Abstract
Fluorescent cell barcoding (FCB) is a cell-based multiplexing technique for high-throughput flow cytometry. Barcoded samples can be stained and acquired collectively, minimizing staining variability and antibody consumption, and decreasing required sample volumes. Combined with functional measurements, FCB can be used for drug screening, signaling profiling, and cytokine detection, but technical issues are present. We optimized the FCB technique for routine utilization using DyLight 350, DyLight 800, Pacific Orange, and CBD500 for barcoding six, nine, or 36 human peripheral blood specimens. Working concentrations of FCB dyes ranging from 0 to 500 μg/ml were tested, and viability dye staining was optimized to increase robustness of data. A five-color staining with surface markers for Vβ usage analysis in CD4+ and CD8+ T cells was achieved in combination with nine sample barcoding. We provide improvements of the FCB technique that should be useful for multiplex drug screening and for lymphocyte characterization and perturbations in the diagnosis and during the course of disease.
Keywords: flow cytometry, fluorescent cell barcoding, phenotyping, viability dye assay
Introduction
Flow cytometry, a laser-based technology enabling simultaneous multiparametric analysis at the single-cell level, is routinely used in research and clinical diagnosis for cell, protein, and functional analysis. Fluorescent cell barcoding (FCB) allows high-throughput multiplexed assays, combining samples from one or more donors, minimizing staining variability, antibody consumption, and decreasing required sample volumes (1,2). FCB is based on the use of an N-hydroxysuccinimide (NHS)-derived reactive form of a fluorophore (FCB dye) which covalently binds the amine functional group of lysine side chains and N-terminus of protein (3). Using different dye concentrations and combinations, each sample acquires a unique fluorescent signature (barcode), based on fluorescence intensity and cytoplasmic complexity. For these reasons, different samples acquired together can be analyzed individually, because every barcoded population has a unique position on dot plot, according to fluorophore intensity and side-scattered light (SSC) (1–4). Others have reported barcoding optimization for four to 96 samples, using DyLight 350, Pacific Orange, DyLight 800, and Pacific Blue and/or AF488 at various working concentrations of individual dyes. These variations are related to the different efficacy to bind the amine functional group by FCB dyes, based on cell types and excitation wavelengths of dyes (1–4).
With the single-cell analysis, attention must be given to quantification of cell-to-cell variation in gene and protein expressions, and standardization efforts are made to model and measure such variability (5,6). FCB has been developed for single-cell phospho-specific flow cytometry (phosphoflow) in order to measure the phosphorylation status of intracellular proteins (7,8) for drug screening (4) and signaling profiling (1,9), but FCB also can be used for detection of intracellular cytokines (2,10). Here, we apply the FCB technique to routine immunophenotyping of human peripheral blood cells, but optimization is required to minimize the potential “spill-over” of one barcoded sample to another by choosing the best combination of dyes according to instrument configuration, the number of samples, and fluorophores (1,11).
Materials and Methods
Human samples
Heparinized and EDTA whole blood was collected from healthy donors (n=18; 10M/8F; mean age, 35 years old) after informed consent was obtained in accordance with the Declaration of Helsinki (12) and protocols approved by the National Heart, Lung, and Blood Institute (NHLBI) Institutional Review Board (National Institutes of Health, Bethesda, MD, USA). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque gradient centrifugation (MP Biomedicals, LLC, Santa Ana, CA, USA), according to manufacturer’s instructions. Cells were frozen in medium containing 50% FCS, 40% RPMI 1640, and 10% dymethyl-sulfoxide (DMSO, Sigma-Aldrich, St. Louis, MO, USA), and stored at −80°C until use.
Reagents
The following FCB dyes were used: CBD500 (BD Biosciences, San Jose, CA, USA); Pacific Orange NHS ester, DyLight 350 NHS ester, and DyLight 800 NHS ester (Thermo Fisher Scientific, Waltham, MA, USA). Antibodies tested for surface staining were: CD3-BV605 (OKT3) (BioLegend, San Diego, CA); CD4-APC (RPA-T4) (BD Biosciences, San Jose, CA, USA); CD8-PE-Cy5 (B9.11), and tube B of IOTest Beta Mark, containing Vβ 9-PE, Vβ 17-PE/FITC, and Vβ 16-FITC (FIN9, E17.5F3, and TAMAYA1.2) (Beckman Coulter, Miami, FL). LIVE/DEAD Fixable Aqua (a viability dye) for 405 nm excitation was used to exclude dead cells from analysis (Thermo Fisher Scientific). Aqua dye was dissolved in DMSO and stored at −80°C, according to the manufacturer’s instructions. Just before use, Aqua dye was diluted 1:16 with PBS and used for staining. All buffers (Phosflow Lyse/Fix Buffer 5X, Phosflow Perm Buffer II, and Phosflow Barcoding Wash Buffer 4X; BD Biosciences) were prepared, according to the manufacturer’s instructions.
Staining with FCB dyes
Each FCB dye was dissolved in DMSO at a final concentration of 500 μg/ml and stored at −80°C. Using the 500 μg/ml stock solution, FCB dyes were diluted with DMSO: 0, 1.56, 13, 50, 250, and 500 μg/ml. For barcoding, a final volume of 40 μl/well was used for each experiment. For single-dye FCB staining, 10 μl of each dye was combined with 6–7.5×105 cells/30 μl/well (final concentrations: 0, 0.39, 3.25, 12.5, 62.5, and 125 μg/ml). Using various combinations of two FCB dyes, 6–7.5×105 cells/30 μl/well were stained with 5 μl of each dye to have a final volume of 40 μl/well at a final concentration of each dye: 0, 0.195, 1.63, 6.25, 31.25, or 62.5 μg/ml.
After thawing, cells were suspended in PBS (3 ml) and centrifuged at 400g for 5 min, followed by aspiration of supernatant and then fixation with BD Phosflow Lyse/Fix Buffer (3 ml) for 15 min at room temperature (RT) (Supplemental Fig. 1). Following centrifugation, cell pellets were resuspended in cold BD Phosflow Perm Buffer II (2 ml), and incubated at 4°C for 20 min. After washing with PBS (2 ml), cells were resuspended in cold BD Phosflow Perm Buffer II to have 6–7.5×105 cells/30 μl. During permeabilization of cells, FCB dyes were diluted in a round-bottom 96-well plate, according to the designed matrix. Permeabilized PBMCs were added to individual wells with FCB dyes prepared in the prior step, and incubated at 4°C for 20 min in the dark. Subsequently, barcoded cells were combined and washed twice with BD Phosflow Barcoding Wash Buffer (3 ml) by centrifugation at 400g for 5 min, followed by resuspension with BD Phosflow Barcoding Wash Buffer (300 μl) (Supplemental Fig. 1). Single-color controls were processed in separate wells/tubes in a similar manner.
