Abstract
PRCC-TFE3 renal cell carcinoma (RCC) is one of the most common types of Xp11.2 translocation renal cell carcinoma (tRCC), of which the diagnosis mainly relies on reverse transcription-polymerase chain reaction (RT-PCR) or chromosomal analysis in fresh frozen samples. Herein, we developed a new dual-fusion fluorescence in situ hybridization (FISH) probe to succinctly identify PRCC-TFE3 RCC in paraffin-embedded tissue. We immunohistochemically analyzed TFE3 and cathepsin K expression in 23 cases of Xp11.2 tRCC which had been confirmed by break-apart TFE3 FISH probe. Next, the dual-fusion FISH assay was performed on these selected cases. Twenty typical cases of clear renal cell carcinoma and 20 cases of papillary renal cell carcinoma were collected as control groups. Seven cases were finally confirmed as PRCC-TFE3 RCC by FISH detection, emerging dual-fusion signals, of which 2 cases were identified as PRCC-TFE3 RCC by RT-PCR previously. All remaining cases were negative for the PRCC-TFE3 rearrangement by FISH. The TFE3 immunohistochemistry was positive in 22/23 cases and the cathepsin K was positive in 16/23 cases. All 7 PRCC-TFE3 RCCs showed positive cathepsin K immunoreactivity. Our results reveal that PRCC-TFE3 dual-fusion FISH probe is an efficient and concise technique for diagnosing PRCC-TFE3 RCC in paraffin-embedded tissue.
Introduction
Xp11.2 translocation renal cell carcinoma (tRCC), a rare subtype of renal cell carcinoma (RCC), result from gene fusions involving the TFE3 transcription factor gene[1], and it is included into the MiT family tRCCs in the recently published World Health Organization (WHO) classification of tumors of the urinary system[2]. Among renal carcinomas, Xp11.2 tRCCs comprise 20% to 75% of paediatric renal neoplasms [3] and about 1.5% of adult RCC cases [4]. Most common gene fusions in Xp11.2 tRCC are TFE3 gene on Xp11.2 with PRCC at 1q21 and TFE3 with ASPL at 17q25, which arise from the translocations t(X; 1) (p11.2; q21)[5] and t(X; 17) (p11.2; q25.3)[6]. Other less recurrent reported TFE3 fusion partners include SFPQ (alias PSF), NonO [7], CLTC [8] and RBM10 [9], resulting from t(X; 1) (p11.2; p34), inv(X) (p11.2q12), t(X; 17) (p11.2; q23), and inv(X) (p11.2p11.23), respectively. Some fusion partners, such as PARP14 [10], KHTFESRP [11], LUC7L3 [11] and DVL2 [12], have only been identified in single patients.
Regarding diagnosis, when fresh frozen samples or viable tumor cells are available, reverse transcription-polymerase chain reaction (RT-PCR) or cytogenetic karyotypic analysis is highly sensitive and specific technique to confirm known gene alteration [13]. However, in archival formalin-fixed paraffin-embedded (FFPE) tissues, morphological characteristics and immunohistochemistry (IHC) are the diagnostic bases of Xp11.2 tRCC. Morphologically, distinctive features of Xp11.2 tRCC are papillary architecture composed of clear or eosinophilic cells and psammoma bodies [1]. However, Xp11.2 tRCCs often present with unusual morphology, which can mimic other types of RCC, and can also be mimicked by some other atypical tumors [14]. In contrast to most RCCs, Xp11.2 tRCC underexpress epithelial markers, but TFE3 and cathepsin K are always high expressed [14]. TFE3 is a ubiquitously expressed transcription factor which contains a basic helix-loop-helix region followed by a leucine zipper (bHLHzip) [15], but its normal level is generally undetectable by IHC. Once TFE3 gene fusion occurs, the partners as strong promoters lead to overexpression of the fusion proteins which can be clearly detected immunohistochemically [14]. IHC with the polyclonal TFE3 antibody was regarded as a very sensitive and specific diagnostic method for Xp11.2 RCC previously[8], but over time increasing evidences have showed false-positive, false-negative, and equivocal results in TFE3 IHC[16, 17]. Studies have found that high sensitivity and specificity is always achieved with a manual overnight labeling, but the automated method with short incubation time creates more false positives. However, in routine, immunohistochemistry in automated immunostainer with a 30 min incubation period is more common [8,18,19]. Cathepsin K is another possible immunohistochemical marker for MiT family tRCC[20]. It plays an important role in osteoclast function, whose expression can be mediated by aberrantly expressed microphthalmia transcription factors [21]. Among Xp11.2 RCCs, approximately 60% label for cathepsin K, while almost all conventional RCCs stain negative [20, 22]. Recently, break-apart fluorescence in situ hybridization (FISH) assay in FFPE archival sections is viewed as the optimal test for diagnosing Xp11.2 RCC for most laboratories [23] (However, the break-apart FISH assay is less useful than TFE3 immunohistochemistry in detecting some Xp11.2 RCCs, like the RBM10-TFE3 RCC which are associated with a subtle chromosome inversion [9].), while it is unable to identify the fusion partner of TFE3 gene.
