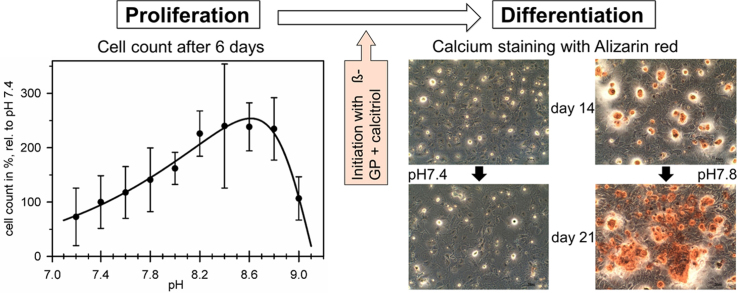
Abstract
We investigated the effects of alkaline pH on developing osteoblasts. Cells of the osteoblast-like cell line MC3T3-E1 were initially cultured for six days in HEPES-buffered media with pH ranging from 7.2 to 9.0. Cell count, cellular WST-1 metabolism, and ATP content were analyzed. The three parameters showed a pH optimum around pH 8.4, exceeding the recommended buffer range of HEPES at the alkaline flank. Therefore, only pH 7.2, 7.4, 7.8, and 8.4 media were used in more elaborate, daily investigations to reduce the effects of pH change within the pH control intervals of 24 h. All parameters exhibited similar pH behaviors, roughly showing increases to 130% and 230% at pH 7.8 and 8.4, as well as decreases to 70% at pH 7.2 when using the pH 7.4 data for reference. To characterize cell differentiation and osteoblastic cell function, cells were cultured at pH 7.4 and under alkaline conditions at pH 7.8 and 8.4 for 14 days. Gene expression and mineralization were evaluated using microarray technology and Alizarin staining. Under alkaline conditions, ATF4, a regulator for terminal differentiation and function as well as DMP1, a potential marker for the transition of osteoblasts into osteocytes, were significantly upregulated, hinting at an accelerated differentiation process. After 21 days, significant mineralization was only detected at alkaline pH. We conclude that elevated pH is beneficial for the cultivation of bone cells and may also provide therapeutic value in bone regeneration therapies.
Keywords: Proliferation, Differentiation, Alkalosis, Calcification, p38 MAPK
Graphical abstract
Highlights
-
•
Cell count, WST-metabolism and ATP-content are increased at severely alkaline pH.
-
•
Alkaline pH enhances osteoblast proliferation.
-
•
Markers for terminal differentiation/function (ATF4/DMP1) are upregulated at pH 7.8.
-
•
Alkaline pH accelerates terminal differentiation and calcification.
-
•
Elevated pH is beneficial for bone cell cultivation.
1. Introduction
It is well known that pH plays a pivotal role in the control of bone remodeling [1]. In situ, low pH stimulates osteoclast function and works as regulating factor in the process of bone matrix production by osteoblasts. The osteoclast-mediated resorption of bone matrix is directly correlated with pH, with a maximum around pH 7.0 and ceasing above pH 7.4 [1], [2]. Systemic acidosis has implications in the pathogenesis of various metabolic diseases, including a negative effect on bone mineralization [3]. Acidosis also stimulates the release of bone mineral in humans [4]. Opposite effects have been observed under metabolic alkalosis, which reduces bone resorption and stimulates osteoblastic collagen synthesis [5]. Both acidosis and alkalosis can be induced by respiration failure, emphasizing the importance of gas exchange for the systemic acid-base balance, which is maintained by systemic carbonate buffer systems that regulate pH in various tissues of an organism.
The in vivo conditions are mimicked by the sodium/hydrogen carbonate buffer systems, which are commonly used in the culture of many cell types. The habitual use of standardized cell culture media may be one of the reasons that pH effects have not yet become a focus of research. Most standardized media are stabilized to pH 7.4, the systemic pH of blood. However, the local pH of the other tissues, such as muscle or connective tissues, may differ strongly [6], [7], [8], which implies that the optimum pH for the various cell types may differ from each other considerably. We believe that a more critical examination of the specific pH needs of different cell types is highly needed.
Different in vitro systems exist for research into bone cell culture. Some groups have used pieces of bone to investigate the pH influence on matrix, mineral, and energy metabolism [5], [9]. Very few studies have been conducted at the cellular level. Leem and Kohn [10], [11] investigated the influence of medium pH on the differentiation of bone mesenchymal stem cells (BMSC) into an osteogenic cell lineage, and Kaysinger and Ramp [12] studied adult human osteoblasts. The latter authors described an increase of osteoblastic activity with increasing pH in the pH 7.0–7.6 range, which was not associated with any elevated glycolytic activity. At pH 7.8, the authors reported reduced osteoblast activity. However, a pH drift of up to 0.4 pH steps in their culture media during a 48-h cultivation sounded a note of caution regarding the interpretation of the data.
To our knowledge, there are no comparable studies of pH effects on the proliferation or long-term cultivation of osteoblastic cell lines, even though such investigations may prove useful for cell culture in 3D scaffolds, where the medium and oxygen supply in the bulk of the scaffolds has become a focus of research [13]. Although current research mainly aims at establishing a dynamic culture to increase oxygen supply [14], we assume that this problem may be less important than the removal of CO2 to avoid acidification by carbonate. With MC3T3 osteoblasts, we showed that a feeding medium-pH of 7.8 helped uphold the pH above 7.0 in confined microfluidic cell culture systems for longer medium exchange periods [15]. We speculate that elevated pH may provide a simple way to avoid adverse effects on cell proliferation and cell vitality during static cell culturing in 3D scaffolds, not only for osteoblasts. Finally, research into the pH conditions of bone cell cultures may provide a new starting point for prospective bone regeneration therapies.
2. Material and methods
2.1. Cell culture
The mouse-osteoblast precursor cell line MC3T3-E1 (referred to below as osteoblast-like cells) was obtained from the German collection of microorganisms and cell culture (DSMZ, Braunschweig, Germany). The cells were cultured in aqueous alpha minimum essential medium (Alpha medium; ord. No. F0925) supplemented with 1% of a penicillin/streptomycin stock solution (100 U/ml penicillin and 100 µg/ml streptomycin) and 10% fetal bovine serum (all purchased from Biochrom AG, Berlin, Germany) in T25 cell-culture flasks (Greiner bio-one, Frickenhausen, Germany). The incubator ensured 95% humidity in a 5% CO2 atmosphere at 37 °C. Cells grown to confluence were trypsinized (0.05% trypsin + EDTA 0.02%, PAN Biotech GmbH, Aidenbach, Germany) and diluted to one million cells per ml before subculture. For pH experiments, carbonate-free alpha medium powder (Ord. no. P03-2510, Pan Biotech, Aidenbach, Germany) was dissolved in deionized water (analytical grade) and buffered with 20 mM 4-(2-hydroxyethyl)−1-piperazineethanesulfonic acid (HEPES). The pH was adjusted in parallel series of up to five different pH values from 7.2 to 9.0 with NaOH and controlled with a pH meter (Mettler-Toledo GmbH, Gießen, Germany).
All media were sterile-filtered and supplemented with 1% penicillin/streptomycin (stock solution: 100 U/ml penicillin/100 µg/ml streptomycin) and 10% fetal bovine serum (FBS). In total, 20,000 cells (20 µl suspension) were seeded per well (500 µl) of a 24-well plate at the desired pH and cultured in an incubator with 95% humidity under normal atmospheric conditions. Proliferation and differentiation experiments were continued for up to 6 and 21 days, respectively. To improve comparability, cells with comparable passage numbers ranging from 10 to 20 were used for each experimental run. The medium pH was measured daily, before half of the medium was exchanged. Selected metabolic parameters and cell proliferation were measured with standard assays. The results of all the consecutive runs were pooled in pH groups. Throughout the paper, the parameter values obtained for the pH 7.4 group were used for reference.
