Abstract
Bladder cancer (BC) is the sixth most common cancer in the United States and is the number one cause of death among patients with urinary system malignancies. This makes the identification of invasive regulator(s)/effector(s) as the potential therapeutic targets for managing BC a high priority. p63 is a member of the p53 family of tumor suppressor genes/proteins, plays a role in the differentiation of epithelial tissues, and is believed to function as a tumor suppressor. However, it remains unclear whether and how p63 functions in BC cell invasion after tumorigenesis. Here, we show that p63α protein levels were much higher in mouse high-invasive BC tissues than in normal tissues. Our results also revealed that p63α is crucial for heat shock protein 70 (Hsp70) expression and subsequently increases the ability of BC invasion. Mechanistic experiments demonstrated that p63α can transcriptionally up-regulate Hsp70 expression, thereby promoting BC cell invasion via the Hsp70/Wasf3/Wave3/MMP-9 axis. We further show that E2F transcription factor 1 (E2F1) mediates p63α overexpression-induced Hsp70 transcription. We also found that p63α overexpression activates E2F1 transcription, which appears to be stimulated by p63α together with E2F1. Collectively, our results demonstrate that p63α is a positive regulator of BC cell invasion after tumorigenesis, providing significant insights into the biological function of p63α in BC and supporting the notion that p63α might be a potential target for invasive BC therapy.
Keywords: 70-kilodalton heat shock protein (Hsp70), deleted in bladder cancer 1 (DBC1), E2F transcription factor, invasion, p63, E2F1, Hsp70, Wasf3/Wave3, bladder cancer cell invasion, p63α
Introduction
Bladder cancer (BC)3 is the sixth most common cancer in the United States and is the number one cause of death among patients with urinary system malignancies (1). The incidence of BC has steadily risen in recent decades. It is estimated that >76,960 Americans will be diagnosed with BC and >16,390 will die of this disease in 2016 (1). Approximately 20∼30% of BCs are muscle-invasive, and 50% of these patients die from metastasis within 2 years of diagnosis (2). The 5-year survival rate for metastatic BC is only 5% (2). The remaining 70–80% of BCs are initially diagnosed as non-muscle-invasive, of which ∼20% later progress to become muscle-invasive bladder cancer. These progressive BCs have a 43% lower 5-year survival rate than non-muscle invasive cancers (3). The depth of invasion of the bladder wall is closely associated with the clinical treatment of bladder cancers (4). High-grade invasive bladder cancers can rapidly progress to life-threatening metastases, and the 5-year overall survival rate in patients with lymph node-only disease was 20.9% (5). For these reasons the identification of key molecules that are responsible for mediating human BC invasion and metastasis is of tremendous importance for improving the clinical outcome of patients with invasive BCs.
Tumor protein p63, or TP63, is a transcription factor belonging to the p53 family (6). All members of the family have highly conserved domains: a transactivation domain (TA), a DNA-binding domain, and an oligomerization domain (6, 8, 9). The p63 protein has been shown to be important in the development of epithelial tissues. p63-deficient mice have several developmental defects, such as the lack of limbs, teeth, and mammary glands (10). Like other members of the p53 family, the TP63 gene is expressed as multiple isoforms arising by either alternative promoter usage or differential splicing events at the C terminus. In particular, The p63 isoforms containing the transactivation TA domain (TAp63) are capable of efficiently transactivating different p53-responsive genes, such as p21, bax, mdm2, and other unique targets (11), thus largely mimicking p53 tumor-suppressive activities (11–13). Accordingly, the ectopic expression of TAp63 is able to induce cell cycle arrest and apoptosis (14). Conversely, the ΔN proteins, which lack the canonical TA and are endowed with an alternative activation domain, promote cancer cell survival and tumor progression (15–17). It has been reported that loss of p63 results in spontaneous tumor formation, although the mechanism underlying the tumorigenesis is not yet fully understood (18). p63α is the longest TA transcript variant of p63 and has been characterized as a tumor suppressor responsible for preventing cancer development (19–23).
Most of the existing studies on p63α have focused on its role as a tumor suppressor gene (11, 24). Our previous studies demonstrate that the XIAP RING domain mediates miR-4295 expression, inhibiting p63α protein translation and promoting malignant transformation of bladder epithelial cells (25). However, much less is known about p63α's role in invasion and metastasis after tumor formation. In our present studies, we attempted to fill this gap in knowledge and define the function of p63α in formed human high invasive BC cells. We found that p63α protein levels were higher in BC tissues than in normal bladder tissues and that p63α was able to regulate BC invasion via transcriptional regulation of E2F1 expression and subsequent increases in heat shock protein 70 (Hsp70) transcription. We further showed that Hsp70 expression mediated by p63α was able to promote BC invasion through the Hsp70/Wasf3/Wave3/MMP-9 axis. Given that our current studies together with our published studies show that p63α inhibits human BC growth, we conclude that p63α could function as a double-edged sword for its promoting BC invasion accompanied with suppression of human BC growth.
Results
p63α was up-regulated in mouse bladder cancer tissues
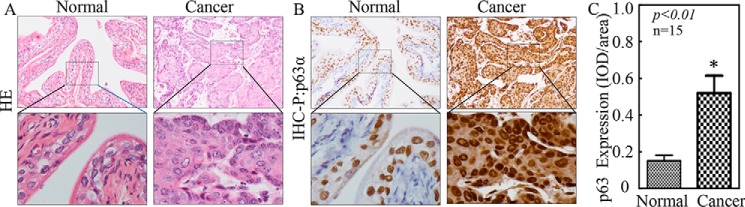
Previous studies have revealed that p63α can serve as a tumor suppressor (19–23). However, its function in invasion after tumorigenesis remains elusive. N-Butyl-N-(4-hydroxybutyl) nitrosamine (BBN) is a well-characterized bladder carcinogen for its induction of 100% invasive BC in mouse model (26). Consistent with the above, exposure of mice to BBN in drinking water led to 100% (15/15) invasive bladder cancer formation, whereas there were no tumors observed in the control group of 15 mice receiving normal drinking water (n = 15) as demonstrated by pathological hematoxylin and eosin staining (Fig. 1A). The expression level of p63α in the mouse invasive bladder cancer were further evaluated by using immunohistochemistry (IHC) staining, and the results showed that p63α protein levels were remarkably increased in BBN-treated mouse bladder cancer tissues in comparison with bladder tissues from mice in control group (Fig. 1, B and C).
Figure 1.
p63α was up-regulated in mouse BC tissues. A, BBN-induced mouse high invasive BC tissues and normal mouse bladder tissue (n = 15) were collected for hematoxylin and eosin (HE) and IHC staining. Hematoxylin and eosin staining was performed, and the representative images of each tissue were captured as described in our previous studies (7). B and C, IHC-P (immunohistochemistry-paraffin staining) was carried out to evaluate p63α protein expression in mouse BC tissues and normal bladder tissues. The optical density was analyzed as described under “Experimental Procedures.” The asterisk (*) indicates a significant increase in comparison with that of normal tissues (p < 0.05). IOD/area, integrated optical density per stained area.
p63α up-regulation promoted BC cell invasion
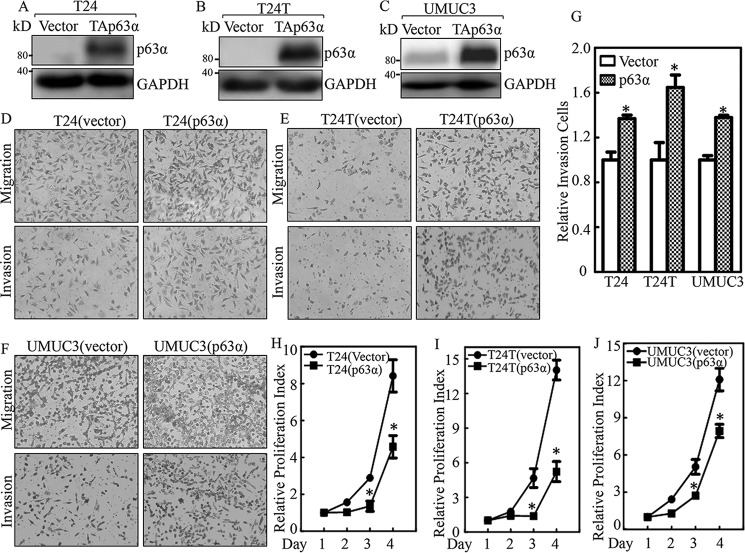
To elucidate the molecular mechanisms underlying the role of p63α in BC development, we constructed p63α overexpression plasmids and stably transfected them into BC cell lines T24, T24T. and UMUC3. The stable transfectants of p63α and its scramble control were established as shown in Fig. 2, A–C. We then utilized a transwell assay to compare the invasion abilities of the stable transfectants. The invasion abilities of p63α overexpression cells was significantly increased compared with those of the vector control transfectants (Fig. 2, D–G). It should be noted that based on the manufacturer's instructions for the “transwell invasion assay,” migrated cells from transwells without Matrigel were used as the internal control to normalize the invasive cells observed in transwells with Matrigel, which should exclude the effect of cell proliferative factor on cell invasion. Given that our most recent studies indicate that p63α is able to inhibit EGF-induced anchorage-independent growth of human urothelial cells, an ATP assay was performed to test whether p63α overexpression inhibited human invasive BC growth. As shown in Fig. 2, H–J, p63α overexpression did inhibit monolayer growth of all three human high-invasive BC cells tested. These results demonstrate that p63α serves as an important positive regulator for BC cell invasion.
