Abstract
High-throughput screening (HTS) of small-molecule libraries accelerates the discovery of chemical leads to serve as starting points for probe or therapeutic development. With this approach, thousands of unique small molecules, representing a diverse chemical space, can be rapidly evaluated by biologically and physiologically relevant assays. The origins of numerous United States Food and Drug Administration–approved cancer drugs are linked to HTS, which emphasizes the value in this methodology. The National Institutes of Health Molecular Libraries Program made HTS accessible to the public sector, enabling the development of chemical probes and drug-repurposing initiatives. In this work, the impact of HTS in the field of oncology is considered among both private and public sectors. Examples are given for the discovery and development of approved cancer drugs. The importance of target validation is discussed, and common assay approaches for screening are reviewed. A rigorous examination of the PubChem database demonstrates that public screening centers are contributing to early-stage drug discovery in oncology by focusing on new targets and developing chemical probes. Several case studies highlight the value of different screening strategies and the potential for drug repurposing.
I. Introduction
The identification of new oncology targets drives the development of novel biologic assays to support discovery programs for experimental therapeutics. Over the past 20 years, high-throughput screening (HTS) has been a major contributor to the discovery of chemical leads. A range of assay technologies is being developed to distinguish biologically relevant hits from HTS, and strategies exist to identify targets that synergize with existing therapies.
A number of Food and Drug Administration (FDA)–approved chemotherapeutics can be traced to small-molecule screens, with examples of cancer drugs shown in Table 1. It takes about 20 years from the initiation of a discovery program to the registration of a clinical agent (Eder et al., 2014). HTS is an efficient means of identifying chemical leads, and its applications for discovery will most likely continue to increase over time (Macarron et al., 2011).
TABLE 1.
Examples of United States FDA-approved cancer drugs that were developed from hit compounds identified in small-molecule screens
Wherever possible, the target of the small-molecule screen and assay type are indicated.
| Drug | Date Approved | US Brand Name | Approved Use in Cancer | Target of Small-Molecule Screen | Assay Type | Reference(s) |
|---|---|---|---|---|---|---|
| Alectinib | 12/11/2015 | Alecensa | Non-small cell lung cancer | Anaplastic lymphoma kinase (ALK) | Not available | Sakamoto et al. (2011) |
| Ixazomib Citrate | 11/20/2015 | Ninlaro | Multiple myeloma | 20S proteasome | Not available | Kupperman et al. (2010) |
| Sonidegib | 7/24/2015 | Odomzo | Basal cell carcinoma | Hedgehog (Hh) signaling | Cell-based: Hh-responsive Gli-luciferase reporter assay in the TM3 mouse cell line | Pan et al. (2010) |
| Olaparib | 12/19/2014 | Lynparza | Ovarian cancer | Poly(ADP-ribose) polymerase- 1 | Biochemical: FlashPlate scintillation proximity assay | Dillon et al. (2003), Loh et al. (2005), Menear et al. (2008) |
| Ceritinib | 4/29/2014 | Zykadia | Non-small cell lung cancer | Anaplastic lymphoma kinase | Cell-based: selective cytotoxicity of Ba/F3 NPM-ALK vs. nontransformed parental Ba/F3 cells | Galkin et al. (2007) |
| Ibrutinib | 11/13/2013 | Imbruvica | Chronic lymphocytic leukemia, mantle cell lymphoma, Waldenstrom macroglobulinemia | Bruton’s tyrosine kinase | Biochemical: Forster resonance energy transfer enzyme assay | Pan et al. (2007), Honigberg et al. (2010) |
| Trametinib | 5/29/2013 | Mekinist | Melanoma | Cyclin-dependent kinase 4 inhibitor B (p15INK4b) | Cell-based: branched-DNA assay with p15INK4b-specific probes in HT-29 cells | Yamaguchi et al. (2007), Abe et al. (2011) |
| Enzalutamide | 8/31/2012 | XTANDI | Prostate cancer | Androgen receptor | Cell-based: human prostate cancer cells | Tran et al. (2009) |
| Vismodegib | 1/30/2012 | Erivedge | Basal cell carcinoma | Hh signaling | Cell-based: Hh-responsive Gli8x-luciferase reporter assay in C3H/10T1/2 cells | Williams et al. (2003), Romer et al. (2004), Zhang et al. (2009) |
| Lapatinib | 3/13/2007 | Tykerb | Breast cancer | Epidermal growth factor receptor | Not available | Fry et al. (1994), Macarron et al. (2011) |
| Dasatinib | 6/28/2006 | Sprycel | Acute lymphoblastic leukemia, chronic myelogenous leukemia | Lymphocyte-specific protein tyrosine kinase | Biochemical: filter-binding assay | Padmanabha et al. (1998), Das et al. (2006) |
| Sorafenib | 12/20/2005 | Nexavar | Hepatocellular carcinoma, renal cell carcinoma, thyroid cancer | Raf1 (C-Raf) kinase | Biochemical: scintillation proximity assay | Wilhelm et al. (2006) |
| Erlotinib | 11/18/2004 | Tarceva | Non-small cell lung cancer, pancreatic cancer | Epidermal growth factor receptor | Not available | Fry et al. (1994), Macarron et al. (2011) |
| Gefitinib | 5/5/2003 | Iressa | Non-small cell lung cancer | Epidermal growth factor receptor | Not available | Fry et al. (1994), Macarron et al. (2011) |
| Zoledronic acid | 8/20/2001 | Zometa | Multiple myeloma, hypercalcemia | Not available | Not available | Green et al. (1994) |
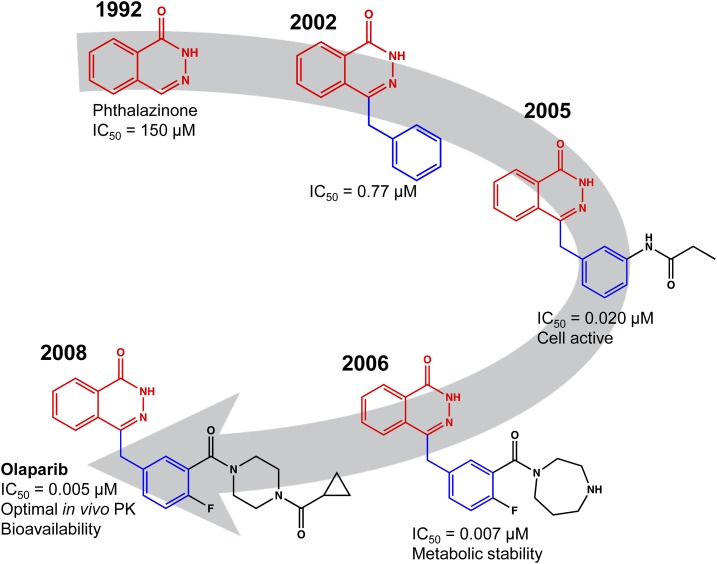
The drugs linked to small-molecule screening can be deceptively circuitous. In many cases, the active constituent is several generations removed from the original HTS hit. The poly(ADP-ribose) polymerase (PARP) inhibitor olaparib (AZD2281, registered in 2014) is a case in point. The recognition that cells with BRCA1/2-inactivating mutations were reliant on PARP for DNA-damage repair led to the discovery of the synthetic lethality of PARP inhibition in BRCA1/2 mutant cells (Bryant et al., 2005; Farmer et al., 2005; Drew, 2015). The development of olaparib (Fig. 1) began with a FlashPlate scintillation proximity assay used for HTS against the Maybridge Screening Collection (Dillon et al., 2003). This work enabled the identification of phthalazinones as leads in 1992 (Banasik et al., 1992). Medicinal chemistry efforts resulted in cell-active PARP inhibitors in 2005 (Loh et al., 2005), with good metabolic stability in 2006 (Cockcroft et al., 2006), and finally the report of a potent inhibitor with single-agent cytotoxicity against BRCA1-deficient cells in 2008 (Menear et al., 2008). At each step of development, the scintillation proximity assay was used to track inhibitor activity along with orthogonal cell–based assays. The legacy of the original HTS continues in a recent high-content screen for molecules that decluster cellular centrosomes. A hit was identified from the AstraZeneca phthalazinone PARP inhibitor library, leading to the PARP 1/2/6 inhibitor AZ0108 (Johannes et al., 2015).
Fig. 1.
The evolution of olaparib. Example structures from each stage of the screening and discovery process are shown. A HTS FlashPlate scintillation proximity assay revealed weak PARP inhibitory activity of phthalazinone (1992, red). Addition of an aryl group (blue) conferred several orders-of-magnitude improved activity (2002, patent WO 2004080976 A1). Subsequent medicinal chemistry optimization identified low nanomolar inhibitors (2005), and reversal of the anilide to an amide retained activity but conferred much-needed metabolic stability (2006). Structural optimization with retention of PARP inhibitory activity yielded olaparib, which was granted accelerated approval by the FDA in 2014. Note that the phthalazinone core (red) from the original screen hit was retained through the discovery process, as was the aryl group (blue) that was identified as part of a focused phthalazinone library screen.
There are many examples of multiple drugs sharing a common target. Nevertheless, the drugs might have been discovered by different strategies. Inhibitors of topoisomerases are good examples of this, and there are FDA-approved inhibitors of topoisomerase I (e.g., topotecan, irinotecan) and II (e.g., etoposide, doxorubicin, daunorubicin), with several more in human clinical trials (Pommier, 2006; Nitiss, 2009). Although describing the discovery pathway for each is beyond the scope of this review, it is instructive to compare an early inhibitor with an advanced compound currently in human clinical trials.
Topoisomerases are required to relax DNA supercoiling during cellular events such as replication, transcription, and remodeling. As part of this process, TOP1 generates single-stranded DNA breaks, whereas TOP2 produces double-stranded DNA breaks. TOP1 inhibitors can trap the topoisomerase–DNA complex during cleavage, causing DNA damage, cell cycle arrest, and cell death. The first inhibitor, camptothecin, was identified as part of the National Cancer Institute (NCI) natural product screening program in the late 1950s and progressed to clinical trials in the 1970s, but was not tolerated (Wall and Wani, 1995). In the meantime, camptothecin stability and solubility properties were improved by the second-generation drugs topotecan and irinotecan (a prodrug of the active inhibitor SN-38). In 1985, it was shown that topoisomerase I is the target of camptothecin (Hsiang et al., 1985), and the on-target activities of newer drugs were confirmed. The camptothecins bind an enzyme–DNA complex, and structural studies of the complex have allowed the rational design of newer-generation interfacial inhibitors (reviewed by Pommier, 2006).
At the contemporary end of the spectrum is vosaroxin (also known as SNS-595, voreloxin), a topoisomerase II inhibitor currently in human clinical trials. Its discovery was driven by a mechanism-informed phenotypic screen (Hawtin et al., 2010). Quinolones are known to cause DNA damage in bacteria by disrupting DNA gyrase and topoisomerase IV, so Tomita et al. (2002) screened antibacterial agents containing quinolone ring structures for cell-killing activity. The lead compound was further optimized for cell killing against murine P388 leukemia cells, resulting in vosaroxin (Tsuzuki et al., 2004). Subsequent mechanistic work showed that vosaroxin intercalates DNA and poisons topoisomerase II, leading to double-stranded DNA breaks (Hawtin et al., 2010). It is interesting that 30 years of topoisomerase drug discovery is bookended by cell-based phenotypic assays, despite knowledge of the biochemical target. Although cell-based and biochemical assays exist to identify interfacial modulators that stabilize the topoisomerase–DNA interaction (Plückthun and Knowles, 1987), cell-based phenotypic screening allowed the discovery of cell-active inhibitors, lowering the barrier to development.