2.2. Cell count determination
To determine cell counts, the medium was removed, and adherent cells were washed with PBS and incubated with trypsin at 37 °C for 5 min. Trypsinization was stopped by adding the same amount of FBS-containing medium to each well before 10 µl of the cell suspensions were transferred to a Neubauer chamber and counted under the microscope.
2.3. ATP measurement (ATP-Lite®assay)
The cellular ATP status was analyzed with a commercial luminescence ATP assay (Perkin Elmer, Waltham, MA, USA) according to the manufacturer´s protocol. In short, the culture medium in each well of the 24-well plates (Sarstedt AG, Nürnbrecht, Germany) was exchanged with aqueous Alpha medium mixed with lysis buffer at a 2:1 ratio without further supplements. Four cell lysate samples per culture well were transferred into intransparent 96-well plates (LumiNunc, Thermo Scientific Nunc, Schwerte, Germany). Cell-free lysis buffer-containing medium served as a negative control. The detected reaction was started by adding 50 µl of the assay's substrate buffer. The plates were incubated in the dark for 10 min followed by luminescence measurement with a plate reader (Lumistar, BMG Labtech GmbH, Ortenberg, Germany).
2.4. WST assay
The WST proliferation assay (Roche Diagnostics GmbH, Mannheim, Germany) uses WST-1 reagent to detect the metabolic activity of the respiratory chain of cultured cells. The assay is based on changes in the light absorbance resulting from the metabolism of WST-1 into formazane by mitochondrial succinate reductase.
For measurements, the medium in the wells of a pH series was replaced by 500 µl of fresh medium of the respective pH supplemented with 1% WST-1 reagent. After another hour in the incubator, the optical absorbance of the supernatants was determined in a plate reader (Polarstar, BMG Labtech GmbH, Ortenberg, Germany). For this, four 100 µl samples from each measuring well were transferred to a 96-well test plate (Sarstedt AG, Nürnbrecht, Germany). The absorbance was determined at 450 nm against a reference wavelength of 620 nm. In all tests, cell-free controls with WTS-1 reagent were prepared in parallel.
Two different tests were carried out to determine whether the WST assay readouts were pH-dependent. For both tests, the cells were seeded at high numbers (approximately 250,000 cells per well of a 12-well plate) and cultured at the standard pH of 7.4 until confluence. The first test was performed to evaluate pH-dependence of mitochondrial succinate reductase, which metabolizes the WTS-1 reagent. The medium in the wells was replaced by 1 ml medium of either pH 7.4, 7.8 or 8.4 containing WST-1 reagent. After the assay incubation time of one hour, the samples were collected as described above. Seven repeats were done at each pH. A linear regression model revealed a statistically significant (p<0.001) pH dependence of the measured absorbance per one million cells. A correlation coefficient of 0.79 (Pearson) proved the strong correlation between the parameters [16].
A second test was performed to exclude a possible pH influence on the photometric measurements (see [16]). In the relevant pH range, no significant differences were measured in the absorbance, excluding a pH effect on the photometric readouts of the WST assay.
2.5. Gene expression analysis
For gene expression analysis, osteoblast-like cells were cultured at pH 7.4, 7.8 or 8.4. After seven days, 10 mg/ml beta-glycerol phosphate and 10 ng/ml calcitriol were added to restrict the proliferation and support the differentiation of the cells [17], [18], [19].
After 14 days, the cells were lysed in RTL Plus buffer, which was included in the RNeasy Plus Kit (Qiagen, Hilden, Germany) and extracted according to the manufacturer's protocol. Total RNA samples were quantified with a spectrophotometer (NanoDrop 1000, Thermo Fisher Scientific, Waltham, MA USA) and their integrity was controlled using the Agilent Bioanalyzer 2100 with the RNA Pico chip kit (both from Agilent Technologies, Waldbronn, Germany). RNA integrity number values between 9.3 and 9.8 were achieved.
The expression profiling was performed according to the manufacturer's instructions with Affymetrix GeneChip Mouse 2.0 ST Arrays (Affymetrix, Santa Clara, CA, USA) using 200 ng RNA. The so-called Whole Transcriptome protocol was started by introducing T7 promoter tags to all RNA molecules using N6 3′-ends for DNA strand synthesis. After RNA strand replacement according to Eberwine (19), non-labeled aRNA was produced by in vitro transcription. To avoid a 3′ bias, all RNA molecules were linearly amplified followed by a purification step. Using the aRNA as template, a new strand-identical single-strand DNA was produced by adding random primers and dNTPs. In addition, a certain amount of dTTP was replaced by dUTP. The following purification included digestion with RNaseH. Endpoint fragmentation was performed with uracil-DNA-glycosylase in combination with apurinic apyrimidinic endonuclease 1. Desoxynucleotidyl-transferase was used to add biotinylated dNTPs to the 3′-ends of the single-stranded DNA fragments. The hybridization was carried out at 45 °C in the GeneChip® Hybridization Oven 645 (Affymetrix) overnight. The prepared microarrays were scanned using the GeneChip Scanner 3000 (Affymetrix) at 0.7 µm resolution.
Primary data analysis was carried out with the Affymetrix Expression Console 1.4.1.46 software including the Robust Multiarray Average module for normalization. Gene expression data were log-transformed. A change was considered significant when the FDR-corrected p-value/q-value thresholds met the criterion q<0.01 at fold changes >|2|, i.e. expression increments or declines larger than two. Our complete microarray data are available in the Gene Expression Omnibus database (GEO accession: GSE84907).
To extract canonical pathways and global functional networks corresponding to those known from mammalian (mouse, rat or human) in vivo and in vitro systems, lists of differentially expressed genes were imported into the Ingenuity program (Ingenuity Pathway Analyses, Ingenuity Systems/Qiagen). Enriched pathways were individually reviewed to exclude pathways that were less meaningful for our cell type, such as sperm motility or neuronal guidance. Significance values were calculated by right-tailed Fisher's exact test. Standard z-scores were used to assess the activation (z>1) or repression (z<1) of certain pathways. No predictions could be made for pathways with z-scores close to zero.
2.6. Alizarin red assay
The mineral deposition ability of osteoblast-like cells was tested with a commercial Alizarin Red S staining assay (AppliChem GmbH, Darmstadt, Germany). The cells were cultured at pH 7.4, 7.8 or 8.4 for 14 or 21 days. To facilitate differentiation, 10 mM beta-glycero-phosphate and 10 nM calcitriol were added when the cells reached confluence after approximately four days [17], [18], [19]. For calcium detection, the cells were washed with PBS and fixed by the addition of 4% paraformaldehyde at 37 °C for 15 min. After another washing step with distilled water, the cells were stained with 10 mM Alizarin-staining solution for 5 min. Excess solution was rinsed off with distilled water, and the staining results were evaluated under a light microscope.
