Abstract
High-risk breast cancer comprises distinct tumor entities such as triple-negative breast cancer (TNBC) which is characterized by lack of estrogen (ER) and progesterone (PR) and the HER2 receptor and breast malignancies which have spread to more than three lymph nodes. For such patients, current (inter)national guidelines recommend anthracycline-based chemotherapy as the standard of care, but not all patients do equally benefit from such a chemotherapy. To further improve therapy decision-making, predictive biomarkers are of high, so far unmet, medical need. In this respect, predictive biomarkers would permit patient selection for a particular kind of chemotherapy and, by this, guide physicians to optimize the treatment plan for each patient individually. Besides DNA mutations, DNA methylation as a patient selection marker has received increasing clinical attention. For instance, significant evidence has accumulated that methylation of the PITX2 (paired-like homeodomain transcription factor 2) gene might serve as a novel predictive and prognostic biomarker, for a variety of cancer diseases. This review highlights the current understanding of treatment modalities of high-risk breast cancer patients with a focus on recommended treatment options, with special attention on the future clinical application of PITX2 as a predictive biomarker to personalize breast cancer management.
1. Introduction
Breast cancer is the most common malignancy in women with >464,000 new cases diagnosed in 2012 in Europe; 131,000 patients have died as a result of this disease [1]. In the Western, one in eight women will experience breast cancer in her lifetime; every fourth is younger than 55 years of age at the time of diagnosis [1]. Therapy decision in breast cancer is mainly based on the established histopathological parameters tumor size, lymph node status, histological type, and histological grade. These factors do provide information about the future clinical course of the disease (exemplified as disease-free survival, DFS, and overall survival, OS) of patients not subjected to any systemic cancer therapy. Different from that, predictive factors are biological markers, which foretell the probability of a cancer patient to respond to a specific anticancer therapy.
At present, these biomarkers include the steroid hormone receptors ER and PR, the oncoprotein HER2, the urokinase-type plasminogen activator (uPA), and the uPA inhibitor PAI-1 [2] (ASCO: http://www.asco.org/practice-guidelines/quality-guidelines/guidelines, AGO: http://www.ago-online.de/en/guidelines-mamma/march-2016). According to the opinion of the St. Gallen breast cancer classification and treatment panel, Ki-67, a cell proliferation marker, should also be considered for therapy decision-making although no standard protocols for this analysis were defined and established so far [3, 4].
By use of molecular classifiers and gene expression signatures, invasive breast carcinomas can be further clinically categorized into at least four molecular subgroups (Table 1), which are associated with different clinical outcomes and are the basis for choosing which kind of systemic therapy should be applied, both for the adjuvant and the neoadjuvant setting [4–8]. Patients in the luminal A/B groups are generally treated with endocrine therapy but, due to biological heterogeneity in this group, quite often combined with chemotherapy (Table 1). It is estimated that 20–40% of those patients are treated cost- and side-effect-rich with chemotherapy, although endocrine therapy would be sufficient. Women with early-stage breast cancer presenting with HER2+ tumors (HER2-type) are generally treated in the adjuvant setting with the humanized antibody Herceptin® plus additional chemotherapy. Since such targeted therapy is not available for the TNBC patients, they are subject to systemic adjuvant or neoadjuvant chemotherapy. Yet avoiding not only overtreatment but also undertreatment of TNBC patients has become a major treatment issue. Therefore, TNBC-specific predictive biomarkers are urgently needed to allow for the identification of TNBC patients who will benefit from a particular systemic therapy. If not so, such TNBC patients should be offered alternative treatment plans to achieve optimal cancer treatment, thereby avoiding potentially toxic side effects.
Table 1.
Traditional classification of breast cancer subgroups.
| Classification | Expression | Distribution (%) |
|---|---|---|
| Luminal A | ER/PR+, HER2− | ~65 |
| Luminal B | ER/PR+, HER2+/− | ~15 |
| HER2-type | ER/PR−, HER2+ | ~5 |
| Triple-negative | ER/PR−, HER2− | ~15 |
According to [3, 5, 8] (http://ww5.komen.org/BreastCancer/SubtypesofBreastCancer.html).
2. Definition of High-Risk Breast Cancer Patients
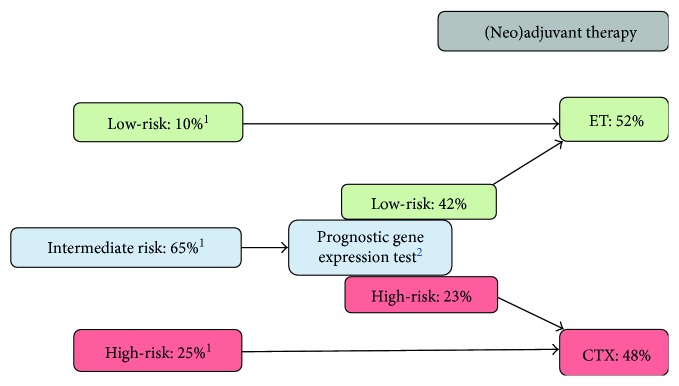
Detailed information for risk stratification of breast cancer patients who would develop metastases is urgently needed to provide effective care early enough to ensure provision of adequate anticancer treatment. In February 1998, the 6th International Conference on “Adjuvant Therapy of Primary Breast Cancer” was held in St. Gallen, Switzerland. In an attempt to solve this problem, at this conference and subsequent St. Gallen meetings, guidelines and recommendations were introduced in the scientific literature on how to select the right breast cancer patient for adequate adjuvant systemic treatment [9, 10]. According to the St. Gallen's patient stratification criteria (Figures 1 and 2), patients with a low risk to progress or to develop metastases (tumor size < 2 cm, node-negative, or up to 3 lymph nodes affected) will receive endocrine therapy if ER and/or PR are expressed. High-risk patients (node-positive with >3 lymph nodes involved, tumor size ≥ 2 cm) are subject to chemotherapy; the same applies to TNBC patients. Intermediate-risk patients, which are defined by pN0-1 [4] or histological grade 2 [11], can be allocated to the high-risk or low-risk group by applying commercially available risk-assessment tests such as MammaPrint® (Agendia), Endopredict® (Myriad Genetics), or Oncotype DX® (Genomic Health) [4, 12].
Figure 1.
Clinicopathological risk stratification of breast cancer patients according to St. Gallen criteria. 1Clinicopathological risk assessment according to St. Gallen consensus panel [9, 20–24]. 2Endopredict®, OncotypeDX®. ET = endocrine therapy; CTX = chemotherapy.
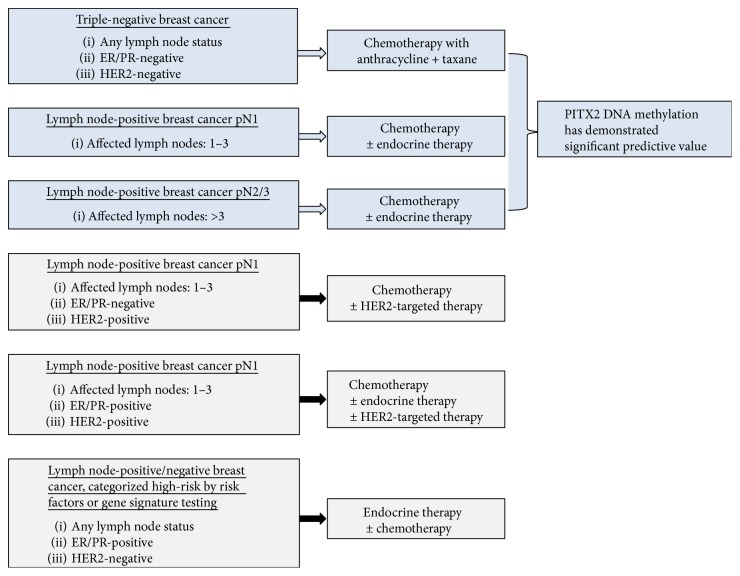
Figure 2.
Classification of the high-risk breast cancer subtypes. PITX2 DNA methylation status can serve as a significant predictive biomarker for anthracycline-based chemotherapy in the two major high-risk breast cancer subtypes [44, 66].
TNBC tumors and tumors with >3 affected lymph nodes are both considered high-risk breast cancers. TNBC is characterized by an increase in tumor size and high histological grade with disease recurrence as early as 3–5 years after the start of systemic cancer therapy [13, 14] and shorter survival time following the first metastatic event [1, 15–17]. Thus, the 5-year survival rate of TNBC patients is significantly reduced compared to other breast cancer subtypes.