Figure 2.
p63α up-regulation promoted invasion in human high invasive BC cells. A–C, Western blotting was used to evaluate the stable ectopic expression of p63α in p63α transfectants as indicated. D–G, the invasion abilities of p63α stable transfectants and their scramble vector transfectants were evaluated by using BD BioCoatTM MatrigelTM Invasion Chamber. The asterisk (*) indicates a significant difference in invasion ability between p63α-overexpressed cells and their scramble vector transfectants (p < 0.05). The bars are presented as the mean ± S.D. from three independent experiments. H–J, cell proliferation was determined in the indicated transfectants using ATP assay. The asterisk (*) indicates a significant inhibition as compared with the scramble vector transfectants.
Hsp70 and Wave3 were up-regulated in p63α-overexpressed BC cells
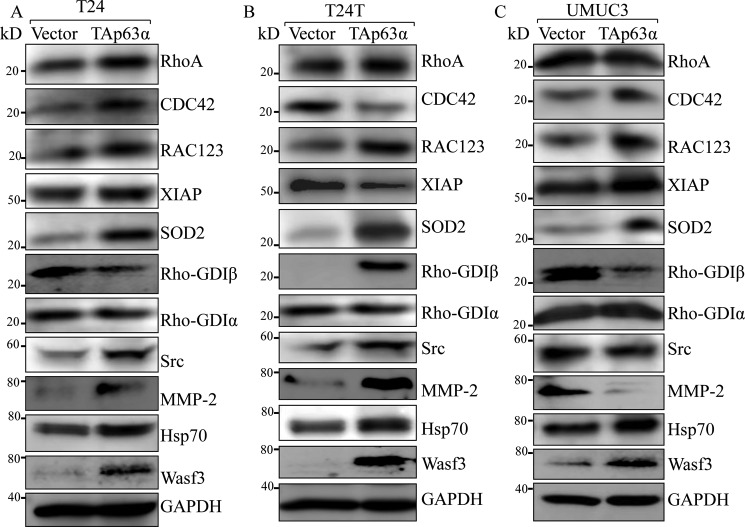
To define the mechanism by which p63α promotes BC cell invasion, we compared the expression levels of key proteins involved in the regulation of BC migration and invasion between scramble vector transfectants and p63α overexpressed T24, T24T, and UMUC3 cells. As shown in Fig. 3, only Hsp70 and Wave3 were consistently up-regulated in all three human high-invasive BC cell lines with ectopic expression of TAp63α in comparison with their related scramble vector transfectants, whereas the expression levels of other proteins, including RhoA, CDC42, RAC123, XIAP, SOD2, RhoGDIα, RhoGDIβ, and SRC did not show consistent alteration in three cell lines (T24, T24T, and UMUC3) after TAp63 overexpression. Our results revealed that Hsp70 and Wave3 may be associated with BC cell invasion.
Figure 3.
Hsp70 and Wave3 were consistently up-regulated in p63α ectopic expressed human BC cells. A–C, the indicated cells were seeded into 6-well plates. The cells were extracted upon the cell density reaching 80–90%, and the cell extracts were subjected to Western blot for determination of protein expression as indicated. GAPDH was used as a protein loading control.
Hsp70 was crucial for the invasive abilities of BC cells
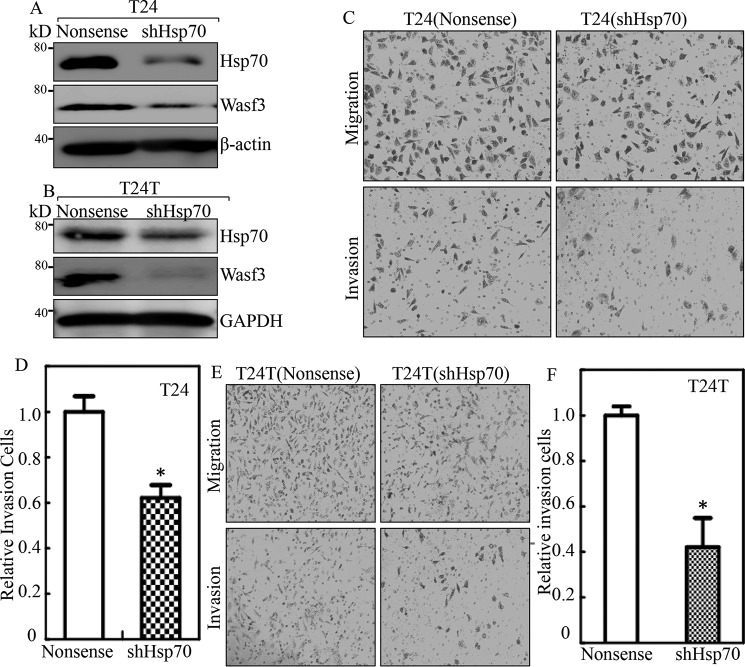
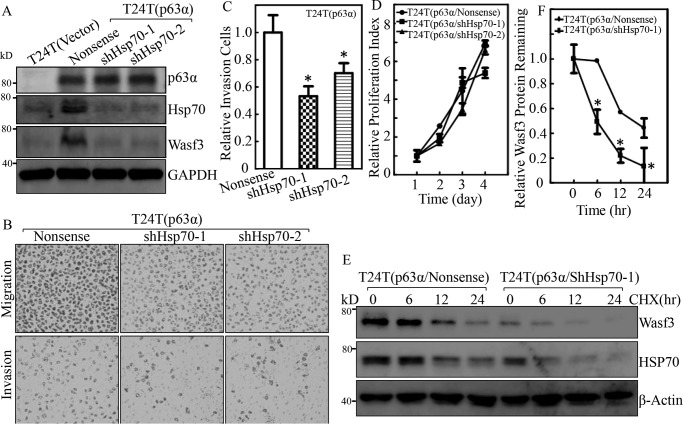
Hsp70 is an important chaperone protein that plays a key regulator for many cell functions, including cell growth and tumor promotion (27). To test whether Hsp70 contributes to increased BC cell invasion, shRNA specifically targeting Hsp70 (shHsp70) was stably transfected into invasive BC cells. The stable transfectant T24(shHsp70), T24T(shHsp70), and their nonsense scramble transfectants were established and identified as shown in Fig. 4, A and B. The knockdown of Hsp70 led to a decreased invasive ability as compared with their scramble Nonsense transfectants in an invasion assay (Fig. 4, C–F). Given that Hsp70 acts as a chaperonin and in turn stabilized Wasf3 protein (28), we tested whether Wasf3 was regulated by Hsp70 in BC cells. The results showed that knockdown of Hsp70 led to a dramatic decrease in Wasf3 protein in T24 and T24T cells as compared with their nonsense scramble transfectants (Fig. 4, A and B), suggesting that Wasf3 might be a downstream effector of Hsp70 responsible for BC invasion.
Figure 4.
Hsp70 was an important player in the invasion of human BC cells. A and B, the cell extracts from the indicated cells were subjected to Western blot for determination of levels of Hsp70 and Wasf3. β-Actin or GAPDH was used as a protein loading control. C–F, invasion abilities of T24(shHsp70) versus T24(Nonsense) cells and T24T(shHsp70) versus T24T(Nonsense) cells were determined using BD BioCoatTM MatrigelTM Invasion Chamber. The asterisk (*) indicates a significant difference of invasion abilities between T24(shHsp70) versus T24(Nonsense) cells or T24T(shHsp70) and T24T(nonsense) cells (p < 0.05). The bars are presented as the mean ± S.D. from three independent experiments.
