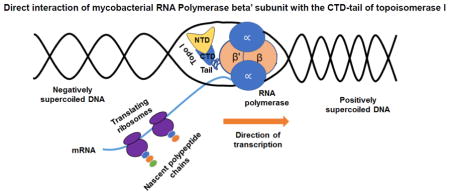
Abstract
We report here a distinct mechanism of interaction between topoisomerase I and RNA polymerase in Mycobacterium tuberculosis and Mycobacterium smegmatis that has evolved independently from the previously characterized interaction between bacterial topoisomerase I and RNA polymerase. Bacterial DNA topoisomerase I is responsible for preventing the hyper-negative supercoiling of genomic DNA. The association of topoisomerase I with RNA polymerase during transcription elongation could efficiently relieve transcription-driven negative supercoiling. Our results demonstrate a direct physical interaction between the C-terminal domains of topoisomerase I (TopoI-CTD) and the β′ subunit of RNA polymerase of M. smegmatis in the absence of DNA. The TopoI-CTD in mycobacteria are evolutionarily unrelated in amino acid sequence and three-dimensional structure to the TopoI-CTD found in the majority of bacterial species outside Actinobacteria, including Escherichia coli. The functional interaction between topoisomerase I and RNA polymerase has evolved independently in mycobacteria and E. coli, with distinctively different structural elements of TopoI-CTD utilized for this protein-protein interaction. Zinc ribbon motifs in E. coli TopoI-CTD are involved in the interaction with RNA polymerase. For M. smegmatis TopoI-CTD, a 27 amino acid tail that is rich in basic residues at the C-terminal end is responsible for the interaction with RNA polymerase. Overexpression of recombinant TopoI-CTD in M. smegmatis competed with the endogenous topoisomerase I for protein-protein interactions with RNA polymerase. The TopoI-CTD overexpression resulted in decreased survival following treatment with antibiotics and hydrogen peroxide, supporting the importance of the protein-protein interaction between topoisomerase I and RNA polymerase during stress response of mycobacteria.
Keywords: TB, evolution, stress response, transcription elongation, antibiotic sensitivity
Graphical Abstract
Introduction
Topoisomerases are essential enzymes that are responsible for controlling DNA topology and facilitating vital cellular processes that include replication, transcription, recombination, and DNA repair 1–5. The active site for DNA cleavage and rejoining by type IA topoisomerases is evolutionarily conserved at the N-terminal domains of bacterial topoisomerase I and topoisomerase III 6, 7. Bacterial topoisomerase I is responsible for relieving the transcription-driven negative supercoiling, generated behind the RNA polymerase complex during transcription elongation 8–10. The absence of topoisomerase I activity in Escherichia coli has been shown to result in increased R-loop formation via the stable association of the nascent transcript with the unwound DNA 11, 12. This could potentially block transcription elongation 13, 14. A direct association between RNA polymerase and topoisomerase I would facilitate gene expression at highly transcribed loci, including genes induced for survival in stress response.
It was previously reported that the E. coli RNA polymerase β′ subunit interacts directly with the zinc ribbon and zinc ribbon-like C-terminal domains (CTD) of topoisomerase I 15, 16. In E. coli and most bacterial species, the CTD of topoisomerase I have multiple zinc ribbon domains, each with a Zn(II) ion coordinated by four cysteines 17–19. In contrast, the topoisomerase I proteins in Actinobacteria, including mycobacteria, evolved with TopoI-CTD that do not have zinc ribbon or zinc ribbon-like domains 20, 21. Structural determination by X-ray crystallography showed that the Mycobacterium tuberculosis topoisomerase I CTD are formed by repeats of a novel protein fold of a four-stranded antiparallel β-sheet, stabilized by a crossing-over α-helix 22. We speculated that the CTD of mycobacterial topoisomerase I could be involved in protein-protein interactions, among its other functions. The protein-protein interactions of topoisomerase I from M. smegmatis were studied by co-immunoprecipitation (Co-IP) and pull-down assays coupled to tandem mass spectrometry 23–26. The assays identified an interaction between DNA-dependent RNA polymerase (RNAP), and topoisomerase I in M. smegmatis. This TopoI-RNAP interaction also employed the CTD of topoisomerase I, even though the mycobacterial topoisomerase I CTD do not have zinc ribbons. The conservation of this interaction in M. tuberculosis with a distinct mode of protein-protein interactions was verified with further studies here.
Results
Identification of RNAP as protein-protein interaction partner of MsmTopoI
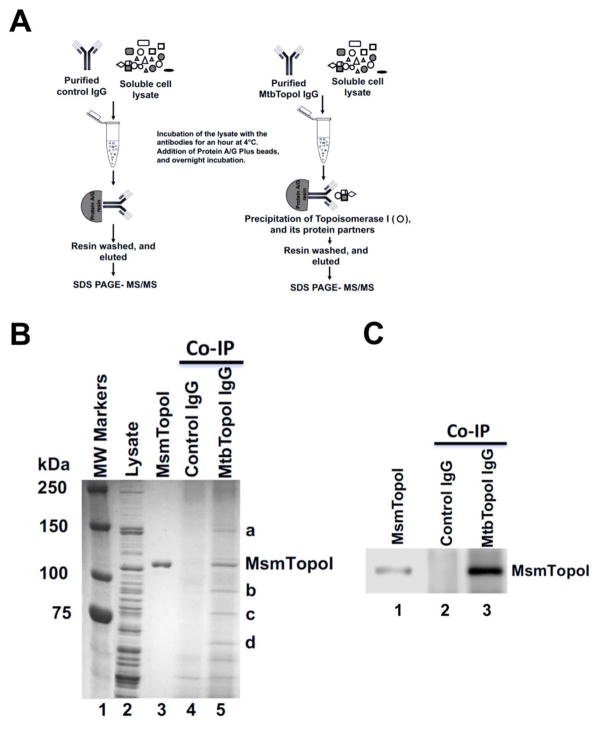
The protein-protein interaction network of the target protein (MsmTopoI) was first analyzed by Co-IP (Figure 1) coupled to mass spectrometry. Antibodies raised against MtbTopoI also recognized MsmTopoI (Figure S1). Following SDS-PAGE of the proteins immunoprecipitated by these antibodies from the soluble lysate of M. smegmatis, coomassie-stained proteins labeled as a-d in lane 5 of Figure 1B were selected. The control purified pre-immune antibodies did not precipitate the proteins corresponding to the bands a-d, as evident in lane 4 (Figure 1B). The ability of the antibodies raised against MtbTopoI to immunoprecipitate MsmTopoI from the cell lysate was confirmed by western blot analysis of the Co-IP eluates (Figure 1C). LC MS/MS analysis identified M. smegmatis RNAP β and β′ subunits in protein band a (Supplementary material, S5).
