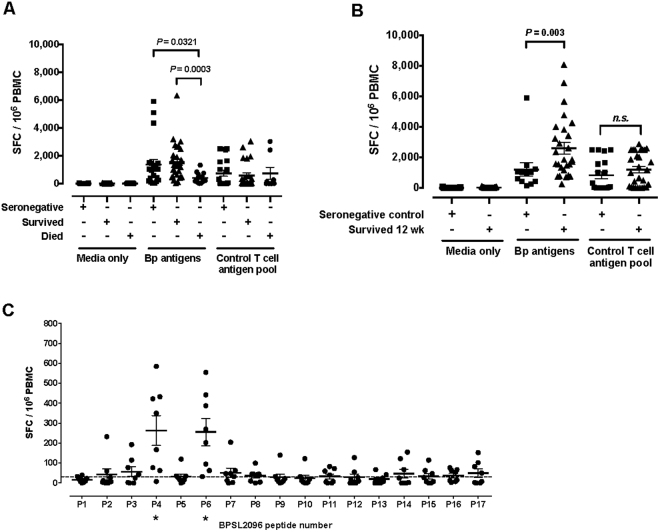
Enhanced immunity 12 weeks after recovery from acute infection is shown in the cumulative antigen responses to all fifteen Bp antigens (Fig. 4). T cell responses at the time of acute infection are higher in survivors than those who died (Fig. 4a), and in recovered individuals 12 weeks after acute infection than in seronegative controls (Fig. 4b). The median, summed IFNу ELIspot response for survivors is 1,301 SFC/106 compared to 234 SFC/106 for fatal cases (p = 0.0003). This is the result of a defect in specific immunity to Bp rather than a wider breakdown of immunity resulting from acute sepsis, since no significant difference was seen for T cell responses to the CEF peptide pool. While some individuals fail to show a response to the CEF peptide pool, it will be noted that in Fig. 4b this non-responsiveness is equally distributed between patients and controls, and that a much smaller number of patients show baseline responses to Bp antigens than to the CEF peptide pool. At 12 weeks there is a higher cumulative frequency of Bp-reactive T cells in convalescent patients (Fig. 4b) than during acute melioidosis.
Figure 4.
Magnitude of T cell responses to Bp antigens in survivors and fatal cases compared to T cell immunity to Bp antigen BPSL2096 is concentrated on 2 peptide epitopes. IFNγ T cell responses to all 15 Bp proteins and to a control T cell antigen pool were summed for each individual in the seronegative control (black squares, n = 18) melioidosis survivor (black triangles, n = 30) and melioidosis fatality (black circles, n = 12) groups. Data are shown for both acute melioidosis (A) and at 12 weeks post acute illness (B). Error bars represent mean + SEM and statistical significance between groups was determined using the Mann Whitney test. IFNγ ELISpot assays were performed for a BPSL2096 20mer peptide panel. A positive response (*) was defined as 2 SD above the mean of the media only control and is represented by a dotted line. Error bars represent mean + SEM (C).
While these IFNγ ELIspot assays were designed primarily to identify CD4 T cell epitopes (exogenous supply antigen-presenting cells [APC] with 20mer peptides that, in the absence of uptake and processing into the class I pathway, are too large for an HLA class I groove) we have previously described Bp antigens presented via class I and II14. CD8 T cell responses to Bp are of interest, this being an intracellular pathogen of APCs where antigen presentation by access to the class I pathway is required for CD8 recognition. We therefore examined the source of the antigen-specific IFNγ signal in responding cells by intracellular cytokine staining (ICS) gating on CD4 or CD8 (Supplementary Fig. 1). Responses were predominantly through CD4 T cells for BPSL2096 AhpC, but either CD4 or CD8 T cells for two of the other antigens tested (BPSL2520 and BPSS1525 (BopE). T cell responses to individual peptide epitopes of BPSL2096 (AhpC) in survivors of melioidosis were evaluated by ex-vivo IFNγ ELIspot assay using 20-mer peptides overlapping by 10 amino acids covering the sequence of BPSL2096 (AhpC) (Supplementary Table 3). Two immunodominant peptides were identified, peptides 4 and 6 of BPSL2096 (AhpC) (Fig. 4c). Peptide 6 from BPSL2096 (AhpC) has previously been reported by us as a strong peptide antigen that can be presented across diverse HLA types14.
Thus, we have identified T cell mediated correlates of protection during acute infection with melioidosis and 12 weeks after recovery.
Antibody recognition of BPSL2096 (AhpC) is a marker of acute melioidosis, while strong antibody responses to BPSS1525 (BopE) are associated with survival
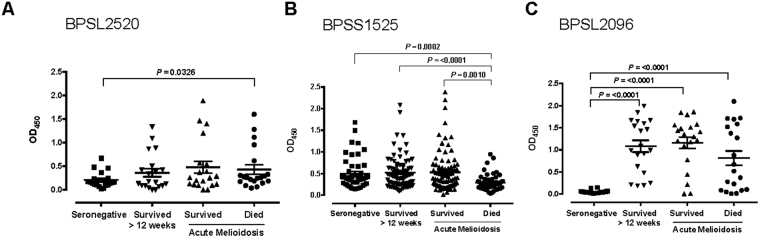
Three of the protein antigens that elicited the strongest T cell responses in survivors, BPSL2520, BPSS1525 (BopE) and BPSL2096 (AhpC), were tested for antibody seroreactivity (Fig. 5a,b,c). Antibody levels were measured for 21 seronegative controls, for 83 surviving melioidosis patients during acute disease and in convalescence at 12 weeks, and for 47 acute melioidosis patients who died. Antibody responses to BPSS1525 (BopE) are associated with survival (p = 0.001) from acute infection, mirroring the differential T cell response to this antigen (Fig. 5b). Strong positive antibody responses to BPSL2096 (AhpC) are associated with Bp infection and a history of melioidosis (Fig. 5c).
Figure 5.