Decreased Hsp70 resulted in invasive ability in p63α-overexpressed BC cells, and Wave3 was a downstream effector of Hsp70
To determine whether Hsp70 is required for overexpressed p63a promoting BC cell invasion, we stably transfected shHsp70 into p63α-overexpressed cells T24T(p63α), and the stable transfectants of T24T(p63α/shHsp70-1) and T24T(p63α/shHsp70–2) as well as their related control transfectants T24T(p63α/Nonsense) and T24T(Vector) were established. As shown in Fig. 5A, Hsp70 was successfully knocked down in T24T(p63α/shHsp70-1) and T24T(p63α/shHsp70-2) cells in comparison with T24T(p63α/Nonsense) cells. T24T(p63α/shHsp70-1) and T24T(p63α/shHsp70–2) cells showed a reduction in their invasive abilities when compared with T24T(p63α/Nonsense) cells (Fig. 5, B and C), suggesting that Hsp70 is an important player in p63α promoting BC invasion. These results indicate that p63α overexpression promotes Hsp70 expression, which plays a critical role in promoting T24T cell invasion. It was noted that knockdown of Hsp70 did not affect BC cell proliferation in p63α-overexpressed T24T cells (Fig. 5D). We next investigated how Hsp70 regulated Wasf3 protein. The T24T(p63α/Nonsense) and T24T(p63α/shHsp70) cells were treated with cycloheximide to inhibit new protein synthesis in order to observe the potential regulation of Hsp70 on Wasf3 protein gradation. As shown in Fig. 5, E and F, Wasf3 protein degradation in T24T(p63α/shHsp70-1) cells was much faster than that in T24T(p63α/Nonsense) cells, suggesting that Hsp70 expression is able to stabilize Wasf3 protein.
Figure 5.
Wave3 was an Hsp70 downstream mediator responsible for p63α promotion of BC invasion. A, T24T(Vector), T24T(p63α/Nonsense), T24T(p63α/shHsp70-1), and T24T(p63α/shHsp70–2) cells were extracted upon the density reaching 80–90%, and the cell extracts were subjected to Western blotting with the specific antibodies as indicated. GAPDH was used as a protein-loading control. B and C, the invasion abilities of T24T(p63α/Nonsense), T24T(p63α/shHsp70-1), and T24T(p63α/shHsp70–2) cells were subjected to a transwell invasion assay. B, the invasion rate was normalized with the insert control according to the manufacturer's instruction, and the results are presented as relative invasion cells. C, the asterisk (*) indicates a significant difference of invasion abilities between T24T(p63α/Nonsense) and T24T(p63α/shHsp70) cells (p < 0.05). D, cell proliferation was evaluated by ATG assay. E, T24T(p63α/Nonsense) and T24T(p63α/shHsp70-1) cells were treated with 50 μg/ml cycloheximide (CHX) for the indicated time periods, and the cell extracts were subjected to Western blotting to analyze Wasf3 and Hsp70 protein degradation rates. β-Actin was used as a protein loading control. F, Wasf3 protein degradations from three independent experiments were analyzed and presented. The asterisk (*) indicates a significant difference between the indicated two transfectants.
p63α promoted Hsp70 transcription by up-regulating E2F1 and Sp1 protein expression
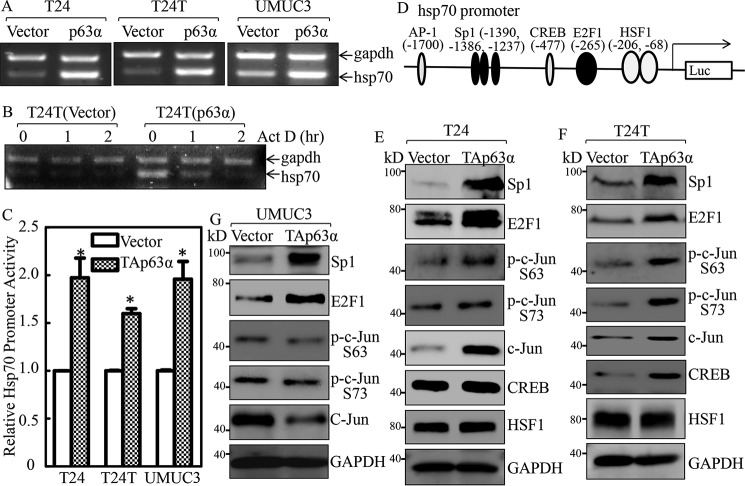
Hsp70 expression is delicately regulated at multiple levels, including transcriptional, post-transcriptional, translational, and post-translational levels (29). Given the above results showing that p63α is important for Hsp70 up-regulation, our subsequent efforts were directed at identifying the mechanisms behind p63α-mediated Hsp70 up-regulation. Hsp70 mRNA levels were markedly increased in p63α-overexpressed BC cells as compared with those observed in their control vector transfectants (Fig. 6A). In contrast, the mRNA degradation in p63α-overexpressed cells was faster than those in control vector transfectants (Fig. 6B). We then evaluated the p63α regulation of Hsp70 transcription by using an Hsp70 promoter-driven luciferase reporter. We found that Hsp70 promoter activity was significantly increased in p63α-overexpressed transfectants (Fig. 6C). These results demonstrate that p63α up-regulates Hsp70 transcription.
Figure 6.
Hsp70 was up-regulated at transcriptional level by p63α in bladder cancer cells. A, the indicated cells were extracted with TRIzol reagent to isolate total RNA upon the density reaching 80–90%. hsp70 mRNA levels were determined with RT-PCR by using the specific primers. GAPDH was used as an internal control. B, T24T(Vector) and T24T(p63α) cells were seeded into 6-well plates. After synchronization, T24T(Vector) and T24T(p63α) cells were treated with actinomycin D (Act D) for the indicated time points, then total RNA was isolated and subjected to RT-PCR analysis to evaluate mRNA levels of hsp70 and GAPDH. C, the indicated cells were transfected with Hsp70 promoter-driven luciferase reporter together with pRL-TK. The transfectants were seeded into 96-well plates and then subjected to determine Hsp70 promoter activity by measuring luciferase activity. pRL-TK was used as an internal control to normalize the transfection efficiency. Each bar indicates the mean ± S.D. from three replicate assays. The asterisk (*) indicates a significant increase in promoter-driven promoter activity in p63-overexpressed cells in comparison with Vector transfectants (p < 0.05). D, potential transcriptional factor-binding sites in the Hsp70 promoter region (−2000 + 1) were analyzed using the TRANSFAC 8.3 engine online. E–G, p63α stable transfectants as indicated were extracted, and the cell extracts were subjected to Western blotting to determine expression of the indicated proteins. GAPDH was used as a protein loading control.
To elucidate the mechanisms underlying p63α up-regulating Hsp70 transcription, TFANSFAC® Transcription Factor Binding Sites Software (Biological Database, Wolfenbüttel, Germany) was applied for Bioinformatics analysis of the Hsp70 promoter region. The results indicated that the Hsp70 gene promoter region contains the putative DNA-binding sites for nuclear factor AP-1, Sp1, cAMP-response element-binding protein (CREB)-binding protein (CBP), E2F1, and activating heat shock factor 1 (HSF1; Fig. 6D). We next determined the expression level of these transcription factors in TAp63-overexpressed cells in comparison with the scramble vector transfectants. As shown in Fig. 6, E–G, Sp1 and E2F1 proteins were consistently elevated in TAp63α-overexpressed BC cells as compared with their scramble vector transfectants, whereas the effect of ectopic expression of Tap63 on expression/phosphorylation of C-Jun, CREB, and HSF1 was not consistent in three type BC cell lines, including T24, T24T, and UMUC3, revealing that Sp1 and E2F1 is modulated by TAp63α and may participate in hsp70 transcriptional regulation by p63α.