PRCC-TFE3 RCC, one of the most common types of Xp11.2 tRCCs, is the first documented case of Xp11.2 tRCCs[24]. The breakpoint of the translocation in PRCC-TFE3 RCC was cloned in 1996, and it was found that this translocation resulted in a fusion of the TFE3 gene on the Xp11.2 to a novel PRCC gene on 1q21.2 [25–27]. The morphologic features of this tumor are slightly different from ASPL-TFE3 RCC; however PRCC-TFE3 RCC cannot be distinguished from other Xp11.2 tRCCs on these criteria alone [28]. Immunohistochemically, the expression of cathepsin K in PRCC-TFE3 RCC is more frequent than ASPL-TFE3 RCC, indicating biologic differences among subtypes of Xp11.2 tRCCs [14]. Although previous studies showed that PRCC-TFE3 RCC presented at lower stage and less metastatic frequency than ASPL-TFE3 RCC, metastatic disease was also found to occur in some PRCC-TFE3 RCCs [14, 29]. Therefore, accurate, concise and early diagnosis of PRCC-TFE3 RCC would benefit clinical management and research.
Up to now, it has been difficult to diagnose PRCC-TFE3 RCC only relying on pathologic morphology, IHC, and break-apart FISH probe in FFPE archival tissue in clinical practice. Clinicopathologic data of PRCC-TFE3 RCC and other subtypes of Xp11.2 tRCC are limited. In a previous study, we have demonstrated the value of a novel ASPL-TFE3 dual-fusion FISH probe in confirming ASPL and TFE3 gene fusion [30]. We report herein a PRCC-TFE3 dual-fusion FISH assay for diagnosing PRCC-TFE3 RCC and collect the clinical, pathological, and IHC features of the cases.
Patients and Methods
Case selection
A total of 23 cases of Xp11.2 tRCC (from January 2007 to February 2015) were included in this study, which were confirmed by break-apart TFE3 FISH probe, and 6 Xp11.2 tRCCs (2 with ASPL-TFE3 gene fusion, 2 with PRCC-TFE3 gene fusion, 1 with PSF-TFE3 gene fusion, and 1 with CLTC-TFE3 gene fusion) were genetically confirmed by RT-PCR previously[30]. FFPE tissues were obtained from resection specimens from the files of the department of Pathology, Nanjing Drum Tower Hospital, Medical School of Nanjing University, and prepared for IHC and FISH assay. The clinicopathologic features including clinical manifestations, treatment strategies, pathological findings, clinical outcomes and follow-up information of Xp11.2 tRCC were recorded and evaluated. Twenty typical cases of clear renal cell carcinoma and 20 cases of papillary renal cell carcinoma were also collected as control groups. This study was approved by the Institutional Review Board of Nanjing Drum Tower Hospital. All the patients have signed informed written consents to have their medical record data used in research.
Immunohistochemistry
Immunoreaction for TFE3 (prediluted, ZSGB-BIO, Beijing, China) and cathepsin K (Abcam, Cambridge, MA, USA) were performed in tumor tissue sections of all RCCs. Briefly, after deparaffinization, four-micrometer-thick paraffin-embedded sections were rehydrated using graded ethanol concentrations and treated in hydrogen peroxidase and absolute alcohol for 10 minutes at room temperature to block endogenous peroxidase activity. The tissue sections were subsequently incubated with the primary antibody against the TFE3 or cathepsin K protein for 40 minutes at 25°C. After tris-buffered saline rinses, the tissue was incubated using the secondary antibody for 30 minutes followed by diaminobenzidine for 5 minutes. Positive and negative controls were stained concurrently and showed appropriate immunostaining.
The immunohistochemical assessment for TFE3 and cathepsin K were evaluated as previously described [31, 32]. We assessed the cathepsin K and TFE3 expression by the proportion or intensity of positive cells. For cathepsin K, a score was assigned to represent the estimated percentage of positive cells as follows: 0 or negative, <5% tumor cell positivity; + or focal, 5% to 10% tumor cell positivity; 2+ or moderate, 11% to 50% tumor cell positivity; and 3+ or strong, >50% tumor cell positivity. Moderately (2+) to strongly (3+) positive staining was considered positive result. For TFE3, we defined nuclear immunoreactivity that was apparent at low-power magnification as positive result. And these cases were subdivided into moderately (2+) and strongly (3+) positive on the basis of intensity.
PRCC-TFE3 dual-fusion FISH assay
FISH on the paraffin-embedded materials were performed with a probe consisting of 2 contigs that covered the entire TFE3 gene on the short arm of the X chromosome and the PRCC gene on the long arm of chromosome 1. The contig on the X chromosome consisted of 7 BAC clones (CTD-2311N12, RP11-416B14, CTD-2522M13, CTD-2516D6, CTD-2312C1, CTD-2248C21 and RP11-959H17) labeled with fluorescein-12-dUTP as green fluorescein, and the contig on chromosome 1 consisted of 4 BAC clones (CTD-2534H6, RP11-1047J23, RP11-730I22 and CTD-2547N15) labeled with tetramethylrhodamine-5-dUTP as red fluorescein.
After deparaffinization and washing, three-micrometer-thick paraffin-embedded sections were rehydrated in 100%, 85%, and 70% ethanol in turn for 3 minutes and digested with 10μL pepsin (4 mg/mL, 0.02M HCl; Sigma-Aldrich, Beijing, China) at 37°C for 3 to 5 minutes, followed by subsequent dehydration. Then, the probe mixture was applied, and the slides were denatured at 85°C for 5 minutes and target DNA simultaneously, followed by hybridization overnight at 37°C. The slides were immersed in 2×SSC for 10 minutes and in 0.1% NP-40/2×SSC for 5 minutes at 37°C. The nuclei were counterstained with 4, 6-diamidino-2-phenylindole (DAPI). After hybridization, all slides were maintained at 4°C in the dark.
