Abstract
Over the past several decades, numerous studies have greatly expanded our knowledge about how microtubule organization and dynamics are controlled in cultured cells in vitro. However, our understanding of microtubule dynamics and functions in vivo, in differentiated cells and tissues, remains under-explored. Recent advances in generating genetic tools and imaging technologies to probe microtubules in situ, coupled with an increased interest in the functions of this cytoskeletal network in differentiated cells, are resulting in a renaissance. Here, we discuss the lessons learned from such approaches, which have revealed that, although some differentiated cells utilize conserved strategies to remodel microtubules, there is considerable diversity in the underlying molecular mechanisms of microtubule reorganization. This highlights a continued need to explore how differentiated cells regulate microtubule geometry in vivo.
KEY WORDS: Centrosome, Differentiation, Microtubule, Nucleation
Summary: This Review summarizes current knowledge of how microtubule organization and dynamics change upon cellular differentiation and gives an overview of the functions of non-centrosomal microtubule arrays in differentiated cells.
Introduction
Cellular differentiation is often accompanied by reorganization of the microtubule cytoskeleton into cell type-specific arrays. It is perhaps surprising that, despite numerous studies focused on the cytoskeleton over the last several decades, we have a very limited understanding of how differentiation-induced microtubule arrays are formed and what their functions are in many cell types. In general, proliferative cells have centrosomal microtubule arrays in which the centrosome acts as the microtubule-organizing center (MTOC). In contrast, most differentiated cells have non-centrosomal arrays that are not centered at the centrosome and are cell type specific. Although a great deal of work has identified the fundamental biochemical principles and molecular factors that regulate microtubule growth, dynamics and organization, there is a substantial need to translate this knowledge to understanding microtubule form and function in differentiated cells in vivo. As recent work has demonstrated, probing microtubules in vivo has the power both to provide new insight into microtubule regulation by microtubule-associated proteins, signaling cascades and transcription factors and to uncover novel functions for microtubules in tissue physiology. These studies also have implications for understanding human disease. Indeed, numerous ciliopathies arise from abnormalities in microtubule-based cilia (reviewed by Hildebrandt et al., 2011), and several human diseases are proposed to involve microtubule array dysfunction (Ballatore et al., 2012; Zempel and Mandelkow, 2014). Additionally, microtubule-targeting drugs – notably those that are being explored for their anti-cancer activities – operate in vivo through mostly unknown mechanisms (reviewed by Stanton et al., 2011; Dalbeth et al., 2014; Leung et al., 2015), highlighting a need for increased research focus on microtubule function in distinct cell types in vivo.
The study of differentiation-induced microtubule reorganization has traditionally been challenging because microtubule arrays formed in differentiated cultured cells often fail to fully recapitulate the organization of in vivo arrays. Recently, however, there has been an increased interest in developing models to study non-centrosomal microtubule arrays in situ. A number of pioneering studies, especially in neurons, muscle and epithelia, have begun to provide answers to long-standing questions about the signals that regulate microtubule reorganization and the functions of non-centrosomal microtubule arrays during development. Here, we highlight what is known about non-centrosomal microtubule organization and dynamics in vivo, with a special emphasis on how in vivo studies have provided unexpected insights into the mechanisms of array formation and their physiological functions. We direct the reader to previous reviews that have thoroughly covered the formation of non-centrosomal microtubule arrays in cultured cells (Bartolini and Gundersen, 2006).
Microtubule organization: centrosomal and non-centrosomal arrays
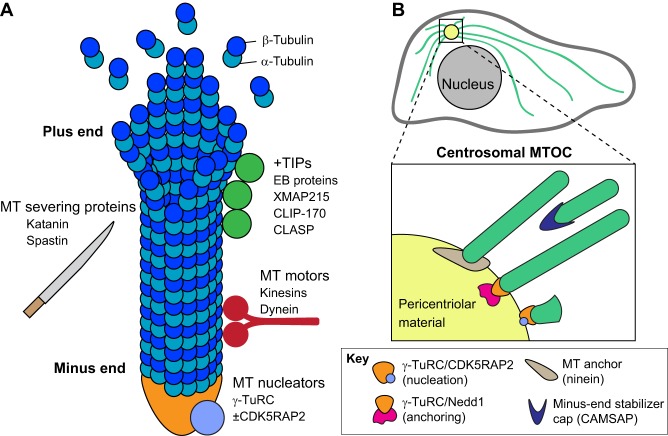
Microtubules are composed of α- and β-tubulin heterodimers that assemble into protofilaments, which associate laterally to form hollow tubes (Fig. 1). They are polar structures that harbor two distinct ends – the plus and minus ends – and their organization within the cell is tightly controlled by a large number of microtubule-associated proteins (MAPs) that promote or suppress dynamic behavior at both of these ends (Fig. 1). Microtubule nucleation, the formation of new microtubule filaments, begins from the minus end and is mostly dependent on γ-tubulin ring complexes (γ-TuRCs) in cells (Moritz and Agard, 2001). Importantly, nucleation by γ-TuRCs can be modulated by activators such as CDK5RAP2 (Choi et al., 2010). The minus end can remain attached to γ-TuRC, which has been shown to bind and cap minus ends of non-centrosomal microtubules (Wiese and Zheng, 2000; Anders and Sawin, 2011) and to anchor microtubules to the centrosome when complexed with Nedd1 (Muroyama et al., 2016). Microtubule minus ends are also colocalized with ninein at both the centrosome and at distal sites, suggesting that ninein mediates microtubule anchoring at MTOCs, although a direct interaction with microtubules has not been reported (Mogensen et al., 2000; Delgehyr et al., 2005). Minus ends can also slowly polymerize in vitro and in vivo when decorated by calmodulin-regulated spectrin-associated protein (CAMSAP) family proteins, which also serve to stabilize and potentially cap minus ends (Goodwin and Vale, 2010; Meng et al., 2008; Jiang et al., 2014; Hendershott and Vale, 2014).
Fig. 1.
Regulators of microtubule dynamics and organization. (A) Numerous microtubule-associated proteins (MAPs) influence microtubule behavior. Many of these, such as EB proteins, XMAP215, CLIP-170 and CLASP proteins, regulate plus-tip dynamics and are collectively known as microtubule plus-end tracking proteins (+TIPs). Only a few proteins are known to bind specifically to the minus end. One of these, the γ-tubulin ring complex (γ-TuRC), is the primary microtubule nucleator in the cell. Nucleation by γ-TuRCs can be modulated by activators such as CDK5RAP2. Microtubule motors can intrinsically influence microtubule dynamics and also regulate microtubule organization by guiding microtubules along existing filaments. Microtubule-severing proteins induce breaks along the length of the filament to impact microtubule organization within the cell. (B) The centrosome is the primary microtubule organizing center (MTOC) in many proliferative cells. However, note that non-centrosomal microtubules and centrosomal microtubules can co-exist within the same cell. MTOC activity is conferred through both microtubule nucleation and anchoring abilities. CDK5RAP2 and Nedd1, acting via γ-TuRC, can promote these activities, respectively, but both basal activity and other activators are also likely to be involved. Ninein colocalizes with microtubule minus ends and may play a role in anchoring. CAMSAP proteins also preferentially localize to microtubule minus ends and serve to stabilize and potentially cap minus ends.
By contrast, microtubule polymerization and depolymerization in cells primarily occur at the highly dynamic plus ends (Desai and Mitchison, 1997). These dynamics are controlled by a host of MAPs that localize to the plus end, such as the EB (end binding) family proteins, CLIP-170 (CLIP1), XMAP215 (CKAP5), and the CLASP family (Mimori-Kiyosue et al., 2000; Perez et al., 1999; Brouhard et al., 2008; reviewed in Akhmanova and Steinmetz, 2008). In addition to the proteins that localize to the plus end, some MAPS, including Tau (MAPT) and MAP4, bind along the lattice and promote microtubule stabilization (Kadavath et al., 2015; Nguyen et al., 1997). Microtubule organization can also be regulated through the microtubule-severing proteins katanin and spastin (reviewed by Roll-Mecak and McNally, 2010) and numerous tubulin post-translation modifications, which can influence polymer dynamics by tuning MAP activity and affinity (reviewed by Song and Brady, 2015; Valenstein and Roll-Mecak, 2016).
Given their key roles, MAPS have served as useful tools to assess and perturb microtubule organization in cells. For example, live-imaging of GFP-tagged EB1 (MAPRE1) and EB3 (MAPRE3) has been invaluable for quantifying microtubule dynamics and visualizing microtubule architecture in a variety of cell types. Genetically controlled overexpression of specific MAPs in vivo has also yielded insight into roles for microtubules in a variety of tissue contexts. Using these genetically encoded microtubule probes has greatly facilitated progress in understanding the dynamics for microtubules in tissue.
