Abstract
Clostridium difficile remains a leading nosocomial pathogen, putting considerable strain on the healthcare system. The ability to form endospores, highly resistant to environmental insults, is key to its persistence and transmission. However, important differences exist between the sporulation pathways of C. difficile and the model Gram-positive organism Bacillus subtilis. Amongst the challenges in studying sporulation in C. difficile is the relatively poor levels of sporulation and high heterogeneity in the sporulation process. To overcome these limitations we placed Ptet regulatory elements upstream of the master regulator of sporulation, spo0A, generating a new strain that can be artificially induced to sporulate by addition of anhydrotetracycline (ATc). We demonstrate that this strain is asporogenous in the absence of ATc, and that ATc can be used to drive faster and more efficient sporulation. Induction of Spo0A is titratable and this can be used in the study of the spo0A regulon both in vitro and in vivo, as demonstrated using a mouse model of C. difficile infection (CDI). Insights into differences between the sporulation pathways in B. subtilis and C. difficile gained by study of the inducible strain are discussed, further highlighting the universal interest of this tool. The Ptet-spo0A strain provides a useful background in which to generate mutations in genes involved in sporulation, therefore providing an exciting new tool to unravel key aspects of sporulation in C. difficile.
Keywords: sporulation, Clostridium difficile, spo0A, spore, inducible expression, allele-coupled exchange, anhydrotetracycline
Introduction
Clostridium difficile is a Gram-positive, anaerobic, spore-forming pathogen and the leading cause of healthcare associated diarrhea worldwide (Rupnik et al., 2009; Smits et al., 2016). C. difficile infection (CDI) occurs most commonly in nosocomial settings, among patients whose natural gut microbiota has been perturbed by prolonged treatment with broad-spectrum antibiotics (Dethlefsen et al., 2008; Lawley and Walker, 2013). Two large cytotoxins secreted by the vegetative cell are responsible for the clinical manifestations of CDI, which range from mild, self-limiting diarrhea to severe, often fatal inflammatory complications, such as pseudomembranous colitis or toxic megacolon (Dubberke and Olsen, 2012). However, it is the spore that is the primary infectious agent, as mutant strains defective in sporulation are unable to efficiently persist in the environment and transmit disease (Deakin et al., 2012). Due to their multi-layered structure, spores are extremely robust and resistant to both chemical and physical insults, enabling C. difficile to survive exposure to heat, oxygen, alcohol, noxious chemicals, and certain disinfectants (Lawley et al., 2010; Vohra and Poxton, 2011). Despite its key relevance in CDI, many aspects of sporulation in C. difficile are still understudied, hampered by low sporulation efficiency and high heterogeneity in the sporulation process.
In order to sporulate, the cell must undergo an ordered program of unicellular differentiation that leads to a dormant, non-replicative state. The decision to enter the developmental program is highly regulated, requiring multiple environmental and metabolic cues (Al-Hinai et al., 2015). It is mediated by Spo0A, a conserved transcription factor that plays a central role in phenotypic differentiation and adaptation to changing environments. The role of Spo0A in orchestrating sporulation has been studied in exquisite detail in Bacillus subtilis (reviewed in Sonenshein, 2000). Spo0A is post-translationally activated via phosphorylation facilitated by a complex phosphorelay consisting of several sensor kinases (KinA-E). These kinases respond to external signals by auto-phosphorylating and subsequently transferring a phosphate residue either directly to Spo0A or via intermediary proteins Spo0F and Spo0B (Jiang et al., 2000b). In contrast to B. subtilis, there are no Spo0F or Spo0B homologs, and therefore presumably no phosphorelay system, in C. difficile; rather, Spo0A is believed to be phosphorylated directly by the sensor kinases (Underwood et al., 2009; Steiner et al., 2011).
The phosphorelay affords opportunity for multiple layers of regulation, such as Rap phosphatases that modulate the level of phosphorylated Spo0A, permitting a degree of fine-tuning and flexibility to the system (Veening et al., 2005). It remains unclear whether this level of control is absent in C. difficile or is achieved by other means (Jiang et al., 2000a; Paredes et al., 2005). Studies published to date have shown that C. difficile initiates sporulation in response to nutrient availability (Edwards et al., 2014; Nawrocki et al., 2016) and that a Rap-like protein, RtsA, acts as a positive regulator of sporulation, although the exact nature of this effect remains unclear (Edwards et al., 2016). Sporulation, initiated by an asymmetric division of the vegetative cell into a smaller forespore and a larger mother cell, then proceeds via an expression cascade controlled by sporulation-specific sigma factors (σE, σF, σG, and σK). Differential expression of sigma factors in each of those compartments allows the sporulation process to be completed.
Recent studies of Spo0A in B. subtilis have demonstrated that the importance of this regulator extends beyond just sporulation, revealing the existence of an intricate Spo0A regulatory network fine-tuned to respond to the specific amount of Spo0A present in the cell (Fujita et al., 2005; Vishnoi et al., 2013). Efficient sporulation requires a progressive increase in the levels of Spo0A, which prompts the gradual activation initially of low-threshold Spo0A-regulated genes and subsequently high-threshold Spo0A-regulated genes (Fujita et al., 2005; Vishnoi et al., 2013). In B. subtilis the Spo0A regulon comprises approximately 120 genes (Molle et al., 2003). At low levels of Spo0A, genes involved in lifestyle changes that might delay the need to sporulate, such as biofilm formation, are activated and only if levels of Spo0A continue to rise until a threshold level is reached does the cell commit to sporulation (Fujita et al., 2005). Thus, Spo0A serves as a focal point for the integration of a number of factors that determine whether the cell needs to enter a dormant state. In C. difficile, Spo0A is thought to control approximately 300 genes (Fimlaid et al., 2013; Pettit et al., 2014), many of which have been linked to biofilm formation (Dawson et al., 2011), swimming motility (Pettit et al., 2014) and toxin production (Underwood et al., 2009), although conflicting results have been reported across different strains and laboratories (Rosenbusch et al., 2012; Mackin et al., 2013), indicating the complex nature of Spo0A signaling.
In an attempt to provide a more robust and tractable way to study sporulation in C. difficile, we have generated an inducible spo0A strain and investigated whether controlled expression of the transcription factor is a viable method for achieving higher sporulation efficiency and homogeneity. We demonstrate that spo0A expression can be induced in a dose-dependent manner and that the level of expression correlates with sporulation efficiency, which reaches values higher than those observed in an isogenic wild type (WT) strain. We also show that spo0A-dependent genes involved in other aspects of the bacterial life cycle can be indirectly controlled by use of the inducing agent. Finally, using the mouse model of CDI, we demonstrate that induction of spo0A can be carried out in vivo, providing a new tool in studying the role of Spo0A-dependent phenotypes in virulence.