II. Target Validation
A critical element of any new cancer therapeutic discovery program is to ensure that the target is validated. At its simplest, the validity of a target or pathway can be achieved through the demonstration that direct modulation will elicit the desired phenotype (Cortés-Cros et al., 2013). Validation increases confidence in a target or pathway prior to significant investments in the development of a molecule. Put simply, the usefulness of any chemical tools that emerge from a discovery program depends on the biologic relevance of the target (Hoelder et al., 2012). Validation of a putative target can be achieved via a range of strategies, such as genetic knockdown or knockout (RNAi, CRISPR/Cas) or overexpression of targets (gain-of-function), using both cell lines and mouse models (including xenografts) (Benson et al., 2006). High-quality chemical probes allow pharmacological validation and can demonstrate exposure at the site of action as well as target engagement (Hughes et al., 2011; Bunnage et al., 2013). Critically, validation of the target alone does not guarantee success, and there remain significant challenges associated with target classes that are considered to be undruggable (Epstein, 2013; Cox et al., 2014; Lazo and Sharlow, 2016). Reasons for failure in drug development are manifold (for both small molecules and biologics) (Waring et al., 2015), but there are examples of failure due to a lack of understanding of the target (Mak et al., 2014). For academic discovery, the barrier to executing a HTS might be lower, but the importance of demonstrating target relevance to oncology is equally critical.
An interesting example of target discovery and validation is that of BCR-ABL and the success of the inhibitor imatinib (also called STI-571 and CGP 57148) (Capdeville et al., 2002). Imatinib’s discovery is somewhat unconventional as it emerged through a medicinal chemistry campaign before being chemically optimized as a potent BCR-ABL inhibitor (Manley et al., 2002). The phenylamino pyridine chemotype that was developed into imatinib was first identified as an inhibitor of protein kinase C by Ciba Pharmaceuticals (Basel, Switzerland) (Zimmermann et al., 1996b). Further optimization yielded an inhibitor of the platelet-derived growth factor receptor tyrosine kinase (Buchdunger et al., 1996; Zimmermann et al., 1996a) that also inhibited Abl kinase. The chemotype was further optimized with a piperazine group to improve solubility and bioavailability (Zimmermann et al., 1997), producing imatinib, a potent Bcr-Abl inhibitor that entered the clinic. In effect, the drug was ultimately the grandchild of a HTS campaign against an alternative kinase.
III. Screening Strategies for Oncology
The discovery strategies used for HTS generally delineate into cell-based phenotypic assays and biochemical target-based assays. These strategies are briefly described in this work, with examples of their benefits. An emphasis is placed on newer screening approaches informed by current treatment strategies, and how the two are being integrated to inform the drug discovery process. The division of biochemical versus phenotypic assays presented in this study is based on practical considerations. A general agreement in the literature is that cell-based phenotypic assays are agnostic to the mechanisms by which phenotypic changes are initiated (Eder et al., 2014). In contrast, target-based discovery aims to modulate a preidentified target directly, using biochemical and/or engineered cell-based assays. It is clear from retrospective analyses of oncology drugs that both target-based and phenotypic HTS play an important role in the drug discovery pipeline (Swinney and Anthony, 2011). Whereas the following sections divide discovery assays by type, a continuum of assays exists from purely biochemical to phenotypic.
A. Target-Based Screening
Where possible, performing HTS with purified protein confers a number of advantages. Where the enzymatic activity or interaction between molecules is well characterized, access to purified recombinant protein(s) can enable screening for compounds that elicit the desired activity on protein function. A range of assay formats is available: biochemical assays can be read directly (e.g., fluorogenic product), coupled to reporters amenable to detection (e.g., luciferase for ATP) (Acker and Auld, 2014), diaphorase for NAD(P)H levels (Davis et al., 2016; Hall et al., 2016), or coupled to a separate spectroscopy for analysis, such as mass spectrometry for quantifying reaction products [such as RapidFire (Haslam et al., 2016)]. Biochemical assays can also be tuned to bias discovery toward inhibitors with a particular mechanism (Copeland, 2013). For an enzyme utilizing a cofactor such as NADPH, inhibitors that bind outside of the cofactor pocket are often desired, and this can be achieved by screening with cofactor concentrations in excess of the KM (Acker and Auld, 2014).
Protein–protein interactions (PPIs) are recognized as difficult targets, but are of great interest, particularly in the context of disrupting protein complex formation and signaling pathways (Arkin and Wells, 2004). For example, venetoclax (ABT-199, GDC-0199) selectively binds BCL-2 to block interactions with proapoptotic proteins and induce programmed cell death in chronic lymphocytic leukemia (CLL) cells (Souers et al., 2013). Modulation of PPIs and other molecular interactions can be measured by proximity assays, where the signal is sensitive to the association of labeled partners. Examples of assay formats include Förster resonance energy transfer, time-resolved fluorescence energy transfer, amplified luminescent proximity homogeneous assay, and fluorescence polarization (Arkin et al., 2004). Although assays using Förster resonance energy transfer, time-resolved fluorescence energy transfer, amplified luminescent proximity homogeneous assay, and fluorescence polarization can be employed with cell lysate, complementary techniques such as the split luciferase assays inform on in-cell activity (Paulmurugan et al., 2002). Many anticancer drugs function as interfacial inhibitors to disrupt PPIs (microtubule inhibitors) or protein–DNA interactions (topoisomerase I and II inhibitors) by forming a ternary complex with their targets and stabilizing a toxic intermediate (Pommier and Marchand, 2011; Pommier et al., 2015). Assays to identify interfacial inhibitors must be sensitive to the stabilization of macromolecular complexes, which is currently less common in HTS and drug discovery (Pommier and Marchand, 2011).
Purified targets also provide ancillary benefits by enabling characterization of chemical leads through biophysical measurements of binding thermodynamics and kinetics as well as structural studies. The implementation of orthogonal cell–based phenotypic assays is necessary for identifying cell-active hits and subsequent analogs.
B. Mechanism-Informed Phenotypic Drug Discovery
The alternative target-based screening route is to engineer a cell-based assay that informs directly on target engagement or modulation (Moffat et al., 2014). This approach, which has been termed mechanism-informed phenotypic drug discovery (MIPDD), has been described as falling between phenotypic screening and true target-based drug discovery (Moffat et al., 2014). A multitude of assays with engineered cell lines can be implemented for MIPDD, including gene reporter assays, high-content approaches, split-protein complementation, and substrate dyes. In many cases, MIPDD platforms have been used in rational design efforts around a second-generation drug, such as epirubicin (derived from doxorubicin) (Giuliani and Kaplan, 1980). Another example is the anti-estrogen, fulvestrant, which resulted from a search for molecules that bind the estrogen receptor without activating the normal transcriptional hormone responses. This was accomplished by screening with a cellular estrogen receptor-binding assay (17β-[3H]estradiol displacement) for compounds that did not elicit downstream estrogen effects (Wakeling et al., 1991). It is unlikely that this mode of action could have been identified via biochemical or true phenotypic screening.
Ultimately, MIPDD carries the limitation that a compound eliciting the desired phenotype might not act on the intended pathway. A major challenge in screening is the development of assays that report on target engagement rather than modulation. Affinity-related techniques can be implemented with purified protein. A high-profile example of this is the utilization of differential scanning fluorimetry to identify (+)-JQ-1 as being able to confer thermal stability among all bromodomains of the BET family (Filippakopoulos et al., 2010). Several techniques were recently described that report on intracellular target engagement by small molecules. The cellular thermal shift assay has used both Western analysis and AlphaScreen technology to detect targets and antitargets (Jafari et al., 2014; Martinez Molina and Nordlund, 2016). Thermal proteome profiling by quantitative mass spectrometry can aid in identifying small-molecule targets (Franken et al., 2015). Recently, Promega scientists (Promega, Fitchberg, WI) reported a bioluminescence resonance energy transfer assay that utilizes a NanoLuc-tagged protein and a cell-permeable quencher-tagged inhibitor to suppress the signal. Competitive binding of test compounds to the target displaces the quencher-tagged inhibitor, resulting in bioluminescence and signifying target engagement independent of protein function (Robers et al., 2015). Although these techniques will require further development to increase throughput, we anticipate that efforts to screen directly for cellular target engagement will enable inaccessible targets to become druggable.
C. Phenotypic-Based Screening
Phenotypic assays can identify active compounds with diverse mechanisms to alter a particular cellular phenotype and have been the backbone of oncology discovery programs. It is important to note that compound activity in cell-based assays suggests cellular target engagement, which is a clear advantage when the target is intracellular. A major argument in favor of phenotypic assays is that the activity observed is more likely to translate to animal models and the clinic. For over 50 years, the predominant cellular phenotype for screening has been differential cell killing or growth arrest. This is perhaps best typified by the NCI Developmental Therapeutics Program that tests small molecules against a 60–cell-line panel (NCI-60), assessing three growth parameters from a single experiment: growth inhibition, total growth inhibition, and lethal concentration (Shoemaker, 2006; Holbeck et al., 2010). The NCI-60 is a publically available drug development resource that has enabled a variety of important investigations. For example, profiling studies of 1429 candidate anticancer drugs with the NCI-60 revealed transporters that are more likely to confer resistance to particular agents (Szakács et al., 2004). Analyses of drug-activity profiles with the NCI-60 also facilitated the discovery of the indenoisoquinoline topoisomerase I inhibitors (LMP400 indotecan, LMP776 idimitecan, and LMP744), which were identified by comparing cytotoxicity–pattern similarities with camptothecins (Kohlhagen et al., 1998; Pommier, 2006). The freely available CellMiner application (http://discover.nci.nih.gov/cellminer) integrates drug activity, gene expression, and microRNA expression datasets for the NCI-60 cell lines and can be used to discover “me-too” compounds (Shankavaram et al., 2009; Reinhold et al., 2012). Another resource that can be mined to develop insights into mechanisms of action for small molecules is the Cancer Therapeutics Response Portal (http://www.broadinstitute.org/ctrp/) (Basu et al., 2013; Seashore-Ludlow et al., 2015; Rees et al., 2016).
There are many examples of pure phenotypic screening (i.e., in the absence of knowledge about the target) leading to drugs, including the histone deacetylase inhibitors vorinostat (Marks and Breslow, 2007) and romidepsin (Nakajima et al., 1998), and the proteasome inhibitor carfilzomib (Kim and Crews, 2013). Romidepsin (FR901228) was originally isolated and identified as the active constituent of a natural product extract from Chromobacterium violaceum that reversed the cellular morphology of ras-transformed cells (Ueda et al., 1994). It was subsequently shown to induce transcription of the simian virus 40 promoter-driven chloramphenicol acetyltransferase reporter gene (Nakajima et al., 1998). Follow-up studies demonstrated profound cytotoxicity and cell cycle block, and the compound was developed by the NCI before approval by the FDA (Piekarz et al., 2001). In the meantime, mechanistic work revealed increased histone acetylation in cells and indicated that the compound acted by inhibiting histone deacetylase (Kim and Crews, 2013). As noted, although phenotypic discovery is mechanism-agnostic, mechanism of action can be determined and provides a basis for future target-based discovery. Taxol is an important example of this discovery pathway, with the identification of tubulin as the target leading to the second-generation docetaxel and further drug discovery (Zhao et al., 2016).
D. Combination Screening
Combination chemotherapy in cancer is almost universally applied, with drugs being administered both simultaneously and sequentially. Clinical trials are often designed to establish the benefit of the new agent introduced with the standard-of-care regimen. Identifying synergistic combinations that might be candidates for clinical trials through screening strategies is an emerging trend. A straightforward strategy, termed the curve-shift assay, involves the incubation of cells with a subtoxic dose of test agent along with library compounds. If cells are sensitized to a library compound in the presence of the test agent, compared with the vehicle control, the combination might be clinically important and worthy of follow-up (Uitdehaag et al., 2015). This strategy is particularly powerful when searching for synergies among a library of agency-registered compounds, such as the National Center for Advancing Translational Sciences Pharmaceutical Collection (Huang et al., 2011), which might accelerate long-term clinical trial approval (Wilkinson and Pritchard, 2015).