Figure 1. Co-Immunoprecipitation of topoisomerase I and interacting proteins from M. smegmatis lysate.
(A) Schematic of the Co-IP. (B) Proteins were stained with coomassie blue following SDS-PAGE. Lane 1: MW standards; Lane 2: 15 μg of total M. smegmatis proteins in soluble lysate; Lane 3: purified recombinant MsmTopoI. Proteins were immunoprecipitated from M. smegmatis lysate (500 μg total proteins) by pre-immune rabbit antibodies (lane 4) or antibodies raised against MtbTopoI (lane 5). Bands a-d were selected for LC MS/MS analysis. (C) A fraction of the eluates from Co-IP reactions were electrophoresed on a SDS-PAGE and immunoblotted with TopoI antibodies. Lane 1: 25 ng of purified MsmTopoI. The efficiency of the Co-IP assay was verified by analyzing a small fraction of eluates from the reaction of M. smegmatis lysate with pre-immune rabbit antibodies (lane 2), or antibodies raised against MtbTopoI (lane 3).
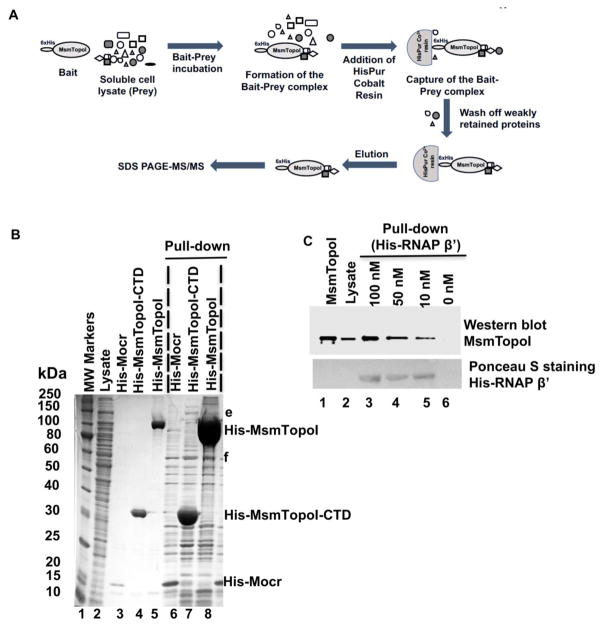
Protein networking of MsmTopoI was further investigated by pull-down assays as described in Figure 2A with His-tagged MsmTopoI and MsmTopoI-CTD. Coomassie-stained proteins (e, f in lanes 7 and 8, Figure 2B) of M. smegmatis pulled down by either His-tagged MsmTopoI or MsmTopoI-CTD, but not control His-Mocr (lane 6, Figure 2B), were analyzed by LC MS/MS. M. smegmatis RNAP β and β′ subunits were again identified in protein band e.
Figure 2. Pull-down assay of MsmTopoI-RNAP interaction with HisPur Cobalt agarose resin.
(A) General scheme of the pull-down assay. (B) Proteins pulled down from M. smemgatis soluble lysate by recombinant N-terminal His-tagged MsmTopoI (lane 8) or MsmTopoI-CTD (lane 7). His-Mocr, a recombinant viral protein, was used as bait in the control reaction (lane 6). Lane 1: MW standards; Lane 2: purified recombinant His-Mocr; Lane 3: purified recombinant His-MsmTopoI-CTD; Lane 4: purified recombinant His-MsmTopoI. (C) Reverse pull-down of TopoI from M. smegmatis soluble lysate with purified recombinant RNAP β′ subunit (N-terminal His-tagged). Following SDS-PAGE, the proteins eluted from the HisPur Cobalt were analyzed by western blot using anti-TopoI antibodies. Lane 1: purified recombinant MsmTopoI (100 ng); Lane 2: 15 μg of total proteins in M. smegmatis soluble lysate; Lanes 3–6: eluates from pull-down assay using 500 μg of total proteins in M. smegmatis soluble lysate with His-tagged RNAP β′ subunit of concentrations 100 nM (lane 3), 50 nM (lane 4), 10 nM (lane 5), and 0 nM (lane 6). The nitrocellulose membrane was also stained with Ponceau S for detection of the recombinant bait (His-tagged RNAP β′) used in the assay.
Protein-protein interaction between RNAP and topoisomerase I in M. smegmatis was further verified by reverse pull-down followed by immunoblotting (Figure 2C). In this assay, recombinant RNAP β′ subunit (N-terminal 6xHis) was used as a bait to capture topoisomerase I from the M. smegmatis soluble cell lysate. The assay confirmed a concentration-dependent binding between the two proteins (lanes 3–5 in Figure 2C). The combined results from Co-IP and pull-down experiments confirmed the networking of topoisomerase I and RNAP with a distinct mode of protein-protein interaction that does not involve the zinc ribbon domains found in other bacterial topoisomerase I.
RNAP-Topoisomerase I interactions are conserved in M. tuberculosis H37Rv
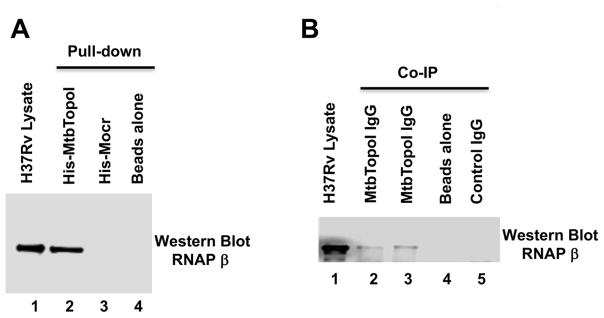
M. smegmatis is a useful model system for the understanding of processes in pathogenic M. tuberculosis 27, 28. The protein-protein interaction between topoisomerase I and RNAP could be conserved among these mycobacteria. The RNAP-topoisomerase I interaction in M. tuberculosis H37Rv was studied by pull-down (Figure 3A) and Co-IP assays (Figure 3B). Results from both approaches (lane 2 in Figure 3A; lanes 2, 3 in Figure 3B) confirmed the conservation of the RNAP-topoisomerase I interaction in M. tuberculosis.
Figure 3. M. tuberculosis H37Rv topoisomerase I and RNAP are protein-protein interaction partners.