Combination staining with Aqua Viability and FCB dyes
On thawing, cells from eight healthy subjects were washed and resuspended in PBS. Aqua viability dye staining was performed by adding 3 μl of a diluted FCB dye to 100 μl of cell suspension and then incubated at RT for 20 min in the dark. After washing twice with PBS, cells stained with Aqua dye were resuspended in an appropriate volume of PBS to have 6–7.5×105 cells/30 μl. For an Aqua dye alone control, cells stained with Aqua dye were kept on ice until use after addition of PBS (300 μl). For combination staining with Aqua and FCB dyes, cells stained with Aqua dye were added to individual wells containing FCB dyes at appropriate concentrations for barcoding in a similar manner as described above. For FCB dye alone controls, PBMCs were directly barcoded without Aqua dye staining.
Combination staining with FCB dyes and antibodies
For combination staining with FCB dyes and antibodies, barcoding was performed using Combo 1 concentrations (0, 13, and 250 μg/ml) of DyLight 350 and DyLight 800. Cells (6–7.5×105/30 μl) from eight healthy subjects were added to each well with FCB dyes, barcoded as described, and stained with antibodies. Manufacturers’ instructions of IOTest Beta Mark were optimized as follows: 5 μl of CD3-BV605, 10 μl of CD4-APC, 10 μl of CD8-PE-Cy5, and 10 μl of Tube B of IOTest Beta Mark were added. Then, cells were washed with BD Phosflow Barcoding Wash Buffer (3 ml). For acquisition, cells were resuspended in 300 μl of the same buffer.
Data acquisition and analysis
Sample acquisition was implemented on a LSR Fortessa cytometer (BD Biosciences) equipped with ultraviolet (UV, 355 nm), violet (407 nm), blue (488 nm), green (532 nm), and red (633 nm) lasers, and BD FACSDiva software (v.8.0.1, BD Biosciences). Compensation was performed using a bead standard for each fluorochrome (anti-Mouse Ig, κ/Negative Control Compensation Particles Set, BD Biosciences) and barcoded cells with the highest concentration of each dye. Samples stained with the same FCB dyes and/or antibody combination were run using the same PMT voltages. A minimum of 10,000–30,000 lymphocytes was recorded. Post-acquisition compensation and flow cytometric analysis were performed using FlowJo software (v.10.0.7b, Treestar, Ashland, OR, USA). Lymphocytes were identified based on forward scatter area (FSC-A) vs SSC-A and doublet exclusion (FSC-A vs FSC-H), and then single cells were gated for FCB dye channels (Supplemental Fig. 2A). Additional gating strategies for viability dye and antibody staining are shown in Supplemental Fig. 2B–D.
Statistical analysis
Data were collected from a computerized database and analyzed using Prism (v.7.02; GraphPad software, Inc., La Jolla, CA). Fluorescence values from each FCB dye combination were reported as mean fluorescence intensity (MFI) and CV (CV = SD/mean of population). To determine FCB dye combinations with the best separation between each population, MFI fold change was calculated as follows: MFI fold increase = [MFIpeak2 − CVpeak2]/[MFIpeak1 + CVpeak1]. For every antibody and dye tested, percentages of positive lymphocyte populations and/or MFI values were reported for each barcoded population, and compared with matched non-barcoded controls by paired or unpaired t-test, and p<0.05 was considered statistically significant. For combination staining with FCB dyes and antibodies, percentages of positive cells and MFI values were converted in a color scale ranging from black (< mean in controls −2SD or > mean in controls +2SD) to yellow (within ±2SD). To check variability between barcoded samples and matched controls, a mean of positive cells or MFI values ±2SD was calculated from all barcoded samples within the same matrix and used as a reference range to define low (within the range) or high (out of the range) variability. In addition, a ratio between the mean of positive cells or MFI values from all barcoded samples within the same matrix and matched controls was used to improve the measurement of sample-to-sample variability. Values between 0.8 and 1.2 were considered within the range of acceptable variation. Each experiment included a minimum of three different healthy subjects.
Results
Single FCB dye staining
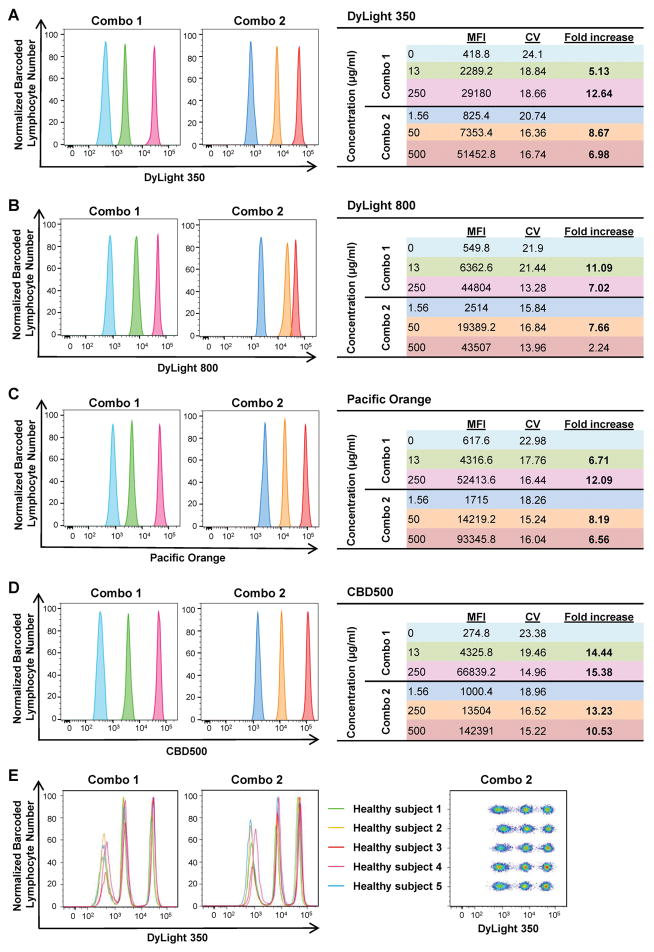
To assess barcoding efficiency using the same six working concentrations (0, 1.56, 13, 50, 250, and 500 μg/ml) for four FCB dyes chosen, six PBMC samples were barcoded individually and combined together in different combinations. Each condition was tested in five healthy donors. As shown in Supplemental Fig. 3, a clear separation between populations was achieved when MFI fold increase was ≥2. Therefore, samples were barcoded using two different FCB dye concentration sets: Combo 1 (0, 13, and 250 μg/ml) and Combo 2 (1.56, 50, and 500 μg/ml). Both dye combinations derived from DyLight 350, DyLight 800, Pacific Orange, or CBD500 displayed three sharp fluorescent peaks in single-parameter histograms, with minimum spill-over of each barcoded sample (MFI fold increase ranging from 2.24 to 15.38). Mean MFIs, CVs, and fold increase for each FCB dye working concentration resulting from all five healthy donors are also shown in Fig. 1.