Results
Generating an Inducible spo0A Strain
Allele-coupled exchange was used to place the inducible Ptet promoter with its regulatory elements 30 bp upstream of the spo0A ORF on the C. difficile 630 chromosome, generating Ptet-spo0A (Figure 1A). PCR and sequencing of the resulting product was used to confirm correct integration of the promoter (Figure 1B). The tet element comprises two divergent promoters, Ptet and PtetR, each overlapping with a tet operator sequence and regulated by tetracycline (Figure 1C). TetR, which represses both Ptet and PtetR, introduces a negative feedback loop to the system that is tightly dose-dependent, allowing an increased level of control (Fagan and Fairweather, 2011). To minimize potential confounding factors resulting from using an antibiotic, the non-antibiotic tetracycline analog anhydrotetracycline (ATc) was used as the inducing agent. To exclude the possibility that ATc was exerting a deleterious effect on normal growth, WT and Ptet-spo0A cultures were incubated in the presence or absence of ATc and their growth rate was monitored over time by measuring optical density (OD600). No appreciable difference in growth rate was observed between WT and Ptet-spo0A, irrespective of the ATc concentration used (Supplementary Figure S1).
FIGURE 1.
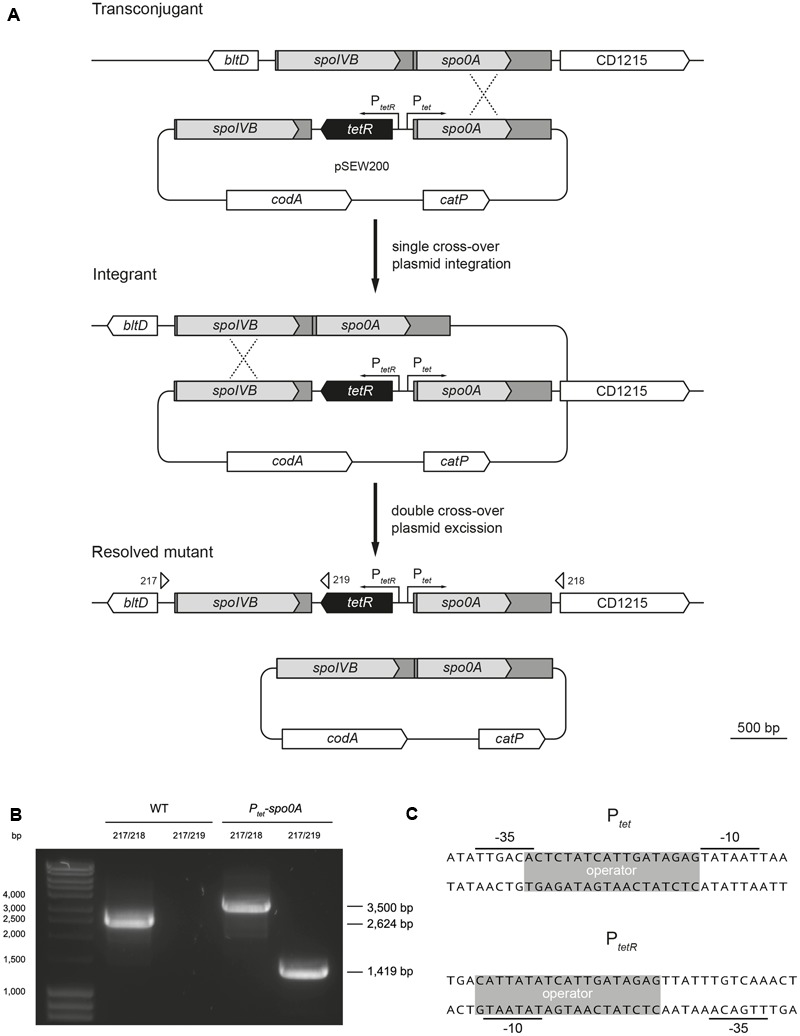
Confirmation of Ptet integration into the C. difficile 630 chromosome. (A) Stepwise diagram detailing the stages of ACE-mediated mutagenesis. Upstream and downstream homology regions spanning the spoIVB and spo0A ORFs are indicated as gray boxes. The Ptet regulatory elements are depicted in black. Binding sites for primers used in PCR screening are indicated with arrows. (B) Agarose gel electrophoresis analysis of PCR products obtained during screening for WT and Ptet-spo0A. Expected products sizes are indicated on the right side of the panel. (C) Detailed representation of the Ptet regulatory elements inserted upstream of spo0A (CD1214).
Expression of spo0A and spo0A-Dependent Genes in Ptet-spo0A Can be Controlled in Response to ATc
To verify whether spo0A expression can be driven by the introduced Ptet promoter upon induction, qRT-PCR was used to measure the level of spo0A transcripts in Ptet-spo0A relative to WT following growth with a range of ATc concentrations spanning 0–250 ng/ml. A clear dose-dependent response in transcript levels was observed (Figure 2A). In the absence of the inducing agent, Ptet-spo0A showed an approximately 40-fold reduction in detectable spo0A mRNA when compared to WT (p < 0.0001), a background level of expression possibly attributable to a small amount of promoter ‘leakage.’ Transcript levels gradually increased with rising ATc concentration, with the largest change observed between 50 and 100 ng/ml, where the levels of spo0A transcripts in Ptet-spo0A were fourfold higher than those found in WT at 100 ng/ml. At the highest concentration of ATc used, 250 ng/ml, spo0A transcript levels were approximately 5.6-fold higher in the mutant relative to WT grown at the same concentration of ATc (p = 0.0161) (Figure 2A).
FIGURE 2.
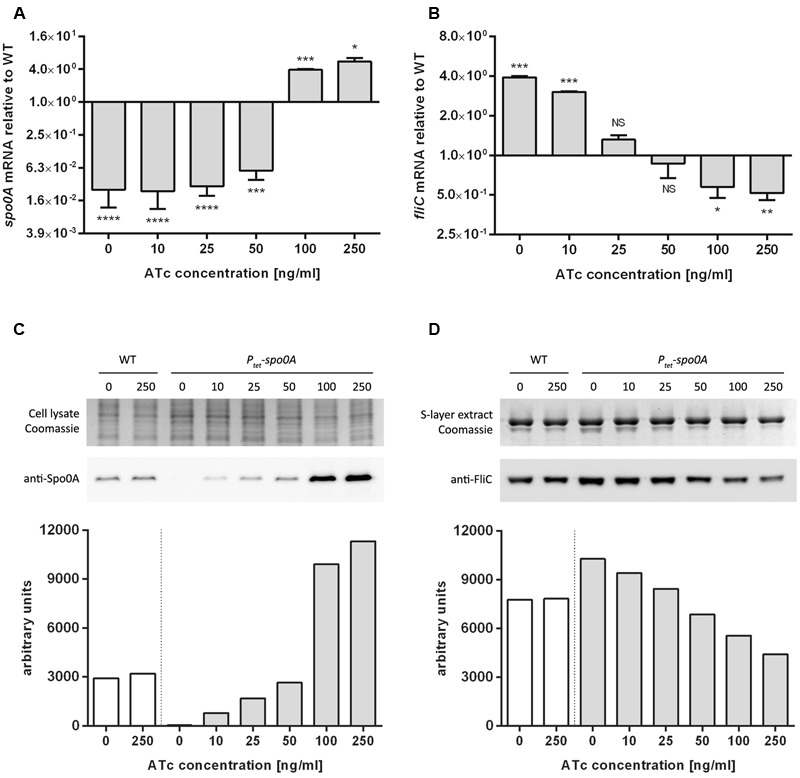
Analysis of dose-dependent expression of spo0A and fliC in Ptet-spo0A. qRT-PCR analysis of spo0A (A) and fliC (B) transcripts isolated from WT or Ptet-spo0A grown with increasing concentrations of ATc. Data displayed is Ptet-spo0A relative to WT (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 by a two-tailed Student’s t-test). Densitometric analysis of anti-Spo0A (C) and anti-FliC (D) immunoblots on protein extracts form WT or Ptet-spo0A cells grown with increasing concentrations of ATc. Coomassie-stained gels are provided as loading control.
