Abstract
The aim of the present study was to identify potential therapeutic target genes and miRNAs for primary myelofibrosis (PMF). The dataset GSE53482 was downloaded from the Gene Expression Omnibus database. The differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs) of peripheral blood (PB) cluster of differentiation (CD)34+ cells from PMF patients (PB-PMF group) and peripheral blood CD34+ cells from healthy individuals (PB-control group) were analyzed using the Linear Models for Microarray Data package in R. The Kyoto Encyclopedia of Genes and Genomes was used for pathway enrichment analysis. MiRNA-gene joint enrichment analysis was performed by ENViz and a miRNAs-gene regulatory network was constructed. A total of 1,182 DEGs (773 upregulated and 109 downregulated) and 48 DEMs (28 upregulated and 20 downregulated) were identified. According to the pathway enrichment analysis, a number of DEGs were enriched in metabolic pathways, including IDH1 and DNMT1. Other DEGs were enriched in the citrate cycle (tricarboxylic acid cycle; IDH1 and IDH3A) and certain DEGs were enriched in pyrimidine metabolism, including CARD8. For downregulated genes, certain DEGs were enriched in the spliceosome, including SF3B1 and CDC40. Furthermore, hsa-miR-127-3p, hsa-miR-140-3p and hsa-miR345 were associated with cell cycle-related biological processes, signal transduction and cell surface receptor signaling pathway. The DEM-DEG regulatory network indicated that hsa-miR-543 regulated 113 genes, including CARD8 and TIFA. The present study identified a number of genes, including IDH1, DNMT1, SF3B1 and CARD8, and miRNAs, including hsa-miR-127-3p and hsa-miR-140-3p, which may be therapeutic targets in the treatment of PMF.
Keywords: primary myelofibrosis, differentially expressed genes, differentially expressed miRNAs, network
Introduction
Primary myelofibrosis (PMF) is a clonal stem cell disorder currently classified as a myeloproliferative neoplasm (MPN), and is typically characterized by megakaryocyte proliferation and a typical morphology, extensive bone marrow fibrosis and splenomegaly secondary to extra-medullary hematopoiesis (1). Ineffective erythropoiesis and extra-medullary hematopoiesis are the primary causes of anemia and organomegaly, respectively (2). PMF is a rare hematological malignancy, with an estimated incidence of 0.3–1.5 cases per 100,000 individuals annually (3). Although hematopoietic stem cell transplant and splenectomy are the conventional treatment options for PMF patients, the 5-year survival rates in the transplant cohorts are 69% for low-risk, 52% for intermediate (int)-1, 50% for int-2, and 32% for high-risk patients (4). The mortality rate and median survival for all patients after splenectomy is 14% and 18.5 months (range 0.1–71.9 months), respectively (5). Thus, there is a need for novel, effective treatment strategies for PMF.
Previously, the genetic underpinnings of MPN were identified and a number of studies have indicated that the Philadelphia chromosome, which has been demonstrated to harbor an oncogenic BCR-ABL1 fusion transcript, is the disease-causing mutation in chronic myelogenous leukemia (6–8). In addition, Janus kinase 2 (JAK2) gain of function (JAK2V617F) and myeloproliferative leukemia virus (MPL) gene mutations have been described in ~50% and 5–10% of PMF patients, respectively (9). Norfo et al (10) identified biomarkers of PMF, including FGR, LCN2, and OLFM4, and using miRNA-gene expression integrative analysis, revealed that miR-155-5p affects the in vitro expansion of the megakaryocyte lineage through the modulation of JARID2 expression. This suggests that the miR-155-5p/JARID2 axis may contribute to the abnormal megakaryopoiesis typical in PMF. Nevertheless, the disease genes and its related miRNA molecular signature of PFM remain to be completely identified and the cause of disease is not clearly understood. Therefore, it is essential to comprehensively describe the alterations in gene expression profiles.
In order to reduce the variables, the present study only investigated the expression microarray data of peripheral blood. The microarray dataset GSE53482 deposited by Norfo (10) was used to identify differentially expressed genes (DEGs) and differentially expressed miRNAs (DEMs). Then miRNAs-target gene network analysis combined with functional enrichment analysis was performed. The aim of the present study was to investigate PMF-associated miRNAs and key genes, which may serve key roles in the treatment of PMF.
Materials and methods
Microarray data
The expression profile dataset of GSE53482 was downloaded from Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (10). A total of 73 chips were available, including 42 peripheral blood (PB) cluster of differentiation (CD) 34+ cells from PMF patients (PB-PMF group), 16 peripheral blood CD34+ cells from healthy individuals (PB-control group), 15 bone marrow (BM) CD34+ cells of healthy individuals (BM-control group). In order to reduce the variables, only the expression microarray data from PB tissue were analyzed. Raw data from mRNA and miRNA profiles were collected using GPL13667 (HG-U219) Affymetrix Human Genome U219 Array and GPL14613 (miRNA-2_0) Affymetrix Multispecies miRNA-2_0 Array (Affymetrix Inc., Santa Clara, CA, USA), respectively.