Figure 1. Single-dye barcoding combinations.
PBMCs (6–7.5×105/30 μl/well) were stained individually in a 96-well plate with various concentrations of single FCB dyes. Two combinations of barcoded cells were prepared by mixing three barcoded lymphocyte populations with various concentrations of one dye: Combo 1 (0, 13, and 250 μg/ml) and Combo 2 (1.56, 50, and 500 μg/ml). Each dye was measured individually in single-parameter histograms, and fluorescent peaks were displayed using different colors based on concentrations. (A) DyLight 350; (B) DyLight 800; (C) Pacific Orange; and (D) CBD500. Mean MFIs, CVs, and fold increase from five different healthy subjects are shown for each concentration, according to the different combinations. Fold increase values ≥3 are reported in bold. (E) Combo 1 and 2 concentrations of DyLight 350 also are shown by concatenating all five subjects.
Nine- and 36-sample FCB staining
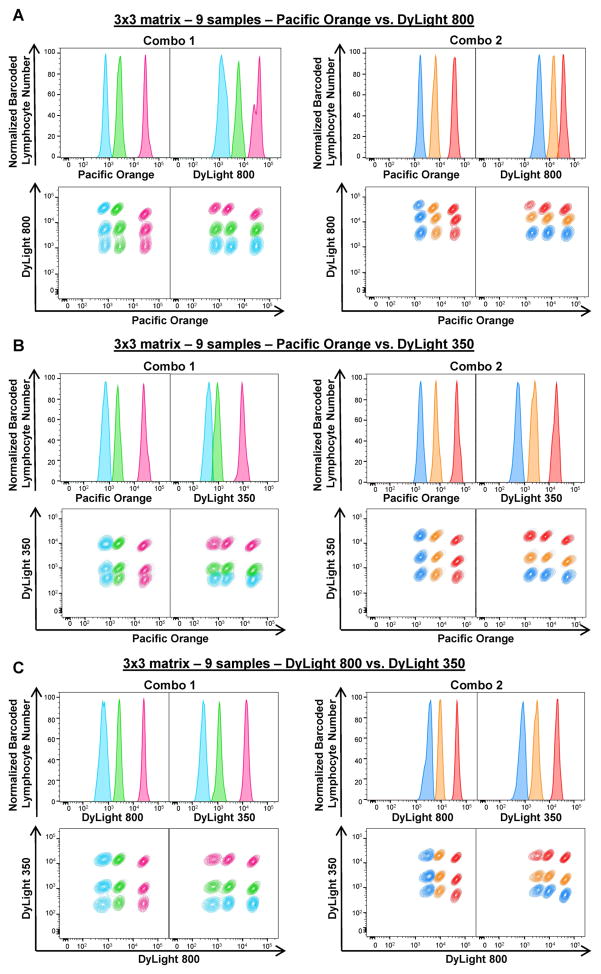
As Combo 1 and Combo 2 concentrations showed minimum spill-over of one barcoded sample into the next one, these combinations were tested to barcode nine or 36 samples, using two or three FCB dyes. To barcode nine samples together, 3×3 matrices were prepared using Combo 1 or Combo 2 concentrations of Pacific Orange plus DyLight 800 (Figs. 2A), Pacific Orange plus DyLight 350 (Figs. 2B), or DyLight 800 plus DyLight 350 (Fig. 2C). PBMCs were barcoded with nine different combinations of FCB dyes. Three sharp fluorescent peaks were observed in single-parameter histograms for each dye, showing apparent separation of cells stained with different FCB concentrations (Figs. 2A–C, top panels). When gated for both FCB dyes in Combo 1 or Combo 2, nine lymphocyte populations were detected (Figs. 2A–C, bottom panels). MFIs, CVs, and MFI fold increase for each combination are represented in Supplemental Fig. 4. Based on our results, Combo 1 and Combo 2 concentrations of DyLight 800 plus DyLight 350 and Combo 2 concentrations of Pacific Orange plus DyLight 350 produced the best deconvolution. These combinations were used for further experiments.
Figure 2. Two-dye barcoding combinations.
Two FCB dyes with different concentrations were combined as indicated and PBMCs were subjected to staining. To prepare a 3×3 matrix (9 samples), PBMCs (6–7.5×105/30 μl/well) were stained with FCB dye combinations: Combo 1 (left panels) or Combo 2 (right panels) concentrations of Pacific Orange plus DyLight 800 (A), Pacific Orange plus DyLight 350 (B), or DyLight 800 and DyLight 350 (C). Single-parameter histograms (Normalized barcoded lymphocyte number vs Dye, top rows) and linear properties (Dye X vs Dye Y, bottom rows) were used to visualize barcoded lymphocyte populations for each matrix. Fluorescent peaks in the histograms and gated lymphocyte populations were displayed in the same colors of the corresponding dye concentration.