Spo0A is linked to many phenotypes in C. difficile. One of these is flagella-mediated motility where Spo0A has been shown to negatively regulate a number of genes, including fliC (Pettit et al., 2014). To further validate the Ptet-spo0A system and to demonstrate that the strain can be used to study spo0A-related phenotypes beyond sporulation, we sought to confirm the link between fliC and spo0A by way of ATc induction. In keeping with previous reports on the link between Spo0A and FliC, qRT-PCR demonstrated that fliC transcription levels decreased with increasing ATc concentrations, dropping from 3.9-fold (p = 0.0003) to 0.5-fold (p = 0.0075) relative to WT (Figure 2B). To confirm that the dose-dependent changes in spo0A and fliC transcript levels observed upon ATc induction correlated with similar changes in protein levels, OD-normalized protein extracts from Ptet-spo0A cultures grown with a range of ATc concentrations were probed for Spo0A and FliC by Western blotting. In line with our qRT-PCR results, levels of Spo0A in whole cell lysates increased with the increase in ATc concentration while the amount of FliC in surface layer (S-layer) extracts decreased. Importantly, the amount of Spo0A and FliC remained constant in WT, irrespective of ATc concentration (Figures 2C,D).
Sporulation Efficiency in Ptet-spo0A Can be Controlled in Response to ATc
Studies reported to date have shown substantial variation in sporulation between different strains of C. difficile (Underwood et al., 2009; Burns et al., 2010; Vohra and Poxton, 2011). Accordingly, making comparisons between strains has proven problematic and is further compounded by a lack of uniform methods for induction and assessment of sporulation (reviewed in Burns and Minton, 2011). Irrespective of the methodologies used, efficiency of sporulation under laboratory conditions appears to be low, particularly when compared to B. subtilis, hampering work on the molecular basis of sporulation in C. difficile. Having shown that spo0A expression can be manipulated in Ptet-spo0A via induction with ATc, we asked whether the observed changes in Spo0A levels would affect the overall sporulation efficiency. We first determined whether the tet element is sufficiently tightly regulated to render the Ptet-spo0A strain asporogenous in the absence of ATc. Supporting this, after 120 h of nutrient starvation in liquid culture, typically regarded as the time after which the sporulation process of a C. difficile culture is complete (Burns and Minton, 2011), no spores could be detected based on heat-resistant CFU counts (Figure 3B). As expected, Ptet-spo0A cultures grown in the presence of increasing concentrations of ATc showed a dose-dependent increase in heat-resistant CFUs within 16 h from induction. Maximum spore titers were observed at 24–48 h post-inoculation, reaching 3 × 103 ± 3.28 × 102 CFU/ml; 2.25 × 104 ± 1.32 × 103 CFU/ml; 1.48 × 105 ± 2.47 × 104 CFU/ml; 5.17 × 105 ± 2.75 × 105 CFU/ml and 9.83 × 105 ± 1.04 × 105 CFU/ml in cultures induced with 10, 25, 50, 100, and 250 ng/ml of ATc respectively (Figures 3A,C), and remained largely constant throughout the duration of the experiment. Taking into account the total CFU observed at t0, these values correspond to a sporulation efficiency of 0.001% (10 ng/ml ATc), 0.006% (25 ng/ml ATc), 0.039% (50 ng/ml ATc), 0.387% (100 ng/ml ATc), and 1.209% (250 ng/ml ATc) showing a clear dose response. Importantly the observed titers were reached faster and, at the maximum concentration of ATc, were significantly higher than in WT (2.58 × 105 ± 1.25 × 104 CFU/ml) (p = 0.0364) (Figure 3B). While the inducing agent had no effect on the final spore titers observed in WT cultures, it did change the dynamics of the sporulation process, resulting in higher spore titers at 16–24 h post-induction (Figure 3B), possibly an effect of additional stress that high concentration of ATc exerts on the cells.
FIGURE 3.
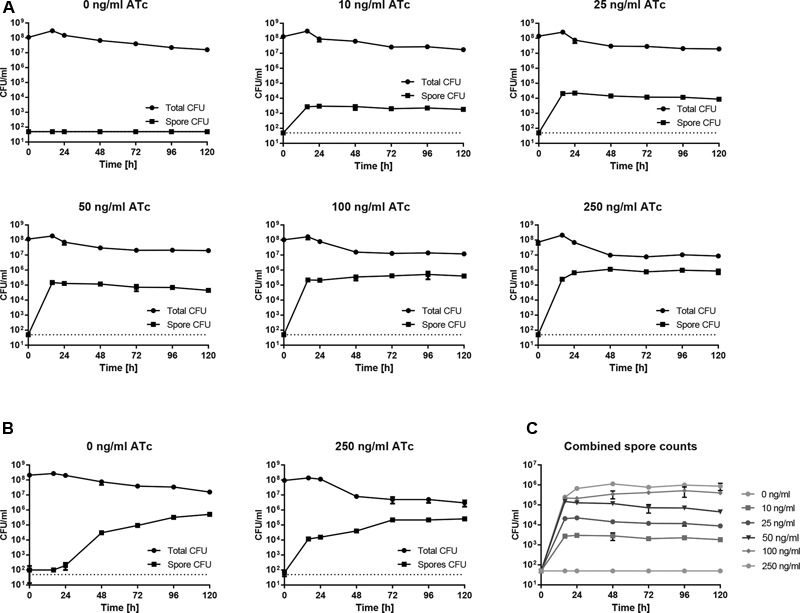
Dose-dependent increase in sporulation efficiency in Ptet-spo0A. Ptet-spo0A (A) or WT (B) were grown to OD ∼0.8 and sub-cultured 1:10,000 in fresh BHIS supplemented with the concentration of ATc indicated, or 1% ethanol (0 ng/ml ATc). Samples were removed at the indicated time points post-inoculation, serially diluted in sterile, pre-reduced PBS and plated onto BHIS agar supplemented with 0.1% Tch with (spore CFU) or without (total CFU) prior heat-treatment at 70°C for 30 min. Colonies were enumerated after 24 h of incubation under anaerobic conditions. (C) Combined Ptet-spo0A counts showing a dose dependent increase in sporulation efficiency. Data presented as means ± SD from three technical replicates. Each result is representative of experiments performed in at least biological triplicate. Dotted line represents the limit of detection.
