Abstract
Available plastomes of the Lauraceae show similar structure and varied size, but there has been no systematic comparison across the family. In order to understand the variation in plastome size and structure in the Lauraceae and related families of magnoliids, we here compare 47 plastomes, 15 newly sequenced, from 27 representative genera. We reveal that the two shortest plastomes are in the parasitic Lauraceae genus Cassytha, with lengths of 114,623 (C. filiformis) and 114,963 bp (C. capillaris), and that they have lost NADH dehydrogenase (ndh) genes in the large single-copy region and one entire copy of the inverted repeat (IR) region. The plastomes of the core Lauraceae group, with lengths from 150,749 bp (Nectandra angustifolia) to 152,739 bp (Actinodaphne trichocarpa), have lost trnI-CAU, rpl23, rpl2, a fragment of ycf2, and their intergenic regions in IRb region, whereas the plastomes of the basal Lauraceae group, with lengths from 157,577 bp (Eusideroxylon zwageri) to 158,530 bp (Beilschmiedia tungfangensis), have lost rpl2 in IRa region. The plastomes of Calycanthus (Calycanthaceae, Laurales) have lost rpl2 in IRb region, but the plastome of Caryodaphnopsis henryi (Lauraceae) remain intact, as do those of the nonLaurales magnoliid genera Piper, Liriodendron, and Magnolia. On the basis of our phylogenetic analysis and structural comparisons, different loss events occurred in different lineages of the Laurales, and fragment loss events in the IR regions have largely driven the contraction of the plastome in the Lauraceae. These results provide new insights into the evolution of the Lauraceae as well as the magnoliids as a whole.
Keywords: Lauraceae, chloroplast, genome, phylogenetic relationship, loss event
Introduction
In land plants, most chloroplast genomes are single, circular, double-stranded DNA sequences 100–220 kb in size, with a quadripartite structure including one large single-copy (LSC) region, one small single-copy (SSC) region, and a pair of inverted repeat (IR) regions (Bock 2007). Together these regions include >30 structural RNA genes and around 80 protein-coding genes, with the latter including genes related to photosynthesis, transcription or translation, and other functions (Gao etal. 2010). Generally, the ribosomal RNA genes are in the IR region, almost all of the photosynthesis related genes in the LSC region, and a number of the NADPH dehydrogenase genes in the SSC region. The plastomes of land plants originated once, from a free-living algal ancestor (Turmel etal. 2006), but the gene contents and order vary considerably among species, and significant structural rearrangements and gene losses have been reported in several unrelated lineages, including ferns (Roper etal. 2007; Karol etal. 2010), gnetophytes (McCoy etal. 2008; Wu etal. 2009), and multiple angiosperm families (Goremykin etal. 2003a; Cai etal. 2006), as well as nonphotosynthetic plants (Wicke etal. 2016).
Comparative analyses of the plastomes of algae and embryophytes show that four genes, tufA, ftsH, odpB, and rpl5, have been lost or transferred to the nucleus and three genes, matK, ycf1, and ycf2, have been gained in charophyte algae and embryophytes (Turmel etal. 2006). For example, the tufA gene, encoding chloroplast protein synthesis elongation factor Tu, is encoded in the plastomes of most algae, but is a pseudogene in Isoetes, fragmented in Anthoceros, cycads, and Gingko, and completely lost in the angiosperms (Karol etal. 2010). Within the angiosperms, three genes, ycf1, ycf2, and accD, have been lost in the Poaceae (Guisinger etal. 2010), whereas rpl22, infA, and accD were lost in the legumes, Lemnoideae, and Acoraceae, respectively (Wang and Messing 2011; Goremykin etal. 2005; Doyle etal. 1995). In plants with a heterotrophic lifestyle, pseudogenization and entire loss events of ndh-genes were detected (Wickett etal. 2008; Barrett etal. 2014; Wicke etal. 2016). However, the ndh-gene loss events have also occurred in autotrophic orchids, gnetophytes, and Pinaceae (Braukmann etal. 2009; Kim etal. 2015; Wakasugi etal. 1994).
In addition to gene losses, large inversions, and other structural rearrangements have been also reported. In ferns and seed plants, a 30-kb fragment flanked by the complete matK and rpoC2 has been identified as an inversion, with gene organization different from that in liverworts, mosses, hornworts, lycophytes, and Chaetosphaeridium (Wickett etal. 2011). In rice, maize, Calamus, and orchids, two identical trnH-rps19 gene clusters were detected as a duplication event before the diversification of extant monocot lineages (Chang etal. 2006; Wang etal. 2008; Luo etal. 2016). In Tetracentron and Trochodendron, a 4-kb extra region containing the five genes rpl22, rps3, rpl16, rpl14, and rps8 was found as evidence for unstable boundaries of the IR region across early-diverging eudicots (Sun etal. 2013, 2016). Interestingly, most of the rearrangements were detected in the boundary regions of IR, suggesting that the IR regions represent hotspots for structural rearrangements within the plastome (Wicke etal. 2011; Zhu etal. 2016).
The IR regions in the plastome of angiosperms have been used as evolutionary markers for elucidating relationships among some taxa, because they are frequently subject to contraction, expansion, or even complete loss (Lavin etal. 1990; Kim and Jansen 1994; Plunkett and Downie 2000; Luo etal. 2016; Sun etal. 2016; Zhu etal. 2016). In the early-diverging eudicots, the IR regions range from 24.3 to 36.4 kb in length and contain from 18 to 33 genes (Sun etal. 2016). In early-diverging monocots, the IR regions range from 25.2 to 33.3 kb in length and contain from 16 to 20 genes (Luo etal. 2016). As extreme examples, loss of one or two IR regions has been detected in Cephalotaxaceae (Yi etal. 2013), Pinaceae (Wu etal. 2011 b), Taxodiaceae (Hirao etal. 2008), Leguminosae (Palmer etal. 1987; Lavin etal. 1990), Geraniaceae (Guisinger etal. 2011), and Cactaceae (Sanderson etal. 2015).
