Abstract
Objective(s)
To determine the association between cytochrome p450 2B6 genotypes and efavirenz-based HIV treatment outcomes.
Design
Observational cohort study of HIV infected adults initiating efavirenz-based regimens in Botswana.
Methods
The primary endpoint was a composite of death or loss to care or HIV RNA>25 copies/ml at 6 months. CYP2B6 516G>T and 983T>C genotyping was done with Taqman Open Array platform. Adverse experiences were measured using the Subject Experience Questionnaire. Metabolism alleles were included in logistic regression models of the composite endpoint.
Results
801 individuals included 406 (51%) males, median age 37 years, median baseline CD4 count 195 cells/mm3 and plasma HIV RNA 4·9 log10 copies/ml. 288 (36%) reached the endpoint including 34 (4%) deaths, 151 (19%) lost to care, 11 (1%) lost to the study, but alive and in care, and 92 (11%) with plasma HIV RNA>25 copies/ml. Metabolism variant alleles were common with 396 (49%) intermediate and 192 (24%) slow metabolizers. There were no statistically significant associations between metabolism and treatment endpoints. However, slower metabolism was associated with fewer adverse experiences.
Conclusions
Slow metabolism alleles were associated with lower efavirenz clearance but not any of the treatment endpoints. Slow efavirenz metabolism did not exacerbate CNS toxicity. These results should allay concern that slow efavirenz metabolism adversely impacts individuals in sub-Saharan African settings in which these alleles are common.
Introduction
The widespread distribution of combination antiretroviral therapy into resource-constrained settings has dramatically improved survival in HIV-infected individuals [1, 2]. The current clinical management approach in such settings focuses on testing as many at risk individuals as can be reached, and prescribing affordable antiretrovirals as soon as feasible regardless of the ability to measure biomarkers of response [3]. In contrast, in resource rich settings, virologic monitoring, resistance testing, and tailoring regimens to patient characteristics are standard of care [4]. As the cost of such a management strategy decreases, it will likely become more widely implemented in resource-constrained settings.
Efavirenz, one of the most commonly used drugs for HIV around the world, is also known to have substantially higher drug concentrations in an identifiable subset of patients, namely, those with polymorphisms in cytochrome P450 2B6 enzyme [5–7]. CYP2B6 516G>T and 983T>C are both associated with a significant decrease in enzyme function. If adverse effects are associated with lower efavirenz clearance and subsequently higher concentrations, as has been described by others [8], it would be clinically important if it results in non-adherence or loss to follow-up [9]. Alternatively, slower clearance could result in improved drug effects in individuals who experience short-term non-adherence yet maintain adequate concentrations of at least one drug adequate to sustain viral suppression. Prior studies based on data from clinical trials participants have failed to demonstrate an impact of the 516 G>T polymorphism on treatment success (i.e., being on the efavirenz-based regimen and in care with an undetectable viral load). [5, 10]. However, there are limited data on this association in routine care settings in sub-Saharan Africa, where efavirenz is widely used and patient attrition in ART programs is common and associated with subsequent death [11].
Recently, efavirenz has been relegated to second line status in the US Department of Health and Human Services guidelines due to concerns of tolerability [12]. We sought to determine if slow efavirenz clearance was associated with better or worse outcomes in an African population with a high prevalence of slow efavirenz metabolism alleles initiating antiretroviral therapy outside of a clinical trial [13].
Methods
Participant Characteristics and Visits
We conducted a prospective observational cohort study of treatment-naïve HIV infected individuals at public HIV clinics in and around Gaborone, Botswana. Participants had to have confirmed HIV infection by standard assays, be of black African origin, age 21 or older, planning to stay in the Gaborone area for the subsequent 6 months, not be pregnant or intending to get pregnant, and initiating a standard 3-drug efavirenz-based regimen including nucleoside or nucleotide reverse transcriptase inhibitors irrespective of CD4 count. The baseline visit included tests for plasma HIV RNA and CD4 count, and blood was obtained for DNA extraction. Follow-up visits occurred at month 1 and month 6, allowing a 3-month window for the month 6 assessment to be completed.
Participants were approached for enrollment in the clinics on the day they were scheduled to initiate antiretroviral therapy. They were then contacted by telephone the day before their scheduled follow-up visits. If participants failed to present to the study visit or did not respond to the telephone call, follow-up calls were made and short message system (SMS) messages were sent over the subsequent 2 weeks. Calls and SMS messages were also sent to their personal contacts requesting that the contacts notify the participant that the study team needed to speak with them. If participants did not show for the final visit by month 9, we searched all available records for evidence of their death or follow-up in clinical care including paper records at the clinic care site and electronic medical and laboratory records for sites that had centralized electronic data capture. Loss to follow-up after active tracing has been associated with death in this setting [14].
