ABSTRACT
Staphylococcus aureus is an important pathogen causing a spectrum of diseases ranging from mild skin and soft tissue infections to life-threatening conditions. Bloodstream infections are particularly important, and the treatment approach is complicated by the presence of methicillin-resistant S. aureus (MRSA) isolates. The emergence of new genetic lineages of MRSA has occurred in Latin America (LA) with the rise and dissemination of the community-associated USA300 Latin American variant (USA300-LV). Here, we prospectively characterized bloodstream MRSA recovered from selected hospitals in 9 Latin American countries. All isolates were typed by pulsed-field gel electrophoresis (PFGE) and subjected to antibiotic susceptibility testing. Whole-genome sequencing was performed on 96 MRSA representatives. MRSA represented 45% of all (1,185 S. aureus) isolates. The majority of MRSA isolates belonged to clonal cluster (CC) 5. In Colombia and Ecuador, most isolates (≥72%) belonged to the USA300-LV lineage (CC8). Phylogenetic reconstructions indicated that MRSA isolates from participating hospitals belonged to three major clades. Clade A grouped isolates with sequence type 5 (ST5), ST105, and ST1011 (mostly staphylococcal chromosomal cassette mec [SCCmec] I and II). Clade B included ST8, ST88, ST97, and ST72 strains (SCCmec IV, subtypes a, b, and c/E), and clade C grouped mostly Argentinian MRSA belonging to ST30. In summary, CC5 MRSA was prevalent in bloodstream infections in LA with the exception of Colombia and Ecuador, where USA300-LV is now the dominant lineage. Clonal replacement appears to be a common phenomenon, and continuous surveillance is crucial to identify changes in the molecular epidemiology of MRSA.
KEYWORDS: bacteremia, Latin America, Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus is a major human pathogen causing a wide range of infections, including bacteremia, infective endocarditis, and osteoarticular, skin and soft tissue, pleuropulmonary, and device-related infections (1). S. aureus is a leading cause of hospital-associated infections and is the most common cause of soft tissue infections requiring visits to emergency services in the United States (2). Bloodstream infection (BSI) is a potentially fatal complication of S. aureus disease, with an estimated incidence of 80 to 190 cases/100,000 inhabitants per year in developed countries (3–5). For example, in the United States and France, it is estimated that ca. 600,000 and 125,000 cases of S. aureus BSIs occur per year, respectively (3–5).
The emergence of methicillin-resistant S. aureus (MRSA) poses important therapeutic challenges (6). Although initially identified as an important nosocomial pathogen, MRSA is now endemic in the community (7). The Centers for Disease Control and Prevention (CDC) estimates that 80,461 invasive MRSA infections and 11,285 related deaths occurred in the United States in 2011 (8). Community-associated MRSA (CA-MRSA) strains were first described among healthy individuals with no health care contact or hospital-associated risk factors (8). In North America (NA), the CA-MRSA epidemic is widely attributed to the spread of a clone designated USA300-ST8 (9, 10). Interestingly, a USA300-related genetic lineage (designated USA-300 Latin American variant [USA300-LV]) has emerged in the northern part of South America (SA) and appears to have become endemic in hospitals in the region (Colombia, Venezuela, and Ecuador) (11–14). Recent phylogenetic analyses of USA300 and USA300-LV MRSA revealed that the two genetic lineages are closely related and that the NA and SA clades segregated geographically. Molecular clock analyses suggested that both clades had a common ancestor that may have emerged in the mid-1970s with individual segregation occurring by 1989 and 1985 for the NA and SA clades, respectively (14). Geographic segregation of these parallel epidemics coincided with the independent acquisition of the arginine catabolic mobile element (ACME) in NA isolates and a copper and mercury resistance (COMER) mobile element in SA isolates (14).
Apart from the introduction of the USA300-LV in the northern countries of SA, important changes in the population structure of MRSA have also occurred in Argentina and Brazil (15–17). However, a comprehensive molecular analysis of S. aureus bacteremia in Latin America has not been attempted before. Here, we sought to characterize the population structure of S. aureus bacteremia and the changing molecular epidemiology of MRSA in a prospective multicenter cohort study of S. aureus bacteremia spanning 3.5 years (2011 to 2014) in selected hospitals from nine Latin-American countries.
RESULTS
MRSA versus MSSA in bloodstream infections from 24 Latin American hospitals.
