Abstract
IMPORTANCE
The family of genes implicated in hereditary spastic paraplegias (HSPs) is quickly expanding, mostly owing to the widespread availability of next-generation DNA sequencing methods. Nevertheless, a genetic diagnosis remains unavailable for many patients.
OBJECTIVE
To identify the genetic cause for a novel form of pure autosomal dominant HSP.
DESIGN, SETTING, AND PARTICIPANTS
We examined and followed up with a family presenting to a tertiary referral center for evaluation of HSP for a decade until August 2014. Whole-exome sequencing was performed in 4 patients from the same family and was integrated with linkage analysis. Sanger sequencing was used to confirm the presence of the candidate variant in the remaining affected and unaffected members of the family and screen the additional patients with HSP. Five affected and 6 unaffected participants from a 3-generation family with pure adult-onset autosomal dominant HSP of unknown genetic origin were included. Additionally, 163 unrelated participants with pure HSP of unknown genetic cause were screened.
MAIN OUTCOME AND MEASURE
Mutation in the neuronal isoform of carnitine palmitoyl-transferase (CPT1C) gene.
RESULTS
We identified the nucleotide substitution c.109C>T in exon 3 of CPT1C, which determined the base substitution of an evolutionarily conserved Cys residue for an Arg in the gene product. This variant strictly cosegregated with the disease phenotype and was absent in online single-nucleotide polymorphism databases and in 712 additional exomes of control participants. We showed that CPT1C, which localizes to the endoplasmic reticulum, is expressed in motor neurons and interacts with atlastin-1, an endoplasmic reticulum protein encoded by the ATL1 gene known to be mutated in pure HSPs. The mutation, as indicated by nuclear magnetic resonance spectroscopy studies, alters the protein conformation and reduces the mean (SD) number (213.0 [46.99] vs 81.9 [14.2]; P < .01) and size (0.29 [0.01] vs 0.26 [0.01]; P < .05) of lipid droplets on overexpression in cells. We also observed a reduction of mean (SD) lipid droplets in primary cortical neurons isolated from Cpt1c−/− mice as compared with wild-type mice (1.0 [0.12] vs 0.44 [0.05]; P < .001), suggesting a dominant negative mechanism for the mutation.
CONCLUSIONS AND RELEVANCE
This study expands the genetics of autosomal dominant HSP and is the first, to our knowledge, to link mutation in CPT1C with a human disease. The association of the CPT1C mutation with changes in lipid droplet biogenesis supports a role for altered lipid-mediated signal transduction in HSP pathogenesis.
Hereditary spastic paraplegias (HSPs) are a group of neurological disorders with progressive spasticity and weakness of the lower limbs, likely a result of distal axonopathy in the corticospinal tract. Hereditary spastic paraplegias have been classified as pure or complicated on the basis of the absence or presence of associated clinical features, such as cognitive dysfunction, ataxia, and peripheral neuropathy.1 With nearly 75 distinct loci and more than 50 disease genes identified to date, the genetics of HSPs provide an opportunity for unraveling the requirements for corticospinal tract axon integrity and identifying convergent themes at the cellular level. The most common presentation of HSP is pure adult-onset autosomal dominant (AD-HSP).2 Mutations in SPAST (SPG4; OMIM #182601),3 ATL1 (SPG3A; OMIM #182600),4 and REEP1 (SPG31; OMIM #610250)5 account for almost 60% of AD-HSPs overall.6 The localization of the encoded proteins to the endoplasmic reticulum (ER), where proteins interact with one another and shape the tubular network, suggests that abnormalities in ER morphogenesis may be a common pathogenic theme.7,8 Which myriad functions of these proteins in the ER are most relevant for the pathogenesis of HSP remains an unanswered question.
In this study, we identified a mutation in the neuronal isoform of the carnitine palmitoyl-transferase (CPT1C) gene as the genetic cause of a pure AD-HSP, termed hereditary spastic paraplegia type 73 (SPG73).
Mammalian CPT1 is encoded by a family of 3 genes that have distinct properties and expression profiles. CPT1A and CPT1B are primarily expressed in the liver and muscle, respectively, where they control the transport of long-chain fatty acids from the cytoplasm to the mitochondrial matrix by catalyzing the conversion of long-chain fatty acyl-CoA into acylcarnitines.9 CPT1C is mainly expressed in neurons.10–12 It localizes to the ER and not to the mitochondria11 and has little or no enzymatic activity in fatty acid oxidation.10,13,14
In this study, we further characterize wild-type and mutant CPT1C by examining its structural properties by solution of nuclear magnetic resonance (NMR) spectroscopy. We show that CPT1C is expressed in motor neurons and interacts with atlastin-1, the protein encoded by the ATL1 gene. Similar to other HSPs,15–18 mutant CPT1C impairs the formation of lipid droplets (LDs), further supporting a role for altered lipid metabolism in HSP pathogenesis.
Methods
A family with hereditary spastic paraplegia of unknown genetic cause was examined and followed up at the Department of Neurosciences, Reproductive Sciences, and Odontostomatology at the University of Naples Federico II in Naples, Italy, for a decade until August 2014. Patients provided written consent for the study according to the Declaration of Helsinki and the study was approved by the Institutional Ethics Review Board of the University of Naples Federico II.