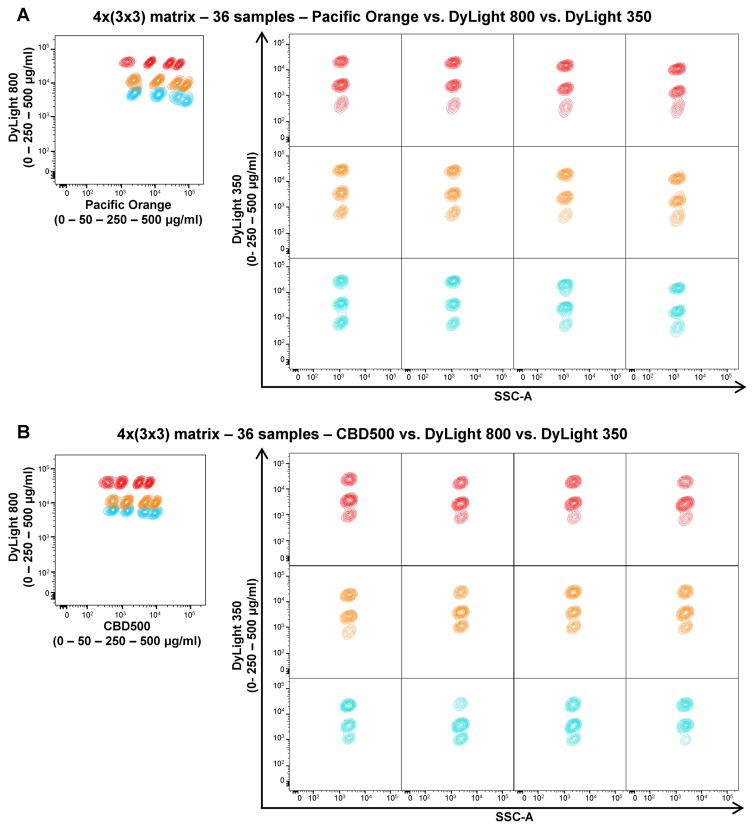
To assess the maximum number of samples that could be detected simultaneously, a 4×(3×3) matrix was designed using three different FCB dyes (CBD500 or Pacific Orange, DyLight 800, and DyLight 350) at various concentrations (Supplemental Fig. 5A). A total of 36 samples were barcoded. Twelve lymphocyte populations were gated using Pacific Orange or CBD500 vs DyLight 800 (Figs. 3A and B, left panels). On each of them, other three populations were displayed using DyLight 350 vs SSC-A for a total of 36 samples analyzed (Figs. 3A and B, right panels). In addition, MFIs, CVs, and MFI fold increase values were calculated for each dye (Supplemental Fig. 5B–C). Even if manual gating allowed the identification of all barcoded populations, MFI fold increase values for Pacific Orange and CBD500 between 250 and 500 μg/ml were lower than the cutoff for a clear separation (1.8 and 1.98, respectively). Also for DyLight 800 between 0 and 250 μg/ml concentrations, fold increase values were lower than 3 in both combinations.
Figure 3. Multiple-dye barcoding combinations for 36 samples.
Each of the following three FCB dyes with different concentrations was combined to make a 4×(3×3) matrix (36 samples): Pacific Orange or CBD500 (0, 50, 250, and 500 μg/ml); DyLight 800, and DyLight 350 (0, 250, and 500 μg/ml). Twelve barcoded populations were identified when gated for DyLight 800 and Pacific Orange (A) or CBD500 (B) (left panels). On each barcoded population, three DyLight 350-barcoded samples are shown (right panels), using the same color of the correspondent DyLight 800 concentration.
Viability dye staining
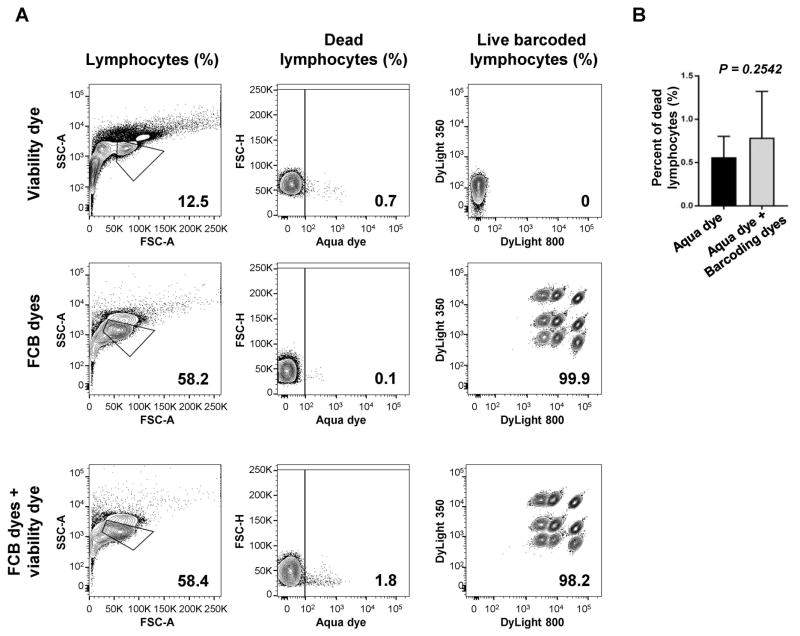
As viability dyes are routinely employed, we next examined whether their use could be combined with FCB dyes without interferences, as both bind the amine functional groups of proteins, and also because they require a different procedure for staining. LIVE/DEAD Fixable Aqua viability dye (405 nm excitation) was tested in combination with Combo 2 concentrations of DyLight 350 plus DyLight 800. Staining with Aqua dye alone or DyLight 350 plus DyLight 800 was also performed as controls (Fig. 4A). When compared to matched controls, no significant differences were observed in percentages of dead lymphocytes (controls [0.56%] vs FCB dyes plus Aqua dye [0.79%], p=0.2542) (Fig. 4B). Similarly, no different frequencies were seen between barcoded lymphocytes stained with Aqua dye plus FCB dyes and FCB dyes alone, resulting in nine clearly separated populations. However, when Aqua dye was used, dead lymphocytes were identified on barcoded populations.
Figure 4. Viability dye and FCB staining.
PBMCs were stained with Aqua viability dye, followed by barcoding with Combo 2 concentrations of DyLight 350 plus DyLight 800. As controls, cells were also stained with either Aqua viability dye or DyLight 350 plus DyLight 800 alone. A representative sample is provided (A). Numbers indicated are percentages of lymphocytes, dead lymphocytes, and barcoded live populations. No differences were described in percentage of dead lymphocytes between controls (n=8) and barcoded samples (n=8) (B). Paired t-test was performed and P<0.05 was considered statistically significant. Data are represented as mean±SD.
Co-staining with FCB dyes and antibodies
FCB has been used in combination with antibody staining for phosphoflow analysis and cytokine detection in T cell subpopulations (1–2,4,7). In our work, we sought to optimize a two-dye FCB staining for routine phenotyping and analyze Vβ usage in CD4+ and CD8+ T cells, as oligoclonal expansion of Vβ groups occurs in autoimmune and malignant diseases (13–15). Antibodies to surface markers were tested using Combo 1 concentrations of DyLight 800 plus DyLight 350 (Fig. 5, Supplemental Fig. 2C–D and 6 for compensation matrix). PBMCs were first barcoded, and then stained with antibodies. A matched sample without barcoding was used as a control for each donor in order to compare percentages of positive cells and confirm the integrity of the staining with and without barcoding (Supplemental Figs. 2C–D, and 7). Lymphocytes were identified as described and gated for CD3 expression. On CD3+ T cells, nine populations were displayed in DyLight 350 and DyLight 800 parameters. For each barcoded population, CD4 and CD8 expression levels were analyzed and subsequently gated by Vβ expression. Results of Vβ expression in CD8+ cells were subjected to further statistical analysis. At least 50,000 lymphocytes were acquired.