M. tuberculosis RNAP was detected by western blot using a monoclonal antibody against E. coli RNAP β subunit that cross-reacts with mycobacterial RNAP. (A) Pull-down assay. Lane 1: total soluble lysate of M. tuberculosis H37Rv (10 μg). Eluates from HisPur cobalt resin incubated with 250 μg of lysate and purified His-tagged MtbTopoI (lane 2), 6xHisMocr (lane 3) or without any His-tagged protein (lane 4). (B) Co-Immunoprecipitation assay. Lane 1: total soluble lysate of M. tuberculosis H37Rv (10 μg). Immunoprecipitates from 250 μg of total soluble lysates incubated with IgG against MtbTopoI (lanes 2, 3), no IgG added (lane 4), and IgG from pre-immune serum (lane 5) were analyzed by western blot with the antibody recognizing RNAP β subunit.
Direct physical interaction between Topoisomerase I and RNAP β′ subunit
The results from the Co-IP and pull-down experiments indicate the presence of mycobacterial topoisomerase I and RNAP in the same complex. However, a direct physical interaction between these two partners in the absence of DNA or other mycobacterial proteins was not established. The direct protein-protein interactions were analyzed with purified recombinant proteins.
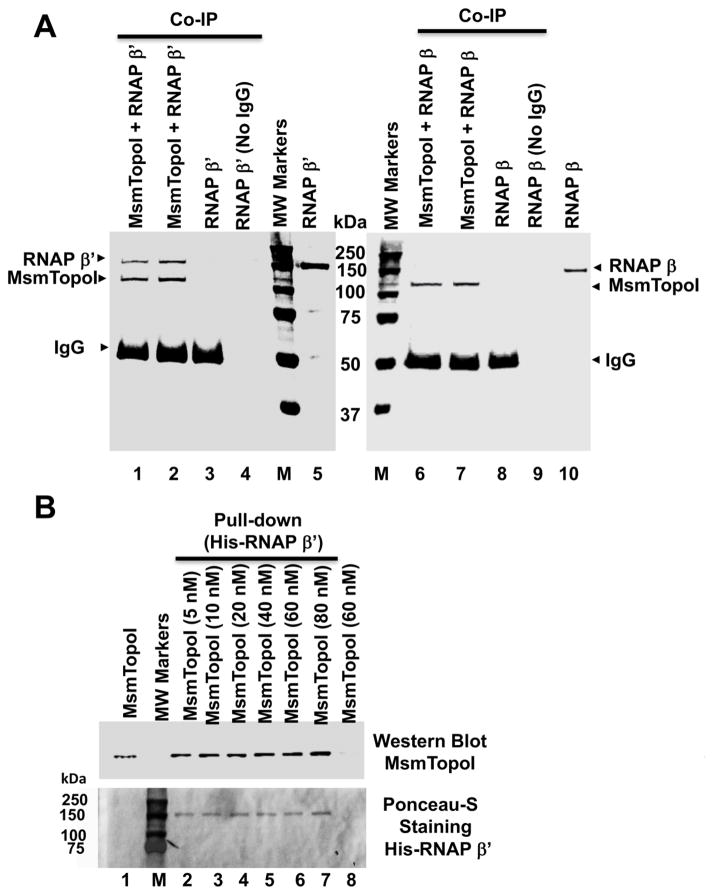
Physical interaction between the M. smegmatis RNAP β′ subunit and topoisomerase I was demonstrated by both co-immunoprecipitation (Figure 4A) and pull-down experiments (Figure 4B). This protein-protein interaction is specific. No interaction with MsmTopoI could be observed for M. smegmatis RNAP β subunit (Figure 4A). E. coli topoisomerase I, with CTD that share no homology with MsmTopoI, also did not interact with the M. smegmatis RNAP β′ subunit (Figure S2).
Figure 4. Direct physical interaction between purified M. smegmatis topoisomerase I and RNAP β′ subunit.
(A) Purified RNAP β′ subunit (lanes 1–4), but not β subunit (lanes 6–9), can be co-immunoprecipitated with MsmTopoI. The co-immunoprecipitation reaction contained 100 nM (lanes 1, 6) or 200 nM of MsmTopo I (lanes 2, 7), along with 125 nM of β′ (lanes 1–4) or β (lanes 6–9) subunit of RNAP. Lanes 3, 8: no MsmTopoI added; Lanes 4, 9: no IgG added; Lanes 5, 10: purified RNAP subunit (1.5 μg). The proteins in the gel were stained with coomassie blue following SDS-PAGE. M: Molecular weight standards. (B) Pull-down assay with HisPur cobalt resin. Lane 1: 25 ng of MsmTopoI. The pull-down reactions contained 15 nM of His-RNAP β′ subunit incubated with 5, 10, 20, 40, 60, 80 nM (lanes 2–7) of MsmTopoI. Lane 8: control of 60 nM MsmTopoI with no RNAP β′ subunit added. MsmTopoI pulled down by His-RNAP β′ subunit was visualized by western blot. The nitrocellulose membrane was then stained with Ponceau S. M: Protein molecular weight standards.
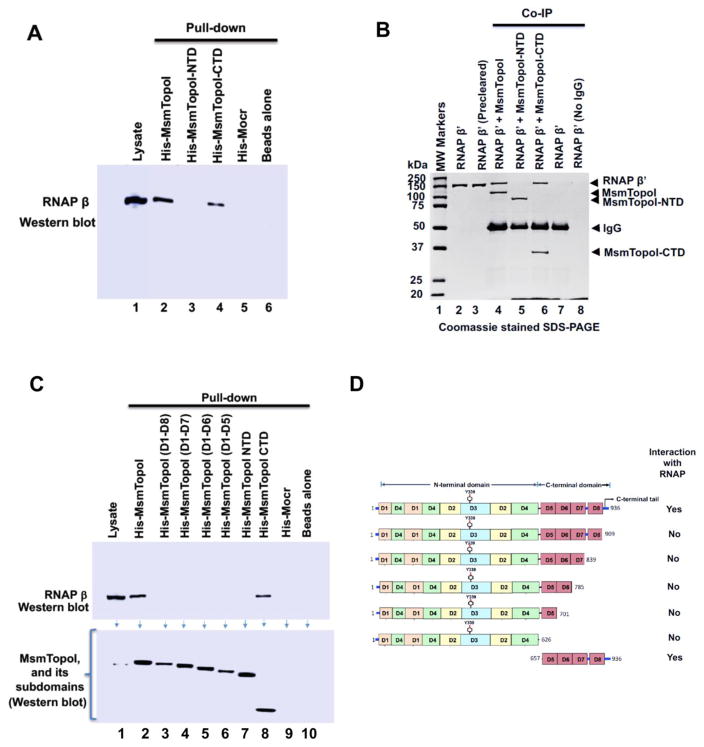
Mapping of MsmTopoI sequence required for interaction with RNAP
Pull-down (Figure 5A) and Co-IP (Figure 5B) assays were first conducted to verify that only the CTD (D5–D8, plus tail at C-terminal end), but not the NTD (D1–D4) of MsmTopoI are involved in the protein-protein interaction with RNAP. Pull-down assays with purified recombinant His-tagged proteins retaining different regions of the CTD of MsmTopoI as bait were then used to further identify the sequence in MsmTopoI-CTD required for pull-down of RNAP (prey) from the soluble cell extract. The pull-down of RNAP (prey) by protein-protein interaction could be observed only when the full-length MsmTopoI, or MsmTopoI-CTD was used as the bait (Figure 5C; lanes 2, 8). Equal molar amounts of the bait protein (subdomains of topoisomerase I) were used in these assays. MsmTopoI-909t (D1–D8) of topoisomerase I is only missing the tail at the C-terminal end (last 27 amino acids), but it still lacks affinity for RNAP. This data indicated that the protein-protein interaction with RNAP is mediated via the C-terminal tail sequence (amino acids 910-936) of MsmTopoI (Figure 5D).