The slides were examined using an Olympus BX51TRF fluorescence microscope (Olympus, Tokyo, Japan) with the following three filters (DAPI/FITC/TexasRed) and the FISH analysis software (Imstar, Paris, France). For each case, a minimum of 100 nonoverlapping tumor cell nuclei were examined under fluorescence microscopy. Cells without the rearrangement presented split green and red signals (negative result), indicating intact Xp11 and 1q21. Dual fusion signal pattern (two fusion signals) was interpreted as the existence of reciprocal translocation of PRCC gene and TFE3 gene (positive result). We considered a yellow or closely approximated green-red signal as a fusion signal. Clear FISH signals should be observed in >100 nonoverlapping nuclei for each case.
Normal renal tissues from 20 cases of non-PRCC-TFE3 renal cell carcinoma were randomly selected to determine the cutoff. We observed the signals of 50 nuclei in each case, and calculated the percentage of the cells with fusion signal. Subsequently, mean and standard deviation of the percentages were calculated. The cutoff is the sum of mean and three times of standard deviation. Using this method, a positive result was reported when >2% of the tumor nuclei showed the fusion-signal pattern. (http://dx.doi.org/10.17504/protocols.io.jdfci3n)
Results
Patients
The total 23 Xp11.2 tRCCs were definitely diagnosed by break-apart TFE3 FISH probe previously, and 6 cases of these Xp11.2 tRCCs were identified as ASPL-TFE3 RCC by dual-fusion FISH probe [30]. Their ages ranged from 3 to 64 years (mean age, 28y; median age, 26y) and the patients were identified as 14 females and 9 males (ratio of female to male patients was 1.6:1).
TFE3 and cathepsin K IHC, Dual-Fusion FISH Probe in FFPE tissues
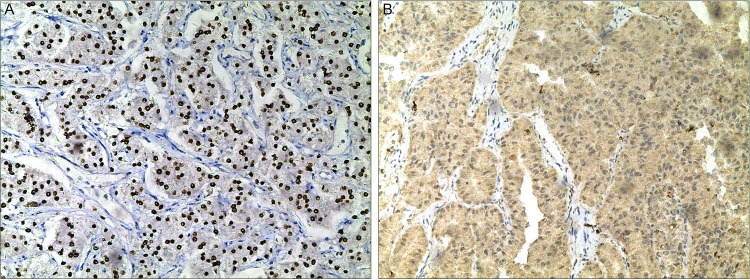
TFE3 and cathepsin K IHC were performed on the 23 Xp11.2 tRCCs and 40 non- Xp11.2 tRCCs. For TFE3, 22/23 Xp11.2 tRCCs and 3/40 non- Xp11.2 tRCCs showed moderately (2+) or strongly (3+) positive staining, and all PRCC-TFE3 and ASPL-TFE3 RCCs were positive (Fig 1A). Of the 23 Xp11.2 tRCCs, 16 cases demonstrated moderately (2+) or strongly (3+) positive staining for cathepsin K (include 7 PRCC-TFE3 RCCs) (Fig 1B) and the remaining 7 cases were negative (include 6 ASPL-TFE3 RCCs). All 40 non- Xp11.2 tRCCs were negative for cathepsin K.
Fig 1. Images of immunohistochemical staining for PRCC-TFE3 RCC.
(A) Strong nuclear immunostaining for TFE3 protein (×100); (B) The neoplastic cells showed diffuse cytoplasmic labeling for cathepsin K (×100). RCC, renal cell carcinoma; TFE3, transcription factor E3; H&E, hematoxylin and eosin.
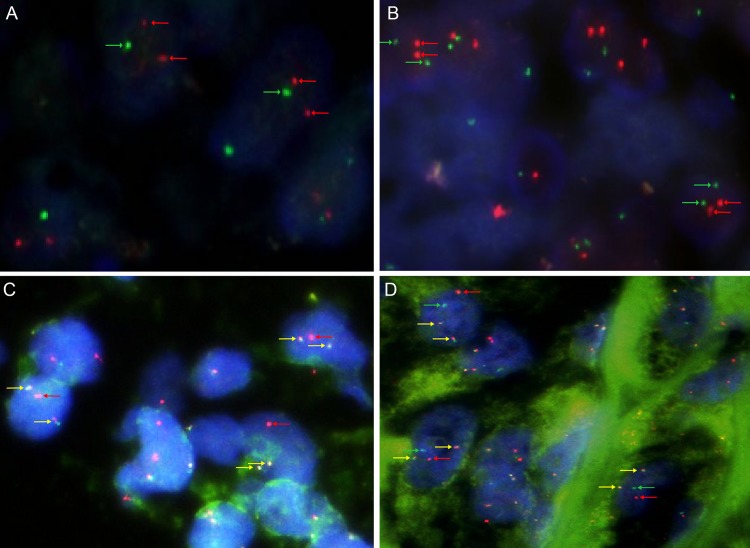
To identify the PRCC-TFE3 fusion gene, PRCC-TFE3 dual-fusion FISH assay was implemented on these Xp11.2 tRCCs, clear renal cell carcinomas and papillary renal cell carcinomas. Seven cases were diagnosed as PRCC-TFE3 RCC, with 2 fusion signals emerged, but the other 16/23 cases and control groups were negative. Two cases (case 1 and 7) of the PRCC-TFE3 RCCs were confirmed by PCR experiment previously [30]. Male and female patients had different signal patterns. In male patients, a positive result included 2 fused and 1 red signals (1R2F) (Fig 2C), but the positive result of female patients showed 2 fused, 1 green and 1red signals (1GR2F) (Fig 2D).