E2F1 was crucial for Hsp70 transcriptional up-regulation and promotion of BC invasion
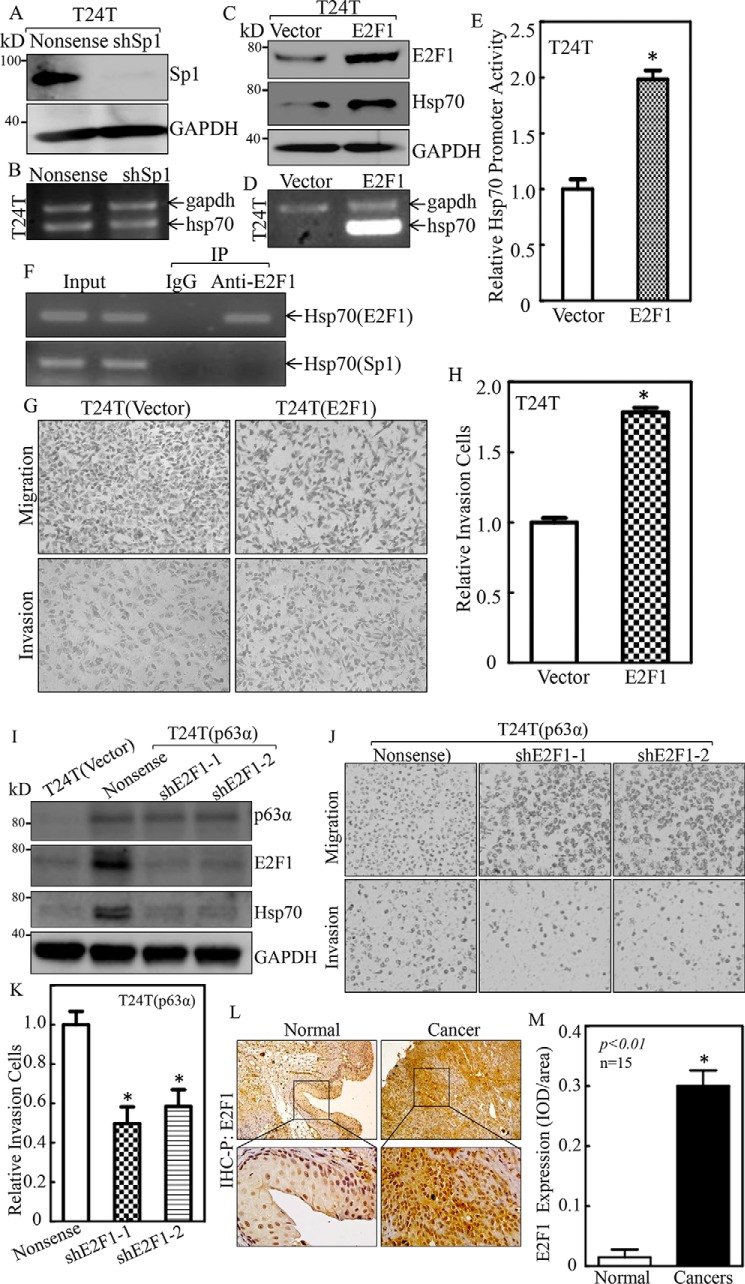
Given that both Sp1 and E2F1 protein were increased in ectopic TAp63-expressed BC cells, we determined which of them was more likely to promote Hsp70 transcription. We transfected shRNA-specific targeting human Sp1 (shSp1) (Fig. 7A) or E2F1 overexpressed plasmid into T24T cells (Fig. 7C) in T24T cells. The stable transfectants T24T(Nonsense) and T24T(shSp1) were used to evaluate the effect of Sp1 knockdown on Hsp70 mRNA levels. The results indicated that knockdown of Sp1 did not show any observable effect on Hsp70 mRNA abundance (Fig. 7B), excluding the participating of Sp1 in Hsp70 modulation. The stable transfectants, T24T(vector) and T24T(E2F1), were further employed to evaluate the effects of E2F1 on Hsp70 expression. As shown in Fig. 7, C and D, Hsp70 mRNA and protein levels were remarkably increased in T24T(E2F1) cells as compared with those in T24T(Vector) cells, revealing the important role of E2F1 in TAp63 promoting Hsp70 mRNA level. To test whether E2F1 regulated Hsp70 mRNA transcription, an Hsp70 promoter-driven luciferase reporter was transfected into T24T(Vector) and T24T(E2F1) cells, and the promoter transcription activities were compared between the two transfectants. As shown in Fig. 7E, Hsp70 promoter-driven luciferase reporter transcription activity was significantly higher in T24T(E2F1) cells than in T24T(Vector) cells, revealing that E2F1 positively regulates Hsp70 mRNA transcription. To test whether E2F1 binds to the Hsp70 promoter region, a chromatin immunoprecipitation (ChIP) assay was carried out in which anti-E2F1 antibodies were used to pull down all E2F1 protein and any DNA with which E2F1 physically interacted. The DNA was then extracted from the precipitated complex, and PCR was performed to detect the presence of the Hsp70 promoter. As shown in Fig. 7F, specific primers of the Hsp70 promoter targeting the E2F1-binding site were used to perform the PCR (top panel of Fig. 7F), whereas the primers of the Hsp70 promoter region that targets the Sp1-binding site was used as a negative control (bottom panel of Fig. 7F). IgG was used as the antibody negative control. The results shown in Fig. 7F strongly indicated that the DNA fragment containing E2F1-binding site, but not Sp1-binding site, was specifically presented in immune-complex pulldown by anti-E2F1 antibodies, revealing the interaction of E2F1 with Hsp70 promoter. In addition, the effect of E2F1 on BC invasion was also evaluated in E2F1-overexpressed T24T cells. As shown in Fig. 7, G and H, overexpression of E2F1 in T24T cells profoundly promoted invasion of T24T cells. To further determine the role of E2F1 in p63α promotion of Hsp70 expression and BC invasion, we stably transfected E2F1 shRNA into T24T(p63a) cells (Fig. 7I). E2F1 knockdown led to an attenuation of Hsp70 expression and invasive abilities of T24T(p63α) cells (Fig. 7, J and K). Moreover, consistent with p63α overexpression in BBN-induced mouse-invasive BC, E2F1 was also remarkably overexpressed in same BBN-induced mouse BC tissues (Fig. 7, L and M). These results demonstrate that E2F1 is critical for p63α promoting Hsp70 transcriptional up-regulation and BC invasion.
Figure 7.
E2F1 was the transcription factor mediating p63α promotion of Hsp70 transcription and cell invasion in human BC cells. A and C, knockdown efficiency of Sp1 or E2F1 ectopic expression in T24T cells was evaluated by Western blotting. GAPDH was used as a protein loading control. B and D, T24T(Nonsense) versus T24T(shSp1) cells or T24T(Vector) versus T24T(E2F1) cells were extracted for total RNA with TRIzol reagent. RT-PCR was used to determine hsp70 mRNA expression, whereas GAPDH was used as an internal control. E, T24T(Vector) and T24T(E2F1) cells were transfected with an hsp70 promoter-driven luciferase reporter together with pRL-TK. The transfectants were used to determine hsp70 promoter activity by measuring luciferase activity. pRL-TK was used as an internal control to normalize the transfection efficiency. Each bar indicates the mean ± S.D. from three replicate assays. F, ChIP assay was carried out as described under “Experimental Procedures,” anti-E2F1 specific antibody was used to pull down its E2F1-bound DNA fragments, and primers for E2F1-binding site and negative control Sp1-binding site were used to carry out PCR. IP, immunoprecipitation. G and H, the invasion abilities of T24T(Vector) and T24T(E2F1) cells were determined using a transwell invasion assay. The results are presented as the number of invasive T24T(E2F1) cells relative to invasive T24T(Vector) transfectants (H). I, the knockdown efficiency of E2F1 in T24T(p63α) cells was determined by Western blotting. GAPDH was used as a protein loading control. J and K, the invasion abilities of T24T(p63α/Nonsense), T24T(p63α/shE2F1–1), and T24T(p63α/shE2F1–2) cells were determined using a transwell invasion assay. L and M, IHC-P was carried out to evaluate E2F1 protein expression in mouse BC tissues and normal bladder tissues. The optical density was analyzed as described under “Experimental Procedures.” The asterisk (*) indicates a significant increase in p63α protein expression in comparison with that observed in normal tissues (p < 0.05).