After the eudicots and monocots, the magnoliids is the third-largest group of Mesangiospermae, and includes four orders, 19 families, and over 9,000 woody species from all over the world (www.theplantlist.org). However, <30 species have assembled chloroplast genome sequences, and there has not been a systemic structural comparison of these plastomes. To improve understanding of the dynamics and evolution of plastome structure in magnoliids, we therefore focused on the plastomes of the important family Lauraceae and the related families Calycanthaceae (Laurales), Chloranthaceae (Chloranthales), Magnoliaceae (Magnoliales), Piperaceae (Piperales), and Winteraceae (Canellales). We included 15 newly sequenced and 33 previously reported plastomes in our study, representing 25 genera from all four orders of magnoliids. The main objectives of this study were 1) to reconstruct the phylogenetic relationships using the sequenced magnoliid plastomes, 2) to reveal plastome structural variations in Lauraceae, 3) to trace the evolutionary pattern of plastome contraction.
Materials and Methods
Plant Material and Plastome Sequencing
Fresh leaves and silica-gel dried materials were sampled from 15 species representing 10 genera of Lauraceae. The voucher specimens for the 15 sampled plants collected from China and Indonesia were deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden (HITBC), Chinese Academy of Sciences (CAS; table 1). Genomic DNA was extracted from 2 g leaves using the CTAB method (Doyle and Dickson 1987), in which 4% CTAB was used, and we added ∼1% polyvinyl polypyrrolidone (PVP) and 0.2% dl-dithiothreitol (DTT). From each purified sample of total DNA, 0.5 μg was fragmented to construct short-insert (500 bp) libraries following the manufacturer‘s manual (Illumina) and then used for sequencing. The DNA samples were indexed by tags and pooled together in one lane of a Genome Analyzer (Illumina HiSeq 2000) for sequencing at BGI-Shenzhen, and >4.0 Gb of reads for each sample were obtained.
Table 1.
Sampled Species of Lauraceae and Their Voucher Specimens Sequenced in This Study
| No | Species | Herbarium | Taxon | Voucher | Geographic Origin | Accession Number in GenBank |
|---|---|---|---|---|---|---|
| 1 | Eusideroxylon zwageri | HITBC-BRG | Eusideroxylon zwageri Teijsm. & Binn. | SY34806 | Sulawesi, Indonesia | MF939351 |
| 2 | Cryptocarya chinensis | HITBC-BRG | Cryptocarya chinensis (Hance) Hemsl. | SY34239 | Jianfenglin, Hainan | MF939349 |
| 3 | Cryptocarya hainanensis | HITBC-BRG | Cryptocarya hainanensis Merr. | SY01426 | Menghai, Yunnan | MF939350 |
| 4 | Beilschmiedia tungfangensis | HITBC-BRG | Beilschmiedia tungfangensis S.K. Lee & L.F. Lau | SY34805 | Wenshan, Yunnan | MF939348 |
| 5 | Beilschmiedia pauciflora | HITBC-BRG | Beilschmiedia pauciflora H.W. Li | SY01364 | Mengla, Yunnan | MF939347 |
| 6 | Cassytha filiformis | HITBC-BRG | Cassytha filiformis Linnaeus | SY34802 | Menghai, Yunnan | MF939337 |
| 7 | Cassytha capillaris | HITBC-BRG | Cassytha capillaris Meisn. | SY34803 | Sulawesi, Indonesia | MF939338 |
| 8 | Neocinnamomum caudatum | HITBC-BRG | Neocinnamomum caudatum (Nees) Merr. | SY01561 | Puer, Yunnan | MF939344 |
| 9 | Neocinnamomum lecomtei | HITBC-BRG | Neocinnamomum lecomtei H. Liu | SY33249 | Wenshan, Yunnan | MF939345 |
| 10 | Caryodaphnopsis henryi | HITBC-BRG | Caryodaphnopsis henryi Airy Shaw | SY01542 | Honghe, Yunnan | MF939346 |
| 11 | Caryodaphnopsis malipoensis | HITBC-BRG | Caryodaphnopsis malipoensis Bing Liu & Y. Yang | SY32618 | Wenshan, Yunnan | MF939343 |
| 12 | Actinodaphne trichocarpa | HITBC-BRG | Actinodaphne trichocarpa C.K. Allen | SY32938 | Emei, Sichuan | MF939342 |
| 13 | Neolitsea sericea | HITBC-BRG | Neolitsea sericea (Blume) koidzumi | SY33307 | Linan, Zhejiang | MF939341 |
| 14 | Nectandra angustifolia | HITBC-BRG | Nectandra angustifolia (Schrad.) Nees & Mart. | SY34804 | Sulawesi, Indonesia | MF939340 |
| 15 | Sassafras tzumu | HITBC-BRG | Sassafras tzumu (Hemsl.) Hemsl. | SY34790 | Anqing, Anhui | MF939339 |
Genome Annotation and Comparison
The paired-end reads were filtered using GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle) to get plastid-like reads, then the filtered reads were assembled using SPAdes version 3.10 (Bankevich etal. 2012). To retain pure chloroplast contigs, the final “fastg” files were filtered using the “slim” script of GetOrganelle. The filtered De Brujin graphs were viewed and edited using Bandage (Wick etal. 2015), then a circular chloroplast genome was generated. The genome was automatically annotated using CpGAVAS (Liu etal. 2012), then adjusted using Geneious version 9.1.7 (Kearse etal. 2012). The annotated chloroplast genomes have been submitted to GenBank (accession number: MF939337 to MF939351). The genome maps of all the 15 plastomes were drawn by OrganellarGenomeDRAW tool (OGDRAW; Lohse etal. 2013) and the gene organization maps were drawn by Gene Structure Display Server (GSDS) version 2.0 (Hu etal. 2015). Mauve version 2.4.0 software was used for alignment and determining the plastome rearrangements among the Magnoliids (Darling etal. 2004).