Fig 2. Images of the PRCC-TFE3 Dual-Fusion FISH assay.
(A) Photomicrograph showed a pair of split red signals and a green signal (negative result, red and green arrows) in each nucleus in male (×1000); (B) Photomicrograph showed a pair of split red signals and two split green signals (negative result, red and green arrows) in each nucleus in female (×1000); (C) A positive result included 2 fused and 1 red signals (yellow and red arrows) in each nucleus in male, indicating that PRCC-TFE3 fusion genes have been formed (×1000); (D) A positive result of female patient showed 2 fused, 1 green and 1red signals (yellow, green and red arrows) in each nucleus (×1000). FISH, fluorescence in situ hybridization.
Clinicopathologic features of PRCC-TFE3 RCC
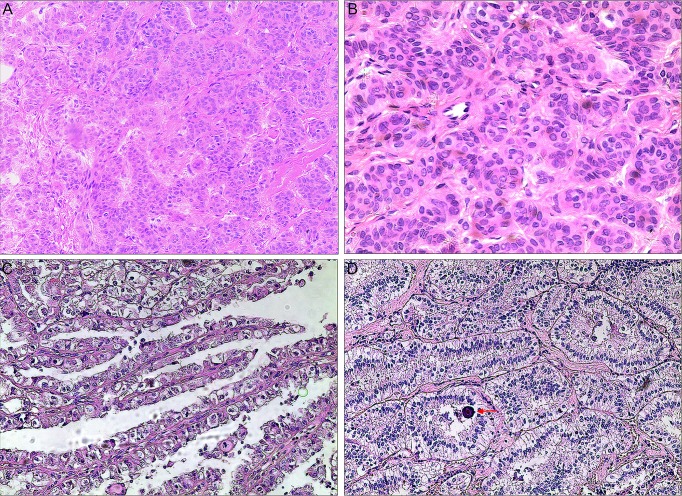
Seven cases were finally confirmed as PRCC-TFE3 RCC by the dual-fusion FISH probe. Of the 7 PRCC-TFE3 RCC patients, 3 were male and 4 female and median age was 30 years (22y to 64y). Two patients (cases 2 and 3) presented with hematuria and the remaining 5 patients were asymptomatic and discovered fortuitously. Nephrectomy was performed in all cases except case 6 who was treated by nephron-sparing surgery (tumor size was 3 cm). Mean size of the tumor was 6.1 cm (3 to 12 cm). Postoperative AJCC staging showed that 5 cases were classified as stage I, while the other 2 cases were classified as stage III. Mean follow-up was 34.6 months (5 to 76 mo). Five of the stage I cases received satisfactory results, except for case 1 who was diagnosed with lung metastasis at the 11th postoperative month. However, the 2 stage III patients (case 4 and 5) who received surgery and postoperative adjuvant molecular-targeted therapy developed local recurrence, and they received secondary surgeries in our hospital to resect recurrent lesions, with no disease progression until the last follow-up. Regarding pathologic morphology, the architecture of this tumor was predominantly nested and focal papillary or acinar, including clear or little eosinophilic cytoplasm, and psammoma bodies could be found in 2 cases (Fig 3). Data of the 7 cases, including the clinicopathologic features, TFE3 and cathepsin K IHC, PRCC-TFE3 dual-fusion FISH assay, are summarized in Table 1.
Fig 3. Images of microscopic morphology for PRCC-TFE3 RCC.
(A) Photomicrograph showed solid nested architecture composed of compactly arranged, eosinophilic cells (H&E,×100); (B) Tumor cells showed irregular nuclei, inconspicuous nucleoli, eosinophilic cytoplasm, and indistinct cell borders (H&E,×200); (C) Photomicrograph showed histopathology of neoplasm containing clear to eosinophilic cells with voluminous cytoplasm arranged in a papillary pattern(H&E,×100); (D) Photomicrograph showed acinar pattern composed of compactly arranged, clear to slightly eosinophilic cells, and a psammoma body(arrow)(H&E,×100); RCC, renal cell carcinoma; H&E, hematoxylin and eosin.
Table 1. The data of clinicopathologic features, TFE3 and cathepsin K IHC, PRCC-TFE3 FISH assay of PRCC-TFE3 renal cell carcinomas.