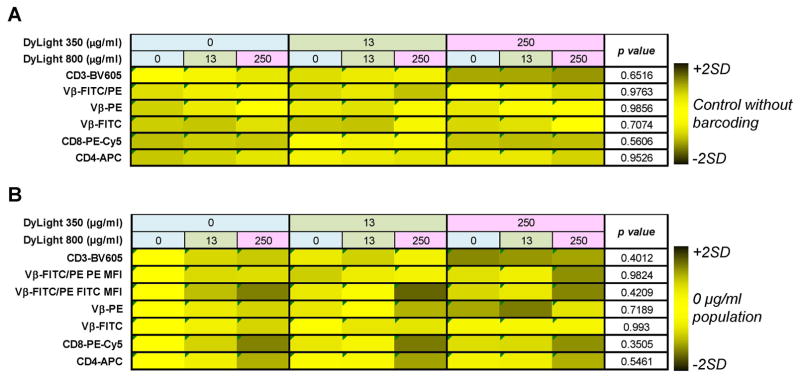
Figure 5. Co-staining cells with FCB dyes and antibodies.
PBMCs were stained with Combo 1 concentrations of DyLight 350 plus DyLight 800. Subsequently, barcoded specimens were stained with various antibodies for cell surface markers as indicated. Barcoded populations are shown according to dye combinations (upper rows). Heatmaps were generated using the mean of percentages of positive cell populations (A) and MFI values (B) for each antibody tested calculated from all barcoded populations. Values were compared to the mean of controls stained using PBS for percentages of positive cells (A) or to the mean of DyLight 350 (0 μg/ml) vs DyLight 800 (0 μg/ml) populations for MFI values (B). Then, values were converted in color scale ranging from black (MFI <mean of controls −2SD or >+2SD) to yellow (MFI within mean of controls ±2SD). Unpaired t-test was performed and p<0.05 was considered statistically significant.
No variations in percentages of positive cells were detected between barcoded samples and matched controls (p=0.6516 for CD3-BV605; p=0.9763 for Vβ-FITC/PE; p=0.9856 for Vβ-PE; p=0.7074 for Vβ-FITC; p=0.5606 for CD8-PE-Cy5; and p=0.9526 for CD4-APC) (Fig, 5A and Supplemental Fig. 7). Variability among barcoded samples within the same matrix was low in 92% of cases, with SD ranging from 0.2% to 11.4% (higher in CD3+ populations). Only in two cases, variations were simultaneously greater than ±2SD and out of the range of acceptable variations (Supplemental Fig. 7). In a similar way, MFIs of individual antibodies were analyzed and compared to MFI values of the DyLight 350 (0 μg/ml) vs DyLight 800 (0 μg/ml) population used as controls (Fig. 5B). No differences were found for BV605-, PE-Cy5-, APC-, FITC-, PE-, and FITC/PE-conjugated antibodies (p=0.4012, p=0.3505, p=0.5461, p=0.993, p=0.7189, p=0.9824, and p=0.4209, respectively). In 90% of cases, variability was low, while it was simultaneously high and out of the range of acceptable variation only in 4% of cases (Supplemental Fig. 8). MFI variations in FITC-, PE-, and APC-conjugated fluorochromes were detected when single barcoded populations were compared with those acquired together (p<0.0001, p=0.0301, and p=0.0133, respectively), likely due to compensation changes on barcoded populations acquired individually for PE- and APC-conjugated antibodies. Nevertheless, variations in MFI values of FITC-conjugated fluorochromes need more investigations, but by using proper matched controls, these variations can be corrected.
Discussion
FCB, a new flow cytometric technique, allows high-throughput multiplexed assays using reactive fluorophores (dyes). However, to implement FCB to routine staining for research and diagnostic purposes, several technical issues have to be considered (1,3). In our study, we optimized the technique for barcoding six, nine, or 36 human PBMC samples using four FCB dyes at the same working concentrations. In addition, the assay was combined with five-color antibody staining for T cell subpopulation detection and Vβ usage analysis.
Others have demonstrated optimization of barcoding for four to 96 samples (1,3). In particular, DyLight 350, Pacific Orange, and DyLight 800 were used for primary cell barcoding at various working concentrations of individual dyes (3). Combination staining of DyLight 800 and Pacific Orange is also optimized for barcoding in mass cytometry using transient partial permeabilization with 0.02% saponin (16). Further, Pacific Orange, Pacific Blue (2,9) and/or AF488 (1,4,9) can be combined for barcoding. However, AF488 does not allow the use of FITC- or AF488-conjugated antibodies, which are commonly chosen for routine staining (1,4,9). FCB dyes with far red wavelengths (DyLight 800 and AF750) can be combined with APC-, AF647-, or AF700-conjugated antibodies for staining (1–2,4,9,16). In the current work, we successfully barcoded six, nine, or 36 human PBMC samples with FCB dyes with UV (DyLight 350), violet (CBD500 and Pacific Orange), and red (DyLight 800) wavelengths. For nine-sample barcoding, three FCB-dye combinations (Pacific Orange plus DyLight 350, Pacific Orange plus DyLight 800, and DyLight 350 plus DyLight 800) were used with two different sets of concentrations. To barcode 36 samples, a combination of three dyes was made with four concentrations of CBD500 or Pacific Orange, and three of DyLight 350 and DyLight 800.