A total of 1,346 isolates were collected from the bloodstreams of patients in 24 hospitals in Latin America. A total of 161 isolates were excluded due to protocol violations, which were commonly due to misidentification or contamination. The final analysis included 1,185 isolates from 1,010 patients. A total of 875, 102, 26, and 7 patients had one, two, three, and four isolates, respectively (see Table S2 in the supplemental material). Table 1 shows the distribution of bloodstream S. aureus isolates for each country. Overall, methicillin resistance was found in 45% of all isolates with important regional variations (Table 1). The hospitals with the highest rates of MRSA were those located in Brazil (62%), Venezuela (57%), Mexico (57%), Peru (54%), and Guatemala (54%). A total of 45% and 40% of S. aureus isolates recovered from hospitals in Chile and Argentina, respectively, exhibited methicillin resistance. Conversely, only a minority of S. aureus isolates obtained from Colombia and Ecuador were methicillin resistant (22% and 29%, respectively). Overall, the majority of MRSA isolates exhibited high rates of resistance to erythromycin (81%), clindamycin (74%), and ciprofloxacin (78%). Chloramphenicol (CHL), tetracycline (TET), rifampin (RIF), and trimethoprim-sulfamethoxazole (SXT) remained active against the majority of MRSA isolates with resistance rates of 15%, 5%, 3%, and 2%, respectively. All S. aureus isolates were susceptible to minocycline (MIN), linezolid (LZD), and vancomycin (VAN) (MIC90, 1 μg/ml) (see Tables S3 and S4). We did not detect any MRSA with reduced susceptibility to vancomycin. Methicillin-susceptible S. aureus (MSSA) isolates were highly susceptible to the majority of antibiotics tested (Table S4). The highest rates of resistance in MSSA were found for erythromycin and tetracycline (13% and 9%, respectively).
TABLE 1.
Distribution of MRSA and MSSA in Latin America
| Country | No. (%) of isolates |
||
|---|---|---|---|
| S. aureus | MRSA | MSSA | |
| Argentina | 149 | 60 (40) | 89 (60) |
| Brazil | 204 | 126 (62) | 78 (38) |
| Chile | 163 | 74 (45) | 89 (13) |
| Colombia | 188 | 41 (22) | 147 (78) |
| Ecuador | 101 | 29 (29) | 72 (71) |
| Guatemala | 138 | 74 (54) | 64 (46) |
| Mexico | 30 | 17 (57) | 13 (43) |
| Peru | 154 | 84 (54) | 70 (46) |
| Venezuela | 58 | 33 (57) | 25 (43) |
| Total | 1,185 | 538 (45) | 647 (55) |
Genetic lineages of MRSA.
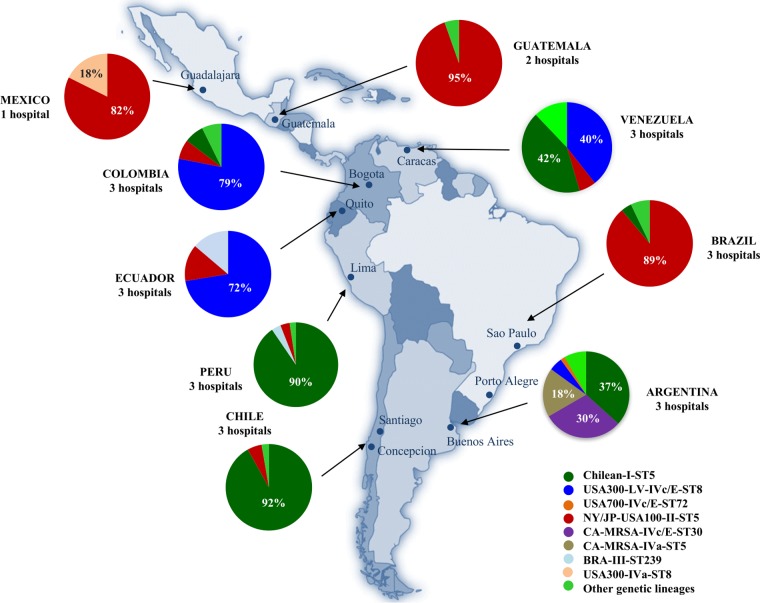
Figure 1 shows the distribution of MRSA clones in Latin America. The most striking result was that the majority of MRSA isolates recovered from participating hospitals in Colombia and Ecuador (79% and 72%, respectively) belonged to the USA300-LV lineage (ST8) (11–14), consistent with the clonal replacement of MRSA described before in hospitals throughout these countries (11–14). All USA300-LV isolates harbored staphylococcal chromosomal cassette mec (SCCmec) IVc/E, and the majority (85% in Colombia and Ecuador) also carried lukS-PV and lukF-PV encoding Panton-Valentine leukocidin (PVL). Of note, isolates belonging to USA300-LV were fully susceptible to RIF, MIN, SXT, LZD, and VAN and only exhibited low resistance rates (3 to 16%) to erythromycin (ERY), clindamycin (CLI), ciprofloxacin (CIP), gentamicin (GEN), TET, and CHL. Multilocus sequence typing (MLST) of USA300-LV indicated that they belonged to sequence type ST8. The remaining MRSA recovered from participating hospitals in Colombia and Ecuador belonged to the Chilean/Cordobes (ST5, SCCmec I, previously prevalent in this region) (11–14), pulsed-field gel electrophoresis (PFGE) pattern USA100 (ST5, SCCmec II, previously designated New York/Japan clone), and Brazilian (ST239, SCCmec III) clones (Fig. 1). These clones exhibited higher rates of resistance than USA300-LV isolates. The clonal distribution of Venezuelan MRSA was more heterogeneous. Indeed, USA300-LV strains represented only 40% of MRSA but harbored more diverse SCCmec subtypes, including IVc/E (62%), IVb (23%), IVa (7.5%), and IVd (7.5%). Moreover, rates of resistance for Venezuelan USA300-LV isolates were higher than those observed in Colombian and Ecuadorian USA300-LV (ERY [23%], TET [15%], CIP [8%], and CLI [8%]). The Chilean/Cordobes clone was represented in a proportion similar to that of USA300-LV (42%).