Fatal cases of acute melioidosis show reduced antibody responses to Bp antigen BPSS1525 compared to seronegative controls. Serum samples from seronegative controls (black squares), acute melioidosis survivors (upward black triangles), acute melioidosis fatal cases (black circles) and melioidosis survivors 12 weeks after acute illness (downward black triangles) (n = 20 per group) were used to measure antibody titres against the Bp antigens BPSL2520 (A), BPSS1525 (B) and BPSL2096 (C). Error bars represent mean + SEM and statistical significance between groups was determined using the Mann Whitney test.
The presence of class I alleles HLA-B*46, and HLA-C*01 are associated with increased mortality following acute melioidosis
In line with a possible role of CD8 (and/or NK cell) immunity, we found an increased risk for death from melioidosis in individuals carrying the class I alleles, HLA-B * 46 or HLA-C * 01 (Supplementary Tables 1 and 2). For individuals carrying an HLA- B * 46 allele, the odds ratio (OR) for death from acute melioidosis was 2.8 (95% confidence interval CI 1.3–5.7, p = 0.005). For individuals carrying an HLA- C * 01 allele, the OR for death from acute melioidosis was 3.1 (95% CI 1.5–6.2, p = 0.002).
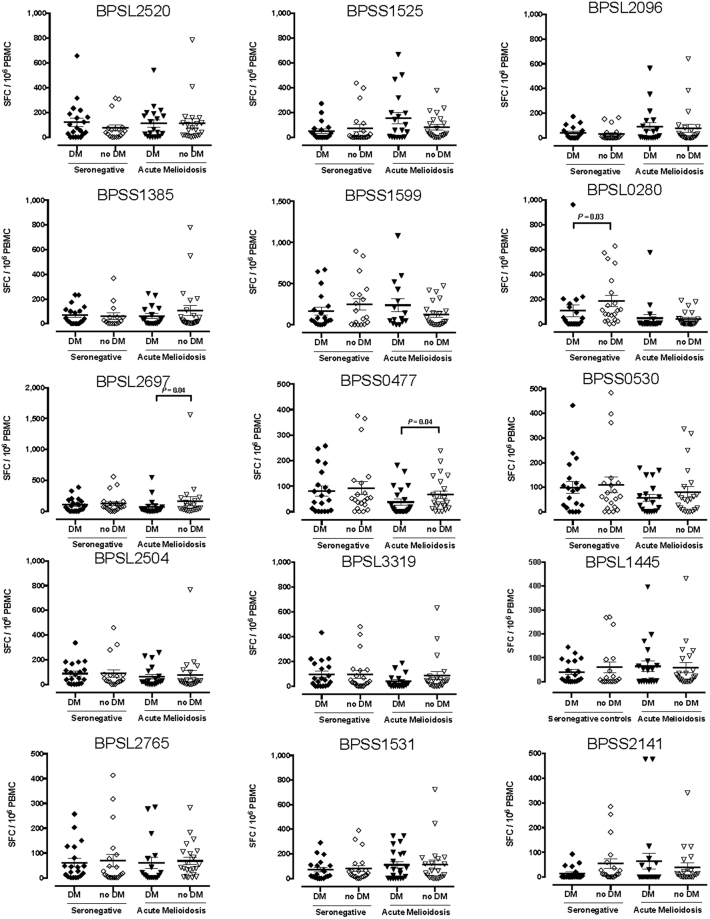
Acute melioidosis in the presence of concurrent diabetes is associated with a reduced T cell response to the GroEL (BPSL2697) and GroEL2 (BPSS0477) stress proteins
Diabetes is a risk factor for melioidosis, although the immune mechanisms underlying susceptibility are not fully understood16. We, therefore, explored functional immune differences between individuals with and without diabetes. Half of the acute melioidosis patients in this cohort had a concurrent diagnosis of diabetes. Diabetic individuals had reduced T cell immunity to two GroEL protein antigens, BPSL2697, GroEL, and BPSS0477, GroEL2 during acute infection compared to non-diabetics (Fig. 6). Altered T cell immunity to the GroEL antigens is of interest, since cross-reactivity between microbial GroEL antigens and mammalian Hsp60 homologues has been implicated in the immunopathogenesis of diabetes17,18. It is hypothetically possible that responsiveness to Bp GroEL antigens was reduced through regulation or exhaustion of responsiveness to the endogenous homologue. To interrogate this premise further, transgenic mice expressing the human HLA-DR1 heterodimer were immunized with recombinant BPSL2697 (GroEL) and T cell peptide epitopes mapped by analyzing recall to a peptide panel comprising BPSL2679 (GroEL) peptides 20aa in length overlapping by 10aa (Supplementary Table 4). Four HLA-DR1 restricted peptide epitopes were identified: p15, p18, p22 and p30 (Supplementary Fig. 2a). HLA-DR1 transgenic mice were then immunized with recombinant BPSL2697 (GroEL) and T cell recall responses analysed against the peptide library for bacterial BPSL2697 (GroEL) or the homologous sequence from human Hsp60 (Supplementary Table 5). There was no evidence to support immunological cross-reactivity at the T cell level in this experiment (Supplementary Fig. 2b,c).
Figure 6.
Diabetic patients with acute melioidosis show a reduced T cell responses to heat shock proteins GroEL and GroEL2 compared to non-diabetic melioidosis patients. IFNγ ELISpot assays for HIA-Bp and one of 15 Bp antigens were performed on cryopreserved PBMC samples from 20 diabetic seronegative controls (black diamonds), 18 non-diabetic seronegative controls (open diamonds), 20 diabetic acute melioidosis patients (black triangles) and 22 non-diabetic acute melioidosis patients (open triangles). Data shown for each antigen is the number of spot forming cells (SFC) per 106 PBMC following subtraction of the number of SFC from culture with media alone. Error bars represent mean + SEM and statistical significance between groups was determined using the Mann Whitney test.