Loss of centrosomal MTOC activity in differentiated cells
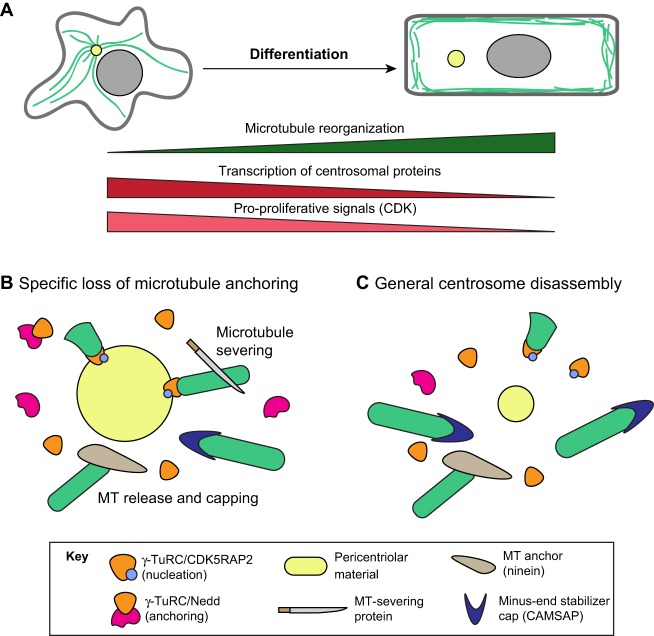
Differentiation is broadly associated with microtubule reorganization from centrosomal into non-centrosomal microtubule arrays (Fig. 2). For this to occur, the centrosome must lose MTOC activity while novel cellular sites are specified and activated to acquire this function. Loss of centrosomal MTOC activity could be driven through various processes, including changes in transcription, RNA splicing, protein localization at the centrosome, and post-translational modifications that alter the ability of the centrosome to nucleate or anchor microtubules. However, most current data suggest that centrosomal protein localization is the predominant form of regulation. For instance, in many tissues, MTOC inactivation is correlated with delocalization of pericentriolar material (PCM). Although the signals that induce this delocalization remain unidentified in many cell types, decreased levels or activity of cell-cycle regulators, in particular cyclin-dependent kinase (CDK) and PLK1 (Fig. 3A), have been shown to result in PCM dispersal in some tissues (Yang and Feldman, 2015; Muroyama et al., 2016; Pimenta-Marques et al., 2016). These cell-cycle regulators presumably control centrosome inactivation through post-translational modifications on centrosomal proteins that lead to PCM shedding, in an extreme form of the inactivation that follows mitotic exit (Fig. 3B,C). Centrosome inactivation upon cell-cycle exit can also be reinforced by transcriptional downregulation of genes encoding centrosomal proteins (Fig. 3A), as occurs during differentiation in the mammalian epidermis (Sen et al., 2010). A recent report described a mechanism in neurons resulting in alternative splicing of the centrosomal protein ninein that removes its centrosome-targeting domain, resulting in ninein dispersal (Zhang et al., 2016). In addition, cells could theoretically inactivate MTOC activity through post-translational modifications of centrosomal proteins without changing protein levels, although examples of this have not been reported. Below, we consider the different mechanisms that specific cell types use to inactivate the centrosome.
Fig. 2.
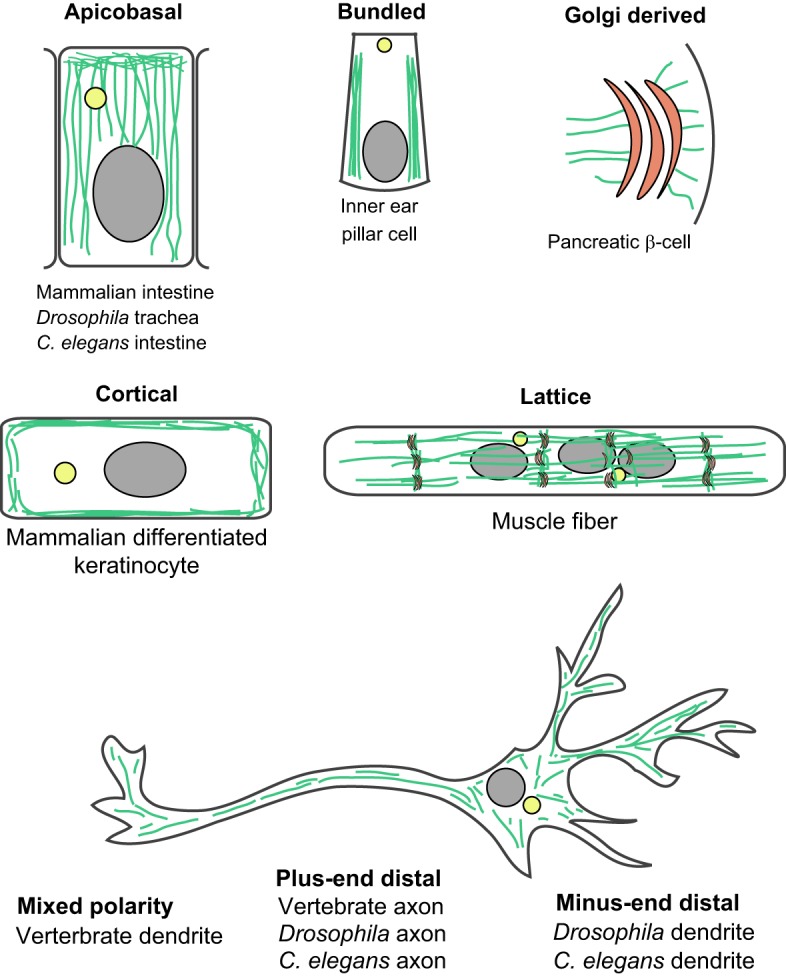
Differentiated animal cells form a variety of non-centrosomal MT arrays. Array geometry is highly cell type specific, but similar cells across species form analogous arrays. Selected representative cell types for different array geometries are illustrated.
Fig. 3.
Models for differentiation-induced centrosome inactivation. (A) Both transcriptional downregulation of centrosomal components and decreased signaling through cell-cycle regulators accompany microtubule reorganization. (B) Specific loss of microtubule anchoring can be the first step in centrosome inactivation; this can be mediated through increased microtubule severing or delocalization/degradation of specific anchoring factors. (C) Centrosome inactivation can also be caused by a general dispersal of pericentriolar material. Note that the mechanisms illustrated in B and C are not mutually exclusive.
Simple epithelia
A number of simple epithelial cells in different species form apicobasal arrays of microtubules, with the minus ends anchored near the apical surface and the plus tips directed towards the basal surface. A comparison of apicobasal arrays in Caenorhabditis elegans, Drosophila and mammals suggests that, although these arrays share common features, distinct mechanisms can be used to generate geometrically similar arrays. Thus, although γ-tubulin is delocalized from centrosomes and subsequently relocalizes to just below the apical surface in all of these cells, the mechanisms that they each use to accomplish this appear to be distinct.
In the C. elegans intestinal epithelium, centrosomal γ-tubulin is actively redistributed to the apical side of the cell in a ‘plume’ of PCM with other known microtubule regulators, including ZYG-9/XMAP215 and GIP1 (Bobinnec et al., 2000; Feldman and Priess, 2012). Coincident with γ-tubulin delocalization, microtubules no longer associate with the centrosome, suggesting that PCM removal directly inactivates the centrosome. The mechanism that induces γ-tubulin release from the centrosome is unknown but is linked to decreased CDK activity (Yang and Feldman, 2015). How γ-tubulin is tethered to the apical surface in these cells and whether it is required for microtubule reorganization is currently unknown.
During Drosophila trachea morphogenesis, the invaginating tracheal epithelium reorganizes its microtubules into apicobasal arrays, and this is coincident with γ-tubulin removal from the centrosome and its subsequent recruitment to the apical surface (Brodu et al., 2010). Subsequently, γ-tubulin is stabilized at the apical side by the transmembrane protein Piopio, which also promotes proper microtubule organization in the pupal wing (Bokel et al., 2005). In the tracheal epithelium, γ-tubulin delocalization is Spastin dependent, although how Spastin-mediated microtubule severing regulates γ-tubulin removal is not known. The transcription factor Trachealess also induces γ-tubulin delocalization, although further work is required to delineate whether it mediates this effect through cell-cycle regulation or a trachea-specific differentiation program. Additionally, whether other PCM components are delocalized remains to be tested.
The differentiated enterocytes that line the villi of the mammalian small intestine also have apicobasal microtubule arrays. In vivo, γ-tubulin is primarily associated with the apical surface of these cells (Salas, 1999; Ameen et al., 2001). Unlike the C. elegans intestine and Drosophila trachea, it is not known whether this apical γ-tubulin is directly trafficked from the centrosome during differentiation or if it is independently derived from cytoplasmic γ-TuRCs. In this case, γ-tubulin is tethered to the apical keratin filament network via the γ-TuRC-specific component GCP6 (TUBGCP6), and disruption of keratin filaments perturbs apical γ-tubulin localization (Ameen et al., 2001; Oriolo et al., 2007). Intriguingly, CDK phosphorylation of GCP6 destabilizes its interaction with keratin filaments, suggesting that decreased CDK activity at differentiation onset could promote γ-tubulin relocalization in these cells (Oriolo et al., 2007).
Taken together, these studies show that centrosome inactivation in simple epithelial cells is associated with γ-tubulin recruitment to the apical surface. The proteins that mediate γ-tubulin release from the centrosome and subsequent stabilization at the apical side appear to be cell type specific but can involve cell-cycle regulators. Future work will be needed to determine whether similar cell-cycle effectors link centrosome inactivation to differentiation onset in other simple epithelial cells.