FIG 1.
Clonal distribution of MRSA isolates in Latin American hospitals included in the study. Specific genetic lineages derived from pulsed-field gel electrophoresis, SCCmec typing, and antimicrobial susceptibility testing are indicated by colors in each corresponding country. ST, sequence type; LV, Latin American variant.
The above findings contrast with those of the other participating hospitals in Latin American countries. In the participating tertiary care centers of Peru and Chile, the clonal distributions of MRSA were almost identical, with the overwhelming majority of isolates (≥90%) belonging to the Chilean/Cordobes clone (Fig. 1). The included hospitals in Mexico, Guatemala, and Brazil had similar MRSA lineages, with the USA100 (ST5, SCCmec II) representing >80% of isolates recovered from bacteremia. Our results are consistent with previous reports of a major clonal replacement of MRSA in Brazil, where the Brazilian clone (ST239, SCCmec III) was the previous predominant clone (16, 17). Interestingly, MRSA from included hospitals in Argentina exhibited the most heterogeneous population structure compared with that from the rest of Latin America. Apart from the Chilean/Cordobes clone, we were able to characterize an important proportion of isolates belonging to three different community-associated genetic lineages, ST30 carrying SCCmec IVc/E (30%), ST5 harboring SCCmec IVa (18%), and USA300-LV (SCCmec IVc/E [7%]) (Fig. 1), consistent with previous local reports (15, 17, 18). In fact, Argentina was the only country in southern Latin America in which USA300-LV isolates were identified from bloodstream infections.
Phylogenetics of MRSA.
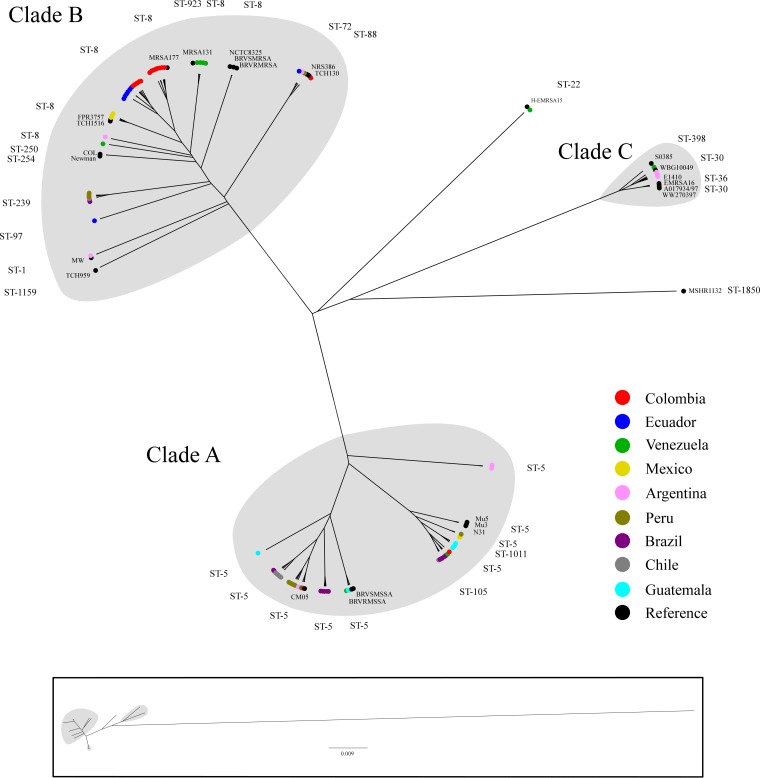
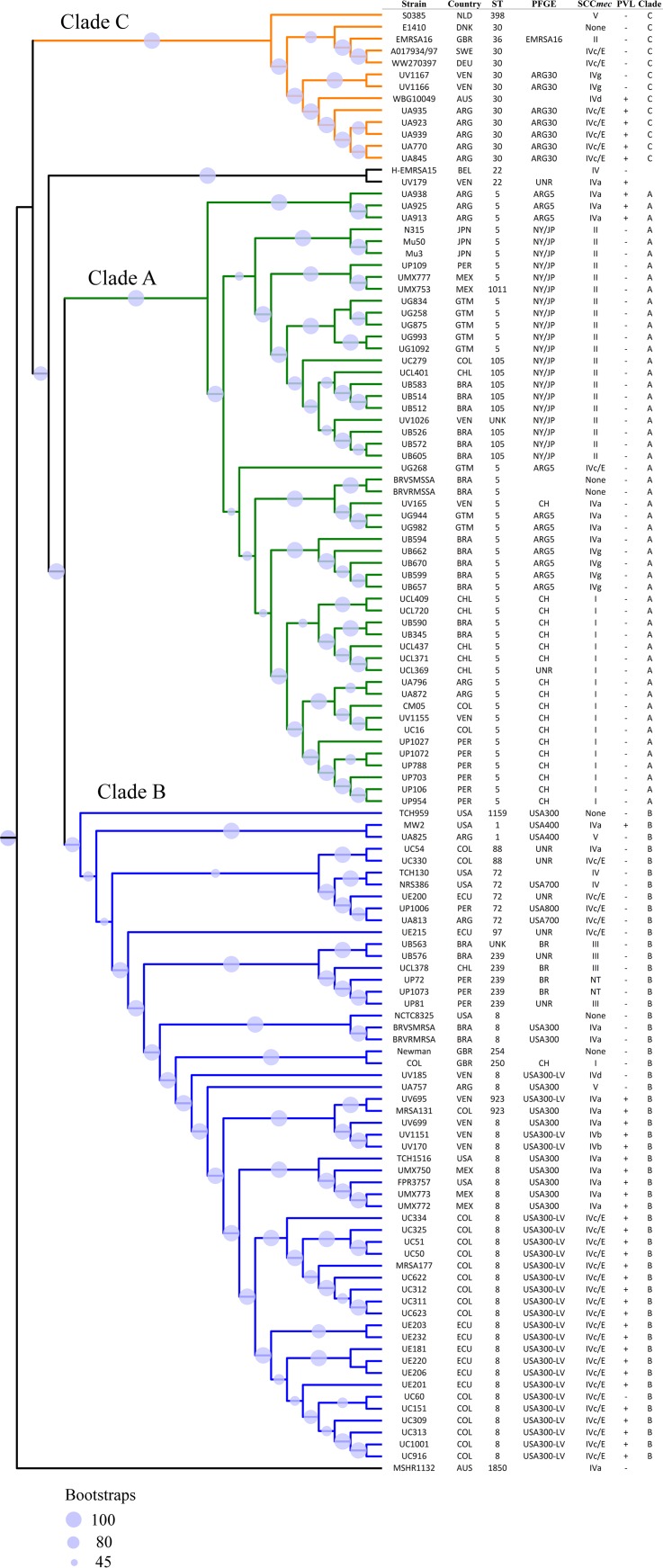
To explore the genetic diversity of MRSA strains circulating in the hospitals included in this study, we constructed a phylogenetic tree that included 96 genomes from representative isolates of the major circulating MRSA clones in the region and isolates with unusual genotypes/phenotypes. Although the selection strategy was driven primarily by budget limitations, we opted to implement this approach (see Materials and Methods) to dissect the population structure of the major genetic lineages circulating in the region and to characterize emerging genotypes. We added 27 S. aureus genomes available on the NCBI database for comparison (see Table S1). The phylogenetic analysis revealed three major clades, denoted A, B, and C (Fig. 2 and 3). Clade A, which included S. aureus N315 (a hospital-associated