Acute melioidosis in the presence of concurrent diabetes is associated with suppressed immune activation
Diabetic individuals with acute melioidosis show differential and suppressed immune activation compared to those without diabetes. PBMC from 24 patients with acute melioidosis showed greater cytokine release of IL-10 and IL-23 from PBMC of non-diabetic compared to diabetic individuals. There were no differences for IL-17A, IL-4, and IL-13 (Supplementary Fig. 3).
Host immunity to Bp in acute melioidosis in diabetic individuals is associated with distinctive transcriptomic changes
To identify further the transcriptomic changes associated with host immunity to Bp, we cultured PBMC from acutely infected diabetic patients for 18 hours in the presence of either heat-killed Bp (HkBp) or one of three Bp antigens (BPSL2520, BPSS1525, or BPSL2096). Since the presence of diabetes is the major comorbidity associated with increased susceptibility to melioidosis (Supplementary Table 6), our initial focus was to characterise the nature of immune responsiveness during acute infection, within this group. Comparison with non-diabetic donors was not undertaken at this stage due to the confounder of the diverse array of other comorbidities, making this a phenotypically heterogeneous group to study.
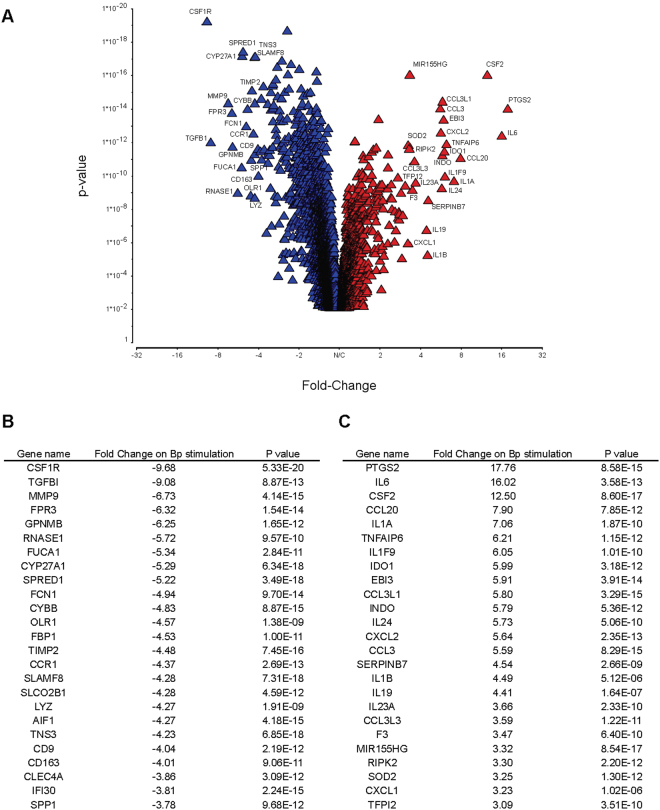
Several of the transcripts up-regulated in response to HkBp were those that might be expected to contribute either to initial innate inflammation or to arming of adaptive immunity and chemotaxis (Fig. 7a,b,c and Supplementary Tables 7 and 8). Within this group are PTGS2/COX2, CSF2, IL-6, IL-1A, IL-23A and CCL20. Up-regulation of MiR-155 is of interest since it is implicated in pro- and anti-inflammatory settings, notably in this context as the regulator of a network that actively orchestrates macrophage survival and T cell immunity in Mtb infection19. The down-regulated transcripts are more functionally diverse, including CSF1R, TGFB1, MMP9 and FPR3.
Figure 7.
Transcriptomic analysis during acute infection in diabetics demonstrates altered regulation of a wide variety of immune related genes following stimulation of PBMC with HIA-Bp. PBMC from acute melioidosis patients were stimulated with either HIA-Bp or media alone for 18 h before extraction of RNA and transcriptomic analysis using an Illumina bead array. Genes significantly up or down-regulated following HIA-Bp stimulation compared to media alone were determined using a paired ANOVA, FDR adjusted with a P value cut off of 0.05. Statistically significant genes are represented in a volcano plot (A) with the top 25 downregulated (red triangle) and 25 upregulated (blue triangle) genes annotated (B,C).
A total of 1,751 genes were differentially expressed in response to each of all three of the Bp antigens tested separately. Pathways identified with high enrichment scores and significant P values included lysosome and phagosome pathways, cytokine and cytokine receptor pathways, Toll-like receptor signalling pathways, and antigen processing and presentation pathways. Notably, there was up-regulation of components of the MHC class I and down regulation of components of MHC class II antigen processing and presentation pathways.
Discussion
Bp is associated with diverse clinical outcomes ranging from asymptomatic seroconversion, to localised infection, sepsis and death. Of around 5,000 proteins encoded in the genome, several hundred are seroreactive. A multi-antigen vaccine platform inducing protective humoral and cellular immunity against Bp likely needs to overcome redundancy in bacterial virulence factors and host evasion strategies. It is important to look beyond reactogenicity and seek host immunity that contributes to protection, as for Salmonella20; hence there is urgent need to characterise those antigen responses involved in protection from a lethal outcome, and generally to define immune correlates of clinical outcome. Target populations for any melioidosis vaccine include individuals with defined risk factors such as diabetes mellitus, making an understanding of the immune deficits in diabetic individuals that predispose to susceptibility to this bacterium extremely important.
Here, we show enhanced T cell immunity to nine specific antigens in individuals who survived the acute infection compared to those who died. Five of these antigens (PilO, ATP binding, AhpC, BopE and the uncharacterised BPSL2520) elicited a memory response in the survivors 12 weeks after acute infection (compared to healthy seronegative controls).