3. Results
3.1. Proliferation parameters: overview in a wide pH range
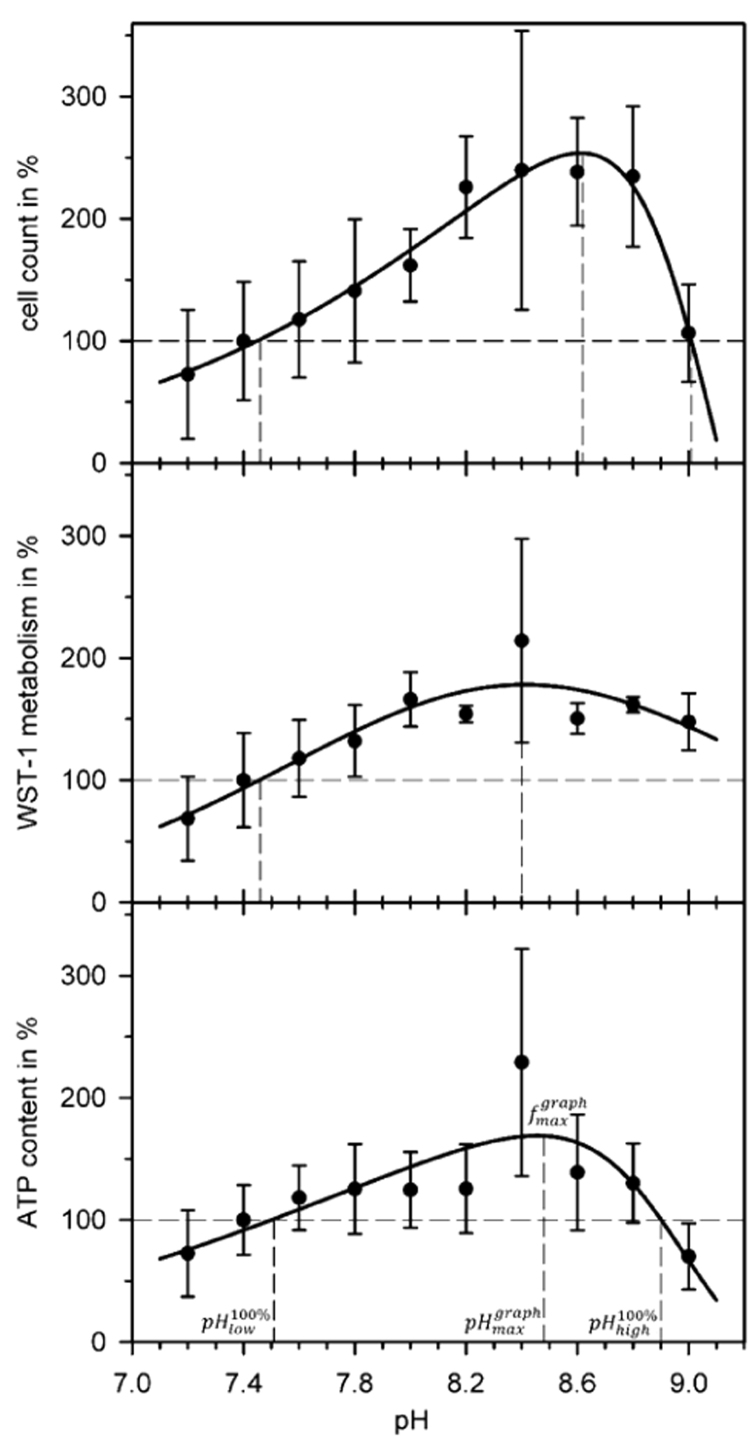
For a first overview, tests of the influence of pH on cell count, WST-1 metabolism, and cellular ATP content were carried out in the 7.2–9.0 pH range after six days of culture. To ensure comparability of the results, HEPES buffer was used in all experiments, even though its recommended buffer range of pH 6.8–8.2 was exceeded [20]. As expected, considerable drops of the initial pH were observed above pH 8.4 within our control interval of 24 h. At pH 9.0, the drop reached up to one pH step, introducing difficulties in the statistical analysis of the results. Despite this problem, the three parameters were plotted over the initial pH values (Fig. 1).
Fig. 1.
pH dependence of the proliferation parameters. The parameters cell count, WST-1 metabolism and cellular ATP content were determined after six days of cell culture. All parameters were normalized to their 100% values at pH 7.4. Each point represents the mean and standard deviation of 10 measurements. The fitting procedure is described in S1 File. The dashed lines illustrate the graphically determined values , , and given in Table 2. , confine the theoretical pH range, in which the fitted curves predict increased parameter values with respect to the standard pH of 7.4.
All three parameters exhibited an optimum pH range with their highest values around pH 8.4. Although the cell count data seemed to exhibit a plateau, ranging from pH 8.2–8.8 with a steep decline toward pH 9.0, the plateau of the WST assay data was even broader, ranging from pH 8.0–9.0. The high WST values seemed to contrast with the reduced cell count, especially at pH 9, whereas the ATP content dependence largely reflected the cell count data, though with a moderately elevated, broad plateau. We were unable to trace back the reasons for the high scatter at pH 8.4.
From a number of different functions, which were tested to fit the pH dependencies of the experimental data, a double-logistic function with five free parameters was found to provide the best fits ([21], see S1 File).
The and the related pH values as well as and , were numerically determined from the plots of the double-logistic functions in Fig. 1. Table 1 summarizes the obtained parameters. All parameters peaked within the pH 8.4–8.6 range.
Table 1.
Graphically determined parameters obtained from the fits of the pH-dependent equation given in S1 File.
| Parameter | Cell count | WST-1 metabolism | ATP content |
|---|---|---|---|
| 7.46/9.01 | 7.46/n.d. | 7.51/8.90 | |
| 8.62 | 8.40 | 8.48 | |
| 251.4 | 177.9 | 168.5 |
These overview results were used to determine a set of four pH values for more elaborate proliferation investigations. We chose pH 7.2, 7.4, 7.8, and 8.4 for a pH below the cell culture standard, the cell culture standard, an intermediate step, and the optimum found in the pretests, respectively. The pH range in which pH could not be kept stable by HEPES-buffering was not further investigated. Long-term experiments on differentiation were carried out only at pH 7.4, 7.8, and 8.4.
3.2. Changes in medium pH during cell culture
In parallel to the investigations on cell proliferation at pH 7.2, 7.4, 7.8, and 8.4, we checked the pH before the daily medium exchanges. For each day, the pH of each group was measured after pooling the media of four randomly selected wells. The observed shifts differed in their directions in the different pH groups (Table 2). Although the pH remained nearly constant in the pH 7.4 group, moderate changes were observed in the pH 7.2 and 7.8 groups. Within 24 h, the pH was increased by approximately 0.07 and reduced by 0.15 in the pH 7.2 and 7.8 groups, respectively, when the measured pH values were averaged over six days. The second column of Table 2 presents means and mean deviations for n≥24. The shift magnitudes were increased with cultivation time, especially after more than three days. The pH shifts observed were particularly large in the pH 8.4 group.
Table 2.
Comparison of adjusted pH and averaged pH values before the daily medium exchanges.
| Adjusted pH | Averaged media pH of 6 consecutive days | pH difference |
|---|---|---|
| 7.2 | 7.27±0.03 | +0.07 |
| 7.4 | 7.41±0.03 | +0.01 |
| 7.8 | 7.65±0.08 | −0.15 |
| 8.4 | 8.03±0.18 | −0.37 |
Importantly, taking the increasing pH shifts into account would compress the abscissae, especially above pH 8.4, leading to even steeper decays of the functions and shifting the optima toward more acidic values. This consideration suggests a slightly shifted optimal pH range with respect to the parameter peak range, rendering the range from pH 8.0–8.4 a sound assumption for the optimal pH.
3.3. Effect of medium pH on proliferation
To characterize the influence of medium pH on the proliferation of osteoblast-like cells, experiments were conducted in carbonate-free Alpha medium of pH 7.2, 7.4, 7.8, and 8.4. A relatively low cell number (20,000) was seeded per well of 24-well plates to obtain a subconfluent cell layer before the cells were cultured at the different pH values. Cell count, WST-1 metabolism, and ATP-content were analyzed over an experimental period of six days.