Genetic Analyses
Blood samples were collected and DNA was extracted using standard methods from peripheral blood lymphocytes from 5 affected individuals (II-6, III-2, III-3, III-4, and IV-1; Figure 1A) and 6 unaffected individuals (III-1, III-5, III-8, III-10, IV-2, and IV-3; Figure 1A). No neurological history was available for participant II-2, who was therefore marked as unknown (Figure 1A). Participant III-9 was not included in the analysis because he developed nonprogressive mild weakness in the lower limbs at 55 years of age, possibly ascribed to spondylosis. Libraries were prepared using NimbleGen SeqCap EZ Exome Library SR per the manufacturer’s protocol. Libraries were diluted in Qiagen Buffer EB (QIAquick PCR Purification Kit) to a concentration of 6.5pM. Exomes were prepared using the SeqCap EZ Human Exome Library version 2.0 and a 50–base pair paired-end run was performed on the HiSeq 2000 (Illumina). Sequence alignment, quality control, and variant calling were performed with BWA, SAMTools, the Genomic Analysis Toolkit, and Picard (http://picard.sourceforge.net/index.shtml). Sequencing was done (50 base pair, paired end) on the HiSeq2000, with each sample exome library on a single lane of a TruSeq version 2 flow cell (Illumina). Sequencing coverage was adequate for these samples (individual II-6, 94.1% of the exome was sequenced at 10 times depth and 89.3% was sequenced at 30 times depth; individual III-2, 93.7% at 10 times depth and 89.3% at 30 times depth; individual III-3, 94.6% at 10 times depth and 90.0% at 30 times depth; and individual IV-1, 94.6% at 10 times depth and 78.9 at 30 times depth). Data analysis was based on the autosomal dominant mode of inheritance and the hypothesis that the underlying mutation was not present in neurologically healthy control individuals or the general population. Variants not shared between all affected individuals in the family studied and variants that were nonautosomal, homozygous, and synonymous were excluded. Any variant reported in the SeattleSeq National Heart, Lung, and Blood Institute Exome Sequencing Project database (http://evs.gs.washington.edu/EVS/) was also excluded. Variants remaining after exome data analysis containing missing data were Sanger sequenced using the BigDye Terminator version 3.1 chemistry (Applied Biosystems), run on an ABI 3730xl analyzer, and analyzed using Sequencher software version 4.2 (Gene Codes). Parametric linkage analysis was performed using MERLIN version 1.1.219 with an autosomal dominant model.
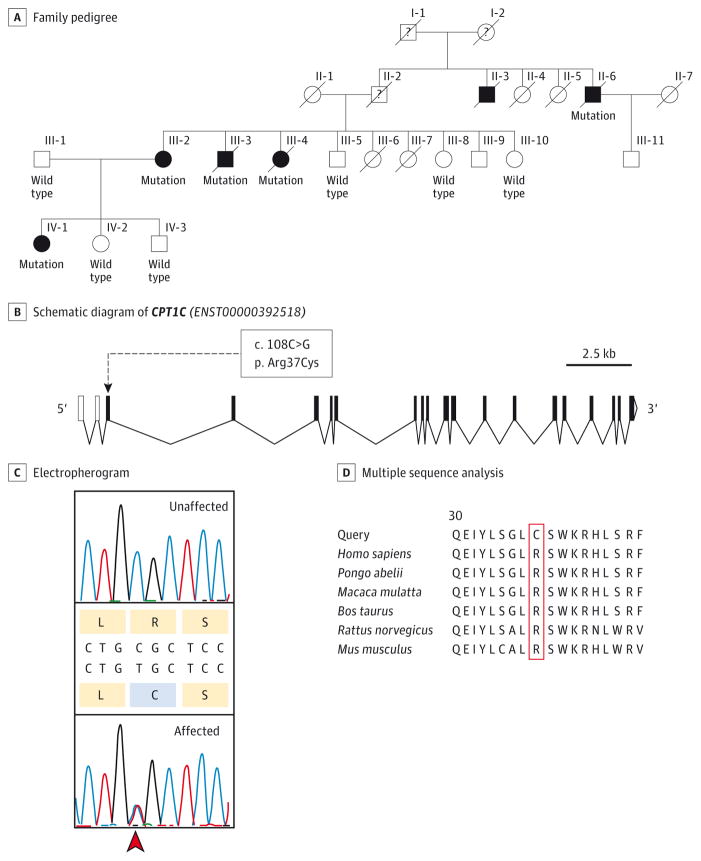
Figure 1. Pedigree and Sequence Analysis of CPT1C.
A, Pedigree of the family. White indicates unaffected individuals and black indicates affected individuals. Presence (mutation) or absence (wild type) of the mutation in CPT1C is indicated below each individual. B, Schematic diagram of CPT1C. Exons are represented as solid black boxes, noncoding exons are depicted as white boxes, and introns are depicted as bent connectors. The c.108C>G variant (p.Arg37Cys) is indicated. C, Electropherograms show the c.109C>T (p.Arg37Cys) variant (arrowhead) in CPT1C in an affected (heterozygote) and a nonaffected individual. D, Multiple sequence analysis of CPT1C in various species.
NMR Spectroscopy
The regulatory domain of human CPT1C (Met1-Phe50), the N domain, incorporated the Arg37Cys substitution and was produced as a fusion to the third IgG-binding domain of protein G within the context of the bacterial pET-44 expression vector.20 Gene expression, 2H/13C/15N labeling, and purification of the N domain (Arg37Cys) were carried out as described for the wild-type N domain elsewhere.21 Two samples, Nα and Nβ, which corresponded to an inhibitory and noninhibitory state in CPT1A, were prepared.20 The Nα results were from 0.2mM N peptide, 25mM HEPES-NaOH (pH, 7.4), and 100mM tetradecyltrimethylammonium chloride. The Nβ results were from 0.5mM N peptide, 25mM MES-NaOH (pH, 5.6), and 150mM dodecyltrimethylammonium chloride. The amide 1 (1HN)/nitrogen 15 (15N)/carbon α 13 (13Cα) backbone assignments of Nα(Arg37Cys) and Nβ(Arg37Cys) were obtained using HNCA and HNCACB experiments acquired on a cryoprobe-equipped Bruker Avance 700 spectrometer.