Clinical Endpoints
Death was determined by documentation in the medical record or by verbal report from the participant’s family. Loss to care was determined by inability to identify any records on the participant up to 9 months after initiation of therapy. Plasma HIV RNA was assessed using NucliSENS Easy Q HIV-1 (Biomerieux) with a lower limit of quantification of 25 copies/ml. We defined a composite primary endpoint as, death, loss to care, or viremia and secondarily assessed each of the component endpoints separately. Participants lost to the study but found to be alive and in care but without available viral load data were classified as having met the primary endpoint. We also assessed adverse experiences on therapy at months 1 and 6 using the 35-item Subject Experience Questionnaire [9]. It was translated into Setswana and then back-translated into English by different bilingual translators and modified until the Setswana version was considered equivalent. Each symptom was rated 0–3 and summed across all items with higher scores representing greater adverse experiences. In secondary analyses, we summed only items pertaining to central nervous system symptoms.
Efavirenz Concentrations
Blood was drawn at month 1 and month 6 for plasma efavirenz concentrations. Participants were queried regarding the date and time of their dose prior to the blood draw and the date and time of the blood draw was also recorded. Plasma efavirenz concentrations were measured at a single laboratory using a validated liquid chromatography/tandem mass spectrometry assay [15].
CYP2B6 Genotyping
DNA was extracted from whole blood samples and individual CYP2B6 genotypes were determined by measuring allele-specific fluorescence using Taqman Genotyper software, version 1.3 (Life Technologies). CYP2B6 516G>T and 983T>C genotypes were jointly considered in defining the metabolism genotype. The SNPs passed a standardized quality control procedure that consisted of a >95% successful call rate and Hardy-Weinberg Equilibrium testing (p>0·01). All genotype-specific major and minor allele frequencies were concordant with individuals of African ancestry from the International HapMap reference panel.
Statistical Analysis
Genotype-Endpoint Association
We compared the association between genotype and composite endpoint using logistic regression models. The primary analysis used the CYP2B6 516G>T and 983T>C genotypes to categorize individuals as extensive, intermediate or slow metabolizers. We included genotype status as a categorical variable in our logistic modeling approach to address the potential concern that the assumption of a linear relationship with outcome might not hold and then as ordinal variables to assess for a dose response relationship between metabolism genotype and the various outcomes.
Multivariable models were constructed by including covariates associated with the endpoints with p values <0.1. Covariates were retained in the model if they changed the magnitude of the association between genotype and endpoint by >15%. Because of its known association with virological response, we included log 10 baseline viral load in all models irrespective of its association with endpoints.
We assessed the relation between CYP2B6 genotypes and change in CD4 count between baseline and month 6 using linear regression models with genotype included as a categorical variable and ordinal variable. The secondary analyses of viremia and change in CD4 count only included individuals who remained in care by 6 months since those out of care did not have available data.
The Subject Experience Questionnaire scores at months 1 and 6 were compared separately between the CYP2B6 genotypes using ANOVA and then linear regression with genotype included as an ordinal variable.
Due to known effects of concomitant isoniazid treatment further decreasing efavirenz clearance in slow metabolizers [16], we conducted additional sensitivity analysis excluding these individuals to assess for evidence of confounding or effect modification.
Population Pharmacokinetic Genotype Analysis
The population pharmacokinetic analysis was conducted using non-linear mixed effects modeling (NONMEM) software version VII, level 2.0 (ADVAN 2, TRANS 2) interfaced with PDx-POP (both from ICON Development Solutions). All models were run with the first order conditional estimation with interaction (FOCE-I) method. We used S-Plus Version 6.2 (Insightful, Inc., Data Analysis Products Division, Seattle, WA) for goodness-of-fit diagnostics and graphical displays. The goodness-of-fit from each NONMEM run was assessed using visual inspection of diagnostic plots and the precision of the parameter estimates (data not shown). Within-subject dosing histories greater than 30 days with no intermittent pharmacokinetic observations were condensed as a single steady-state dosing record. Given the limited sampling, only a one-compartment model was considered. Models were parameterized by apparent oral clearance (CL/F, mL/min), apparent central volume of distribution (V1/F, liters) and the first-order absorption rate constant (ka, hr−1). We used an exponential variance model to describe the unexplained random variability of parameters across individuals. Models were explored using various inter-individual random effect covariance structures. Inter-individual variability was initially estimated for CL/F, and then subsequently evaluated for the remaining pharmacokinetic parameters. A combined additive and proportional error model was used to describe random residual variability. The impact of weight on all pharmacokinetic parameters was investigated using an allometric model and fixed at 0·75 for CL/F, and 1 for V1/F [17–20] and a reference weight of 70 kg. Finally, the impact of genotypes on CL/F based on polymorphisms for CYP2B6 516G>T and 983T>C jointly was estimated. A total of 13 individuals had efavirenz concentrations below the lower limit of quantification and were not included in the pharmacokinetic analysis.