Figure 5. Identification of MsmTopoI sequence required for interaction with RNAP.
(A) M. smegmatis soluble lysate was incubated with the His-tagged recombinant MsmTopoI or its fragment, and the eluates from the reaction were analyzed by western blotting. TopoI-CTD, but not TopoI-NTD, can interact with RNAP in M. smegmatis cell lysate. (B) Direct physical interaction between TopoI-CTD and RNAP β′ subunit was verified by Co-IP assay. Purified RNAP β′ subunit was incubated with MsmTopoI or its fragment, and the proteins immunoprecipiated by TopoI antibodies from the reactions were analyzed by SDS-PAGE/coomassie staining. The assay confirms a physical interaction of RNAP β′ with MsmTopoI-CTD (lane 6), and not MsmTopoI-NTD (lane 5). Purified RNAP β′, by itself, did not bind to the antibody (lane 7) or the beads (lane 8). (C) Pull-down assay with MsmTopoI truncation mutants lacking different segments of the TopoI-CTD. M. smegmatis soluble lysate was incubated with different constructs of the recombinant MsmTopoI, and the eluates were probed for the presence of RNAP. The full-length recombinant MsmTopoI (lane 2), and CTD-MsmTopoI (lane 8) can interact with RNAP. (D) Domain arrangement of MsmTopoI is shown here. The results from pull-down and Co-IP assays, taken together, demonstrate that the tail at the C-terminal end of MsmTopoI interacts with RNAP.
Increased sensitivity to antibiotics and oxidative stress from overexpression of MsmTopoI-CTD
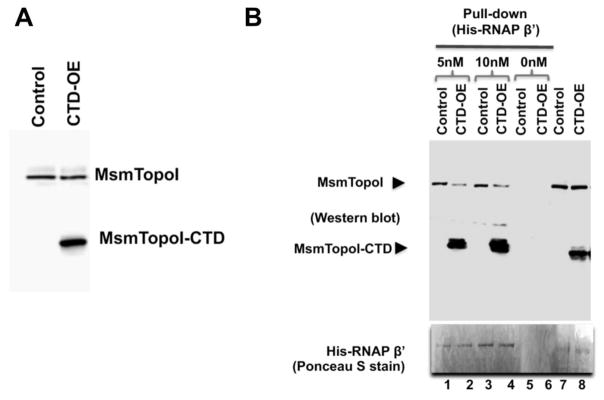
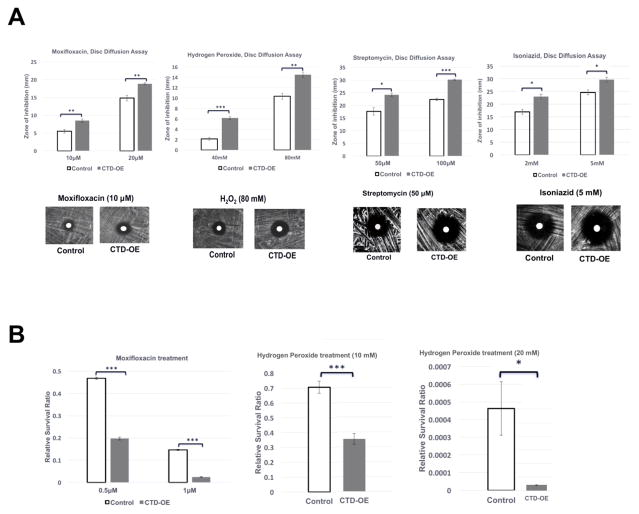
Overexpression of the MsmTopoI-CTD is expected to inhibit the interaction between native MsmTopoI and RNAP due to its competition for binding. Following tetracycline-induced overexpression of recombinant MsmTopoI-CTD in M. smegmatis mc2 155 (Figure 6A), less full-length MsmTopoI was pulled down by the His-tagged RNAP β′ subunit (Figure 6B). The MsmTopoI-CTD overexpression did not affect the growth rate in 7H9 media in the absence of any stress challenge, however, growth rate was reduced by the MsmTopoI-CTD overexpression in the presence of sublethal concentration of moxifloxacin (Figure S3). Increased sensitivity to stress challenges from antibiotics (moxifloxacin, streptomycin, isoniazid) and hydrogen peroxide resulted from the MsmTopoI-CTD overexpression, as seen from the comparison of the diameters of the zones of inhibition between the control strain and MsmTopoI-CTD overexpressing strain (Figure 7A). No significant effect from MsmTopoI-CTD overexpression was observed for sensitivity to rifampicin (data not shown). This is likely because stress gene transcription is already inhibited by rifampicin. Following treatment with either moxifloxacin (0.5 μM, 1 μM) or hydrogen peroxide (10 mM, 20 mM), loss of viability was significantly greater for the MsmTopoI-CTD overexpressing strain when compared to the control strain (Figure 7B). These results showed that inhibition of the MsmTopoI-CTD protein-protein interactions can be correlated to enhanced sensitivity to antibiotics and oxidative stress.
Figure 6. Inhibition of MsmTopoI-RNAP interaction with overexpression of recombinant MsmTopoI-CTD.