Currently, there is no routinely available targeted therapy established for TNBC patients and no predictive factors have been introduced into the clinic which could provide information about the response of a TNBC patient to a specific cancer therapy [2]. Primary breast cancer patients with ≤3 affected lymph nodes but ER/PR negative status or HER2 positive are considered high-risk breast cancer patients as well [18]. For those patients, anthracycline-based adjuvant chemotherapy has become the standard of care. Several studies showed that the addition of taxanes to this protocol may further improve clinical outcome. Still, not all of these high-risk patients will benefit from such an intensified therapy plan [19].
3. Treatment Options for High-Risk Breast Cancer Patients
In addition to the established traditional histopathological parameters (lymph node status, tumor size, histological type, and histological grade), the decision who to treat and how to treat is also based on whether ER/PR and HER2 are expressed by the primary breast cancer tumor cells or not (Table 2). The biomarkers uPA and PAI-1 are indicative factors to predict response to adjuvant chemotherapy but in node-negative breast cancer patients only [18, 25].
Table 2.
Current therapeutic regimens for breast cancers contain.
| (i) For patients with luminal A/B tumors (ER/PR+, HER2−): endocrine therapy or chemotherapy followed by endocrine therapy. (ii) For patients with HER2+ tumors: chemotherapy plus anti-HER2 therapy (e.g., with trastuzumab/pertuzumab) and, if appropriate, endocrine therapy. The combination with chemotherapy is currently recommended since by this therapy a survival benefit for the patients has been reported [4]. (iii) For TNBC patients: chemotherapy (anthracycline plus taxane) (AGO guidelines 2016: http://www.ago-online.de/en/guidelines-mamma/march-2016). |
Originally, treatment in the neoadjuvant setting was advised for the treatment of larger sometimes inoperable breast cancers, but various considerations support the implementation of neoadjuvant systemic chemotherapy in operable breast cancer patients as well [4, 26]. Benefits of such a systemic therapy procedure are higher rates of breast-conserving surgery and the opportunity to assess early in vivo response to systemic treatment prior to primary surgery. In addition, this approach might improve survival of breast cancer patients by early elimination of occult metastases. Neoadjuvant chemotherapy is particularly recommended for TNBC patients and patients afflicted with HER2+ breast cancer [4]. Current adjuvant chemotherapy standards for early breast cancer patients involve an anthracycline plus taxane. These regimens generally do not differ for the neoadjuvant and the adjuvant settings [4].
Standard treatment for breast cancer patients with more than three affected lymph nodes is systemic therapy employing an anthracycline plus a taxane (Table 3), administered in the adjuvant or neoadjuvant setting. In addition, anti-HER2 therapy for HER2+ tumors and/or endocrine therapy for ER/PR+ tumors can be applied. Results of several trials addressing N+ high-risk breast cancer patients have shown that dose-dense chemotherapy improves outcome compared to standard interval chemotherapy [4]. Patients are treated by adjuvant anthracycline-based chemotherapy; additional treatment with adjuvant endocrine therapy will be administered when tumor cells are ER/PR+. In patients with more than three affected lymph nodes, dose-dense and dose-intensified epirubicin, paclitaxel, and cyclophosphamide led not only to a significant reduction in disease recurrence and mortality but also to higher toxicities [4].
Table 3.
Treatment options for high-risk breast cancer patients.
| TNBC | >3 lymph nodes affected |
|---|---|
| Therapy: CTX | Therapy: CTX, anti-HER2, or ET |
| (i) Anthracycline plus taxane (ii) Neoadjuvant treatment (presurgical) (iii) Adjuvant treatment (postsurgical) (iv) Addition of carboplatin may improve pCR and event-free survival |
(i) Adjuvant treatment with anthracycline plus taxane (ii) Anti-HER2 treatment if tumor HER2+, and/or endocrine therapy if tumor ER/PR+ (iii) Regimens may also include platinum salts (cisplatin, carboplatin) (iv) Dose-dense or dose-intensified CTX improves outcome in N+ patients |
pCR: pathological complete response; CTX: chemotherapy; anthracyclines: doxorubicin, epirubicin; Taxanes: paclitaxel, docetaxel; N+: node-positive. According to AGO guidelines (http://www.ago-online.de/en/guidelines-mamma/march-2016).
In general, in Germany, for the presurgical neoadjuvant setting, TNBC patients are treated following the AGO guidelines (http://www.ago-online.de/en/guidelines-mamma/march-2016), which are characterized by core biopsies taken to allow histological diagnosis, followed by neoadjuvant chemotherapy, primary surgery, and, if applicable, further postsurgical chemotherapy. Anthracycline-based chemotherapy is hereby the standard therapy option. For high-risk N+ TNBC patients, a definite benefit for the patients was reported when anthracyclines were applied in combination with taxanes [27]. In patients with highly proliferating TNBC, in the adjuvant setting, anthracycline-containing regimes improve disease-free and overall survival when compared to treatment with CMF (combination of cyclophosphamide, methotrexate, and 5-fluorouracile [28]).
In TNBC patients, a favorable prognosis is predicted by an effective response to (neo)adjuvant chemotherapy, which is defined by high rate of pCR (pathological complete remission) which may serve as a surrogate endpoint for estimation of long-term clinical outcomes of the patients [29]. Those patients who will not achieve a pCR are at high risk of disease recurrence within the next two years following surgery [29]. Until now, predictors of benefit from anthracyclines, or a test which predicts response to anthracycline-based treatment, are not commercially available [28]. The addition of carboplatin or cisplatin can significantly improve the rate of pCR [13]. Platinum salts do effect DNA cross-link strand breaks and, thus, prevent DNA replication [7]. Staudacher et al. [30] reported that overall survival was improved in metastatic breast cancer patients responding to platinum-based chemotherapy. Since 2014, results from several clinical trials have indicated that addition of platinum salts to a neoadjuvant anthracycline-taxane combination or sequence does improve pCR [4].
4. Predictive Biomarkers in High-Risk Breast Cancer
Breast cancer is a genetically and phenotypically very heterogeneous disease equipped with molecular diversity. It may exhibit with distinct clinical appearance, regarding varying response to treatment and clinical outcome. Consequently, precise breast cancer stratification for and forecasting of effectiveness of therapeutic modalities is a crucial step toward a more beneficial treatment design. For many years, breast cancer patients have been stratified according to their histopathological parameters such as histological type and grade, tumor size, lymph node status, and the ER/PR/HER2− status.
In recent years, significant advances have been made in characterizing the molecular characteristics of TNBC [5, 8, 14]. This has led to the identification of biomarkers that potentially could be used for diagnostic purpose to assess patient's prognosis or as therapeutic targets. Irrespective of the fact that today for TNBC there are no effective targeted therapies available. About one-third of the patients will achieve pCR through standard-of-care anthracycline/taxane chemotherapy; but how are these responders classified? So far, the broad molecular heterogeneity of TNBC tumors has hindered the discovery of effective biomarkers to identify such patients in order to tailor chemotherapy at an individual level [13, 31, 32]. To further characterize TNBC on a molecular level, various subtypes of TNBC have been proposed [33] (Table 4).
Table 4.
Subclassification of TNBC based on gene expression analysis.
| Characteristics | |
|---|---|
| BL1 (basal-like 1) | Increased expression of cell cycle and DNA repair genes |
| BL2 (basal-like 2) | Increased expression of growth factor signaling and myoepithelial markers |
| M | Increased expression of genes involved in epithelial-mesenchymal-like transition and growth factor pathways |
| MSL | Decreased expression of genes involved in proliferation (mesenchymal stem cell-like) |
| IM | Immune cell processes, expression of genes involved in cytokine signal immunomodulatory transduction pathway |
| LAR | Luminal gene expression and androgen receptor signaling genes |
Eighty to ninety percent of TNBCs belong to the BL subtype. Despite some differences in the number of subtypes or the classifying methods, regarding this issue, all of the published studies suggested that TNBC consists of several subtypes and thus does require subtype-specific therapy based on its biological characteristics [34]. The clinical relevance of the TNBC subtypes listed in Table 4, however, is not yet defined. Cancer biomarkers may be useful as diagnostic, prognostic, or predictive indicators and may represent potential targets for cancer therapy [34, 35]. Although there are no clinically meaningful prognostic or predictive cancer biomarkers available, a series of potential biomarkers have been identified, for example, in the blood (VEGF), on the cell surface (epithelial growth factor receptor 1 (EGFR)), and in the cell nucleus (BRCA1, BRCA2) [32]. VEGF (vascular epithelial growth factor receptor) causes proliferation of endothelium cells and regulates vascular permeability and migration of endothelial stem cells from the bone marrow [34]. In TNBC patients, elevated VEGF levels were observed to be associated with disease recurrence and survival probability [34], making VEGF a potential therapy target in TNBC.