Each dye differs in efficacy in binding amine functional groups, depending on cell types and excitation wavelengths of dyes (1–4,9,16). Because higher concentrations of dyes increase the background fluorescence of unstained populations, likely due to residual non-reactive dye after washing (1), the use of small amounts of dyes (from 0 to 5 μg in our case) and further extensive washing of cells after staining are required (1–4,9). Also important for FCB optimization is the number of cells required for staining robustness (1). Krutzik and Nolan have demonstrated robust staining using 5×106 or 2×107 primary cells for six- or 96-sample barcoding, respectively. Stam et al. have reported optimization of the technique, based on blood volume (100 μl of whole blood), regardless of cell number (2). Considering various factors affecting barcoding procedures, we sought to develop standardized barcoding protocols using the same six concentrations (0, 1.56, 13, 50, 250, and 500 μg/ml) of four FCB dyes and further in combination with antibodies. In our study, these six concentrations were divided in two combination sets (Combo 1 and Combo 2), and 3.6–4.5×106 or 5.4–6.7×106 human PBMCs were used for six or nine sample barcoding, respectively. Thus, we successfully achieved optimization of our FCB staining, resulting in good separation of barcoded populations, with minimum spill-over between barcoded samples. Purity of deconvolution is defined as the distance between MFIs and CVs to obtain 70% of the respective barcoded population with 95% purity, and MFIs should be separated by a three-fold increase (1). In our work, human PBMCs displayed CVs of 8 – 28% using the same six concentrations for four FCB dyes tested. Barcoded populations were identified with high resolution when MFIs were separated by a three- or more fold- increase, using only three concentrations of FCB dyes, similarly to previous reports.
For barcoding, cells are fixed, permeabilized, and then combined, so that live and dead cells are barcoded and included in post-acquisition analysis. Dead cells can be removed with viability dye staining, as they acquire a more intense fluorescence, compared to live cells, due to exposure of surface and intracellular epitopes caused by damaged membranes (17). However, permeabilization is not required for viability dye staining, because permeabilization buffers impair membrane integrity in both live and dead cells. In our work, we optimized staining with viability dye prior barcoding to ensure high quality data. No significant variations were observed in percentages of live and barcoded populations, compared to matched controls, indicating that the viability dye staining did not interfere with the activity of FCB dyes. Therefore, combination staining with FCB and viability dyes allowed the exclusion of dead cells from gating strategy, minimizing non-specific binding.
The FCB technique has been developed for phosphoflow assay (1,3–4,9,18), but it can be also applied for immunophenotyping and intracellular cytokine detection (1–2,9), and in computational and system biology analyses (19,20). For staining optimization, there are several technical considerations. First, saponin-based buffer is preferred in epitopes known to be disrupted by methanol permeabilization (16). Second, the choice of FCB dyes should be adequate to avoid spill-over into neighboring fluorescent channels (3). Based on these considerations, five-color antibody staining for routine phenotyping and Vβ usage analysis in CD4+ and CD8+ T cells was tested with one set of concentrations of DyLight 350 plus DyLight 800, allowing the use of BV605-, FITC-, PE-, PE-Cy5, and APC- conjugated antibodies. When compared to matched not-barcoded controls, we did not observe any variations in percentages of positive cells and in MFI values as displayed by ratios and variabilities (Supplemental Figs. 7 and 8). However, some slight differences were seen between donors (donor HC-8 showed high variability and a higher ratio for CD4 and Vβ staining). However, FCB technique minimizes staining variability, but the quantification of technical and biological variations is important for better evaluation of gene and protein expression at single-cell levels (5). The variability described for some fluorochromes should be considered when experiments are run, for example, to not use the FITC channel for markers of interest or when results are reported as MFI values.
After optimization of FCB dye staining, we could perform combination staining with FCB dyes and antibodies for surface phenotyping or viability dye assays in as many as six samples from one or different donors. Moreover, our FCB methods in combination with viability dye and further antibody staining should be useful in various fields, such as in assessing diagnosis and prognosis of patients and in multiplexed drug screening or high-throughput technologies for system biology analysis.
Supplementary Material
Supplemental Figure 1. Graphical FCB protocol. After thawing, PBMCs were washed with PBS (3 ml) (w/wash), centrifuged at 400g for 5 min (
 ), and fixed with Phosflow Lyse/Fix Buffer (3 ml) for 15 min at room temperature (RT). Following centrifugation without (w/o) washing, cell pellets were permeabilized with cold Phosflow Perm Buffer II (2ml) and incubated at 4°C for 20 min. After washing and centrifugation, cells (6–7.5×105 cells/30 μl) were barcoded with 10 μl of FCB dye alone or in combination at various concentrations, followed by incubation at 4°C for 20 min in the dark. Subsequently, barcoded cells were combined and washed twice with Phosflow Barcoding Wash Buffer (3 ml) (w/FCB wash). After addition of antibodies, cells were incubated for 30 min at RT, washed with FCB wash, and centrifuged, followed by resuspension in 300 μl of the same buffer for acquisition.
), and fixed with Phosflow Lyse/Fix Buffer (3 ml) for 15 min at room temperature (RT). Following centrifugation without (w/o) washing, cell pellets were permeabilized with cold Phosflow Perm Buffer II (2ml) and incubated at 4°C for 20 min. After washing and centrifugation, cells (6–7.5×105 cells/30 μl) were barcoded with 10 μl of FCB dye alone or in combination at various concentrations, followed by incubation at 4°C for 20 min in the dark. Subsequently, barcoded cells were combined and washed twice with Phosflow Barcoding Wash Buffer (3 ml) (w/FCB wash). After addition of antibodies, cells were incubated for 30 min at RT, washed with FCB wash, and centrifuged, followed by resuspension in 300 μl of the same buffer for acquisition.
Supplemental Figure 2. Gating strategy and FCB matrix design. (A) Gating strategy used to identify barcoded lymphocytes populations. Lymphocytes were identified based on FSC-A vs SSC-A and doublet exclusion (FSC-A vs FSC-H), and then single cells were gated for FCB dye channels. (B) Gating strategy for dead cell discrimination. Lymphocytes were identified based on FSC-A vs SSC-A and then gated for Aqua dye vs FSC-H. Live cells were then displayed in FCB dye channels (DyLight 800 vs DyLight 350). Total dead cells were also gated on total events (Aqua dye vs FSC-H). Gating strategy for Vβ usage in CD4+ and CD8+ T cells in samples stained using PBS and without barcoding (C), and in barcoded samples (D). Lymphocytes were identified based on FSC-A and SSC-A and doublet exclusion (FSC-H vs FSC-A). For not barcoded samples, single cells were gated directly for CD3 expression (SSC-A vs CD3-BV605), otherwise, barcoded samples were first identified using FCB dye channels (DyLight 350 vs DyLight 800). On CD3+ cells, CD4+ and CD8+ lymphocytes were gated (CD8-PE-Cy5 vs CD4-APC), and then, on each population, Vβ usage was studied (Vβ-PE vs Vβ-FITC).