MRSA strain) grouped isolates with ST5, ST105, and ST1011 harboring SCCmec types I and II, but also type IV (more related to community-associated strains) with subtypes IVa, IVc/E, and IVg (Fig. 2 and 3). Clade A included the most multidrug-resistant MRSA strains (Fig. 4), including strains with fluoroquinolone resistance harboring substitutions in GyrA and GrlA (coding for large subunits of DNA gyrase and topoisomerase IV, respectively) in most isolates. Of note, five genes encoding different aminoglycoside-modifying enzymes, including the gene encoding the bifunctional enzyme AAC(6′)-APH(2″) conferring high-level resistance to GEN, amikacin, and tobramycin, were identified (Fig. 4). Additionally, ermA, encoding a ribosomal methylase (macrolide-lincosamide-streptogramin B [MLSB]-type resistance), was found in a high proportion of isolates of this clade. Interestingly, within the clade A, isolates that harbored SCCmec IV were fluoroquinolone susceptible and had fewer aminoglycoside and MLSB-type resistance determinants (Fig. 4).
FIG 2.
Whole-genome maximum likelihood phylogeny of MRSA isolates. The circular representations in the phylogenetic tree show the three main clades designated A, B, and C (highlighted in gray). Major clades (A, B, and C) are composed mainly of strains from CC5, CC8, and CC30, respectively (sequence types [ST] are indicated). Each branch tip is labeled by a colored circle according to the country of isolation or if it was considered a reference strain (black). The box at the bottom shows the representation of the true distance of the phylogenetic tree.
FIG 3.
Site of isolation and molecular characteristics of MRSA lineages. The rectangular transformed representation shows the three main clades (A, B, and C) highlighted in green, blue, and orange, respectively. Molecular typing characteristics of the evaluated strains are shown, including multi locus sequence type (ST), pulse field gel electrophoresis type (PFGE), staphylococcal chromosomal cassette mec type (SCCmec), and presence (+) or absence (−) of the Panton-Valentine leukocidin (PVL). ARG, Argentina; AUS, Australia; BRA, Brazil; CHL, Chile; COL, Colombia; DEU, Germany; DNK, Denmark; ECU, Ecuador; GBR, Great Britain; GTM, Guatemala; JPN, Japan; MEX, Mexico; NDL, The Netherlands; PER, Peru; SWE, Sweden; USA, United States of America; VEN, Venezuela; ARG30, Argentinian clone ST30; ARG5, Argentinian clone ST5; CH, Chilean clone; NY/JP, New York/Japan-USA100 clone; BR, Brazilian clone; NT, nontypeable; UNR, unrelated by PFGE.