Activation of EGFR causes transcription of genes thus inducing cell proliferation, angiogenesis, metastatic spread, and inhibition of apoptosis. EGFR was found to be overexpressed in many TNBC tumors and was shown to be a prognostic factor for disease recurrence by univariate and multivariate analysis [16, 29, 32]. Currently, tyrosine kinase inhibitors (e.g., lapatinib and gefitinib) are applied in breast cancer patients to block EGF-stimulated growth signal transduction and that of HER2 and the combination of these drugs with carboplatin or docetaxel synergistically may enhance the treatment effect in TNBC patients [29, 34, 36–38].
BRCA1 and BRCA2 germline mutations are more frequent in TNBC than in other types of breast cancer and affect ~30% of TNBC patients [13, 39]. There is increasing evidence that BRCA1 germline mutant breast cancers present with above-average platinum sensitivity and increased sensitivity to poly-ADP ribose polymerase (PARP) inhibitors [13]. Germline mutations in the BRCA genes (BRCA1, BRCA2) predispose individuals to develop several kinds of cancer, including that of the breast [32]. Both genes play a crucial role in DNA repair processes, and the lack of functional BRCA1/2 proteins leads to loss of repair of DNA double-strand breaks and subsequently increases the risk of cancer. Mutated BRCA1-related breast cancers share pathological features with TNBC, including ER, PR, HER2, EGFR, TP53 expression, and genomic instability [34, 40].
The link between TNBC and germline BRCA1 mutations has led to the investigation of PARP inhibitors in TNBC. PARPs are cell signaling enzymes which catalyze the poly(ADP-ribosylation) of DNA binding proteins. The main function is to act as a nick sensor for DNA damage, which plays a vital role in DNA repair through the base excision repair pathway: the base excision pathway fails when PARP1 is inhibited. In BRCA1-deficient cells, inhibition of PARP1 leads to cell death through apoptosis [29]. Trials of PARP inhibitors (e.g. iniparib, ocaparib, veliparib, talazoparib, and rucaparib) in TNBCs have shown improved rates of response and progression-free survival [6, 15, 29, 41]; however, these studies are inconclusive [7].
Ki-67 is a marker for cell proliferation activity. In TNBC patients, Ki-67 levels were related to tumor size and histological grade, and it was associated with increased pCR when responding to chemotherapy but with poor disease-free and overall survival [34]. Ki-67 is one of the biomarkers which is recommended to be included in breast cancer therapy decision-making. However, so far, no standards for this analysis are defined [3].
In TNBC, the prevalence of the androgen receptor (AR) is ~10–20% [13, 42]. The LAR subtype of TNBC (Table 4) is characterized by luminal gene expression, enriched for AR expression and its target genes [33]. Clinically, individuals with AR-TNBC have a higher likelihood of achieving a pCR by treatment with neoadjuvant chemotherapy than those with AR+ TNBC [42]. Next-generation sequencing activities have identified further genes recurrently mutated in TNBC, including TP53, PIK3CA, PTEN, RB1, RAS, and ERBB3, but none of these genes have any predictive or prognostic relevance [31].
According to the St. Gallen risk stratification scheme (Figure 1), the intermediate-risk group (pN0-1) can be further classified into low- or high-risk by using multigene expression assays, for example, Endopredict (Myriad Genetics), Mammaprint (Agendia), OncotypeDX (Genomic Health), and others. OncotypeDX delivers a 21-gene recurrence score, which gives information about the likelihood of chemotherapy benefit as well as the risk of disease recurrence in early-stage breast cancer. This test is recommended for breast cancer patients with tumors classified as N0/N+, ER+ [12]. Mammaprint (70-gene signature) gives information about a breast cancer patient's risk for disease recurrence and identifies those patients that may safely forgo chemotherapy. Mammaprint is recommended for pN0 and pN+ breast cancer patients. Endopredict provides information how to devise personalized treatment plans for breast cancer patients. It detects the likelihood of late metastases (>5 years) and can thus guide treatment decision for CTX and predicts disease recurrence. This test is recommended for pN0-1, ER+, HER2− breast cancer patients [4, 36].
All these multigene assays, however, deliver prognostic information only but have no predictive value, also not for TNBC patients [2]. In summary, none of these tests can currently be recommended for predicting the response to a specific form of chemotherapy or to be prognostic for any kind of high-risk breast cancer patients [36]. In addition to tumor size and nodal status, the ER/PR/HER2 status is currently the most important prognostic and predictive marker in high-risk breast cancer. There is still a so far unmet need for individualized systemic treatment of breast cancer to predict the necessity, efficacy, and potentially toxic side effects of anticancer drugs and clinical outcome of breast cancer patients under therapy. Thus, molecular tests, which can predict response or failure to a certain therapy, are highly needed to provide a tailored therapy for the appropriate patient.
5. Epigenetics and Breast Cancer
The term epigenetics describes dynamic alterations in a cell that switch genes on and off without changes in the DNA sequence. Epigenetic modifications are reversible. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters gene expression. Many cellular processes are influenced by epigenetic changes, including gene expression, cellular differentiation, genomic imprinting, and embryogenesis [43]. DNA methylation is a chemical process that adds a methyl group to DNA on the 5th position of the pyrimidine ring of cytosine. It is highly specific and frequently happens in a region in which a cytosine nucleotide is located next to a guanine nucleotide that is linked by a phosphate: a so-called CpG site. A region of several hundred CpG sites is called CpG island [41, 43–45]. Many human gene promoters are associated with CpG islands and are usually unmethylated; a few become methylated during cell development or differentiation [41, 43]. DNA methylation can inhibit the binding of transcription factors to the promoter, and consequently, methylation in the promoter region is associated with silencing of the adjacent gene due to the abrogation of transcription [43, 46].
DNA methylation plays a crucial role in the development of a variety of cancers, including breast cancer [46]. DNA hypomethylation can lead to oncogene activation and chromosome instability in tumor development [41]. The loss of methylation in CpG-depleted regions, where CpG-dinucleotides are expected to be methylated, may be associated with aberrant or inappropriate expression of some genes that could contribute to neoplastic transformation, tumorigenesis, or cancer progression [47]. Conversely, hypermethylation has been shown to inhibit tumor suppressor genes, thereby releasing cells from their normal physiological control [41, 43].
Multiple reports have suggested that determination of the methylation status of specific promoter regions can provide important information for early detection of cancer, determine prognosis, and predict the response of a cancer patient to anticancer drugs [46]. In cancer, many tumor suppressor genes and various other cancer-related genes have been found to be hypermethylated. Their biological function includes cell-cycle regulation, apoptosis, DNA repair, cellular homeostasis, cell adhesion, and cell invasion [43, 45]. Examples for hypermethylated breast cancer-associated genes are BRCA1, CCND2 (cyclin D2), ER, PR, CDH1 (E-cadherin), and many others [43–49]. The ubiquity of such epigenetic changes in cancer events through DNA methylation has led to a variety of innovative diagnostic and therapeutic strategies.
The most recent technical advances have demonstrated the great potential of DNA methylation markers as valuable tools for decision-making in the treatment of cancer patients [43, 50]. In breast cancer, DNA methylation has shown promise as a potential biomarker for early detection, therapy monitoring, assessment of prognosis, and prediction of therapy response [46]. DNA methylation markers predicting response to endocrine therapy with tamoxifen in early and metastatic breast cancer have been described [45, 50, 51]. Furthermore, the DNA methylation status of the ER gene has been suggested as a marker for treatment response in breast cancer patients receiving antihormonal therapy [52]. Association between DNA methylation levels and clinicopathological parameters was reported, confirming complex relationships between DNA methylation and TP53 status or the ER status [43].
There have been only a few studies focusing on the investigation of the DNA methylation of certain genes in TNBC tumor tissues. For example, a DNA methylation signature relevant for TNBC patients was identified by Stirzaker et al. exploring Cancer Genome Atlas data [53]. They showed that TNBC patients with low levels of tumor DNA methylation in their gene signature had the best prognosis, and by their gene methylation signature, TNBC could be separated from non-TNBC tumors. Hafez et al. observed that p16 (cyclin-dependent kinase inhibitor 2A), a tumor suppressor gene, which has a central function in the regulation of cell cycle activation, was frequently hypermethylated in TNBC cases, and p16 hypermethylation was significantly increased in TNBC compared to non-TNBC [54].
Several studies have focused on the methylation status of the BRCA1 gene, a key player in breast cancer including TNBC. TNBC breast cancer cell lines with BRCA1 DNA methylation were more sensitive to PARP inhibitors when BRCA1 gene was methylated [55]. TNBC patients with pCR to adjuvant chemotherapy expressed higher BRCA1 methylation values than nonresponders [56], and the study from Xu et al. demonstrated an increased 10-year disease-free survival of 78% in TNBC patients with BRCA1 methylation compared to 55% in patients without BRCA1 methylation [57].