Cell Culture and Plasmid Preparation
The COS7 cell line was maintained in Dulbecco modified Eagle medium and supplemented with 10% fetal bovine serum, 2mM glutamine, 100 U/mL of penicillin, and 100 μg/mL of streptomycin. Cells were incubated at 37°C with 5% carbon dioxide in a humidified incubator. Human complementary DNA for CPT1C was purchased by OriGene Technologies (catalog number RG226726) and subcloned into pCMV6-AC-HA (OriGene, catalog number PS100004). The mutation was introduced using a directed mutagenesis kit (QuickChange II Site; Agilent Technologies, catalog number 200521). The cells were transiently transfected with the indicated plasmids using Lipofectamine 2000 (Life Technologies), following the manufacturer’s instructions. The induced pluripotent stem cell culture and differentiation into motor neurons were performed as previously described.22 Primary cultures of mouse cortical neurons were prepared from E15–E16 C57BL/6J wild-type or Cpt1c−/− embryos.23 All animal procedures followed the guidelines of the European Community Directive and met National Institutes of Health standards for use of laboratory animals. The cortex was enzymatically and mechanically dissociated as described previously.24 The neuronal cells were seeded in 48-well plates at a density of 5 × 104 cells/cm2 and maintained in a humidified incubator at 37°C with 5% carbon dioxide and 95% air. The cells were used for experiments at 7 days in vitro.
Immunohistochemistry
After fixation with 4% paraformaldehyde, slides were placed in blocking solution (10% normal goat serum and 0.3% Triton X-100 in phosphate-buffered saline [PBS]) for 45 minutes at room temperature. Primary antibody staining was done at 4°C overnight in PBS with 5% normal goat serum and 0.1% Triton X-100 (The Dow Chemical Company). The slides were then washed 3 times with PBS (0.1% Triton X-100 in PBS), incubated with secondary antibody (Invitrogen, 1:500) for 1 hour at room temperature in the dark, and then washed 3 times before drying and adding Vectashield/4′,6-diamidino-2-phenylinodole stain (Vector Laboratories). Antibody staining was performed in PBS with 3% bovine serum albumin and 0.1% Tween with 0.1% Tween/PBS used for all washes. Coverslips were mounted with Permount (Fisher Scientific). The cells were imaged using a confocal microscope (TCS SP5 II; Leica Microsystems) and image acquisition was done using the Application Suite Advanced Fluorescence for Windows (Leica Microsystems).
Immunoblotting and Coimmunoprecipitation
Cell pellets were lysed with radioimmunoprecipitation assay buffer (150mM sodium chloride, 0.5% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50mM TRIS pH 7.5, 2mM EDTA pH 8, and protease inhibitor cocktail [Roche]). Protein levels were measured using Bio-Rad protein assay. For immunoblotting, proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then electrophoretically transferred to nitrocellulose. After blocking with nonfat milk/0.1% Tween 20/TRIS-buffered saline (pH, 7.5), antibodies (1–5 μg/mL) were added overnight at 4°C. For coimmunoprecipitation, COS7 cells were transfected with the indicated constructs, lysed in coimmunoprecipitation buffer (50mM TRIS hydrogen chloride pH 7.5, 150mM sodium chloride, 1mM EDTA, 0.5% NP-40, and 10% glycerol and protease inhibitor cocktail [Roche]) and subjected to immunoprecipitation using G-coupled Sepharose beads (Sigma-Aldrich) followed by immunoblotting with the indicated antibodies.
Results
Identification of CPT1C Mutation in a Family With Pure AD-HSP
We identified a 3-generation family from southern Italy with dominantly inherited, adult-onset, and pure spastic paraplegia of unknown genetic cause (Figure 1A). The disease was slowly progressive, leading to impairment or loss of ambulation 10 to 15 years after the onset of symptoms. Cognitive functions were normal in all participants. Neurophysiologic examination showed prolonged central motor conduction time and delayed sensory evoked potentials, indicating abnormalities of the corticospinal tract and the dorsal column. Peripheral nerve conduction velocities were normal in all participants examined (Table).
Table.
Clinical Characteristics
| Characteristic | Outcome |
|---|---|
| No. of affected individuals | 6 |
| No. of affected individuals clinically examined | 5 |
| Age, mean (SD) [range], y | |
| Onset | 36 (12) [19–48] |
| Death | 70 (4) [67–75] |
| Spasticity | Moderate/severe phenotype |
| Weakness | Mild phenotype, proximal |
| Muscle wasting | Mild phenotype |
| Hyperreflexia | Moderate phenotype |
| Plantar reflex | Extensor |
| Vibration sensation at ankles | Decreased |
| Urinary dysfunction | Mild phenotype |
| Additional features (No. of individuals/total individuals) | Foot deformity (2/5) |
| Brain MRI | Normal |
| Neurophysiology | CMT delayed, normal NCS and EMG |
Abbreviations: CMT, central motor conduction time; EMG, electromyography; MRI, magnetic resonance imaging; NCS, nerve conduction studies.
We performed whole-exome sequencing on genomic DNA from the affected individuals II-6, III-2, III-3, and IV-1 (Figure 1A). Genetic analysis provided adequate coverage (94.1%, 93.7%, 94.6%, and 94.6% greater than or equal to 10×). After alignment and filtering, 13 heterozygous single-nucleotide variants were identified that were not described in online databases and were shared by all 4 individuals (eFigure 1 in the Supplement). Sanger sequencing of these variants in an additional affected member (III-4; list reduced to 8 variant candidates) and the unaffected members older than 70 years (III-5, III-8, and III-10) allowed us to reduce the list to the single-nucleotide substitution c.109C>T (RefSeq NM_001136052; GRCh37/hg19 chromosome 19: 50 195 618C>T) in exon 3 of CPT1C (OMIM *608846) (Figure 1B). This variant was also absent in the other unaffected participants (III-1, IV-2, and IV-3), thus strictly cosegregating with the disease phenotype. In addition to the online databases dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and the National Heart, Lung, and Blood Institute (http://evs.gs.washington.edu/EVS/), this variant was absent in 712 exomes of control individuals from the ClinSeq cohort.25 To examine the robustness of our results, we performed linkage analysis using the variant as a single marker and a logarithm of odds score of 2.48 was obtained. The base change substituted a Cys residue for an Arg in the gene product. This Cys residue was evolutionarily conserved (p.Arg37Cys; Figure 1C and D) and was considered to be damaging by several bioinformatics prediction tools (PolyPhen2 score, 0.995; PMut score, 0.927; and PROVEAN score, −4.08). No mutation in CPT1C was found in 163 other unrelated participants with pure HSP of unknown genetic cause.