(A) The tetracycline-induced overexpression of MsmTopoI-CTD was confirmed by western blot analysis with rabbit polyclonal antibodies against TopoI. Lane 1: lysate (10 μg) of M. smegmatis transformed with control vector. Lane 2: lysate (10 μg) of M. smegmatis transformed with pMsmTopoI-CTD. (B) Pull-down of MsmTopoI from M. smegmatis lysate by His-tagged RNAP β′ subunit is reduced by the competing overexpressed MsmTopoI-CTD. Pull-down of MsmTopoI and MsmtopoI-CTD from the lysate (350 μg) by His-RNAP β′ (5 nM or 10 nM) was analyzed by western blot using antibodies against MtbTopoI (upper panel). Lanes 1, 3: pull-down reactions with lysate from the strain transformed with the control vector (control) in the presence of 5 nM His-RNAP β′ or 10 nM His-RNAP β′. Lanes 2, 4: pull-down reactions with lysate from the strain overexpressing the MsmTopoI-CTD (CTD-OE) in the presence of 5 nM His-RNAP β′ or 10 nM His-RNAP β′. Lanes 5, 6: lysates from either the control strain or the overexpression strain were incubated with the beads as a negative control for the pull-down assays. Lane 7: lysate (10 μg) from M. smegmatis transformed with the control vector. Lane 8: lysate (10 μg) from M. smegmatis overexpressing the MsmTopoI-CTD. The nitrocellulose membrane was stained with Poneau S (bottom panel) for the detection of the bait (His-RNAP β′).
Figure 7. Effect of TopoI-CTD overexpression on sensitivity of M. smegmatis to stress challenge.
(A) The tetracycline-induced cultures of the vector control strain (control), and the MsmTopoI CTD-overexpression strain (CTD-OE) were spread on LB plates, and a blank paper disc was placed on the plate. 20 μL of the antibiotic solution or hydrogen peroxide was added to the disc. The plates were incubated at 37°C for 60 hours, and the zone of inhibition was measured. Error bars represent the standard deviation (n=3). Student’s t-test was used to calculate the p-values (* p<0.05; ** p<0.005, ***p<0.0005). (B) Following treatment with moxifloxacin or hydrogen peroxide for 12 hours, the untreated and treated cultures of the tetracycline-induced vector control strain (control), and MsmTopoI CTD-overexpression (CTD-OE) strain were serially diluted and spread on LB plates. The viable colony counts (CFU/ml) were determined to calculate the relative survival ratios as the colony counts of the treated cultures divided by the colony counts of the untreated cultures.
Discussion
Most bacterial species have at least two type IA topoisomerases encoded in their genome 29, 30 to carry out specific associated functions in replication, transcription, and recombination that require the passage of DNA through a break in a single strand of DNA 3. TopoI is the only type IA topoisomerase encoded in the genomes of mycobacteria, and has been shown to be essential for the viability of both M. tuberculosis 31, 32 and M. smegmatis33. The identification of the protein-protein interaction partners of mycobacterial TopoI is likely to provide insights into the in vivo functions and regulation of this potential antibacterial target. It has been reported that the relaxation activity of MtbTopoI and MsmTopoI can be inhibited by interaction with a MazF homolog 34, as well as D-ribokinase 35. The catalytic activity of MtbTopoI can also be modulated by interaction with the nucleoid associated protein HU 36. These previously identified protein-protein interactions may be relevant for the regulation of mycobacterial TopoI activity. However, experimental data on protein-protein interactions of mycobacterial TopoI that may inform on the physiological setting of its function 37 is currently not available. In this study, we tried to identify such potential partners present in the total soluble proteins of M. smegmatis using the approaches of Co-IP and pull-down assays coupled to mass spectrometry. RNAP was identified as a protein-protein interaction partner for MsmTopoI by both approaches. A recent ChIP-Seq study showed that M. tuberculosis TopoI and gyrase are recruited to genomic loci with high transcriptional activity, and MtbTopoI was localized behind RNAP to be in position for relaxing the negative supercoils generated during transcription 38. Our studies here demonstrated that M. tuberculosis RNAP can recruit TopoI via protein-protein interaction to facilitate the co-localization during transcription elongation.
While protein-protein interaction with the RNAP β′ subunit has been previously reported for the CTD of E. coli TopoI 15, it should be noted that the CTD of mycobacterial TopoI share no sequence and structural similarity with the CTD of bacteria outside the Actinobacteria phylum 39, 21. The CTD of E. coli TopoI are formed by zinc ribbon motifs stabilized by Zn(II) coordinated with four cysteines in each motif (D5–D7), or zinc ribbon-like motifs of similar structures (D8–D9) 17, 18. Interaction sites for RNAP have been mapped to E. coli TopoI CTD regions of D5–D7 as well as D8–D9 15. Analysis of the MsmTopoI protein sequence 40 and determination of the D1–D5 structure of MtbTopoI 22 have shown that the CTD of mycobacteria are formed by repeats of a distinctively different structural motif containing a β-sheet and α-helix, with no Zn(II) present. The results reported here demonstrate that while the different bacterial species have evolved to have distinct sequences and structures of the CTD in TopoI, potentially through acquiring different duplicated gene segments during evolution 41, the protein-protein interaction with the β′ subunit of RNAP has simultaneously co-evolved through divergent mechanisms in different bacterial phylum.
Molecular simulations predicted that salt bridges and hydrogen bonds formed by basic residues positioned over a large molecular surface formed by the zinc ribbon motifs of E. coli TopoI are responsible for interactions with acidic residues in RNAP 16. In contrast, a distinct mode of protein-protein interaction utilizing a short stretch of C-terminal tail is employed instead for the TopoI-RNAP interaction in bacteria that do not have Zn(II) binding TopoI-CTD. As shown in Figure S4, the amino acid sequence of this C-terminal tail is highly conserved in mycobacterial TopoI and is rich in basic residues. The basic region represented by the C-terminal tail of MsmTopoI and MtbTopoI has been proposed to participate in the binding of DNA to promote strand passage during catalysis 20, 42. Direct interaction of the C-terminal tail with RNAP would facilitate the rewinding of single-stranded DNA following its exit from the RNAP elongation complex to prevent stabilization of R-loop structures and inhibition of transcription elongation. The TopoI-RNAP interaction may be particularly important for efficient transcriptional response to stress conditions to achieve maximal survival following antibiotics or oxidative challenge 14, 43. Competition for RNAP interaction by overexpressed recombinant MsmTopoI-CTD may have contributed to the increase in sensitivity to antibiotics and oxidative stress challenge observed here. Inhibition of the SOS response induction following moxifloaxcin treatment would result in the decreased survival for the MsmTopoI-CTD overexpressing strain. Nevertheless, we cannot rule out that other protein-protein interactions inhibited by overexpressed MsmTopoI-CTD may also contribute to the increased sensitivity. Other potential protein-protein interaction partners identified in our Co-IP and pull down experiments are currently being investigated in further studies. Mycobacterial topoisomerase I may have functional roles associated with topoisomerase III in bacteria that have this additional type IA topoisomerase. The actinobacterium Streptomyces coelicolor also does not have topoisomerase III, and it has been hypothesized that S. coelicolor topoisomerase I is recruited to ParB complexes because the topoisomerase activity is needed to resolve segregating chromosomes 44.