Being aware that DNA methylation is altered in breast cancer cells compared to normal breast cells and new assays that determine these changes and thus provide information about the patient's response to anti-cancer drugs need to be introduced into clinical practice in the near future. Although some genes altered by DNA methylation have been associated with response to adjuvant therapy in breast cancer patients in small exploratory studies using laboratory-developed tests, currently, for breast cancer testing, no predictive DNA methylation test is commercially available yet. This is remarkable since, in contrast to RNA and proteins, DNA is a very stable biological material that can be extracted from the same archived clinical tissue sample that is subjected to inspection by the pathologist for routine malignancy assessment.
6. Pathobiology of PITX2 in Breast Cancer
PITX2 (paired-like homeodomain transcription factor 2, also known as pituitary homeobox 2) is a transcription factor, which is involved in the morphogenesis of anterior structures, such as eyes and teeth. PITX2 is involved in pituitary-specific gene regulation and left-right patterning during embryonic and organogenic development [58, 59]. PITX2 has three different isoforms, leading to three different proteins (PITX2A, PITX2B, and PITX2C), which differentially regulate transcription of their target genes [60]. Two promoters (P1 and P2) are operational: P2 drives transcription of two mRNA variants, leading to the PITX2A and B proteins; P1 drives the third transcript variant encoding the C protein [58, 61, 62]. Expression of isoforms 1 and 2 is regulated by the Wnt/β-catenin pathway, and expression of isoform 3 is regulated by TGF-β family members [48, 58]. Both the Wnt/β-catenin pathway and the TGF-β pathway play important roles in carcinogenesis, but the results from Pillai et al. support the notion that PITX2 plays a role in mediating invasiveness of cancer cells through the Wnt/β-catenin pathway [58]. PITX2 functions in the Wnt-signaling pathway by recruiting and interacting with cytosolic β-catenin, the central molecule in the canonical Wnt pathway. This leads to stabilization of β-catenin, which then enters the nucleus and associates with transcription factors leading to transcription of cell cycle regulatory and proliferation genes (e.g., cyclin D1, c-Myc, and MMP7), and subsequently enhances cell proliferation [17].
Even today, the role of PITX2 in breast carcinogenesis is unclear; however, there is a role of PITX2 DNA methylation for the prognosis of the course of the breast cancer disease. Nimmrich et al. reported that PITX2 DNA methylation assessed in breast cancer tissue is a high-risk indicator of disease recurrence in N0 ER/PR+ patients [48]. Another study, employing immunohistochemistry for PITX2 determination, revealed a significant association between PITX2 protein and ER/PR expression, indicating that PITX2 and ER/PR protein expression may be useful prognostic markers in invasive breast cancer [63]. Jezkova et al. observed hypermethylation of PITX2 in about 50% of invasive breast cancers [64]; an association of PITX2 expression with established parameters such as ER, PR, and HER2 was described by Rahman et al. [63]. Further, PITX2 DNA methylation rates were found to be higher in tumors with elevated ER levels [49]. Jezkova et al. found that PITX2 DNA methylation status was associated with high tumor grade and clinical tumor stage of breast cancer patients [64].
A further breast cancer study (ER/PR+, N0) was conducted by Nimmrich et al. [48]. The authors reported that PITX2 DNA methylation acts as a statistically independent prognostic marker for these untreated breast cancer patients, implying that tumors with a hypermethylated PITX2 status are more aggressive. In a univariate survival analysis, the authors did show that PITX2 DNA methylation is associated with early distant metastases and poor overall survival. In a multivariate analysis, PITX2 retained its statistical significance, together with the established prognostic factors age, tumor size, and nuclear grade, plus ER/PR [48]. This study also confirmed that in clinical samples PITX2 DNA hypermethylation is positively associated with breast cancer disease progression [45, 48].
Results from the scientific literature provide further evidence that PITX2 DNA methylation analysis may allow clinically relevant risk assessment in tamoxifen-treated primary breast cancers [51, 65]. In the study performed by Maier et al., PITX2 DNA methylation showed the strongest correlation with metastasis-free survival in N0 ER/PR+, tamoxifen-treated breast cancer patients, with high PITX2 DNA methylation representing poor metastasis-free survival [51]. Harbeck et al. could show that PITX2 methylation in N0 ER/PR+, tamoxifen-treated patients added significant information to the histopathological factors tumor size, histological grade, and patient age [65]. Both studies underline that PITX2 DNA methylation may be a potential biomarker for predicting outcome in patients with N0 ER/PR+ breast cancer patients treated with tamoxifen monotherapy [51, 65]. Furthermore, a strong correlation between PITX2 DNA methylation and disease recurrence was found: 86% of patients with low PITX2 DNA methylation were metastasis-free after 10 years, compared to only 69% with elevated PITX2 DNA methylation [51]. In survival analyses, PITX2 DNA methylation added statistically significant independent prognostic value to the clinical impact of the established clinical and histomorphological factors.
7. Clinical Implication of PITX2 in High-Risk Breast Cancer Patients
According to the St. Gallen risk stratification panel, breast cancer patients with >3 affected axillary lymph nodes and tumor size > 2 cm are classified high-risk and treated with chemotherapy; the same applies to TNBC patients. Anthracycline-based chemotherapy has become the standard of care for these patients.
7.1. High-Risk Breast Cancer of the Triple-Negative Subtype (TNBC)
In a recent study, PITX2 DNA methylation status and its clinical impact for TNBC patients were investigated [66]. PITX2 DNA methylation was determined in primary tumor tissues obtained from TNBC patients before treatment with adjuvant anthracycline-based chemotherapy. In this retrospective study, PITX2 DNA methylation was the only significant factor as assessed by univariate and multivariate survival analysis; in combination with PITX2 DNA methylation status; none of the established clinical and histomorphological parameters (age, histological grade, tumor size, and lymph node status) showed statistical significance for predicting the rate of disease-free or overall survival [66].
In contrast to the above-cited studies involving PITX2 DNA methylation status in ER/PR+ breast cancer patients, the results obtained by Absmaier et al. revealed evidence that for TNBC patients treated with adjuvant anthracycline-based chemotherapy, a low PITX2 DNA methylation status is associated with a decrease in the progression-free interval [66]. Kaplan-Meier analyses demonstrated that a high PITX2 DNA methylation status is associated with a favorable prognosis for both disease-free and overall survivals. 5-year observation rates (disease-free survival) differ significantly in favor of the high PITX2 DNA methylation group (Figure 3). For patients who did not receive any chemotherapy or who received chemotherapy that was not based on anthracyclines, this risk group separation was not apparent. These results indicate that for TNBC patients treated with adjuvant anthracycline-based chemotherapy, assessment of the PITX2 DNA methylation status may serve as a marker that predicts response to anthracycline-based chemotherapy.
Figure 3.
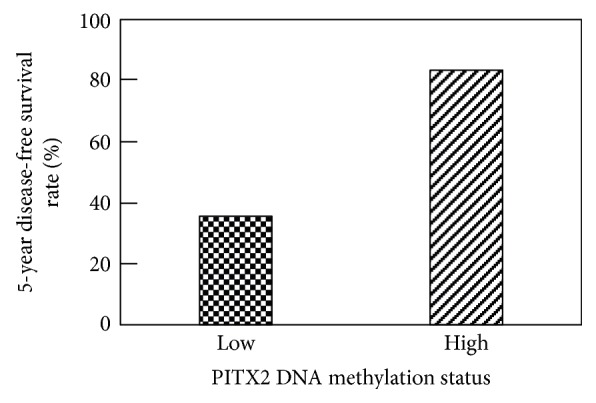
5-year disease-free survival rate analysis of TNBC patients treated with anthracycline-based adjuvant chemotherapy. At 5 years of follow up, the TNBC patients were grouped according to their PITX2 DNA methylation value with a cut-off of 6.35 percent methylation ratio (PMR). Low PITX2 DNA methylation status (n = 18) shows a poor disease-free survival rate at 5 years (35.6%); high PITX2 DNA methylation status (n = 38) is associated at 5-year observation time with favorable disease-free survival (83.5%). (p values: log-rank test, p = 0.006; Wilcoxon test, p = 0.003) [66].