Stratified epithelia
The mammalian epidermis is an example of a stratified epithelium that is composed of multiple layers of progressively differentiated epithelial cells. Coincident with differentiation in the mammalian epidermis, microtubules reorganize from radial to cortical arrays (Lechler and Fuchs, 2007). Recently, a two-step mechanism for centrosome inactivation involving two functionally distinct γ-TuRCs – Nedd1/γ-TuRC and CDK5RAP2/γ-TuRC – was proposed for differentiating keratinocytes (Muroyama et al., 2016). This work revealed that Nedd1/γ-TuRC complexes are specific for microtubule anchoring to the centrosome and are lost from centrosomes at initial differentiation commitment. Cell-cycle exit and decreased CDK activity are sufficient to promote Nedd1 degradation, γ-TuRC delocalization and centrosome inactivation in the epidermis, linking quiescence with MTOC status. A host of additional proteins, including ninein, Ndel1 and Lis1 (Pafah1b1), are delocalized from the centrosome and recruited to the cell cortex to form a non-centrosomal MTOC (Lechler and Fuchs, 2007; Sumigray et al., 2011). As epidermal cells then continue down the differentiation pathway, additional centrosomal proteins including CDK5RAP2/γ-TuRCs, which are active microtubule nucleators, are lost from the centrosome. Therefore, during epidermal differentiation, PCM is generally dispersed following cell-cycle exit, but distinct protein sub-complexes are delocalized with different kinetics.
Muscle
As myoblasts fuse to form myotubes, microtubules form longitudinal arrays and then a stationary lattice; the microtubule minus ends in this lattice are associated with both the Golgi and nuclei whereas the dynamic plus ends track along existing microtubules (Tassin et al., 1985; Musa et al., 2003; Oddoux et al., 2013). During myoblast differentiation, centrosomal proteins such as pericentrin, ninein and γ-tubulin are delocalized from centrosomes (Bugnard et al., 2005; Srsen et al., 2009). Some of these are subsequently recruited both to the nuclear envelope and to Golgi outposts (Musa et al., 2003; Bugnard et al., 2005; Oddoux et al., 2013). Microtubule reorganization has also been studied in vivo in the context of cardiac muscle cells (cardiomyocytes), which become post-mitotic shortly after birth. Cardiomyocytes have both longitudinal arrays of microtubules within the cell and orthogonal networks beneath the cell cortex (Watkins et al., 1987; Kerr et al., 2015). In mammalian cardiomyocytes in vivo, cell-cycle exit is correlated with centrosomal inactivation (Zebrowski et al., 2015), although the mechanisms for PCM dispersal in these cells are yet to be determined. Interestingly, zebrafish cardiomyoctes that retain proliferative potential do not inactivate their centrosomes (Zebrowski et al., 2015).
Neurons
Mammalian, Drosophila and C. elegans neurons organize their axonal microtubules with their plus ends distal to the cell body (Baas et al., 1988; Stone et al., 2008; Yan et al., 2013). By contrast, microtubules within the dendrites of mammalian neurons have mixed polarity, whereas Drosophila and C. elegans dendritic microtubules have their minus ends oriented away from the cell body (Stone et al., 2008; Hill et al., 2012; Yau et al., 2016; Goodwin et al., 2012). Several studies have demonstrated that as neurons mature both in culture and also in vivo, the centrosome is inactivated and γ-tubulin is gradually delocalized from the centrosome coincident with complete centrosome inactivation (Leask et al., 1997; Stiess et al., 2010). In cultured rat hippocampal neurons, which likely represent a more immature state, γ-tubulin is maintained at the centrosome, and centrosomal MTOC inactivation is initially controlled through a loss of microtubule anchoring (Baas and Joshi, 1992; Yu et al., 1993). Intriguingly, in maturing neurons, Nedd1 protein is lost but total γ-tubulin levels within the neuron remain unchanged. The specific loss of the microtubule-anchoring Nedd1/γ-TuRC is reminiscent of what has been shown in the mammalian epidermis, suggesting that a conserved control point centered on Nedd1 might regulate centrosome inactivation and differentiation-dependent centrosomal γ-TuRC delocalization in various mammalian cell types (Stiess et al., 2010). Additionally, as discussed earlier, differential splicing of the ninein transcript, which eliminates its centrosome targeting domain, might be a second, parallel mechanism to inactivate the centrosome during mammalian neuronal maturation (Zhang et al., 2016).
Centrosome maintenance
Although the studies described above have highlighted mechanisms that are used by various cells to inactivate the MTOC activity of centrosomes, it is not clear whether centrosomes have to be inactivated to render the cell competent to generate non-centrosomal microtubule arrays. In theory, maintenance of an intact centrosome could prevent non-centrosomal microtubule array formation. In the C. elegans intestine, cell fusion experiments have demonstrated that the centrosome MTOC state is dominant (Yang and Feldman, 2015). Therefore, to form apicobasal microtubule arrays properly, the centrosome must be shut off. In the future, it will be important to assess whether forced maintenance of centrosome composition interferes with non-centrosomal microtubule array formation during differentiation in other tissues.
More recently, several groups have begun to explore how centrosome inactivation might in fact reinforce differentiation or cell-cycle status independently of non-centrosomal microtubule array formation, suggesting that the centrosome acts as a scaffold for cellular signaling in post-mitotic cells. One study found that the delocalization of pericentriolar proteins is required to induce centriole disassembly in the female Drosophila germline; forced maintenance of PCM resulted in abnormal divisions and sterility (Pimenta-Marques et al., 2016). In mammalian cardiomyoctes, centrosome inactivation is required to keep cardiomyocytes in a post-mitotic, G0/G1 cell-cycle state (Zebrowski et al., 2015). Finally, it was shown that the expression of a differentiated splice isoform of ninein in neuronal progenitor cells is sufficient to promote their differentiation (Zhang et al., 2016). Therefore, future work should continue to explore how centrosome MTOC inactivation can influence the differentiation status of the cell.
Microtubule nucleation in differentiated cells
As mentioned above, cellular differentiation is associated with a loss of centrosomal arrays and the reorganization of microtubules into non-centrosomal arrays. Such non-centrosomal arrays can be formed through several distinct, although not mutually exclusive, mechanisms (Fig. 4). The centrosome can continue to nucleate microtubules, which can then be released through uncapping or severing and stabilized at a novel cellular site. Nucleation activity can also be relocated to a non-centrosomal MTOC. Alternatively, non-centrosomal microtubule arrays can be formed by maintaining and reconfiguring existing microtubules independently of any requirement for new nucleation. Importantly, microtubule nucleation can also proceed from multiple sites in the cell to establish local, distinct microtubule configurations. As we discuss below, studies using a variety of approaches to analyze microtubule nucleation (see Box 1) have revealed that all of these mechanisms contribute to the formation of non-centrosomal arrays of microtubules as cells undergo differentiation.
Fig. 4.
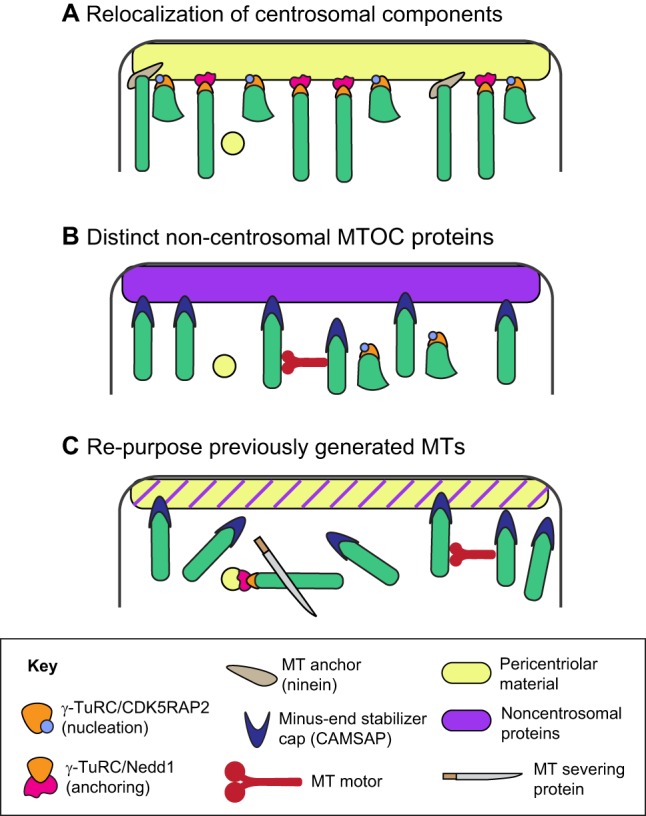
Potential models of non-centrosomal MTOC activation. (A) Centrosomal proteins can be relocalized to a novel cellular site to re-specify that site for microtubule nucleation and/or anchoring. (B) A distinct set of non-centrosomal proteins can be utilized to generate non-centrosomal MTOCs. (C) Non-centrosomal arrays can be generated through microtubule severing and subsequent reorganization independently of new nucleation. These models are not mutually exclusive.