FIG 4.
Genomic characteristics of Latin American MRSA strains. (A) Presence (dark) and absence (light) of 2,132 antibiotic resistance genes from the ResFinder 2.1 database, and (B) the presence (black) and absence (white) of the genes in the genetic locus “copper and mercury resistance” (COMER) and “arginine catabolic mobile element” (ACME). The strains are organized according to the phylogenetic reconstruction of these strains, and the three main clades (A, B, and C) are highlighted in green, blue, and orange, respectively. (A) The genes are grouped according to the type antibiotic they confer resistance to: aminoglycosides, glycopeptides, macrolides-lincosamides-streptogramin B (MLSB), trimethoprim (TMP), tetracyclines, beta-lactams, phenicols-lincosamides-oxazolidinones-pleuromutilins-streptogramin A (PhLOPSA), and fluoroquinolones. (B) The genetic organization of the loci COMER and ACME is shown; reference sequences were obtained from the strains CA12 (accession no. CP007672.1, regions 53520 to 77705) and USA300FPR3757 (accession no. CP000255.1, regions 63100 to 88681), respectively. TR, transcriptional regulator; Cc, cytochrome c.
Clade B included ST8, ST88, ST97, and ST72 strains, mostly harboring SCCmec IV (subtypes a, b, c/E, and d) and were less enriched in antibiotic-resistance genes (Fig. 4). Within this clade, a small group (four strains) was ST239, which harbored SCCmec III and a high number of resistance genes, including those encoding aminoglycoside, tetracycline, MLSB, and fluoroquinolone resistance (Fig. 4). Since the ST8 USA300-LV lineage (included within this clade) typically harbors the novel COMER mobile element and lacks the ACME island (typical of NA-USA300), we specifically analyzed the contents of both COMER and ACME within strains of clade B. As shown in Fig. 4, both islands were enriched in strains belonging to this clade and were almost absent in isolates belonging to clades A and C. Most of the COMER-containing strains were recovered in Ecuador (ECU) and Colombia (COL), in agreement with the geographic distribution of the USA300-LV genetic lineage. ACME was found only in Mexican strains with the profile typical of NA-USA300. Of interest, some isolates belonging to ST72, ST88, and ST97 harbored some fragments of the COMER island, supporting the notion that the COMER element was mobile and might represent remnants of this DNA fragment, reinforcing the genetic relationship of strains in clade B. Finally, clade C grouped mostly Argentinian MRSA isolates with ST30 and SCCmec IVc/E, a finding previously reported for this country (15, 17, 18).
DISCUSSION
In this work, we have assembled the largest cohort of patients with S. aureus bacteremia in Latin America to date. The study included patients enrolled in selected hospitals from nine countries, from Mexico to Argentina. Our main objective was to understand the clinical and molecular characteristics of S. aureus bacteremia on a large scale. Although we understand that the hospitals enrolled may not represent the situation of other hospitals in the countries included in this study, it is important to note that our findings are supported by many previous local studies (11–19). Thus, our results may actually reflect an accurate picture of circulating S. aureus lineages causing bloodstream infections in the region.
One of the most interesting findings of our study is that the proportion of MRSA strains remains high in the region but exhibits important regional variations. Indeed, rates of MRSA higher than 40% were found in the majority of countries, with the highest prevalence in the participating hospitals in Brazil (62%), a finding that is consistent with those of recently reported surveillance studies (11–19). A notable exception to the above appears to be in the included hospitals from both Colombia and Ecuador. In these centers, the rates of MRSA appear to have markedly decreased compared with those from previous studies. Indeed, in a multicenter surveillance carried out by our group in Colombia from 2006 to 2008 (which included the three hospitals of the current study), methicillin resistance in S. aureus was documented in ca. 50% of hospital-associated isolates (13). By contrast, in the present study, <30% of S. aureus isolates from these hospitals were MRSA. Although the former study included isolates from clinical samples other than blood, the decrease in the rates of MRSA seems significant compared with previous reports of hospitals in Peru and Venezuela (19). Although we cannot exclude that such reductions in MRSA prevalence could be due to major changes in infection control practices in the participating hospitals, the lower rates of MRSA seem to be associated with the shift in the population structure.