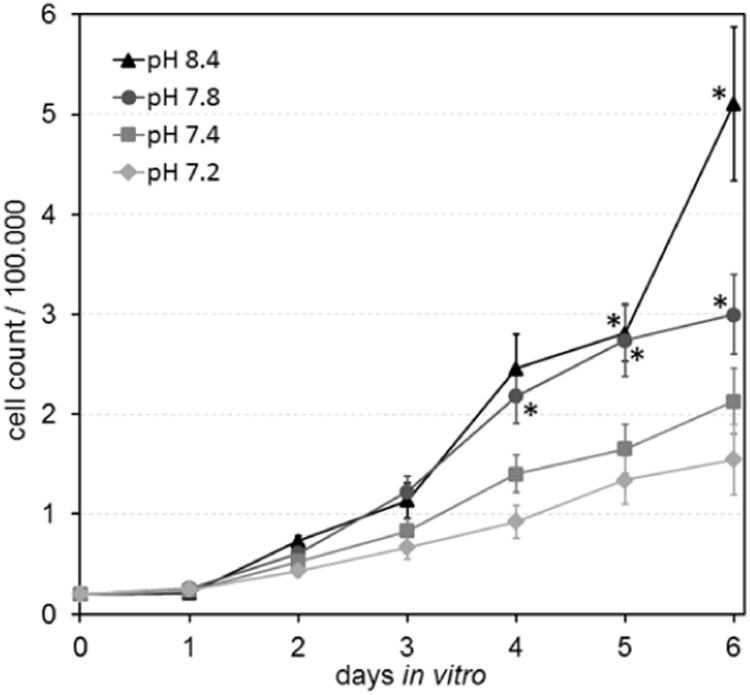
Cell counts increasing with pH were already apparent on day three (Fig. 2). On day four, the difference between pH 7.8 and 7.4 (control) became statistically significant. The cell count differences remained considerable at the end of the experiment. Compared to the standard culture conditions at pH 7.4, the mean cell counts were approximately 1.4-fold and 2.4-fold higher in the pH 7.8 and 8.4 groups, respectively. The pH 7.2 group showed the lowest cell counts throughout the entire experimental period, with the approximately 0.8-fold values of the pH 7.4 control group at the end of the experiment.
Fig. 2.
Cell counts over six days in dependence on medium pH. Each data point represents the mean of ten wells. A RM ANOVA revealed significantly increased cell counts at alkaline pH (p<0.05, indicated by asterisks) with respect to the pH 7.4 group, starting on day four for pH 7.8. On day six, the count peaked for pH 8.4 with a factor of approx. 2.4 with respect to the pH 7.4 group.
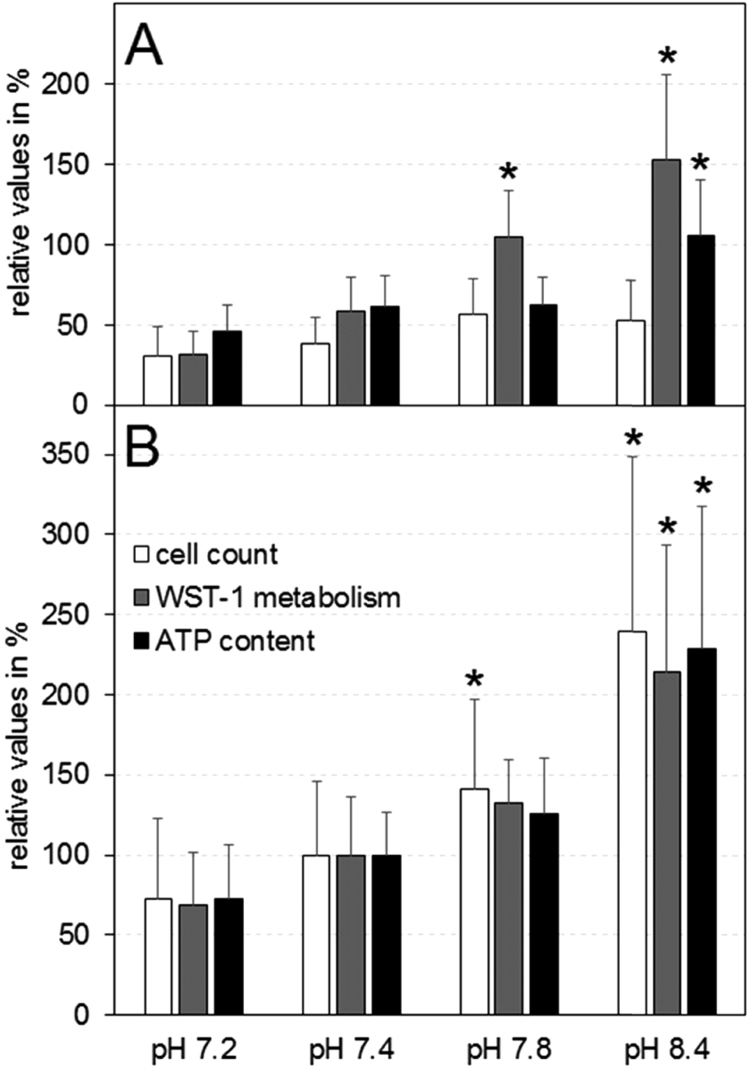
Fig. 3 compares cell count, WST-1 metabolism, and ATP content on days 3 and 6. All values were normalized to their pH 7.4 means on day 6. The cell count data correspond to those in Fig. 2.
Fig. 3.
pH dependence of proliferation parameters. Relative cell count (white bars), WST-1 metabolism (gray bars) and ATP content (black bars) are given for 3 (A) and 6 (B) days in culture. All data were normalized to their means of pH 7.4 on day 6. All parameters were increased with culture time and more alkaline medium pH. Each column represents the mean of ten wells collected from different culture plates. The data were analyzed by a RM ANOVA. Significant differences (p<0.05), referring to the pH 7.4 (control) values at their corresponding days are indicated by asterisks. A strong increase at alkaline pH was found in the WST-1 metabolism already on day 3, whereas the other parameters showed a delayed increase.
WST-1 metabolism was increasing over the experimental period of six days, suggesting cell proliferation in all pH groups. The assay showed the same tendencies as the cell counts, even though it was more sensitive, detecting significant differences after only two days of culture. The statistical significance was verified by a Repeated Measures (RM) ANOVA using the Holm-Sidak method. For clarity, significant differences were marked only with respect to the pH 7.4 group and not for all possible pairs of the tested pH groups. The pH 8.4 group showed a significantly increased WST-1 metabolism with respect to the pH 7.4 group. Although the pH 7.8 group also displayed considerably elevated values, the RM ANOVA failed to detect these differences. Nevertheless, a pairwise comparison for significance using Student's t-test led to significant differences for this group, as well.
The overview on metabolism and proliferation was completed by the ATP-Lite®assay. The ATP content was measured on six consecutive days. For more alkaline medium pH a tendency toward higher ATP content, and thus enhanced proliferation, was observed. In the first phase of the experiment, the results were insignificant. However, from day 4 onward, the ATP content of the pH 7.2 group became significantly lower than that of the pH 7.8 and 8.4 groups. At this point the pH 8.4 group also showed significant differences compared to the standard, peaking in ATP content (119.45±48.44 µM) on day 6. As in the WST assay, the ATP values appeared to be considerably elevated in the pH 7.8 group on day 6. Although a pairwise comparison with Student's t-test revealed a significantly higher ATP content at pH 7.8 (65.34±19.17 µM) than at pH 7.4 (52.13±14.87 µM), the RM ANOVA did not show a significant difference.
3.4. Transcriptional response to pH
To examine whether osteoblast-like cells retained their ability to differentiate under our experimental conditions, long-term experiments were conducted. The cells were cultured either under standard conditions (pH 7.4) or under alkaline pH (pH 7.8 or pH 8.4). To support differentiation, beta-glycerophosphate and calcitriol were supplemented as soon as the cells reached confluence after 4 days.