Box 1. Experimental approaches to analyze microtubule nucleation.
Three assays are commonly used to determine sites of microtubule nucleation, each with their own sets of strengths and caveats. First, γ-tubulin localization is often used to identify microtubule-nucleation sites indirectly. However, it was recently demonstrated that not all γ-TuRCs in the cell are nucleation competent, suggesting that γ-tubulin localization may not provide a faithful read-out of nucleation sites in all cases (Muroyama et al., 2016). Second, microtubule regrowth following washout of nocodazole (a chemical that induces microtubule depolymerization) has been used to identify sites of microtubule nucleation in vivo. Nocodazole washout is a robust assay that can often be used to identify nucleation sites in many cells at a time. The major caveats, however, are that it does not identify where microtubules are nucleated during steady-state conditions and high levels of cytoplasmic tubulin dimer following microtubule depolymerization could induce nucleation at sites where it does not typically occur. Third, live-imaging of end-binding (EB) family proteins is useful to visualize sites of microtubule growth, although caution must be applied when interpreting whether the appearance of an EB puncta (known as a ‘comet’) marks a true nucleation event. Such methods to visualize EB dynamics directly in vivo have begun to settle persistent questions about non-centrosomal microtubule array formation, especially when combined with concurrent visualization of microtubules.
Nucleation from the centrosome
In several cell types, centrosomal nucleation continues during the initial stages of non-centrosomal microtubule array establishment. For example, although the centrosome is inactivated as the primary MTOC as neurons reach maturity, during their initial differentiation the centrosome continues to nucleate microtubules, which are then severed by katanin and trafficked out of the cell body (Yu et al., 1993; Ahmad et al., 1999). Although a transgenic mouse has been recently generated to track EB3-GFP in neurons (Kleele et al., 2014), no group has directly assessed centrosomal nucleation in mammalian neurons in vivo. The centrosome is also considered to be the primary microtubule nucleator in the inner pillar cells of the mammalian cochlea; in this context, γ-tubulin remains enriched at the centrosome during differentiation and cytoplasmic microtubules can be seen transiting away from the centrosome to the non-centrosomal MTOC (Mogensen et al., 1997; Tucker et al., 1998). In primary cultures of epidermal cells, too, the centrosome retains nucleation capacity during early differentiation (Lechler and Fuchs, 2007; Muroyama et al., 2016). However, sites of nucleation in terminally differentiated epidermal cells, which have cortical microtubules, are still unknown; γ-tubulin is not associated with either the centrosome or the cell cortex in these cells, suggesting that microtubule nucleation occurs primarily in the cytoplasm. Together, these findings highlight that cells that maintain centrosomal nucleation do so during their initial stages of differentiation. There is little evidence to suggest that the centrosome retains nucleation capacity in fully mature, differentiated cells.
An outstanding question is how microtubules are released and subsequently trafficked to their ultimate stabilization site. Microtubule release from centrosomes has been directly visualized in some cultured cells (Keating et al., 1997; Abal et al., 2002). However, few data exist for how this occurs in vivo. Microtubule release could be mediated through loss of attachment with the γ-TuRC or through severing; however, the free minus end would presumably have to be rapidly capped to prevent depolymerization of the microtubule. The mechanisms controlling subsequent transport of microtubules to the non-centrosomal MTOC also remain largely unexplored with some notable exceptions. In neurons, dynein plays a key role in trafficking microtubules and/or tubulin to their ultimate locations (He et al., 2005; del Castillo et al., 2015). Microtubule-dependent microtubule trafficking has also been proposed to establish bundled microtubules in the inner ear pillar cells (Moss et al., 2007). Clearly, more studies are required to define the mechanisms coordinating centrosomal nucleation with minus-end release and transport during non-centrosomal microtubule array formation.
Nucleation from non-centrosomal sites
A number of intracellular sites have been identified as potential sites of microtubule nucleation during cellular differentiation. Because γ-tubulin is highly enriched at the apical surface of many simple epithelial cells, current models suggest that the apical surface is the primary microtubule nucleator. In both the C. elegans intestine and Drosophila trachea, microtubules regrow from the apical surface after nocodazole washout, consistent with a role for γ-tubulin in nucleation, although other possibilities remain (Yang and Feldman, 2015; Brodu et al., 2010). A similar role for plasma membrane-associated γ-tubulin has been proposed for the C. elegans gonad (Zhou et al., 2009). Whether the apical γ-tubulin in the mammalian intestine is nucleation competent is still unknown. Experiments conducted in cultured simple epithelial cell lines (e.g. MDCK and Caco-2 cells) suggest that the centrosome is still competent for microtubule nucleation following nocodazole washout; however, whether this reflects the natural state in vivo has not been determined (Bre et al., 1987; Meads and Schroer, 1995). Importantly, centrosomes in these cultured cells retain γ-tubulin in contrast to their fully differentiated counterparts in vivo. To date, no group has visualized microtubule growth in apicobasal arrays in vivo under homeostatic conditions to definitively address whether nucleation and anchoring are coordinated at the apical surface.
In mature neurons, the centrosome may no longer be the active nucleator. Laser ablation of the centrosome in well-differentiated neurons in culture does not cause detectable microtubule-organization defects (Stiess et al., 2010). Similarly, in Drosophila dendritic arborization (da) sensory neurons, centrosome ablation either by laser ablation or mutation of the essential centriole component SAS-4 does not cause defects in microtubule organization (Nguyen et al., 2011). The Golgi apparatus has also been demonstrated to be a site for microtubule nucleation in several settings (Efimov et al., 2007; Miller et al., 2009; Rivero et al., 2009; Zhu et al., 2015). In Drosophila da sensory neurons, for instance, EB1-GFP comets grow from Golgi outposts in dendrites, suggesting that these may function as nucleation sites (Ori-McKenney et al., 2012). That Centrosomin, the Drosophila homolog of CDK5RAP2, is localized to the Golgi in these cells supports the idea that they are microtubule nucleators (Yalgin et al., 2015). However, Golgi-mediated microtubule nucleation might not be necessary in all Drosophila neurons, as forced sequestration of the Golgi out of dendrites in several da neuron subtypes does not cause changes in γ-tubulin localization (Nguyen et al., 2014). Interestingly, a recent report demonstrated that microtubules in murine hippocampal neurons are nucleated by γ-TuRCs tethered to existing microtubules through augmin, reinforcing proper non-centrosomal microtubule polarity (Sánchez-Huertas et al., 2016). Augmin-dependent microtubule polymerization of existing microtubules is an attractive model for maintaining uniform microtubule polarity in non-centrosomal arrays. Future work will be needed to identify whether similar augmin-dependent mechanisms are required for non-centrosomal array formation in other cell types.
A number of studies using cultured myotubes have suggested that microtubules can also be nucleated from the nuclear envelope and cytoplasm (Tassin et al., 1985; Musa et al., 2003; Bugnard et al., 2005). More recently, using a remarkable setup, Oddoux et al. performed live imaging of microtubule growth under steady-state conditions in situ in living mouse skeletal muscle (Oddoux et al., 2013). Their visualization of EB3-GFP and GFP-tubulin in the digitorum brevis muscle of the mouse foot directly confirmed that microtubule nucleation occurs from both the nuclear envelope but also, unexpectedly, from Golgi elements at the intersection of microtubule tracks (Oddoux et al., 2013). This work highlights the value of developing systems to visualize microtubule behavior in vivo.
The stabilization and anchoring of microtubule minus ends
Non-centrosomal microtubule arrays, by definition, have minus ends anchored to sites that are distinct from the centrosome, and these sites may or may not be the same sites where microtubules are nucleated. This process of microtubule anchoring can use either the same or distinct proteins from the ones utilized to mediate microtubule anchoring at the centrosome (Fig. 4A,B).
To date, few bona fide minus-end binding proteins have been identified. As mentioned above, γ-TuRCs can cap and anchor microtubules both in the cytoplasm and at the centrosome and may perform a similar function at non-centrosomal sites (Anders and Sawin, 2011; Muroyama et al., 2016). In Arabidopsis and the C. elegans epidermis, cortical microtubules can be both nucleated and anchored by γ-TuRC (Walia et al., 2014; Wang et al., 2015). Similar mechanisms could operate in many cell types where γ-tubulin is relocalized to the non-centrosomal MTOC, although this has yet to be explicitly tested. In addition, although it has never been shown to bind directly to minus ends, ninein has been linked to microtubule anchoring at the centrosome and has been proposed to capture microtubule minus ends at non-centrosomal sites (Mogensen et al., 2000; Moss et al., 2007; Goldspink et al., 2017). In the mammalian epidermis, ninein relocalizes to the cell cortex in differentiated keratinocytes, suggesting that it captures microtubules to form cortical microtubule arrays (Lechler and Fuchs, 2007). Ninein is also deployed to the apical surface in the inner pillar cells of the mammalian cochlea, where it associates with the minus ends of bundled microtubules (Tucker et al., 1998; Mogensen et al., 2000). The relocalization of ninein to cell junctions during epithelial differentiation has also been proposed to play a role in non-centrosomal microtubule formation in simple epithelial cells (Goldspink et al., 2017). Additionally, a recently described putative ninein homolog in C. elegans, NOCA-1, helps to establish microtubule arrays in the C. elegans epidermis (Wang et al., 2015).