NMR Spectroscopy Shows Structural Changes of Mutant CPT1C
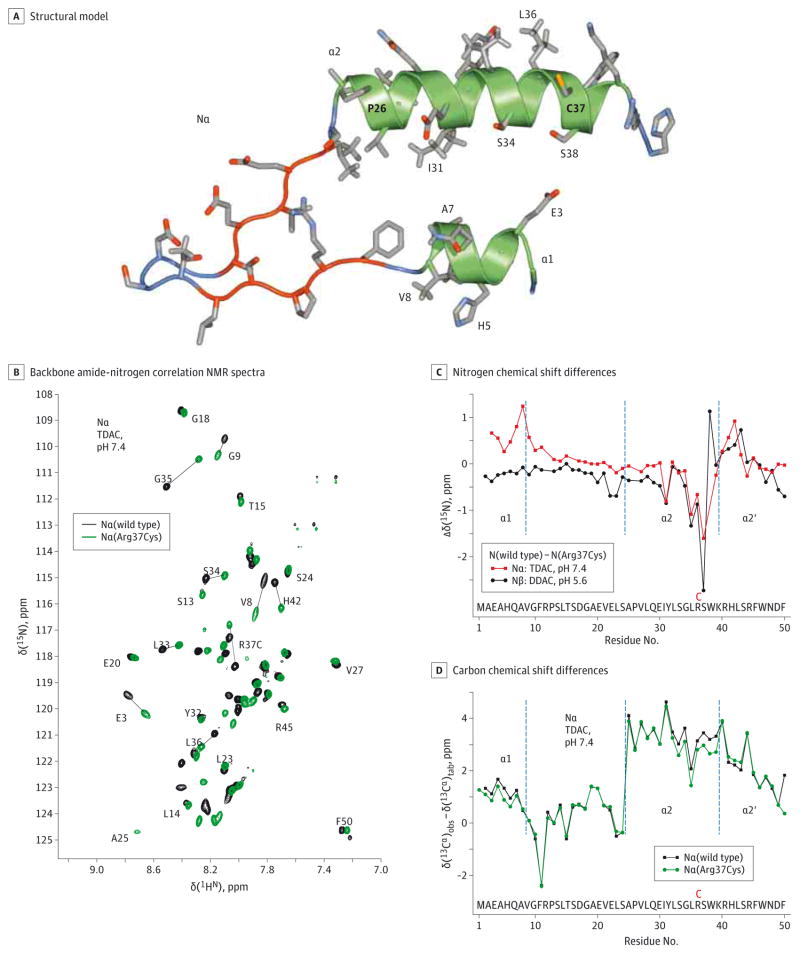
Members of the CPT1 protein family have an N-terminal regulatory domain (N) and a C-terminal catalytic domain, which are separated by 2 transmembrane helices. Arg37 is located in helix α2 of the N domain (Figure 2A). Using NMR spectroscopy, we evaluated the effects of Arg37Cys on structural properties of the inhibitory and noninhibitory N states, designated Nα and Nβ, respectively.20 In lieu of the membrane and catalytic domain surface, the Nα and Nβ states were obtained in the presence of micelle folding scaffolds.20,21 The NMR spectra showed that Arg37Cys resulted in a shift of many backbone hydrogen-nitrogen resonances relative to those of the wild-type protein (Figure 2B; eFigure 2A in the Supplement). The magnitudes of the observed chemical shift changes mapped the Arg37Cys-induced structural perturbations. In both Nα and Nβ, the largest changes in 1HN and 15N chemical shifts were observed around the mutation site (Figure 2C; eFigure 2B in the Supplement). This was accompanied by decreased secondary 13Cα shifts relative to wild-type protein around residue 37 in both the Nα and Nβ states (Figure 2D and eFigure 2C in the Supplement). Unlike 1HN and 15N shifts, which reflected a mix of their surrounding chemical environments and local protein conformations, 13Cα shifts correlated exclusively with the protein backbone conformation,26 indicating a destabilization of the helical structure near Cys 37. This effect may have occurred from the reduced amphiphilic character of helix α2, resulting from the loss of the positive charge of Arg37 (Figure 2A). In contrast to the strictly local effects of Arg37Cys in Nβ (Figure 2C; eFigure 2B and C in the Supplement), in Nα, the mutation created perturbations in helix α1 far from the mutation site (Figure 2B and C; eFigure 2B in the Supplement). This was accompanied by a small decrease in secondary 13Cα shifts in helix α1 relative to wild-type Nα (Figure 2D) indicative of its destabilization.
Figure 2. Structural Model and Nuclear Magnetic Resonance (NMR) Spectroscopy of CPT1C.
A, Structural model of the inhibitory Nα state of CPT1C. This model is based on the Nα state of CPT1A10 and looks onto the hydrophilic faces of amphiphilic helices α1 and α2. The model depicts Cys 37, which replaces Arg 37. In CPT1C, Glu26 of CPT1A is instead Pro26.13 B, Superposition of backbone amide-nitrogen correlation NMR spectra of wild-type Nα and Nα(Arg37Cys) of CPT1C. The spectra were recorded in the presence of tetradecyltrimethylammonium chloride (TDAC) at pH 7.4, 25°C, and a hydrogen 1 (1H) frequency of 700 MHz. C, Nitrogen 15 (15N) chemical shift differences, Dd(15N) between Nα and Nα(Arg37Cys), and Nβ and Nβ(Arg37Cys). The borders of the secondary structure elements of Nα, helices α1, α2, and α2′, are indicated. D, Comparison of Nα and Nα(Arg37Cys) secondary carbon α 13 (13Cα) chemical shifts, d(13Cα), defined as the difference between observed and tabulated random coil 13Cα shifts. Positive and negative shifts indicate helical and extended backbone conformations, respectively.14 DDAC indicates didecyldimethylammonium chloride.