| Case | Age(years)/Sex | Symptom | Operation | Tumor size(cm) | ACJJ stage | TFE3 IHC | cathepsin K IHC | PRCC-TFE3 dual-fusion FISH | Follow-up(months) and outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 35/M | Symptomless | LRN | 6 | pT1bN0M0,Ⅰ | ++ | ++ | 1R2F | 52, Lung metastasis in 11 months, stable now. |
| 2 | 22/F | Gross hematuria | LRN | 5 | pT1bN0M0,Ⅰ | +++ | ++ | 1G1R2F | 40,Normal |
| 3 | 25/F | Gross hematuria | LRN | 3.5 | pT1aNxM0,Ⅰ | ++ | +++ | 1G1R2F | 15,Normal |
| 4 | 45/F | Symptomless | ORN | 12 | pT3aN0M0,Ⅲ | +++ | ++ | 1G1R2F | 32, Recur in 12 months |
| 5 | 30/F | Symptomless | LRN | 9.5 | pT3aN0M0,Ⅲ | +++ | +++ | 1G1R2F | 22, Recur in 14 months |
| 6 | 64/M | Symptomless | LNSS | 3 | pT1aN0M0,Ⅰ | ++ | +++ | 1R2F | 5,Normal |
| 7 | 26/M | Symptomless | ORN | 3.7 | pT1aN0M0,Ⅰ | +++ | +++ | 1R2F | 76,Normal |
LRN: Laparoscopic radical nephrectomy; ORN: Open radical nephrectomy; LNSS: laparoscopic nephron-sparing surgery; IHC: immunohistochemistry; FISH: fluorescence in situ hybridization; TFE3, transcription factor E3.
Discussion
Although PRCC-TFE3 RCC is the first reported Xp11.2 tRCC, because of less clinicopathologic data, distinctive histopathologic characterization, recommended diagnostic criteria, and standardized therapeutic strategy of PRCC-TFE3 RCC has yet to be established. As is well-known, accurate and concise diagnosis is crucial for treatment, and also for basic research. Relying only upon pathologic morphology and IHC, the subtypes of Xp11.2 tRCC cannot be discriminated. Genetic approaches, such as RT-PCR and FISH assay, are available to identify the type of genetic changes in tumor cells[13], but RT-PCR usually requires high quality RNA (fresh frozen samples). For FFPE archival sections, it has been demonstrated that FISH assay is an effective method for identifying gene translocation. Therefore, we performed this FISH probe on paraffin-embedded tissue to distinguish PRCC-TFE3 RCC.
The design strategy in this assay imitates the ASPL-TFE3 dual-fusion probe: BACs cover the entire PRCC and TFE3 genes [30]. Two fusion signals emerge simultaneously in 1 nucleus, indicating that PRCC-TFE3 fusion genes have been formed. In this study, 7 cases were diagnosed as PRCC-TFE3 RCC using the probe. The result of FISH assay on 2 PRCC-TFE3 RCCs was in accordance with the PCR result, which validated the efficacy of the PRCC-TFE3 dual-fusion probe. Argani et al also analyzed PRCC-TFE3 RCC by FISH assay to establish their TFE3 fusion gene partner, but what they employed were the TFE3 and PRCC break-apart FISH probes [11]. They detected the separation of TFE3 gene to diagnose Xp11.2 tRCC first, followed by using PRCC break-apart probe to identify PRCC-TFE3 RCC.
Morphologically, PRCC-TFE3 RCC may be associated with differing features. Argani et al and subsequently Rao et al’s studies found that this tumor always had a nested(solid, alveolar, acinar, or tubular) pattern with foci of papillary architecture consisted by compact cells with less abundant cytoplasm and fewer psammoma bodies compared with ASPL-TFE3 RCC[28, 33]. In our series, 7 cases had nested and focal papillary or acinar architecture and tumor cells included clear or little eosinophilic cytoplasm with only 2 cases to be found psammoma body, which was consistent with published data. However, this type of gene fusion-associated neoplasm cannot be validated by this distinction, which is not suitable for all cases. Immunohistochemically, except for molecular genetic analysis, nuclear reactivity of TFE3 protein is the most sensitive and specific method for TFE3 rearrangement-associated neoplasm [8]. But TFE3 IHC in diagnosing Xp11.2 tRCC may lead to false-positive or false-negative results [34, 35]. Nuclear TFE3 labeling can also appear in ASPL-TFE3 RCC, PSF-TFE3 RCC and other subtypes [8]. Our data showed that 22/23 Xp11.2 tRCCs, all 7 PRCC-TFE3 RCCs and 3/40 non- Xp11.2 tRCCs were positive for TFE3 IHC, which indicated the meaninglessness of TFE3 IHC in diagnosing PRCC-TFE3 RCC. Cathepsin K, a lysosomal papain-like cysteine protease which affects the bone reabsorption and remodeling [21], is always detected by IHC in MiT family tRCCs but not in other more common RCC subtypes. Interestingly, cathepsin K is distinguishingly expressed in Xp11.2 tRCC relying upon the fusion partner of the TFE3 gene. For example, in Martignoni et al’s study [22], 8 ASPL-TFE3 RCCs entirely did not express cathepsin K. Nevertheless, 12 of 14 PRCC-TFE3 RCCs and all 18 alveolar soft part sarcomas were positive for this protein. Argani et al found that the Xp11.2 tRCCs with the SFPQ-TFE3, NONO-TFE3, DVL2-TFE3, and ASPL-TFE3 gene fusions were almost cathepsin K negative by IHC, whereas a majority of PRCC-TFE3 RCCs and 6 Xp11.2 tRCCs with unknown fusion partner were cathepsin K positive [11].The result of our study showed that seven PRCC-TFE3 RCCs exhibited positive staining for cathepsin K but all of the ASPL-TFE3 RCCs were negative, which was consistent with the findings of Martignoni et al. This diversity could be due to the functional differences between the fusion variants. However, the detailed molecular mechanism has not been well defined, which need further research.