Data preprocessing
The downloaded raw gene expression data of mRNA and miRNA were preprocessed. Background correction, quantile normalization, probe summarization and log2-transformation were performed using a Robust microarray analysis package (version 3.3.2; Bioconductor, Buffalo, NY, USA) (11). If one probe corresponded to multiple genes, the expression value of this probe was removed. However, if multiple probes corresponded to one given gene, the mean expression value was defined as the final expression value of the gene.
DEGs and DEMs screening
DEGs and DEMs of the PB-PMF and PB-control groups were analyzed by the Linear Models for Microarray Data (LIMMA) package (R) (version: 3.30.3; Bioconductor) (12). In order to circumvent the multiple testing problems, which may induce false-positive results, the Benjamini-Hochberg procedure was used to control false discovery rate (FDR) by adjusting the raw P-values (13). log2FC (fold change)≥1 and FDR<0.05 were set as the screening criteria for DEGs. However, log2FC≥0.5 and FDR<0.05 were set as the screening criteria for DEM (12).
Functional enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted with R package clusterProfiler (version: 3.2.14; Bioconductor) (14). KEGG is a database resource for understanding high-level functions and utilities of the biological system, including the cell, organism and ecosystem, from molecular level information (15). The P-values were adjusted using the Benjamini-Hochberg procedure. P<0.05 was considered to indicate a statistically significant difference.
MiRNA-gene joint enrichment analysis
In order to investigate the biological function of DEMs, miRNA-gene joint enrichment analysis was performed by Enrichment Analysis and Visualization (ENViz) in Cytoscape (version 3.4.0; Cytoscape Consortium, San Diego, CA, USA) which was driven by three input matrices: the primary data matrix, the annotation matrix and the pivot data matrix (16). The multiple types of data were integrated in the same sample and the graphical results were presented (16). Primary data (gene expression profile), pivot data (miRNA expression profile) and annotation data were entered into the procedure. The enrichment analysis for the primary element, which was most associated with the pivot element, was performed through ENViz to annotate the biological function of the pivot element.
Construction of miRNAs-target gene regulatory network
MiRNAs are single-stranded RNA molecules of 21–23 nucleotides in length that regulate gene expression generally by inducing cleavage of mRNAs or inhibiting translation in a sequence-specific manner (17). miRanda (www.microrna.org) (18), MirTarget (http://cbit.snu.ac.kr/~miTarget/) (19), PicTar (http://pictar.bio.nyu.edu/) (20), TargetScan (www.targetscan.org) (21), miRecords (http://mirecords.biolead.org/) (22) and miRwalk (www.umm.uni-heidelberg.de/apps/zmf/mirwalk) (23) were used for the miRNA target gene prediction. MiRNA-genes present in at least three of the aforementioned databases were used to construct the miRNA-gene regulatory network.
Statistical analysis
The expression levels of genes are presented as mean ± standard deviation. Statistical analysis was performed using SPSS 20.0 (IBM SPSS, Armonk, NY, USA). Student's t-test was used to analyze differences between PB-PMF and PB-control groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Screening of DEGs and DEMs
A total of 1,182 DEGs including 773 upregulated and 109 downregulated genes were selected from the PMF samples (Table I). Following DEMs screening, 48 DEMs were identified, including 28 upregulated and 20 downregulated DEMs (Table II).
Table I.
Number of differentially expressed genes and microRNAs in the PB-PMF group compared with PB-control group.
| Category | Genes | MicroRNAs |
|---|---|---|
| Upregulated | 773 | 28 |
| Downregulated | 109 | 20 |
| Total | 1,182 | 48 |
PMF, primary myelofibrosis; CD, cluster of differentiation; PB-PMF group, peripheral blood CD34+ cells from PMF patients; PB-control group, peripheral blood CD34+ cells from healthy individuals.
Table II.
Differentially expressed microRNAs in the peripheral blood cluster of differentiation (CD)34+ cells from primary myelofibrosis patients.