In 2005, we characterized the first strains of CA-MRSA reported in Colombia and found that, in contrast to in other parts of the world, the majority of these infections were caused by MRSA isolates that belonged to a genetic lineage closely related to the USA300 strain prevalent in NA (we designated this lineage USA300-LV) (11–14). Both USA300 genetic lineages encompass ST8 isolates harboring SCCmec IV and genes encoding the PVL toxin. However, the most important genetic distinction between these two is the absence of the ACME island in USA300-LV, which is replaced by a gene cluster encoding proteins involved in the metabolism of copper and mercury (14). The results of our study, which are supported by several other local findings, indicate that in stark contrast with USA300, USA300-LV strains have been able to completely replace previously prevalent hospital-associated clones in Colombia and Ecuador (11–19). The reasons for such remarkable clonal switch in a relatively short time span are not known. Thus, the understanding of the pathogenic properties of this genetic lineage and the role of the COMER island in virulence and the ability to disseminate has become a priority of our future studies.
To dissect the population genetics of isolates from our studies, we selected isolates for whole-genome sequencing (WGS) based on our initial molecular characterization to offer a more detailed picture of the molecular epidemiology and population structure of MRSA. Our genomic analyses indicate that three major clades are likely to circulate in the Latin American centers included in this study, which in general, clustered in previous MLST analyses. The most multidrug-resistant of these clades is clade A, which encompasses the majority of MRSA strains of the region (except in Colombia and Ecuador). The strains are grouped within the CC5 (Chilean/Cordobes and NY/Japan-USA100 clones) and exhibit high rates of resistance to quinolones, MLSB, and aminoglycosides associated with the presence of SCCmec I and II. An exception to the above characteristics is a cluster of ST5 isolates recovered in Argentina (ARG) and Brazil (BRA) that carry SCCmec IVa and IVg and have been associated with community infections in these countries (15, 16), suggesting that a novel ST5 CA-MRSA lineage is in its ascendency in the southern parts of SA.
Clade B encompasses isolates from CC8 and CC239, including those belonging to USA300-LV and NA-USA300. In general, the CC8 isolates harbor the PVL genes, carry SCCmec IV, and are less enriched in antibiotic resistance genes than those of clade A, with the exception of isolates belonging to the Brazilian clone which harbor SCCmec III and harbor resistance profiles similar to those of clade A. Of note, 18% of Mexican MRSA isolates from a tertiary hospital (grouped in clade B) display the pattern typical of that of NA-USA300, suggesting that this strain is likely to be circulating in Mexico. Finally, clade C groups most of ST30 CA strains, which are found mainly in the ARG hospitals and are related to the Oceania-Pacific ST30 clone initially described in Australia. These strains also carry SCCmec IVc/E, with genes encoding PVL present in 61% of them. Of note, the Argentinian hospitals harbored the most highly diverse population of MRSA, and the reasons for such heterogeneity in the population genetics of MRSA are unclear.
As mentioned above, an important limitation of our study is that we only included a few participating centers in each country, and our results may not reflect the situation of the entire region. However, this is an inherent limitation of studies similar to ours, and a more comprehensive inclusion of hospitals with such diversity and different social and economic realities is challenging. Moreover, the results from many local studies seem to support our findings, making them more generalizable (11–19). Also, we did not have the support to perform WGS on the entire collection, which would have provided a deeper understanding of the population structure of the MRSA isolates. Nonetheless, our genomic selection strategy was robust (96 isolates), with the objective of including isolates that may accurately represent the phylogenetic picture of MRSA bacteremia in the participating centers. Indeed, our genomic findings are also supported by results from other studies and point to specific clonal changes that may contribute to the understanding of the dissemination and dynamics of bacteremic S. aureus infections in the region.
In summary, we presented a comprehensive characterization of S. aureus bacteremia in Latin America, providing strong evidence that clonal replacement is frequent and that the population structure is in continuous evolution. The identification of newly emerged genetic lineages would help in defining specific therapeutic approaches for MRSA infections in the region.
MATERIALS AND METHODS
Study design, inclusion criteria, and collection of isolates.
We included patients in 24 tertiary care hospitals from ARG (3 hospitals in Buenos Aires), BRA (3 hospitals in Sao Paulo and Porto Alegre), Chile ([CHI] 3 hospitals in Santiago and Concepcion), COL (3 hospitals in Bogota), ECU (3 hospitals in Quito), Guatemala ([GTM] 2 hospitals in Guatemala City), Mexico ([MEX] 1 hospital in Guadalajara), Peru ([PER] 3 hospitals in Lima), and Venezuela ([VEN] 3 hospitals in Caracas) between January 2011 and July 2014. These hospitals were chosen because (i) they had infrastructures for clinical research that included personnel for data collection, (ii) the investigators were experienced in clinical studies, and (iii) they possessed microbiology laboratory infrastructures. In this prospective observational cohort study, we included patients older than 18 years who had an episode of S. aureus bacteremia (detailed inclusion and exclusion criteria are specified in the supplemental material). Isolates were identified by standard microbiological techniques at the local hospitals and were subsequently sent to a reference laboratory (Universidad El Bosque, Bogota, Colombia) for additional characterization (see the supplemental material). All participating hospitals had local Institutional Review Board (IRB) approval for the study.
Identification of bacterial isolates and antimicrobial susceptibility testing.
The identification at the species level of all S. aureus isolates was performed by a multiplex PCR assay as described previously (20). Antimicrobial susceptibility testing was performed using the agar dilution method (see the supplemental material) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (21).