Phylogenetic Analysis
To estimate phylogenetic relationships within the magnoliids, 47 taxa with available complete plastomes were compared, including one taxon each from Canellales and Chloranthales, four from Piperales, six from Magnoliales, and 35 from Laurales. The 35 taxa included the 15 new plastomes and 20 complete plastomes which have been published elsewhere or adopted from NCBI (Song etal. 2015, 2016; Wu etal. 2017). Amborella trichopoda (AJ506156) was treated as the outgroup. For the species tree, maximum likelihood (ML) analyses were performed on data sets of 48 plastome sequences with single IR, SSC, and LSC regions. The whole genome matrix was aligned using MAFFT version 3.73 (Katoh and Standley 2013), then manually edited using Geneious version 9.1.7 (Kearse etal. 2012). ML analysis was conducted using RAxML version 7.2.6 with the GTR + G model to search the best-scoring ML tree (Tamura etal. 2011). One thousand bootstrap replicates were performed to obtain the confidence support. Bayesian inference (BI) was performed using MrBayes version 3.2.6 (Ronquist and Huelsenbeck 2003). The best-fit DNA substitution model of the Bayesian information criterion (BIC) was evaluated by using jModeltest version 2.1.10 (Darriba etal. 2012; Guindon etal. 2003). Markov Chain Monte Carlo (MCMC) analyses were run in MrBayes for 10,000,000 generations. The BI analysis started with a random tree and sampled every 1,000 generations. The first 25% of the trees was discarded as burn-in, and the remaining trees were used to generate a majority-rule consensus tree (supplementary fig. S1, Supplementary Material online). The trees were viewed and edited with the Fig tree version 1.4.0 software (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Overall Structure and Gene Pool
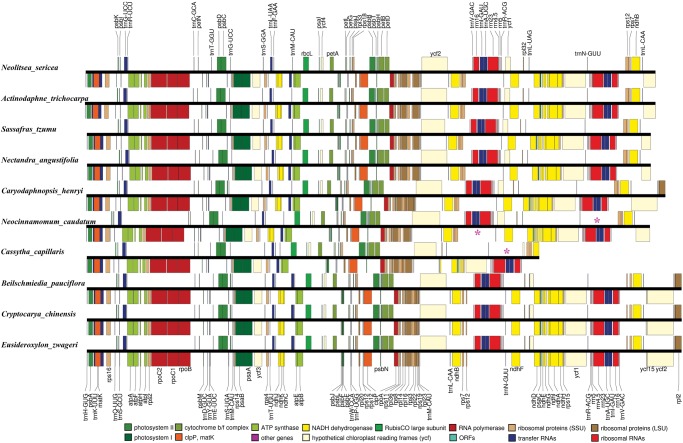
Thirteen of the 15 newly sequenced Lauraceae plastomes displayed the typical quadripartite structure of angiosperms, including LSC, SSC, and a pair of IR regions, whereas the two plastomes from Cassytha, a genus of parasitic vines, have lost one copy of the IR (fig. 1). The complete plastome of Cassytha filiformis is 114,623 bp in length, 340 bp shorter than that of Cassytha capillaris (114,963 bp), and 42,954 bp shorter than that of Eusideroxylon zwageri (157,577 bp; table 2). Among the other 13 plastomes, genome size ranged from 150,749 bp (Nectandra angustifolia) to 158,530 bp (Beilschmiedia tungfangensis). In the LSC region, the length varied from 86,035 (Caryodaphnopsis henryi) to 93,803 bp (Neolitsea sericea), in the SSC region from 15,751 bp (Caryodaphnopsis malipoensis) to 19,222 bp (Cryptocarya chinensis), and in the IR region from 19,292 (N. angustifolia) to 25,601 bp (C. henryi). The plastomes of Eusideroxylon, Cryptocarya, Beilschmiedia, and C. henryi shared identical complements of coding genes; a total of 130 genes, including 8 rRNA genes, 37 tRNA, and 85 protein-coding genes, of which 17 are duplicated in IR regions. A total of 128 genes were detected on the plastomes of Neocinnamomum, Nectandra, Sassafras, Actinodaphne, Neolitsea, and Caryodaphnopsis malipoensis, 113 of which are single copy, while 15 are duplicated in IR regions. The different gene numbers reflect the duplication of rpl23 and trnI-CAU in the first group. The plastomes of Cassytha have not only lost the duplicated genes in the IR region, but also six NADH dehydrogenase (ndh) genes, ndhA, ndhC, ndhG, ndhI, ndhJ, and ndhK, and their five ndh genes are pseudogenes.