7.2. High-Risk Breast Cancer with >3 Axillary Lymph Nodes
PITX2 DNA methylation was also analyzed in high-risk (N+, ER/PR+, and HER2−) breast cancer patients treated with adjuvant anthracycline-based chemotherapy, which improved clinical outcome of the patients. In this study, the authors could show that PITX2 DNA methylation status significantly predicts the outcome of the patients [44]. The finding was that PITX2 DNA methylation status and that of fourteen other methylated genes predicted clinical outcome in these patients. PITX2 DNA hypermethylation was associated with a high risk of developing metastases in this group of patients (Figure 4). In multivariate analysis, PITX2 hypermethylation evolved as a significant marker to predict outcome [44], when assessed together with the parameters age at the time of surgery, tumor stage, nuclear grade, progesterone receptor status, and adjuvant endocrine therapy. This study provided additional strong evidence that PITX2 DNA methylation status may serve as a useful biomarker in high-risk N+ breast cancer to predict response to anthracycline-based chemotherapy (Figure 4).
Figure 4.
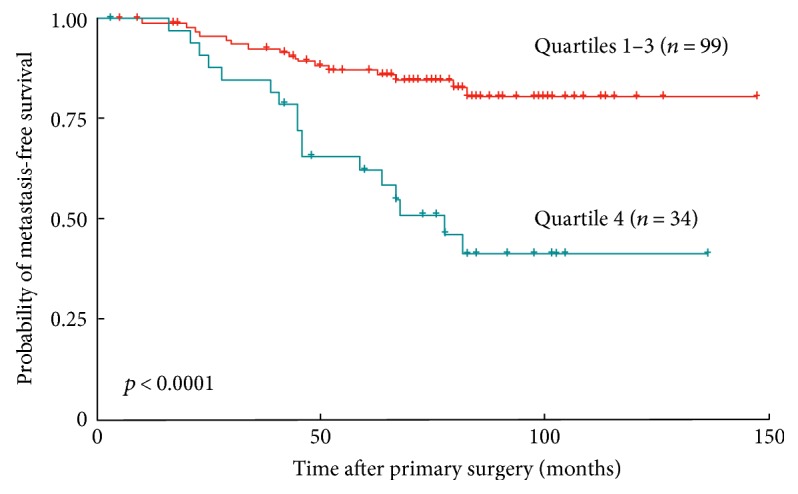
Kaplan-Meier survival curves demonstrating metastasis-free survival probability of high-risk breast cancer patients (n = 133). ER+ breast cancer patients with >3 lymph nodes affected were treated with anthracycline-based chemotherapy plus endocrine therapy. Patients were grouped according to their PITX2 DNA methylation score. PITX2 high gene methylation (quartile 4) predicts poor survival and PITX2 low gene methylation (quartiles 1–3) favorable survival (data reanalyzed from [44]).
Anthracyclines can cause severe side effects; therefore, a marker which could predict sensitivity to anthracyclines in high-risk breast cancers would be highly valuable. Several studies described a positive association of PITX2 DNA methylation status with aggressiveness of the disease and with clinical outcome. Summarizing the clinical impact of PITX2 DNA methylation in high-risk breast cancer patients, there is evidence that PITX2 DNA methylation may serve as a valuable predictive marker to distinguish between responding and nonresponding patients (Figure 2).
8. Hypothesis to Explain the Controversial PITX2 DNA Methylation Status in Predicting Therapy Response in Breast Cancer Patients
The data above raise the question how we can explain that hypomethylation of PITX2 predicts nonresponders in TNBC and hypermethylation of the PITX2 gene predicts nonresponders in ER+ breast cancer. The canonical Wnt-signaling pathway is activated in several tumor types including breast cancer (Figure 5). The major effector of this pathway is β-catenin, which is stabilized in the cytoplasm, translocates to the nucleus, and controls gene expression [17]. PITX2 and β-catenin pathways upregulate the ABC transporter system, especially ABCG2, which is also known as BCRP (breast cancer resistant protein). ABCG2, which belongs to the family of membrane proteins, possesses an ATP-binding cassette and transports specific substrates actively through cellular membranes. It mediates the efflux of drugs and contributes to multidrug resistance in cancer. Substrates of ABCG2 include anticancer drugs such as anthracyclines, topoisomerase inhibitors, tyrosine kinase inhibitors, and antimetabolites [67].
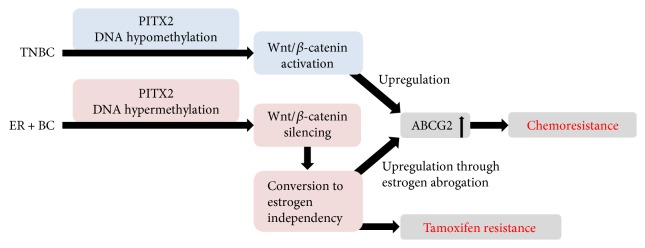
Figure 5.
Predictive significance of PITX2 gene methylation in high-risk breast cancer patients. TNBC and ER+ BC are two biologically different high-risk breast cancer entities. Yet, for both, the PITX2 DNA methylation status has been shown to predict response or failure to anthracycline-based chemotherapy [44, 66]. The controversial results of the favorable clinical significance of PITX2 gene hypermethylation in TNBC patients versus PITX2 gene hypomethylation in ER+ BC patients are outlined. The hypothesis presented is based on published evidence; other, so far unknown, mechanisms may be involved as well. ER+ BC = estrogen receptor-positive breast cancer.
The increased expression of such transporters on plasma membranes results in an increased efflux and decreased intracellular accumulation of many anticancer drugs, ultimately leading to multidrug resistance [67]. Several transcription factors regulate ABCG2, including, but not limited to, the steroid hormone receptors ER/PR and estrogen/progesterone response elements [67]. In TNBC, low methylation (hypomethylation) of the PITX2 gene predicts poor disease-free and/or overall survival in nonresponders to neoadjuvant chemotherapy [58]. PITX2 regulates the Wnt/β-catenin pathway [60], and β-catenin is required for the tumorigenic behavior of TNBC [21]. PITX2 and the β-catenin pathway upregulate the ABCB1 transporter [68, 69], another efflux transporter of the same gene family involved in multidrug resistance. ABCB1 is responsible to efflux small drugs such as anthracyclines and therefore mediates chemoresistance [70]. In summary, hypomethylation of PITX2 may lead to the activation of the Wnt/β-catenin pathway with subsequent upregulation of the ABCG2 or ABCB1 transporter triggering resistance to chemotherapy.
In high-risk ER+ BC, high PITX2 gene methylation (hypermethylation) predicts poor disease-free and overall survival [44, 71]. The Wnt/β-catenin pathway is a positive regulator of ER in breast cancer. Hypermethylation of PITX2 results in silencing of the Wnt/β-catenin pathway with subsequent downregulation of estrogen and its receptor ER, causing estrogen deprivation/independency in ER-positive cancer cells [72]. The ABCG2 transporter is downregulated by estrogen [73]; that is, estrogen deprivation leads to increased ABCG2 expression. If PITX2 is hypermethylated, the Wnt/β-catenin pathway is silenced and subsequent ER downregulation leads to (a) overexpression of the ABCG2 transporter and thereby to anthracycline resistance and (b) tamoxifen (ER antagonist) resistance because of estrogen independency.
9. Prospects and Perspectives for Clinical Use of PITX2 DNA Methylation Status in Cancer Patient Management
Determination of the DNA methylation status of certain genes reflects an emerging field of cancer biomarkers. Promising recent results highlight its potential for early detection, assessment of prognosis, prediction of therapy response, and therapy monitoring in various cancer diseases, which can be performed on tissue samples and on cell-free-circulating DNA in body fluids [88]. For optimum management of cancer patients, however, accurate and highly significant prognostic and predictive factors are of eminent medical need. In particular, predictive cancer biomarkers are needed to determine the right systemic therapy for patients afflicted with any of the heterogeneous high-risk breast cancer subgroups, characterized by varying outcomes, since certain systemic adjuvant therapies may be beneficial for a subgroup of patients only, while others will suffer from potentially toxic and unnecessary therapy-related side effects [41].
In order to be of clinical use, a cancer biomarker should be detectable in biological samples readily available without interrupting the routine clinical workflow. A suitable assay for routine diagnostics must be robust, simple to use, standardized, evaluated in external quality assurance schemes, and made available at affordable costs. Evaluation of such a marker should adhere to the REMARK/BRISQ criteria [79, 80]. In this respect, assessment of the PITX2 DNA methylation status in primary tumor tissues has demonstrated its prognostic and predictive value for several cancer indications (Table 5). A cancer patient's PITX2 DNA methylation status can be determined reliably by real-time PCR technology employing DNA extracted from formalin-fixed, paraffin-embedded (FFPE) tissue [46]. For this purpose, a validated PITX2 test kit is required to expedite its clinical utility. Once these requirements are met, determination of PITX2 DNA methylation status might become an important measure to aid clinicians in advising the optimal cancer therapy to their patients.
Table 5.