Sample Size Considerations
We targeted enrollment of 940 individuals to achieve 80% power to detect a relative risk of 2·0 for the association between slow metabolism genotype and composite endpoint at 6 months. We assumed a prevalence of the variant allele of the SNP with the more common polymorphism (i.e., CYP2B6 516G>T) of 40% [13] and a composite endpoint rate of 20% [14]. We increased the target sample size by 50% [21] to account for the presence of potential effect modifiers of the relation between genotype and endpoint.
Ethical Considerations and Funding
All participants gave written consent to participate after the study was explained to them in English or Setswana, depending on their preference. Participants were compensated with ~5 US dollars per visit for three visits based upon guidelines from the Botswana Ministry of Health. The assays for CYP2B6 genotypes and efavirenz concentrations were performed in laboratories blinded to the endpoint data. Likewise, the assessment of loss to care was performed by staff blinded to the CYP2B6 genotypes. The study was approved by the Ethics Board of the Health Research and Development Committee of the Botswana Ministry of Health and by the Committee on Human Subjects Research of the University of Pennsylvania.
Results
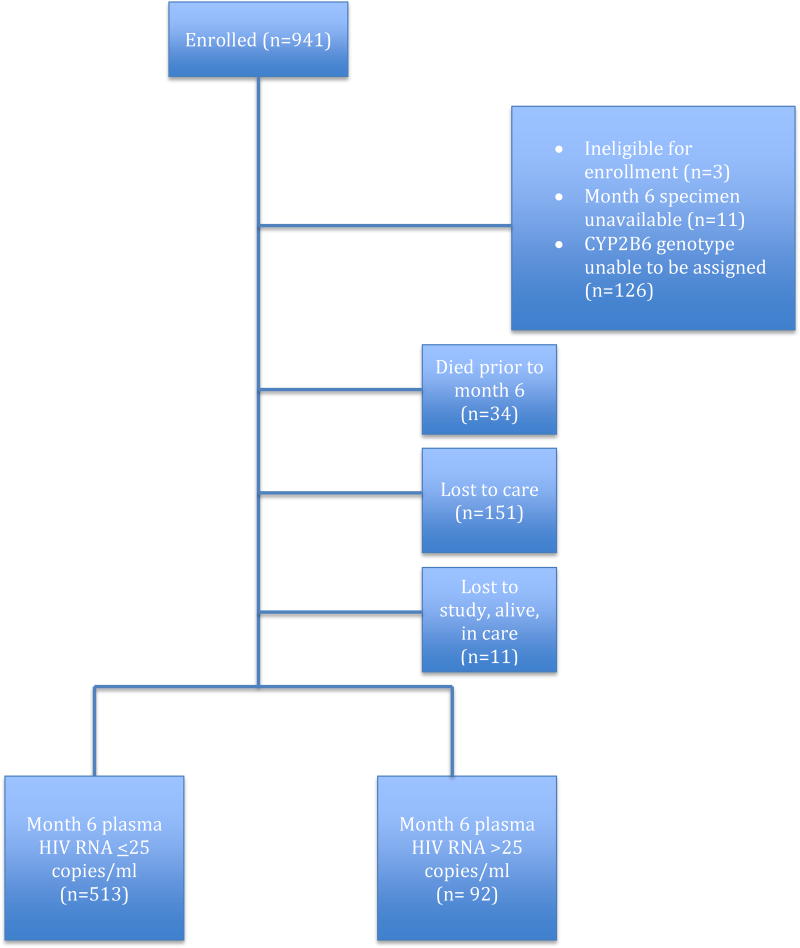
Beginning in June 2009 and ending in February 2013, we enrolled 941 individuals with follow-up ending in November 2013. Figure 1 displays the disposition of the participants. We successfully extracted DNA on all participants, but full exposure and outcome data were available in 801 individuals. Of the 128 participants excluded from all analyses due to inability to reliably classify CYP2B6 metabolism genotype, 45 samples showed no target amplification and in the remaining samples, there was insufficient allelic discrimination for the genotype to be determined by the automatic call algorithm. There were no apparent differences in baseline characteristics between those who were and were not successfully genotyped (see Supplemental Table 1). Plasma HIV RNA was unavailable in 11 participants who were excluded from the analysis of the composite endpoint and the secondary endpoint of viremia, but included in the secondary analyses of death and loss to care. The 801 individuals included 406 (51%) males, median age 37 years (ranging from 21 to 67), median baseline CD4 count 195 cells/mm3 (IQR 112, 256) and plasma HIV RNA 4.9 log10 copies/ml (IQR 4·2, 5·4). Table 1 displays the baseline characteristics of the 801 participants included in the primary analyses. Males were at higher risk of treatment failure. Median baseline CD4 counts were lower and plasma HIV RNA were higher in those meeting the endpoint than those in study and suppressed at 6 months. Tenofovir disoproxil fumarate (TDF) and emtricitabine were the nucleos(t)ide analog drugs in 769 (96%), with 24 (3%) on zidovudine and lamivudine, 2 (<1%) on stavudine and lamivudine, and 6 (1%) on an unknown combination of nucleos(t)ide analogs.