Transcriptome analysis with the Affymetrix platform revealed several hundred genes to be differentially regulated at pH 7.8 and pH 8.4 compared to the standard conditions. Even though the effects were similar in the two alkaline groups, the existing differences not only hinted at the amplification of effects at pH 8.4 that were already induced at pH 7.8 but also at the induction of different signaling pathways. In our analysis, this led to a more ambiguous picture at pH 8.4, which was based on the data bases used by the IPA software, probably also because of the limited existing knowledge in the alkaline pH range. For a more stringent discussion of the alkalizing effects, we focus at pH 7.8 below. Additional reasons are that i) the medium pH was more stable throughout the culturing time at pH 7.8 (Table 3); and ii) we consider pH 7.8 a stepping stone towards pH 8.4. All data including the pH 8.4 data will be summarized and made accessible online.
Table 3.
Gene symbols of the Top5s of the upregulated (top) and downregulated (bottom) genes at pH 7.8. Fold changes compare the transcript numbers at pH 7.8 with those at pH 7.4.
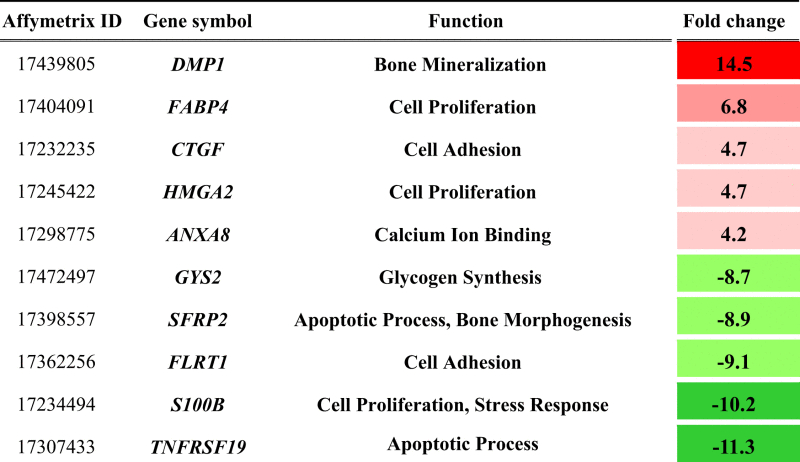 |
The Top5 of the upregulated and the Top5 of the down-regulated genes are involved in mineralization (DMP1, ANXA8), proliferation and cell adhesion (FABP4, HMGA2, CTGF, FLRT1, S100B), cell differentiation (SFRP2, TNFRSF19) as well as metabolism (GYS2) (Table 4). Above all, the transcription of DMP1 was strongly enhanced. These regulations reflect the profound effects of the alkaline medium on cell physiology. Surprisingly, the gene expression of typical early osteoblastic transcription factors such as Runx2 and Osx (SP7) was either unaltered or even downregulated (fold changes: −1.57 and −3.14), resulting in a reduced gene expression of the early osteoblastic markers ALP and BGLAP. At the same time, the transcription factor ATF4, which is considered a critical regulator for terminal differentiation and function [22], was upregulated (fold change: 2.22).
Table 4.
Selected canonical pathways activated or repressed at pH 7.8.
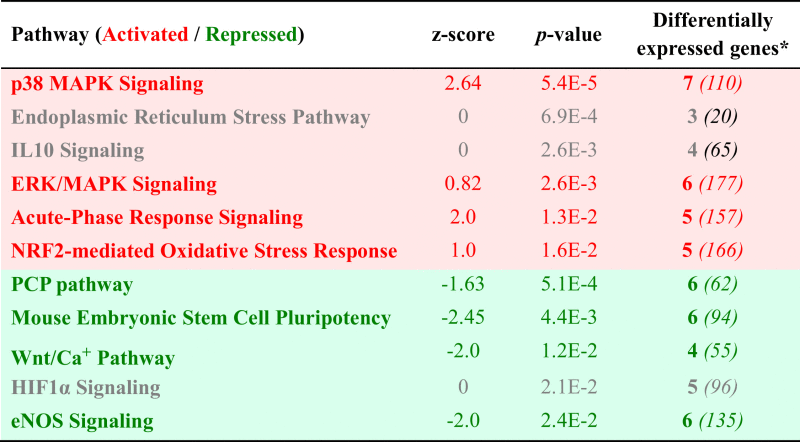 |
*The known numbers of genes within each pathway are shown in parentheses.
The complete set of differentially expressed genes was assigned to functional pathways and networks. Amongst these pathways many were related to stress, differentiation and bone cell function (Table 4). For example, p38 MAPK is known to be involved in the responses to extracellular stress stimuli and to inflammatory cytokines, such as TNF, IL1 and IL18 [23], [24]. We found the receptors of these cytokines significantly upregulated, while the cytokine expression itself was unaltered or slightly downregulated. Even though the p38 MAPK itself was not significantly regulated, its signaling network was subject to various regulation processes, resulting in a generally enhanced activity of the network (z-score: 2.64).
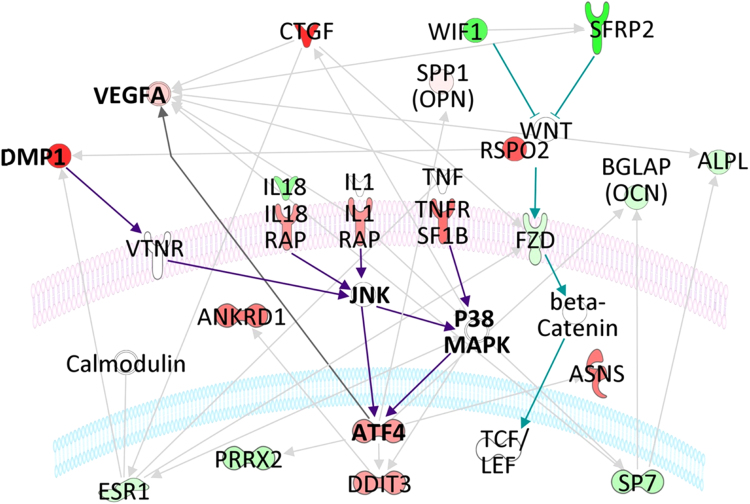
The WNT β-catenin pathway was influenced by a severe downregulation of the cytoplasmic inhibitors WIF1 and SFRP2, accompanied by negligible regulations in the WNT family (Fig. 4).
Fig. 4.
Schematic network of relevant pathways regulated at pH 7.8. Upregulated genes, down-regulated genes, and insignificantly regulated genes are colored in red, green, and white, respectively. To identify relevant pathways, we manually screened all the global functional networks that were extracted from lists of differentially expressed genes by the Ingenuity Pathway Analyses program.
The condensed scheme in Fig. 4 summarizes the interaction of genes that we consider relevant for the response to alkaline pH. The genes were selected from the extended picture presented in S3 File.
3.5. Effect of medium pH on mineralization
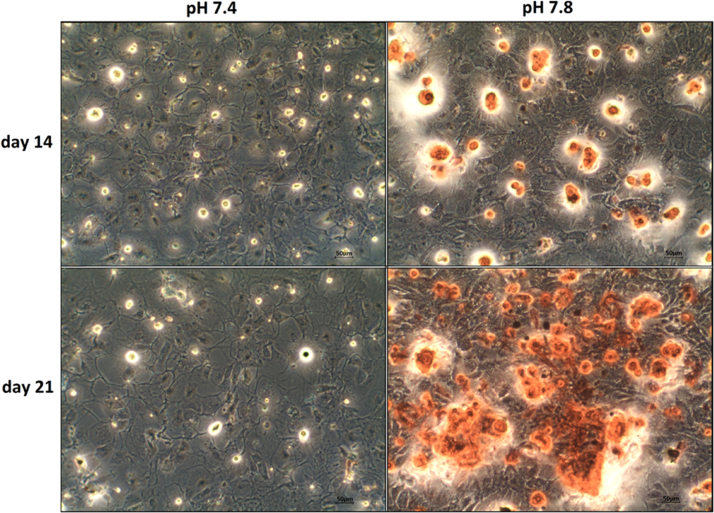
The cellular potential for mineral deposition was determined by calcium staining in Alizarin Red assays. Staining was performed after 14 and 21 days in culture. Without supplementing beta-glycerophosphate and calcitriol at the time of confluence, the cells lost their osteoblastic morphology, gaining a rather fibroblastic phenotype independent of pH after several days (data not shown). As expected, no calcium deposition could be detected in these cultures. In the supplemented cultures, the confluent cells retained their osteoblastic morphology, though at considerably lower cell counts, with some apoptotic cells being recognizable independently of pH. The deposition of calcium was pH-dependent. Calcium was not detectable at pH 7.4 up to day 21. At pH 7.8, cells could first be stained on day 14, and large calcium deposits were detectable after 21 days. They were located in an optically cell-free matrix layer atop the cells, which we interpreted as a well-developed extracellular matrix. Fig. 5 shows representative images. Separate, prolonged tests revealed calcium deposition also at pH 7.4, though this was undetectable before day 28 (data not shown).