It was reported that Xp11.2 tRCC typically had a predominance of females and affects young adults less than 45 years of age [36]. However, in PRCC-TFE3 RCC, the gender diversity has not been proven and the frequency in adults is underestimated. In the current study, the sex ratio was three to four and case 6 was confirmed in a 64-year-old. There had been reported a series of cases who were older than 45 years [28, 37, 38]. We showed 2 patients presented with hematuria, whose tumor size was not the largest. ASPL-TFE3 RCC was previously found to present at more advanced stage, poor prognosis and more likely to present with regional lymph node metastasis than the PRCC-TFE3 RCC [29, 39]. However, we reported here one case in low stage developed metastasis (lung) 11month after resection, whereas the condition of this patient was fortunately stable with 52 month of follow up. Another two cases with large tumor size (stage III) developed local recurrence approximately 1 year after surgery. According to Argani et al’s studies, some cases of PRCC-TFE3 RCC presented with metastatic disease and recurrence [11, 28]. Ellis et al observed that node-positive but non-metastatic cases with PRCC-TFE3 RCC tended to have a worse outcome than those with ASPL-TFE3 RCC [29]. But regional nodal metastasis was not found in these PRCC-TFE3 RCC cases of our study. Differentiating the PRCC-TFE3 RCC from other types is necessary for patient management.
At the molecular level, oncogenic mechanism of TFE3 in Xp11.2 tRCC is driven by the different gene fusion partners, whereas it has not been well defined. It has been reported that the TFE3 gene fusion partners owning strong promoters cause overexpression of the chimeric protein. Thereinto, PRCC-TFE3 acts as an aberrant transcription factor leading to strong nuclear labeling for TFE3 by IHC. About the function of PRCC, Skalsky et al’s results suggest that it was a component of the pre-mRNA splicing complex [40]. When the PRCC gene is fused to TFE3, the function of splicing factor is disordered. PRCC-TFE3 may also influence the transactivation capacity of the transcription factor and perturb mitotic checkpoint control. It has been reported that PRCC-TFE3 was an approximately threefold stronger transactivator than native TFE3, causing the change of genetic regulation [41]. Weterman et al found that PRCC-TFE3 fusion protein could impair the interaction of PRCC and MAD2B, sequentially disrupting the mitotic checkpoint in RCC [42]. The carcinogenic mechanism of PRCC-TFE3 gene translocation in RCC is not yet clear, but it is crucial for the therapy of this carcinoma.
Conclusions
In summary, PRCC-TFE3 RCC is a rare tumor belonging to Xp11.2 tRCC. We developed a dual-fusion FISH assay as a relatively rapid and precise method to identify this type carcinoma in paraffin-embedded tissue. Furthermore, our study also characterized the morphologic, immunohistochemical and clinical features of the 7 PRCC-TFE3 RCCs.
Supporting information
(DOCX)
A targeted PRCC-TFE3 transcript was in lane 1; lanes 2 was negative RT–PCR results of ASPL-TFE3; lanes 3 was clear cell RCC.
(DOCX)
Acknowledgments
The authors greatly thank Jun Yang and Ming Chen (the department of pathology, Nanjing Drum Tower Hospital) for providing technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Natural Science Foundation of China (ID: 81572512) and Natural Science Foundation of Jiangsu Province (ID: BK20131281). The websites of the program are http://www.nsfc.gov.cn/ and http://www.jstd.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Argani P, Ladanyi M. Distinctive neoplasms characterised by specific chromosomal translocations comprise a significant proportion of paediatric renal cell carcinomas. Pathology. 2003;35(6):492–8. doi: 10.1080/00313020310001619901 [DOI] [PubMed] [Google Scholar]
- 2.Argani P. MiT family translocation renal cell carcinomas In: Moch H, Humphrey PA, Ulbright TM, Reuter VE. WHO Classification of Tumours of the Urinary System and Male Genital Organs. 4thed. Lyon: IARC Press, 2016; 8: 33–4. [Google Scholar]
- 3.Ross H, Argani P. Xp11 translocation renal cell carcinoma. Pathology. 2010;42(4):369–73. doi: 10.3109/00313021003767348 [DOI] [PubMed] [Google Scholar]
- 4.Komai Y, Fujiwara M, Fujii Y, Mukai H, Yonese J, Kawakami S, et al. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin Cancer Res. 2009;15(4):1170–6. doi: 10.1158/1078-0432.CCR-08-1183 [DOI] [PubMed] [Google Scholar]
- 5.Shipley JM, Birdsall S, Clark J, Crew J, Gill S, Linehan M, et al. Mapping the X chromosome breakpoint in two papillary renal cell carcinoma cell lines with a t(X;1)(p11.2;q21.2) and the first report of a female case. Cytogenet Cell Genet. 1995;71(3):280–4. [DOI] [PubMed] [Google Scholar]