The recent identification and characterization of CAMSAP/Patronin/Nezha family proteins has provided fresh insight into how microtubule minus ends are stabilized. These proteins associate with microtubule minus ends in all species in which they have been identified (Baines et al., 2009; Goodwin and Vale, 2010; Meng et al., 2008). CAMSAP homologs have different minus-end protection properties (reviewed by Akhmanova and Hoogenraad, 2015) and their functions and localizations in vivo are beginning to be probed in various cell types. For example, Nezha/CAMSAP3 was originally identified as an adherens junction-associated protein in Caco-2 cells that regulates apicobasal microtubule organization (Meng et al., 2008), and more recently it was reported that microtubules are disorganized in the small intestines of a Camsap3 mutant mouse in which the microtubule-binding domain (the CKK domain) was deleted (Toya et al., 2016). CAMSAP2 has been demonstrated to organize microtubules in mammalian hippocampal neurons independently of γ-tubulin (Yau et al., 2014), and the C. elegans homolog of vertebrate CAMSAP proteins, PTRN-1, is important for microtubule organization in neurons and also in the epidermis (Richardson et al., 2014; Wang et al., 2015). How CAMSAP proteins recognize the minus end is still unknown. Several recent reports have identified proteins that bridge between CAMSAP/Patronin/Nezha and a non-centrosomal docking site. For example, the adherens junction protein PLEKHA7 can bind directly to Nezha to localize to cortical sites in cultured Caco-2 cells (Meng et al., 2008). CAMSAP targeting via various spectraplakins has also been demonstrated in several contexts; ACF7 (MACF7) can link CAMSAP3/Nezha to actin filaments in intestinal epithelial cells (Noordstra et al., 2016) and in migrating cultured cells (Ning et al., 2016), and the Drosophila spectraplakin Short stop (Shot) has been shown to localize Patronin in the Drosophila oocyte (Khanal et al., 2016; Nashchekin et al., 2016). Further studies are needed to examine whether similar mechanisms operate in differentiated cells in vivo to regulate localization of CAMSAP family members.
Microtubule dynamics in differentiated cells
Microtubule dynamics include growth rates and lifetimes at both the microtubule plus and minus ends as well as catastrophe, pause and rescue frequencies, which in addition to microtubule organization, control the functions of microtubules. Although the studies described above have highlighted how microtubule arrays become reorganized as cells undergo differentiation, it is not clear if or how microtubule dynamics are altered to facilitate microtubule reorganization. Although fluorescently tagged EB proteins have been used to live-image microtubule growth, the visualization of microtubule dynamics in distinct cell types over the differentiation process has only been reported in a few studies. EB1-GFP has been used to quantify microtubule growth speeds and clarify microtubule organization in Drosophila neurons (Ori-McKenney et al., 2012; Mattie et al., 2010; Hill et al., 2012), although, to our knowledge, these tools have not been used to analyze microtubule dynamics comprehensively during differentiation. Similarly, a number of transgenic zebrafish lines have been generated and used to visualize microtubules in vivo, although no data exist about how differentiation impacts their dynamics (Distel et al., 2010; Yoo et al., 2012). Recently, using confocal imaging of GFP-tubulin in the C. elegans egg-laying apparatus, Lacroix et al. demonstrated that microtubule dynamics do indeed change during differentiation (Lacroix et al., 2014). Furthermore, by performing an RNA interference screen, they showed that different sets of MAPs are required for distinct microtubule behaviors during differentiation. Additionally, the authors demonstrated that alteration of microtubule dynamics perturbs muscle function in vivo. Another recent study described roles for specific MAPs in regulating both proper microtubule organization and cargo trafficking in the axons of C. elegans DA9 motor neurons (Yogev et al., 2016). How alterations in microtubule dynamics impact cell function in other tissues remains unexplored. Several mammalian EB1/EB3-GFP systems have been developed, but they are currently restricted to specific cell types and have not been used to track changes in microtubule dynamics during differentiation in vivo (Oddoux et al., 2013; Muroyama et al., 2016; Kleele et al., 2014).
Although few studies have tracked how microtubule dynamics change during differentiation, several lines of evidence suggest that regulation of microtubule behavior may be crucial for proper non-centrosomal microtubule array formation. In many differentiated tissues, microtubules are stabilized, as inferred through increased microtubule post-translational modifications and upregulation of MAPs that promote microtubule stability (Sumigray et al., 2012; Brodu et al., 2010; Bre et al., 1987). However, during microtubule reorganization, microtubules may transiently be more dynamic. An intriguing hypothesis is that formation of non-centrosomal microtubules requires microtubules to be more dynamic, and once the final architecture has been established, microtubules are stabilized. In support of this, it has been shown that suppressing microtubule dynamics impairs proper differentiation of the egg-laying apparatus in C. elegans (Lacroix et al., 2014). Furthermore, during myotube differentiation, microtubules are transiently destabilized before being ultimately stabilized (Mian et al., 2012; Mogessie et al., 2015). Similarly, EB3 is required for proper myoblast differentiation in culture, suggesting that appropriate regulation of microtubule dynamics is crucial for non-centrosomal microtubule formation (Straube and Merdes, 2007). Finally, EB2 has been shown to regulate apicobasal microtubule formation, once again linking dynamic microtubules to microtubule reorganization (Goldspink et al., 2013). Intriguingly, injury increases microtubule dynamics in multiple neuronal types, suggesting that microtubule dynamics might revert to a more plastic state during wound healing or periods of regeneration (Kleele et al., 2014; Lu et al., 2015; Stone et al., 2010). However, it should be noted that these studies have primarily used cultured cell models to assess microtubule dynamics, and much work lies ahead to define how microtubule dynamics change during differentiation in distinct tissues. We propose that special focus should be paid to developing novel tools and setups to visualize these parameters in vivo in order to clearly delineate how microtubule dynamics are altered during differentiation in native settings.
Functions for non-centrosomal microtubule arrays in vivo
What purposes do cell type-specific non-centrosomal microtubule arrays serve? In recent years, increased efforts have been made to understand the functions of non-centrosomal microtubule arrays in vivo, but the genetic dissection of these networks has traditionally been challenging because: (1) many MAPs are required for mitosis, rendering global knockout/knockdown impractical; (2) many MAPs have redundant functions, particularly in mammals; and (3) with a limited understanding of the mechanisms regulating non-centrosomal microtubule array formation, there are few clear candidates to target for disruption. In some cases, microtubule-perturbing drugs have been successfully used to probe microtubule function in tissues (Achler et al., 1989; Zhu et al., 2015). Below, we highlight some of the recently developed genetic tools that have been used successfully to understand the functions of non-centrosomal microtubule arrays in tissue development and physiology.
Several groups have used the GAL4/UAS system in Drosophila to overexpress the microtubule-severing protein Spastin in a tissue-specific manner to grossly perturb microtubule organization (Sherwood et al., 2004). Using this approach, it was shown that non-centrosomal microtubule arrays are important for zippering of the epithelium during Drosophila dorsal closure (Jankovics and Brunner, 2006), for maintaining proper cortical levels of adherens junction proteins (Le Droguen et al., 2015) and for denticle spacing in the Drosophila embryo (Spencer et al., 2017). A similar spastin overexpression strategy was used to perturb non-centrosomal microtubules in the C. elegans epidermis and led to a mild elongation defect (Quintin et al., 2016).
More recently, perturbation of CAMSAP family members has been useful in delineating the role of non-centrosomal microtubules in various organisms (Wang et al., 2015). Perturbation of PTRN-1 negatively affects neurite morphology and axon regeneration in C. elegans (Richardson et al., 2014; Chuang et al., 2014). In the Drosophila oocyte, non-centrosomal microtubules stabilized through Patronin are important for polarity and tissue architecture (Nashchekin et al., 2016). Global disruption of the microtubule-binding domain of CAMSAP3 in mice results in disorganized microtubule arrays in the small intestine and an accompanying perturbation of nuclear position and organelle placement (Toya et al., 2016), and CAMSAP2 disruption in the mouse brain results in cell migration defects and axon extension defects (Yau et al., 2014). Moving forward, it will be important to compare how well in vitro phenotypes of microtubule disruption reflect the in vivo functions of microtubules. From the studies that have been performed thus far, it is becoming clear that cells in vivo have distinct mechanisms to tolerate microtubule disruption. In at least one case, Drosophila motor neurons, we have a molecular framework for how this may occur; in these cells, microtubule disruption leads to downregulation of a transcription factor, FoxO, that normally promotes microtubule destabilization (Nechipurenko and Broihier, 2012). Because cells appear remarkably robust to microtubule loss in vivo, and because compensatory mechanisms to tolerate microtubule disruption may operate at the tissue level, canonical roles for microtubules in processes such as polarity establishment and cell-cell adhesion should be further examined in in vivo studies.