One limitation of curve-shift studies is that the single dose of test agent might not be adequate to identify synergistic or inhibitory combinations. Systematically testing the combination of two agents at multiple concentrations (for example, a matrix of six concentrations for each compound = 36 pairwise combinations) can reveal synergies that would not be detected by curve-shift studies (six combinations) (Borisy et al., 2003). A major challenge in the implementation of this strategy is that it requires a large number of multiwell plates per experiment. Mathews Griner et al. (2014) confronted this challenge by developing the following: 1) an oncology-focused library of mechanistically annotated agents (called MIPE), 2) an automated dispensing protocol that utilizes 35 6 × 6 matrices per assay plate with test agents delivered by acoustic dispensing, and 3) robust data analysis algorithms. A combinatorial screen of 495 compounds and the Bruton’s tyrosine kinase inhibitor ibrutinib, against activated B cell-like subtype of diffuse large B cell lymphoma, revealed strong synergy with the phosphatidylinositol 3-kinase inhibitor, BKM-120, that is currently in clinical trials. Additionally, a number of clinical agents, including etoposide, doxorubicin, and dexamethasone, also showed synergy (Mathews Griner et al., 2014). Follow-up combination screening with the same malignancy demonstrated that BET protein inhibitors synergize with ibrutinib, and further xenograft studies demonstrated efficacy of the combination (Ceribelli et al., 2014). An advantage of utilizing a mechanistically annotated library is that efficacious combinations can be readily linked to likely targets and further validated genetically to inform on the cancer biology.
Recently, therapeutic activities were evaluated for over 5000 pairs of FDA-approved cancer drugs against the NCI-60 cell lines to identify synergistic combinations. The results were compiled into a database called the NCI-ALMANAC (A Large Matrix of Anti-Neoplastic Agent Combinations) (https://dtp.cancer.gov/ncialmanac) to enable hypothesis-generating assessments of oncology drug combinations. Two phase 1 clinical trials were initiated at the NCI to evaluate combinations identified by the NCI-ALMANAC (Holbeck et al., 2017).
E. Patient-Derived Cells for Personalized Medicine and Discovery
Cancer cell lines maintained in laboratories for many years are recognized to be limited in their relationship to the clinical cancer from which they were derived (Masters, 2000). There are many causes for these differences, including but not limited to clonality, genetic drift, misdiagnosis of the originating malignancy, cross-contamination, and a lack of physiologic complexity (Wilding and Bodmer, 2014). Examples emerge regularly in the literature—such as the recent report that the most commonly-used ovarian cancer cell lines are the least similar (with respect to copy-number changes, mutations, and gene expression) to the high-grade serous ovarian carcinomas they were reported to be derived from (Domcke et al., 2013). Worse, it was reported that all six adenoid cystic carcinoma cell lines used for oncology studies are contaminated by other common cell lines, rendering a significant amount of work in the field redundant (Phuchareon et al., 2009). These issues emphasize the need for patient-derived primary cells in screening, both from the perspective of clinical relevance and that individualized therapeutic strategies might be divined through screening against oncology drugs. Culturing of primary tumor cells from any malignancy presents unique challenges, but blood and solid tumors have been used. A screen of cancer drugs against 174 peritoneal carcinomatosis patient samples indicated oxaliplatin is the preferable agent (Cashin et al., 2013), and it was subsequently implemented in the clinic for this purpose. Additionally, a case study below describes a phenotypic screen with chemotherapy-naive CLL patient samples that led to the identification of auranofin as a clinical candidate based on sensitivity (Shen et al., 2013).
One strategy to preserve human tumor features is to transplant a patient’s primary tumor directly into an immunodeficient mouse and subsequently passage the tumor from animal to animal (i.e., never culturing on plastic), which are termed patient-derived tumor xenografts (PDXs) (Choi et al., 2014). This is in some ways a renaissance, as, prior to tissue culture screening, the primary tools available for identifying drug activity were transplantable syngeneic rodent tumors, and many FDA-registered chemotherapeutics were identified using these models (Fiebig and Burger, 2011). PDX cells can be carried into tissue culture as three-dimensional models (see below) for immediate drug-response evaluation without a dramatic loss of phenotype/genotype (Fong et al., 2014). A recent study from Novartis (Basel, Switzerland) reported HTS using PDXs in vivo after testing mice carrying 277 different PDXs from a range of malignancies with 62 different treatment conditions (Gao et al., 2015). As with cell line studies, sensitivity could be correlated with genomic information to identify the mechanisms of resistance and discover potential new targets. An obvious limitation to screening with mice is that it is not a readily accessible (or affordable) high-throughput strategy. Certainly targets can be validated or identified, but the power is largely in screening across multiple tumor types for sensitivity. Additionally, the compounds screened in the Novartis example were largely medicinal chemistry products and possessed reasonable drug metabolism and pharmacokinetics properties, so the strategy lends itself to mechanistic discovery of probes rather than phenotypic screening of library compounds.
F. Tumor Models: Spheroids, Organoids, and Other Disease-Specific Models
Two-dimensional tissue culturing is not reflective of the physiologic tumor environment. A common model used in oncology is the multicellular tumor spheroid, achieved by culturing tumor cells in low-adherent environments, such as hanging drops, agar-coated plates, or spinner flasks (Martinez et al., 2015). Tumor spheroids recapitulate aspects of tumor heterogeneity, such as a hypoxic core, high interstitial pressure, and acidification related to the accumulation of metabolites. Spheroids also mirror the limited penetration into tumors observed with many drugs (Trédan et al., 2007). Mixed-cell spheroids utilizing supporting cells, such as fibroblasts, or spheroids created from patient samples or PDXs (termed organoids), offer the potential for greater clinical relevance (Unger et al., 2014).
There are challenges with adapting spheroids to screening platforms, including the development of physiologically and biologically reproducible spheroids on a well-by-well basis in HTS multiwell plates. Also, the physical dimensions of spheroids generally limit their use to 96- and 384-well plates (Martinez et al., 2015). Detailed protocols for 96- and 384-well HTS spheroid viability have optimized cell line utility, assay setup, viability readout, and analysis (Kijanska and Kelm, 2004; Friedrich et al., 2009). Screening has revealed selective killing of three-dimensional spheroids versus monolayer culture (Tung et al., 2011), and screens of OVCAR-3 spheroids against the NCI Mechanistic Set (875 compounds) identified compounds selective for cancer stem cell–enriched spheroids (Mezencev et al., 2012). High-content imaging allows specific readouts, including diameter measurement, live- and dead-cell assessment, apoptosis activation, and direct observation of fluorescent test agent penetration (Sirenko et al., 2015).
Cell migration and invasion present separate challenges for assay development. Recently, a multilayered culture (rather than spheroidal) that mimics the ovarian cancer metastatic microenvironment, containing primary human mesothelial cells, fibroblasts, and extracellular matrix, was developed for 384- and 1536-well plates (Kenny et al., 2015). Fluorescent-labeled ovarian cancer cell lines were applied to the multilayered culture, and their invasion was analyzed by high-content imaging. This work reflects the potential for implementation of emerging advanced oncology models to HTS screening platforms (Tanner and Gottesman, 2015).
IV. The Impact of Screening Centers in Cancer
The NCI Cancer Chemotherapy National Service Center was initiated in 1955 to evaluate novel chemicals as cancer therapeutic agents, and in 1976 the functions of the NCI Cancer Chemotherapy National Service Center were incorporated into the Developmental Therapeutics Program (DTP) (Monga and Sausville, 2002). The discovery or development of many United States–licensed chemotherapeutic agents has been supported by the DTP (Sausville and Feigal, 1999). In the late 1980s, the 60 human tumor cell line anticancer drug screen (NCI-60) was developed by the DTP as an in vitro drug-discovery tool, which evolved into a service screen to support cancer research (Shoemaker, 2006). The DTP also maintains a repository of synthetic compounds and pure natural products that are made available to extramural investigators for anticancer evaluation. Before 2005, applications of high-throughput screening and medicinal chemistry to small-molecule discovery were predominately carried out within pharmaceutical companies. To advance the understanding of biologic processes and accelerate the discovery of novel therapeutics, the Molecular Libraries Program was initiated in 2005 by the National Institutes of Health (Austin et al., 2004; Schreiber et al., 2015). The Molecular Libraries Probe Production Centers Network included screening centers located throughout the United States with capabilities to screen a small-molecule library that grew to nearly 390,000 compounds (Schreiber et al., 2015). Active molecules identified from screens were optimized by medicinal chemistry with the goal of developing high-quality chemical probes (Oprea et al., 2009; Frye, 2010; Workman and Collins, 2010).
Datasets from the Molecular Libraries Probe Production Centers Network were made available to researchers worldwide through the PubChem database, for use in studying biology and disease. Overall screening projects in the database are assigned a unique summary assay identification number (AID) and the summary might reference multiple BioAssay records (descriptions of individual assays) and molecular targets. To examine the impact of public screening centers within the field of cancer biology, we performed a rigorous examination of the database entries. An initial list of 401 unique summary AIDs was obtained by searching the PubChem database on December 29, 2015 with the following terms: angiogenesis, cancer, carcinoma, chemotherapy, epigenetic, hematologic, leukemia, oncology, oncogene, malignant, malignancy, metastasis, metastases, mutation, and tumor. The list was manually curated to retain only unique AIDs with clear cancer relevance, resulting in a final list of 285. Collectively, the 285 summary AIDs include 405 cancer-associated targets that were applied to small-molecule screens. This work resulted in 135 probes that modulate diverse targets that can be organized into 96 classifications by the Panther Classification System (Mi et al., 2013, 2016). Information about primary screens was also evaluated, including the assay technology and whether each screen used a biochemical, cell-based, or in vivo assay approach.
To provide a comparison for targets investigated by academic screening centers, we generated a separate list with the targets for 121 FDA-approved small-molecule cancer drugs. An initial list of drugs was compiled from single-agent small molecules included in the “A to Z List of Cancer Drugs” by NCI (http://www.cancer.gov/about-cancer/treatment/drugs) on January 5, 2016. Drugs indicated for chemotherapy-induced nausea and vomiting (palonosetron hydrochloride, ondansetron hydrochloride, and rolapitant hydrochloride) were removed from the list to retain only anticancer molecules. Additionally, 2-mercaptoethanesulfonic acid, leucovorin, aminolevulinic acid, and dexrazoxane were eliminated from the list, because the mechanisms of action do not involve a cancer-related target. Finally, hydroxyurea was added to the list.
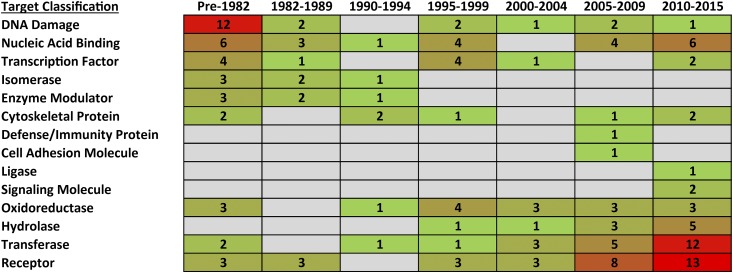
Figure 2 shows the targets of small-molecule cancer drugs organized by the FDA-approval date. Over 31% of the drugs approved before 1982 are associated with a general mechanism of inducing DNA damage. Approval of drugs with a similar mechanism of action has diminished over time, whereas the number of specific protein targets has increased. The approval of drugs targeting classes such as nucleic acid–binding proteins, transcription factors, cytoskeletal proteins, and oxidoreductases has remained relatively consistent over time. In contrast, no drugs were approved during the past 20 years that target the isomerase and enzyme modulator protein classes. Conversely, drugs targeting defense/immunity proteins, cell adhesion molecules, ligases, and signaling molecules have been approved for the first time during the past decade. Drugs targeting hydrolases have steadily increased since the mid-1990s. Also, there has been a major increase during the past 15 years in the approval of drugs that target transferases, which include kinases. Similarly, drugs that target receptors have increased substantially over the past decade. Contributions of small-molecule screening, which began in the early 1990s, to approved drugs would be most prominent from 2000 to 2015.