Controlled Expression of spo0A Has No Effect on Overall Homogeneity of Sporulating Cultures
The lack of synchronicity observed in sporulating C. difficile cultures has been a significant barrier to population-wide studies as any measurements have to be interpreted as an average value across cells at different stages of the bacterial life cycle. Since sporulation in Ptet-spo0A under ATc induction was capable of proceeding faster than in WT, we asked whether this would be reflected in a more synchronous process. To investigate this, we monitored sporulating Ptet-spo0A cultures grown in the presence of increasing ATc concentrations by fluorescence microscopy following staining with FM4-64 and Hoechst 33258 to visualize the membrane and DNA, respectively. In order to overcome the high limit of detection of the technique, cultures were sporulated on a 70:30 mixture of sporulation media (SM) and BHIS agar, previously used in sporulation studies by several groups as it seems to provide faster and more efficient sporulation (Fimlaid et al., 2013; Putnam et al., 2013; Edwards et al., 2016). In line with our previous experiments conducted in liquid culture, a clear dose response in sporulation efficiency was observed, although a twofold higher concentration of ATc had to be used to reach saturation, presumably due to limited ATc availability in plated culture when compared to liquid culture. Mean values of sporulation efficiency, calculated as the percentage ratio of cells that had undergone asymmetric cell division to total cells counted, reached 0.47 ± 1%; 12.37 ± 2.35%; 50.44 ± 6.17%, and 58.99 ± 6.02% in cultures induced with 50, 100, 250, and 500 ng/ml ATc, respectively. No sporulation was observed in cultures induced with 25 ng/ml ATc or in uninduced cultures. As expected, ATc had no significant effect on sporulation efficiency in WT (Figure 4A), reaching 55.6 ± 3.04% in the uninduced culture and 55.93 ± 6.59% in cultures induced with 500 ng/ml ATc. In order to assess whether controlled expression of spo0A has an effect on the overall homogeneity of sporulation, we examined the composition of sporulating cultures by counting cells that had reached one of three defined morphological stages: (i) cells with flat, asymmetric septa; (ii) cells undergoing engulfment and (iii) cells with fully engulfed sporangia (including mature spores) (Figure 4B). While the distribution of cells at stage 1, 2, or 3 did change with increasing ATc concentration, the overall homogeneity of the culture did not improve when compared to WT, particularly at higher concentrations of ATc required to match WT sporulation efficiency, suggesting that controlled expression of spo0A on its own does not increase the synchronicity of the sporulating culture (Figures 4A,C).
FIGURE 4.
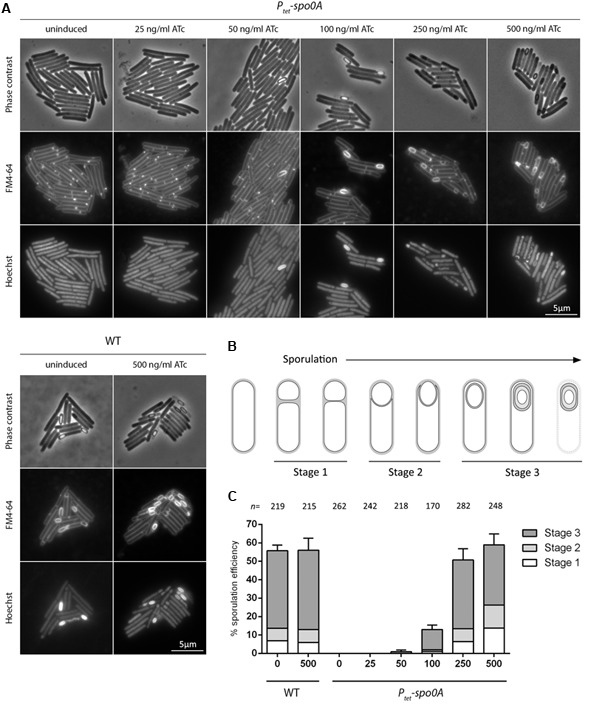
Analysis of Ptet-spo0A sporulation dynamics. (A) WT and Ptet-spo0A cultures were harvested after 14 h of growth on 70:30 agar supplemented with ATc as indicated. Cells were stained with FM4-64 and Hoechst 33258 and analyzed by phase contrast and fluorescence microscopy. Scale bar corresponds to 5 μm. (B) Schematic representation of the sporulation process indicating stages corresponding to asymmetric cell division (stage 1), engulfment (stage 2) and spore maturation (stage 3). (C) Sporulation efficiency and distribution of cells at different stages of sporulation in WT and Ptet-spo0A grown for 14 h on 70:30 agar supplemented with ATc as indicated. Sporulation efficiency is expressed as the percentage ratio of cells at a given stage of sporulation to total cells counted. Data reported as means ± SD from 10 random fields of view. The total number of cells counted (n) is indicated above each column on the graph.
Use of Ptet-spo0A as a Background Strain in Studies of Sporulation-Related Phenotypes
One of the main benefits of using allele-coupled exchange to genetically engineer C. difficile is the possibility of creating strains carrying multiple mutations through subsequent rounds of mutagenesis since any changes to the chromosome are left unmarked and therefore not reliant on the limited availability of suitable antibiotic resistance markers (Cartman et al., 2012; Heap et al., 2012). Having shown that inducible expression of spo0A is a viable strategy for controlling sporulation in C. difficile, we set out to investigate whether Ptet-spo0A could be used as a parental strain in studies of sporulation-related genes. As ‘proof of concept’ we have created sigE, sigF, and sigG mutants in the Ptet-spo0A background by introducing in-frame deletions into the respective genes by allele-coupled exchange. The resulting strains designated Ptet-spo0AΔsigE, Ptet-spo0AΔsigF, and Ptet-spo0AΔsigG were sporulated on solid media in the presence or absence of ATc and analyzed by microscopy following membrane and DNA staining (Figure 5). When induced, both the sigE and sigF mutants were blocked at the asymmetric division stage, revealing abortive disporic cells and occasionally multiple, closely located polar septa. Given the apparent lack of DNA staining in compartments created by these multiple septa, the observed staining pattern could equally be the result of membrane delamination between the forespore and the mother cell. Additionally, sigF mutant forespores showed distinct bulging. While cells of the sigG mutant were able to complete engulfment, they did not proceed further in morphogenesis and the resulting forespores were often misshapen and dislodged, resting at an angle to the longitudinal axis of the cell. As expected, strains grown in the absence of ATc did not initiate sporulation (Figure 5). These observations are largely consistent with previous work characterizing the morphology of sporulation-specific sigma factor mutants created using ClosTron insertional mutagenesis (Pereira et al., 2013) and demonstrate the potential of using Ptet-spo0A in studies that would benefit from more control over the sporulation process.
FIGURE 5.