Figure 1.
Disposition of Study Participants for Primary Analysis
Table 1.
Demographic and Clinical Characteristics at Baseline by Outcome Group
| Characteristic | Confirmed HIV suppression at 6 months n=513 |
Composite failure by 6 months n=288 |
p value |
|---|---|---|---|
| Median age (range) | 37 (21, 67) | 38(23, 65) | <0·02 |
| Male sex | 233 (45%) | 173 (60%) | <0·001 |
| Median baseline CD4 count (cells/mm3) (interquartile range) | 207 (131, 269) | 169 (89, 243) | <0·001 |
| Median baseline plasma HIV RNA (log10 copies/ml) (interquartile range) | 4·8 (4·1, 5·3) | 5·0 (4·3, 5·5) | <0·001 |
| History of tuberculosis | 19 (4%) | 13 (5%) | >0·5 |
Of the 801 included in the primary analyses, 288 (36%) met the composite endpoint with 34 (4%) who died, 151 (19%) lost to care, 11 (1%) lost to the study, but alive and in care, and 92 (11%) with detectable plasma viremia. In the individuals with viremia, the level was low with a median of 85 copies/ml (interquartile range 45, 160 copies/ml). Eleven (12%) of those with viremia had plasma HIV RNA>1000 copies/ml.
CYP2B6 Polymorphisms and Efavirenz Clearance
788 participants had 1 and 633 had 2 plasma efavirenz specimens available at either month 1 or month 6 and were included in the efavirenz population pharmacokinetic analysis. All models were developed using FOCE-I estimation, and the final model was minimized with successful execution of the covariance step. Final parameter estimates and inter-individual variability are represented in Table 3. There was a dose-response relationship between the joint genotype categories and clearance and individuals were characterized as extensive metabolizers if they were homozygous for the CYP2B6 native allele at both sites (516 G,G and 983 T,T) and intermediate metabolizers if they were heterozygous for one of the SNPs, but not both (516 G,T or 983 T,C). All others were categorized as slow metabolizers. Of the 13 individuals with efavirenz concentrations below the limit of quantification, 10 were successfully genotyped with a distribution of 2 extensive, 6 intermediate, and 2 slow metabolizers.
Table 3.
Efavirenz Population Pharmacokinetic Model Parameters
| a. Fixed-Effect Parameters | ||
|---|---|---|
| Parameter* | Point Estimate | Standard Error % |
| θCL/F (L/hr) | 11.2 | 2·17 |
| θVd (L) | 150 | NA |
| θQ (hr−1) | 0·3 | NA |
| Impact of 516GT variant on CL/F | 0.697 | 1.9 |
| Impact of 516TT variant on CL/F | 0.354 | 1.5 |
| Impact of 983TC variant on CL/F | 0.587 | 2.3 |
| Impact of 983TT variant on CL/F | 0.180 | 1.4 |
| b. Variability Parameters | |
|---|---|
| Parameter | Estimate |
| Inter-individual (ω2CL) | 40.5 %CV |
| Residual, proportional (σ2proportional) | 0·0831 %CV |
| Residual, additive (σ2additive) | 0·0699 SD |
PK parameters are reported for a typical 70kg individual with no slow metabolism variants
CYP2B6 Polymorphisms and Endpoints
Metabolism variant alleles were common with 396 (49%) intermediate and 192 (24%) slow metabolizers. In unadjusted analyses, there were no statistically significant associations between metabolism and either the composite or secondary endpoints (Table 2). There was no apparent confounding of any of the associations but, analyses adjusted for log10 baseline HIV viral load are included in Table 2. Only 13 individuals were treated with isoniazid during the study and excluding them had no substantive effect on the results (data not shown). Of the 11 individuals with plasma HIV RNA>1000 copies/ml, the genotypes were 1 (9%) extensive, 7 (64%) intermediate, and 3 (27%) slow.