Fig. 2.
Targets of FDA-approved small-molecule cancer drugs organized by approval date. Values indicate the number of drugs for a particular target, and the plot is colored as a heat map with high values in red and low values in green. Approved small-molecule drugs were collected from the National Cancer Institute “A to Z List of Cancer Drugs,” and the targets were defined according to the Panther Classification System (Mi et al., 2013, 2016). Some drugs, including cisplatin, induce general DNA damage and are included under the classification “DNA Damage.”
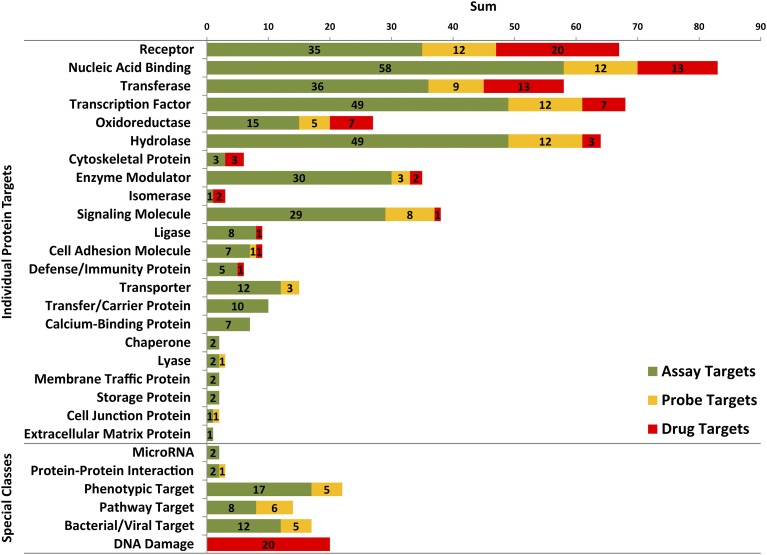
To examine the contributions of public screening centers, the oncology-relevant targets of screens and chemical probes were examined. Unique targets rather than total targets were evaluated to assess the overall diversity. Figure 3 shows the distribution of unique targets for FDA-approved small-molecule cancer drugs (red bars), which defines the current standard of care for cancer treatment. Also shown are the unique cancer-relevant targets of HTS assays (green bars) and chemical probes (yellow bars) from public screening centers. The largest categories of protein targets among drugs include the following: receptors (21%), nucleic acid–binding proteins (14%), transferases (14%), transcription factors (7%), and oxidoreductases (7%). Table 2 expands these general target classifications for HTS assays, chemical probes, and approved drugs to more specific designations, such as kinase and G protein–coupled receptor. Well-represented targets of HTS assays also include the following: nucleic acid–binding proteins (14%), transcription factors (12%), receptors (9%), transferases (9%), and oxidoreductases (4%). Similarly, the number of targets modulated by chemical probes includes the following: receptors (13%), nucleic acid–binding proteins (13%), transcription factors (13%), transferases (9%), and oxidoreductases (5%).
Fig. 3.
Bar chart showing the distributions for unique targets of FDA-approved cancer drugs, cancer-relevant HTS assays from PubChem, and chemical probes. The individual protein target classifications (above) are organized from top to bottom by decreasing number of unique targets for FDA-approved drugs (Drug Targets, red), with the numbers of unique targets for HTS assays and chemical probes indicated by green (Assay Targets) and yellow (Probe Targets), respectively. The bottom of the chart shows special classifications, such as targets that are not defined by the Panther Classification System (MicroRNA and Bacterial/Viral Target) or not easily classified (Protein-Protein Interaction, Phenotypic Target, and Pathway Target). Additionally, many FDA-approved cancer drugs, including cisplatin, induce general DNA damage and are grouped together under the classification “DNA Damage.” All numbers reported in the special classes section represent a total count.
TABLE 2.
Primary and secondary target classifications
Primary and secondary classifications for the unique targets of cancer-relevant HTS assays from PubChem (assay targets), chemical probes (probe targets), and FDA-approved small-molecule cancer drugs (drug targets) shown in Fig. 2. Approved small-molecule drugs were collected from the NCI “A to Z List of Cancer Drugs” (http://www.cancer.gov/about-cancer/treatment/drugs), and the two levels of target classes were defined according to the Panther Classification System (Mi et al., 2013, 2016). Numbers signify the sum of unique targets at or below the indicated target classification level. If a given target was only annotated at the primary level in the database, the sum of primary targets in the table might exceed the sum of secondary targets. Alternatively, the sum of secondary targets can exceed that of primary targets if a given target is linked to multiple secondary Panther protein classifications. An example is thioredoxin reductase 1, cytoplasmic (Uniprot: Q16881), which has a primary classification of oxidoreductase and the secondary classifications reductase and oxidase, due to its potential to catalyze both reactions. Finally, some targets belong to multiple primary classifications, such as nuclear hormone receptors, which fall under both transcription factor and receptor primary classes. The bottom six rows list total numbers for targets that are not currently defined by the Panther Classification System (MicroRNA and bacterial/viral target) or not easily classified (protein–protein interactions, phenotypic target, and pathway target). A total of 20 approved chemotherapeutics is associated with general DNA damage and grouped together under the primary classification of DNA damage.
| Primary Classification | Assay Targets | Probe Targets | Drug Targets | Secondary Classification | Assay Targets | Probe Targets | Drug Targets |
|---|---|---|---|---|---|---|---|
| Nucleic acid binding | 58 | 12 | 13 | DNA-binding protein | 22 | 4 | 5 |
| Helicase | 5 | 1 | |||||
| RNA-binding protein | 6 | 2 | 1 | ||||
| Nuclease | 4 | ||||||
| Transcription factor | 49 | 12 | 7 | Nuclear hormone receptor | 15 | 3 | 7 |
| Transcription cofactor | 5 | ||||||
| Basic helix-loop-helix transcription factor | 6 | 2 | |||||
| Helix-turn-helix transcription factor | 2 | 1 | |||||
| Zinc finger transcription factor | 4 | 2 | |||||
| Hydrolase | 49 | 12 | 4 | Protease | 20 | 5 | 2 |
| Phosphatase | 9 | 3 | 1 | ||||
| Esterase | 6 | 4 | |||||
| Lipase | 6 | 1 | |||||
| Deacetylase | 1 | 1 | |||||
| Deaminase | 1 | ||||||
| Phosphodiesterase | 2 | ||||||
| Receptor | 35 | 12 | 20 | Nuclear hormone receptor | 15 | 3 | 7 |
| G protein–coupled receptor | 11 | 7 | 5 | ||||
| Cytokine receptor | 3 | 2 | |||||
| Protein kinase receptor | 1 | 7 | |||||
| Enzyme modulator | 30 | 3 | 2 | G protein modulator | 9 | ||
| G protein | 8 | ||||||
| Protease inhibitor | 6 | 2 | |||||
| Kinase modulator | 3 | ||||||
| Transferase | 36 | 9 | 13 | Kinase | 21 | 6 | 8 |
| Methyltransferase | 6 | 1 | 2 | ||||
| Acetyltransferase | 5 | 1 | |||||
| Acyltransferase | 1 | 1 | |||||
| Glycosyltransferase | 1 | ||||||
| Signaling molecule | 29 | 8 | 1 | Peptide hormone | 3 | ||
| Cytokine | 2 | 1 | |||||
| Membrane-bound signaling molecule | 2 | ||||||
| Growth factor | 1 | ||||||
| Oxidoreductase | 15 | 5 | 7 | Dehydrogenase | 6 | 1 | 2 |
| Reductase | 5 | 2 | 5 | ||||
| Oxidase | 4 | 1 | 1 | ||||
| Oxygenase | 4 | 2 | 2 | ||||
| Transporter | 12 | 3 | ATP-binding cassette transporter | 3 | 2 | ||
| Ion channel | 3 | 1 | |||||
| Amino acid transporter | 2 | ||||||
| Mitochondrial carrier protein | 1 | ||||||
| Transfer/Carrier protein | 10 | Apolipoprotein | 2 | ||||
| Calcium-binding protein | 7 | Intracellular calcium-sensing protein | 4 | ||||
| Annexin | 3 | ||||||
| Ligase | 8 | 1 | Ubiquitin-protein ligase | 3 | |||
| Cell adhesion molecule | 7 | 1 | 1 | Immunoglobulin superfamily cell adhesion molecule | 2 | 1 | |
| Cadherin | 1 | 1 | |||||
| Integrin | 1 | ||||||
| Cytoskeletal protein | 3 | 3 | Microtubule family cytoskeletal protein | 1 | 3 | ||
| Defense/Immunity protein | 5 | 1 | Immunoglobulin receptor superfamily | 2 | 1 | ||
| Complement component | 1 | ||||||
| Extracellular matrix protein | 1 | Extracellular matrix linker protein | 1 | ||||
| Membrane traffic protein | 2 | Membrane trafficking regulatory protein | 2 | ||||
| Chaperone | 2 | Hsp90 family chaperone | 1 | ||||
| Isomerase | 1 | 2 | Epimerase/Racemase | ||||
| Lyase | 2 | 1 | |||||
| Cell junction protein | 1 | 1 | |||||
| Storage protein | 2 | ||||||
| MicroRNA | 2 | ||||||
| Protein–protein interaction | 2 | 1 | |||||
| Phenotypic target | 17 | 5 | |||||
| Pathway target | 8 | 6 | |||||
| Bacterial/Viral target | 12 | 5 | |||||
| DNA damage | 20 |
Transcription factors, which have been described as “undruggable” (Lazo and Sharlow, 2016), are the second most common target of public screening centers. Moreover, probes have been developed to modulate 12 unique transcription factor targets. Hydrolases (12%) are another major target of public screening centers, and 18% of the unique hydrolase targets are phosphatases (Table 2), which have also been described as “undruggable” (Lazo and Sharlow, 2016). Two probes, ML119 and ML120, target protein tyrosine phosphatase nonreceptor type 7 isoform 2 (Sergienko et al., 2010a,b), and the probe ML113 targets dual specificity phosphatase 3 (Tautz et al., 2010). Also, the probe ML174 is a selective and potent inhibitor of protein phosphatase methylesterase-1 that regulates the activity of protein phosphatase 2A indirectly through posttranslational modifications (Bachovchin et al., 2010). Both enzyme modulator (7%) and signaling molecule (7%) classes have been substantial targets of public screening centers. No drugs that target the enzyme modulator class have been approved for at least 20 years (Fig. 2), although probes were developed against three enzyme modulator targets. Eight unique signaling molecule targets are modulated by probes (Fig. 3), whereas just one target in that class is modulated by two drugs, vismodegib and sonidegib, which were both approved within the past 5 years to treat basal cell carcinoma (Fig. 2). Only four target classes modulated by approved drugs are without probes: cytoskeletal protein, isomerase, ligase, and defense/immunity protein. In the case of isomerases, no drugs have been approved for at least 20 years (Fig. 2), and that class has been a minor focus of screens (0.2%).