Fig. 5.
Comparison of calcium secretion at pH 7.4 and 7.8. Secretion was assayed with Alizarin (red stained clusters) in media supplemented with 10 nM calcitriol and 10 mM β-glycerophosphate after 14 and 21 days. At pH 7.8, stained cells and huge calcium deposits were clearly visible on days 14 and 21, respectively. At pH 7.4, no calcium could be detected until day 21.
Analogous experiments could not be completed at pH 8.4 because pH control would have required the repetitive replacement of larger amounts of culture medium. Medium replacement would have removed secerned molecules and prevented the conditioning of the medium by the cells themselves.
4. Discussion
4.1. The setup
Osteoblasts undergo a developmental sequence with three principal phases: cell proliferation, extracellular matrix maturation, and mineralization [25], [26]. Only mature, differentiated osteoblasts are able to produce and modulate the extracellular matrix that is finally mineralized. Here, we used three parameters, which reflect different aspects of proliferation, namely cell count, cellular metabolism, and ATP content, to show that alkaline culture media facilitate the proliferation of osteoblast-like cells. Moreover, alkaline pH was shown to foster the development of characteristic cell properties of mature osteoblasts, such as the expression of typical late osteoblastic transcription factors and the ability to mineralize.
Most literature studies have reported pH effects for culture times of up to 72 h or shorter. Often, different extracellular pH was adjusted only after the cells reached confluence [9], [12]. We believe that these approaches provide only limited information on the pH effects on proliferation and differentiation processes in osteoblasts. To our knowledge, we are the first to report beneficial effects of alkaline pH on osteoblast-like cell proliferation during the initial log-phase and their differentiation in long-term cultures.
4.2. Results on proliferation parameters
The tested proliferation parameters showed similar pH dependencies, with optima in the alkaline range (Fig. 1). Of the three parameters, WST-1 metabolism was the fastest to detect increased proliferation at alkaline pH after only two days in culture. This increase was corroborated by cell count, which showed significant differences than the standard after three days at pH 8.4 and later at lower pH (Fig. 2).
However, another aspect must be taken into account for this comparison. Tests with equal cell numbers at different pH during the incubation times of the WST-1 assay revealed increased absorbances for increased pH values in the WST medium [16]. In other words, the increased WST-1 turnover detected with alkaline culture media resulted from both higher cell numbers and pH dependence of the WST-1 turnover during the incubation time of the WST-1 assay.
Assuming that the abundance of the WST-1-metabolizing succinate reductase was similar in all pH groups, the enzyme's pH-dependence would lead to increased absorbance values per cell at more alkaline pH. In fact, detected absorbance was increased by a factor of approximately 1.7 when comparing pH 8.4 and 7.4 on day 3 (see Fig. 3) but not on day 6. We attributed the disappearance of the increase on day 6 to a downregulation of enzyme copy number as a result of its enhanced efficiency [16].
Enhanced activity of succinate reductase in the respiratory chain will inevitably increase ATP synthesis. Indeed, we found significantly higher ATP contents per well in the pH 8.4 group starting on day 3. On day 6, ATP content peaked at pH 8.4. This value was more than twice the value found at pH 7.4. Nevertheless, when the ATP content was normalized to cell count, very similar values were obtained in all pH groups (see S2 Table). For longer incubation times, we found insignificant reductions in the ATP content per cell in all pH groups. This finding corresponds to results of Komarova et al. [27], who reported decreased cellular ATP levels between days 3 and 7 when the cells were cultured in alpha minimum essential medium without beta-glycerophosphate. However, we could not exclude the possibility that the reduction in our experiments was caused by the test itself, e.g., a hampered lysis of cell layers with gradually developed cellular matrix.
We expected proliferation to approach the plateau phases within six days, resulting in pH-independent converging cell counts. Nevertheless, on day six, cell counts were clearly increased at elevated pH, with strikingly high counts at pH 8.4 (Fig. 2). In parallel experiments, we found ATP content and cell count per well to be strongly correlated for at least the first six days of culture. The corresponding constant ATP content per cell suggests a stable balance between ATP synthesis and energy consumption during proliferation [28]. The cellular ATP content was also constant for the high cell counts at pH 8.4 on day six, suggesting that this measuring point was not an outlier (Fig. 2).
Higher final cell counts might have resulted from multilayer formation by osteoblasts, which would require their differentiation [27]. In this case, elevated pH would have supported both proliferation and differentiation. In our continuative differentiation experiments, the addition of supplements diverted the cell fate from those in the proliferation experiments at approximately day 4. Nevertheless, a more detailed investigation of the interrelationship between proliferation and differentiation was beyond the scope of this manuscript.
4.3. Results on the transcriptional response to alkaline pH
The pH dependence of differentiation was investigated in long-term experiments over 14 or 21 days. In our experiments, osteoblast-like cells did not keep their typical osteoblastic morphology but developed a rather fibroblastic phenotype without the addition of beta-glycerophosphate and calcitriol on approximately day 4. This observation is in accordance with recent findings of a positive selection of fast-growing cells from populations of the heterogeneous MC3T3 cell line by continuous passaging [29]. These fast-growing cells gradually lost their capacity to form mineralized-nodules, an effect that is in accordance with our observation of a missing calcium deposition in the long-term mineralization experiments. In our experiments, the typical morphology of mature osteoblasts was stabilized by the addition of the supplements, an effect that was likely based on the antiproliferative properties of calcitriol. Cessation of proliferation is a known prerequisite for the initiation of osteoblast-specific gene expression [30]. Our differentiation experiments suggest that this mechanism also applies to the immortalized MC3T3 cell line.
Ongoing differentiation processes can be revealed by gene expression profiling at a relevant timepoint. After 14 days in culture at pH 7.8, we found the expression of genes encoding the early osteoblastic transcription factor Runx2 and Osx to be either unaltered or even downregulated while the late transcription factor ATF4 was significantly upregulated. ATF4 is a crucial factor for osteoblast differentiation and the major determinant of osteoblast function [22]. ATF4-deficient mice displayed a delayed skeletal development and developed a severe low-bone-mass phenotype due to decreased bone formation. Proper type I collagen synthesis required ATF4, even though ATF4 is known not to regulate the transcription of type I collagen [31].
The p38 MAPK network, which also controls ATF4, showed an enhanced activity at pH 7.8. The network is involved in responses to extracellular stress stimuli as well as to inflammatory cytokines such as TNF, IL-1 and IL-18 [23], [24], which all had upregulated receptors in our experiments. Beyond its role in the stress response, p38 MAPK itself is also involved in regulating the differentiation and proliferation of bone progenitors. The phosphorylation activity of p38 MAPK promotes progression in osteogenesis by enhancing the expression or the activity of genes of osteoblast-specific transcription factors. Deletion of p38 MAPK hampers the terminal differentiation of osteoblasts and the appearance of osteocytes, thus directly affecting bone composition and maintenance in vivo [32]. The activation of the p38 MAPK network in our experiments points at the predestined differentiation of the osteoblast-like cells into osteocytes.