Table 2.
Association between Efavirenz Metabolism Genotypes* and Treatment Endpoints
| Endpoint (# observations) | Unadjusted OR (95% CI) |
p value |
† Adjusted OR (95% CI) |
p value |
|---|---|---|---|---|
| Composite Failure (n=801) |
1·13 (0·92, 1·39) | 0·23 | 1·13 (0.92, 1.39) | 0·25 |
| Virologic Failure (n=615) |
1·21 (0·88, 1·67) | 0·25 | 1·24 (0·89, 1·73) | 0·20 |
| Loss to Follow-Up (n=767) |
1·02 (0·79, 1·31) | >0·5 | 1·01 (0·79, 1·31) | >0·5 |
| Death (n=801) |
1·35 (0·83, 2·20) | 0·23 | 1·35 (0·83, 2·20) | 0·22 |
Metabolism included as ordinal variable at three levels: extensive (base case), intermediate and slow.
Adjusted for log10 baseline HIV RNA
Of the 450 (56%) participants who had both baseline and month 6 CD4 counts available, there were neither statistically nor clinically significant differences in the magnitude of CD4 increase by genotype. The extensive metabolizers had a median of 95 cells/mm3 increase (IQR 33, 189), intermediate metabolizers had a median of 92 cells/mm3 increase (IQR 42, 165), and the slow metabolizers had a median of 88 cells/mm3 increase (IQR 29, 168) with p values for all pairwise comparisons >0.5.
CYP2B6 Genotypes and Adverse Experiences
The results of the adverse experiences are displayed in Table 4. At month 1 of therapy, slower metabolism was associated with statistically significantly fewer adverse experiences. Scores on the Subject Experience Questionnaire were 1.16 points lower (95% CI 1.9 points lower, 0.40 points lower) and 0.66 points lower (95% CI 1.2 points lower, 0.14 points lower) for each slower metabolism category on the overall questionnaire and the CNS questions only, respectively at month 1. At month 6 of therapy, among those remaining in care, this association was of a lesser degree and no longer statistically significant. Scores on the Subject Experience Questionnaire 0.35 points lower (95% CI 0.84 points lower, 0.13 points higher) and 0.16 points lower (95% CI 0.47 points lower, 0.15 points higher) for each slower metabolism category on the overall questionnaire and the CNS questions only, respectively at month 6.
Table 4.
Subject Experience Questionnaire by Metabolism Genotype
| Total Adverse Experience Score, Mean (95% CI) | CNS Adverse Experience Score, Mean (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|
| Extensive | Inter-mediate | Slow | p value* | Extensive | Inter-mediate | Slow | P value* | |
| Month 1 | 5.2 (4.1, 6.2) |
4.6 (3.9, 5.4) |
2.8 (1.7, 3.9) |
<0.01 | 3.0 (2.3, 3.7) |
2.8 (2.3, 3.3) |
1.6 (0.9, 2.4) |
<0.02 |
| Month 6 | 2.3 (1.7, 3.0) |
2.1, (1.6, 2.5) |
1.6 (0.9, 2.3) |
0.34 | 1.3 (0.9, 1.7) |
1.2 (0.9, 1.5) |
0.9 0.5, 1.4) |
0.55 |
for differences between groups
Discussion
In this large population of HIV-infected adults initiating ART in routine clinical practice in Botswana, we found that polymorphisms in CYP2B6 516G>T and 983T>C were associated with decreased efavirenz clearance and less efavirenz-related toxicity, but not differences in composite virologic endpoints. In addition, we established that the additive allele-dose relationship between CYP2B6 516G>T and 983T>C and efavirenz clearance seen in other settings [7, 22] holds in this population. The genotype-specific clearance estimates as presented here are in line with other studies in Africa [23, 24], but in contrast to findings in African-Americans.