| Neocinnamomum caudatum | Neocinnamomum lecomtei | Caryodaphnopsis henryi | Caryodaphnopsis malipoensis | Actinodaphne trichocarpa | Neolitsea sericea | Nectandra angustifolia | Sassafras tzumu |
|---|---|---|---|---|---|---|---|
| 150,842 | 150,838 | 154,938 | 149,239 | 152,739 | 152,442 | 150,749 | 151,798 |
| 91,881 | 91,912 | 86,035 | 91,901 | 93,783 | 93,803 | 93,783 | 92,752 |
| 20,257 | 20,257 | 25,601 | 20,036 | 20,078 | 20,067 | 19,292 | 20,096 |
| 18,447 | 18,412 | 17,701 | 17,266 | 18,800 | 18,505 | 18,382 | 18,854 |
| 38.80% | 38.80% | 39.00% | 39.00% | 39.20% | 39.20% | 39.20% | 39.20% |
| 128 (113) | 128 (113) | 131 (113) | 128 (113) | 128 (113) | 128 (113) | 128 (113) | 128 (113) |
| 84 | 84 | 86 | 84 | 84 | 84 | 84 | 84 |
| 36 | 36 | 37 | 36 | 36 | 36 | 36 | 36 |
| 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| 5,517 | 5,517 | 5,526 | 5,526 | 5,574 | 5,568 | 5,535 | 5,586 |
| 928 | 928 | 1,473 | 1,473 | 1,378 | 1,372 | 1,372 | 1,419 |
| 6,831 | 6,831 | 6,894 | 6,894 | 6,876 | 6,846 | 6,909 | 6,294 |
| 3,110 | 3,110 | 6,894 | 3,186 | 3,168 | 3,162 | 2,478 | 3,168 |
Fig. 1.
—Gene maps of the plastomes of Cassytha, Eusideroxylon, Cryptocarya, Beilschmiedia, Caryodaphnopsis, Neocinnamomum, Nectandra, Sassafras, Neolitsea, and Actinodaphne in the Lauraceae. The pink asterisks indicate the structural differences of IR loss.
Table 2.
Summary of 15 Complete Plastomes of Lauraceae
| Eusideroxylon zwageri | Cryptocarya chinensis | Cryptocarya hainanensis | Beilschmiedia tungfangensis | Beilschmiedia pauciflora | Cassytha filiformis | Cassytha capillaris | |
|---|---|---|---|---|---|---|---|
| Total cpDNA size (bp) | 157,577 | 157,675 | 157,145 | 158,530 | 157,901 | 114,623 | 114,963 |
| Length of LSC region (bp) | 89,231 | 89,199 | 89,002 | 89,351 | 88,673 | – | – |
| Length of IR region (bp) | 24,717 | 24,627 | 24,621 | 25,473 | 25,496 | – | – |
| Length of SSC region (bp) | 18,912 | 19,222 | 18,901 | 18,233 | 18,236 | – | – |
| Total GC content | 39.10% | 39.10% | 39.10% | 39.00% | 39.00% | 36.90% | 36.90% |
| Total number of genes (unique) | 130 (113) | 130 (113) | 130 (113) | 130 (113) | 130 (113) | 107 (107) | 107 (107) |
| protein encoding | 85 | 85 | 85 | 85 | 85 | 73 | 73 |
| tRNA | 37 | 37 | 37 | 37 | 37 | 30 | 30 |
| rRNA | 8 | 8 | 8 | 8 | 8 | 4 | 4 |
| Length of ycf1 (bp) | 5,493 | 5,460 | 5,436 | 5,436 | 5,460 | 5,211 | 5,211 |
| Length of truncated ycf1(bp) | 971 | 977 | 974 | 1,863 | 1,863 | – | – |
| Length of ycf2(bp) | 6,882 | 6,885 | 6,885 | 6,843 | 6,849 | 5,583 | 5,583 |
| Length of complete or truncated ycf2(bp) | 6,882 | 6,885 | 6,885 | 6,843 | 6,849 | – | – |
Phylogenomic Analysis
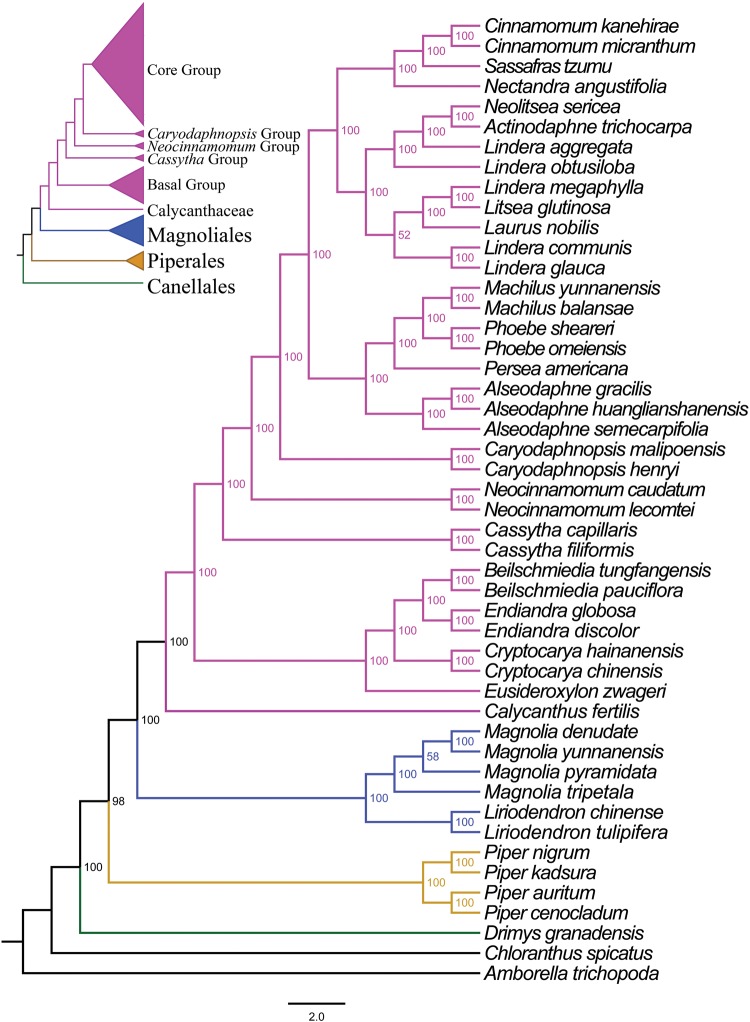
The matrix of complete plastomes was used to reconstruct a phylogenetic tree of magnoliids (fig. 2). Magnoliids are divided into five main clades (ML-BS = 100%) corresponding to five orders: Canellales, Chloranthales, Laurales, Magnoliales, and Piperales. Sisterhood of Laurales and Magnoliales, with Piperales and Canellales being the next sister groups, was highly supported. Two major clades, including Calycanthaceae and Lauraceae, were recognized within the Laurales. There was 100% support for the monophyly of Lauraceae family. Five well-supported groups were recovered within the Lauraceae (ML-BS = 100%). The basal group (ML-BS = 100%), including the genera Eusideroxylon, Cryptocarya, Beilschmiedia, and Endiandra, the Cassytha group (ML-BS = 100%), the Neocinnamomum group (ML-BS = 100%), the Caryodaphnopsis group (ML-BS = 100%), and the core group (ML-BS = 100%), including Alseodaphne, Persea, Phoebe, Machilus, Lindera, Laurus, Actinodaphne, Neolitsea, Litsea, Nectandra, Sassafras, and Cinnamomum.