The most strongly regulated gene codes for DMP1 (Table 4), which is a critical factor for the proper mineralization of bone. DMP1 is a member of the small integrin-binding ligand N-linked glycoprotein (SIBLING) family of extracellular matrix proteins. Other prominent members of the SIBLING family are osteopontin and bone sialoprotein, which were only modestly regulated in our experiments. In primary osteoblast cultures, DMP1 is expressed very late and coincides with the formation of the mineralized matrix and the presence of cells with an osteocytic phenotype as described by Kalajzic et al. [33]. These authors suggested DMP1 was a potential marker for the transition of osteoblasts into osteocytes. In our experiments, the upregulation of DMP1 and the late transcription factor ATF4 together with the downregulation of typical early osteoblastic markers, such as ALP and BGLAP imply an accelerated transition from the osteoblastic phenotype into a more osteocytic phenotype under alkaline conditions.
4.4. Results on calcification
A late indicator of osteoblast differentiation is matrix mineralization. At pH 7.4, no calcium was detectable in our experiments at up to 21 days in culture. At pH 7.8, Alizarin-stained osteoblasts and some calcium deposition appeared on day 14. Seven days later, huge Alizarin-stained calcium deposits appearing in the extracellular matrix outshined stained osteoblasts (Fig. 5).
A comparable sequence of mineral deposition was described by Sudo et al. [34], who established the MC3T3 cell line. Using a constant standard pH, these authors found that calcium vesicles appeared in the cells after 24 days, and they observed calcified spherules from day 30 onward. To compare our experiments with these findings, we prolonged several experiments up to 30 days. At pH 7.4, we observed Alizarin-stained osteoblasts in the presence of the appropriate supplements only on day 28. When the experiment was terminated on day 30, there were still no detectable calcified spherules or calcium deposits. Nevertheless, at pH 7.8, the sequence of calcium verifiability in our experiments was accelerated with respect to the results of Sudo et al. [34] (Fig. 5).
In summary, our results show that even though alkaline pH is not sufficient to retain the osteoblast-like phenotype, it clearly supports osteoblastic properties. Our view is supported by reports on the favorable influence of elevated pH on the differentiation of BMSCs into an osteogenic cell lineage [11].
4.5. On the cellular role in the acid-base homeostasis
Our results support the ideas of Brandao et al. [35], who showed that bone cell function is regulated by pH. These and other authors have postulated a major role for bone as an alkaline buffer reserve against systemic acidification, which ensures the acid–base homeostasis even under pathophysiological acidic conditions [4]. During acidosis, the osteoblast-induced deposition of alkaline minerals in bone is reduced, and the resorptive activity of the osteoclasts is enhanced [1]. As a result, the availability of dissolved hydroxyl ions, which buffer surplus protons, is maximized. In contrast, the process of mineralization involves the deposition of alkaline hydroxyapatite, a process that inevitably releases protons into the extracellular fluid.
In our experiments, all pH-shifts tended toward pH 7.4. Nevertheless, this does not imply that pH 7.4 is optimal for cell viability in different tissues. It is more likely that the mechanisms behind our observation are part of homeostatic pH regulation, which would make blood capillaries in bone the interface for the systemic acid-base homeostasis and pH 7.4 the “lingua franca” for inter-systemic fluid exchange between tissues. pH 7.4 is actually the physiologically buffered value of blood and other systemic fluid reservoirs, such as liquor and lymph. In contrast, the local pH inside tissues may largely differ from pH 7.4.
We believe that the observed enhanced proliferation and differentiation of osteoblast-like cells at elevated pH in vitro, together with the observed medium acidification, reflects the cellular contribution to the in vivo feedback mechanism of homeostasis. Under alkaline conditions, osteoblast-like cells are more efficient at mineralization, a process that would drive the pH toward 7.4 under alkaline in vivo conditions. Even the alkalization observed in our pH 7.2 group did not contradict this picture. Nevertheless, in our in vitro experiments, alkalization may have been partly caused by the FBS of our medium. It introduces plasma proteins, which contribute to the blood buffer system under in vivo conditions. The cells themselves may have also contributed to the alkalization of the pH 7.2 medium by the exchange of intracellular HCO3− for extracellular Cl- by the Cl-/HCO3− antiporter [36].
Nevertheless, literature on the pH in the interstitia of tissues is scarce. In most interstitia, the pH is probably lower than 7.4, depending on the metabolic activity of the cells and their distance to the nearest blood capillary. For osteoblasts, our results suggest that within the range up to pH 8.4, any increase above pH 7.2 is beneficial for proliferation and mineralization. Alkalization of the extracellular milieu is most likely part of the physiological processes occurring during bone regeneration. For example, Chakkalakal et al. [37] reported a pH increase up to pH 7.55 following an initial pH drop in the surrounding bone tissue during bone healing. The pH rise was accompanied by rapidly increasing calcium content in the tissue.
5. Conclusions
In bone, osteoclast and osteoblast activities are decreased and increased at elevated pH, respectively [1]. Surprisingly, literature on the influence of alkaline pH on bone cell proliferation and differentiation is still scarce. Taking pH drifts of the culture media into account, we found the maximum proliferation of osteoblast-like cells in the pH range from 8.0 to 8.4. Our results hint at the importance of research deep into the alkaline range to complete the picture. In view of these findings, an elevated pH would be beneficial for the in vitro or ex vivo cultivation of bone cells or bone tissue and might also open new avenues for bone regeneration therapies.
Acknowledgments
The authors are grateful for a DFG grant (German Research Council, graduate school GRK1505/2 "WELISA"), which covered the expenses for consumables as well as the PhD positions of AMG and SMB.