- 6.Heimann P, El Housni H, Ogur G, Weterman MA, Petty EM, V G. Fusion of a novel gene, RCC17, to the TFE3 gene in t(X;17)(p11.2;q25.3)-bearing papillary renal cell carcinomas. Cancer Res. 2001;61(10):4130–5. [PubMed] [Google Scholar]
- 7.Clark J, Lu Y-J, Sidhar SK, Parker C, Gill S, Smedley D, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9. doi: 10.1038/sj.onc.1201394 [DOI] [PubMed] [Google Scholar]
- 8.Argani P, Lal P, Hutchinson B, Lui MY, Reuter VE, M L. Aberrant Nuclear Immunoreactivity for TFE3 in Neoplasms With TFE3 Gene Fusions: A Sensitive and Specific Immunohistochemical Assay. Am J Surg Pathol. 2003;27(6):750–61. [DOI] [PubMed] [Google Scholar]
- 9.Argani P, Zhang L, Reuter VE, Tickoo SK, Antonescu CR. RBM10-TFE3 Renal Cell Carcinoma: A Potential Diagnostic Pitfall Due to Cryptic Intrachromosomal Xp11.2 Inversion Resulting in False-negative TFE3 FISH. Am J Surg Pathol. 2017;41:655–62. doi: 10.1097/PAS.0000000000000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argani P, Olgac S, Tickoo SK, Goldfischer M, Moch H, et al. Xp11 translocation renal cell carcinoma in adults: Expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–60. doi: 10.1097/PAS.0b013e318031ffff [DOI] [PubMed] [Google Scholar]
- 11.Malouf GG, Su X, Yao H, Gao J, Xiong L, He Q, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res. 2014;20(15):4129–40. doi: 10.1158/1078-0432.CCR-13-3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argani P, Zhong M, Reuter VE, Fallon JT, Epstein JI, Netto GJ, et al. TFE3-Fusion Variant Analysis Defines Specific Clinicopathologic Associations Among Xp11 Translocation Cancers. Am J Surg Pathol. 2016;40:723–37. doi: 10.1097/PAS.0000000000000631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong M, De Angelo P, Osborne L, Paniz-Mondolfi AE, Geller M, Yang Y, et al. Translocation renal cell carcinomas in adults: a single-institution experience. Am J Surg Pathol. 2012;36(5):654–62. doi: 10.1097/PAS.0b013e31824f24a6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Argani P. MiT family translocation renal cell carcinoma. Semin Diagn Pathol. 2015;32(2):103–13. doi: 10.1053/j.semdp.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 15.Artandi SE, Merrell K, Avitahl N, Wong KK, C K. TFE3 contains two activation domains, one acidic and the other proline-rich, that synergistically activate transcription. Nucleic Acids Res. 1995;23:3865–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macher-Goeppinger S, Roth W, Wagener N, Hohenfellner M, Penzel R, Axel Haferkamp, et al. Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Modern Pathology. 2012;2012(25):308–15. [DOI] [PubMed] [Google Scholar]
- 17.Camparo P, Vasiliu V, Molinie V, Couturier J, Dykema KJ, Petillo D, et al. Renal translocation carcinomas:clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol. 2008;32:656–70. doi: 10.1097/PAS.0b013e3181609914 [DOI] [PubMed] [Google Scholar]
- 18.Argani P, Aulmann S, Illei PB, Netto GJ, Ro J, Cho H, et al. A Distinctive Subset of PEComas Harbors TFE3 Gene Fusions. Am J Surg Pathol. 2010;34:1395–406. doi: 10.1097/PAS.0b013e3181f17ac0 [DOI] [PubMed] [Google Scholar]
- 19.Gaillot-Duranda L, Chevallierb M, Colombelc M, Couturierd J, Pierrond G, Scoazeca JY, et al. Diagnosis of Xp11 translocation renal cell carcinomas in adult patients under 50 years: Interest and pitfalls of automated immunohistochemical detection of TFE3 protein. Pathol Res Pract. 2013; 209:83–9. doi: 10.1016/j.prp.2012.10.013 [DOI] [PubMed] [Google Scholar]
- 20.Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22(8):1016–22. doi: 10.1038/modpathol.2009.58 [DOI] [PubMed] [Google Scholar]
- 21.Motyckova G, Weilbaecher KN, Horstmann M, Rieman DJ, Fisher DZ, F DE. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci U S A. 2001;98(10):5798–803. doi: 10.1073/pnas.091479298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martignoni G, Gobbo S, Camparo P, Brunelli M, Munari E, Segala D, et al. Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod Pathol. 2011;24(10):1313–9. doi: 10.1038/modpathol.2011.93 [DOI] [PubMed] [Google Scholar]
- 23.Green WM, Yonescu R, Morsberger L, Morris K, Netto GJ, Epstein JI, et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol. 2013;37:1150–63. doi: 10.1097/PAS.0b013e31828a69ae [DOI] [PubMed] [Google Scholar]
- 24.de Jong B, Molenaar IM, Leeuw JA, Idenberg VJ, Oosterhuis JW. Cytogenetics of a renal adenocarcinoma in a 2-year-old child. Cancer Genet Cytogenet. 1986;21:165–9. [DOI] [PubMed] [Google Scholar]
- 25.Sidhar SK, Clark J, Gill S, Hamoudi R, Crew AJ, Gwilliam R, et al. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet. 1996;5:1333–8. [DOI] [PubMed] [Google Scholar]
- 26.Weterman MA, Wilbrink M, A GvK. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1)(p11;q21)-positive papillary renal cell carcinomas. Proc Natl Acad Sci U S A. 1996;93:15294–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weterman MA, Wilbrink M, Janssen I, Janssen HA, van den Berg E, Fisher SE, et al. Molecular cloning of the papillary renal cell carcinoma-associated translocation (X;1)(p11;q21) breakpoint. Cytogenet Cell Genet. 1996;75:2–6. [DOI] [PubMed] [Google Scholar]