Currently, no FDA-approved cancer drugs target nine of the protein classes that were pursued by public screening centers: transporter, transfer/carrier protein, calcium-binding protein, chaperone, lyase, membrane traffic protein, storage protein, cell junction protein, and extracellular matrix protein (Fig. 3). However, probes were developed that target three of these protein classes: transporter, lyase, and cell junction protein. Three unique transporter targets are modulated by probes, including two ATP-binding cassette transporter proteins by ML230 (Strouse et al., 2010), and the potassium channel subfamily K member 9 by ML308 (Miller et al., 2010). The lyase protein, human apurinic/apyrimidinic endonuclease 1, is inhibited by the probe ML199 (Rai et al., 2010). Expression of the cell junction protein, E-cadherin, is increased by the probe ML327 (Stoops et al., 2010). Continued efforts toward developing chemical probes to modulate new and underrepresented target classes will likely further the understanding of cancer biology, provide starting points for therapeutic development, and help to push the limits of what are considered to be “druggable” targets (Lazo and Sharlow, 2016).
Two screening center projects targeted protein–protein interactions, as indicated in the special classes section of Fig. 3. One project enabled the development of the probe, ML223, which disrupts the interaction of the transcription factor, RunX1, with its activator, CBFβ (Marugan et al., 2010). Protein–protein interactions are also considered to be tough targets for small molecules (Thiel et al., 2012), and the tumor-suppressive activity of two probes, ML203 and ML265, was shown to result from stabilization of protein–protein interactions within the pyruvate kinase isoform M2 tetramer (Boxer et al., 2010; Anastasiou et al., 2012). Mechanistically, 20 of 121 FDA-approved small-molecule cancer drugs act by inducing DNA damage and were grouped together in the special classes section of Fig. 3. Correlates of the “DNA Damage” target among approved drugs might exist for chemical probes in the categories “Phenotypic Target” and “Pathway Target,” which lack a defined molecular target. These two categories combined account for only 25 of the 405 cancer-relevant primary target classifications (6%) (Table 2), which indicates a major focus on target-based approaches. Nevertheless, the approaches were fruitful, as 11 of the 25 screens (44%) resulted in probes. Additionally, some cancer-relevant small-molecule screens were directed at bacterial and viral targets (3%), which resulted in the development of five probes.
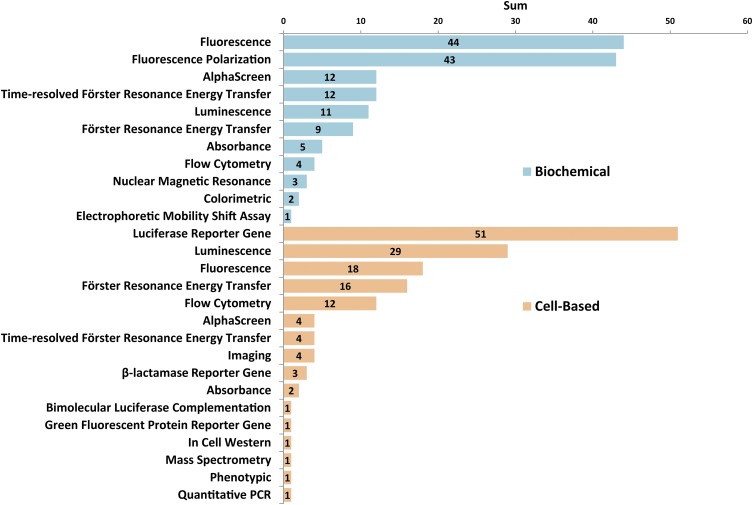
There was a near-perfect split between biochemical-based (146) and cell-based (149) assays among the HTS assays from the PubChem database (Fig. 4). Additionally, one screen was performed in vivo with a fluorescence-based angiogenesis assay using zebrafish (Cross et al., 2003). The distribution of technologies applied to biochemical assays for oncology targets shows that general fluorescence-based methods (30%) and fluorescence polarization (29%) have been the most frequently used. Technologies such as electrophoretic mobility shift (1%) and nuclear magnetic resonance (2%) are challenging to apply in HTS, but provide valuable information. The distribution of technologies implemented for screening with cell-based assays indicates that luciferase reporter genes are the most common (34%), followed by luminescence-based (19%) and fluorescence-based (12%) approaches. Interestingly, methodologies such as mass spectrometry (1%), Western analysis (1%), and quantitative polymerase chain reaction (1%) are used, which are powerful experimental approaches that are difficult to adapt for screening applications.
Fig. 4.
Distribution of assay technologies applied to cancer-relevant HTS assays reported in the PubChem database. Of 295 annotated assays, 146 were biochemical (top) and 149 were cell-based (bottom). A fluorescence-based angiogenesis assay was performed with zebrafish in vivo and is not included in this figure (Cross et al., 2003).
To summarize, a range of oncology-relevant targets was pursued by public screening centers of the Molecular Libraries Program. Both biochemical- and cell-based assays were used for small-molecule screens and critical for the development of probes. Perhaps not surprisingly, the most common primary protein target classes for cancer drugs were also a major focus of screening centers. This does not imply that screening centers have focused on the same molecular targets of approved drugs. For instance, recent genomic studies identified 91 protein kinases as “cancer drivers,” only 22% of which are targeted by FDA-approved therapies (Fleuren et al., 2016). Indeed a substantial focus by screening centers on the primary target class, transferase, is apparent, and kinases represent the majority of targets in that class (Table 2). In general, classes containing greater numbers of unique assay targets are associated with more probes. Interestingly, many of the screens among the special classes of targets resulted in the development of chemical probes. Particularly, two protein–protein interaction screens resulted in 1 probe, 17 phenotypic screens resulted in 5 probes, 8 screens for pathway targets resulted in 6 probes, and 12 screens for bacterial/viral targets resulted in 5 probes. As described above, assays designed to reveal modulators of pathways and phenotypes might allow a greater number of targets to be discovered. Additionally, the approach provides opportunities to discover unanticipated biologic relationships and open new areas of basic research. Nine of the 22 primary protein target classes pursued by screening centers are not associated with approved drugs, demonstrating an emphasis on novel targets. Chemical probes were reported for three of these nine classes, which increases the number of cancer-relevant targets that can be pharmacologically modulated. Altogether, these results indicate that screening centers are contributing overall to early stage drug discovery in the field of oncology by exploring new targets and developing chemical probes.
V. Case Studies
Two examples of screening strategies that played a critical role in the development of a drug program are described in this work. The first conveys the importance of biochemical assays and target-based screening to support the rapid pursuit of a newly identified oncology-related genetic mutation as a potential drug target. The second highlights the use of patient-derived primary cells for phenotypic screening, with a focus on the potential for drug repurposing.
A. Case Study 1: Mutant Isocitrate Dehydrogenases 1 and 2
A contemporary example of the speed at which researchers can mobilize on a putative target is that of mutant isocitrate dehydrogenase (IDH) 1 and 2 (Cairns and Mak, 2013). Wild-type IDH1 is a homodimer that converts isocitrate to α-ketoglutarate (α-KG) using NADP+ as a cofactor. Between 2008 and 2009, separate studies demonstrated that acute myeloid leukemia (AML) and glioblastoma samples carried heterozygous mutations in IDH1 at a single amino acid position (R132) (Parsons et al., 2008; Mardis et al., 2009). Further research revealed that over 75% of some brain cancers and 20% of AML carry such an IDH1/2 mutation, and the mutations occur in a range of other solid tumors, including chondrosarcoma, cholangiocarcinoma, colorectal, and prostate cancer (Cairns and Mak, 2013). Although the somatic mutation results in a loss of enzymatic efficiency, it was also demonstrated by Agios scientists to confer a gain-of-function (neomorphic) activity. Metabolic profiling led to the discovery that the wild-type IDH1 product, α-KG, was converted by mutant IDH1 to 2-hydroxyglutarate (2-HG), utilizing NADPH (Dang et al., 2010). The 2-HG has no known biochemical role and was shown to accumulate in patients with mutant IDH1/2 tumors. This led to a body of work demonstrating that 2-HG plays a role in tumorigenesis and growth by processes such as DNA and histone methylation and NADPH depletion (Turcan et al., 2012; Losman et al., 2013). As a target, mutant IDH1/2 represented a discrete genetic modification amenable to personalized medicine via both tumor gene sequencing and metabolite (2-HG) detection, which appeared to have a gain-of-function oncogenic role. Furthermore, it was anticipated that specific targeting of mutant IDH1/2 would have few clinical side effects, as no endogenous biochemistry would be disrupted by pharmacologic modulation. Mutant IDH1 and IDH2 were therefore pursued as candidates for the development of small-molecule inhibitors.
Multiple pharmaceutical and academic centers rapidly adopted mutant IDH1/2 for inhibitor discovery campaigns. Given that the underlying biochemistry of IDH1/2 as part of the citric acid cycle was well established, the barrier to mutant IDH1 recombinant protein production was relatively low. However, the biochemical reaction was new and required the development of an assay amenable to HTS. The first report of a mutant IDH1 inhibitor (AG-5198, Agios) resulted from a biochemical HTS using recombinant R132H IDH1 homodimer (the most common cellular product would be a mutant/wild-type heterodimer) (Popovici-Muller et al., 2012). The biochemical assay (α-KG to 2-HG) coupled NADPH consumption to diaphorase/resazurin, which uses NADPH to convert resazurin to fluorescent resorufin. With this assay setup, inhibition of mutant IDH1 results in an elevated signal (Hall et al., 2016). The phenyl-glycine hit was chemically optimized into the lead AG-5198 using biochemical assays to assess selectivity against wild-type IDH1 and cell-based assays to monitor the reduction of 2-HG levels (Popovici-Muller et al., 2012). This lead was demonstrated to affect growth and promote differentiation in mutant IDH1 glioma cells, confirming the target relevance (Rohle et al., 2013). A similar medicinal chemistry and assay approach was used by the National Center for Advancing Translational Sciences and Agios to generate the phenyl-glycine analog ML309 (Davis et al., 2014). The Agios efforts have progressed to yield mutant IDH1 (AG-120) (2015), mutant IDH2 (AG-221) (Stein, 2015), and pan-mutant IDH1/2 (AG-881, brain-permeable) inhibitors. Although the development of these inhibitors has not been disclosed in the literature to date, all three are currently in clinical trials for patients with AML and a range of solid tumors (where the tumors were confirmed to carry an appropriate IDH1 mutation). All of this occurred in a remarkable 7-year window from the first report of the mutation to the present.
A number of other discovery programs have been reported against this class of enzymes. Separate discovery efforts around mutant IDH1 led by Sanofi (Cambridge, MA) (Deng et al., 2015) and GlaxoSmithKline (Collegeville, PA) (Okoye-Okafor et al., 2015) used a similar biochemical screen, although they prepared wild-type-IDH1/mutant-IDH1 heterodimer protein (physiologic state) using the two most common IDH1 mutations (R132C and R132H) for their screening campaigns. In each case, they pursued distinct chemical scaffolds, and the primary in-cell confirmation of activity was demonstrated through inhibition of 2-HG production. Agios also pursued mutant IDH2 inhibitors using an analogous biochemical approach to develop the heterocyclic urea sulfonamide, AGI-6780 (Wang et al., 2013). An alternative cell-based HTS strategy used Escherichia coli 3-phosphoglycerate dehydrogenase to convert extracellular 2-HG to α-KG. The NADH produced by the 3-phosphoglycerate dehydrogenase reaction was coupled to the diaphorase/resazurin system, although the hit zaprinast (a PDE5 inhibitor) was found to inhibit 2-HG production by an off-target mechanism, highlighting the risks associated even with cell-based assays that rely on a specific biochemical readout (Elhammali et al., 2014).