| MicroRNAs | logFC | FDR |
|---|---|---|
| hsa-miR-127-3p | 3.387003333 | 1.59E-11 |
| hsa-miR-487b | 2.898958717 | 4.77E-09 |
| hsa-miR-409-3p | 2.38774578 | 1.90E-06 |
| hsa-miR-1246 | 2.117570289 | 0.001769255 |
| hsa-miR-486-5p | 1.987400714 | 0.000180305 |
| hsa-miR-1308 | 1.918602857 | 1.74E-09 |
| hsa-miR-494 | 1.770673137 | 7.10E-06 |
| hsa-miR-134 | 1.503354598 | 3.53E-05 |
| hsa-miR-382 | 1.503337152 | 0.000110142 |
| hsa-miR-503 | 1.425249491 | 8.13E-05 |
| hsa-miR-486-3p | 1.392454196 | 0.047462229 |
| hsa-miR-18a | 1.270815722 | 4.58E-08 |
| hsa-miR-1979 | 1.267982054 | 0.012984921 |
| hsa-miR-92a-1 | 1.184540893 | 0.003833481 |
| hsa-miR-152 | 1.141451113 | 0.011362506 |
| hsa-miR-379 | 1.042055815 | 9.74E-07 |
| hsa-miR-432 | 0.957774043 | 9.27E-05 |
| hsa-miR-19b | 0.940775327 | 1.48E-05 |
| hsa-miR-195 | 0.940193085 | 1.37E-05 |
| hsa-miR-376c | 0.849398149 | 0.007550278 |
| hsa-miR-485-5p | 0.811446693 | 0.000110142 |
| hsa-miR-543 | 0.754953378 | 0.003583896 |
| hsa-miR-34a | 0.754130661 | 0.000852361 |
| hsa-miR-21 | 0.710161631 | 0.023095868 |
| hsa-miR-18b | 0.633363635 | 0.000111584 |
| hsa-miR-370 | 0.604671741 | 0.016171501 |
| hsa-miR-20b | 0.510119307 | 0.002615483 |
| hsa-miR-146b-5p | 0.500727976 | 0.023667101 |
| hsa-miR-744 | −0.53118132 | 1.74E-05 |
| hsa-miR-1915 | −0.55023808 | 0.000708478 |
| hsa-miR-423-5p | −0.55960845 | 0.000997801 |
| hsa-miR-3196 | −0.59688598 | 0.028223221 |
| hsa-miR-140-3p | −0.62122708 | 0.001454615 |
| hsa-miR-23a | −0.63403583 | 0.003583896 |
| hsa-miR-345 | −0.64862991 | 0.01242366 |
| hsa-miR-1275 | −0.66625937 | 0.003833481 |
| hsa-miR-638 | −0.74490649 | 0.005061941 |
| hsa-miR-4281 | −0.76486024 | 0.007550278 |
| hsa-miR-2861 | −0.78904119 | 0.010372911 |
| hsa-miR-324-3p | −0.81814792 | 0.009448033 |
| hsa-miR-27a | −0.85423333 | 0.007550278 |
| hsa-miR-197 | −0.9544247 | 0.000228802 |
| hsa-miR-762 | −0.95937914 | 0.003304652 |
| hsa-miR-766 | −1.04226524 | 0.000725484 |
| hsa-miR-1469 | −1.04372292 | 9.21E-05 |
| hsa-miR-181a-2 | −1.23940304 | 0.003054099 |
| hsa-miR-3185 | −1.31621408 | 0.007373289 |
| hsa-miR-663 | −1.55496702 | 3.62E-07 |
FC, fold-change; FDR, false discovery rate.
Functional enrichment analysis results
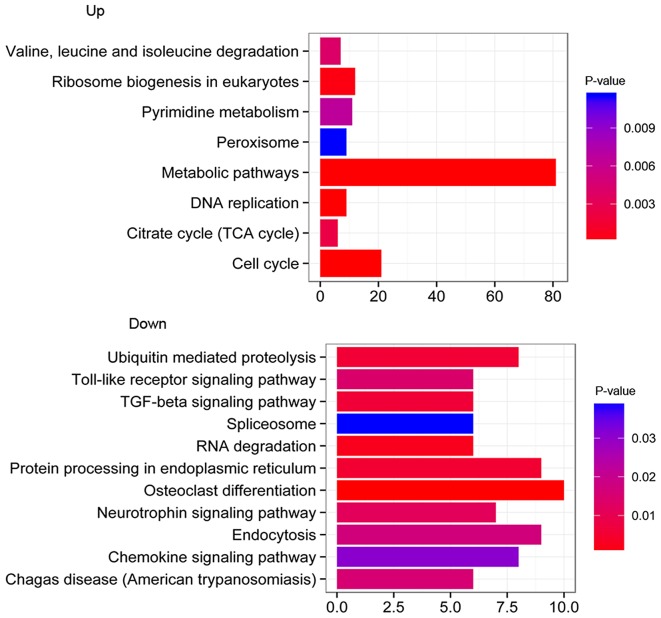
According to KEGG pathway enrichment analysis, the upregulated DEGs were significantly enriched in metabolic pathways (P=3.21E-04), cell cycle (P=1.31E-05), ribosome biogenesis in eukaryotes (P=2.83E-03) and DNA replication (P=3.21E-04). The downregulated DEGs were significantly enriched in the Toll-like receptor signaling pathway (P=0.025), osteoclast differentiation (P=0.002), protein processing in the endoplasmic reticulum (P=0.016) and the transforming growth factor-β signaling pathway (P=0.016) (Fig. 1). As presented in Table III, for upregulated genes a number of DEGs were significantly enriched in metabolic pathways (IDH1 and DNMT1; P=3.21E-04), the citrate cycle (TCA cycle; IDH1 and IDH3A; P=1.47E-02) and within the pyrimidine metabolism (CARD8; P=2.92E-02). However, for downregulated genes, a number of DEGs were significantly enriched in the spliceosome (SF3B1 and CDC40; P=4.54E-02).