- 28.Argani P, Antonescu CR, Couturier J, Fournet JC, Sciot R, Debiec-Rychter M, et al. PRCC TFE3 renal carcinoma: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1) (p11.2;q21). Am J Surg Pathol. 2002;26(12):1553–66. [DOI] [PubMed] [Google Scholar]
- 29.Ellis CL, Eble JN, Subhawong AP, Martignoni G, Zhong M, Ladanyi M, et al. Clinical heterogeneity of Xp11 translocation renal cell carcinoma: impact of fusion subtype, age, and stage. Mod Pathol. 2014;27(6):875–86. doi: 10.1038/modpathol.2013.208 [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Yang Y, Gan W, Xu L, Ye Q, Guo H. Newly designed break-apart and ASPL-TFE3 dual-fusion FISH assay are useful in diagnosing Xp11.2 translocation renal cell carcinoma and ASPL-TFE3 renal cell carcinoma: a STARD-compliant article. Medicine (Baltimore). 2015;94(19):e873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao Q, Liu B, Cheng L, Zhu Y, Shi QL, Wu B, et al. Renal cell carcinomas with t(6;11)(p21;q12): a clinicopathologic study emphasizing unusual morphology, novel alpha-TFEB gene fusion point, immunobiomarkers, and ultrastructural features, as well as detection of the gene fusion by fluorescence in situ hybridization. Am J Surg Pathol. 2012;36:1327–38. doi: 10.1097/PAS.0b013e31825aafb5 [DOI] [PubMed] [Google Scholar]
- 32.Rao Q, Williamson SR, Zhang S, Eble JN, Grignon DJ, Wang M, et al. TFE3 Break-apart FISH Has a Higher Sensitivity for Xp11.2 Translocation–associated Renal Cell Carcinoma Compared With TFE3 or Cathepsin K Immunohistochemical Staining Alone. Am J Surg Pathol. 2013;37:804–15. doi: 10.1097/PAS.0b013e31827e17cb [DOI] [PubMed] [Google Scholar]
- 33.Rao Q, Chen JY, Wang JD, Ma HH, Zhou HB, Lu ZF, et al. Renal cell carcinoma in children and young adults: clinicopathological, immunohistochemical, and VHL gene analysis of 46 cases with follow-up. Int J Surg Pathol. 2011;19(2):170–9. doi: 10.1177/1066896909354337 [DOI] [PubMed] [Google Scholar]
- 34.Klatte T, Streubel B, Wrba F, Remzi M, Krammer B, de Martino M, et al. Renal cell carcinoma associated with transcription factor E3 expression and Xp11.2 translocation: incidence, characteristics, and prognosis. Am J Clin Pathol. 2012;137(5):761–8. doi: 10.1309/AJCPQ6LLFMC4OXGC [DOI] [PubMed] [Google Scholar]
- 35.Camparo P, Vasiliu V, Molinie V, Couturier J, Dykema KJ, Petillo D, et al. Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol. 2008;32(5):656–70. doi: 10.1097/PAS.0b013e3181609914 [DOI] [PubMed] [Google Scholar]
- 36.Hirobe M, Masumori N, Tanaka T, Kitamura H, Tonooka A, Hasegawa T, et al. Clinicopathological characteristics of Xp11.2 translocation renal cell carcinoma in adolescents and adults: Diagnosis using immunostaining of transcription factor E3 and fluorescence in situ hybridization analysis. Int J Urol. 2016;23(2):140–5. doi: 10.1111/iju.13007 [DOI] [PubMed] [Google Scholar]
- 37.Meloni AM, Dobbs RM, Pontes JE, Sandberg AA. Translocation (X;1) in papillary renal cell carcinoma. A new cytogenetic subtype. Cancer Genet Cytogenet. 1993;65(1):1–6. [DOI] [PubMed] [Google Scholar]
- 38.Dijkhuizen T, van den Berg E, Storkel S, Terpe HI, Burger H, de Jong B. Distinct features for chromophilic renal cell cancer with Xp11.2 breakpoints. Cancer Genet Cytogenet. 1998;104(1):74–6. [DOI] [PubMed] [Google Scholar]
- 39.Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, et al. Primary Renal Neoplasms with the ASPL-TFE3 Gene Fusion of Alveolar Soft Part Sarcoma. A Distinctive Tumor Entity Previously Included among Renal Cell Carcinomas of Children and Adolescents. Am J Pathol. 2001;2001(159):179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skalsky YM, Ajuh PM, Parker C, Lamond AI, Goodwin G, C CS. PRCC, the commonest TFE3 fusion partner in papillary renal carcinoma is associated with pre-mRNA splicing factors. Oncogene. 2001;20:178–87. doi: 10.1038/sj.onc.1204056 [DOI] [PubMed] [Google Scholar]
- 41.Weterman MAJ, van Groningen JJM, Jansen A, vK AG. Nuclear localization and transactivating capacities of the papillary renal cell carcinoma-associated TFE3 and PRCC (fusion) proteins. Oncogene. 2000;19:69–74. doi: 10.1038/sj.onc.1203255 [DOI] [PubMed] [Google Scholar]
- 42.Weterman MA, van Groningen JJ, Tertoolen L, vK AG. Impairment of MAD2B-PRCC interaction in mitotic checkpoint defective t(X;1)-positive renal cell carcinomas. Proc Natl Acad Sci U S A. 2001;98(24):13808–13. doi: 10.1073/pnas.241304198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
A targeted PRCC-TFE3 transcript was in lane 1; lanes 2 was negative RT–PCR results of ASPL-TFE3; lanes 3 was clear cell RCC.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.