Molecular typing and detection of Panton-Valentine leukocidin.
SCCmec typing (I to V) and SCCmec IV subtyping were performed in all MRSA isolates by multiplex PCR according to previously reported methods (see the supplemental material) (22–25). PFGE was performed on all MRSA isolates with some modifications of a previously described method (26). Patterns were analyzed with the GelCompar II software (version 6.5; Applied Maths, Sint-Martens-Latem, Belgium) (see the supplemental material). The presence of lukS-PV and lukF-PV encoding PVL was investigated using PCR assays and primers described previously (27).
WGS and phylogenetic analysis.
WGS was used to dissect the genetic diversity of MRSA. Due to funding limitations, we chose to sequence representatives of the most prevalent clones defined by PFGE (banding patterns), susceptibility profiles, and SCCmec typing. Additionally, representative isolates with unusual PFGE patterns and SCCmec typing were also selected for WGS to provide a broad view of the population structure of S. aureus in Latin America. For all assembled genomes, we performed MLST, detection of antibiotic resistance determinants, and detection of ACME and COMER (14, 28, 29) (see the supplemental material). All sequenced genomes from this study were included in the phylogenetic reconstruction and tree generation (see Table S1 in the supplemental material) (30–33).
Supplementary Material
ACKNOWLEDGMENTS
We dedicate the manuscript to the memory of Carlos Mejia-Villatoro. We thank Juan David Garavito, Natalia Rojas, and Paola Porras for technical assistance in the characterization of the isolates. We also thank the following members of the Latin-America Working Group on Bacterial Resistance. From Argentina, Didier Bruno, Hospital de Clinicas, Buenos Aires; Ernesto Efron, Hospital Británico, Buenos Aires; and Marcelo Del Castillo, Sanatorio Mater Dei, Buenos Aires. From Brazil, Thaís Guimarães, Hospital do Servidor Publico Estadual de São Paulo, São Paulo. From Chile, María Elena Ceballos, Escuela de Medicina, Pontificia Universidad Católica de Chile, Santiago; Isabel Domínguez and Daniela Beltrán, Hospital Sótero del Río, Santiago; and Gisela Riedel, Hospital Guillermo Grant Benavente, Concepción. From Colombia, Sandra Liliana Valderrama, Unidad de Enfermedades Infecciosas, Hospital Universitario San Ignacio, Pontificia Universidad Javeriana, Bogota; Sandra Milena Gualtero, Hospital Universitario San Ignacio, Clínica Shaio; and Carlos Humberto Saavedra, Unidad Infectologia, Hospital Universitario Clínica San Rafael, Facultad de Medicina, Universidad Nacional de Colombia, Bogotá. From Ecuador, Betzabé Tello, Hospital Vozandes, Quito; Juan Carlos Aragón, Hospital General de las Fuerzas Armadas, Quito; and Fausto Guerrero, Hospital Carlos Andrade Marín, Quito. From Guatemala, María Mónica Silvestre, Hospital Roosevelt. From Mexico, Rayo Morfin-Otero, Hospital Civil de Guadalajara, Fray Antonio Alcalde, Centro Universitario Ciencias de la Salud, Universidad de Guadalajara, Guadalajara. From Peru, Jose Hidalgo, Hospital Guillermo Almenara, Lima; and Luis Hercilla, Hospital Alberto Sabogal, Lima. From Venezuela, Ana María Cáceres Hernández, Clinica La Floresta, Caracas; Marisela Silva, Hospital Universitario de Caracas, Caracas; and Alfonso José Guzmán, Centro Médico de Caracas, Caracas.
This work was supported by an independent investigator-initiated grant to E.G. and C.S. C.A.A. is supported by the National Institutes of Health–National Institute of Allergy and Infectious Diseases (NIH/NIAID) (grants K24-AI114818, R01-AI093749, R21-AI114961, and R21/R33 AI121519). DNA sequencing was supported in part by Departamento de Ciencia, Tecnologia e Innovacion (COLCIENCIAS) (grant 130871250417/906-2015 to J.R.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
All authors have completed and submitted the ICMJE form for the disclosure of potential conflicts of interest. Cesar A. Arias has received grant support, consulted for or provided lectures for Pfizer, Merck, Bayer, Allergan Pharmaceuticals, Novartis, Theravance, and the Medecins Company. Mauro J. Salles has received grant support from Pfizer and Novartis, and lecture and consulting fees from Pfizer, Novartis, Bayer, MSD, Sanofi-Aventis, and Astra-Zeneca. Paul J. Planet and Eduardo Rodríguez-Noriega have received lecture and consulting fees from Pfizer. Carlos Seas has received fees and grant support from Pfizer and grant support from Glaxo Smith Kline and Bristol Myers Squibb. No other conflict of interest was reported.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00816-17.