CPT1C Is Expressed in Motor Neurons and Interacts With Atlastin-1
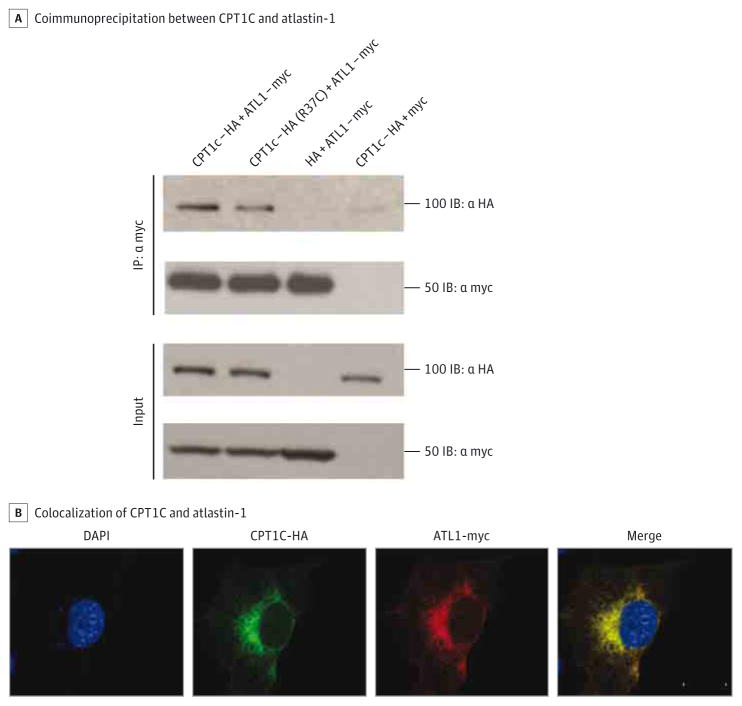
We looked for CPT1C expression in motor neurons derived from human-induced pluripotent stem cells and found localization in the soma and dendritic and axonal projections (eFigure 3 in the Supplement). By Western blot analysis, we showed CPT1C expression in the ventral horn from spinal cords of wild-type mice, further suggesting CPT1C enrichment in motor neurons (eFigure 4 in the Supplement). We also confirmed that CPT1C localized to the ER and not the mitochondria (eFigure 5 in the Supplement), supporting the hypothesis that CPT1C has a different role compared with its isozymes. To determine whether CPT1C is a member of the ER interaction network, we examined the interaction between CPT1C and atlastin-1 (SPG3A). SPG3A is the second most common cause of AD-HSP, accounting for approximately 10% of cases.4,6 We completed coimmunoprecipitation experiments using hemagglutinin-tagged CPT1C and avian myelocytomatosis–tagged atlastin-1 in COS7 cells and found that atlastin-1 coimmunoprecipitated with CPT1C (Figure 3A). Consistent with this finding, we also found strong colocalization of avian myelocytomatosis–tagged atlastin-1 with hemagglutinin-tagged CPT1C (Figure 3B). The Arg37Cys mutation did not affect the ability of CPT1C to bind atlastin-1 (Figure 3A). This interaction was specific because CPT1C does not interact with the ER protein Sec61 (eFigure 6 in the Supplement).
Figure 3. CPT1C Interaction With Atlastin-1.
A, COS7 cells cotransfected with 1.5 μg of avian myelocytomatosis (myc)–tagged atlastin-1 and 1.5 μg of HA-tagged CPT1C. Coimmunoprecipitation using an anti-myc antibody (C3956; Sigma) was performed and interactions were detected by Western blot analysis using an anti-hemagglutinin (HA) antibody (clone 16B12, 1:5000; Covance).
B, COS7 cells were cotransfected and signal was detected using anti-HA (clone 16B12, 1:5000; Covance) and anti-myc antibody (C3956, 1:5000; Sigma). The nuclear 4′,6-diamidino-2-phenylinodole stain is blue. Scale bar = 20 μm.
Mutation in CPT1C Decreases Number and Size of Lipid Droplets by a Dominant Negative Mechanism
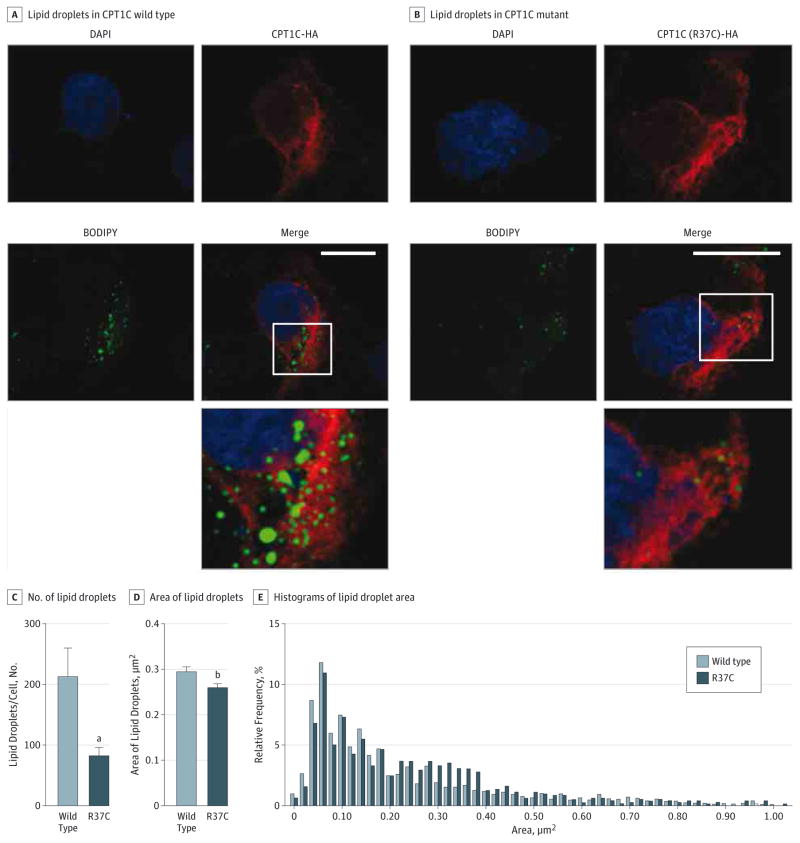
To investigate the effects of CPT1C on LD morphology, we immunostained COS7 cells transfected with either wild-type or mutant CPT1C with BODIPY 493/503 (Figure 4A and B). We found that the average number of LDs per cell was dramatically reduced when mutant CPT1C was overexpressed compared with the wild-type protein (P < .01; Figure 4C). The average area of LDs was also reduced (P < .05; Figure 4D), most likely because of a shift in the size distribution toward LDs with small to medium areas (0.30–0.40 μm2) (Figure 4E).