Figure 1.
Pathway enrichment analysis for up- and downregulated genes. The abscissa represents P-value following log2-transformation, the ordinate represents the enriched biological pathways with log2FC≥1 and P<0.05, and the x-axis represents the number of genes. TGF-β, transforming growth factor-β; TCA, tricarboxylic acid cycle (citric acid cycle).
Table III.
Functional annotation for differentially expressed genes in primary myelofibrosis.
| Category | ID | Description | Count | Gene symbol | P-value |
|---|---|---|---|---|---|
| Upregulated genes | hsa04110 | Cell cycle | 21 | BUB1, BUB1B, CCNA2, CCNB1, CCNB2,CDC6, CDK1, CHEK1, CUL1, E2F5 | 1.31E-05 |
| hsa01100 | Metabolic pathways | 81 | ACADM, ACO1, B3GALNT1, B3GALT6, BCAT2, IDH1, CYP2R1, DNMT1, EXTL2, GALK2 | 3.21E-04 | |
| hsa03030 | DNA replication | 9 | FEN1, MCM2, MCM3, MCM4, MCM6, PCNA, POLE2, RNASEH2A, RPA2 | 3.21E-04 | |
| hsa03008 | Ribosome biogenesis in eukaryotes | 12 | EFTUD1, GNL3L, IMP3, MPHOSPH10, POP5, POP7, PWP2, REXO2, RPP40, UTP14C | 2.83E-03 | |
| hsa00020 | Citrate cycle (TCA cycle) | 6 | ACO1, DLAT, IDH1, IDH3A, PDHB, SUCLA2 | 1.47E-02 | |
| hsa00280 | Valine, leucine and isoleucine degradation | 7 | ACADM, ALDH3A2, BCAT2, HIBADH, HIBCH, HMGCL, MUT | 2.12E-02 | |
| hsa00240 | Pyrimidine metabolism | 11 | CARD8, PNP, PNPT1, POLE2, POLR1B, POLR2H, POLR3B, RRM2, TK1, TYMS, UMPS | 2.92E-02 | |
| hsa04146 | Peroxisome | 9 | ACSL3, ACSL5, FAR2, HMGCL, HSD17B4, IDH1, PEX3, PRDX1, SLC27A2 | 4.46E-02 | |
| Downregulated genes | hsa04380 | Osteoclast differentiation | 10 | CTSK, CYLD, FOSL2, IFNGR1, IL1A, JUND, NFKBIA, PIK3CA, RELB, SQSTM1 | 2.31E-03 |
| hsa03018 | RNA degradation | 6 | BTG1, BTG3, CNOT8, DCP1A, DDX6, XRN1 | 1.57E-02 | |
| hsa04141 | Protein processing in endoplasmic reticulum | 9 | DDIT3, EIF2AK3, ERO1L, ERO1LB, SEC24A, SEC24B, UBE2G1, UBE2J1, YOD1 | 1.57E-02 | |
| hsa04120 | Ubiquitin-mediated proteolysis | 8 | BIRC3, ITCH, RNF7, SIAH1, SMURF1, UBE2G1, UBE2H, UBE2J1 | 1.57E-02 | |
| hsa04350 | TGF-β signaling pathway | 6 | EP300, ID1, ID2, ID3, SMAD7, SMURF1 | 1.57E-02 | |
| hsa04722 | Neurotrophin signaling pathway | 7 | CDC42, FOXO3, IRS2, MAPKAPK2, NFKBIA, NFKBIE, PIK3CA | 2.48E-02 | |
| hsa04620 | Toll-like receptor signaling pathway | 6 | CCL3, CTSK, IL8, NFKBIA, PIK3CA, TLR1 | 2.48E-02 | |
| hsa05142 | Chagas disease (American trypanosomiasis) | 6 | CCL3, GNA15, IFNGR1, IL8, NFKBIA, PIK3CA | 2.48E-02 | |
| hsa04144 | Endocytosis | 9 | ADRBK1, CDC42, CHMP2B, CHMP4B, ITCH, SMAD7, SMURF1, STAM, VPS37B | 2.48E-02 | |
| hsa04062 | Chemokine signaling pathway | 8 | ADRBK1, CCL20, CCL3, CDC42, FOXO3, IL8, NFKBIA, PIK3CA | 4.00E-02 | |
| hsa03040 | Spliceosome | 6 | CDC40, RBMXL1, SF3B1, SLU7, SYF2, WBP11 | 4.54E-02 |
TGF-β, transforming growth factor-β; TCA cycle, tricarboxylic acid cycle (citric acid cycle).