Based on studies in HLA transgenic mice, we have previously highlighted BPSL2096 (AhpC) as a highly immunogenic Bp antigen14. Across several intracellular bacterial pathogens, the alkyl hydroperoxide reductases are up-regulated in response to host oxidative stress, yet this up-regulation exposes them as prominent targets for adaptive immunity. A 40-fold up-regulation of AhpC has been implicated in resistance to high concentrations of reactive oxygen species by B. cenocepacia strains from biofilms, making it a virulence factor for Burkholderia21. AhpC was highly immunogenic for T cells both in our cohort and in a separate cohort of exposed individuals in Khon Kaen, northeast Thailand14. Enhanced T cell immunity to AhpC was observed in survivors compared to fatal cases during acute melioidosis, and in survivors compared to healthy seronegative controls. The pattern of immunodominant T cell epitopes from AhpC was similar to epitope mapping results in HLA-DR and -DQ transgenics14, with p6 immunodominant in human populations and in HLA transgenics. While the case for host-dependent selection pressure is a difficult one to make for Bp, it is noteworthy that p6, a T cell epitope highlighted in the present study, as in our earlier work, was found to have been deleted from a pathogenic, clinical isolate from Cambodia14. Although the Bp immunome is clearly large and complex, the definition of highly immunodominant, common, peptide-HLA complexes offers the potential for developments in future immune monitoring of immune status12–14,22,23. A new tool developed from analysis of Bp pMHC and Pseudomonas aerguniosa 24 pMHC complexes in responder datasets is BIITE, a means of elucidating likely pMHC combinations out of bulk PBMC responses in HLA-heterozygous individuals who will be expressing multiple HLA class II heterodimers to present peptide23. AhpC, and specifically, the p6 epitope from this antigen, stand out as a part of the Bp immunome that is highly visible to the T cell repertoire.
While those individuals who succumbed to fatal infection showed almost complete shutdown of specific T cell responses to Bp antigens, this was pathogen-specific rather than a function of more general immune shutdown in sepsis25, with no differential impairment in responses to control antigens. Thus, while this case is generally argued in terms of systemic immune susceptibility, partly through Treg function, our findings offer an alternate scenario whereby there is a specifically impaired response to Bp antigens, more compatible with the notion of tolerogenic presentation to effector T cells. Individuals with diabetes have altered innate and adaptive immunity to infection, with differences described in several cellular processes including neutrophil, macrophage, NK and lymphocyte function16. There is considerable healthcare concern about the ‘double burden’ of increased risk from the convergence of non communicable diseases (NCDs) and infection: in this case the prevalence of diabetes increasing in Asia in countries which also carry a burden of intracellular bacterial infections such as Burkholderia and tuberculosis26. In this cohort, half of acute melioidosis patients had concurrent diabetes. Diabetes in Northern Thailand has not been extensively characterized, but is typically adult onset, not initially requiring insulin, and frequently occurring in individuals of normal BMI. Improved understanding of the enhanced susceptibility to intracellular infections in diabetics is urgently needed. Here, diabetic individuals showed lower T cell immunity to GroEL proteins during acute melioidosis compared to non-diabetic patients. GroEL proteins, which have been explored as vaccine candidates in Shigella27 are molecular chaperones involved in folding of newly synthesised proteins and maintenance of protein homeostasis28. The eukaryote protein Hsp60 is structurally and functionally highly similar to GroEL. Presumably as a consequence of this homology, autoimmunity to self-Hsp60 following infection can be a feature of autoimmune diseases, including diabetes29; it is interesting in this context that we observed specifically down-regulated responses to Bp GroEL proteins in diabetic individuals. This may indicate immune regulation or exhaustion of this repertoire as a consequence of diabetogenesis, leaving a deficit also in the anti-bacterial response. T cell responses to heat-killed Bp are suppressed in diabetics with acute melioidosis compared to non-diabetics9, and lower responses to GroEL could be part of this mechanism.
Our study showed reduced IL-23 and IL-10 cytokine responses in diabetic individuals, suggesting dysregulated bacterial immunity. Cytokine production by macrophages in response to infection has previously been reported as lower in diabetic mice30 and diabetic people31, with reduced macrophage migration and phagocytosis32. We found decreased responses in healthy diabetics compared to healthy non-diabetic controls to another flagellar subunit protein, BPSL0280 FlgK (Fig. 5), the flagellar hook-associated protein. Antibody responses to FlgK have been noted as significantly raised in recovered melioidosis patients; FlgK is intrinsically cytotoxic to macrophages, though it remains to be seen whether this might facilitate or impair subsequent FlgK presentation33.
HLA genotyping of this acute melioidosis cohort identified increased mortality associated with the presence of HLA-B46 and HLA-C * 01. HLA-B46 and HLA-C * 01 are in linkage disequilibrium. Both of these alleles could function in this context either to present peptide to CD8 T cells or to interact with inhibitory killer immunoglobulin-like receptors (KIR) of NK cells. The presence of these alleles is associated with a lethal outcome, suggesting highly impaired host immunity. HLA-B46 is one of the most common HLA-B alleles in SE Asia. However, it is associated with several negative disease outcomes, including enhanced risk of cerebral malaria34. It has been proposed that HLA-B46 has a novel peptide binding groove, presenting a highly constrained peptide pool35.
Transcriptomic studies of murine infection have been reported36, and an analysis of unstimulated whole blood from patients with acute melioidosis has previously demonstrated a gene expression signature dominated by interferon-signalling pathways similar to tuberculosis infection37. Our analysis considered Bp specific immunity during acute melioidosis in diabetics. There is a strong picture of early integration and up-regulation of innate and adaptive immune pathways. This includes innate cytokines such as IL-1A, TNFA1 and IL-6, as well as cytokines polarising adaptive Th17 immunity such as IL-23A. Bp infection of primary human monocytes has been shown to activate gene expression of IL-2338,39. This is somewhat different from a recent report of cytokine transcription in unstimulated PBMC of melioidosis cases, which emphasised global up-regulation of Th17 and Th2 pathways39. Interestingly, transcripts associated with regulation of adaptive immunity, such as IDO-1 and IL-24, are activated. Several key changes relate to chemotaxis or mobilisation of appropriate subsets – CSF2, CCL20, CXCL2 and CCL3. Further studies will be required to interrogate the specific functional implications of these pathways for acute immune responses to Bp. The strong up-regulation of the immunomodulatory microRNA, MiR-155, is reminiscent of observations in M. tuberculosis infection of macrophages19. In that setting a dual activity of MiR-155 was proposed, both maintaining the survival of infected macrophages and promoting survival of responding T cells. It is also noteworthy that PTGS2/COX2 was so strongly up-regulated, since two studies have shown impaired host defence against Bp through COX2/PGE240,41. Our new transcriptomic findings offer a mechanistic rationale for the efficacy of COX-2 inhibitors, tolfenamic acid (TA) and NS398, in the post-exposure protection of Bp challenged mice, and support further efforts to develop clinical therapeutics targeting the COX2 pathway.