Fig. 2.
—Molecular phylogenetic tree of 47 taxa of Magnoliids based on complete plastome sequences using unpartitioned ML. Numbers at each node are bootstrap support value.
Plastome Comparisons
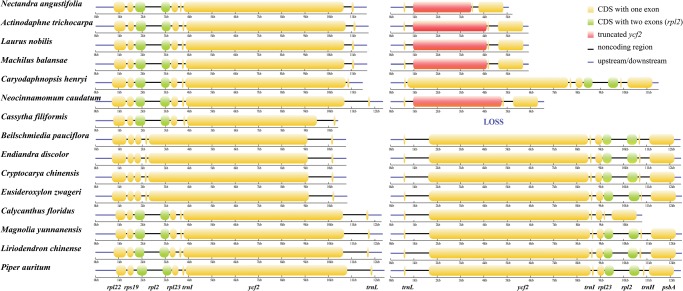
Synteny and rearrangements were detected in ten plastomes of Lauraceae. A significant degree of synteny was found within the basal group, including E. zwageri and B. tungfangensis, and the core group, including N. angustifolia, Laurus nobilis, Lindera communis, Machilus balansae, Alseodaphne semecarpifolia, Neocinnamomum caudatum, and C. capillaris. However, the two groups differ in the orientation of a 13.7-kb fragment flanked by rps7 and rpl2 (fig. 3). In the basal group, the rps7-ndhB-trnL-ycf2-trnI-rpl23-rpl2 segment has been combined with trnH-GUG, whereas the segment of the core group species has been combined with rps19 (fig. 4), indicating that a rearrangement event occurred in Lauraceae plastome evolution. In the plastomes of C. henryi and in the basal group species, two unbroken protein-coding copies of ycf2 were detected, suggesting that fragmentation of ycf2 has occurred in other species of Lauraceae. Moreover, upstream of rps19 adjoining the IR region, we detected one copy of a protein-coding gene rpl23 and a tRNA gene trnM-CAU in the plastome of C. henryi and the basal group species, but not in the plastomes of other species, indicating that significant IR boundary changes occurred in Lauraceae plastome evolution.
Fig. 3.
—Synteny and rearrangements detected in Lauraceae plastomes using the Mauve multiple-genome alignment program. The yellow asterisks 1 indicate the varied gene locus of rpl2, the blue asterisks 2 indicate a rearrangement of the fragment flanked by rpl2 and rps7, and the green asterisks 3 indicate an insert of intergenic region rpl2–rpl23.
Fig. 4.
—Comparisons of gene loci in the fragments flanked by rps19 and trnL in IRa and trnL and trnH in IRb among 15 plastomes of Magnoliids.
IR Expansion and Contraction
In the sequenced plastomes of Lauraceae, two complete or fragmented copies of ycf1and ycf2 were located at the boundaries between the IR regions and the LSC or SSC regions. The full lengths of ycf2 and ycf1 ranged from 5,583 bp in Cassytha filiformis to 6,894 bp in Caryodaphnopsis malipoensis and from 5,211 bp in Cassytha filiformis to 5,586 bp in Sassafras tzumu, respectively (table 2). Double complete copies of the ycf2 genes were detected in the seven sequenced Lauraceae plastomes of the basal group species, but only one complete copy and one fragment in the 24 plastomes of C. malipoensis, Neocinnamomum, and the core group species, except those of C. henryi and both Cassytha species. The length of the fragment of ycf2 ranged from 2,478 bp in N. angustifolia to 3,168 bp in Actinodaphne trichocarpa. In contrast, all 32 sequenced Lauraceae plastomes, except the two species of Cassytha, had one complete copy and a fragment of ycf1. The length of the fragment of ycf1 ranged from 971 bp in E. zwageri to 1, 863 bp in Beilschmiedia pauciflora. Neither Cassytha plastome had fragments of ycf1 and ycf2, but only one complete copy of each due to the IR loss.