Conclusions
A recent resurgence of interest in the formation of non-centrosomal microtubule arrays has led to a number of discoveries uncovering the mechanisms controlling microtubule reorganization. However, we believe that several outstanding questions warrant special focus in the future. Clearly, differentiation induces microtubule reorganization in many cell types and in many species, so it will be interesting to explore whether conserved mechanisms link the cell cycle to centrosome inactivation in different tissues. Second, further development of live-imaging tools to visualize microtubules in vivo will help to settle long-standing questions about microtubule nucleation and reorganization. We are intrigued by the suggestion that reorganization of microtubules requires a transient increase in dynamics that are ultimately suppressed. Future work on the functions for non-centrosomal microtubules in vivo is also required. Moving forward, the development of novel tools to perturb microtubule dynamics, organization and polymer levels in the organism will be needed to answer these questions and to go from simple descriptions of phenotypes to a mechanistic understanding of microtubule function. It should also be emphasized that proper microtubule function in vivo might depend not only on the spatial position of filaments but also on the complex interplay of regulated microtubule dynamics, post-translational modifications and cell-specific MAPs. How all of these distinct elements are collectively organized within the cell to generate functional non-centrosomal arrays is a major outstanding question.
Acknowledgements
We thank the many creative researchers who have contributed to our understanding of microtubule dynamics, organization and function, and regret that length constraints prevent us from discussing all the works in the detail they deserve.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The Lechler Lab is supported by grants from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH/NIAMS) (R01 AR055926 and AR067203), and the National Institute of General Medical Sciences (GM111336). A.M. was supported by a National Science Foundation predoctoral grant. Deposited in PMC for release after 12 months.
References
- Abal M., Piel M., Bouckson-Castaing V., Mogensen M., Sibarita J. B. and Bornens M. (2002). Microtubule release from the centrosome in migrating cells. J. Cell Biol. 159, 731-737. 10.1083/jcb.200207076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achler C., Filmer D., Merte C. and Drenckhahn D. (1989). Role of microtubules in polarized delivery of apical membrane proteins to the brush border of the intestinal epithelium. J. Cell Biol. 109, 179-189. 10.1083/jcb.109.1.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F. J., Yu W., McNally F. J. and Baas P. W. (1999). An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 145, 305-315. 10.1083/jcb.145.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A. and Hoogenraad C. C. (2015). Microtubule minus-end-targeting proteins. Curr. Biol. 25, R162-R171. 10.1016/j.cub.2014.12.027 [DOI] [PubMed] [Google Scholar]
- Akhmanova A. and Steinmetz M. O. (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309-322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Ameen N. A., Figueroa Y. and Salas P. J. (2001). Anomalous apical plasma membrane phenotype in CK8-deficient mice indicates a novel role for intermediate filaments in the polarization of simple epithelia. J. Cell Sci. 114, 563-575. [DOI] [PubMed] [Google Scholar]
- Anders A. and Sawin K. E. (2011). Microtubule stabilization in vivo by nucleation-incompetent gamma-tubulin complex. J. Cell Sci. 124, 1207-1213. 10.1242/jcs.083741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. W. and Joshi H. C. (1992). Gamma-tubulin distribution in the neuron: implications for the origins of neuritic microtubules. J. Cell Biol. 119, 171-178. 10.1083/jcb.119.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. W., Deitch J. S., Black M. M. and Banker G. A. (1988). Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc. Natl. Acad. Sci. USA 85, 8335-8339. 10.1073/pnas.85.21.8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines A. J., Bignone P. A., King M. D. A., Maggs A. M., Bennett P. M., Pinder J. C. and Phillips G. W. (2009). The CKK domain (DUF1781) binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol. Biol. Evol. 26, 2005-2014. 10.1093/molbev/msp115 [DOI] [PubMed] [Google Scholar]
- Ballatore C., Brunden K. R., Huryn D. M., Trojanowski J. Q., Lee V. M.-Y. and Smith A. B. III (2012). Microtubule stabilizing agents as potential treatment for Alzheimer's disease and related neurodegenerative tauopathies. J. Med. Chem. 55, 8979-8996. 10.1021/jm301079z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F. and Gundersen G. G. (2006). Generation of noncentrosomal microtubule arrays. J. Cell Sci. 119, 4155-4163. 10.1242/jcs.03227 [DOI] [PubMed] [Google Scholar]
- Bobinnec Y., Fukuda M. and Nishida E. (2000). Identification and characterization of Caenorhabditis elegans gamma-tubulin in dividing cells and differentiated tissues. J. Cell Sci. 113, 3747-3759. [DOI] [PubMed] [Google Scholar]
- Bokel C., Prokop A. and Brown N. H. (2005). Papillote and Piopio: Drosophila ZP-domain proteins required for cell adhesion to the apical extracellular matrix and microtubule organization. J. Cell Sci. 118, 633-642. 10.1242/jcs.01619 [DOI] [PubMed] [Google Scholar]
- Bre M. H., Kreis T. E. and Karsenti E. (1987). Control of microtubule nucleation and stability in Madin-Darby canine kidney cells: the occurrence of noncentrosomal, stable detyrosinated microtubules. J. Cell Biol. 105, 1283-1296. 10.1083/jcb.105.3.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodu V., Baffet A. D., Le Droguen P. M., Casanova J. and Guichet A. (2010). A developmentally regulated two-step process generates a noncentrosomal microtubule network in Drosophila tracheal cells. Dev. Cell 18, 790-801. 10.1016/j.devcel.2010.03.015 [DOI] [PubMed] [Google Scholar]
- Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J. and Hyman A. A. (2008). XMAP215 is a processive microtubule polymerase. Cell 132, 79-88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnard E., Zaal K. J. M. and Ralston E. (2005). Reorganization of microtubule nucleation during muscle differentiation. Cell Motil. Cytoskelet. 60, 1-13. 10.1002/cm.20042 [DOI] [PubMed] [Google Scholar]
- Choi Y. K., Liu P., Sze S. K., Dai C. and Qi R. Z. (2010). CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191, 1089-1095. 10.1083/jcb.201007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang M., Goncharov A., Wang S., Oegema K., Jin Y. and Chisholm A. D. (2014). The microtubule minus-end-binding protein patronin/PTRN-1 is required for axon regeneration in C. elegans. Cell Rep. 9, 874-883. 10.1016/j.celrep.2014.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbeth N., Lauterio T. J. and Wolfe H. R. (2014). Mechanism of action of colchicine in the treatment of gout. Clin. Ther. 36, 1465-1479. 10.1016/j.clinthera.2014.07.017 [DOI] [PubMed] [Google Scholar]
- del Castillo U., Winding M., Lu W. and Gelfand V. I. (2015). Interplay between kinesin-1 and cortical dynein during axonal outgrowth and microtubule organization in Drosophila neurons. eLife 4, e10140 10.7554/eLife.10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J. and Bornens M. (2005). Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118, 1565-1575. 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Desai A. and Mitchison T. J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117. 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- Distel M., Hocking J. C., Volkmann K. and Köster R. W. (2010). The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J. Cell Biol. 191, 875-890. 10.1083/jcb.201004154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P. M., Andreyeva N., Gleeson P., Galjart N., Maia A. R., McLeod I. X. et al. (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12, 917-930. 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J. L. and Priess J. R. (2012). A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr. Biol. 22, 575-582. 10.1016/j.cub.2012.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink D. A., Gadsby J. R., Bellett G., Keynton J., Tyrrell B. J., Lund E. K., Powell P. P., Thomas P. and Mogensen M. M. (2013). The microtubule end-binding protein EB2 is a central regulator of microtubule reorganisation in apico-basal epithelial differentiation. J. Cell Sci. 126, 4000-4014. 10.1242/jcs.129759 [DOI] [PubMed] [Google Scholar]