p63α promoted E2F1 transcription
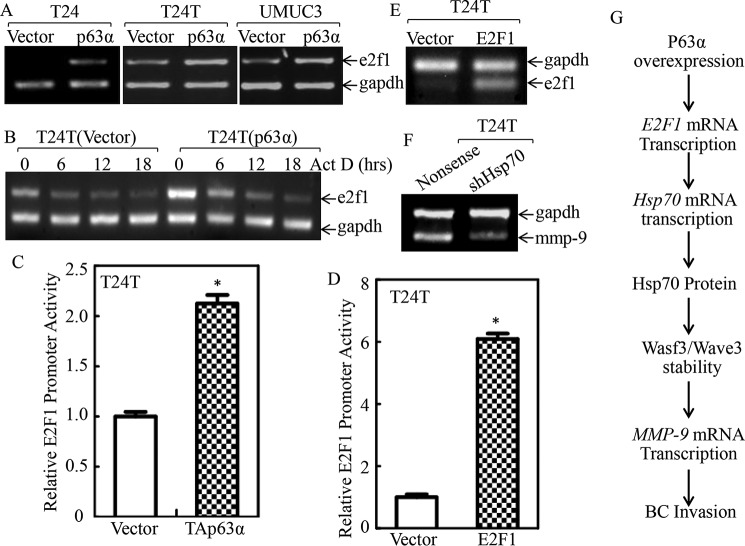
Given our results showing that E2F1 is important for p63α up-regulation of Hsp70 expression and BC invasion, our subsequent efforts were directed at elucidating the mechanisms responsible for p63α up-regulation of E2F1. Therefore, we first examined E2F1 mRNA levels in p63α-overexpressed cells versus its scramble vector transfectants. The results indicated that E2F1 mRNA levels were significantly elevated in p63α ectopically expressed cells than those in the scramble vector transfectants in all T24, T24T, and UMUC3 cells (Fig. 8A). The results obtained from evaluation of E2F1 mRNA stability revealed that E2F1 mRNA degradation rate in p63α transfectants was comparable with their scramble vector transfectants (Fig. 8B). Thus, E2F1 promoter-driven luciferase reporter transcription activities were evaluated in T24T(Vector) and T24T(p63α) cells. As exhibited in Fig. 8C, E2F1 promoter activity was remarkably up-regulated in T24T(p63α) cells. E2F1 has been reported to be capable of promoting its own transcription (30), and we examined E2F1 promoter activity in T24T(Vector) and T24T(E2F1) cells. As expected, E2F1 promoter activity was markedly increased in T24T(E2F1) cells in comparison with its scramble vector transfectants (Fig. 8D), strongly suggesting that E2F1 promotes its own mRNA transcription. This notion was greatly supported by the observation of promotion of endogenous E2F1 mRNA expression in T24T(E2F1) cells (Fig. 8E). MMP-9 has been reported to hydrolyze type IV collagen, a key component of the extracellular matrix, and promotes BC invasion (31). Thus, reverse transcription-PCR (RT-PCR) was employed to determine mmp-9 expressions in T24T(shHsp70) and T24T(Nonsense) cells. As shown in Fig. 8F, mmp-9 mRNA was remarkably attenuated in T24T(shHsp70) cells, revealing that MMP-9 might act as a downstream effector of p63α/E2F1/Hsp70 axis responsible for p63α promoting BC invasion. Taken together, we demonstrated that p63α overexpression initiated E2F1 mRNA transcription/protein expression through E2F1 self-promoting mechanisms, which also promoted Hsp70 transcription and in turn stabilized Wasf3/Wave3 protein, consequently leading to mmp-9 transcription and BC cell invasion (Fig. 8G).
Figure 8.
p63α promoted E2F1 transcription. A, the indicated stable transfectants were extracted with TRIzol reagent for total RNA isolation. e2f1 mRNA was determined with RT-PCR using the specific primers. GAPDH was used as an internal control. B, the T24T(Vector) and T24T(p63α) cells were treated with actinomycin D (Act D) for the indicated time points, then total RNA was isolated and subjected to RT-PCR for analysis of e2f1 mRNA degradation. GAPDH was used as a loading control. C and D, T24T(Vector) versus T24T(p63α) cells (C) and T24T(Vector) versus T24T(E2F1) cells (D) were transfected with an E2F1 promoter-driven luciferase reporter together with pRL-TK. The transfectants were seeded into 96-well plates to determine E2F1 promoter transcriptional activity. pRL-TK was used as an internal control to normalize the transfection efficiency. Each bar indicates the mean ± S.D. from three replicate assays. The asterisk indicates significant increases in E2F1 promoter-driven reporter activity in p63α-overexpressed cells (C) or in E2F1-overexpressed cells (D) in comparison with Vector transfectants (p < 0.05). E and F, T24T(Vector) and T24T(E2F1) cells (E) or T24T(Nonsense) and T24T(shHsp70) cells (F) were extracted for total RNA with TRIzol reagent. RT-PCR was used to determine endogenous e2f1 mRNA expression, whereas GAPDH was used as an internal control. G, illustration of proposed molecular mechanisms underlying overexpressed p63α promoting human BC invasion.
Discussion
Muscular invasive BC is fatal and represents a major therapeutic challenge of this disease. The key to developing future medications is in understanding the mechanisms underlying the affirmative effect on BC invasion. The p63 gene was discovered as the tumor suppressor function along with p73, upon the discovery of the p53 (32). TPp63 contains a transactivation domain (TAD) and can initiate transcription of p53-regulated genes (11). p63α is the longest TA transcript variant of p63 and has been characterized as a tumor suppressor responsible for preventing cancer development. In this study we found that p63α was overexpressed in mouse invasive BC tissues as compared with normal bladder tissues and that overexpressed p63α could promote BC cell invasion. This positive effect on invasion is regulated by E2F1-dependent increases of Hsp70 mRNA transcription and protein expression. Further studies found that Hsp70 is crucial for BC cell invasion through the activation of the Hsp70/Wasf3/Wave3/MMP-9 axis. Our findings that p63α up-regulates Hsp70 mRNA transcription and subsequently activates the Hsp70/Wasf3/Wave3/MMP-9 axis and promotes BC cell invasion via initiating the transcription factor E2F1 makes a new discovery of p63α in promotion of BC invasion, which is distinct from its acting as tumor suppression and further provides new insight into the understanding the double faces of p63α in regulation of BC cell growth and progression/invasion in cancer development.
The stress-inducible Hsp70, also known as HSPA1A, Hsp70-1, Hsp72, or HspA1, is produced at low or undetectable levels in unstressed, healthy cells (33). Upon a variety of stresses, its expression is rapidly induced through activating HSFs via mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK)- and stress-activated protein kinase (SAPK)-signaling cascades (34, 35). Hsp70 restores the balance of the cell proteome by normalizing the level of unfolded and denatured proteins. As a molecular chaperone, Hsp70 is an important regulator in cellular networks, including transcriptional, signaling, membrane, and organelle networks (36). Hsp70 exerts a dual role in cancer, promoting survival and dissemination of tumor cells while simultaneously contributing to antitumor immunity (37). Hsp70 overexpression correlates with an aggressive phenotype in several cancer types. Elevated expression of Hsp70 has been found to correlate with lymph-node metastasis in breast cancer cells (38) and with vascular invasion in gastric cancers (28). In cervical and BC cells, knockdown of Hsp70 by its shRNA has been shown to suppress invasion and migration (39). The results from Rohde et al. (40) also reveal a role of Hsp70 in regulation of cancer cell adhesion as depletion of Hsp70 resulted in cell detachment. In the current studies we discovered that p63α is able to promote Hsp70 transcription and BC invasion, which provides a new insight into the understanding of new Hsp70 function in promotion of high invasive BC progression.
Wasf3, also known as Wave3, is a member of the Wiskott-Aldrich family of proteins (41) and is an effector for cancer cell motility, invasion, and metastasis (42–45). Wasf3 is critical for the regulation of actin cytoskeleton dynamics via activating Arp2/3 complex. This process leads to reorganization of the actin cytoskeleton, mediates cell movement through increasing lamellipodia formation (46), and promotes invasion through activating matrix metalloproteinases (MMPs) (42). Activation of Wasf3 is achieved to some extent through its phosphorylation by ABL kinase and the resulting lamellipodia formation and invasion in breast cancer cells (43). Compared with lower stage tumors and normal tissue, advanced breast and prostate cancer tumors have increased WASF3 expression (47, 48). Down-regulation of WASF3 has been found to inhibit the invasion and metastasis of breast cancer cells, and it has thus been proposed as a metastasis promoter gene (45). Hsp70 was found in the WASF3 immuno-complex, and inactivation of Hsp70 results in destabilization of WASF3 through proteasome degradation (39); therefore, the influence chaperone proteins such as Hsp70 show the effect on invasion, at least in part, through their controlling Wasf3 protein (39). Our studies showed that Hsp70 and Wasf3/Wave3 were up-regulated in p63α-overexpressed BC cells, and WASF3/Wave3 was a downstream effector of Hsp70 that mediated the increased invasion abilities in p63α-overexpressed BC cells. In addition, our results indicated that knockdown of Hsp70 led to reduction of MMP-9 mRNA expression, suggesting that MMP-9 might also be an Hsp70/WASF3 downstream effector for mediating BC cell invasion.