In summary, we have characterised immune correlates of protection in a cohort of acute melioidosis patients. HLA-B46 and HLA-C*01, were associated with increased mortality. We identified antigens and epitopes, immunodominant in survivors compared to endemic controls: BPSS1525 (BopE) – a Type III secreted protein, BPSL2096 (AhpC) - the alkyl hydroperoxide reductase, BPSL2520 – an uncharacterised secreted protein, BPSS1385 (ATP binding protein) a Type III effector protein, and BPSS1599 (PilO) – a Type IV pilus biosynthesis protein and peptides 4 and 6 from BPSL2096 (AhpC). Antibody responses to BPSL2096 (AhpC) were associated with a history of acute melioidosis, while strong antibody responses to BPSS1525 (BopE) were associated with survival. Impaired responses to GroEL proteins were demonstrated in diabetics during acute infection. Transcriptomic analysis of acute infection in diabetics illuminates the interplay of pathways controlling chemotaxis, antigen presentation, innate and adaptive immunity, as well as their regulation. Overall, differential host immunity to these antigens offers insights into correlates of protection, making them attractive candidates for development of a vaccine platform.
Materials and Methods
Ethics statement
Mouse experiments were performed in accordance with UK Home Office legislation under the terms of Home Office Project License Project License PPL 70/7708, Imperial College London UK granted for this work under the “Animals (Scientific Procedures) Act 1986”. Local ethical review and formal approval was also obtained through the Imperial College Ethical Review Process Committee.
Human studies and consent forms were approved by the ethics committees of Faculty of Tropical Medicine, Mahidol University, of Sunpasitthiprasong Hospital, Ubon Ratchathani and the Oxford Tropical Research Ethics Committee. The study was conducted according to the principles of the Declaration of Helsinki (2008) and the International Conference on Harmonization (ICH) Good Clinical Practice (GCP) guidelines. Written informed consent was obtained for all patients enrolled in the study.
HLA genotyping
Genomic DNA was isolated from peripheral blood by high salt extraction. HLA‐A, B, and C loci were typed by SSOP PCR.
Subjects
For human immunology studies, blood samples were collected from 183 patients over 18 years of age with cultured-confirmed melioidosis at Sunpasitthiprasong Hospital, Ubon Ratchathani at a median of 5 days after admission to hospital (IQR 4–6 days), and from 83 of these patients after recovery 12 weeks later (Supplementary Table 1). 44/183 patients (24%) died within 28 days. This is in line with a larger published cohort from the same hospital9. Healthy control subjects were recruited from the blood donation clinic at Sunpasitthiprasong Hospital, and 21 subjects were selected as seronegative controls for melioidosis if their indirect hemagglutination assay (IHA) titre was <1:4015,42. Of the 183 patients with a diagnosis of acute melioidosis studied on enrolment (week 0), 123 subjects (67%) had a diagnosis of diabetes (defined for the purpose of this study as a past medical history of diabetes and / or a blood glycated haemoglobin (HbA1c) of ≥7%. For patients who did not attend follow-up, their 28-day survival status was determined using hospital mortality records and contact by telephone. Here, we analyse immune correlates of host immunity to Bp in a total cohort of 189 acute melioidosis patients, 44 of whom died (Table S1). Immune correlates of mortality were characterised at the level of HLA class I alleles (n = 189), T cell IFNγ response to fifteen immunodominant Bp antigens (n = 44), T cell epitope mapping (n = 26), and antibody responses (n = 130). Cytokine response profiles (n = 44) and comprehensive transcriptomics of antigen-stimulated PBMC from infected diabetic individuals (n = 13) were also studied.
Antigens and peptides
Fifteen antigens from B. pseudomallei strain K96243 were selected for investigation (Fig. 2). Protein sequences were codon optimised, sub-cloned into an E-Coli expression vector, and purified using a His-Tag. Proteins were checked by SDS-PAGE and western blot, using an anti 6-His antibody. All proteins were produced by Biomatik (Biomatik USA, Delaware, USA). BPSS1599 was produced as previously described (Lassaux et al. 2014 PMID: 24728008). Unless otherwise stated, Bp antigens were used individually in all assays at a final concentration of 20 µg/ml. For peptide epitope mapping of BPSL2096, BPSL2697 and Hsp60, synthetic peptides of 20 amino acids in length and overlapping by 10 amino acids were synthesized (GL Biochem Ltd, Shanghai) and used individually at a final concentration of 25 µg/ml (Supplementary Tables 3–5). Whole heat-inactivated Bp (HIA-Bp) was prepared from two Thai patient isolates 199a and 207a as previously described9 and used at a dilution of 1:60 (20 µg/ml by Bicinchoninic Acid Assay, Sigma) in ELIspot assays. The CEF T-cell epitope peptide pool (Mabtech, AB, Sweden) was used at a final concentration of 1 µg/mL.