MiRNA gene joint enrichment analysis
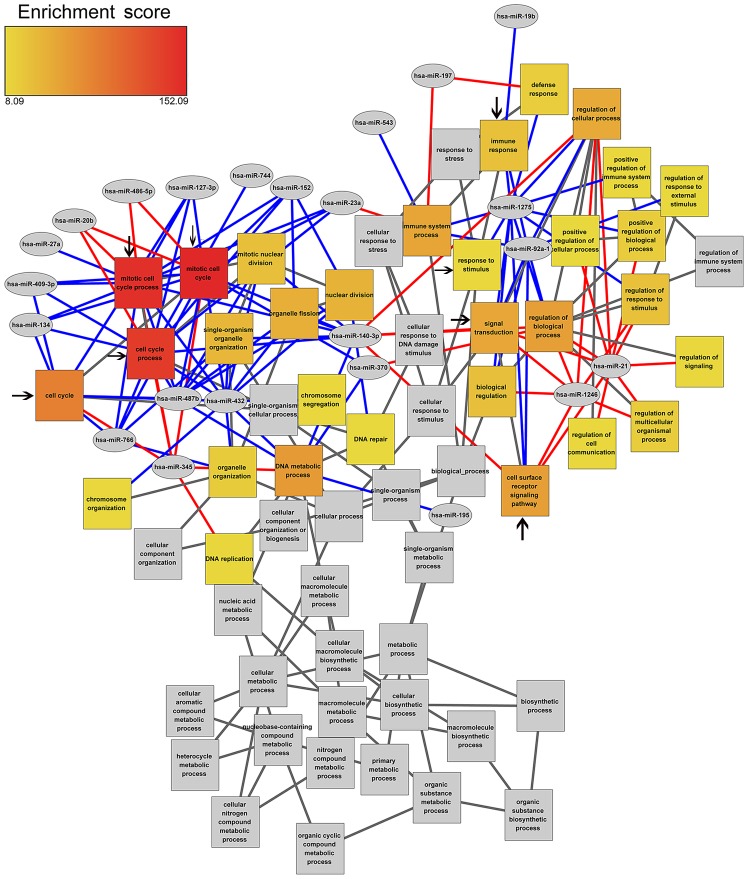
In order to study the effect of DEMs and DEGs in different biological processes (BP), DEMs and DEGs joint enrichment analysis was performed. DEMs were primarily enriched in the biological processes associated with the cell cycle, immune response and response to stimulus. It was also demonstrated that the expression of hsa-miR-127-3p, which exhibited a significantly higher expression, was negatively associated with four cell cycle-related BP terms, including cell cycle, mitotic cell cycle, mitotic cell cycle process and cell cycle process. Notably, hsa-miR-140-3p was negatively associated with the cell cycle and positively associated with different biological pathways, including signal transduction and cell surface receptor signaling pathway (Fig. 2).
Figure 2.
miRNA-GO enrichment network. Cumulative enrichment score for each GO term is the sum of enrichment scores for all edges connected to the GO term node. Unconnected parents are colored gray. Pivot nodes (miRNAs) are colored gray. Edges are color-coded by direction of gene-pivot association. Red edges indicate that the genes related to the enriched GO term are positively regulated by the pivot miRNA, and blue edges indicate that the genes related to the GO term are negatively regulated by the pivot miRNA. The colour of boxes (yellow to red) indicates the enrichment score from 8.09 to 152.09. Black arrows indicate the important GO terms. GO, gene ontology; miRNA, microRNA.
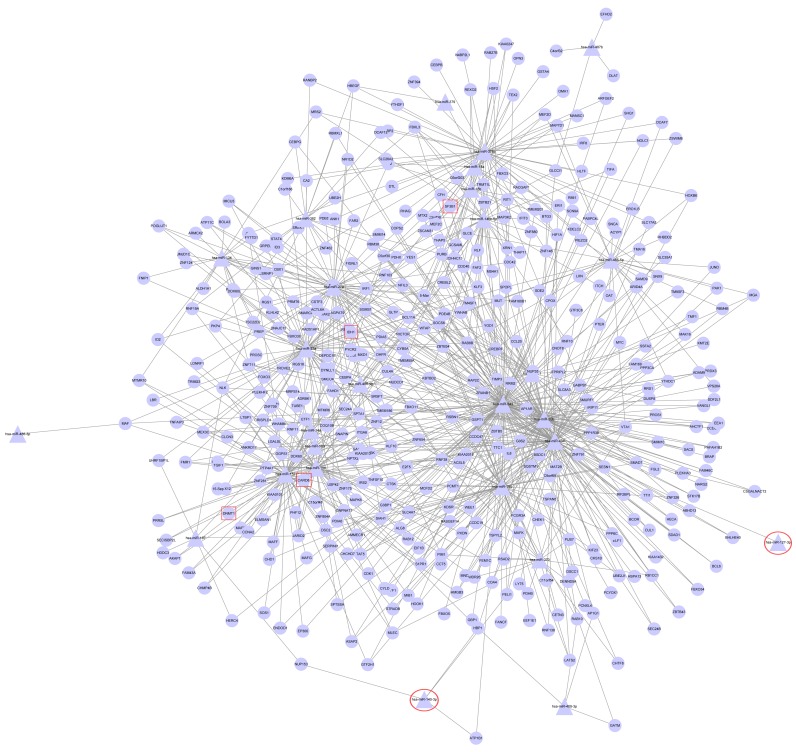
DEM-DEG regulatory network analysis
Based on the aforementioned 7 miRNA target gene prediction methods, the regulation associated between DEMs and DEGs was identified (Fig. 3). The regulatory network included 27 miRNAs, 427 genes and 820 edges. The top 5 miRNAs with the highest connective degree in the DEM-DEG regulatory network included hsa-miR-543, hsa-miR-494, hsa-miR-20b, hsa-miR-23a and hsa-miR-152 (Table IV). For example, hsa-miR-543 regulated 113 genes, including CARD8, PELI1 and TIFA (Fig. 3).