- Goldspink D. A., Rookyard C., Tyrrell B. J., Gadsby J., Perkins J., Lund E. K., Galjart N., Thomas P., Wileman T. and Mogensen M. M. (2017). Ninein is essential for apico-basal microtubule formation and CLIP-170 facilitates its redeployment to non-centrosomal microtubule organizing centres. Open Biol. 10.1098/rsob.160274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S. S. and Vale R. D. (2010). Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143, 263-274. 10.1016/j.cell.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin P. R., Sasaki J. M. and Juo P. (2012). Cyclin-dependent kinase 5 regulates the polarized trafficking of neuropeptide-containing dense-core vesicles in Caenorhabditis elegans motor neurons. J. Neurosci. 32, 8158-8172. 10.1523/JNEUROSCI.0251-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Francis F., Myers K. A., Yu W., Black M. M. and Baas P. W. (2005). Role of cytoplasmic dynein in the axonal transport of microtubules and neurofilaments. J. Cell Biol. 168, 697-703. 10.1083/jcb.200407191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershott M. C. and Vale R. D. (2014). Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc. Natl. Acad. Sci. USA 111, 5860-5865. 10.1073/pnas.1404133111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., Benzing T. and Katsanis N. (2011). Ciliopathies. N Engl. J. Med. 364, 1533-1543. 10.1056/NEJMra1010172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. E., Parmar M., Gheres K. W., Guignet M. A., Huang Y., Jackson F. R. and Rolls M. M. (2012). Development of dendrite polarity in Drosophila neurons. Neural Dev. 7, 34 10.1186/1749-8104-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovics F. and Brunner D. (2006). Transiently reorganized microtubules are essential for zippering during dorsal closure in Drosophila melanogaster. Dev. Cell 11, 375-385. 10.1016/j.devcel.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Jiang K., Hua S., Mohan R., Grigoriev I., Yau K. W., Liu Q., Katrukha E. A., Altelaar A. F. M., Heck A. J., Hoogenraad C. C. et al. (2014). Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell 28, 295-309. 10.1016/j.devcel.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Kadavath H., Hofele R. V., Biernat J., Kumar S., Tepper K., Urlaub H., Mandelkow E. and Zweckstetter M. (2015). Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. USA 112, 7501-7506. 10.1073/pnas.1504081112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating T. J., Peloquin J. G., Rodionov V. I., Momcilovic D. and Borisy G. G. (1997). Microtubule release from the centrosome. Proc. Natl. Acad. Sci. USA 94, 5078-5083. 10.1073/pnas.94.10.5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. P., Robison P., Shi G., Bogush A. I., Kempema A. M., Hexum J. K., Becerra N., Harki D. A., Martin S. S., Raiteri R. et al. (2015). Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat. Commun. 6, 8526 10.1038/ncomms9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal I., Elbediwy A., Diaz de la Loza Mdel C., Fletcher G. C. and Thompson B. J. (2016). Shot and Patronin polarise microtubules to direct membrane traffic and biogenesis of microvilli in epithelia. J. Cell Sci. 129, 2651-2659. 10.1242/jcs.189076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleele T., Marinkovic P., Williams P. R., Stern S., Weigand E. E., Engerer P., Naumann R., Hartmann J., Karl R. M., Bradke F. et al. (2014). An assay to image neuronal microtubule dynamics in mice. Nat. Commun. 5, 4827 10.1038/ncomms5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B., Bourdages K. G., Dorn J. F., Ihara S., Sherwood D. R., Maddox P. S. and Maddox A. S. (2014). In situ imaging in C. elegans reveals developmental regulation of microtubule dynamics. Dev. Cell 29, 203-216. 10.1016/j.devcel.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A., Obrietan K. and Stearns T. (1997). Synaptically coupled central nervous system neurons lack centrosomal gamma-tubulin. Neurosci. Lett. 229, 17-20. 10.1016/S0304-3940(97)00412-6 [DOI] [PubMed] [Google Scholar]
- Lechler T. and Fuchs E. (2007). Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J. Cell Biol. 176, 147-154. 10.1083/jcb.200609109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Droguen P. M., Claret S., Guichet A. and Brodu V. (2015). Microtubule-dependent apical restriction of recycling endosomes sustains adherens junctions during morphogenesis of the Drosophila tracheal system. Development 142, 363-374. 10.1242/dev.113472 [DOI] [PubMed] [Google Scholar]
- Leung Y. Y., Yao Hui L. L. and Kraus V. B. (2015). Colchicine--Update on mechanisms of action and therapeutic uses. Semin. Arthritis Rheum. 45, 341-350. 10.1016/j.semarthrit.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Lakonishok M. and Gelfand V. I. (2015). Kinesin-1-powered microtubule sliding initiates axonal regeneration in Drosophila cultured neurons. Mol. Biol. Cell 26, 1296-1307. 10.1091/mbc.E14-10-1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie F. J., Stackpole M. M., Stone M. C., Clippard J. R., Rudnick D. A., Qiu Y., Tao J., Allender D. L., Parmar M. and Rolls M. M. (2010). Directed microtubule growth, +TIPs, and kinesin-2 are required for uniform microtubule polarity in dendrites. Curr. Biol. 20, 2169-2177. 10.1016/j.cub.2010.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meads T. and Schroer T. A. (1995). Polarity and nucleation of microtubules in polarized epithelial cells. Cell Motil. Cytoskelet. 32, 273-288. 10.1002/cm.970320404 [DOI] [PubMed] [Google Scholar]
- Meng W., Mushika Y., Ichii T. and Takeichi M. (2008). Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135, 948-959. 10.1016/j.cell.2008.09.040 [DOI] [PubMed] [Google Scholar]
- Mian I., Pierre-Louis W. S., Dole N., Gilberti R. M., Dodge-Kafka K. and Tirnauer J. S. (2012). LKB1 destabilizes microtubules in myoblasts and contributes to myoblast differentiation. PLoS ONE 7, e31583 10.1371/journal.pone.0031583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P. M., Folkmann A. W., Maia A. R. R., Efimova N., Efimov A. and Kaverina I. (2009). Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 11, 1069-1080. 10.1038/ncb1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N. and Tsukita S. (2000). The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10, 865-868. 10.1016/S0960-9822(00)00600-X [DOI] [PubMed] [Google Scholar]
- Mogensen M. M., Mackie J. B., Doxsey S. J., Stearns T. and Tucker J. B. (1997). Centrosomal deployment of gamma-tubulin and pericentrin: evidence for a microtubule-nucleating domain and a minus-end docking domain in certain mouse epithelial cells. Cell Motil. Cytoskelet. 36, 276-290. [DOI] [PubMed] [Google Scholar]
- Mogensen M. M., Malik A., Piel M., Bouckson-Castaing V. and Bornens M. (2000). Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113, 3013-3023. [DOI] [PubMed] [Google Scholar]
- Mogessie B., Roth D., Rahil Z. and Straube A. (2015). A novel isoform of MAP4 organises the paraxial microtubule array required for muscle cell differentiation. eLife 4, e05697 10.7554/eLife.05697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M. and Agard D. A. (2001). Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11, 174-181. 10.1016/S0959-440X(00)00187-1 [DOI] [PubMed] [Google Scholar]
- Moss D. K., Bellett G., Carter J. M., Liovic M., Keynton J., Prescott A. R., Lane E. B. and Mogensen M. M. (2007). Ninein is released from the centrosome and moves bi-directionally along microtubules. J. Cell Sci. 120, 3064-3074. 10.1242/jcs.010322 [DOI] [PubMed] [Google Scholar]
- Muroyama A., Seldin L. and Lechler T. (2016). Divergent regulation of functionally distinct gamma-tubulin complexes during differentiation. J. Cell Biol. 213, 679-692. 10.1083/jcb.201601099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa H., Orton C., Morrison E. E. and Peckham M. (2003). Microtubule assembly in cultured myoblasts and myotubes following nocodazole induced microtubule depolymerisation. J. Muscle Res. Cell Motil. 24, 301-308. 10.1023/A:1025477807393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashchekin D., Fernandes A. R. and St Johnston D. (2016). Patronin/shot cortical foci assemble the noncentrosomal microtubule array that specifies the drosophila anterior-posterior axis. Dev. Cell 38, 61-72. 10.1016/j.devcel.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko I. V. and Broihier H. T. (2012). FoxO limits microtubule stability and is itself negatively regulated by microtubule disruption. J. Cell Biol. 196, 345-362. 10.1083/jcb.201105154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. L., Chari S., Gruber D., Lue C. M., Chapin S. J. and Bulinski J. C. (1997). Overexpression of full- or partial-length MAP4 stabilizes microtubules and alters cell growth. J. Cell Sci. 110, 281-294. [DOI] [PubMed] [Google Scholar]
- Nguyen M. M., Stone M. C. and Rolls M. M. (2011). Microtubules are organized independently from the centrosome in Drosophila neurons. Neural Dev. 6, 38 10.1186/1749-8104-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. M., McCracken C. J., Milner E. S., Goetschius D. J., Weiner A. T., Long M. K., Michael N. L., Munro S. and Rolls M. M. (2014). Gamma-tubulin controls neuronal microtubule polarity independently of Golgi outposts. Mol. Biol. Cell 25, 2039-2050. 10.1091/mbc.E13-09-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning W., Yu Y., Xu H., Liu X., Wang D., Wang J., Wang Y. and Meng W. (2016). The CAMSAP3-ACF7 complex couples noncentrosomal microtubules with actin filaments to coordinate their dynamics. Dev. Cell 39, 61-74. 10.1016/j.devcel.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Noordstra I., Liu Q., Nijenhuis W., Hua S., Jiang K., Baars M., Remmelzwaal S., Martin M., Kapitein L. C. and Akhmanova A. (2016). Control of apico-basal epithelial polarity by the microtubule minus-end-binding protein CAMSAP3 and spectraplakin ACF7. J. Cell Sci. 129, 4278-4288. 10.1242/jcs.194878 [DOI] [PubMed] [Google Scholar]
- Oddoux S., Zaal K. J., Tate V., Kenea A., Nandkeolyar S. A., Reid E., Liu W. and Ralston E. (2013). Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J. Cell Biol. 203, 205-213. 10.1083/jcb.201304063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori-McKenney K. M., Jan L. Y. and Jan Y.-N. (2012). Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron 76, 921-930. 10.1016/j.neuron.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriolo A. S., Wald F. A., Canessa G. and Salas P. J. I. (2007). GCP6 binds to intermediate filaments: a novel function of keratins in the organization of microtubules in epithelial cells. Mol. Biol. Cell 18, 781-794. 10.1091/mbc.E06-03-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Diamantopoulos G. S., Stalder R. and Kreis T. E. (1999). CLIP-170 highlights growing microtubule ends in vivo. Cell 96, 517-527. 10.1016/S0092-8674(00)80656-X [DOI] [PubMed] [Google Scholar]