Discussion
Relationships in Lauraceae
This study included 47 complete chloroplast genomes for plants from all five orders (Canellales, Chloranthales, Laurales, Magnoliales, and Piperales) of the magnoliids. All of these complete plastome sequences of Lauraceae and related families yielded a fully resolved tree, consistent with the Angiosperm Phylogeny Group’s most recent phylogeny, APG IV (Byng etal. 2016). Relationships among the five orders of the magnoliids are clarified as sisterhood of Laurales and Magnoliales, with Piperales and Canellales being the next sister groups, and Chloranthales the most basal group. Calycanthaceae and Lauraceae were recognized within the Laurales. All of these clades were recognized by Renner (Renner 1999).
The deep relationships of 34 Lauraceae taxa are separated into the following groups in our study. Eusideroxylon, Cryptocarya, Beilschmiedia, and Endiandra form the first group in the phylogeny. Cassytha, Neocinnamomum, and Caryodaphnopsis form the second, third, and fourth groups, respectively. The fifth group includes Alseodaphne, Persea, Phoebe, and Machilus. The sixth group includes Nectandra, Sassafras, and Cinnamomum. And the last group includes Lindera, Laurus, Litsea, Actinodaphne, and Neolitsea. The phylogenetic placements of the first, fourth, fifth, and sixth groups are consistent with previously published phylogenetic relationships (Chanderbali etal. 2001; Rohwer and Rudolph 2005). The position of Cassytha, considered as a ‘jumping genus’ by Rohwer and Rudolph (2005), was settled here in the way predicted from morphology (Chanderbali etal. 2001). The seventh group, equivalent to the tribe Laureae (Chanderbali etal. 2001), was confirmed as sister to the sixth group, tribe Cinnamomeae (including Sassafras), which has always been assumed based on morphological characters, although previous molecular analyses failed to prove it convincingly (Chanderbali etal. 2001; Rohwer and Rudolph 2005).
Unusual Structure of the Cassytha Plastomes
The sizes of the fifteen newly sequenced Lauraceae plastomes differed greatly, from 114,623 bp in the hemiparasitic vine, C. capillaris, to 158,530 bp in B. tungfangensis, as a result of the loss of one IR copy and six ndh genes in Cassytha. Cassytha is the only stem hemiparasitic genus with reduced leaves and roots in the magnoliids, and the only nonwoody member of the Lauraceae. We show that it is also unique in the Lauraceae the loss of one IR copy in its plastome, although similar losses have occurred independently in the Leguminosae (Cai etal. 2008), Pinaceae (Raubeson and Jansen 1992), Cephalotaxaceae (Yi etal. 2013), and cupressophytes (Wu etal. 2011a). In addition, six ndh genes, ndhA, ndhC, ndhG, ndhI, ndhJ, and ndhK, have been lost, and the other five, ndhB, ndhD, ndhE, ndhF, and ndhH, are clearly pseudogenes in both Cassytha taxa sequenced in this study. All eleven ndh genes encode independent subunits of a plastid NADPH-dehydrogenase complex (Ndh 1-complex) which carries out one of the recycled electron pathways around Photosystem I (Casano etal. 2000). Cyclic electron flow is vital for maintenance of efficient photosynthesis and enablement of photoprotection under environmental stresses in higher plants (Wang etal. 2006). The ndh genes are frequently pseudogenized or lost in plant groups with a degree of heterotrophy, such as Aneura, Cuscuta, Epifagus, Hydnora, and nonphotosynthetic orchid species, and in some autotrophic gymnosperms and ferns (dePamphilis and Palmer 1990; Wicke etal. 2011; Wickett etal. 2008; McNeal etal. 2007; Kim etal. 2015; Naumann etal. 2016), but this is first report for Cassytha, the only hemiparasitic genus in the Laurales. This adds to the evidence that the Ndh1-complex is not essential for plant survival, while the ndh-independent antimycin-A-sensitive pathway, which functions in cyclic electron flow as another choice, could be more important under most conditions (Shikanai 2014).
Loss Events in the Laurales
Comparative genomic analysis indicated that missing segments of DNA in Lauraceae plastids mainly drive the genome contraction events. A fragment flanked by rps7 and rpl2 was detected as a rearrangement event between the basal group species and the other species except C. henryi. However, it looks more like two or more independent loss events when we choose the plastomes of C. henryi or nonLaurales species as reference. Double IR fragments with the gene order of trnL-ycf2-trnI-rpl23-rpl2 are highly conserved in the plastomes of C. henryi (fig. 4) and nonLaurales genera such as Drimys, Piper, Liriodendron, and Magnolia (Cai etal. 2006; Zhu etal. 2016; Yang etal. 2014), indicating the plastome of C. henryi is evolutionarily conserved. In Calycanthus (Laurales) plastome (Goremykin etal. 2003a), one copy of rpl2 with the length of 1,480 bp disappeared from the trnL-rpl2 fragment in IRb, but all of the sequenced Lauraceae plastomes of the basal group, including Endiandra, Beilschmiedia, Cryptocarya, and Eusideroxylon, lost another copy of rpl2 from the trnL-rpl2 fragment in the IRa region (fig. 4). More interesting are the sequenced Lauraceae plastomes of the core group, including Alseodaphne, Persea (Song etal. 2016), Phoebe, Machilus (Song etal. 2015), Lindera, Laurus, Litsea, Nectandra, Sassafras, Cinnamomum (Wu etal. 2017), Actinodaphne, and Neolitsea, which have further lost a segment of at least 4,500 bp which contains a fragment of ycf2 and one copy of rpl23 and trnI-CAU in IRb of Calycanthus. This segment was also lost in the plastomes of Neocinnamomum species and C. malipoensis. Taken together, these independent loss events show that in the Lauraceae the plastomes of Neocinnamomum, Cassytha, the core group, and the basal group could share a common ancestral genome structure like that of C. henryi, but have subsequently evolved independently with different loss patterns.