Figure 4. p.Arg37Cys CPT1C Is Associated With Reduced Lipid Droplet (LD) Number in COS7 Cells.
A and B, COS7 cells were transfected with wild-type or mutant hemagglutinin (HA)-tagged CPT1C and stained with HA antibody (clone 16B12, 1:5000; Covance) and BODIPY 493/503 for LDs (0.1 μg/mL; Invitrogen). The bottom panel shows an enlarged view of the boxed area. Scale bar = 20 μm. C and D, Numbers and areas of LDs were counted blindly in an automated fashion and results were derived from a total of 2949 LDs. Results are given as mean (SEM). E, Relative distribution of LD areas in wild-type and mutant CPT1C. Counting of LDs was performed in a blinded and automated fashion using the software Volocity 3D Image Analysis Software (PerkinElmer). The data were analyzed using GraphPad Prism software version 6 (http://www.graphpad.com) and P ≤ .05 was considered significant. DAPI indicates 4′,6-diamidino-2-phenylinodole.
aP < .01.
bP < .05.
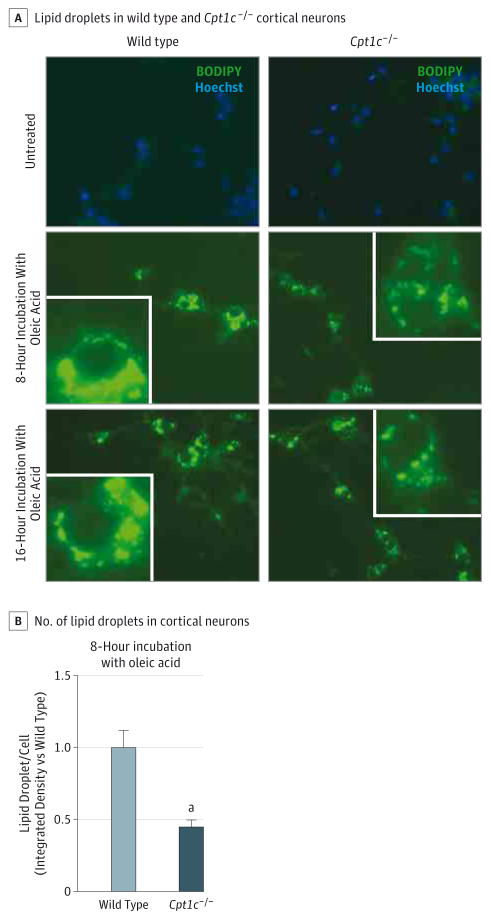
Furthermore, cortical neurons derived from Cpt1c−/− mice23,27 similarly exhibited a marked reduction in the number of LDs, suggesting a dominant negative mechanism for the Arg37Cys mutation (Figure 5).
Figure 5. Lipid Droplet Number Is Decreased in Cpt1c−/− Cortical Neurons.
A, Cells were obtained from the cortex of wild-type or Cpt1c−/− E16 embryos and seeded in 48-well plates during 7 days in vitro. For lipid loading, 300μM oleic acid was added for 8 or 16 hours before fixation. Representative images are shown. Images of neural lipids and the nucleus were acquired on a Nikon Eclipse TE-2000E optic microscope using a plan apochromat objective (BODIPY 493/503 and Hoechst dyes [Life Technologies], respectively; original magnification ×20). For quantification, sets of cells were cultured and stained simultaneously and imaged using identical settings. The region of interest was randomly selected using nucleic staining. B, For quantification, the Fiji image processing package was used to determine the integrated density stain per soma in at least 80 cells per condition. The results are shown as the mean of 2 independent experiments ± SEM.
aP < .001.
Discussion
The list of genes involved in HSPs is rapidly expanding. Even as their numbers increase, the functions of the encoded proteins are converging on a small number of common themes, such as intracellular trafficking and the shaping of membrane compartments.28 In particular, the genetic heterogeneity of pure HSPs offers a great opportunity to understand the actual requirements for axonal integrity and cell survival in motor neurons. Several of these proteins have been shown to localize to the ER where they interact with one another to assist in the formation and maintenance of long cellular processes,29 suggesting that the ER plays a central role in axonal health of corticospinal neurons. For example, it has been shown that the SPG4 protein spastin interacts with the SPG3A protein atlastin-1,30 the SPG31 protein REEP1 interacts with both atlastin-1 and spastin,7 and the SPG12 protein reticulon-2 (RTN2; OMIM #604805) interacts with spastin8 and likely atlastin-1 as well.31
In this study, we showed that the mutation Arg37Cys in the ER protein CPT1C causes a pure form of AD-HSP (SPG73). The protein is expressed in the soma and dendritic and axonal projections of motor neurons and interacts with atlastin-1, thus taking part in the network of ER proteins involved in HSP pathogenesis. The molecular functions of CPT1C are not yet understood. CPT1C shares high levels of homology with its related isozymes, the liver isoform CPT1A and the muscle isoform CPT1B10; nevertheless, it localizes to the ER and not the mitochondria,11 has little or no enzymatic activity in fatty acid oxidation when overexpressed in yeast or mammalian cells,10,13,14 and is mainly expressed in neurons, which have a limited capacity for lipid β-oxidation.10–12 CPT1A mutagenesis data indicated that inhibitory Nα catalytic domain interactions rely on electrostatic interaction between Glu26 and Lys561.32 Glu26 and Arg37 are both located in helix α2 of the N domain, which is conserved between CPT1A and CPT1C.21 Therefore, we hypothesized that the Arg37Cys mutation alters the interaction between the regulatory and catalytic domains in CPT1C. The NMR studies indicated that the mutation altered the protein conformation and disturbed a likely interaction between the regulatory and catalytic domains. The destabilizing effects of Arg37Cys on the Nα state and the putative loss of Arg37-mediated inhibitory helix α2 catalytic domain contacts pointed to a possible increase in an activity of mutant CPT1C that, to our knowledge, has not been characterized to date.