B. Case Study 2: Drug-Repurposing Efforts with Patient-Derived Primary Cells Identify Auranofin for CLL
An emerging application of small-molecule screening is aimed at repurposing approved drugs for new indications (Weir et al., 2012). The National Center for Advancing Translational Sciences Pharmaceutical Collection was assembled for this purpose and contains approximately 2800 clinically approved and pharmacologically active small molecules, including drugs approved by the FDA for human or animal use, molecules approved for human use in other countries, and investigational compounds that have been tested in clinical trials (Huang et al., 2011). A recent example that highlights the great potential in repurposing is the identification of multiple therapeutic candidates for the treatment of CLL among compounds within the National Center for Advancing Translational Sciences Pharmaceutical Collection (Shen et al., 2013). New treatment options for CLL are critically needed because current approaches are often not curative and relapse of the disease is frequently observed after treatment with available chemotherapeutics (Chiorazzi et al., 2005). A luminescent cell viability assay was used to compare cytotoxicity of library compounds among CLL cells donated by six individual patients compared against cells donated by five unaffected donors. This phenotypic assay approach effectively casts a wide net in terms of the molecular target or mechanism of action, with the focus on revealing a therapeutic window between the susceptibility of leukemic versus normal cells, which is the desired therapeutic endpoint. A comparison of responses among CLL cells from six individual patients was critical to identify the best compounds because substantial variations in the sensitivities of cells to different drugs were observed. Responses of the most sensitive patient cells resulted in 356 active compounds (13% hit rate), whereas 117 actives were identified with the least sensitive cells (4% hit rate). Moreover, among compounds that were pan-active across CLL cells from all six patients, substantial variations in potencies were observed. Notably, vinblastine showed a >100-fold potency shift between CLL cells from two patients. Whereas the physiologic relevance of screening primary patient cells is a clear advantage, these studies demonstrate the potential bias introduced by only screening one cell source and emphasize the importance of screening multiple cell populations. Remarkably, five compounds were identified that were efficacious against all six patient-derived primary CLL cells, with little or no activity against lymphocytes from healthy donors. In particular, auranofin (Ridaura) exhibited a 30-fold shift in potency between CLL (IC50 ∼70 nM) and normal donor cells, which suggested a potential therapeutic window for CLL patients. Notably, the screening data did not indicate a therapeutic window for drugs in current clinical use, including fludarabine, chlorambucil, bendamustine, mitoxantrone, and vincristine. Auranofin is a gold-containing compound approved for treatment of rheumatoid arthritis, and had been previously reported to inhibit thioredoxin reductase (Kean et al., 1997; Gandin et al., 2010; Debnath et al., 2012). Further studies with primary CLL cells supported auranofin-induced inhibition of thioredoxin reductase and demonstrated that auranofin reduced the leukemia cell burden and increased the survival of TCL-1 transgenic mice, an in vivo model of CLL (Fiskus et al., 2014). Auranofin was subsequently advanced to a phase II clinical study in CLL patients (Shen et al., 2013). Recently, auranofin was included in the CUSP9* treatment protocol for a revolutionary glioblastoma trial consisting of a nine-drug cocktail (Kast et al., 2014). This trial was targeted to inhibit multiple escape pathways to enable the effectiveness of temozolomide, the current standard of care for primary glioblastoma.
VI. Conclusion
Since small-molecule screening was first implemented in the pharmaceutical industry during the early 1990s, it has proven to be a powerful methodology that enabled the development of numerous drugs, including those indicated for cancer (Table 1) (Macarron et al., 2011). Subsequently, the National Institutes of Health Molecular Libraries Program established designated screening centers throughout the United States, which applied small-molecule screening toward a variety of cancer-relevant targets. These efforts resulted in the development of 135 chemical probes that modulate diverse target classes. Technological advancements within a variety of fields have improved the process of small-molecule screening, such as increasing throughput, reducing the burden of assay artifacts, new methods for signal detection, and improved reagents. Assays that are more biologically and physiologically relevant will continue to result from recent advancements in three-dimensional tissue culturing, genomic editing, stem cell biology, and access to primary patient cells. Additionally, accumulating knowledge (within both public databases and the literature) about compound activity (Wassermann et al., 2015), reactivity (Dahlin et al., 2004), and interference (Simeonov et al., 2008; Baell and Walters, 2014) informs the selection of hit compounds from primary screens and influences the assembly and composition of small-molecule libraries. As always, the utility of all these advancements toward therapeutic development depends critically upon the continued efforts of basic researchers to unravel the complicated molecular details that drive human diseases. An exciting and emerging trend is increasing public–private partnerships, which optimally leverages resources and capabilities in both sectors to catalyze advancements toward oncology drug discovery (Stock et al., 2015). Although there is little doubt that small-molecule screens will continue to drive advancements in research and medicine, the utilization of this powerful technology in translational science is expanding.
Abbreviations
- 2-HG
2-hydroxyglutarate
- α-KG
α-ketoglutarate
- AID
assay identification number
- AML
acute myeloid leukemia
- CLL
chronic lymphocytic leukemia
- DTP
Developmental Therapeutics Program
- FDA
Food and Drug Administration
- HTS
high-throughput screening
- IDH
isocitrate dehydrogenase
- MIPDD
mechanism-informed phenotypic drug discovery
- NCI
National Cancer Institute
- PARP
poly(ADP-ribose) polymerase
- PDX
patient-derived tumor xenograft
- PPI
protein–protein interaction
Footnotes
This work was supported by the Intramural Research Program of the National Institutes of Health, National Center for Advancing Translational Sciences.
References
- (2015) IDH1 inhibitor shows promising early results. Cancer Discov 5:4. [DOI] [PubMed] [Google Scholar]
- Abe H, Kikuchi S, Hayakawa K, Iida T, Nagahashi N, Maeda K, Sakamoto J, Matsumoto N, Miura T, Matsumura K, et al. (2011) Discovery of a highly potent and selective MEK inhibitor: GSK1120212 (JTP-74057 DMSO solvate). ACS Med Chem Lett 2:320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker MG, Auld DS. (2014) Considerations for the design and reporting of enzyme assays in high-throughput screening applications. Perspect Sci 1:56–73. [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, et al. (2012) Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol 8:839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin MR, Glicksman MA, Fu H, Havel JJ, Du Y. (2004) Inhibition of protein-protein interactions: non-cellular assay formats, in Assay Guidance Manual (Sittampalam GS, Coussens NP, Nelson H, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Iversen PW, et al., eds) Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD. [PubMed] [Google Scholar]
- Austin CP, Brady LS, Insel TR, Collins FS. (2004) NIH molecular libraries initiative. Science 306:1138–1139. [DOI] [PubMed] [Google Scholar]
- Bachovchin DA, Speers AE, Brown SJ, Spicer TP, Fernandez V, Ferguson J, Mohr JT, Murphy J, Fu GC, Cravatt BF, et al. (2010) Probe development efforts to identify novel inhibitors of protein phosphatase methylesterase-1 (PME-1), in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Baell J, Walters MA. (2014) Chemistry: chemical con artists foil drug discovery. Nature 513:481–483. [DOI] [PubMed] [Google Scholar]
- Banasik M, Komura H, Shimoyama M, Ueda K. (1992) Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J Biol Chem 267:1569–1575. [PubMed] [Google Scholar]
- Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al. (2013) An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson JD, Chen YN, Cornell-Kennon SA, Dorsch M, Kim S, Leszczyniecka M, Sellers WR, Lengauer C. (2006) Validating cancer drug targets. Nature 441:451–456. [DOI] [PubMed] [Google Scholar]
- Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehar J, Price ER, Serbedzija G, Zimmermann GR, Foley MA, Stockwell BR, et al. (2003) Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci USA 100:7977–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, et al. (2010) Evaluation of substituted N,N′-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. J Med Chem 53:1048–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913–917. [DOI] [PubMed] [Google Scholar]
- Buchdunger E, Zimmermann J, Mett H, Meyer T, Müller M, Druker BJ, Lydon NB. (1996) Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res 56:100–104. [PubMed] [Google Scholar]
- Bunnage ME, Chekler EL, Jones LH. (2013) Target validation using chemical probes. Nat Chem Biol 9:195–199. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Mak TW. (2013) Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov 3:730–741. [DOI] [PubMed] [Google Scholar]
- Capdeville R, Buchdunger E, Zimmermann J, Matter A. (2002) Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov 1:493–502. [DOI] [PubMed] [Google Scholar]
- Cashin PH, Mahteme H, Graf W, Karlsson H, Larsson R, Nygren P. (2013) Activity ex vivo of cytotoxic drugs in patient samples of peritoneal carcinomatosis with special focus on colorectal cancer. BMC Cancer 13:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y, Mathews Griner LA, Guha R, Shinn P, Keller JM, et al. (2014) Blockade of oncogenic IκB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc Natl Acad Sci USA 111:11365–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiorazzi N, Rai KR, Ferrarini M. (2005) Chronic lymphocytic leukemia. N Engl J Med 352:804–815. [DOI] [PubMed] [Google Scholar]
- Choi SY, Lin D, Gout PW, Collins CC, Xu Y, Wang Y. (2014) Lessons from patient-derived xenografts for better in vitro modeling of human cancer. Adv Drug Deliv Rev 79–80:222–237. [DOI] [PubMed] [Google Scholar]
- Cockcroft XL, Dillon KJ, Dixon L, Drzewiecki J, Kerrigan F, Loh VM, Jr, Martin NM, Menear KA, Smith GC. (2006) Phthalazinones 2: optimisation and synthesis of novel potent inhibitors of poly(ADP-ribose)polymerase. Bioorg Med Chem Lett 16:1040–1044. [DOI] [PubMed] [Google Scholar]
- Copeland RA. (2013) Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists, John Wiley & Sons, Hoboken, NJ. [PubMed] [Google Scholar]
- Cortés-Cros M, Schmelzle T, Stucke VM, Hofmann F. (2013) The path to oncology drug target validation: an industry perspective. Methods Mol Biol 986:3–13. [DOI] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. (2014) Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 13:828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross LM, Cook MA, Lin S, Chen JN, Rubinstein AL. (2003) Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol 23:911–912. [DOI] [PubMed] [Google Scholar]
- Dahlin JL, Baell J, Walters MA. (2004) Assay interference by chemical reactivity, in Assay Guidance Manual (Sittampalam GS, Coussens NP, Nelson H, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Iversen PW, et al., eds) Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD. [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. (2010) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 465:966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J, Chen P, Norris D, Padmanabha R, Lin J, Moquin RV, Shen Z, Cook LS, Doweyko AM, Pitt S, et al. (2006) 2-Aminothiazole as a novel kinase inhibitor template: structure-activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase inhibitor. J Med Chem 49:6819–6832. [DOI] [PubMed] [Google Scholar]