We observed an unexpected inverse relationship between efavirenz metabolism and efavirenz-related adverse effects. Evidence in the literature regarding the association between efavirenz metabolism alleles, efavirenz concentrations, and toxicities has been conflicting. In contrast to our observation, the earliest studies of efavirenz clearance and CNS toxicity by Haas et al and Rotger et al reported that slow efavirenz metabolizing genotypes were associated with increased incidence of CNS toxicity [22, 25]. Notably, these studies were conducted predominantly in white patients, whereas most studies in patients of African ancestry have failed to substantiate this association. For example, Mukonzo found that CYP2B6 516/983 genotype was not associated with sleep disorders or hallucinations by the second week of therapy in 197 Ugandans [24]. Ribaudo found a similar lack of association between CYP2B6 genotype and toxicity in people of African origin [5]. Sarfo et al showed in 299 Ghanaian patients that had a very high prevalence of 516G>T genotype, a particularly low incidence of neuropsychiatric toxicities (9·4%)[26]. In contrast, Gounden et al however did report CYP2B6 516G>T genotype to be associated with adverse events in a study of 80 patients of African origin in South Africa [27]. A potential explanation for the conflicting findings include polymorphisms in genes other than CYP450 mitigating CNS toxicity in people of African origin [28, 29]. Another potential explanation may be that 8-hydroxy efavirenz generated by CYP2B6 is the primary contributor to efavirenz’s known CNS adverse effects [30]. Thus, slow efavirenz metabolizers of African origin may have lower levels of downstream toxic metabolites. Consistent with this, levels of this metabolite have been found to be lower in the plasma [31] and cerebrospinal fluid of slow metabolizers [32] although other evidence does not support this hypothesis [33].
The lack of effect of efavirenz metabolism on virologic outcome we observed adds to an existing body of conflicting evidence. For example, Ribaudo et al. found the 516G>T slow metabolism allele to be protective against virologic failure in African American patients, but not in Caucasians and Hispanics whereas Haas’s et al. study lacked any associations between CYP2B6 genotype and virologic failure in Haiti [5, 10]. Our results should help allay concerns that efavirenz might not be a good drug choice in places where slow metabolism genotype is common due to concerns for abandonment of therapy by slow metabolizers.
Despite its strengths of large size, a racially homogenous group, and being conducted in a routine care setting, this study has limitations. Although we expended great effort at finding individuals lost to care, we do not know their ultimate disposition. Yet, other studies have demonstrated that many of these individuals have poor outcomes [34]. Because we lacked a precise measure of adherence, such as microelectronic monitors [35], we were unable to determine whether adherence was impacted by the CNS toxicity in the extensive metabolizers. We also lacked data on cognitive functioning in relation to the CNS adverse effects reported. Finally, this study is limited to the impact of efavirenz metabolism on relatively short-term outcomes despite antiretroviral therapy remaining lifelong for the foreseeable future.
In conclusion, we found that variants in CYP2B6 SNPs at positions 516 and 983 resulted in differences in the rate of efavirenz clearance with slower metabolism conferring lower rates of adverse effects. While it did not impact short term virological outcomes, identification of the cause of increased toxicity in the setting of greater clearance may provide novel insights into the mechanism by which drugs and their metabolites cause CNS adverse effects.
Supplementary Material
Acknowledgments
Financial Support
We thank the medical staff at the Bontleng, Broadhurst 3, Broadhurst Traditional Area, Morwa, Nkoyaphiri, Phase II, and Village Infectious Diseases Care Clinics for their assistance with carrying out this study. We thank the Ministry of Health of Botswana for supporting the project and the patients who participated. We also thank Marc Gastonguay PhD for his advice on the pharmacokinetics analyses.
The study was funded by the U.S. National Institute of Mental Health (R01 MH080701 and the Penn Mental Health AIDS Research Center P30 MH097488) with supplemental funding by the National Institute of Allergy and Infectious Diseases (Penn Center for AIDS Research P30 AI045008). The funding agencies had no input on the analysis or interpretation of results or writing or editing of the manuscript.
Footnotes
The authors declare no conflicts of interest.