HLA transgenic mouse studies
This study used HLA class II transgenic mouse lines for the alleles HLA-DR1 (DRB1 * 0101) which were all maintained in the context of a homozygous knockout for murine H2-Aβ, as described previously12–14,43–45. Mice were maintained in individually ventilated cages and used in experiments as young adults. For T cell epitope mapping studies, mice were primed sub-cutaneously in one hind footpad with 25 μg of recombinant protein emulsified in Hunters Titermax Gold adjuvant (Sigma-Aldrich). Ten days post immunisation, the draining popliteal lymph node was removed and disaggregated into a single-cell suspension for ELISpot assays. The frequency of cells producing IFNγ in response to antigen was quantified by ELISpot (Diaclone; 2B Scientific, Oxon, U.K). Briefly, 2 × 105 cells plus 25 μg/ml of protein or peptide were added to wells in HL-1 serum free medium (Lonza, Slough, U.K), supplemented with L-glutamine and penicillin-streptomycin (ThermoFisher Scientific, U.K). Plates were incubated for 72 h at 37 °C with 5% CO2. Following assay development, spots were counted on an automated ELISpot reader (Autoimmun Diagnostika, Strasbourg, France). Response frequencies were expressed as Δ spot forming cells (SFC) per 106 cells, with an epitope confirmed when the majority of immunised mice responded with a magnitude greater than the mean SFCs in the absence of any antigen + 2 SD. Mean + 2 SD background SFC for each ELISpot is indicated by a dotted line.
Human interferon gamma (IFNγ) enzyme-linked immunosorbent spot-forming cell assay (ELIspot)
Human cellular responses to protein antigens and peptide were assessed by ex-vivo IFNγ ELISpot assay using cryopreserved PBMC as described previously9. Briefly, 96-well Multiscreen-I plates (Millipore, UK) were coated overnight with 1D1K anti-human IFNγ (Mabtech, AB, Sweden) at 4 °C. PBMCs were added in triplicate wells at 2 × 105 PBMCs/well and antigens added at a final concentration of 20 µg/ml. After 18 hours, secreted IFNγ was detected (Mabtech, AB, Sweden) using a CTL ELIspot reader. Results are expressed as IFNγ spot-forming cells (SFC) per million PBMC. Background (media only) responses in unstimulated control wells were typically less than 20 spots, and were subtracted from those measured in antigen-stimulated wells. Responses were considered positive if they were above the mean background response plus 2 standard deviations and above 20 SFC/106 PBMC. PHA (phytohaemagglutinin) was used as a positive control to ensure T cell responsiveness, with a minimum of 100 SFC/106 PBMC required to be used in the assay. All PBMC samples tested were randomised independently and ELISpot testing was performed and analysed by laboratory staff blinded to the patient group and survival status.
Flow cytometry
PBMC were stimulated with protein (0.4 µg /well) or media only for 18 h. Brefeldin A (ebioscience, USA) was added at 10 µg/ml and following 4 h further incubation, staining for intracellular APC-IFNγ and for the immune cell surface markers PCP-anti-CD3, FITC-anti-CD4, APCH7-anti-CD8, PE-anti-CD56 and V450-CD14 (BD Biosciences, USA) was performed. Samples were analysed using a MACSQuant Analyzer 10 (Miltenyi Biotec, Germany) with Flowjo software (Treestar Inc, USA). Responses were quantitated for the negative control-subtracted percentage of IFNγ secreted CD4+ or CD8+ cells from the gated lymphocyte population.
Enzyme-Linked Immunosorbent Assay (ELISA)
Three Bp antigens (BPSL2520, BPSS1525 or BPSS2096) or coating buffer alone were added to wells of MaxiSorp U-bottom 96-Well immunoplates (Thermo Scientific, Denmark) and incubated overnight at 4 °C. Immunoplates were blocked with 5% skimmed milk/PBS for 2 h at 37 °C. Sera from melioidosis patients or healthy control were diluted and added to the immunoplate in duplicate, then further incubated for 1 h. HRP-conjugated goat anti-human IgG (Sigma) was then added to the ELISA plate, and further incubated for 1 h. ELISAs were developed using TMB substrate. Results were measured using a Multiskan™ GO Microplate Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and expressed as absorbance value (OD450).
Cytokine analysis
Levels of 23 cytokines were measured in the supernatants of PBMC stimulated for 18 hours with Bp antigens (BPSL2520, BPSS1525 or BPSS2096) or media alone, using the Milliplex MAP Human High sensitivity T-cell panel kit according to the manufacturer’s instructions.
Transcriptomic analysis
PBMC from thirteen diabetic subjects with acute melioidosis were stimulated for 18 hours with either HIA-Bp, media alone or one of three Bp antigens (BPSL2520, BPSS1525, BPSL2096) for 18 h and RNA was extracted using Qiagen RNeasy minikits (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Biotinylated and amplified RNA was generated using Illumina TotalPrep RNA Amplification Kit followed by Thermo Scientific GeneJET RNA Cleanup and Concentration Micro Kit (both ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions to generate a median of 4.9 µg RNA (range 1.6–12.0 µg), and hybridized to HumanHT-12 v4 Expression BeadChips (Illumina, San Diego, CA, USA). Arrays were scanned with an Illumina bead array reader confocal scanner, according to the manufacturer’s instructions. Array data processing and preliminary analysis with background subtraction was performed using Illumina BeadStudio software. Transcriptomics data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6093.
Data management and statistical analysis
Statistical analysis was performed using Graphpad Prism version 6 (San Diego, CA, USA) and IBM SPSS Statistics for Windows version 22.0 (Armonk, NY, USA). Non-parametric tests were used (Mann Whitney, paired Wilcoxon, Kruskal-Wallis and Spearman’s rank correlation tests) and HLA frequency comparisons were made by Chi-square, or 2-tailed Fisher’s exact test where <5 in a group. A P value (2-tailed) of <0.05 was considered significant. Transcriptomics data underwent variance stabilization and normalization in R using the vsn package from Bioconductor46. Further transcriptomics analysis and visual representation was performed using Partek Genomics suite with Pathway version 6.6 (St Louis, MO, USA). Differential gene expression between HIA-Bp stimulated and media alone culture conditions was performed using ANOVA paired analysis with a false discovery rate adjusted P value of <0.05 accepted as significant, with results represented graphically by volcano plots.