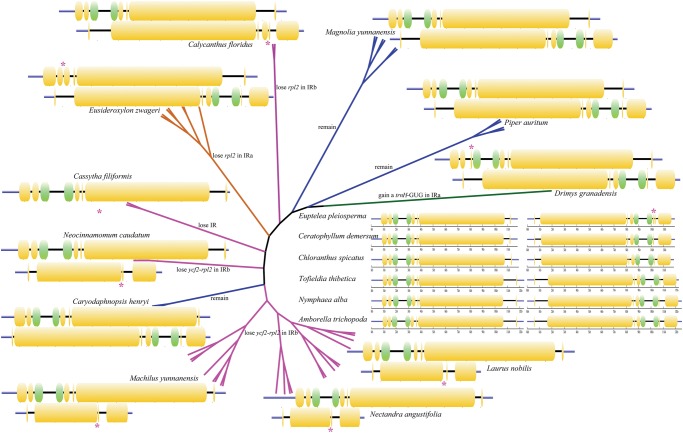
Evolutionary Pattern in Angiosperms
To put these results in a wider phylogenetic context, we traced the fragments flanked by trnL-CAA and rps19 in the IRa region and by trnL-CAA and trnH-GUG in the IRb region in the six major groups of the angiosperms and found that the gene backbone and order are conserved (fig. 5). In the early-diverging angiosperm species, A. trichopoda and Nymphaea alba, of the ANITA group (Qiu etal. 1999), the gene orders of the fragments are rps19-rpl2-rpl23-trnI-ycf2-trnL and trnL-ycf2-trnI-rpl23-rpl2-trnH (Goremykin etal. 2003b, 2004). These orders are retained in the early diverging monocot Tofieldia thibetica (Luo etal. 2016) and Ceratophyllum demersum in the Ceratophyllaceae (Moore etal. 2007). In the early diverging eudicot Euptelea pleiosperma (Sun etal. 2016), the only change in the gene order is a new insertion of a fragment of rps19. In the magnoliids, the same gene order for both fragments is retained in the sequenced species of Choranthaceae (Hansen etal. 2007), Piperales (Cai etal. 2006), and Magnoliales (Zhu etal. 2016), but a new copy of trnH has been inserted between rps19 and rpl2 in the IRa fragment of Drimys granadensis in the Canellales (Cai etal. 2006) and the copy of rpl2 has been lost between rps19 and rps23 in the IRa region of Endiandra, Beilschmiedia, Cryptocarya, and Eusideroxylon species in Lauraceae, and in IRb of Calycanthus in Calycanthaceae (Goremykin etal. 2003a). Nevertheless, our comparative genomic analysis concluded that the regions encompassing the ycf2 and the adjoined trnH-GUG or trnL-CAA gene in the plastomes of C. henryi and other early-diverging angiosperms are the retained IRs, corresponding to either IRa or IRb in the basal and core groups of Lauraceae.
Fig. 5.
—Model of the origin and variation of the fragments flanked by rps19 and trnL in IRa and trnL and trnH in IRb among plastomes of angiosperms. The pink asterisks indicate the varied gene loci.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Jing Yang, Juan-Hong Zhang, Zheng-Shan He, Chun-Yan Lin, and Ji-Xiong Yang at the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy Sciences, for sequencing technology. They sincerely thank the anonymous referees and Prof. Giovanni Vendramin for their critical and invaluable comments that greatly improved our manuscript. This work was supported by the National Natural Science Foundation of China (No. 31600531), a grant of the Large-scale Scientific Facilities, CAS (No.2017-LSF-GBOWS-02), the CAS “Light of West China” Program (Y7XB061B01), the 1000 Talents Program (WQ20110491035), and the project of the Southeast Asia Biodiversity Research Institute, CAS.