Figure 3.
Differentially expressed microRNAs-differentially expressed gene regulatory network. The triangle nodes represent miRNAs and the circle nodes represent genes. Red squares and circles represent the key DEGs and miRNAs, respectively.
Table IV.
Top 5 microRNAs with the highest connective degree in the differentially expressed microRNAs-differentially expressed genes regulatory network.
| MicroRNA | Connective degree |
|---|---|
| hsa-miR-543 | 113 |
| hsa-miRt-494 | 87 |
| hsa-miR-20b | 73 |
| hsa-miR-23a | 62 |
| hsa-miR-195 | 54 |
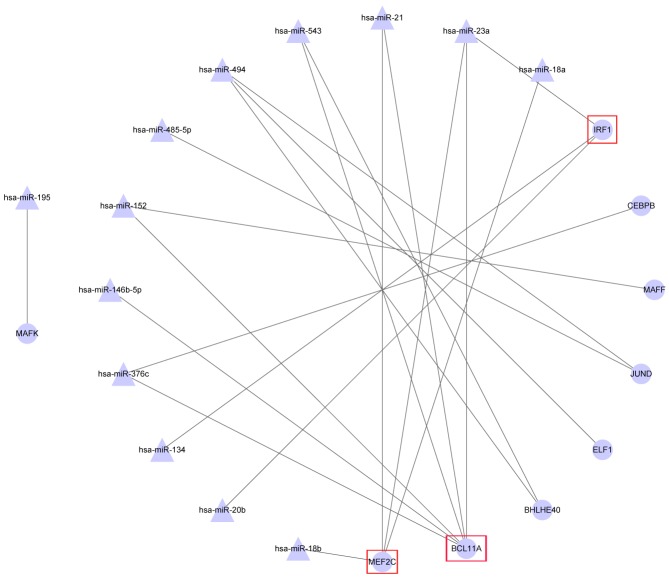
There was a regulatory association between miRNA and transcription factors (TF) in the DEM-DEG regulatory network (Fig. 4), which included 13 miRNAs and 9 TFs. Of these, MEF2C and BCL11A were regulated by >2 DEMs; for example MEF2C connected with hsa-miR-21, hsa-miR-23a, hsa-miR-18a and hsa-miR-18b.
Figure 4.
Differentially expressed microRNA-transcription factor regulatory network. The triangle nodes represent microRNAs, and the circle nodes represent transcription factors. Red squares indicate the transcription factors that are regulated by more than two differentially expressed microRNAs.
Discussion
PMF is a Philadelphia-negative myeloproliferative neoplasm with a heterogeneous clinical presentation (24,25). In the present study, a total of 1,182 DEGs and 48 DEMs were identified from PB CD34+ cells of patients with PMF. Among them, a number of upregulated DEGs were significantly enriched in metabolic pathways, including IDH1 and DNMT1. Certain DEGs were markedly enriched in pyrimidine metabolism, including CARD8. A number of downregulated DEGs were enriched in the spliceosome, including SF3B1.
IDH1 is located on chromosome 2q33.3 and encodes isocitrate dehydrogenase 1, which catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (26). IDH1 serves an important role in cytoplasmic micotinamide adenine dinucleotide phosphate production. IDH mutations are frequent in blast-phase myeloproliferative neoplasms and therefore, may contribute to leukemic transformation. Previous findings have reported higher frequencies of IDH1/IDH2 and LNK mutations in blast-phase PMF, suggesting a pathogenetic contribution to disease progression (27). The mutant IDH1 was associated with a worse prognosis in cytogenetically normal acute myeloid leukemia with a NPM1+/FLT3− molecular profile (28,29). The phenotypic and prognostic effects of the IDH1 mutation among patients with PMF have been previously shown (27). Clinical observations underscore the potential relevance of investigating other mutations or epigenetic abnormalities that functionally mimic the mutant IDH, in JAK2-mutated PMF (30).