Authors’ Contributions
Study Design: RG, SLB, RA, MM, APS, BLS, GPB
Data Collection: BR
Data Analysis: RG, SLB, XH, MV, RA, GM, AFZ
Interpretation of Results: RG, SLB, MM, MV, RA, APS, GM, AFZ, BLS, GPB
Drafting of Manuscript: RG
Editing of Manuscript: SLB, BR, XH, MM, MV, RA, APS, GM, AFZ, BLS, GPB
References
- 1.Johnson LF, Mossong J, Dorrington RE, Schomaker M, Hoffmann CJ, Keiser O, et al. Life expectancies of South African adults starting antiretroviral treatment: collaborative analysis of cohort studies. PLoS Med. 2013;10:e1001418. doi: 10.1371/journal.pmed.1001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills EJ, Bakanda C, Birungi J, Chan K, Ford N, Cooper CL, et al. Life expectancy of persons receiving combination antiretroviral therapy in low-income countries: a cohort analysis from Uganda. Ann Intern Med. 2011;155:209–216. doi: 10.7326/0003-4819-155-4-201108160-00358. [DOI] [PubMed] [Google Scholar]
- 3.Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. Geneva: World Health Organization; 2015. [PubMed] [Google Scholar]
- 4.Gunthard HF, Aberg JA, Eron JJ, Hoy JF, Telenti A, Benson CA, et al. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312:410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 5.Ribaudo HJ, Liu H, Schwab M, Schaeffeler E, Eichelbaum M, Motsinger-Reif AA, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010;202:717–722. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leger P, Dillingham R, Beauharnais CA, Kashuba AD, Rezk NL, Fitzgerald DW, et al. CYP2B6 variants and plasma efavirenz concentrations during antiretroviral therapy in Port-au-Prince, Haiti. J Infect Dis. 2009;200:955–964. doi: 10.1086/605126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas DW, Gebretsadik T, Mayo G, Menon UN, Acosta EP, Shintani A, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African americans. J Infect Dis. 2009;199:872–880. doi: 10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez Martin A, Cabrera Figueroa S, Cruz Guerrero R, Hurtado LP, Hurle AD, Carracedo Alvarez A. Impact of pharmacogenetics on CNS side effects related to efavirenz. Pharmacogenomics. 2013;14:1167–1178. doi: 10.2217/pgs.13.111. [DOI] [PubMed] [Google Scholar]
- 9.Clifford DB, Evans S, Yang Y, Acosta EP, Goodkin K, Tashima K, et al. Impact of efavirenz on neuropsychological performance and symptoms in HIV-infected individuals. Ann Intern Med. 2005;143:714–721. doi: 10.7326/0003-4819-143-10-200511150-00008. [DOI] [PubMed] [Google Scholar]
- 10.Haas DW, Severe P, Jean Juste MA, Pape JW, Fitzgerald DW. Functional CYP2B6 variants and virologic response to an efavirenz-containing regimen in Port-au-Prince, Haiti. J Antimicrob Chemother. 2014;69:2187–2190. doi: 10.1093/jac/dku088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger M, Spycher BD, Sidle J, Weigel R, Geng EH, Fox MP, et al. Correcting mortality for loss to follow-up: a nomogram applied to antiretroviral treatment programmes in sub-Saharan Africa. PLoS Med. 2011;8:e1000390. doi: 10.1371/journal.pmed.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.What to Start: Initial Combination Regimens for the Antiretroviral-Naive Patient. US Department of Health and Human Services; 2016. Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. [Google Scholar]
- 13.Gross R, Aplenc R, Tenhave T, Foulkes AS, Thakur R, Mosepele M, et al. Slow efavirenz metabolism genotype is common in Botswana. J Acquir Immune Defic Syndr. 2008;49:336–337. doi: 10.1097/QAI.0b013e31817c1ed0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bisson GP, Gaolathe T, Gross R, Rollins C, Bellamy S, Mogorosi M, et al. Overestimates of survival after HAART: implications for global scale-up efforts. PLoS One. 2008;3:e1725. doi: 10.1371/journal.pone.0001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava P, Moorthy GS, Gross R, Barrett JS. A sensitive and selective liquid chromatography/tandem mass spectrometry method for quantitative analysis of efavirenz in human plasma. PLoS One. 2013;8:e63305. doi: 10.1371/journal.pone.0063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand J, Verstuyft C, Chou M, Borand L, Chea P, Nay KH, et al. Dependence of efavirenz- and rifampicin-isoniazid-based antituberculosis treatment drug-drug interaction on CYP2B6 and NAT2 genetic polymorphisms: ANRS 12154 study in Cambodia. J Infect Dis. 2014;209:399–408. doi: 10.1093/infdis/jit466. [DOI] [PubMed] [Google Scholar]