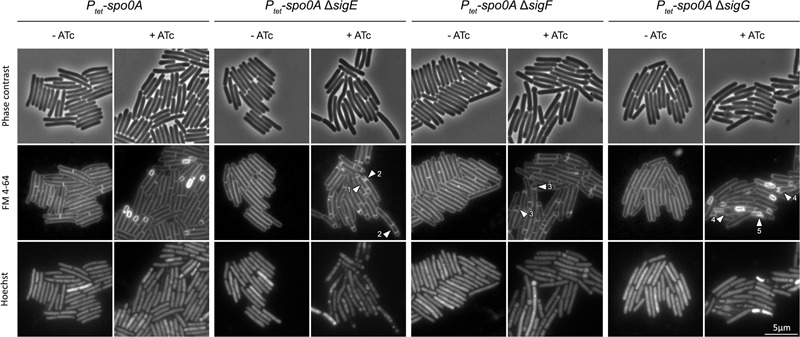
Morphological characterisation of Ptet-spo0AΔsigE, Ptet-spo0A ΔsigF, and Ptet-spo0A ΔsigG mutants. Sporulating cultures of sigE, sigF, and sigG mutants created in the Ptet-spo0A background were harvested after 14 h of growth on 70:30 agar supplemented with ATc as indicated. Cells were stained with FM4-64 and Hoechst 33258 and analyzed by phase contrast and fluorescence microscopy. Arrows indicate: (1) abortive diasporic forms, (2) multiple polar septa, (3) forespore bulging, (4) mother cell bulging and (5) dislodged sporangia resting at an angle to the longitudinal axis of the cell. Scale bar corresponds to 5 μm.
Use of Ptet-spo0A in the Mouse Model of Infection
The spo0A regulatory network co-ordinates multiple aspects of the C. difficile life cycle, including toxin production and the resulting virulence of a given strain. Deakin et al. (2012) reported that a R20291 Δspo0A strain (ribotype 027) produced more toxin in vitro and caused more severe disease than WT in a murine model of infection, while being deficient in persistence and transmission. More recent studies however, suggest that this effect is not consistent across all C. difficile strains (Mackin et al., 2013). While Spo0A was confirmed to be a negative regulator of toxin production in ribotype 027 strains, no such effect was observed in ribotype 012, suggesting that Spo0A could differentially regulate toxin production between phylogenetically distinct strains of C. difficile (Mackin et al., 2013). The exact mechanisms of the increased virulence of some spo0A mutants are not yet known, but the explanation is most likely to be multi-faceted. In B. subtilis, Spo0A does not have an equal affinity for all genes within the spo0A regulon, leading to a programmed change in gene expression with sporulation-specific genes not activated until high levels of Spo0A have accumulated (Fujita et al., 2005). An inducible system that offers titratable control over Spo0A levels provides an opportunity to ask more nuanced questions about the role of Spo0A in the infectious process in a manner that the Δspo0A mutant does not. Previous studies have demonstrated the potential for ATc induction of bacterial gene expression in mouse models (Ji et al., 2001; Kotula et al., 2014). Therefore, having characterized the Ptet-spo0A strain in vitro, we asked whether the inducible system would also work in vivo.
In our study, 12 mice pre-treated with clindamycin were infected with 1 × 107 CFU from an overnight culture of Ptet-spo0A grown without ATc. Six mice received 0.1 mg/ml ATc in their drinking water, with the remaining six receiving 1% (v/v) ethanol as a negative control. Feces were collected at 24, 72, and 120 h post-infection, and total and heat-resistant CFUs measured (Figure 6). No statistically significant difference was found for total cell counts between the ATc- and ATc+ groups until 120 h after infection, indicating successful infection of both groups. For the group of mice that did not receive ATc in their drinking water, no spores could be detected by enumeration of heat resistant colonies, indicating a failure to sporulate within the mouse. In contrast, for the group of mice that received 0.1 mg/ml ATc in their drinking water, heat-resistant CFUs were detected 24 h post-infection, steadily increasing until the final time-point taken 120 h post-infection. To confirm the identity of the isolated strains post-infection, five colonies from each group were selected at random and screened for Ptet-spo0A by PCR. As shown in Supplementary Figure S2, all strains were found to carry Ptet-spo0A. Thus, the Ptet promoter does not become activated in vivo, rendering Ptet-spo0A asporogenous. Furthermore, ATc can be used to control the Ptet system in vivo. Therefore, this approach will permit the study of the role of Spo0A and sporulation/germination kinetics in infection with a level of control not possible with previously used systems.
FIGURE 6.
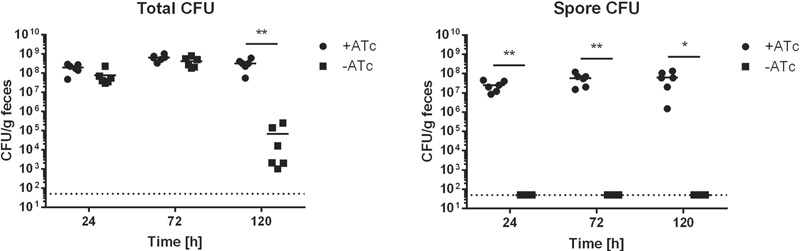
Use of the Ptet inducible system in vivo. 12 mice were treated with clindamycin to disrupt the natural flora. All mice were subsequently infected with the same culture of Ptet-spo0A grown overnight without ATc (infectious dose = 1 × 107 CFU). The mice were then split into two groups of 6, one of which received 1% ethanol in their drinking water, and the other 100 μg/ml ATc. Feces were collected at the time indicated post-infection and enumerated for total and heat-resistant CFUs. Data plotted for each animal individually with mean indicated (∗p ≤ 0.05; ∗∗p ≤ 0.01 by a multiple Student’s t-test using the Holm-Sidak method). Dotted line represents the limit of detection.
Discussion
Sporulation is a key contributing factor in establishing C. difficile as a leading nosocomial pathogen in the developed world. Studies carried out in B. subtilis have undoubtedly provided a useful basis for study of sporulation in Clostridial species, but it has become clear that there are several key differences in the sporulation process which make extrapolating results difficult. These differences are most apparent at the stage of entry into sporulation, governed in Bacilli by a phosphorelay system that is absent in Clostridial species. It is presumed that the phosphorelay system provides tighter regulation over the commitment to sporulate, but this premise remains unproven and the consequences of the lack of such a system remain unclear. Another aspect of the C. difficile life cycle that is uniquely affected by Spo0A is toxin production, a phenotype that cannot be studied in a non-toxigenic species such as B. subtilis. Additionally, Spo0A regulates other lifestyle changes in a manner that can vary from species to species and even between strains. Thus, although Spo0A is the master regulator of sporulation, it is also an important instigator of transcriptional changes that might help evade the need to sporulate, such as cannibalism or biofilm formation.