Literature Cited
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Bock R. 2007. Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr Opin Biotechnol. 18(2):100–106. [DOI] [PubMed] [Google Scholar]
- Braukmann TWA, Kuzmina M, Stefanovic S.. 2009. Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr Genet. 55(3):323–337. [DOI] [PubMed] [Google Scholar]
- Byng JW, et al. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20. [Google Scholar]
- Cai Z, et al. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6:77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai ZQ, et al. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 67(6):696–704. [DOI] [PubMed] [Google Scholar]
- Casano LM, Zapata JM, Martin M, Sabater B.. 2000. Chlororespiration and poising of cyclic electron transport. Plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. J Biol Chem. 275(2):942–948. [DOI] [PubMed] [Google Scholar]
- Chanderbali AS, van der Werff H, Renner SS.. 2001. Phylogeny and historical biogeography of Lauraceae: evidence from the chloroplast and nuclear genomes. Ann Missouri Bot Gard. 88(1):104–134. [Google Scholar]
- Chang CC, et al. 2006. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 23(2):279–291. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT.. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePamphilis CW, Palmer JD.. 1990. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348(6299):337–339. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Dickson EE.. 1987. Preservation of plant-samples for DNA restriction endonuclease analysis. Taxon 36(4):715–722. [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD.. 1995. Multiple independent losses of 2 genes and one intron from legume chloroplast genomes. Syst Bot. 20(3):272–294. [Google Scholar]
- Gao L, Su Y-J, Wang T.. 2010. Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J Syst Evol. 48(2):77–93. [Google Scholar]
- Goremykin V, Hirsch-Ernst K, Wlfl S, Hellwig F.. 2003a. The chloroplast genome of the “basal” angiosperm Calycanthus fertilis – structural and phylogenetic analyses. Plant Syst Evol. 242(1):119–135. [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH.. 2003. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that amborella is not a basal angiosperm. Mol Biol Evol. 20(9):1499–1505. [pii] [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH.. 2004. The chloroplast genome of Nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm. Mol Biol Evol. 21(7):1445–1454. [pii] [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH.. 2005. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol. 22(9):1813–1822. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, Rannala B.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Guisinger M, Chumley T, Kuehl J, Boore J, Jansen R.. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 70(2):149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK.. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28(1):583–600. [DOI] [PubMed] [Google Scholar]
- Hansen DR, et al. 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 45(2):547–563. [DOI] [PubMed] [Google Scholar]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K.. 2008. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 8:70.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, et al. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol KG, et al. 2010. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol. 10(1):321.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, et al. 2015. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE. 10(11):e0142215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YD, Jansen RK.. 1994. Characterization and phylogenetic distribution of a chloroplast DNA rearrangement in the Berberidaceae. Plant Syst Evol. 193(1–4):107–114. [Google Scholar]
- Lavin M, Doyle JJ, Palmer JD.. 1990. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the leguminosae subfamily Papilionoideae. Evolution 44(2):390–402. [DOI] [PubMed] [Google Scholar]
- Liu C, et al. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R.. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, et al. 2016. Plastid phylogenomic analyses resolve Tofieldiaceae as the root of the early diverging monocot order Alismatales. Genome Biol Evol. 8(3):932–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy SR, Kuehl JV, Boore JL, Raubeson LA.. 2008. The complete plastid genome sequence of Welwitschia mirabilis: an unusually compact plastome with accelerated divergence rates. BMC Evol Biol. 8:130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW.. 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE.. 2007. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Nat Acad Sci U S A. 104(49):19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann J, et al. 2016. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol Evol. 8(2):345–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Osorio B, Aldrich J, Thompson WF.. 1987. Chloroplast DNA evolution among legumes – loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr Genet. 11(4):275–286. [Google Scholar]
- Plunkett GM, Downie SR.. 2000. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst Bot. 25(4):648–667. [Google Scholar]
- Qiu YL, et al. 1999. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature 402(6760):404–407. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK.. 1992. A rare chloroplast-DNA structural mutation is shared by all conifers. Biochem Syst Ecol. 20(1):17–24. [Google Scholar]
- Renner SS. 1999. Circumscription and phylogeny of the Laurales: evidence from molecular and morphological data. Am J Bot. 86(9):1301–1315. [PubMed] [Google Scholar]
- Rohwer JG, Rudolph B.. 2005. Jumping genera: the phylogenetic positions of Cassytha, Hypodaphnis, and Neocinnamomum (Lauraceae) based on different analyses of trnK intron sequences. Ann Missouri Bot Gard. 92(2):153–178. [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Roper JM, et al. 2007. The complete plastid genome sequence of Angiopteris evecta (G. Forst.) Hoffm. (Marattiaceae). Am Fern J. 97(2):95–106. [Google Scholar]
- Sanderson MJ, et al. 2015. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am J Bot. 102(7):1115–1127. [DOI] [PubMed] [Google Scholar]
- Shikanai T. 2014. Central role of cyclic electron transport around photosystem I in the regulation of photosynthesis. Curr Opin Biotechnol. 26:25–30. [DOI] [PubMed] [Google Scholar]
- Song Y, et al. 2015. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front Plant Sci. 6:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yao X, Tan YH, Gan Y, Corlett RT.. 2016. Complete chloroplast genome sequence of the avocado: gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can J For Res. 46(11):1293–1301. [Google Scholar]
- Sun Y, et al. 2016. Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol Phylogenet Evol. 96:93–101. [DOI] [PubMed] [Google Scholar]
- Sun YX, et al. 2013. Complete plastid genome sequencing of trochodendraceae reveals a significant expansion of the inverted repeat and suggests a paleogene divergence between the two extant species. PLoS ONE. 8(4):e60429.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C.. 2006. The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol. 23(6):1324–1338. [DOI] [PubMed] [Google Scholar]
- Wakasugi T, et al. 1994. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci U S A. 91(21):9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, et al. 2006. Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 141(2):465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R-J, et al. 2008. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 8(1):36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Messing J.. 2011. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS ONE. 6(9):e24670.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE.. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Nat Acad Sci U S A. 113(32):9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D.. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Forrest LL, Budke JM, Shaw B, Goffinet B.. 2011. Frequent pseudogenization and loss of the plastid-encoded sulfate-transport gene cysA throughout the evolution of liverworts. Am J Bot. 98(8):1263–1275. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, et al. 2008. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 25(2):393–401. [DOI] [PubMed] [Google Scholar]
- Wu CC, Chu FH, Ho CK, Sung CH, Chang SH.. 2017. Comparative analysis of the complete chloroplast genomic sequence and chemical components of Cinnamomum micranthum and Cinnamomum kanehirae. Holzforschung 71(3):189–197. [Google Scholar]
- Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM.. 2009. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol Phylogenet Evol. 52(1):115–124. [DOI] [PubMed] [Google Scholar]
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM.. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM.. 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 3:1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JB, Li DZ, Li HT.. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031. [DOI] [PubMed] [Google Scholar]
- Yi X, Gao L, Wang B, Su YJ, Wang T.. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol Evol. 5(4):688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AD, Guo WH, Gupta S, Fan WS, Mower JP.. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209(4):1747–1756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.