- Pimenta-Marques A., Bento I., Lopes C. A., Duarte P., Jana S. C. and Bettencourt-Dias M. (2016). A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science 353, aaf4866 10.1126/science.aaf4866 [DOI] [PubMed] [Google Scholar]
- Quintin S., Wang S., Pontabry J., Bender A., Robin F., Hyenne V., Landmann F., Gally C., Oegema K. and Labouesse M. (2016). Non-centrosomal epidermal microtubules act in parallel to LET-502/ROCK to promote C. elegans elongation. Development 143, 160-173. 10.1242/jcs.185371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. E., Spilker K. A., Cueva J. G., Perrino J., Goodman M. B. and Shen K. (2014). PTRN-1, a microtubule minus end-binding CAMSAP homolog, promotes microtubule function in Caenorhabditis elegans neurons. eLife 3, e01498 10.7554/eLife.01498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S., Cardenas J., Bornens M. and Rios R. M. (2009). Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28, 1016-1028. 10.1038/emboj.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A. and McNally F. J. (2010). Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22, 96-103. 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas P. J. I. (1999). Insoluble gamma-tubulin-containing structures are anchored to the apical network of intermediate filaments in polarized CACO-2 epithelial cells. J. Cell Biol. 146, 645-658. 10.1083/jcb.146.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Huertas C., Freixo F., Viais R., Lacasa C., Soriano E. and Lüders J. (2016). Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat. Commun. 7, 12187 10.1038/ncomms12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G. L., Reuter J. A., Webster D. E., Zhu L. and Khavari P. A. (2010). DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463, 563-567. 10.1038/nature08683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood N. T., Sun Q., Xue M., Zhang B. and Zinn K. (2004). Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2, e429 10.1371/journal.pbio.0020429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. and Brady S. T. (2015). Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol. 25, 125-136. 10.1016/j.tcb.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A. K., Schaumberg A. J. and Zallen J. A. (2017). Scaling of cytoskeletal organization with cell size in Drosophila. Mol. Biol. Cell 28, 1519-1529. 10.1091/mbc.E16-10-0691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srsen V., Fant X., Heald R., Rabouille C. and Merdes A. (2009). Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 10, 28 10.1186/1471-2121-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R. A., Gernert K. M., Nettles J. H. and Aneja R. (2011). Drugs that target dynamic microtubules: a new molecular perspective. Med. Res. Rev. 31, 443-481. 10.1002/med.20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiess M., Maghelli N., Kapitein L. C., Gomis-Ruth S., Wilsch-Brauninger M., Hoogenraad C. C., Tolic-Norrelykke I. M. and Bradke F. (2010). Axon extension occurs independently of centrosomal microtubule nucleation. Science 327, 704-707. 10.1126/science.1182179 [DOI] [PubMed] [Google Scholar]
- Stone M. C., Roegiers F. and Rolls M. M. (2008). Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol. Biol. Cell 19, 4122-4129. 10.1091/mbc.E07-10-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. C., Nguyen M. M., Tao J., Allender D. L. and Rolls M. M. (2010). Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol. Biol. Cell 21, 767-777. 10.1091/mbc.E09-11-0967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A. and Merdes A. (2007). EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr. Biol. 17, 1318-1325. 10.1016/j.cub.2007.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray K. D., Chen H. and Lechler T. (2011). Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J. Cell Biol. 194, 631-642. 10.1083/jcb.201104009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray K. D., Foote H. P. and Lechler T. (2012). Noncentrosomal microtubules and type II myosins potentiate epidermal cell adhesion and barrier formation. J. Cell Biol. 199, 513-525. 10.1083/jcb.201206143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A. M., Maro B. and Bornens M. (1985). Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 100, 35-46. 10.1083/jcb.100.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M., Kobayashi S., Kawasaki M., Shioi G., Kaneko M., Ishiuchi T., Misaki K., Meng W. and Takeichi M. (2016). CAMSAP3 orients the apical-to-basal polarity of microtubule arrays in epithelial cells. Proc. Natl. Acad. Sci. USA 113, 332-337. 10.1073/pnas.1520638113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker J. B., Mogensen M. M., Henderson C. G., Doxsey S. J., Wright M. and Stearns T. (1998). Nucleation and capture of large cell surface-associated microtubule arrays that are not located near centrosomes in certain cochlear epithelial cells. J. Anat. 192, 119-130. 10.1046/j.1469-7580.1998.19210119.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenstein M. L. and Roll-Mecak A. (2016). Graded control of microtubule severing by tubulin Glutamylation. Cell 164, 911-921. 10.1016/j.cell.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia A., Nakamura M., Moss D., Kirik V., Hashimoto T. and Ehrhardt D. W. (2014). GCP-WD mediates gamma-TuRC recruitment and the geometry of microtubule nucleation in interphase arrays of Arabidopsis. Current Biol. 24, 2548-2555. 10.1016/j.cub.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Wang S., Wu D., Quintin S., Green R. A., Cheerambathur D. K., Ochoa S. D., Desai A. and Oegema K. (2015). NOCA-1 functions with gamma-tubulin and in parallel to Patronin to assemble non-centrosomal microtubule arrays in C. elegans. eLife 4, e08649 10.7554/eLife.08649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins S. C., Samuel J. L., Marotte F., Bertier-Savalle B. and Rappaport L. (1987). Microtubules and desmin filaments during onset of heart hypertrophy in rat: a double immunoelectron microscope study. Circ. Res. 60, 327-336. 10.1161/01.RES.60.3.327 [DOI] [PubMed] [Google Scholar]
- Wiese C. and Zheng Y. (2000). A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2, 358-364. 10.1038/35014051 [DOI] [PubMed] [Google Scholar]
- Yalgin C., Ebrahimi S., Delandre C., Yoong L. F., Akimoto S., Tran H., Amikura R., Spokony R., Torben-Nielsen B., White K. P. et al. (2015). Centrosomin represses dendrite branching by orienting microtubule nucleation. Nat. Neurosci. 18, 1437-1445. 10.1038/nn.4099 [DOI] [PubMed] [Google Scholar]
- Yan J., Chao D. L., Toba S., Koyasako K., Yasunaga T., Hirotsune S. and Shen K. (2013). Kinesin-1 regulates dendrite microtubule polarity in Caenorhabditis elegans. eLife 2, e00133 10.7554/eLife.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R. and Feldman J. L. (2015). SPD-2/CEP192 and CDK are limiting for microtubule-organizing center function at the centrosome. Curr. Biol. 25, 1924-1931. 10.1016/j.cub.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Yau K. W., van Beuningen S. F., Cunha-Ferreira I., Cloin B. M. C., van Battum E. Y., Will L., Schätzle P., Tas R. P., van Krugten J., Katrukha E. A. et al. (2014). Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron 82, 1058-1073. 10.1016/j.neuron.2014.04.019 [DOI] [PubMed] [Google Scholar]
- Yau K. W., Schatzle P., Tortosa E., Pages S., Holtmaat A., Kapitein L. C. and Hoogenraad C. C. (2016). Dendrites in vitro and in vivo contain microtubules of opposite polarity and axon formation correlates with uniform plus-end-out microtubule orientation. J. Neurosci. 36, 1071-1085. 10.1523/JNEUROSCI.2430-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev S., Cooper R., Fetter R., Horowitz M. and Shen K. (2016). Microtubule organization determines axonal transport dynamics. Neuron 92, 449-460. 10.1016/j.neuron.2016.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Lam P.-Y., Eichelberg M. R., Zasadil L., Bement W. M. and Huttenlocher A. (2012). The role of microtubules in neutrophil polarity and migration in live zebrafish. J. Cell Sci. 125, 5702-5710. 10.1242/jcs.108324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Centonze V. E., Ahmad F. J. and Baas P. W. (1993). Microtubule nucleation and release from the neuronal centrosome. J. Cell Biol. 122, 349-359. 10.1083/jcb.122.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebrowski D. C., Vergarajauregui S., Wu C. C., Piatkowski T., Becker R., Leone M., Hirth S., Ricciardi F., Falk N., Giessl A. et al. (2015). Developmental alterations in centrosome integrity contribute to the post-mitotic state of mammalian cardiomyocytes. eLife 4, e05563 10.7554/eLife.05563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempel H. and Mandelkow E. (2014). Lost after translation: missorting of Tau protein and consequences for Alzheimer disease. Trends Neurosci. 37, 721-732. 10.1016/j.tins.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Zhang X., Chen M. H., Wu X., Kodani A., Fan J., Doan R., Ozawa M., Ma J., Yoshida N., Reiter J. F. et al. (2016). Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166, 1147-1162 e1115. 10.1016/j.cell.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Rolls M. M., Hall D. H., Malone C. J. and Hanna-Rose W. (2009). A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 186, 229-241. 10.1083/jcb.200902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Hu R., Brissova M., Stein R. W., Powers A. C., Gu G. and Kaverina I. (2015). Microtubules negatively regulate insulin secretion in pancreatic beta cells. Dev. Cell 34, 656-668. 10.1016/j.devcel.2015.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]