In contrast to the embryonic lethality of complete deficiency of Cpt1a and Cpt1b,33,34 Cpt1c knockout mice (Cpt1c−/−) survive but have decreased body weight and higher susceptibility to obesity induced by a high-fat diet,13,27,35 suggesting that CPT1C may act as a metabolic sensor. Interestingly, Cpt1c−/− mice also display motor coordination and gait abnormalities, hypoactivity, and muscle weakness,23 supporting a role of CPT1C in mammalian motor function. This evidence of a role of CPT1 in lipid metabolism prompted us to test the hypothesis that CPT1C affects LD biogenesis.
By storing fatty acids in the form of neutral triacylglycerol, LDs are thought to help cells to cope with many types of insults. They bud off as lipid globules and are enveloped by the cytoplasmic-facing lipid monolayer of the ER membrane.36 Numerous proteins have been implicated in the synthesis and degradation of neutral lipids; nevertheless, the mechanisms of their regulation and turnover remain elusive. The functional significance of LDs in neuronal physiology has attracted growing interest, especially because several LD-associated proteins are abundantly expressed in the brain and neurons are particularly vulnerable to the oxidative stress induced by lipid peroxidation. Corticospinal neurons, which have an intrinsically low neutral lipid content, may be particularly susceptible to an impairment of LD metabolism, perhaps owing to their extended membranes and highly specialized nature. Moreover, it has been recently shown that the depletion of atlastin-1 or expression of a dominant-negative mutant results in a reduction in LD size, whereas overexpression of at-lastin-1 has the opposite effect.15 Spartin, the protein mutated in Troyer syndrome (SPG20; OMIM #275900),16 associates with the surface of LDs and, when depleted, triggers the accumulation and coalescence of LDs into perinuclear clusters.17 Mutations in seipin, a protein that regulates LD formation,18 have been associated with several neurological disorders, with a phenotypic spectrum ranging from pure hereditary spastic paraplegia (SPG17; OMIM #270685)37 to distal hereditary motor neuropathy type V (OMIM #600794).37
We showed that mutant CPT1C markedly reduces the number and size of LDs in transfected cells. We also observed a reduction of LDs in primary cortical neurons isolated from Cpt1c−/− mice as compared with wild-type mice. This finding, in combination with the recent evidence that Cpt1c−/− mice also display motor abnormalities,23 suggests that CPT1C mutation causes the disease through a dominant negative mechanism. Additional work using animal models expressing the Arg37Cys change in CPT1C is needed to confirm these findings.
Conclusions
This association of CPT1C with an HSP phenotype highlights the importance of lipid metabolism as a theme in the pathogenesis of HSPs and perhaps other motor neuron diseases and deserves further study.
Supplementary Material
Acknowledgments
Funding/Support: This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Neurological Disorders and Stroke, grant Z01-AG000949-02 from the National Institute on Aging (Drs Traynor and Johnson), the European Union’s Seventh Framework Programme for Research (Dr Brice), grant ANR-10-IAIHU-06 from the Investissements d’avenir (Drs Brice and Dürr), and the French Agency for Research (Dr Stevanin). Dr Lee is supported by funds from the Division of Intramural Research of the National Institute of Child Health and Human Development.
Footnotes
Conflict of Interest Disclosures: Dr Biesecker receives royalties from Amgen and Genentech and is an uncompensated advisor to the Illumina Corporation. No other disclosures were reported.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: We thank the DNA bank of the Institut du Cerveau et de la Moelle Épinière (coordinated by Sylvie Forlani, PhD, Neuroscience Research Center, Groupe Hospitalier Pitié-Salpêtrière, Paris, France) for technical assistance. No financial compensation was received.
Author Contributions: Dr Fischbeck had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Rinaldi, Renvoise, De Michele, Santorelli, Filla, Casals, Traynor, Blackstone, Fischbeck.
Acquisition, analysis, or interpretation of data: Rinaldi, Schmidt, Situ, Johnson, Lee, Chen, Bott, Fadó, Harmison, Parodi, Grunseich, Biesecker, Stevanin, Dürr, Brice, Casals, Traynor, Fischbeck.
Drafting of the manuscript: Rinaldi, Casals, Blackstone, Fischbeck.
Critical revision of the manuscript for important intellectual content: Rinaldi, Schmidt, Situ, Johnson, Lee, Chen, Bott, Fadó, Harmison, Parodi, Grunseich, Renvoise, Biesecker, De Michele, Santorelli, Filla, Stevanin, Dürr, Brice, Casals, Traynor, Blackstone, Fischbeck.
Statistical analysis: Rinaldi, Schmidt, Situ, Johnson, Fadó, Casals, Traynor.
Obtained funding: Stevanin, Fischbeck.
Administrative, technical, or material support: Chen, Harmison, Renvoise, Biesecker, Fischbeck.
Study supervision: Casals, Fischbeck.