Supplemental Figure 3. Single-dye FCB staining for six samples. PBMCs (6–7.5×105/30 μl/well) were stained individually in a 96-well plate with six concentrations (0, 1.56, 13, 50, 250, and 500 μg/ml) of single FCB dyes, and then combined together. Each dye was measured individually in single-parameter histograms, and fluorescent peaks were displayed using different colors based on concentrations. (A) DyLight 350; (B) DyLight 800; (C) Pacific Orange; and (D) CBD500. Mean MFIs, CVs and fold increase are shown for each dye concentration, according to different combinations. Fold increase values ≥3 are reported in bold.
Supplemental Figure 4. MFIs and CVs from two-dye FCB combinations. PBMCs were combined with different concentrations of two dyes to barcode nine samples. Each dye was measured individually in single-parameter histograms, and mean MFIs and CVs were reported for each peak. Combo 1 (left panels) and Combo 2 (right panels) concentrations of Pacific Orange plus DyLight 800 (A), Pacific Orange plus DyLight 350 (B), or DyLight 800 plus DyLight 350 (C). Fold increase values ≥3 are reported in bold.
Supplemental Figure 5. MFIs, CVs and MFI fold increase values from three-dye FCB combinations. (A) Layout for designing a 4×(3×3) matrix for 36 sample acquisition. Four Pacific Orange-3×3 matrices (0, 50, 250, and 500 μg/ml) were prepared by adding each of four concentrations of Pacific Orange into 3 columns, respectively. Subsequently, combination of three concentrations (0, 250, and 500 μg/mL) of DyLight 800 and DyLight 350 were added into three rows, generating the 4×(3×3) matrix. Each dye was measured individually in single-parameter histograms, and mean MFIs, CVs and MFI fold increase values were reported for each peak of DyLight 350 plus DyLight 800 plus Pacific Orange (B), or DyLight 350 plus DyLight 800 plus CBD500 (C). Fold increase values ≥3 are reported in bold.
Supplemental Figure 6. Compensation matrix for five-color antibody staining made by FlowJo. Compensated sample is shown in black, while uncompensated in light blue.
Supplemental Figure 7. Percentages of positive cells from five-color antibody staining experiment were provided for each antibody and subject. For each table, barcoded populations are shown according to FCB dye concentrations (upper rows) and donor identification number (from HC-1 to HC-8, first column). Control values were obtained from matched controls of the same donor in the row, processed without FCB buffers and dyes (Control w/o FCB). Variability was measured as described in the text (Variability and Ratio columns). To test the efficiency of FCB, some samples were run with missing populations (missing values in the tables). Contour plots with correspondent populations are shown (left).
Supplemental Figure 8. MFIs from all data of five-color antibody staining experiment are provided for each antibody, in a similar manner shown in Supplemental Fig. 7, using DyLight 350 (0 μg/ml) vs DyLight 800 (0 μg/ml) population as controls.
Acknowledgments
The authors would like to thank Fernandez Ibanez M.D.P., Desierto M., Keyvanfar K., and Belkina N. for technical assistance and Broder K. for assistance in obtaining healthy volunteer samples. This research was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 2.Stam J, Abdulahad W, Huitema MG, Roozendaal C, Limburg PC, van Stuijvenberg M, Schölvinck EH. Fluorescent cell barcoding as a tool to assess the age-related development of intracellular cytokine production in small amounts of blood from infants. PLoS One. 2011;6:e25690. doi: 10.1371/journal.pone.0025690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krutzik PO, Clutter MR, Trejo A, Nolan GP. Fluorescent cell barcoding for multiplex flow cytometry. Curr Protoc Cytom. 2011;Chapter 6(Unit 6.31) doi: 10.1002/0471142956.cy0631s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spurgeon BE, Aburima A, Oberprieler NG, Taskén K, Naseem KM. Multiplexed phosphospecific flow cytometry enables large-scale signaling profiling and drug screening in blood platelets. J Thromb Haemost. 2014;12:1733–1743. doi: 10.1111/jth.12670. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Biancotto A, Cheung F, Remmers E, Shah N, McCoy JP, Tsang JS. Systematic Analysis of Cell-to-Cell Expression Variation of T Lymphocytes in a Human Cohort Identifies Aging and Genetic Associations. Immunity. 2016;45(5):1162–1175. doi: 10.1016/j.immuni.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Curr Opin Chem Biol. 2009;13(5–6):556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- 8.Krutzik PO, Crane JM, Clutter MR, Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 9.Kalland ME, Oberprieler NG, Vang T, Taskén K, Torgersen KM. T cell-signaling network analysis reveals distinct differences between CD28 and CD2 costimulation responses in various subsets and in the MAPK pathway between resting and activated regulatory T cells. J Immunol. 2011;187:5233–5245. doi: 10.4049/jimmunol.1101804. [DOI] [PubMed] [Google Scholar]
- 10.Prussin C, Metcalfe DD. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J Immunol Methods. 1995;188:117–128. doi: 10.1016/0022-1759(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 11.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 13.Risitano AM, Maciejewski JP, Green S, Plasilova M, Zeng W, Young NS. In-vivo dominant immune responses in aplastic anaemia: molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing. Lancet. 2004;364(9431):355–364. doi: 10.1016/S0140-6736(04)16724-X. [DOI] [PubMed] [Google Scholar]
- 14.Clemente MJ, Przychodzen B, Jerez A, Dienes BE, Afable MG, Husseinzadeh H, Rajala HL, Wlodarski MW, Mustjoki S, Maciejewski JP. Deep sequencing of the T-cell receptor repertoire in CD8+ T-large granular lymphocyte leukemia identifies signature landscapes. Blood. 2013;122(25):4077–4085. doi: 10.1182/blood-2013-05-506386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman CG, Yamaguchi R, Tamura K, Weidner J, Imoto S, Kwon J, Fang H, Yew PY, Marino SR, Miyano S, Nakamura Y, Kiyotani K. Characterization of T-cell Receptor Repertoire in Inflamed Tissues of Patients with Crohn’s Disease Through Deep Sequencing. Inflamm Bowel Dis. 2016;22(6):1275–1285. doi: 10.1097/MIB.0000000000000752. [DOI] [PubMed] [Google Scholar]
- 16.Behbehani GK, Thom C, Zunder ER, Finck R, Gaudilliere B, Fragiadakis GK, Fantl WJ, Nolan GP. Transient partial permeabilization with saponin enables cellular barcoding prior to surface marker staining. Cytometry A. 2014;85:1011–1019. doi: 10.1002/cyto.a.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M. Amine-reactive dyes for dead cell discrimination in fixed samples. Curr Protoc Cytom. 2010;Chapter 9(Unit 9.34) doi: 10.1002/0471142956.cy0934s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krutzik PO, Clutter MR, Nolan GP. Coordinate analysis of murine immune cell surface markers and intracellular phosphoproteins by flow cytometry. J Immunol. 2005;175:2357–2365. doi: 10.4049/jimmunol.175.4.2357. [DOI] [PubMed] [Google Scholar]
- 19.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, Moir S, Dickler HB, Perl S, Cheung F Baylor HIPC Center; CHI Consortium. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diggins KE, Ferrell PB, Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. doi: 10.1016/j.ymeth.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Graphical FCB protocol. After thawing, PBMCs were washed with PBS (3 ml) (w/wash), centrifuged at 400g for 5 min (
 ), and fixed with Phosflow Lyse/Fix Buffer (3 ml) for 15 min at room temperature (RT). Following centrifugation without (w/o) washing, cell pellets were permeabilized with cold Phosflow Perm Buffer II (2ml) and incubated at 4°C for 20 min. After washing and centrifugation, cells (6–7.5×105 cells/30 μl) were barcoded with 10 μl of FCB dye alone or in combination at various concentrations, followed by incubation at 4°C for 20 min in the dark. Subsequently, barcoded cells were combined and washed twice with Phosflow Barcoding Wash Buffer (3 ml) (w/FCB wash). After addition of antibodies, cells were incubated for 30 min at RT, washed with FCB wash, and centrifuged, followed by resuspension in 300 μl of the same buffer for acquisition.