- Davis MI, Gross S, Shen M, Straley KS, Pragani R, Lea WA, Popovici-Muller J, DeLaBarre B, Artin E, Thorne N, et al. (2014) Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate dehydrogenase IDH1. J Biol Chem 289:13717–13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MI, Shen M, Simeonov A, Hall MD. (2016) Diaphorase coupling protocols for red-shifting dehydrogenase assays. Assay Drug Dev Technol 14:207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath A, Parsonage D, Andrade RM, He C, Cobo ER, Hirata K, Chen S, García-Rivera G, Orozco E, Martínez MB, et al. (2012) A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat Med 18:956–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G, Shen J, Yin M, McManus J, Mathieu M, Gee P, He T, Shi C, Bedel O, McLean LR, et al. (2015) Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by an allosteric small molecule. J Biol Chem 290:762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon KJ, Smith GC, Martin NM. (2003) A FlashPlate assay for the identification of PARP-1 inhibitors. J Biomol Screen 8:347–352. [DOI] [PubMed] [Google Scholar]
- Domcke S, Sinha R, Levine DA, Sander C, Schultz N. (2013) Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew Y. (2015) The development of PARP inhibitors in ovarian cancer: from bench to bedside. Br J Cancer 113 (Suppl 1):S3–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder J, Sedrani R, Wiesmann C. (2014) The discovery of first-in-class drugs: origins and evolution. Nat Rev Drug Discov 13:577–587. [DOI] [PubMed] [Google Scholar]
- Elhammali A, Ippolito JE, Collins L, Crowley J, Marasa J, Piwnica-Worms D. (2014) A high-throughput fluorimetric assay for 2-hydroxyglutarate identifies Zaprinast as a glutaminase inhibitor. Cancer Discov 4:828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein RJ. (2013) The unpluggable in pursuit of the undruggable: tackling the dark matter of the cancer therapeutics universe. Front Oncol 3:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917–921. [DOI] [PubMed] [Google Scholar]
- Fiebig H-H, Burger AM. (2011) Patient-derived tumor models and explants, in Tumor Models in Cancer Research (Teicher B. ed), pp 167–193, Springer, New York, NY. [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010) Selective inhibition of BET bromodomains. Nature 468:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Saba N, Shen M, Ghias M, Liu J, Gupta SD, Chauhan L, Rao R, Gunewardena S, Schorno K, et al. (2014) Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res 74:2520–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuren ED, Zhang L, Wu J, Daly RJ. (2016) The kinome ‘at large’ in cancer. Nat Rev Cancer 16:83–98. [DOI] [PubMed] [Google Scholar]
- Fong EL, Martinez M, Yang J, Mikos AG, Navone NM, Harrington DA, Farach-Carson MC. (2014) Hydrogel-based 3D model of patient-derived prostate xenograft tumors suitable for drug screening. Mol Pharm 11:2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken H, Mathieson T, Childs D, Sweetman GM, Werner T, Tögel I, Doce C, Gade S, Bantscheff M, Drewes G, et al. (2015) Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc 10:1567–1593. [DOI] [PubMed] [Google Scholar]
- Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. (2009) Spheroid-based drug screen: considerations and practical approach. Nat Protoc 4:309–324. [DOI] [PubMed] [Google Scholar]
- Fry DW, Kraker AJ, McMichael A, Ambroso LA, Nelson JM, Leopold WR, Connors RW, Bridges AJ. (1994) A specific inhibitor of the epidermal growth factor receptor tyrosine kinase. Science 265:1093–1095. [DOI] [PubMed] [Google Scholar]
- Frye SV. (2010) The art of the chemical probe. Nat Chem Biol 6:159–161. [DOI] [PubMed] [Google Scholar]
- Galkin AV, Melnick JS, Kim S, Hood TL, Li N, Li L, Xia G, Steensma R, Chopiuk G, Jiang J, et al. (2007) Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci USA 104:270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Fernandes AP, Rigobello MP, Dani B, Sorrentino F, Tisato F, Björnstedt M, Bindoli A, Sturaro A, Rella R, et al. (2010) Cancer cell death induced by phosphine gold(I) compounds targeting thioredoxin reductase. Biochem Pharmacol 79:90–101. [DOI] [PubMed] [Google Scholar]
- Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, et al. (2015) High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 21:1318–1325. [DOI] [PubMed] [Google Scholar]
- Giuliani FC, Kaplan NO. (1980) New doxorubicin analogs active against doxorubicin-resistant colon tumor xenografts in the nude mouse. Cancer Res 40:4682–4687. [PubMed] [Google Scholar]
- Green JR, Müller K, Jaeggi KA. (1994) Preclinical pharmacology of CGP 42′446, a new, potent, heterocyclic bisphosphonate compound. J Bone Miner Res 9:745–751. [DOI] [PubMed] [Google Scholar]
- Hall MD, Simeonov A, Davis MI. (2016) Avoiding fluorescence assay interference: the case for diaphorase. Assay Drug Dev Technol 14:175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam C, Hellicar J, Dunn A, Fuetterer A, Hardy N, Marshall P, Paape R, Pemberton M, Resemannand A, Leveridge M. (2016) The evolution of MALDI-TOF mass spectrometry toward ultra-high-throughput screening: 1536-well format and beyond. J Biomol Screen 21:176–186. [DOI] [PubMed] [Google Scholar]
- Hawtin RE, Stockett DE, Byl JA, McDowell RS, Nguyen T, Arkin MR, Conroy A, Yang W, Osheroff N, Fox JA. (2010) Voreloxin is an anticancer quinolone derivative that intercalates DNA and poisons topoisomerase II. PLoS One 5:e10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelder S, Clarke PA, Workman P. (2012) Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol Oncol 6:155–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck SL, Camalier R, Crowell JA, Govindharajulu JP, Hollingshead M, Anderson LW, Polley E, Rubinstein L, Srivastava A, Wilsker D, et al. (2017) The National Cancer Institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res 77:3564–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck SL, Collins JM, Doroshow JH. (2010) Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther 9:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, Li S, Pan Z, Thamm DH, Miller RA, et al. (2010) The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci USA 107:13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang YH, Hertzberg R, Hecht S, Liu LF. (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem 260:14873–14878. [PubMed] [Google Scholar]
- Huang R, Southall N, Wang Y, Yasgar A, Shinn P, Jadhav A, Nguyen DT, Austin CP. (2011) The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med 3:80ps16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JP, Rees S, Kalindjian SB, Philpott KL. (2011) Principles of early drug discovery. Br J Pharmacol 162:1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundbäck T, Nordlund P, Martinez Molina D. (2014) The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9:2100–2122. [DOI] [PubMed] [Google Scholar]
- Johannes JW, Almeida L, Daly K, Ferguson AD, Grosskurth SE, Guan H, Howard T, Ioannidis S, Kazmirski S, Lamb ML, et al. (2015) Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering. Bioorg Med Chem Lett 25:5743–5747. [DOI] [PubMed] [Google Scholar]
- Kast RE, Karpel-Massler G, Halatsch ME. (2014) CUSP9* treatment protocol for recurrent glioblastoma: aprepitant, artesunate, auranofin, captopril, celecoxib, disulfiram, itraconazole, ritonavir, sertraline augmenting continuous low dose temozolomide. Oncotarget 5:8052–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean WF, Hart L, Buchanan WW. (1997) Auranofin. Br J Rheumatol 36:560–572. [DOI] [PubMed] [Google Scholar]
- Kenny HA, Lal-Nag M, White EA, Shen M, Chiang CY, Mitra AK, Zhang Y, Curtis M, Schryver EM, Bettis S, et al. (2015) Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat Commun 6:6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M, Kelm J. (2004) In vitro 3D spheroids and microtissues: ATP-based cell viability and toxicity assays, in Assay Guidance Manual (Sittampalam GS, Coussens NP, Nelson H, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Iversen PW, et al., eds) Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD. [PubMed] [Google Scholar]
- Kim KB, Crews CM. (2013) From epoxomicin to carfilzomib: chemistry, biology, and medical outcomes. Nat Prod Rep 30:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhagen G, Paull KD, Cushman M, Nagafuji P, Pommier Y. (1998) Protein-linked DNA strand breaks induced by NSC 314622, a novel noncamptothecin topoisomerase I poison. Mol Pharmacol 54:50–58. [DOI] [PubMed] [Google Scholar]
- Kupperman E, Lee EC, Cao Y, Bannerman B, Fitzgerald M, Berger A, Yu J, Yang Y, Hales P, Bruzzese F, et al. (2010) Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res 70:1970–1980. [DOI] [PubMed] [Google Scholar]
- Lazo JS, Sharlow ER. (2016) Drugging undruggable molecular cancer targets. Annu Rev Pharmacol Toxicol 56:23–40. [DOI] [PubMed] [Google Scholar]
- Loh VM, Jr, Cockcroft XL, Dillon KJ, Dixon L, Drzewiecki J, Eversley PJ, Gomez S, Hoare J, Kerrigan F, Matthews IT, et al. (2005) Phthalazinones. Part 1: the design and synthesis of a novel series of potent inhibitors of poly(ADP-ribose)polymerase. Bioorg Med Chem Lett 15:2235–2238. [DOI] [PubMed] [Google Scholar]
- Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG., Jr (2013) (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339:1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DV, Hertzberg RP, Janzen WP, Paslay JW, et al. (2011) Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov 10:188–195. [DOI] [PubMed] [Google Scholar]
- Mak IW, Evaniew N, Ghert M. (2014) Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res 6:114–118. [PMC free article] [PubMed] [Google Scholar]
- Manley PW, Cowan-Jacob SW, Buchdunger E, Fabbro D, Fendrich G, Furet P, Meyer T, Zimmermann J. (2002) Imatinib: a selective tyrosine kinase inhibitor. Eur J Cancer 38 (Suppl 5):S19–S27. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361:1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Breslow R. (2007) Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol 25:84–90. [DOI] [PubMed] [Google Scholar]
- Martinez Molina D, Nordlund P. (2016) The cellular thermal shift assay: a novel biophysical assay for in situ drug target engagement and mechanistic biomarker studies. Annu Rev Pharmacol Toxicol 56:141–161. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Titus SA, Wagner AK, Simeonov A. (2015) High-throughput fluorescence imaging approaches for drug discovery using in vitro and in vivo three-dimensional models. Expert Opin Drug Discov 10:1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugan JJ, Xiao J, Zheng W, Southall N, Dehdashti S, Cunningham L, Liu PP. (2010) ML223: a small molecule probe with in vivo activity against acute myeloid leukemia subtype M4Eo, in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Masters JR. (2000) Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol 1:233–236. [DOI] [PubMed] [Google Scholar]
- Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, et al. (2014) High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci USA 111:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, Dillon KJ, Drzewiecki J, Garman S, Gomez S, et al. (2008) 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem 51:6581–6591. [DOI] [PubMed] [Google Scholar]
- Mezencev R, Wang L, McDonald JF. (2012) Identification of inhibitors of ovarian cancer stem-like cells by high-throughput screening. J Ovarian Res 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. (2016) PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44 (D1):D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Zou B, Shi J, Flaherty DP, Simpson DS, Yao T, Maki BE, Day VW, Douglas JT, Wu M, et al. (2010) Development of a selective chemical inhibitor for the two-pore potassium channel, KCNK9, in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Moffat JG, Rudolph J, Bailey D. (2014) Phenotypic screening in cancer drug discovery: past, present and future. Nat Rev Drug Discov 13:588–602. [DOI] [PubMed] [Google Scholar]
- Monga M, Sausville EA. (2002) Developmental therapeutics program at the NCI: molecular target and drug discovery process. Leukemia 16:520–526. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S. (1998) FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res 241:126–133. [DOI] [PubMed] [Google Scholar]
- Nitiss JL. (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye-Okafor UC, Bartholdy B, Cartier J, Gao EN, Pietrak B, Rendina AR, Rominger C, Quinn C, Smallwood A, Wiggall KJ, et al. (2015) New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol 11:878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea TI, Bologa CG, Boyer S, Curpan RF, Glen RC, Hopkins AL, Lipinski CA, Marshall GR, Martin YC, Ostopovici-Halip L, et al. (2009) A crowdsourcing evaluation of the NIH chemical probes. Nat Chem Biol 5:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabha R, Shu YZ, Cook LS, Veitch JA, Donovan M, Lowe S, Huang S, Pirnik D, Manly SP. (1998) 1-Methoxy-agroclavine from Penicillium sp. WC75209, a novel inhibitor of the Lck tyrosine kinase. Bioorg Med Chem Lett 8:569–574. [DOI] [PubMed] [Google Scholar]