Electronic supplementary material
Acknowledgements
The authors wish to thank the patients and staff at Sunpasitthiprasong Hospital, Ubon Ratchathani, Thailand and Matthew Holden for helpful discussion about the BPSL2520 protein. The authors acknowledge the support of the NIHR Imperial BRC multi-parameter flow cytometry and confocal imaging facility, Hammersmith Campus, Imperial College London. We thank Dr Dilair Baban and the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z) for the generation of the transcriptomic data, and the Transplant Immunology Laboratory, Oxford for assistance with HLA typing. S.J.D is grateful for the support of a Wellcome Trust Intermediate Clinical Fellowship award ref WT100174/Z/12/Z, an Academy of Medical Sciences Starter Grant for Clinical Fellows (2011–2013) and an Oxford University Clinical Academic Graduate School Academic Lectureship (2009–2013). Mahidol-Oxford Tropical Medicine Research Unit is supported by the Wellcome Trust. L.G. and P.L. were supported by Fondazione CARIPLO contract number 2009–3577. This work was supported by National Institutes of Health–National Institute of Allergy and Infectious Diseases Large Scale T Cell Epitope Discovery Program Contract HHSN27220090046C (to R.J.B. and D.M.A.), the Welton Foundation (to R.J.B.), the National Institute for Health Research Biomedical Research funding scheme (to R.B. and D.A.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
K.J., M.S., P.C., S.C., C.J.R., K.J.Q., and R.S. performed the experiments. S.J.D., K.J., M.S., P.C., C.J.R., D.L., and R.J.B. analysed the data. S.J.D., K.J., S.C., P.A., C.P., P.T., D.L. and N.P.J.D. conducted the clinical study. R.S did the HLA typing. K.J. collected the clinical samples, oversaw the laboratory assays and curated the database. L.G. and P.L. made the BPSS1599 protein antigen. K.J., and C.J.R. helped prepare the manuscript. S.J.D., D.M.A., and R.J.B. conceived the research, designed experiments, interpreted data and wrote the manuscript. All authors read, commented on and agreed the content of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12331-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Susanna J. Dunachie, Email: susie.dunachie@ndm.ox.ac.uk
Rosemary J. Boyton, Email: r.boyton@imperial.ac.uk
References
- 1.Peacock SJ, et al. Melioidosis vaccines: a systematic review and appraisal of the potential to exploit biodefense vaccines for public health purposes. PLoS neglected tropical diseases. 2012;6:e1488. doi: 10.1371/journal.pntd.0001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limmathurotsakul, D. et al. Predicted global distribution of and burden of melioidosis. Nature microbiology1, doi:10.1038/nmicrobiol.2015.8 (2016). [DOI] [PubMed]
- 3.Limmathurotsakul D, et al. Increasing incidence of human melioidosis in Northeast Thailand. Am J Trop Med Hyg. 2010;82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS neglected tropical diseases. 2010;4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones AL, Beveridge TJ, Woods DE. Intracellular survival of Burkholderia pseudomallei. Infect Immun. 1996;64:782–790. doi: 10.1128/iai.64.3.782-790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes JL, et al. Adaptive immunity in melioidosis: a possible role for T cells in determining outcome of infection with Burkholderia pseudomallei. Clinical immunology. 2004;113:22–28. doi: 10.1016/j.clim.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Ketheesan N, et al. Demonstration of a cell-mediated immune response in melioidosis. J Infect Dis. 2002;186:286–289. doi: 10.1086/341222. [DOI] [PubMed] [Google Scholar]
- 8.Tippayawat P, et al. Phenotypic and functional characterization of human memory T cell responses to Burkholderia pseudomallei. PLoS neglected tropical diseases. 2009;3:e407. doi: 10.1371/journal.pntd.0000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenjaroen K, et al. T-Cell Responses Are Associated with Survival in Acute Melioidosis Patients. PLoS neglected tropical diseases. 2015;9:e0004152. doi: 10.1371/journal.pntd.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holden MT, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felgner PL, et al. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci USA. 2009;106:13499–13504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu KK, et al. CD4+ T-cell immunity to the Burkholderia pseudomallei ABC transporter LolC in melioidosis. European journal of immunology. 2011;41:107–115. doi: 10.1002/eji.201040881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musson JA, et al. CD4+ T cell epitopes of FliC conserved between strains of Burkholderia: implications for vaccines against melioidosis and cepacia complex in cystic fibrosis. J Immunol. 2014;193:6041–6049. doi: 10.4049/jimmunol.1402273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds C, et al. T Cell Immunity to the Alkyl Hydroperoxide Reductase of Burkholderia pseudomallei: A Correlate of Disease Outcome in Acute Melioidosis. J Immunol. 2015;194:4814–4824. doi: 10.4049/jimmunol.1402862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng AC, O’Brien M, Freeman K, Lum G, Currie BJ. Indirect hemagglutination assay in patients with melioidosis in northern Australia. Am J Trop Med Hyg. 2006;74:330–334. [PubMed] [Google Scholar]
- 16.Hodgson K, et al. Immunological mechanisms contributing to the double burden of diabetes and intracellular bacterial infections. Immunology. 2015;144:171–185. doi: 10.1111/imm.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen IR. Autoimmunity to chaperonins in the pathogenesis of arthritis and diabetes. Annu Rev Immunol. 1991;9:567–589. doi: 10.1146/annurev.iy.09.040191.003031. [DOI] [PubMed] [Google Scholar]