Based on the essentiality of topoisomerase I for the viability of M. tuberculosis, MtbTopoI is a validated target for discovery of novel TB drugs 32, 45. The SOS response has been shown to be involved in the induction of mycobacterial resistance to fluoroquinolones 46. Due to its involvement in transcriptional response to stress challenges, mycobacterial topoisomerase I inhibitors could be especially useful for use in combination with other antibiotics to increase the efficacy, and decrease the induction of drug resistance.
Materials and methods
Bacterial strains, plasmids, and cloning
M. smegmatis mc2 155 was obtained from ATCC and cultured in 7H9 broth. M. tuberculosis H37Rv genomic DNA and whole cell lysates were obtained from BEI Resources. A plasmid pMsmTopoI-CTD for overexpression of topoisomerase I-CTD in M. smegmatis was constructed by first placing M. smegmatis topA coding sequence (amplified with primers shown in Table S1) under the control of the tetracycline inducible TetRO promoter in pKW08 vector 47 via Gibson Assembly cloning 48. Plasmid pMsmTopoI-CTD was then constructed for overexpression of the C-terminal domains D5–D8 plus tail (residues 627-936) by deleting the N-terminal domains D1–D4 (residues 1-626) via site-directed mutagenesis. Overexpression of the various domains of M. smegmatis topoisomerase I (MsmTopoI) in E. coli BL21 STAR DE3 (Invitrogen) was achieved with pET His6 Mocr TEV LIC cloning vector (2O-T, Addgene Plasmid# 29710). Cloning of recombinant M. tuberculosis topoisomerase I (MtbTopoI) and its CTD in vector pLIC-HK 49 for expression in E. coli BL21- CodonPlus(DE3)-RP (Agilent) was described previously 50. Coding sequences for M. smegmatis RNAP β (RpoB) and β′ (RpoC) subunits were inserted by Gibson cloning into the SspI site of pLIC-HK, and transformed into E. coli SoluBL21 (Genlantis) cells for protein expression and purification. The PCR primers used in the cloning and site-directed mutagenesis are listed in the Supplementary Information (Table S1 and Table S2).
Protein expression and purification
DNA topoisomerase I, sub-domains of topoisomerase I, RNAP β, and RNAP β′ subunits were purified as recombinant proteins for protein-protein interaction studies. Overexpression of these recombinant proteins in the E. coli hosts was induced with 1 mM IPTG. Following the overexpression and cell lysis, recombinant proteins were purified to near homogeneity by affinity chromatography using Ni-Sepharose 6 Fast flow beads (GE Healthcare Life Sciences), followed by size-exclusion chromatography.
Co-Immunoprecipitation (Co-IP)/Tandem Mass spectrometry
The complex of M. smegmatis topoisomerase I (MsmTopoI) and its potential protein partners was isolated from the M. smegmatis soluble cell lysate by Co-IP 23. M. smegmatis mc2 155 was grown to stationary phase in LB-Tween 80 (0.1%). The pelleted cells were resupsended in lysis/wash buffer (50 mM NaCl, 50 mM NaH2PO4, pH 8.0, 0.1% NP-40), and then subjected to 5 pulses of sonication (10 seconds/pulse at medium intensity). The lysate was spun at 16000×g for an hour, and the supernatant soluble lysate was used as prey in the Co-IP assays. The soluble lysate was pre-incubated (pre-cleared) with protein A/G plus agarose beads to remove proteins that bind non-specifically to the beads. The pre-cleared lysate (total protein=500 μg) was first mixed with IgG (10 μg) purified from serum of a rabbit inoculated with MtbTopoI or control pre-immune serum. These rabbit polyclonal antibodies cross react with MsmTopoI (Figure S1). The protein A/G plus agarose beads (Santa Cruz Biotechnology), previously equilibrated in wash buffer, was then added to the lysate-antibody reaction and left overnight at 4°C. On the following day, the reaction mixture was spun at 700×g, and the supernatant was discarded. The bead pellet with the bound proteins was washed three times in the wash buffer. The elution of the proteins bound to the beads was carried out by suspending the bead pellet in 4X SDS sample buffer (240 mM Tris-HCl, 8% SDS, 40% glycerol, 0.04% bromophenol blue), and heating at 95°C for 2 minutes. The eluted proteins (MsmtopoI, and its potential protein partners) were stained with coomassie blue following SDS-PAGE. The protein bands of interest were excised for characterization by tryptic digest and mass spectrometry (nano LC/MS/MS) at the Proteomics and Mass Spectrometry Facility of University of Massachusetts Medical School. The MS/MS results were searched using Mascot (Matrix Science, London, UK; version 2.5.0) against the UniProt_MSmegmatis_071714 database for trypsin digestion products.
Pull-down assay/Tandem Mass spectrometry
For the pull-down assay 24, the target protein (MsmtopoI) with a fusion tag (N-terminal 6xHistidine) was mixed with M. smegmatis mc2 155 soluble cell extract for 2 hours at 4°C, and then immobilized on a HisPur Cobalt agarose resin (Thermofisher). The resin with bound target proteins was washed three times in a pull-down wash buffer (10 mM HEPES, pH 7.5, 10 mM imidazole, 0.005% Tween-20) to remove the weakly retained proteins. The proteins that remained bound to the resin were eluted with pull-down elution buffer (10 mM HEPES, pH 7.5, 350 mM imidazole), separated by SDS-PAGE, and stained with coomassie blue. The protein bands of interest were excised for identification by tryptic digest and nano LC/MS/MS.
Reverse pull-down assay/Immunoblotting
A reverse pull-down assay was used to validate the protein interactions identified by pull-down assays. In this assay, a recombinant M. smegmatis RNAP β′ subunit (N-terminal 6xHistidine) was used as bait to pull down topoisomerase I from the soluble cell extract. The pull-down protocol was similar to that described in the previous section. Western blot analysis with rabbit polyclonal antibodies against MtbTopoI was used to detect the presence of topoisomerase I in the eluate.