Under laboratory conditions, C. difficile sporulation efficiency is low compared to B. subtilis (Burns and Minton, 2011), a technical hurdle to its study that is exacerbated by the high heterogeneity and the current inability to synchronize a sporulating culture. Furthermore, as illustrated in studies of B. subtilis, the study of spo0A-null mutants does not permit recognition of the importance of incremental increases in the level of Spo0A, leading to more nuanced Spo0A-mediated phenotypes to be overlooked. An over-simplification of the intricate Spo0A network is perhaps one reason for conflicting results regarding Spo0A-mediated phenotypes in the literature. In this study, we report the generation of an inducible spo0A system, as schematically detailed in Figure 1A, in an attempt to improve sporulation efficiency, investigate the possibility of achieving a more homogenous culture, and test spo0A-titratable phenotypes.
In WT C. difficile cultures, heat-resistant CFUs are typically observed 8–12 h after reaching stationary phase (Pereira et al., 2013) and the sporulation process can take up to 5 days to reach equilibrium (Burns and Minton, 2011). We have demonstrated that inducing spo0A expression significantly reduces the amount of time necessary to reach maximum spore titers. Analysis of the cultures by fluorescence microscopy indicated that sporulation proceeds similarly to WT even when induction leads to faster completion of sporulation, but transmission electron microscopy would be required to rule out any changes to the fine structure of the spore. These results demonstrates that induction of Spo0A is sufficient to drive sporulation and, at least to some degree, overrides the need for external stimulus. Therefore, the Ptet-spo0A strain is a useful background for generating mutants in genes involved in other stages of the sporulation pathway, as illustrated by construction of sporulation-specific sigma factor mutants. Furthermore, the Ptet promotor removes spo0A from its normal regulatory network; this allows the maintenance of high levels of spo0A expression when mutation of a gene in the sporulation network might otherwise serve to downregulate spo0A and potentially mask phenotypes of interest.
Our finding that accelerated accumulation of Spo0A correlates with more rapid sporulation contrasts with studies performed in B. subtilis, where accelerated accumulation is detrimental to sporulation (Vishnoi et al., 2013). We found no evidence that fast accumulation to elevated levels of Spo0A has any detrimental effect on sporulation, indicating that a gradual build-up of Spo0A is not important to the sporulation process itself in C. difficile. In B. subtilis, it has been suggested that the reduction in sporulation efficiency upon artificial induction of Spo0A resulted from premature repression of DivIVA, which is involved in chromosome segregation during the earlier stages of sporulation but downregulated via Spo0A in the later stages of sporulation (Vishnoi et al., 2013). Existing transcriptomic and proteomic studies of C. difficile Δspo0A have not found a statistically significant regulation of DivIVA by Spo0A (Pettit et al., 2014; Charlton et al., 2015), but the Ptet-spo0A strain will allow a more in-depth study of this connection, as well as investigations into the potential existence of high and low spo0A regulons more generally. It has been previously proposed that the phosphorelay system present in B. subtilis is responsible for the gradual build-up of Spo0A (Fujita and Losick, 2005) and our finding would be in line with this notion.
Finally, we demonstrated that the inducible system works in a mouse model of infection, with Ptet-spo0A remaining asporogenous in the absence of ATc in drinking water but sporulating in its presence. Although beyond the scope of this work, Ptet-spo0A could be used to investigate germination rates and efficiency in vivo, following infection with spores produced by ATc induction in vitro which will be unable to re-sporulate in vivo. Furthermore, we envisage that administration of ATc at different doses relative to mouse weight will allow dose-dependent phenotypes to be studied during infection. This paves the way for more detailed study into the role of Spo0A in virulence, and to delineate the relative importance of the many Spo0A related factors involved. The Ptet-spo0A strain described here is therefore a novel tool to study sporulation in C. difficile in a much more robust and controllable way, that will allow us to gain unique insights into the mechanisms involved in spore formation and related phenotypes, as well as the role of Spo0A in pathways leading to different lifecycle choices.
Materials and Methods
Bacterial Strains and Growth Conditions
Escherichia coli strains were grown in LB-Miller supplemented where necessary with chloramphenicol (15 μg/ml) and/or kanamycin (50 μg/ml). C. difficile strains were cultured statically in BHIS (37 g brain heart infusion, 5 g yeast extract and 1 g L-cysteine per liter), SM (90 g Bacto peptone, 5 g proteose peptone, 1 g NH4SO4, 1.5 g Tris base) or a 70:30 mixture of SM and BHIS as indicated, supplemented where necessary with thiamphenicol (15 μg/ml), D-cycloserine (250 μg/ml), 5-fluorocytosine (FC) (50 μg/ml) or anhydrotetracycline (ATc) (5–500 ng/ml). For plate cultures, media were solidified with 1.5% (w/v) agar. All C. difficile strains were grown and maintained at 37°C in a Don Whitley DG250 anaerobic workstation under anaerobic conditions (10% H2, 10% CO2, 80% N2) (Don Whitley Scientific). A detailed list of strains used in this study is provided in Supplementary Table S1.
Construction of Ptet-spo0A Mutant
To generate plasmid pSEW200, the following DNA fragments were PCR-amplified using KOD Hot Start polymerase (Novagen). The pMTL-SC7315 plasmid backbone (Cartman et al., 2012) was linearized by inverse PCR using primers 103 and 104. 1,200 bp upstream and downstream spo0A homology arms were amplified from C. difficile 630 genomic DNA, using primer pairs 155–156 and 159–160, respectively. Tet regulatory elements were amplified from pRPF185 (Fagan and Fairweather, 2011) using primer pair 157–158. The resulting fragments were ligated using Gibson assembly (NEB) as per manufacturer’s instructions. pSEW200 was transformed into E. coli CA434 and then conjugated into either C. difficile 630 or C. difficile 630Δerm as described previously (Purdy et al., 2002). Resulting colonies were restreaked twice onto fresh BHIS agar plates supplemented with thiamphenicol and D-cycloserine to counter-select for E. coli. Allelic exchange was performed as described previously (Cartman et al., 2012), with the slight modification of colonies being restreaked three times onto C. difficile Defined Medium (CDDM) (Karasawa et al., 1995) agar plates supplemented with FC (50 μg/ml) prior to patch-plating on BHIS agar plates supplemented with thiamphenicol to screen for plasmid excision. FC-resistant, thiamphenicol-sensitive clones were screened by PCR to separate mutants from WT revertants using primer pair 217–218. A detailed list of plasmids and primers used in this study is provided in Supplementary Tables S2 and S3, respectively.
Construction of sigEFG Mutants
sigEFG mutants were created in the 630Δerm Ptet-spo0A background via allele-coupled exchange as described previously (Cartman et al., 2012). Briefly, the pMTL-SC7315 plasmid backbone was linearized by inverse PCR using primers 103 and 104. 900 bp upstream and downstream homology arms were PCR amplified from C. difficile 630 genomic DNA, using primer pairs 161–162 and 163–164 (sigE); 167–168 and 169–170 (sigF); 174–175 and 176–177 (sigG) respectively. The resulting fragments were ligated using Gibson assembly (NEB) as per manufacturer’s instructions yielding pMLD126 (sigE), pMLD127 (sigF), and pMLD128 (sigG). All plasmids were transformed into E. coli CA434 and conjugated into C. difficile 630Δerm Ptet-spo0A as described previously (Purdy et al., 2002). Resulting colonies were restreaked twice onto fresh BHIS agar plates supplemented with thiamphenicol and D-cycloserine to counter-select for E. coli. These were then restreaked onto CDMM agar plates supplemented with FC (50 μg/ml) before patch-plating on BHIS agar supplemented with thiamphenicol to screen for plasmid excision. FC-resistant, thiamphenicol-sensitive clones were screened by PCR to separate mutants from WT revertants using primer pairs 165–166 (sigE), 171–172 (sigF), and 177–178 (sigG).