- Pan S, Wu X, Jiang J, Gao W, Wan Y, Cheng D, Han D, Liu J, Englund NP, Wang Y, et al. (2010) Discovery of NVP-LDE225, a potent and selective smoothened antagonist. ACS Med Chem Lett 1:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Scheerens H, Li SJ, Schultz BE, Sprengeler PA, Burrill LC, Mendonca RV, Sweeney MD, Scott KC, Grothaus PG, et al. (2007) Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem 2:58–61. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R, Umezawa Y, Gambhir SS. (2002) Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA 99:15608–15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuchareon J, Ohta Y, Woo JM, Eisele DW, Tetsu O. (2009) Genetic profiling reveals cross-contamination and misidentification of 6 adenoid cystic carcinoma cell lines: ACC2, ACC3, ACCM, ACCNS, ACCS and CAC2. PLoS One 4:e6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarz RL, Robey R, Sandor V, Bakke S, Wilson WH, Dahmoush L, Kingma DM, Turner ML, Altemus R, Bates SE. (2001) Inhibitor of histone deacetylation, depsipeptide (FR901228), in the treatment of peripheral and cutaneous T-cell lymphoma: a case report. Blood 98:2865–2868. [DOI] [PubMed] [Google Scholar]
- Plückthun A, Knowles JR. (1987) The consequences of stepwise deletions from the signal-processing site of beta-lactamase. J Biol Chem 262:3951–3957. [PubMed] [Google Scholar]
- Pommier Y. (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 6:789–802. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Kiselev E, Marchand C. (2015) Interfacial inhibitors. Bioorg Med Chem Lett 25:3961–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan S, Zhao F, Gross S, Dang L, Yen KE, Yang H, et al. (2012) Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett 3:850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai G, Vyjayanti VN, Dorjsuren D, Simeonov A, Jadhav A, Wilson DM, Maloney DJ. (2010) Small molecule inhibitors of the human apurinic/apyrimidinic endonuclease 1 (APE1), in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL, Bodycombe NE, et al. (2016) Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 12:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, Doroshow J, Pommier Y. (2012) CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res 72:3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robers MB, Dart ML, Woodroofe CC, Zimprich CA, Kirkland TA, Machleidt T, Kupcho KR, Levin S, Hartnett JR, Zimmerman K, et al. (2015) Target engagement and drug residence time can be observed in living cells with BRET. Nat Commun 6:10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. (2013) An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340:626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, et al. (2004) Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/-)p53(-/-) mice. Cancer Cell 6:229–240. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami TA, Oikawa N, Tsukuda T, Ishii N, Aoki Y. (2011) CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 19:679–690. [DOI] [PubMed] [Google Scholar]
- Sausville EA, Feigal E. (1999) Evolving approaches to cancer drug discovery and development at the National Cancer Institute, USA. Ann Oncol 10:1287–1291. [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Kotz JD, Li M, Aubé J, Austin CP, Reed JC, Rosen H, White EL, Sklar LA, Lindsley CW, et al. NIH Molecular Libraries Project Team (2015) Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell 161:1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, Jones V, Bodycombe NE, Soule CK, Gould J, et al. (2015) Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov 5:1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergienko E, Bobkova E, Colayco S, Rascon J, Chan X, Vasile S, Su Y, Dahl R, Chung TDY, Dad S, et al. (2010a) Selective HePTP inhibitors: probe 2, in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Sergienko E, Bobkova E, Vasile S, Yuan H, Su Y, Dahl R, Yang L, Chung TDY, Dad S, Mustelin T, et al. (2010b) Selective HePTP inhibitors: probe 1, in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, Pommier Y, Weinstein JN. (2009) CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics 10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Zhang Y, Saba N, Austin CP, Wiestner A, Auld DS. (2013) Identification of therapeutic candidates for chronic lymphocytic leukemia from a library of approved drugs. PLoS One 8:e75252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH. (2006) The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 6:813–823. [DOI] [PubMed] [Google Scholar]
- Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, et al. (2008) Fluorescence spectroscopic profiling of compound libraries. J Med Chem 51:2363–2371. [DOI] [PubMed] [Google Scholar]
- Sirenko O, Mitlo T, Hesley J, Luke S, Owens W, Cromwell EF. (2015) High-content assays for characterizing the viability and morphology of 3D cancer spheroid cultures. Assay Drug Dev Technol 13:402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, et al. (2013) ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19:202–208. [DOI] [PubMed] [Google Scholar]
- Stein EM. (2015) IDH2 inhibition in AML: finally progress? Best Pract Res Clin Haematol 28:112–115. [DOI] [PubMed] [Google Scholar]
- Stock JK, Jones NP, Hammonds T, Roffey J, Dillon C. (2015) Addressing the right targets in oncology: challenges and alternative approaches. J Biomol Screen 20:305–317. [DOI] [PubMed] [Google Scholar]
- Stoops SL, Waterson AG, An H, Deane N, Daniels JS, Morrison R, Engers JL, Beauchamp D, Lindsley CW. (2010) Discovery and characterization of a small molecule that restores E-cadherin expression in cancer cell lines via a new mechanism, in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Strouse JJ, Ivnitski-Steele I, Njus HM, Foutz TD, Yao T, Weiner WS, Schroeder CE, Simpson DS, Maki BE, Li K, et al. (2010) Selective efflux inhibition of ATP-binding cassette sub-family G member 2, in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Swinney DC, Anthony J. (2011) How were new medicines discovered? Nat Rev Drug Discov 10:507–519. [DOI] [PubMed] [Google Scholar]
- Szakács G, Annereau JP, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, Reinhold W, Guo Y, Kruh GD, Reimers M, et al. (2004) Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell 6:129–137. [DOI] [PubMed] [Google Scholar]
- Tanner K, Gottesman MM. (2015) Beyond 3D culture models of cancer. Sci Transl Med 7:283ps9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz L, Mustelin T, Wu S, Vossius S, Rahmouni S, Vasile S, Sergienko E, Stonich D, Yuan H, Su Y, et al. (2010) Small-molecule inhibitors of vaccinia-H1-related phosphatase VHR, in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information (US), Bethesda, MD. [PubMed] [Google Scholar]
- Thiel P, Kaiser M, Ottmann C. (2012) Small-molecule stabilization of protein-protein interactions: an underestimated concept in drug discovery? Angew Chem Int Ed Engl 51:2012–2018. [DOI] [PubMed] [Google Scholar]
- Tomita K, Tsuzuki Y, Shibamori K, Tashima M, Kajikawa F, Sato Y, Kashimoto S, Chiba K, Hino K. (2002) Synthesis and structure-activity relationships of novel 7-substituted 1,4-dihydro-4-oxo-1-(2-thiazolyl)-1,8-naphthyridine-3-carboxylic acids as antitumor agents. Part 1. J Med Chem 45:5564–5575. [DOI] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. (2009) Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324:787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trédan O, Galmarini CM, Patel K, Tannock IF. (2007) Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst 99:1441–1454. [DOI] [PubMed] [Google Scholar]
- Tsuzuki Y, Tomita K, Shibamori K, Sato Y, Kashimoto S, Chiba K. (2004) Synthesis and structure-activity relationships of novel 7-substituted 1,4-dihydro-4-oxo-1-(2-thiazolyl)-1,8-naphthyridine-3-carboxylic acids as antitumor agents. Part 2. J Med Chem 47:2097–2109. [DOI] [PubMed] [Google Scholar]
- Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. (2011) High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst (Lond) 136:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Nakajima H, Hori Y, Fujita T, Nishimura M, Goto T, Okuhara M. (1994) FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. I. Taxonomy, fermentation, isolation, physico-chemical and biological properties, and antitumor activity. J Antibiot (Tokyo) 47:301–310. [DOI] [PubMed] [Google Scholar]
- Uitdehaag JC, de Roos JA, van Doornmalen AM, Prinsen MB, Spijkers-Hagelstein JA, de Vetter JR, de Man J, Buijsman RC, Zaman GJ. (2015) Selective targeting of CTNBB1-, KRAS- or MYC-driven cell growth by combinations of existing drugs. PLoS One 10:e0125021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger C, Kramer N, Walzl A, Scherzer M, Hengstschläger M, Dolznig H. (2014) Modeling human carcinomas: physiologically relevant 3D models to improve anti-cancer drug development. Adv Drug Deliv Rev 79–80:50–67. [DOI] [PubMed] [Google Scholar]
- Wakeling AE, Dukes M, Bowler J. (1991) A potent specific pure antiestrogen with clinical potential. Cancer Res 51:3867–3873. [PubMed] [Google Scholar]
- Wall ME, Wani MC. (1995) Camptothecin and taxol: discovery to clinic--thirteenth Bruce F. Cain Memorial Award Lecture. Cancer Res 55:753–760. [PubMed] [Google Scholar]
- Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, et al. (2013) Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340:622–626. [DOI] [PubMed] [Google Scholar]
- Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, Pairaudeau G, Pennie WD, Pickett SD, Wang J, et al. (2015) An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov 14:475–486. [DOI] [PubMed] [Google Scholar]
- Wassermann AM, Lounkine E, Hoepfner D, Le Goff G, King FJ, Studer C, Peltier JM, Grippo ML, Prindle V, Tao J, et al. (2015) Dark chemical matter as a promising starting point for drug lead discovery. Nat Chem Biol 11:958–966. [DOI] [PubMed] [Google Scholar]
- Weir SJ, DeGennaro LJ, Austin CP. (2012) Repurposing approved and abandoned drugs for the treatment and prevention of cancer through public-private partnership. Cancer Res 72:1055–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding JL, Bodmer WF. (2014) Cancer cell lines for drug discovery and development. Cancer Res 74:2377–2384. [DOI] [PubMed] [Google Scholar]
- Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. (2006) Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov 5:835–844. [DOI] [PubMed] [Google Scholar]
- Wilkinson GF, Pritchard K. (2015) In vitro screening for drug repositioning. J Biomol Screen 20:167–179. [DOI] [PubMed] [Google Scholar]
- Williams JA, Guicherit OM, Zaharian BI, Xu Y, Chai L, Wichterle H, Kon C, Gatchalian C, Porter JA, Rubin LL, et al. (2003) Identification of a small molecule inhibitor of the hedgehog signaling pathway: effects on basal cell carcinoma-like lesions. Proc Natl Acad Sci USA 100:4616–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P, Collins I. (2010) Probing the probes: fitness factors for small molecule tools. Chem Biol 17:561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Yoshida T, Kurachi R, Kakegawa J, Hori Y, Nanayama T, Hayakawa K, Abe H, Takagi K, Matsuzaki Y, et al. (2007) Identification of JTP-70902, a p15(INK4b)-inductive compound, as a novel MEK1/2 inhibitor. Cancer Sci 98:1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Laterra J, Pomper MG. (2009) Hedgehog pathway inhibitor HhAntag691 is a potent inhibitor of ABCG2/BCRP and ABCB1/Pgp. Neoplasia 11:96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Mu X, Du G. (2016) Microtubule-stabilizing agents: new drug discovery and cancer therapy. Pharmacol Ther 162:134–143. [DOI] [PubMed] [Google Scholar]
- Zimmermann J, Buchdunger E, Mett H, Meyer T, Lydon NB, Traxler P. (1996a) Phenylamino-pyrimidine (PAP) - derivatives: a new class of potent and highly selective PDGF-receptor autophosphorylation inhibitors. Bioorg Med Chem Lett 6:1221–1226. [Google Scholar]
- Zimmermann J, Buchdunger E, Mett H, Meyer T, Lydon NB. (1997) Potent and selective inhibitors of the Abl-kinase: phenylamino-pyrimidine (PAP) derivatives. Bioorg Med Chem Lett 7:187–192. [Google Scholar]
- Zimmermann J, Caravatti G, Mett H, Meyer T, Müller M, Lydon NB, Fabbro D. (1996b) Phenylamino-pyrimidine (PAP) derivatives: a new class of potent and selective inhibitors of protein kinase C (PKC). Arch Pharm (Weinheim) 329:371–376. [DOI] [PubMed] [Google Scholar]