References
- 1.Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;1(8334):1151–1155. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- 2.Reid E, Dearlove AM, Whiteford ML, Rhodes M, Rubinsztein DC. Autosomal dominant spastic paraplegia: refined SPG8 locus and additional genetic heterogeneity. Neurology. 1999;53(8):1844–1849. doi: 10.1212/wnl.53.8.1844. [DOI] [PubMed] [Google Scholar]
- 3.Hazan J, Fonknechten N, Mavel D, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999;23(3):296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 4.Zhao X, Alvarado D, Rainier S, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001;29(3):326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- 5.Züchner S, Wang G, Tran-Viet KN, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79(2):365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7(12):1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 7.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montenegro G, Rebelo AP, Connell J, et al. Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J Clin Invest. 2012;122(2):538–544. doi: 10.1172/JCI60560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergstrom JD, Reitz RC. Studies on carnitine palmitoyl transferase: the similar nature of CPTi (inner form) and CPTo (outer form) Arch Biochem Biophys. 1980;204(1):71–79. doi: 10.1016/0003-9861(80)90008-9. [DOI] [PubMed] [Google Scholar]
- 10.Price N, van der Leij F, Jackson V, et al. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics. 2002;80(4):433–442. doi: 10.1006/geno.2002.6845. [DOI] [PubMed] [Google Scholar]
- 11.Sierra AY, Gratacós E, Carrasco P, et al. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J Biol Chem. 2008;283(11):6878–6885. doi: 10.1074/jbc.M707965200. [DOI] [PubMed] [Google Scholar]
- 12.Carrasco P, Sahún I, McDonald J, et al. Ceramide levels regulated by carnitine palmitoyltransferase 1C control dendritic spine maturation and cognition. J Biol Chem. 2012;287(25):21224–21232. doi: 10.1074/jbc.M111.337493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfgang MJ, Kurama T, Dai Y, et al. The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc Natl Acad Sci U S A. 2006;103(19):7282–7287. doi: 10.1073/pnas.0602205103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hada T, Yamamoto T, Yamamoto A, et al. Comparison of the catalytic activities of three isozymes of carnitine palmitoyltransferase 1 expressed in COS7 cells. Appl Biochem Biotechnol. 2014;172(3):1486–1496. doi: 10.1007/s12010-013-0619-y. [DOI] [PubMed] [Google Scholar]
- 15.Klemm RW, Norton JP, Cole RA, et al. A conserved role for atlastin GTPases in regulating lipid droplet size. Cell Rep. 2013;3(5):1465–1475. doi: 10.1016/j.celrep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel H, Cross H, Proukakis C, et al. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet. 2002;31(4):347–348. doi: 10.1038/ng937. [DOI] [PubMed] [Google Scholar]
- 17.Renvoisé B, Stadler J, Singh R, Bakowska JC, Blackstone C. Spg20−/− mice reveal multimodal functions for Troyer syndrome protein spartin in lipid droplet maintenance, cytokinesis and BMP signaling. Hum Mol Genet. 2012;21(16):3604–3618. doi: 10.1093/hmg/dds191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szymanski KM, Binns D, Bartz R, et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104(52):20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 20.Rao JN, Warren GZ, Estolt-Povedano S, Zammit VA, Ulmer TS. An environment-dependent structural switch underlies the regulation of carnitine palmitoyltransferase 1A. J Biol Chem. 2011;286(49):42545–42554. doi: 10.1074/jbc.M111.306951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samanta S, Situ AJ, Ulmer TS. Structural characterization of the regulatory domain of brain carnitine palmitoyltransferase 1. Biopolymers. 2014;101(4):398–405. doi: 10.1002/bip.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grunseich C, Zukosky K, Kats IR, et al. Stem cell-derived motor neurons from spinal and bulbar muscular atrophy patients. Neurobiol Dis. 2014;70:12–20. doi: 10.1016/j.nbd.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrasco P, Jacas J, Sahún I, et al. Carnitine palmitoyltransferase 1C deficiency causes motor impairment and hypoactivity. Behav Brain Res. 2013;256:291–297. doi: 10.1016/j.bbr.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Fadó R, Moubarak RS, Miñano-Molina AJ, et al. X-linked inhibitor of apoptosis protein negatively regulates neuronal differentiation through interaction with cRAF and Trk. Sci Rep. 2013;3:2397. doi: 10.1038/srep02397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biesecker LG, Mullikin JC, Facio FM, et al. NISC Comparative Sequencing Program. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19(9):1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. In: James T, Dotsch V, Schmitz U, editors. Nuclear Magnetic Resonance Of Biological Macromolecules, Pt A. Waltham, MA: Academic Press; 2001. pp. 3–34. [DOI] [PubMed] [Google Scholar]
- 27.Gao XF, Chen W, Kong XP, et al. Enhanced susceptibility of Cpt1c knockout mice to glucose intolerance induced by a high-fat diet involves elevated hepatic gluconeogenesis and decreased skeletal muscle glucose uptake. Diabetologia. 2009;52(5):912–920. doi: 10.1007/s00125-009-1284-0. [DOI] [PubMed] [Google Scholar]
- 28.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12(1):31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanderson CM, Connell JW, Edwards TL, et al. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet. 2006;15(2):307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J, Shibata Y, Zhu PP, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138(3):549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-Viñas E, Bentebibel A, Gurunathan C, et al. Definition by functional and structural analysis of two malonyl-CoA sites in carnitine palmitoyltransferase 1A. J Biol Chem. 2007;282(25):18212–18224. doi: 10.1074/jbc.M700885200. [DOI] [PubMed] [Google Scholar]
- 33.Ji S, You Y, Kerner J, et al. Homozygous carnitine palmitoyltransferase 1b (muscle isoform) deficiency is lethal in the mouse. Mol Genet Metab. 2008;93(3):314–322. doi: 10.1016/j.ymgme.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyman LR, Cox KB, Hoppel CL, et al. Homozygous carnitine palmitoyltransferase 1a (liver isoform) deficiency is lethal in the mouse. Mol Genet Metab. 2005;86(1–2):179–187. doi: 10.1016/j.ymgme.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Gao S, Zhu G, Gao X, et al. Important roles of brain-specific carnitine palmitoyltransferase and ceramide metabolism in leptin hypothalamic control of feeding. Proc Natl Acad Sci U S A. 2011;108(23):9691–9696. doi: 10.1073/pnas.1103267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24(3):109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- 37.Windpassinger C, Auer-Grumbach M, Irobi J, et al. Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat Genet. 2004;36(3):271–276. doi: 10.1038/ng1313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.