Importance and clinical relevance of PITX2 DNA methylation status for various types of cancer.
| Cancer type | Adjuvant treatment | Clinical endpoint | Clinical impact analyzed | Reference |
|---|---|---|---|---|
| Breast cancer (n = 345) LN−, ER/PR+ |
Tamoxifen | MFS | Therapy response prediction | Maier et al., 2007 [51] |
| Breast cancer (n = 412) LN−, ER/PR+ |
None | TDM, OS | Prognosis | Nimmrich et al., 2008 [48] |
| Breast cancer (n = 399) LN−, HER2±, ER/PR+ |
Tamoxifen | TDM | Therapy response prediction | Harbeck et al., 2008 [65] |
| Breast cancer (n = 241) LN+, ER/PR+ |
Anthracycline | DFS, MFS, OS | Therapy response prediction | Hartmann et al., 2009 [44] |
| Breast cancer (n = 100) LN±, ER/PR±, HER2± |
Chemotherapy: 65% of the patients. Endocrine therapy: 39% of patients | OS, DFS, DSS | Prognosis | Buhmeida et al., 2011 [74] |
| Breast cancer (n = 149) LN±, ER/PR±, HER2± |
Not disclosed | No follow-up | Association of PITX2 with RASSF1A and clinicopathological parameters | Jezkova et al., 2016 [64] |
| Breast cancer (n = 56) LN±, ER/PR/HER2− |
Anthracycline | DFS | Therapy response prediction | Absmaier et al., 2017 [66] |
|
| ||||
| Prostate cancer (n = 719) | Observation, untreated | OS | Prognosis | Ahmad et al., 2016 [75] |
| Prostate cancer (n = 476) | Observation, untreated | Biochemical recurrence | Prognosis | Bañez et al., 2010 [76] |
| Prostate cancer (n = 356) | Not disclosed | Biochemical recurrence | Prognosis | Uhl et al., 2017 [77] |
| Prostate cancer (n = 680) | Observation, untreated | Biochemical recurrence | Prognosis | Dietrich et al., 2013 [78] |
| Prostate cancer (n = 367) | Observation, untreated | Biochemical recurrence | Prognosis | Vasiljević et al., 2014 [79] |
| Prostate cancer (n = 71) | Observation, untreated | Biochemical recurrence | Prognosis | Litovkin et al., 2014 [80] |
| Prostate cancer (n = 45) | Not disclosed | Biochemical recurrence | Prognosis | Vinarskaja et al., 2013 [81] |
|
| ||||
| Biliary cancer (n = 80) | Not disclosed | OS | Prognosis | Uhl et al., 2016 [82] |
|
| ||||
| Esophageal cancer (n = 305) | Chemoradiotherapy including cisplatin plus 5-fluorouracil | DFS | Prognosis Therapy response prediction |
Zhang et al., 2013 [83] |
|
| ||||
| Pancreatic cancer (n = 256) | Not disclosed | OS | Prognosis | Wang et al., 2016 [84] |
|
| ||||
| Head & neck cancer (n = 399) | Radiotherapy ± chemotherapy | OS, DFS | Prognosis | Sailer et al., 2016 [85] |
| Head & neck cancer (n = 528) | Not disclosed | OS | Prognosis | Sailer et al., 2017 [86] |
|
| ||||
| Lung cancer (NSCLC) (n = 474) | ± radiotherapy/chemotherapy | PFS | Prognosis | Dietrich et al., 2012 [87] |
DFS: disease-free survival; DSS: disease-specific survival; OS: overall survival; PFS: progression-free survival; TDM: time to distant metastasis; MFS: metastasis-free survival; LN: lymph node status; ER/PR: receptors for estrogen and/or progesterone; HER2: human epidermal growth factor receptor 2; TNBC: triple-negative breast cancer.
Ethical Approval
Any experiments on humans cited which were conducted in cooperation with the Department of Obstetrics and Gynecology (Frauenklinik) of the Technical University of Munich, Germany, were done in accordance with the Declaration of Helsinki (1964). The laboratory experiments were carried out with the human subjects' understanding, who provided written informed consent for using the respective tissue specimens. The various experiments were approved by the local Ethical Committee.
Conflicts of Interest
Michaela Aubele, Manfred Schmitt, Rudolf Napieralski, and Olaf G. Wilhelm are coworkers of Therawis Diagnostics GmbH. The other authors reported no conflict of interest.
Authors' Contributions
All authors participated in the conception and design of this manuscript. They also have read and approved it.
References
- 1.Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. European Journal of Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Harris L. N., Ismaila N., McShane L. M., et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. Journal of Clinical Oncology. 2016;34:1134–1150. doi: 10.1200/JCO.2015.65.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldhirsch A., Wood W. C., Coates A. S., et al. Strategies for subtypes - dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Annals of Oncology. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389(10074):1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 5.Perou C. M., Sùrlie T., Eisen M. B., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Lukong K. E. Understanding breast cancer-the long and winding road. BBA Clinical. 2017;7:64–77. doi: 10.1016/j.bbacli.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao H., He G., Yan S., et al. Triple-negative breast cancer: is there a treatment on the horizon? Oncotarget. 2017;8:1913–1924. doi: 10.18632/oncotarget.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer F. C., Lopez-Garcia M. A., Lambros M. B., Reis-Filho J. S. Genetic characterization of breast cancer and implications for clinical management. Journal of Cellular and Molecular Medicine. 2009;13:4090–4103. doi: 10.1111/j.1582-4934.2009.00906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldhirsch A., Glick J. H., Gelber R. D., Senn H. J. International consensus panel on the treatment of primary breast cancer. V: update 1998. Recent Results in Cancer Research. 1998;152:481–497. doi: 10.1007/978-3-642-45769-2_46. [DOI] [PubMed] [Google Scholar]
- 10.Gnant M., Harbeck N., Thomssen C. St. Gallen/Vienna 2017: a brief summary of the consensus discussion about escalation and de-escalation of primary breast cancer treatment. Breast Care. 2017;12:102–107. doi: 10.1159/000475698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kantelhardt E. J., Vetter M., Schmidt M., et al. Prospective evaluation of prognostic factors uPA/PAI-1 in node-negative breast cancer: phase III NNBC3-Europe trial (AGO, GBG, EORTC-PBG) comparing 6xFEC versus 3xFEC/3xDocetaxel. BMC Cancer. 2011;11:p. 140. doi: 10.1186/1471-2407-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markopoulos C. Overview of the use of Oncotype DX® as an additional treatment decision tool in early breast cancer. Expert Review of Anticancer Therapy. 2013;13:179–194. doi: 10.1586/era.12.174. [DOI] [PubMed] [Google Scholar]
- 13.Székely B., Silber A. L. M., Pusztai L. New therapeutic strategies for triple-negative breast cancer. Oncology. 2017;31:130–137. [PubMed] [Google Scholar]
- 14.Ahn S. G., Kim S. G., Kim C., Jeong J. Molecular classification of triple-negative breast cancer. Journal of Breast Cancer. 2016;19:223–230. doi: 10.4048/jbc.2016.19.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Pinilla S. M., Sarrio D., Moreno-Bueno G., et al. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Modern Pathology. 2007;20:474–481. doi: 10.1038/modpathol.3800760. [DOI] [PubMed] [Google Scholar]
- 16.Dawood S., Broglio K., Esteva F. J., et al. Survival among women with triple receptor-negative breast cancer and brain metastases. Annals of Oncology. 2009;20:621–627. doi: 10.1093/annonc/mdn682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J., Prosperi J. R., Choudhury N., Olopade O. I., Goss K. H. β-Catenin is required for the tumorigenic behavior of triple-negative breast cancer cells. PLoS One. 2015;10, article e0117097 doi: 10.1371/journal.pone.0117097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy M. J., McGowan P. M., Harbeck N., Thomssen C., Schmitt M. uPA and PAI-1 as biomarkers in breast cancer: validated for clinical use in level-of-evidence-1 studies. Breast Cancer Research. 2014;16:428–438. doi: 10.1186/s13058-014-0428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M., Pienkowski T., Mackey J., et al. Adjuvant docetaxel for node-positive breast cancer. The New England Journal of Medicine. 2005;352:2302–2313. doi: 10.1056/NEJMoa043681. [DOI] [PubMed] [Google Scholar]
- 20.Kiechle M. Genexpressionstests zur Prognoseabschätzung beim Mammakarzinom. Frauenarzt. 2014;55:231–237. [Google Scholar]
- 21.Ettl J. Genexpressionstests zur Diagnose und Prädiktion. Gynaekologe. 2015;48:871–875. [Google Scholar]
- 22.Goldhirsch A., Wood W. C., Gelber R. D., et al. Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Annals of Oncology. 2007;18:1133–1144. doi: 10.1093/annonc/mdm271. [DOI] [PubMed] [Google Scholar]