DNMT1 encodes DNA (cytosine-5-)-methyltransferase 1, which belongs to a family of DNA methyltransferases (DNMT) including DNMT1, DNMT3A and DNMT3B, and catalyzes the addition of methyl groups to cytosine residues of CpG nucleotides (31). DNA methyltransferase is among the most common occurring mutation identified in myeloid malignancies (32). DNA methylation of DNMT1 consists of an enzymatic addition of a methyl group at the carbon 5 position of cytosine in the context of the sequence 5-cytosine-guanosine (CpG), mediated by DNMTs (33). Although the mechanisms leading to aberrant DNA hypermethylation remain to be fully elucidated, increased levels of DNMT1 are observed in malignant myeloid blasts compared with normal bone marrow mononuclear cells, suggesting that DNMT overexpression contributes to gene promoter hypermethylation and in turn, to leukemogenesis (34). Furthermore, DNMT family genes were analyzed in PMF, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms (35).
SF3B1 is a critical component of the splicing machinery, which catalyzes the removal of introns from precursor mRNA and it was observed to be enriched in spliceosome in the present study. One of the most notable findings arising from the recent next-generation sequencing data has been the identification of mutated splicing factor 3b1 (SF3B1) as a putative candidate driver gene of the common lymphoid malignancy, chronic lymphocytic leukemia (36). A clinical, histopathological and genetic association study of 155 patients indicated that SF3B1 mutations in PMF may encode a key component of the mRNA splicing machinery (37). SF3B1 mutations were initially identified in myelodysplastic syndromes and other solid tumors suggesting it is serving an important role in cancer biology (38). SF3B1 mutations are associated with disease subtype (38), progression (39,40), chemotherapy resistance (41) and longer overall patient survival (42).
CARD8 is the role of caspase activating and recruitment domain 8 and encodes a protein whose exact function remains unclear (43). In the current study, it was observed that CARD8 was upregulated and enriched in the pyrimidine metabolism. CARD proteins are generally involved in signal transduction pathways, important for apoptosis, inflammation, immune function and nuclear factor-κB activation (1). Furthermore, a previous study demonstrated that the presence of somatic mutations and expression analysis of CARD family gene may be potentially relevant to PMF pathogenesis, at least in rare cases (1). In the present study, the involvement of CARD8 was predicted by the bioinformatics methods mentioned. This, combined with previous studies, indicates that it may be a therapeutic target for PMF.
In the present study, hsa-miR-127-3p was negatively associated with the four cell cycle-related BP terms, which may suggest that the high expression of hsa-miR-127-3p inhibits the proliferation of stem cells in peripheral blood (44). However, hsa-miR-140-3p was identified to be the most active miRNA in the current study. While it was negatively associated with cell cycle pathways, it was positively associated with signal transduction and cell surface receptor signaling pathway. A previous study demonstrated that miR-140-3p is differentially expressed in hematopoietic lineages, and it may be associated with platelet production and activation (45). Hsa-miR345, hsa-miR20b and hsa-miR486-5p were positively associated with cell cycle-related terms such as mitotic cell cycle process and mitotic cell cycle. In particular, hsa-miR345 was positively associated with DNA replication and metabolic pathways (46). This suggested that these miRNAs play key roles in PMF. A clinical study demonstrated that in patients with PMF, the number of peripheral blood CD34+ cells was significantly higher than normal (47). Therefore, the abnormal expression of these miRNAs in patients with PMF affected the CD34+ cell cycle in peripheral blood, thus affecting the occurrence and development of PMF. Although the target genes and miRNAs for PMF were identified, validation of the results is required in the future.
In conclusion, the DEGs and DEMs were obtained in PMF. A number of significant genes, including IDH1, DNMT1, SF3B1 and CARD8 and miRNAs, including hsa-miR-127-3p and hsa-miR-140-3p, may serve pivotal roles in the development of PMF. These genes and miRNAs may be used as specific therapeutic targets in the treatment of PMF, which may be validated by experiments in future studies.