- 17.Allegaert K, Anderson BJ. Interindividual variability of aminoglycoside pharmacokinetics in preterm neonates at birth. Eur J Clin Pharmacol. 2006;62:1011–1012. doi: 10.1007/s00228-006-0204-1. [DOI] [PubMed] [Google Scholar]
- 18.Holford N, Heo YA, Anderson B. A pharmacokinetic standard for babies and adults. J Pharm Sci. 2013;102:2941–2952. doi: 10.1002/jps.23574. [DOI] [PubMed] [Google Scholar]
- 19.Anderson BJ, McKee AD, Holford NH. Size, myths and the clinical pharmacokinetics of analgesia in paediatric patients. Clin Pharmacokinet. 1997;33:313–327. doi: 10.2165/00003088-199733050-00001. [DOI] [PubMed] [Google Scholar]
- 20.West GB, Brown JH, Enquist BJ. The fourth dimension of life: fractal geometry and allometric scaling of organisms. Science. 1999;284:1677–1679. doi: 10.1126/science.284.5420.1677. [DOI] [PubMed] [Google Scholar]
- 21.Byar DP, Piantadosi S. Factorial designs for randomized clinical trials. Cancer Treat Rep. 1985;69:1055–1063. [PubMed] [Google Scholar]
- 22.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics. 2005;15:1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Nyakutira C, Roshammar D, Chigutsa E, Chonzi P, Ashton M, Nhachi C, et al. High prevalence of the CYP2B6 516G–>T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur J Clin Pharmacol. 2008;64:357–365. doi: 10.1007/s00228-007-0412-3. [DOI] [PubMed] [Google Scholar]
- 24.Mukonzo JK, Okwera A, Nakasujja N, Luzze H, Sebuwufu D, Ogwal-Okeng J, et al. Influence of efavirenz pharmacokinetics and pharmacogenetics on neuropsychological disorders in Ugandan HIV-positive patients with or without tuberculosis: a prospective cohort study. BMC Infect Dis. 2013;13:261. doi: 10.1186/1471-2334-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–2400. [PubMed] [Google Scholar]
- 26.Sarfo FS, Zhang Y, Egan D, Tetteh LA, Phillips R, Bedu-Addo G, et al. Pharmacogenetic associations with plasma efavirenz concentrations and clinical correlates in a retrospective cohort of Ghanaian HIV-infected patients. J Antimicrob Chemother. 2014;69:491–499. doi: 10.1093/jac/dkt372. [DOI] [PubMed] [Google Scholar]
- 27.Gounden V, van Niekerk C, Snyman T, George JA. Presence of the CYP2B6 516G> T polymorphism, increased plasma Efavirenz concentrations and early neuropsychiatric side effects in South African HIV-infected patients. AIDS Res Ther. 2010;7:32. doi: 10.1186/1742-6405-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS. 2009;23:2101–2106. doi: 10.1097/QAD.0b013e3283319908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cummins NW, Neuhaus J, Chu H, Neaton J, Wyen C, Rockstroh JK, et al. Investigation of Efavirenz Discontinuation in Multi-ethnic Populations of HIV-positive Individuals by Genetic Analysis. EBioMedicine. 2015;2:706–712. doi: 10.1016/j.ebiom.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tovar-y-Romo LB, Bumpus NN, Pomerantz D, Avery LB, Sacktor N, McArthur JC, et al. Dendritic spine injury induced by the 8-hydroxy metabolite of efavirenz. J Pharmacol Exp Ther. 2012;343:696–703. doi: 10.1124/jpet.112.195701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngaimisi E, Mugusi S, Minzi O, Sasi P, Riedel KD, Suda A, et al. Effect of rifampicin and CYP2B6 genotype on long-term efavirenz autoinduction and plasma exposure in HIV patients with or without tuberculosis. Clin Pharmacol Ther. 2011;90:406–413. doi: 10.1038/clpt.2011.129. [DOI] [PubMed] [Google Scholar]
- 32.Winston A, Amin J, Clarke A, Else L, Amara A, Owen A, et al. Cerebrospinal fluid exposure of efavirenz and its major metabolites when dosed at 400 mg and 600 mg once daily: a randomized controlled trial. Clin Infect Dis. 2015;60:1026–1032. doi: 10.1093/cid/ciu976. [DOI] [PubMed] [Google Scholar]
- 33.Aouri M, Barcelo C, Ternon B, Cavassini M, Anagnostopoulos A, Yerly S, et al. In Vivo Profiling and Distribution of Known and Novel Phase I and Phase II Metabolites of Efavirenz in Plasma, Urine, and Cerebrospinal Fluid. Drug Metab Dispos. 2016;44:151–161. doi: 10.1124/dmd.115.065839. [DOI] [PubMed] [Google Scholar]
- 34.Brinkhof MW, Pujades-Rodriguez M, Egger M. Mortality of patients lost to follow-up in antiretroviral treatment programmes in resource-limited settings: systematic review and meta-analysis. PLoS One. 2009;4:e5790. doi: 10.1371/journal.pone.0005790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross R, Bilker WB, Friedman HM, Strom BL. Effect of adherence to newly initiated antiretroviral therapy on plasma viral load. AIDS. 2001;15:2109–2117. doi: 10.1097/00002030-200111090-00006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.