Pull-down, Co-IP assays on M. tuberculosis (H37Rv) cell extract
The conservation of RNAP-topoisomerase I protein-protein interactions in M. tuberculosis was verified by pull-down and Co-IP assays. Whole cell extract of M. tuberculosis H37Rv was provided by BEI Resources. The extract was spun at 16000×g for 20 min at 4°C, and the soluble protein fraction was used for the assays. Recombinant MtbTopoI with N-terminal 6x-Histidine was used in the pull-down assay, while antibodies against topoisomerase I were used in the Co-IP assay as bait to determine if RNAP could be captured from the cell extract. Following SDS-PAGE, the proteins in the eluates from the assays were immunoblotted with a monoclonal antibody against RNAP β (Biolegend) which can recognize the RNAP β subunits 51 across multiple species, including M. tuberculosis and M. smegmatis.
Effect of TopoI-CTD overexpression on sensitivity to antibiotics or hydrogen peroxide
The M. smegmatis strain overexpressing the CTD of topoisomerase I from the plasmid pMsmTopA-CTD, or a control M. smegmatis strain transformed with the cloning vector, were cultured to stationary phase in 7H9 medium containing Hygromycin (100μg/ml) and tetracycline (25ng/ml). The cultures were adjusted to OD600=1.0 in 7H9 medium, and 200μl of the OD-adjusted culture was spread on LB plates. Antibiotic or H2O2 was applied to paper discs placed at the center of the plates, followed by incubation at 37°C for 60 hrs. The diameter of the zone of inhibition was measured.
For comparison of viable colony counts following stress challenge, the M. smegmatis strains were grown in Luria broth-Tween 80 (0.1%) containing Hygromycin (100μg/ml) and tetracycline (25ng/ml). At the exponential phase, the cultures were treated with moxifloxacin (0.5 μM or 1 μM) for 12 hours at 37°C. The treated cultures and untreated cultures were serially diluted and spread on LB plates. The viable colonies were counted, and the relative viability ratio (treated versus untreated) was calculated. Similarly, viable colony counts following treatment with hydrogen peroxide (10 mM or 20 mM) was analyzed with the strains cultured in 7H9 medium containing Hygromycin (100 μg/ml) and tetracycline (25 ng/ml).
Supplementary Material
Highlights.
Mycobacteria topoisomerase I interacting protein partner of functional significance
Mycobacteria topoisomerase I interacts directly with RNA polymerase beta’ subunit
Interaction via short C-terminal tail instead of Zn ribbon motifs in other bacteria
Antibiotics hypersensitivity from topoisomerase I C-terminal domain overexpression
Convergent evolution of topoisomerase I-RNAP interaction via distinct mechanism
Acknowledgments
Funding
This work was supported by the National Institute of Health grants GM054226 and AI069313 to YT.
We are grateful to BEI Resources for providing the M. tuberculosis H37Rv genomic DNA and whole cell lysate. We thank Shayna Sandhaus for proof-reading of manuscript. Plasmid pKW08-Lx was a gift from Brian Robertson (Addgene plasmid # 25012). We thank Shayna Sandhaus for proof-reading of manuscript.
Abbreviations
- CTD
C-terminal domains
- TopoI-CTD
Topoisomerase I C-terminal domains
- MsmTopoI
Mycobacterium smegmatis topoisomerase I
- MtbTopo1
Mycobacterium tuberculosis topoisomerase I
- Co-IP
Co-immunoprecipitaton
- RNAP
RNA polymerase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen SH, Chan NL, Hsieh TS. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 2.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [doi]; nrm831 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Baranello L, Levens D, Gupta A, Kouzine F. The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochim Biophys Acta. 2012;1819:632–638. doi: 10.1016/j.bbagrm.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorman CJ, Dorman MJ. DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys Rev. 2016;8:89–100. doi: 10.1007/s12551-016-0238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capranico G, Marinello J, Chillemi G. Type I DNA Topoisomerases. J Med Chem. 2017;60:2169–2192. doi: 10.1021/acs.jmedchem.6b00966. [DOI] [PubMed] [Google Scholar]
- 8.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masse E, Drolet M. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J Biol Chem. 1999;274:16654–16658. doi: 10.1074/jbc.274.23.16654. [DOI] [PubMed] [Google Scholar]
- 10.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. 0092-8674(88)90163-8 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masse E, Drolet M. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J Mol Biol. 1999;294:321–332. doi: 10.1006/jmbi.1999.3264. [doi]; S0022-2836(99)93264-3 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Hraiky C, Raymond MA, Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem. 2000;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 14.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. MMI5006 [pii]; [DOI] [PubMed] [Google Scholar]
- 15.Cheng B, Zhu CX, Ji C, Ahumada A, Tse-Dinh YC. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J Biol Chem. 2003;278:30705–30710. doi: 10.1074/jbc.M303403200. [doi]; M303403200 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Tiwari PB, Chapagain PP, Banda S, Darici Y, Uren A, Tse-Dinh YC. Characterization of molecular interactions between Escherichia coli RNA polymerase and topoisomerase I by molecular simulations. FEBS Lett. 2016;590:2844–2851. doi: 10.1002/1873-3468.12321. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grishin NV. C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J Mol Biol. 2000;299:1165–1177. doi: 10.1006/jmbi.2000.3841. [DOI] [PubMed] [Google Scholar]
- 18.Tse-Dinh YC, Beran-Steed RK. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc-binding domains. J Biol Chem. 1988;263:15857–15859. [PubMed] [Google Scholar]