RNA Extraction and Quantitative Reverse Transcription PCR Analysis (qRT-PCR)
RNA was extracted from C. difficile cultures grown in BHIS to early stationary phase (OD600 ∼ 0.8) using the Total RNA Purification Kit (Norgen Biotek, Corp.) according to the manufacturer’s instructions. Purified RNA was DNase-treated using the Turbo DNA-free Kit (Life Technologies). First strand synthesis was performed on 1 μg of RNA using the SuperScript III VILO cDNA Synthesis Kit (Invitrogen) according to the manufacturer’s instructions. 50 ng of cDNA per reaction was used to perform qRT-PCR analysis using the SensiFast SYBR No-ROX Kit (Bioline) on a RotorGene 6000 instrument (Corbett Research). cDNA synthesis reactions containing no reverse transcriptase were included to control for genomic contamination. Primers for qRT-PCR analysis were designed using PrimerQuest (Integrated DNA Technologies), and primer efficiencies were calculated for each primer set prior to use. qRT-PCR was performed in technical triplicate for each cDNA sample and primer pair and on cDNA isolated from a minimum of two biological replicates. Results were calculated by the comparative cycle threshold method (Schmittgen and Livak, 2008) and normalized to the rpoC transcript. Results are presented as the means and standard deviations of the means and a two-tailed Student’s t-test was performed to determine statistical significance using GraphPad Prism 6.
Sporulation Assays
C. difficile cultures were grown in BHIS broth to logarithmic growth phase (OD600 ∼ 0.6), diluted 1:10,000 in fresh broth, and grown overnight to stationary phase. This allowed synchronization of growth and minimized the carry-over of spores from initial cultures. Total and heat-resistant CFUs were then enumerated at 24 h intervals for 5 days. To this end, at each time point serial 10-fold dilutions were prepared in pre-reduced PBS and 20 μl were spotted in triplicate onto BHIS agar supplemented with 0.1% (w/v) sodium taurocholate. To determine the number of spores, samples were incubated at 70°C for 30 min before spotting onto plates. Colonies were enumerated after 24 h incubation in an anaerobic cabinet.
Microscopy
0.5 ml samples from cultures grown in SM broth were harvested by centrifugation (2 min at 4,000 × g) 14 h after inoculation. Cells were washed with 1 ml of PBS, resuspended in 100 μl of PBS and spotted onto 1.5% (w/v) agarose pads supplemented with a lipophilic steryl membrane dye: N-(3-triethylammoniumprpyl)24-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide (FM4-64; Invitrogen; 1 μg/ml), a DNA dye: 2′-(4-hydroxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5′-bi-1H-benzimidazole trihydrochloride hydrate, bisBenzimide (Hoechst 33258, Thermo Fisher; 1 μg/ml) and 0.5% (v/v) di-methylsulfoxide (DMSO). Images were captured using the Metamorph software package on a Nikon Ti microscope and were processed using ImageJ (Schindelin et al., 2012; Schneider et al., 2012).
Western Blotting
To prepare C. difficile whole cell lysates, 0.5 ml samples from cultures grown overnight in BHIS to stationary phase were harvested by centrifugation (2 min at 4,000 × g), washed with PBS, resuspended to OD600 = 10 in sample buffer and boiled for 10 min. S-layer extracts were prepared using low pH glycine as previously described (Calabi et al., 2001). SDS-PAGE was carried out according to standard protocols. For immunoblot analysis, proteins were transferred to nitrocellulose membranes using the Trans-blot Turbo system (Bio-rad) according to the manufacturer’s instructions. Membranes were blocked in 3% (w/v) skimmed milk in PBS before probing with rabbit anti-Spo0A antibody at 1:5,000 or chicken anti-FliC at 1:100,000. Primary antibodies were detected using horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L) antibody at 1:2,500 (Promega) or goat anti-chicken IgY antibody at 1:10,000 (Novex, Life Technologies) and Clarity Western ECL substrate (Bio-rad).
Animal Studies
12 C57BL/6 mice aged 6–7 weeks (Charles River) were pre-treated with drinking water containing 250 mg/ml clindamycin for 7 days. Clindamycin treatment was then removed for 24 h, after which the mice were infected by oral gavage with 1 × 107 CFU Ptet-spo0A grown for 16 h in TYG (3% w/v Bacto tryptose, 2% w/v yeast, 0.5% w/v glucose). The mice were split into two groups of 6, each receiving either 0.1 mg/ml ATc or 1% (v/v) ethanol in drinking water, refreshed every 24 h. Fresh feces were collected and suspended in sterilized distilled water to 0.2 g/ml. For heat-treated counts, the suspension was treated at 70°C for 30 min. 20 μl of suspension was plated in technical triplicate on Chrome ID plates (BioMerieux) and CFU/g fecal matter enumerated following 16 h growth.
Ethics Statement
All animal work was performed under the United Kingdom Home Office project license PPL 70/8276 and with approval from the Royal Holloway University of London Ethics Committee.
Author Contributions
MD and SW designed the study, developed the methodology, collected and analyzed the data, wrote and revised the manuscript; SH and HH collected the data; PS and SC designed the study, analyzed the data, revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Glen D. Armstrong for providing the anti-FliC antibodies and Dr. Aimee Shen for providing the anti-Spo0A antibodies.