Transcription factor E2F plays critical roles in cell cycle regulation, apoptosis, and senescence (49, 50). E2F1 and its maximum level of expression is observed in the late G1/S phase of the cell cycle during which it induces transcription of the genes required for DNA synthesis (50). E2F1 protein is regulated by retinoblastoma protein Rb. When Rb is not in its hyperphosphorylated form, it binds to E2F1 via its pocket domain and inhibits sequence-specific E2F1 DNA-binding, therefore limiting E2F1-mediated transcriptional induction (51). Although E2F1 as a transcription factor induces transcription of many genes, nothing is known about its effect on Hsp70 expression and potential regulatory effect of p63 on E2F1 expression. Our studies here indicate that p63α can transcriptionally activate E2F1. Interestingly, E2F1 expression could further promote E2F1 transcription in self-regulation manner as well as mediate Hsp70 mRNA transcription. These studies demonstrate novel mechanisms related to regulation of E2F1 and Hsp70.
In summary, our current studies show for the first time that in addition to acting as a tumor suppressor of BC cell growth, p63α also promotes BC cell invasion after tumor formation. p63α up-regulates Hsp70 by transcriptionally activating E2F1 and subsequently promoting BC cell invasion via the WASF3/WAVE3/MMP-9 axis as illustrated in Fig. 8G. This novel mechanism of p63α highlights its downstream regulators/effectors as potential biomarkers for diagnosis and/or targets for therapy of invasive and metastatic human BC patients.
Experimental procedures
Reagents, antibodies, and plasmids
Actinomycin D was purchased from Santa Cruz (Dallas, TX). The dual luciferase assay kit was from Promega (Madison, WI). TRIzol reagent and the SuperScript™ First-Strand Synthesis system were from Invitrogen. The shRNA set specifically targeting human Hsp70 (RHS4533-EG3303) was from Open Biosystems (GE Healthcare). The shRNA-specific knockdown of human E2F1 was constructed as pSGLV/H1/Green/puro Vector by targeting 5′-TTG ATC ACC ATA ACC ATC T-3′ and 5′-TCA GTG AGG TCT CAT AGC G-3′. The p63α expression plasmid a gift from Dr. Zhaohui Feng (State University of New Jersey, New Brunswick, NJ). The E2F1 expression plasmid was a gift from Dr. Daniel S. Peeper, The Netherlands Cancer Institute (52). The E2F1 promoter-driven luciferase reporter was obtained from Addgene (Cambridge, MA). The Hsp70 promoter-driven luciferase reporters were described in our previous study (53). The specific antibodies against p63α (GTX102425) and GAPDH (GTX100118) were purchased from Genetex (Irvine, CA); anti-XIAP (610763) antibodies were from BD Biosciences; antibodies specific against RhoA (2117S), CDC42 (2466S), Rac123 (2465S), Src (2109S), Hsp70 (4872S), WAVE3 (2806S), p-c-Jun Ser-63 (2361S), p-c-Jun Ser-73 (3270S), c-Jun (9165S), and CREB (9197S) were from Cell Signaling (Beverly, MA); antibodies specific for RhoGDIα (sc-360), RhoGDIβ (sc-11359), Sp1 (sc-59P), E2F1 (sc-193), MMP-2 (sc-13594), and HSF1 (sc-17757) were from Santa Cruz (Dallas, TX).
Cell culture and transfection
T24 and T24T cells were described in our previous publications (31) and cultured in DMEM:F-12 (1:1) with 5% FBS (Atlanta Biologicals, Flowery Branch, GA). UMUC3 was described in our previous studies (54, 55) and cultured in DMEM with 10% FBS (Atlanta, Flowery Branch, GA). Cell transfections were performed with PolyJetTM DNA (SL10068) in Vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD) according to the manufacturer's instructions. For stable transfection, T24, T24T, and UMUC3 cells were subjected to selection with hygromycin B (200 μg/ml) (31282-4-9, Goldbio, Ashby Rd., St. Louis, MO), G418 (1000 μg/ml) (sc-29065, Dallas, TX), or puromycin (0.3 μg/ml) (sc-205821, Dallas, TX) depending on the antibiotic resistance of different plasmids transfected. The cells surviving from the antibiotics selection were pooled as mass stable transfectants.
Luciferase reporter assay
Bladder cancer cells were co-transfected with either the Hsp70 or E2F1 promoter-driven luciferase reporter constructs together with the Renilla luciferase vector pRL-TK (Promega). The luciferase activities were determined by using the dual-luciferase assay kit (Promega) together with a luminometer (Lumat LB9507, Berthold Tech., Bad Wildbad, Germany). The firefly luciferase signal was normalized to the Renilla luciferase signal for each individual analysis to eliminate the variations of transfection efficiencies as previously described (56).
N-butyl-N-(4-hydroxybutyl) nitrosamine-induced high invasive mouse BC and immunohistochemistry staining-paraffin (IHC-P)
All animal procedures were approved by the Committee on Animal Resources of the New York University School of Medicine and in accordance with NIH guidelines. C57BL/6 male mice (n = 15/group) at ages of 3–4 weeks were supplied ad libitum with tap water containing 0.05% BBN (B0938, TCI America, Portland, OR) in opaque bottles for 23 weeks, whereas negative control mice received regular tap water. The drinking water was prepared freshly twice a week, and consumption was recorded to estimate BBN intake. Mice were sacrificed at 23 weeks after the experiment began, and bladders were harvested and preserved in paraffin for pathological analysis and immunohistochemistry staining (IHC).
Bladder tissues obtained from the sacrificed mice specimens were formalin-fixed and paraffin-embedded as described in our previous studies (26). Immunohistochemistry staining (IHC) was performed to evaluate p63α expression between BBN-induced invasive bladder cancer tissues and negative control bladder tissues. IHC was performed using antibodies specific against p63α (Genetex) together with the IHC kit following the protocol described in our previous studies (57). The resultant immunostaining images were captured using the AxioVision Rel.4.6 computerized image analysis system (Carl Zeiss, Oberkochen, Germany). p63α protein expression levels were analyzed by calculating the integrated optical density per stained area (IOD/area) using Image-Pro Plus version 6.0 (Media Cybernetics). The IHC-stained sections were evaluated at 400-fold magnifications. At least five representative staining fields of each section were analyzed to calculate the optical density based on typical photographs that had been captured.
RT-PCR
Total RNA was extracted with TRIzol reagent, and 5 μg of total RNA was used for first-strand cDNA synthesis with oligo(dT) primer by SuperScriptTM First-Strand Synthesis system (Invitrogen). Specific primer pairs were designed for amplifying human hsp70 (forward, 5′-CAA CAC GGC AAG GTG GAG TCA-3′; reverse, 5′-TCA GCC GCT TCG CGT CAA ACA-3′), e2f1 (forward, 5′-GAG GTG CTG AAG GTG CAG AA-3′; reverse, 5′-GTT TGC TCT TAA GGG AGA TCT G-3′), endogenous e2f1 (forward, 5′-AGT TCA TCA GCC TTT CCC CAC C-3′; reverse, 5′-GAC AAG GTG AGC ATC TCT GGA AAC-3′), and GAPDH (forward, 5′-AGA AGG CTG GGG CTC ATT TG-3′; reverse, 5′-AGG GGC CAT CCA CAG TCT TC-3′). The PCR products were separated onto 2% agarose gels and stained with ethidium bromide, and the images were scanned with a UV light as described previously (58).
Immunoblotting assay
Whole cells were washed with ice-cold PBS, and then extracted with cell lysis buffer (10 mm, pH 7.4, Tris-HCl, 1% SDS, 1 mm Na3VO4, and proteasome inhibitor). The protein concentration was determined using Nano Drop 2000 (Thermo Scientific, Holtsville, NY). The cell extracts were subjected to Western blotting with each of the antibodies as indicated. The protein bands specifically bound to the primary antibodies were detected using an alkaline phosphatase-linked secondary antibody and ECF (enhanced chemifluorescence) Western blotting analysis system (Amersham Biosciences) as previously described (59). The results shown are representative one from at least three independent experiments.
Cell invasion assay
The invasion kit was purchased from BD Falcon (354480) (Franklin Lakes, NJ). The invasion assay was performed according to the manufacturer's instructions in normal cell culture serum. Upon incubation of the transwell, the cells on both the inside and outside of the chamber were fixed with 3.7% formalin for 2 min, washed twice with PBS, transferred to 100% methanol for 20 min, washed twice again, and finally stained by Giemsa (1:20 diluted with PBS) at room temperature for 15 min in the dark. After staining, the cells were washed twice again with PBS, and the non-invaded cells were scraped off with a cotton swab (PBS-wetted) four times. The cell invasion ability was calculated by invasion normalized to migrated cells. The migration was determined in transwells without Matrigel. The images were captured under an Olympus DP71, and the number of cells was calculated by the software ImageJ as described in previous papers (26, 31).