- 19.Suerbaum S, Brauer-Steppkes T, Labigne A, Cameron B, Drlica K. Topoisomerase I of Helicobacter pylori: juxtaposition with a flagellin gene (flaB) and functional requirement of a fourth zinc finger motif. Gene. 1998;210:151–161. doi: 10.1016/s0378-1119(98)00065-1. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed W, Bhat AG, Leelaram MN, Menon S, Nagaraja V. Carboxyl terminal domain basic amino acids of mycobacterial topoisomerase I bind DNA to promote strand passage. Nucleic Acids Res. 2013;41:7462–7471. doi: 10.1093/nar/gkt506. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikder D, Nagaraja V. Determination of the recognition sequence of Mycobacterium smegmatis topoisomerase I on mycobacterial genomic sequences. Nucleic Acids Res. 2000;28:1830–1837. doi: 10.1093/nar/28.8.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan K, Cao N, Cheng B, Joachimiak A, Tse-Dinh YC. Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold. J Mol Biol. 2016;428:182–193. doi: 10.1016/j.jmb.2015.11.024. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Free RB, Hazelwood LA, Sibley DR. Identifying novel protein-protein interactions using co-immunoprecipitation and mass spectroscopy. Curr Protoc Neurosci. 2009;Chapter 5(Unit 5.28) doi: 10.1002/0471142301.ns0528s46. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brymora A, Valova VA, Robinson PJ. Protein-protein interactions identified by pull-down experiments and mass spectrometry. Curr Protoc Cell Biol. 2004;Chapter 17(Unit 17.5) doi: 10.1002/0471143030.cb1705s22. [doi] [DOI] [PubMed] [Google Scholar]
- 25.Ning Z, Hawley B, Chiang CK, Seebun D, Figeys D. Detecting protein-protein interactions/complex components using mass spectrometry coupled techniques. Methods Mol Biol. 2014;1164:1–13. doi: 10.1007/978-1-4939-0805-9_1. [doi] [DOI] [PubMed] [Google Scholar]
- 26.Speth C, Toledo-Filho LA, Laubinger S. Immunoprecipitation-based analysis of protein-protein interactions. Methods Mol Biol. 2014;1158:175–185. doi: 10.1007/978-1-4939-0700-7_11. [doi] [DOI] [PubMed] [Google Scholar]
- 27.Shiloh MU, Champion PA. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol. 2010;13:86–92. doi: 10.1016/j.mib.2009.11.006. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agrawal P, Miryala S, Varshney U. Use of Mycobacterium smegmatis deficient in ADP-ribosyltransferase as surrogate for Mycobacterium tuberculosis in drug testing and mutation analysis. PLoS One. 2015;10:e0122076. doi: 10.1371/journal.pone.0122076. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed W, Menon S, Godbole AA, Karthik PV, Nagaraja V. Conditional silencing of topoisomerase I gene of Mycobacterium tuberculosis validates its essentiality for cell survival. FEMS Microbiol Lett. 2014;353:116–123. doi: 10.1111/1574-6968.12412. [doi] [DOI] [PubMed] [Google Scholar]
- 32.Ravishankar S, Ambady A, Awasthy D, Mudugal NV, Menasinakai S, Jatheendranath S, Guptha S, Sharma S, Balakrishnan G, Nandishaiah R, Ramachandran V, Eyermann CJ, Reck F, Rudrapatna S, Sambandamurthy VK, Sharma UK. Genetic and chemical validation identifies Mycobacterium tuberculosis topoisomerase I as an attractive anti-tubercular target. Tuberculosis (Edinb) 2015;95:589–598. doi: 10.1016/j.tube.2015.05.004. [doi] [DOI] [PubMed] [Google Scholar]
- 33.Ahmed W, Menon S, Karthik PV, Nagaraja V. Reduction in DNA topoisomerase I level affects growth, phenotype and nucleoid architecture of Mycobacterium smegmatis. Microbiology. 2015;161:341–353. doi: 10.1099/mic.0.000014. [doi] [DOI] [PubMed] [Google Scholar]
- 34.Huang F, He ZG. Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Res. 2010;38:8219–8230. doi: 10.1093/nar/gkq737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Q, Liu Y, Huang F, He ZG. Physical and functional interaction between D-ribokinase and topoisomerase I has opposite effects on their respective activity in Mycobacterium smegmatis and Mycobacterium tuberculosis. Arch Biochem Biophys. 2011;512:135–142. doi: 10.1016/j.abb.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh S, Mallick B, Nagaraja V. Direct regulation of topoisomerase activity by a nucleoid-associated protein. Nucleic Acids Res. 2014;42:11156–11165. doi: 10.1093/nar/gku804. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Freddolino PL, Zhang Y. COFACTOR: improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx366. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmed W, Sala C, Hegde SR, Jha RK, Cole ST, Nagaraja V. Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet. 2017;13:e1006754. doi: 10.1371/journal.pgen.1006754. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szafran MJ, Strick T, Strzalka A, Zakrzewska-Czerwinska J, Jakimowicz D. A highly processive topoisomerase I: studies at the single-molecule level. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku494. gku494 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhaduri T, Bagui TK, Sikder D, Nagaraja V. DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J Biol Chem. 1998;273:13925–13932. doi: 10.1074/jbc.273.22.13925. [DOI] [PubMed] [Google Scholar]
- 41.Viard T, de la Tour CB. Type IA topoisomerases: a simple puzzle? Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Jain P, Nagaraja V. Indispensable, functionally complementing N and C-terminal domains constitute site-specific topoisomerase I. J Mol Biol. 2006;357:1409–1421. doi: 10.1016/j.jmb.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 43.Rui S, Tse-Dinh YC. Topoisomerase function during bacterial responses to environmental challenge. Front Biosci. 2003;8:d256–63. doi: 10.2741/984. [DOI] [PubMed] [Google Scholar]
- 44.Szafran M, Skut P, Ditkowski B, Ginda K, Chandra G, Zakrzewska-Czerwinska J, Jakimowicz D. Topoisomerase I (TopA) is recruited to ParB complexes and is required for proper chromosome organization during Streptomyces coelicolor sporulation. J Bacteriol. 2013 doi: 10.1128/JB.00798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagaraja V, Godbole AA, Henderson SR, Maxwell A. DNA topoisomerase I and DNA gyrase as targets for TB therapy. Drug Discov Today. 2017;22:510–518. doi: 10.1016/j.drudis.2016.11.006. S1359-6446(16)30422-6 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Malik M, Chavda K, Zhao X, Shah N, Hussain S, Kurepina N, Kreiswirth BN, Kerns RJ, Drlica K. Induction of mycobacterial resistance to quinolone class antimicrobials. Antimicrob Agents Chemother. 2012;56:3879–3887. doi: 10.1128/AAC.00474-12. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams KJ, Joyce G, Robertson BD. Improved mycobacterial tetracycline inducible vectors. Plasmid. 2010;64:69–73. doi: 10.1016/j.plasmid.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle SA. High-throughput cloning for proteomics research. Methods Mol Biol. 2005;310:107–113. doi: 10.1007/978-1-59259-948-6_7. [DOI] [PubMed] [Google Scholar]
- 50.Annamalai T, Dani N, Cheng B, Tse-Dinh YC. Analysis of DNA relaxation and cleavage activities of recombinant Mycobacterium tuberculosis DNA topoisomerase I from a new expression and purification protocol. BMC Biochem. 2009;10:18. doi: 10.1186/1471-2091-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess RR, Thompson NE. Advances in gentle immunoaffinity chromatography. Curr Opin Biotechnol. 2002;13:304–308. doi: 10.1016/s0958-1669(02)00340-3. S0958166902003403 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.