), and fixed with Phosflow Lyse/Fix Buffer (3 ml) for 15 min at room temperature (RT). Following centrifugation without (w/o) washing, cell pellets were permeabilized with cold Phosflow Perm Buffer II (2ml) and incubated at 4°C for 20 min. After washing and centrifugation, cells (6–7.5×105 cells/30 μl) were barcoded with 10 μl of FCB dye alone or in combination at various concentrations, followed by incubation at 4°C for 20 min in the dark. Subsequently, barcoded cells were combined and washed twice with Phosflow Barcoding Wash Buffer (3 ml) (w/FCB wash). After addition of antibodies, cells were incubated for 30 min at RT, washed with FCB wash, and centrifuged, followed by resuspension in 300 μl of the same buffer for acquisition.
Supplemental Figure 2. Gating strategy and FCB matrix design. (A) Gating strategy used to identify barcoded lymphocytes populations. Lymphocytes were identified based on FSC-A vs SSC-A and doublet exclusion (FSC-A vs FSC-H), and then single cells were gated for FCB dye channels. (B) Gating strategy for dead cell discrimination. Lymphocytes were identified based on FSC-A vs SSC-A and then gated for Aqua dye vs FSC-H. Live cells were then displayed in FCB dye channels (DyLight 800 vs DyLight 350). Total dead cells were also gated on total events (Aqua dye vs FSC-H). Gating strategy for Vβ usage in CD4+ and CD8+ T cells in samples stained using PBS and without barcoding (C), and in barcoded samples (D). Lymphocytes were identified based on FSC-A and SSC-A and doublet exclusion (FSC-H vs FSC-A). For not barcoded samples, single cells were gated directly for CD3 expression (SSC-A vs CD3-BV605), otherwise, barcoded samples were first identified using FCB dye channels (DyLight 350 vs DyLight 800). On CD3+ cells, CD4+ and CD8+ lymphocytes were gated (CD8-PE-Cy5 vs CD4-APC), and then, on each population, Vβ usage was studied (Vβ-PE vs Vβ-FITC).
Supplemental Figure 3. Single-dye FCB staining for six samples. PBMCs (6–7.5×105/30 μl/well) were stained individually in a 96-well plate with six concentrations (0, 1.56, 13, 50, 250, and 500 μg/ml) of single FCB dyes, and then combined together. Each dye was measured individually in single-parameter histograms, and fluorescent peaks were displayed using different colors based on concentrations. (A) DyLight 350; (B) DyLight 800; (C) Pacific Orange; and (D) CBD500. Mean MFIs, CVs and fold increase are shown for each dye concentration, according to different combinations. Fold increase values ≥3 are reported in bold.
Supplemental Figure 4. MFIs and CVs from two-dye FCB combinations. PBMCs were combined with different concentrations of two dyes to barcode nine samples. Each dye was measured individually in single-parameter histograms, and mean MFIs and CVs were reported for each peak. Combo 1 (left panels) and Combo 2 (right panels) concentrations of Pacific Orange plus DyLight 800 (A), Pacific Orange plus DyLight 350 (B), or DyLight 800 plus DyLight 350 (C). Fold increase values ≥3 are reported in bold.
Supplemental Figure 5. MFIs, CVs and MFI fold increase values from three-dye FCB combinations. (A) Layout for designing a 4×(3×3) matrix for 36 sample acquisition. Four Pacific Orange-3×3 matrices (0, 50, 250, and 500 μg/ml) were prepared by adding each of four concentrations of Pacific Orange into 3 columns, respectively. Subsequently, combination of three concentrations (0, 250, and 500 μg/mL) of DyLight 800 and DyLight 350 were added into three rows, generating the 4×(3×3) matrix. Each dye was measured individually in single-parameter histograms, and mean MFIs, CVs and MFI fold increase values were reported for each peak of DyLight 350 plus DyLight 800 plus Pacific Orange (B), or DyLight 350 plus DyLight 800 plus CBD500 (C). Fold increase values ≥3 are reported in bold.
Supplemental Figure 6. Compensation matrix for five-color antibody staining made by FlowJo. Compensated sample is shown in black, while uncompensated in light blue.
Supplemental Figure 7. Percentages of positive cells from five-color antibody staining experiment were provided for each antibody and subject. For each table, barcoded populations are shown according to FCB dye concentrations (upper rows) and donor identification number (from HC-1 to HC-8, first column). Control values were obtained from matched controls of the same donor in the row, processed without FCB buffers and dyes (Control w/o FCB). Variability was measured as described in the text (Variability and Ratio columns). To test the efficiency of FCB, some samples were run with missing populations (missing values in the tables). Contour plots with correspondent populations are shown (left).
Supplemental Figure 8. MFIs from all data of five-color antibody staining experiment are provided for each antibody, in a similar manner shown in Supplemental Fig. 7, using DyLight 350 (0 μg/ml) vs DyLight 800 (0 μg/ml) population as controls.