ChIP assay
The EZ-ChIP kit (Millipore Technologies) was used to carryout the ChIP assay according to the manufacturer's instructions and as described previously (60). Briefly, T24T cells were treated with 1% formaldehyde for 10 min at room temperature. Cells were then pelleted, resuspended in lysis buffer, and sonicated to generate 200–400 bp if chromatin DNA fragments. After centrifugation (13,000 × g at 4 °C) for 10 min, the supernatants were incubated with an anti-E2F1 antibody or the control rabbit IgG at 4 °C overnight. The immune complex was captured with Protein G-agarose-saturated beads with salmon sperm DNA and then eluted with elution buffer. The reverse cross-linking of protein-DNA complexes to free DNA was conducted by incubating at 65 °C overnight. The DNA was extracted and subjected to PCR analysis. To specifically amplify the region containing the E2F1-binding sites on the human Hsp70 promoter, PCR was performed with the pair of primers specific for E2F1 bind site, 5′-TCC ATT GTA ACG TGG CCG G-3′ and 5′-CTA ATT GAC AGG AAG GGT CGG C-3′, whereas the primers specific for the Sp1-binding site were 5′-TTT CTG GAG CCA ATA ACT GA-3′ and 5′-TGT TAG CCA GGA TGG TAG CC-3′, used as negative control. PCR products were separated on 2% agarose gels and stained with ethidium bromide. The gels were scanned under UV light as described previously.
Statistical analysis
Student's t test was used to determine the significance between treated and untreated groups. The results are expressed as the mean ± S.D. from at least three independent experiments. p < 0.05 was considered a significant difference between compared groups (61).
Author contributions
H. H., X. R. W., and C. H. designed the studies. H. J., Q. X., and A. W. drafted the manuscript, J. X. carried out the IHC assay, H. J. and X. G. conducted the real-time PCR assays, H. J. and J. X. performed the Western blotting assays and luciferase reporter assays. H. J., H. H., and C. H. performed the statistical analysis, J. L. helped to acquire the experimental data, and J. Z. detected the cells' biological function. All authors read and approved the final manuscript.
Acknowledgments
We thank Dr. Zhaohui Feng (State University of New Jersey, New Brunswick, NJ) for providing the p63α expression vector. We thank Dr. Daniel S. Peeper from the Netherlands Cancer Institute for the generous gifts of E2F1 expression plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants CA112557, CA165980, and CA177665 (NCI) and ES000260 (NIEHS). This work was also supported by the Natural Science Foundation of China (NSFC81773391, NSFC81702530, NSFC81372946, and NSFC81601849) and the Key Project of Science and Technology Innovation Team of Zhejiang Province (2013TD10). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- BC
- bladder cancer
- Hsp70
- heat shock protein 70
- TA
- transactivation domain
- BBN
- N-butyl-N-(4-hydroxybutyl) nitrosamine
- IHC
- immunohistochemistry
- HSF1
- heat shock factor 1
- MMP
- matrix metalloproteinase
- CREB
- cAMP-response element-binding protein
- IHC-P
- immunohistochemistry staining-paraffin
- HSF
- heat shock factor.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2016) Cancer statistics. 2016. CA Cancer J. Clin. 66, 7–30 [DOI] [PubMed] [Google Scholar]
- 2. Knowles M. A., and Hurst C. D. (2015) Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 15, 25–41 [DOI] [PubMed] [Google Scholar]
- 3. Murphy W. M., Soloway M. S., Jukkola A. F., Crabtree W. N., and Ford K. S. (1984) Urinary cytology and bladder cancer: the cellular features of transitional cell neoplasms. Cancer 53, 1555–1565 [DOI] [PubMed] [Google Scholar]
- 4. Cowan N. C., and Crew J. P. (2010) Imaging bladder cancer. Curr. Opin. Urol. 20, 409–413 [DOI] [PubMed] [Google Scholar]
- 5. von der Maase H., Sengelov L., Roberts J. T., Ricci S., Dogliotti L., Oliver T., Moore M. J., Zimmermann A., and Arning M. (2005) Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J. Clin. Oncol. 23, 4602–4608 [DOI] [PubMed] [Google Scholar]
- 6. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., and McKeon F. (1998) p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 7. Huang H., Pan X., Jin H., Li Y., Zhang L., Yang C., Liu P., Liu Y., Chen L., Li J., Zhu J., Zeng X., Fu K., Chen G., Gao J., and Huang C. (2015) PHLPP2 Downregulation Contributes to Lung Carcinogenesis Following B[a]P/B[a]PDE Exposure. Clin Cancer Res. 21, 3783–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang A., and McKeon F. (2000) P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 1, 199–207 [DOI] [PubMed] [Google Scholar]
- 9. Levine A. J., Tomasini R., McKeon F. D., Mak T. W., and Melino G. (2011) The p53 family: guardians of maternal reproduction. Nat. Rev. Mol. Cell Biol. 12, 259–265 [DOI] [PubMed] [Google Scholar]
- 10. Crum C. P., and McKeon F. D. (2010) p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu. Rev. Pathol. 5, 349–371 [DOI] [PubMed] [Google Scholar]
- 11. Flores E. R. (2007) The roles of p63 in cancer. Cell Cycle 6, 300–304 [DOI] [PubMed] [Google Scholar]
- 12. Giacobbe A., Bongiorno-Borbone L., Bernassola F., Terrinoni A., Markert E. K., Levine A. J., Feng Z., Agostini M., Zolla L., Agrò A. F., Notterman D. A., Melino G., and Peschiaroli A. (2013) p63 regulates glutaminase 2 expression. Cell Cycle 12, 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Candi E., Dinsdale D., Rufini A., Salomoni P., Knight R. A., Mueller M., Krammer P. H., and Melino G. (2007) TAp63 and ΔNp63 in cancer and epidermal development. Cell Cycle 6, 274–285 [DOI] [PubMed] [Google Scholar]
- 14. Gressner O., Schilling T., Lorenz K., Schulze Schleithoff E., Koch A., Schulze-Bergkamen H., Lena A. M., Candi E., Terrinoni A., Catani M. V., Oren M., Melino G., Krammer P. H., Stremmel W., and Müller M. (2005) TAp63α induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 24, 2458–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rocco J. W., Leong C.-O., Kuperwasser N., DeYoung M. P., and Ellisen L. W. (2006) p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer cell 9, 45–56 [DOI] [PubMed] [Google Scholar]
- 16. Trink B., Osada M., Ratovitski E., and Sidransky D. (2007) p63 transcriptional regulation of epithelial integrity and cancer. Cell Cycle 6, 240–245 [DOI] [PubMed] [Google Scholar]
- 17. Candi E., Agostini M., Melino G., and Bernassola F. (2014) How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum. Mutat. 35, 702–714 [DOI] [PubMed] [Google Scholar]
- 18. Flores E. R., Sengupta S., Miller J. B., Newman J. J., Bronson R., Crowley D., Yang A., McKeon F., and Jacks T. (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7, 363–373 [DOI] [PubMed] [Google Scholar]
- 19. Dohn M., Zhang S., and Chen X. (2001) p63a and DNp63a can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20, 3193–3205 [DOI] [PubMed] [Google Scholar]
- 20. Urist M. J., Di Como C. J., Lu M.-L., Charytonowicz E., Verbel D., Crum C. P., Ince T. A., McKeon F. D., and Cordon-Cardo C. (2002) Loss of p63 expression is associated with tumor progression in bladder cancer. Am. J. Pathol. 161, 1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Su X., Chakravarti D., Cho M. S., Liu L., Gi Y. J., Lin Y.-L., Leung M. L., El-Naggar A., Creighton C. J., Suraokar M. B., Wistuba I., and Flores E. R. (2010) TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melino G. (2011) p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 18, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park B. J., Lee S. J., Kim J. I., Lee S. J., Lee C. H., Chang S. G., Park J. H., and Chi S. G. (2000) Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 60, 3370–3374 [PubMed] [Google Scholar]