We thank R. Modrozynski and Dr. M. Stubbe for their help with the experiments and the theoretical data interpretations.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.02.001.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Arnett T.R. Extracellular pH regulates bone cell function. J. Nutr. 2008;138:415S–418S. doi: 10.1093/jn/138.2.415S. [DOI] [PubMed] [Google Scholar]
- 2.Arnett T.R., Dempster D.W. Effect of pH on bone resorption by rat osteoclasts in vitro. Endocrinology. 1986;119:119–124. doi: 10.1210/endo-119-1-119. [DOI] [PubMed] [Google Scholar]
- 3.Bushinsky D.A. Net calcium efflux from live bone during chronic metabolic, but not respiratory, acidosis. Am. J. Physiol. 1989;256:F836–F842. doi: 10.1152/ajprenal.1989.256.5.F836. [DOI] [PubMed] [Google Scholar]
- 4.Bushinsky D.A. Acid-base imbalance and the skeleton. Eur. J. Nutr. 2001;40:238–244. doi: 10.1007/s394-001-8351-5. [DOI] [PubMed] [Google Scholar]
- 5.Bushinsky D.A. Metabolic alkalosis decreases bone calcium efflux by suppressing osteoclasts and stimulating osteoblasts. Am. J. Physiol. -Ren. Fluid Electrolyte Physiol. 1996;40:F216. doi: 10.1152/ajprenal.1996.271.1.F216. [DOI] [PubMed] [Google Scholar]
- 6.Hermansen L., Osnes J.B. Blood and muscle pH after maximal exercise in man. J. Appl. Physiol. 1972;32:304–308. doi: 10.1152/jappl.1972.32.3.304. [DOI] [PubMed] [Google Scholar]
- 7.Schade H. Die Bedeutung der H-Ionenkonzentration in der Pathologie. Colloid Polym. Sci. 1926;40:252–258. [Google Scholar]
- 8.Martin G.R., Jain R.K. Noninvasive measurement of interstitial pH profiles in normal and neoplastic tissue using fluorescence ratio imaging microscopy. Cancer Res. 1994;54:5670–5674. [PubMed] [Google Scholar]
- 9.Ramp W.K., Lenz L.G., Kaysinger K.K. Medium pH modulates matrix, mineral, and energy metabolism in cultured chick bones and osteoblast-like cells. Bone Miner. 1994;24:59–73. doi: 10.1016/s0169-6009(08)80131-6. [DOI] [PubMed] [Google Scholar]
- 10.Leem Y.H., Nam T.S., Kim J.H., Lee K.S., Lee D.H., Yun J., Chang J.S. The effects of extracellular pH on proliferation and differentiation of human bone marrow stem cells. Korean J. Bone Metab. 2012;19:35. [Google Scholar]
- 11.Kohn D.H., Sarmadi M., Helman J.I., Krebsbach P.H. Effects of pH on human bone marrow stromal cells in vitro: implications for tissue engineering of bone. J. Biomed. Mater. Res. 2002;60:292–299. doi: 10.1002/jbm.10050. [DOI] [PubMed] [Google Scholar]
- 12.Kaysinger K.K., Ramp W.K. Extracellular pH modulates the activity of cultured human osteoblasts. J. Cell. Biochem. 1998;68:83–89. doi: 10.1002/(sici)1097-4644(19980101)68:1<83::aid-jcb8>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 13.Ou K.-L., Hosseinkhani H. Development of 3D in vitro technology for medical applications. Int. J. Mol. Sci. 2014;15:17938–17962. doi: 10.3390/ijms151017938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volkmer E., Drosse I., Otto S., Stangelmayer A., Stengele M., Kallukalam B.C., Mutschler W., Schieker M. Hypoxia in static and dynamic 3D culture systems for tissue engineering of bone. Tissue Eng. Part A. 2008;14:1331–1340. doi: 10.1089/ten.tea.2007.0231. [DOI] [PubMed] [Google Scholar]
- 15.Bonk S.M., Stubbe M., Buehler S.M., Tautorat C., Baumann W., Klinkenberg E.-D., Gimsa J. Design and characterization of a sensorized microfluidic cell-culture system with electro-thermal micro-pumps and sensors for cell adhesion, oxygen, and pH on a glass chip. Biosensors. 2015;5:513–536. doi: 10.3390/bios5030513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galow A., Gimsa J. WST-assay data hint at an increased activity of the succinate reductase in osteoblast-like cells at alkaline pH. Biochem. Biophys. Rep. 2017 doi: 10.1016/j.dib.2017.04.037. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quarles L.D., Yohay D.A., Lever L.W., Caton R., Wenstrup R.J. Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development. J. Bone Miner. Res. 1992;7:683–692. doi: 10.1002/jbmr.5650070613. [DOI] [PubMed] [Google Scholar]
- 18.Fratzl-Zelman N., Fratzl P., Hörandner H., Grabner B., Varga F., Ellinger A., Klaushofer K. Matrix mineralization in MC3T3-E1 cell cultures initiated by β-glycerophosphate pulse. Bone. 1998;23:511–520. doi: 10.1016/s8756-3282(98)00139-2. [DOI] [PubMed] [Google Scholar]
- 19.Jørgensen N.R., Henriksen Z., Sørensen O.H., Civitelli R. Dexamethasone, BMP-2, and 1,25-dihydroxyvitamin D enhance a more differentiated osteoblast phenotype: validation of an in vitro model for human bone marrow-derived primary osteoblasts. Steroids. 2004;69:219–226. doi: 10.1016/j.steroids.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Biological Buffer: AppliChem – BioChemica.Chemica.Synthesis.Service, (n.d.). 〈https://www.applichem.com/en/products/laboratory-biochemicals/biological-buffer/〉 (accessed 04.09.15).
- 21.Buehler S., Stubbe M., Gimsa U., Baumann W., Gimsa J. A decrease of intracellular ATP is compensated by increased respiration and acidification at sub-lethal parathion concentrations in murine embryonic neuronal cells: measurements in metabolic cell-culture chips. Toxicol. Lett. 2011;207:182–190. doi: 10.1016/j.toxlet.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Karsenty G. Transcriptional control of skeletogenesis. Annu. Rev. Genom. Hum. Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 23.Raingeaud J., Gupta S., Rogers J.S., Dickens M., Han J., Ulevitch R.J., Davis R.J. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 24.Freshney N.W., Rawlinson L., Guesdon F., Jones E., Cowley S., Hsuan J., Saklatvala J. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of hsp27. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 25.Owen T.A., Aronow M.S., Barone L.M., Bettencourt B., Stein G.S., Lian J.B. Pleiotropic effects of vitamin D on osteoblast gene expression are related to the proliferative and differentiated state of the bone cell phenotype: dependency upon basal levels of gene expression, duration of exposure, and bone matrix competency in normal rat osteoblast cultures. Endocrinology. 1991;128:1496–1504. doi: 10.1210/endo-128-3-1496. [DOI] [PubMed] [Google Scholar]
- 26.Stein G.S., Lian J.B., Owen T.A. Relationship of cell growth to the regulation of tissue-specific gene expression during osteoblast differentiation. FASEB J. 1990;4:3111–3123. doi: 10.1096/fasebj.4.13.2210157. [DOI] [PubMed] [Google Scholar]
- 27.Komarova S.V., Ataullakhanov F.I., Globus R.K. Bioenergetics and mitochondrial transmembrane potential during differentiation of cultured osteoblasts. Am. J. Physiol. – Cell Physiol. 2000;279:C1220–C1229. doi: 10.1152/ajpcell.2000.279.4.C1220. [DOI] [PubMed] [Google Scholar]
- 28.Ataullakhanov F.I., Vitvitsky V.M. What determines the intracellular ATP concentration. Biosci. Rep. 2002;22:501–511. doi: 10.1023/a:1022069718709. [DOI] [PubMed] [Google Scholar]
- 29.Yan X.-Z., Yang W., Yang F., Kersten-Niessen M., Jansen J.A., Both S.K. Effects of continuous passaging on mineralization of MC3T3-E1 cells with improved osteogenic culture protocol. Tissue Eng. Part C. Methods. 2014;20:198–204. doi: 10.1089/ten.tec.2012.0412. [DOI] [PubMed] [Google Scholar]
- 30.Owen T.A., Aronow M., Shalhoub V., Barone L.M., Wilming L., Tassinari M.S., Kennedy M.B., Pockwinse S., Lian J.B., Stein G.S. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J. Cell. Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 31.Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T.M., Hanauer A., Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Carballo E., Gámez B., Ventura F. p38 MAPK signaling in osteoblast differentiation. Front. Cell Dev. Biol. 2016;4 doi: 10.3389/fcell.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalajzic I., Braut A., Guo D., Jiang X., Kronenberg M.S., Mina M., Harris M.A., Harris S.E., Rowe D.W. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Sudo H., Kodama H.A., Amagai Y., Yamamoto S., Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J. Cell Biol. 1983;96:191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandao-Burch A., Utting J.C., Orriss I.R., Arnett T.R. Acidosis inhibits bone formation by osteoblasts in vitro by preventing mineralization. Calcif. Tissue Int. 2005;77:167–174. doi: 10.1007/s00223-004-0285-8. [DOI] [PubMed] [Google Scholar]
- 36.Madshus I.H. Regulation of intracellular pH in eukaryotic cells. Biochem. J. 1988;250:1. doi: 10.1042/bj2500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakkalakal D.A., Mashoof A.A., Novak J., Strates B.S., McGuire M.H. Mineralization and pH relationships in healing skeletal defects grafted with demineralized bone matrix. J. Biomed. Mater. Res. 1994;28:1439–1443. doi: 10.1002/jbm.820281209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material