REFERENCES
- 1.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talan DA, Krishnadasan A, Gorwitz RJ, Fosheim GE, Limbago B, Albrecht V, Moran GJ, EMERGEncy ID Net Study Group. 2011. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis 53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- 3.Laupland KB. 2013. Incidence of bloodstream infection: a review of population-based studies. Clin Microbiol Infect 19:492–500. doi: 10.1111/1469-0691.12144. [DOI] [PubMed] [Google Scholar]
- 4.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 5.Le Moing V, Alla F, Doco-Lecompte T, Delahaye F, Piroth L, Chirouze C, Tattevin P, Lavigne JP, Erpelding ML, Hoen B, Vandenesch F, Duval X, VIRSTA study group. 2015. Staphylococcus aureus bloodstream infection and endocarditis-a prospective cohort study. PLoS One 10:e0127385. doi: 10.1371/journal.pone.0127385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naber CK. 2009. Staphylococcus aureus bacteremia: epidemiology, pathophysiology, and management strategies. Clin Infect Dis 48:S231–S237. doi: 10.1086/598189. [DOI] [PubMed] [Google Scholar]
- 7.Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 9.Klein E, Smith DL, Laxminarayan R. 2009. Community-associated methicillin-resistant Staphylococcus aureus in outpatients, United States, 1999–2006. Emerg Infect Dis 15:1925–1930. doi: 10.3201/eid1512.081341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez CA, Barrientes OJ, Leal AL, Contreras GA, Barrero L, Rincón S, Diaz L, Vanegas N, Arias CA. 2006. Community-associated methicillin-resistant Staphylococcus aureus, Colombia. Emerg Infect Dis 12:2000–2001. doi: 10.3201/eid1212.060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arias CA, Rincon S, Chowdhury S, Martínez E, Coronell W, Reyes J, Nallapareddy SR, Murray BE. 2008. MRSA USA300 clone and VREF–a U.S.-Colombian connection? N Engl J Med 359:2177–2179. doi: 10.1056/NEJMc0804021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes J, Rincón S, Díaz L, Panesso D, Contreras GA, Zurita J, Carrillo C, Rizzi A, Guzmán M, Adachi J, Chowdhury S, Murray BE, Arias CA. 2009. Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin Infect Dis 49:1861–1867. doi: 10.1086/648426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planet PJ, Diaz L, Kolokotronis SO, Narechania A, Reyes J, Xing G, Rincon S, Smith H, Panesso D, Ryan C, Smith DP, Guzman M, Zurita J, Sebra R, Deikus G, Nolan RL, Tenover FC, Weinstock GM, Robinson DA, Arias CA. 2015. Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J Infect Dis 212:1874–1882. doi: 10.1093/infdis/jiv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egea AL, Gagetti P, Lamberghini R, Faccone D, Lucero C, Vindel A, Tosoroni D, Garnero A, Saka HA, Galas M; S. aureus Study Group-Argentina, Bocco JL, Corso A, Sola C. 2014. New patterns of methicillin-resistant Staphylococcus aureus (MRSA) clones, community-associated MRSA genotypes behave like healthcare-associated MRSA genotypes within hospitals, Argentina. Int J Med Microbiol 304:1086–1099. doi: 10.1016/j.ijmm.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Gelatti LC, Bonamigo RR, Inoue FM, Carmo MS, Becker AP, Castrucci FM, Pignatari AC, D'Azevedo PA. 2013. Community-acquired methicillin-resistant Staphylococcus aureus carrying SCCmec type IV in southern Brazil. Rev Soc Bras Med Trop 46:34–38. doi: 10.1590/0037-868213022013. [DOI] [PubMed] [Google Scholar]
- 17.Aires De Sousa M, Miragaia M, Sanches IS, Avila S, Adamson I, Casagrande ST, Brandileone MC, Palacio R, Dell'Acqua L, Hortal M, Camou T, Rossi A, Velazquez-Meza ME, Echaniz-Aviles G, Solorzano-Santos F, Heitmann I, de Lencastre H. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J Clin Microbiol 39:2197–2205. doi: 10.1128/JCM.39.6.2197-2205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez S, Murzicato S, Sandoval O, Fernández-Canigia L, Mollerach M. 2015. Community-acquired necrotizing pneumonia caused by methicillin-resistant Staphylococcus aureus ST30-SCCmecIVc-spat019-PVL positive in San Antonio de Areco, Argentina. Rev Argent Microbiol 47:50–53. [DOI] [PubMed] [Google Scholar]
- 19.Jones RN, Guzman-Blanco M, Gales AC, Gallegos B, Castro AL, Martino MD, Vega S, Zurita J, Cepparulo M, Castanheira M. 2013. Susceptibility rates in Latin American nations: report from a regional resistance surveillance program (2011). Braz J Infect Dis 17:672–681. doi: 10.1016/j.bjid.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martineau F, Picard FJ, Lansac N, Ménard C, Roy PH, Ouellette M, Bergeron MG. 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother 44:231–238. doi: 10.1128/AAC.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 21st informational supplement CLSI document MS100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milheiriço C, Oliveira DC, de Lencastre H. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother 60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist Updat 6:41–52. doi: 10.1016/S1368-7646(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 26.Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol 28:2059–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 28.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.