- 23.Curigliano G., Burstein H. J., Winer E. P., et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Annals of Oncology. 2017;28:1700–1712. doi: 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morigi C. Highlights from the 15th St Gallen International Breast Cancer Conference 15-18 March, 2017, Vienna: tailored treatments for patients with early breast cancer. Ecancermedicalscience. 2017;11:p. 732. doi: 10.3332/ecancer.2017.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jänicke F., Prechtl A., Thomssen C., et al. Randomized adjuvant chemotherapy trial in high-risk, lymph node-negative breast cancer patients identified by urokinase-type plasminogen activator and plasminogen activator inhibitor type 1. Journal of the National Cancer Institute. 2001;93:913–920. doi: 10.1093/jnci/93.12.913. [DOI] [PubMed] [Google Scholar]
- 26.Untch M., Konecny G. E., Paepke S., von Minckwitz G. Current and future role of neoadjuvant therapy for breast cancer. Breast. 2014;23:526–537. doi: 10.1016/j.breast.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Esserman L. J., Donald B., DeMichele A., et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL—CALGB 150007/150012, ACRIN 6657. Journal of Clinical Oncology. 2012;30:3242–3249. doi: 10.1200/JCO.2011.39.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocca A., Bravaccini S., Scarpi E., et al. Benefit from anthracyclines in relation to biological profiles in early breast cancer. Breast Cancer Research and Treatment. 2014;144:307–318. doi: 10.1007/s10549-013-2819-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J.-F., Liu J., Wang Y., Zhang B. Novel therapeutic strategies for patients with triple-negative breast cancer. OncoTargets and Therapy. 2016;9:6519–6528. doi: 10.2147/OTT.S105716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staudacher L., Cottu P. H., Diéras V., et al. Platinum-based chemotherapy in metastatic triple negative breast cancer: The Institut Curie experience. Annals of Oncology. 2011;22:848–856. doi: 10.1093/annonc/mdq461. [DOI] [PubMed] [Google Scholar]
- 31.Jiang T., Shi W., Wali V. B., et al. Predictors of chemosensitivity in triple negative breast cancer: an integrated genomic analysis. PLoS Medicine. 2016;13, article e1002193 doi: 10.1371/journal.pmed.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleisher B., Clarke C., Ait-Oudhia S. Current advances in biomarkers for targeted therapy in triple-negative breast cancer. Breast Cancer. 2016;8:183–197. doi: 10.2147/BCTT.S114659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann B. D., Bauer J. A., Chen X., et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of Clinical Investigation. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yadav B. S., Chanana P., Jhamb S. Biomarkers in triple negative breast cancer. World Journal of Clinical Oncology. 2015;6:252–263. doi: 10.5306/wjco.v6.i6.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrohl A. S., Holten-Andersen M., Sweep F., et al. Tumor markers: from laboratory to clinical utility. Molecular & Cellular Proteomics. 2003;2:378–387. doi: 10.1074/mcp.R300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Duffy M. J., Harbeck N., Nap M., et al. Clinical use of biomarkers in breast cancer: updated guidelines from the European Group on Tumor Markers. (EGTM) European Journal of Cancer. 2017;75:284–298. doi: 10.1016/j.ejca.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Yadav M., Singh A. K., Kumar H., et al. Epidermal growth factor receptor inhibitor cancer drug gefitinib modulates cell growth and differentiation of acute myeloid leukemia cells via histamine receptors. Biochimica et Biophysica Acta (BBA) - General Subjects. 2016;1860:2178–2190. doi: 10.1016/j.bbagen.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 38.Corkery B., Crown J., Clynes M., O'Donovan N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Annals of Oncology. 2009;20:862–867. doi: 10.1093/annonc/mdn710. [DOI] [PubMed] [Google Scholar]
- 39.Mirzania A. M. Approach to the triple negative breast cancer in new drugs area. International Journal of Hematology-Oncology and Stem Cell Research. 2016;10:115–119. [PMC free article] [PubMed] [Google Scholar]
- 40.Gross E., van Tinteren H., Li Z., et al. Identification of BRCA1-like triple-negative breast cancers by quantitative multiplex ligation-dependent probe amplification (MLPA) analysis of BRCA1-associated chromosomal regions: a validation study. BMC Cancer. 2016;16:p. 811. doi: 10.1186/s12885-016-2848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathe A., Rodney J., Scott R. J., Avery-Kiejda K. A. miRNAs and other epigenetic changes as biomarkers in triple negative breast cancer. International Journal of Molecular Sciences. 2015;16:28347–28376. doi: 10.3390/ijms161226090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saha P., Nanda R. Concepts and targets in triple-negative breast cancer: recent results and clinical implications. Therapeutic Advances in Medical Oncology. 2016;8:351–359. doi: 10.1177/1758834016657071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jovanovic J., Rønneberg J. A., Tost J., Kristensen V. The epigenetics of breast cancer. Molecular Oncology. 2010;4:242–254. doi: 10.1016/j.molonc.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartmann O., Spyratos F., Harbeck N., et al. DNA-methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clinical Cancer Research. 2009;15:315–323. doi: 10.1158/1078-0432.CCR-08-0166. [DOI] [PubMed] [Google Scholar]
- 45.Martens J. W. M., Margossian A. L., Schmitt M., Foekens J., Harbeck N. DNA-methylation as a biomarker in breast cancer. Future Oncology. 2009;5:1245–1256. doi: 10.2217/fon.09.89. [DOI] [PubMed] [Google Scholar]
- 46.Duffy M. J., Napieralski R., Martens J. W. M., et al. Methylated genes as new cancer biomarkers. European Journal of Cancer. 2009;45:335–346. doi: 10.1016/j.ejca.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Sandhu R., Roll J. D., Rivenbark A. G., Coleman W. B. Dysregulation of the epigenome in human breast cancer, contributions of gene-specific DNA hypermethylation to breast cancer pathobiology and targeting the breast cancer methylome for improved therapy. The American Journal of Pathology. 2015;185:282–292. doi: 10.1016/j.ajpath.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Nimmrich I., Sieuwerts A. M., Meijer-van Gelder M. E., et al. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Research and Treatment. 2008;111:429–437. doi: 10.1007/s10549-007-9800-8. [DOI] [PubMed] [Google Scholar]
- 49.Widschwendter M., Apostolidou S., Raum E., et al. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS One. 2008;3, article e2656 doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stefansson O. A., Manel E. M. Epigenetic modifications in breast cancer and their role in personalized medicine. The American Journal of Pathology. 2013;183:1052–1063. doi: 10.1016/j.ajpath.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 51.Maier S., Nimmrich I., Koenig T., et al. DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients—technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. European Journal of Cancer. 2007;43:1679–1686. doi: 10.1016/j.ejca.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 52.Widschwendter M., Siegmund K. D., Müller H. M., et al. Association of breast cancer DNA-methylation profiles with hormone receptor status and response to tamoxifen. Cancer Research. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 53.Stirzaker C., Zotenko E., Song J. Z., et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nature Communications. 2015;6, article 5899 doi: 10.1038/ncomms6899. [DOI] [PubMed] [Google Scholar]
- 54.Hafez M. M., Al-Shabanah O. A., Al-Rejaie S. S., et al. Increased hypermethylation of glutathione s-transferase P1, DNA-binding protein inhibitor, death associated protein kinase and paired box protein-5 genes in triple-negative breast cancer Saudi females. Asian Pacific Journal of Cancer Prevention. 2015;16:541–549. doi: 10.7314/apjcp.2015.16.2.541. [DOI] [PubMed] [Google Scholar]
- 55.Veeck J., Ropero S., Setien F., et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. Journal of Clinical Oncology. 2010;28:e563–e564. doi: 10.1200/JCO.2010.30.1010. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe Y., Maeda I., Oikawa R., et al. Aberrant DNA methylation status of DNA repair genes in breast cancer treated with neoadjuvant chemotherapy. Genes to Cells. 2013;18:1120–1130. doi: 10.1111/gtc.12100. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y., Diao L., Chen Y., et al. Promotor methylation of BRCA1 in triple-negative breast cancer predicts sensitivity to adjuvant chemotherapy. Annals of Oncology. 2013;24:1498–1505. doi: 10.1093/annonc/mdt011. [DOI] [PubMed] [Google Scholar]