- 24. Armstrong S. R., Wu H., Wang B., Abuetabh Y., Sergi C., and Leng R. P. (2016) The regulation of tumor suppressor p63 by the ubiquitin-proteasome system. Int. J. Mol. Sci. 17, E2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin H., Xu J., Guo X., Huang H., Li J., Peng M., Zhu J., Tian Z., Wu X. R., Tang M. S., and Huang C. (2016) XIAP RING domain mediates miR-4295 expression and subsequently inhibiting p63α protein translation and promoting transformation of bladder epithelial cells. Oncotarget 7, 56540–56557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang G., Wu A. D., Huang C., Gu J., Zhang L., Huang H., Liao X., Li J., Zhang D., Zeng X., Jin H., Huang H., and Huang C. (2016) Isorhapontigenin (ISO) Inhibits invasive bladder cancer formation in vivo and human bladder cancer invasion in vitro by targeting STAT1/FOXO1 Axis. Cancer Prevention Research 9, 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang D., Li J., Costa M., Gao J., and Huang C. (2010) JNK1 mediates degradation HIF-1α by a VHL-independent mechanism that involves the chaperones Hsp90/Hsp70. Cancer Res. 70, 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Canöz O., Belenli O., and Patiroglu T. E. (2002) General features of gastric carcinomas and comparison of Hsp70 and NK cell immunoreactivity with prognostic factors. Pathol. Oncol. Res. 8, 262–269 [DOI] [PubMed] [Google Scholar]
- 29. Silver J. T., and Noble E. G. (2012) Regulation of survival gene hsp70. Cell Stress Chaperones 17, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crowe D. L., Nguyen D. C., Tsang K. J., and Kyo S. (2001) E2F-1 represses transcription of the human telomerase reverse transcriptase gene. Nucleic Acids Res. 29, 2789–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin H., Yu Y., Hu Y., Lu C., Li J., Gu J., Zhang L., Huang H., Zhang D., Wu X. R., Gao J., and Huang C. (2015) Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget 6, 522–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu G., Nomoto S., Hoque M. O., Dracheva T., Osada M., Lee C.-C., Dong S. M., Guo Z., Benoit N., Cohen Y., Rechthand P., Califano J., Moon C. S., Ratovitski E., Jen J., Sidransky D., and Trink B. (2003) ΔNp63α and TAp63α regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 63, 2351–2357 [PubMed] [Google Scholar]
- 33. Kampinga H. H., Hageman J., Vos M. J., Kubota H., Tanguay R. M., Bruford E. A., Cheetham M. E., Chen B., and Hightower L. E. (2009) Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morimoto R. I. (1993) Cells in stress: transcriptional activation of heat shock genes. Science 259, 1409–1410 [DOI] [PubMed] [Google Scholar]
- 35. Adler V., Schaffer A., Kim J., Dolan L., and Ronai Z. (1995) UV irradiation and heat shock mediate JNK activation via alternate pathways. J. Biol. Chem. 270, 26071–26077 [DOI] [PubMed] [Google Scholar]
- 36. Soti C., Pál C., Papp B., and Csermely P. (2005) Molecular chaperones as regulatory elements of cellular networks. Curr Opin Cell Biol. 17, 210–215 [DOI] [PubMed] [Google Scholar]
- 37. Juhasz K., Lipp A. M., Nimmervoll B., Sonnleitner A., Hesse J., Haselgruebler T., and Balogi Z. (2013) The complex function of hsp70 in metastatic cancer. Cancers 6, 42–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kluger H. M., Chelouche Lev D., Kluger Y., McCarthy M. M., Kiriakova G., Camp R. L., Rimm D. L., and Price J. E. (2005) Using a xenograft model of human breast cancer metastasis to find genes associated with clinically aggressive disease. Cancer Res. 65, 5578–5587 [DOI] [PubMed] [Google Scholar]
- 39. Teng Y., Ngoka L., Mei Y., Lesoon L., and Cowell J. K. (2012) HSP90 and Hsp70 proteins are essential for stabilization and activation of WASF3 metastasis-promoting protein. J. Biol. Chem. 287, 10051–10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rohde M., Daugaard M., Jensen M. H., Helin K., Nylandsted J., and Jäättelä M. (2005) Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 19, 570–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sossey-Alaoui K., Su G., Malaj E., Roe B., and Cowell J. K. (2002) WAVE3, an actin-polymerization gene, is truncated and inactivated as a result of a constitutional t(1;13)(q21;q12) chromosome translocation in a patient with ganglioneuroblastoma. Oncogene 21, 5967–5974 [DOI] [PubMed] [Google Scholar]
- 42. Sossey-Alaoui K., Ranalli T. A., Li X., Bakin A. V., and Cowell J. K. (2005) WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp. Cell Res. 308, 135–145 [DOI] [PubMed] [Google Scholar]
- 43. Sossey-Alaoui K., Li X., and Cowell J. K. (2007) c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J. Biol. Chem. 282, 26257–26265 [DOI] [PubMed] [Google Scholar]
- 44. Sossey-Alaoui K., Downs-Kelly E., Das M., Izem L., Tubbs R., and Plow E. F. (2011) WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int. J. Cancer 129, 1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sossey-Alaoui K., Safina A., Li X., Vaughan M. M., Hicks D. G., Bakin A. V., and Cowell J. K. (2007) Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am. J. Pathol. 170, 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sossey-Alaoui K., Li X., Ranalli T. A., and Cowell J. K. (2005) WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J. Biol. Chem. 280, 21748–21755 [DOI] [PubMed] [Google Scholar]
- 47. Teng Y., Ren M. Q., Cheney R., Sharma S., and Cowell J. K. (2010) Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br. J. Cancer 103, 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teng Y., Liu M., and Cowell J. K. (2011) Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. Int. J. Cancer 129, 2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matsumura I., Tanaka H., and Kanakura Y. (2003) E2F1 and c-Myc in cell growth and death. Cell Cycle 2, 333–338 [PubMed] [Google Scholar]
- 50. Dimri G. P., Itahana K., Acosta M., and Campisi J. (2000) Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14ARF tumor suppressor. Mol. Cell. Biol. 20, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Helin K., Harlow E., and Fattaey A. (1993) Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 13, 6501–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rowland B. D., Denissov S. G., Douma S., Stunnenberg H. G., Bernards R., and Peeper D. S. (2002) E2F transcriptional repressor complexes are critical downstream targets of p19 ARF/p53-induced proliferative arrest. Cancer cell 2, 55–65 [DOI] [PubMed] [Google Scholar]
- 53. Liu J., Zhang D., Mi X., Xia Q., Yu Y., Zuo Z., Guo W., Zhao X., Cao J., Yang Q., Zhu A., Yang W., Shi X., Li J., and Huang C. (2010) p27 suppresses arsenite-induced Hsp27/Hsp70 expression through inhibiting JNK2/c-Jun- and HSF-1-dependent pathways. J. Biol. Chem. 285, 26058–26065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang H. Y., Shariat S. F., Sun T. T., Lepor H., Shapiro E., Hsieh J. T., Ashfaq R., Lotan Y., and Wu X. R. (2007) Persistent uroplakin expression in advanced urothelial carcinomas: implications in urothelial tumor progression and clinical outcome. Hum. Pathol. 38, 1703–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fang Y., Cao Z., Hou Q., Ma C., Yao C., Li J., Wu X. R., and Huang C. (2013) Cyclin d1 downregulation contributes to anticancer effect of isorhapontigenin on human bladder cancer cells. Mol Cancer Ther. 12, 1492–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang D., Li J., Zhang M., Gao G., Zuo Z., Yu Y., Zhu L., Gao J., and Huang C. (2012) The requirement of c-Jun N-terminal kinase 2 in regulation of hypoxia-inducing factor-1α mRNA stability. J. Biol. Chem. 287, 34361–34371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu J., Wang Y., Hua X., Xu J., Tian Z., Jin H., Li J., Wu X. R., and Huang C. (2016) Inhibition of PHLPP2/cyclin D1 protein translation contributes to the tumor suppressive effect of NFκB2 (p100). Oncotarget 7, 34112–34130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu Y., Zhang D., Huang H., Li J., Zhang M., Wan Y., Gao J., and Huang C. (2014) NF-κB1 p50 promotes p53 protein translation through miR-190 downregulation of PHLPP1. Oncogene 33, 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xie Q., Guo X., Gu J., Zhang L., Jin H., Huang H., Li J., and Huang C. (2016) p85α promotes nucleolin transcription and subsequently enhances EGFR mRNA stability and EGF-induced malignant cellular transformation. Oncotarget 7, 16636–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang D., Liang Y., Xie Q., Gao G., Wei J., Huang H., Li J., Gao J., and Huang C. (2015) A novel post-translational modification of nucleolin, SUMOylation at Lys-294, mediates arsenite-induced cell death by regulating gadd45α mRNA stability. J. Biol. Chem. 290, 4784–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pawson T., and Gish G. D. (1992) SH2 and SH3 domains: from structure to function. Cell 71, 359–362 [DOI] [PubMed] [Google Scholar]