Footnotes
Funding. This work was supported by grants from the MRC (DPFS MR/K015354/1) and European Union (HEALTH-F3-2013-601810) to SC. MD is supported by an MRC NIRG (MR/M000923/1) awarded to PS.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01793/full#supplementary-material
References
- Al-Hinai M. A., Jones S. W., Papoutsakis E. T. (2015). The Clostridium sporulation programs: diversity and preservation of endospore differentiation. Microbiol. Mol. Biol. Rev. 79 19–37. 10.1128/MMBR.00025-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns D. A., Heap J. T., Minton N. P. (2010). The diverse sporulation characteristics of Clostridium difficile clinical isolates are not associated with type. Anaerobe 16 618–622. 10.1016/j.anaerobe.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Burns D. A., Minton N. P. (2011). Sporulation studies in Clostridium difficile. J. Microbiol. Methods 87 133–138. 10.1016/j.mimet.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Calabi E., Ward S., Wren B., Paxton T., Panico M., Morris H., et al. (2001). Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40 1187–1199. 10.1046/j.1365-2958.2001.02461.x [DOI] [PubMed] [Google Scholar]
- Cartman S. T., Kelly M. L., Heeg D., Heap J. T., Minton N. P. (2012). Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between tcdC genotype and toxin production. Appl. Environ. Microbiol. 78 4683–4690. 10.1128/AEM.00249-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton T. M., Kovacs-Simon A., Michell S. L., Fairweather N. F., Tate E. W. (2015). Quantitative lipoproteomics in Clostridium difficile reveals a role for lipoproteins in sporulation. Chem. Biol. 22 1562–1573. 10.1016/j.chembiol.2015.10.006 [DOI] [PubMed] [Google Scholar]
- Dawson L. F., Valiente E., Donahue E. H., Birchenough G., Wren B. W. (2011). Hypervirulent Clostridium difficile PCR-ribotypes exhibit resistance to widely used disinfectants. PLOS ONE 6:e25754 10.1371/journal.pone.0025754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin L. J., Clare S., Fagan R. P., Dawson L. F., Pickard D. J., West M. R., et al. (2012). The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 80 2704–2711. 10.1128/IAI.00147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Huse S., Sogin M. L., Relman D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLOS Biol. 6:e280 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubberke E. R., Olsen M. A. (2012). Burden of Clostridium difficile on the healthcare system. Clin. Infect. Dis. 55(Suppl. 2) S88–S92. 10.1093/cid/cis335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. N., Nawrocki K. L., Mcbride S. M. (2014). Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect. Immun. 82 4276–4291. 10.1128/IAI.02323-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. N., Tamayo R., Mcbride S. M. (2016). A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol. Microbiol. 100 954–971. 10.1111/mmi.13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan R. P., Fairweather N. F. (2011). Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286 27483–27493. 10.1074/jbc.M111.263889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimlaid K. A., Bond J. P., Schutz K. C., Putnam E. E., Leung J. M., Lawley T. D., et al. (2013). Global analysis of the sporulation pathway of Clostridium difficile. PLOS Genet. 9:e1003660 10.1371/journal.pgen.1003660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Gonzalez-Pastor J. E., Losick R. (2005). High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187 1357–1368. 10.1128/JB.187.4.1357-1368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Losick R. (2005). Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19 2236–2244. 10.1101/gad.1335705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap J. T., Ehsaan M., Cooksley C. M., Ng Y. K., Cartman S. T., Winzer K., et al. (2012). Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 40:e59 10.1093/nar/gkr1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Zhang B., Van Horn S. F., Warren P., Woodnutt G., Burnham M. K., et al. (2001). Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293 2266–2269. 10.1126/science.1063566 [DOI] [PubMed] [Google Scholar]
- Jiang M., Grau R., Perego M. (2000a). Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J. Bacteriol. 182 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Shao W., Perego M., Hoch J. A. (2000b). Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38 535–542. [DOI] [PubMed] [Google Scholar]
- Karasawa T., Ikoma S., Yamakawa K., Nakamura S. (1995). A defined growth medium for Clostridium difficile. Microbiology 141(Pt 2) 371–375. 10.1099/13500872-141-2-371 [DOI] [PubMed] [Google Scholar]
- Kotula J. W., Kerns S. J., Shaket L. A., Siraj L., Collins J. J., Way J. C., et al. (2014). Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U.S.A. 111 4838–4843. 10.1073/pnas.1321321111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D., Clare S., Deakin L. J., Goulding D., Yen J. L., Raisen C., et al. (2010). Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl. Environ. Microbiol. 76 6895–6900. 10.1128/AEM.00718-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D., Walker A. W. (2013). Intestinal colonization resistance. Immunology 138 1–11. 10.1111/j.1365-2567.2012.03616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackin K. E., Carter G. P., Howarth P., Rood J. I., Lyras D. (2013). Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PLOS ONE 8:e79666 10.1371/journal.pone.0079666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V., Fujita M., Jensen S. T., Eichenberger P., Gonzalez-Pastor J. E., Liu J. S., et al. (2003). The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50 1683–1701. 10.1046/j.1365-2958.2003.03818.x [DOI] [PubMed] [Google Scholar]
- Nawrocki K. L., Edwards A. N., Daou N., Bouillaut L., Mcbride S. M. (2016). CodY-dependent regulation of sporulation in Clostridium difficile. J. Bacteriol. 198 2113–2130. 10.1128/JB.00220-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes C. J., Alsaker K. V., Papoutsakis E. T. (2005). A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3 969–978. 10.1038/nrmicro1288 [DOI] [PubMed] [Google Scholar]
- Pereira F. C., Saujet L., Tome A. R., Serrano M., Monot M., Couture-Tosi E., et al. (2013). The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLOS Genet. 9:e1003782 10.1371/journal.pgen.1003782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit L. J., Browne H. P., Yu L., Smits W. K., Fagan R. P., Barquist L., et al. (2014). Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics 15:160 10.1186/1471-2164-15-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy D., O’keeffe T. A., Elmore M., Herbert M., Mcleod A., Bokori-Brown M., et al. (2002). Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46 439–452. 10.1046/j.1365-2958.2002.03134.x [DOI] [PubMed] [Google Scholar]
- Putnam E. E., Nock A. M., Lawley T. D., Shen A. (2013). SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J. Bacteriol. 195 1214–1225. 10.1128/JB.02181-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbusch K. E., Bakker D., Kuijper E. J., Smits W. K. (2012). C. difficile 630Deltaerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLOS ONE 7:e48608 10.1371/journal.pone.0048608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik M., Wilcox M. H., Gerding D. N. (2009). Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7 526–536. 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits W. K., Lyras D., Lacy D. B., Wilcox M. H., Kuijper E. J. (2016). Clostridium difficile infection. Nat. Rev. Dis. Primers 2:16020 10.1038/nrdp.2016.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L. (2000). Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3 561–566. 10.1016/S1369-5274(00)00141-7 [DOI] [PubMed] [Google Scholar]
- Steiner E., Dago A. E., Young D. I., Heap J. T., Minton N. P., Hoch J. A., et al. (2011). Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol. Microbiol. 80 641–654. 10.1111/j.1365-2958.2011.07608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood S., Guan S., Vijayasubhash V., Baines S. D., Graham L., Lewis R. J., et al. (2009). Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 191 7296–7305. 10.1128/JB.00882-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J. W., Hamoen L. W., Kuipers O. P. (2005). Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol. Microbiol. 56 1481–1494. 10.1111/j.1365-2958.2005.04659.x [DOI] [PubMed] [Google Scholar]
- Vishnoi M., Narula J., Devi S. N., Dao H. A., Igoshin O. A., Fujita M. (2013). Triggering sporulation in Bacillus subtilis with artificial two-component systems reveals the importance of proper Spo0A activation dynamics. Mol. Microbiol. 90 181–194. 10.1111/mmi.12357 [DOI] [PubMed] [Google Scholar]
- Vohra P., Poxton I. R. (2011). Efficacy of decontaminants and disinfectants against Clostridium difficile. J. Med. Microbiol. 60 1218–1224. 10.1099/jmm.0.030288-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