- 18.Birk OS, et al. A role of Hsp60 in autoimmune diabetes: analysis in a transgenic model. Proc Natl Acad Sci USA. 1996;93:1032–1037. doi: 10.1073/pnas.93.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothchild AC, et al. MiR-155-regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2016;113:E6172–E6181. doi: 10.1073/pnas.1608255113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barat S, et al. Immunity to intracellular Salmonella depends on surface-associated antigens. PLoS Pathog. 2012;8:e1002966. doi: 10.1371/journal.ppat.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zlosnik JE, Speert DP. The role of mucoidy in virulence of bacteria from the Burkholderia cepacia complex: a systematic proteomic and transcriptomic analysis. J Infect Dis. 2010;202:770–781. doi: 10.1086/655663. [DOI] [PubMed] [Google Scholar]
- 22.Swetha RG, Sandhya M, Ramaiah S, Anbarasu A. Identification of CD4+ T-cell epitope and investigation of HLA distribution for the immunogenic proteins of Burkholderia pseudomallei using in silico approaches - A key vaccine development strategy for melioidosis. J Theor Biol. 2016;400:11–8. doi: 10.1016/j.jtbi.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Boelen L, et al. BIITE: A Tool to Determine HLA Class II Epitopes from T Cell ELISpot Data. PLoS Comput Biol. 2016;12(3):e1004796. doi: 10.1371/journal.pcbi.1004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quigley K, et al. Chronic Infection by Mucoid Pseudomonas aeruginosa Associated with Dysregulation in T-Cell Immunity to Outer Membrane Porin F. Am J Respir Crit Care Med. 2015;191(11):1250–64. doi: 10.1164/rccm.201411-1995OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nature reviews. Immunology. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh GC, Peacock SJ, van der Poll T, Wiersinga WJ. The impact of diabetes on the pathogenesis of sepsis. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2012;31:379–388. doi: 10.1007/s10096-011-1337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chitradevi ST, Kaur G, Sivaramakrishna U, Singh D, Bansal A. Development of recombinant vaccine candidate molecule against Shigella infection. Vaccine. 2016;34:5376–5383. doi: 10.1016/j.vaccine.2016.08.034. [DOI] [PubMed] [Google Scholar]
- 28.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 29.Elias D, et al. Vaccination against autoimmune mouse diabetes with a T-cell epitope of the human 65-kDa heat shock protein. Proc Natl Acad Sci USA. 1991;88:3088–3091. doi: 10.1073/pnas.88.8.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallerskog T, Martens GW, Kornfeld H. Diabetic mice display a delayed adaptive immune response to Mycobacterium tuberculosis. J Immunol. 2010;184:6275–6282. doi: 10.4049/jimmunol.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stew SS, Martinez PJ, Schlesinger LS, Restrepo BI. Differential expression of monocyte surface markers among TB patients with diabetes co-morbidity. Tuberculosis. 2013;93(Suppl):S78–82. doi: 10.1016/S1472-9792(13)70015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodgson KA, Morris JL, Feterl ML, Govan BL, Ketheesan N. Altered macrophage function is associated with severe Burkholderia pseudomallei infection in a murine model of type 2 diabetes. Microbes and infection / Institut Pasteur. 2011;13:1177–1184. doi: 10.1016/j.micinf.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Gourlay LJ, et al. From crystal structure to in silico epitope discovery in the Burkholderia pseudomallei flagellar hook-associated protein FlgK. FEBS J. 2105;282(7):1319–33. doi: 10.1111/febs.13223. [DOI] [PubMed] [Google Scholar]
- 34.Hananantachai H, et al. Polymorphisms of the HLA-B and HLA-DRB1 genes in Thai malaria patients. Jpn J Infect Dis. 2005;58:25–28. [PubMed] [Google Scholar]
- 35.Hilton, H. G. et al. The intergenic recombinant HLA-B*46:01 has a distinctive peptidome which includes KIR2DL3 ligands. Cell Reports. (in press) (2017). [DOI] [PMC free article] [PubMed]
- 36.Conejero L, et al. The Blood Transcriptome of Experimental Melioidosis Reflects Disease Severity and Shows Considerable Similarity with the Human Disease. J Immunol. 2015;195:3248–3261. doi: 10.4049/jimmunol.1500641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh GC, et al. Host responses to melioidosis and tuberculosis are both dominated by interferon-mediated signaling. PLoS One. 2013;8:e54961. doi: 10.1371/journal.pone.0054961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulsantiwong, P. et al. Burkholderia pseudomallei induces IL-23 production in primary human monocytes. Med Microbiol Immunol, doi:10.1007/s00430-015-0440-z (2015). [DOI] [PubMed]
- 39.Krishnananthasivam S, et al. Gene Expression Profile of Human Cytokines in Response to Burkholderia pseudomallei Infection. mSphere. 2(2) (2017). [DOI] [PMC free article] [PubMed]
- 40.Wilson WJ et al. Immune Modulation as an Effective Adjunct Post-exposure Therapeutic for B. pseudomallei. PLoS Negl Trop Dis. 10(10), e0005065 (2016). [DOI] [PMC free article] [PubMed]
- 41.Asakrah S, et al. Post-exposure therapeutic efficacy of COX-2 inhibition against Burkholderia pseudomallei. PLoS Negl Trop Dis. 2106;7(5):e2212. doi: 10.1371/journal.pntd.0002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wuthiekanun V, et al. Short report: Melioidosis in Myanmar: forgotten but not gone? Am J Trop Med Hyg. 2006;75:945–946. [PubMed] [Google Scholar]
- 43.Reynolds CJ, et al. The serodominant secreted effector protein of Salmonella, SseB, is a strong CD4 antigen containing an immunodominant epitope presented by diverse HLA class II alleles. Immunology. 2014;143:438–446. doi: 10.1111/imm.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ascough S, et al. Anthrax lethal factor as an immune target in humans and transgenic mice and the impact of HLA polymorphism on CD4+ T cell immunity. PLoS Pathog. 2014;10:e1004085. doi: 10.1371/journal.ppat.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Till SJ, et al. Peptide-induced immune regulation by a promiscuous and immunodominant CD4T-cell epitope of Timothy grass pollen: a role of Cbl-b and Itch in regulation. Thorax. 2014;69:335–345. doi: 10.1136/thoraxjnl-2013-204324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.