- 58.Pillai S. G., Dasgupta N., Siddappa C. M., et al. Paired-like homeodomain transcription factor 2 expression by breast cancer bone marrow disseminated tumor cells is associated with early recurrent disease development. Breast Cancer Research and Treatment. 2015;153:507–517. doi: 10.1007/s10549-015-3576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilting J., Hagedorn M. Left-right asymmetry in embryonic development and breast cancer: common molecular determinants? Current Medicinal Chemistry. 2011;18:5519–5527. doi: 10.2174/092986711798347252. [DOI] [PubMed] [Google Scholar]
- 60.Basu M., Roy S. S. Wnt/β-catenin pathway is regulated by PITX2 homeodomain protein and thus contributes to the proliferation of human ovarian adenocarcinoma cell, SKOV-3. The Journal of Biological Chemistry. 2013;288:4355–4367. doi: 10.1074/jbc.M112.409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franco D., Christoffels V. M., Campione M. Homeobox transcription factor PITX2: the rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends in Cardiovascular Medicine. 2014;24:23–31. doi: 10.1016/j.tcm.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Lamba P., Hjalt T. A., Bernard D. J. Novel forms of paired-like homeodomain transcription factor 2 (PITX2): generation by alternative translation initiation and mRNA splicing. BMC Molecular Biology. 2008;9:p. 31. doi: 10.1186/1471-2199-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rahman W. F. W. A., Fauzi M. H., Jaafar H. Expression of DNA-methylation marker of paired-like homeodomain transcription factor 2 and growth receptors in invasive ductal carcinoma of the breast. Asian Pacific Journal of Cancer Prevention. 2014;15:8441–8445. doi: 10.7314/apjcp.2014.15.19.8441. [DOI] [PubMed] [Google Scholar]
- 64.Jezkova E., Kajo K., Zubor P., et al. Methylation in promoter regions of PITX2 and RASSF1A genes in association with clinicopathological features in breast cancer patients. Tumour Biology. 2016;37(12):15707–15718. doi: 10.1007/s13277-016-5324-3. [DOI] [PubMed] [Google Scholar]
- 65.Harbeck N., Nimmrich I., Hartmann A., et al. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA-methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. Journal of Clinical Oncology. 2008;26:5036–5042. doi: 10.1200/JCO.2007.14.1697. [DOI] [PubMed] [Google Scholar]
- 66.Absmaier M., Napieralski R., Schuster T., et al. PITX2 DNA-methylation predicts response to anthracycline-based adjuvant chemotherapy in triple-negative breast cancer patients. International Journal of Oncology. 2017 doi: 10.3892/ijo.2018.4241. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mo W., Zhang J.-T. Human ABCG2: structure, function, and its role in multidrug resistance. International Journal of Biochemistry and Molecular Biology. 2012;3:1–27. [PMC free article] [PubMed] [Google Scholar]
- 68.Lee W.-K., Chakraborty P. K., Thevenod F. Pituitary homeobox 2 (PITX2) protects renal cancer cell lines against doxorubicin toxicity by transcriptional activation of the multidrug transporter ABCB1. International Journal of Cancer. 2013;133:556–568. doi: 10.1002/ijc.28060. [DOI] [PubMed] [Google Scholar]
- 69.Lee W. K., Thévenod F. Upregulation of the multidrug resistance p-glycoprotein abcb1 by transcription factor pituitary homeobox 2 (PITX2) in human colon and kidney cancers. The FASEB Journal. 2016;30(Supplement 1):439–432. [Google Scholar]
- 70.Edwardson D., Chewchuk S., Parissenti A. M. Resistance to anthracyclines and taxanes in breast cancer. In: Ahmad A., editor. Breast Cancer Metastasis and Drug Resistance. New York: Springer; 2013. pp. 227–247. [DOI] [Google Scholar]
- 71.Lesche R., Martens J. W. M., Maier S., et al. Identification of novel DNA-methylation markers predicting outcome in node-positive, anthracycline-treated breast cancer patients. Breast Cancer Research and Treatment. 2009;100, article A6009 [Google Scholar]
- 72.Gupta N., Schmitt F., Grebhardt S., Mayer D. β-Catenin is a positive regulator of estrogen receptor-α function in breast cancer cells. Cancer. 2011;3:2990–3001. doi: 10.3390/cancers3032990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imai Y., Ishikawa E., Asada S., Sugimoto Y. Estrogen-mediated post transcriptional down-regulation of breast cancer resistance protein/ABCG2. Cancer Research. 2005;65:596–604. [PubMed] [Google Scholar]
- 74.Buhmeida A., Merdad A., Al-Maghrabi J., et al. RASSF1A methylation is predictive of poor prognosis in female breast cancer in a background of overall low methylation frequency. Anticancer Research. 2011;31:2975–2981. [PubMed] [Google Scholar]
- 75.Ahmad A. S., Vasiljević N., Carter P., et al. A novel DNA methylation score accurately predicts death from prostate cancer in men with low to intermediate clinical risk factors. Oncotarget. 2016;7:71833–71840. doi: 10.18632/oncotarget.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bañez L. L., Sun L., van Leenders G. J., et al. Multicenter clinical validation of PITX2 methylation as a prostate specific antigen recurrence predictor in patients with post-radical prostatectomy prostate cancer. The Journal of Urology. 2010;184:149–156. doi: 10.1016/j.juro.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 77.Uhl B., Gevensleben H., Tolkach Y., et al. PITX2 DNA-methylation as biomarker for individualized risk assessment of prostate cancer in core biopsies. The Journal of Molecular Diagnostics. 2017;19:107–114. doi: 10.1016/j.jmoldx.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 78.Dietrich D., Hasinger O., Bañez L. L., et al. Development and clinical validation of a real-time PCR assay for PITX2DNA-methylation to predict prostate-specific antigen recurrence in prostate cancer patients following radical prostatectomy. The Journal of Molecular Diagnostics. 2013;15:270–279. doi: 10.1016/j.jmoldx.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vasiljević N., Ahmad A. S., Carter P. D., et al. DNA methylation of PITX2 predicts poor survival in men with prostate cancer. Biomarkers in Medicine. 2014;8:1143–1150. doi: 10.2217/bmm.14.41. [DOI] [PubMed] [Google Scholar]
- 80.Litovkin K., Joniau S., Lerut E., et al. Methylation of PITX2, HOXD3, RASSF1 and TDRD1 predicts biochemical recurrence in high-risk prostate cancer. Journal of Cancer Research and Clinical Oncology. 2014;140:1849–1861. doi: 10.1007/s00432-014-1738-8. [DOI] [PubMed] [Google Scholar]
- 81.Vinarskaja A., Schulz W. A., Ingenwerth M., Hader C., Arsov C. Association of PITX2 mRNA down-regulation in prostate cancer with promotor hypermethylation and poor prognosis. Urologic Oncology. 2013;31:622–627. doi: 10.1016/j.urolonc.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 82.Uhl B., Dietrich D., Branchi V., et al. DNA-methylation of PITX2 and PANCR is prognostic for overall survival in patients with resected adenocarcinomas of the biliary tract. PLoS One. 2016;11, article e0165769 doi: 10.1371/journal.pone.0165769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang J. X., Tong Z.-T., Yang L., et al. PITX2: a promising predictive biomarker of patients’ prognosis and chemoradioresistance in esophageal squamous cell carcinoma. International Journal of Cancer. 2013;132:2567–2577. doi: 10.1002/ijc.27930. [DOI] [PubMed] [Google Scholar]
- 84.Wang Q., Li J., Wu W., et al. Smad4-dependent suppressor pituitary homeobox 2 promotes PPP2R2A-mediated inhibition of Akt pathway in pancreatic cancer. Oncotarget. 2016;7:11208–11222. doi: 10.18632/oncotarget.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sailer V., Holmes E. E., Gevensleben H., et al. PITX2 and PANCR DNA-methylation predicts overall survival in patients with head and neck squamous cell carcinoma. Oncotarget. 2016;7:75827–75838. doi: 10.18632/oncotarget.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sailer V., Gevensleben H., Dietrich J., et al. Clinical performance validation of PITX2 DNA methylation as prognostic biomarker in patients with head and neck squamous cell carcinoma. PLoS One. 2017;12, article e0179412 doi: 10.1371/journal.pone.0179412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dietrich D., Hasinger O., Liebenberg V., Field J. K., Kristiansen G., Soltermann A. DNA-methylation of the homeobox genes PITX2 and SHOX2 predicts outcome in non–small-cell lung cancer patients. Diagnostic Molecular Pathology. 2012;21:93–104. doi: 10.1097/PDM.0b013e318240503b. [DOI] [PubMed] [Google Scholar]
- 88.He W. S., Bishop K. S. The potential use of cell-free-circulating-tumor DNA as a biomarker for prostate cancer. Expert Review of Molecular Diagnostics. 2016;16:839–852. doi: 10.1080/14737159.2016.1197121. [DOI] [PubMed] [Google Scholar]