References
- 1.Merker JD, Roskin KM, Ng D, Pan C, Fisk DG, King JJ, Hoh R, Stadler M, Okumoto LM, Abidi P, et al. Comprehensive whole-genome sequencing of an early-stage primary myelofibrosis patient defines low mutational burden and non-recurrent candidate genes. Haematologica. 2013;98:1689–1696. doi: 10.3324/haematol.2013.092379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benites BD, Lima CS, Lorand-Metze I, Delamain MT, Oliveira GB, Almeida Dd, Souza CA, Vassallo J, Pagnano KB. Primary myelofibrosis: Risk stratification by IPSS identifies patients with poor clinical outcome. Clinics (Sao Paulo) 2013;68:339–343. doi: 10.6061/clinics/2013(03)OA09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed A, Chang CC. Chronic idiopathic myelofibrosis: Clinicopathologic features, pathogenesis, and prognosis. Arch Pathol Lab Med. 2006;130:1133–1143. doi: 10.5858/2006-130-1133-CIM. [DOI] [PubMed] [Google Scholar]
- 4.Kröger N, Giorgino T, Scott BL, Ditschkowski M, Alchalby H, Cervantes F, Vannucchi A, Cazzola M, Morra E, Zabelina T, et al. Impact of allogeneic stem cell transplantation on survival of patients less than 65 years with primary myelofibrosis. Blood. 2015;125:3347–4350. doi: 10.1182/blood-2014-10-608315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitanaka A, Takenaka K, Shide K, Miyamoto T, Kondo T, Ozawa K, Kurokawa M, Akashi K, Shimoda K. Splenic irradiation provides transient palliation for symptomatic splenomegaly associated with primary myelofibrosis: A report on 14 patients. Int J Hematol. 2016;103:423–428. doi: 10.1007/s12185-016-1940-3. [DOI] [PubMed] [Google Scholar]
- 6.Schnittger S, Bacher U, Eder C, Dicker F, Alpermann T, Grossmann V, Kohlmann A, Kern W, Haferlach C, Haferlach T. Molecular analyses of 15,542 patients with suspected BCR-ABL1-negative myeloproliferative disorders allow to develop a stepwise diagnostic workflow. Haematologica. 2012;97:1582–1585. doi: 10.3324/haematol.2012.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mughal TI, Radich JP, Van Etten RA, Quintás-Cardama A, Skorski T, Ravandi F, DeAngelo DJ, Gambacorti-Passerini C, Martinelli G, Tefferi A. Chronic myeloid leukemia 2011: Successes, challenges, and strategies-proceedings of the 5th annual BCR-ABL1 positive and BCR-ABL1 negative myeloproliferative neoplasms workshop. Am J Hematol. 2011;86:811–819. doi: 10.1002/ajh.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, Zaleskas VM, Van Etten RA. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2(V617F) in mice. Blood. 2012;119:3550–3560. doi: 10.1182/blood-2011-12-397554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vardiman JW, Brunning RD, Arber DA, Le Beau MM, Porwit A, Tefferi A, Bloomfield CD, Thiele J. Introduction and overview of the classification of the myeloid neoplasms. In: Jaffe ES, Harris NL, Swerdlow SH, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: 2008. pp. 18–30. [Google Scholar]
- 10.Norfo R, Zini R, Pennucci V, Bianchi E, Salati S, Guglielmelli P, Bogani C, Fanelli T, Mannarelli C, Rosti V, et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: Role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 2014;124:e21–e32. doi: 10.1182/blood-2013-12-544197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 12.Smyth GK. Limma: linear models for microarray data Bioinformatics and computational biology solutions using R and Bioconductor. Springer; 2005. pp. 397–420. [Google Scholar]
- 13.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altman T, Travers M, Kothari A, Caspi R, Karp PD. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinformatics. 2013;14:112. doi: 10.1186/1471-2105-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, Mäkelä R, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. MicroRNAs: Molecular features and role in cancer. Front Biosci (Landmark Ed) 2012;17:2508–2540. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. microRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SK, Nam JW, Rhee JK, Lee WJ, Zhang BT. MiTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics. 2006;7:411. doi: 10.1186/1471-2105-7-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 21.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian miscoRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 22.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Barosi G, Rosti V, Bonetti E, Campanelli R, Carolei A, Catarsi P, Isgrò AM, Lupo L, Massa M, Poletto V, Viarengo G, et al. Evidence that prefibrotic myelofibrosis is aligned along a clinical and biological continuum featuring primary myelofibrosis. PLoS One. 2012;7:e35631. doi: 10.1371/journal.pone.0035631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mascarenhas JO, Orazi A, Bhalla KN, Champlin RE, Harrison C, Hoffman R. Advances in myelofibrosis: A clinical case approach. Haematologica. 2013;98:1499–1509. doi: 10.3324/haematol.2013.086348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tefferi A, Lasho TL, Abdel-Wahab O, Guglielmelli P, Patel J, Caramazza D, Pieri L, Finke CM, Kilpivaara O, Wadleigh M, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tefferi A, Jimma T, Sulai NH, Lasho TL, Finke CM, Knudson RA, McClure RF, Pardanani A. IDH mutations in primary myelofibrosis predict leukemic transformation and shortened survival: Clinical evidence for leukemogenic collaboration with JAK2V617F. Leukemia. 2012;26:475–480. doi: 10.1038/leu.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boissel N, Nibourel O, Renneville A, Gardin C, Reman O, Contentin N, Bordessoule D, Pautas C, de Revel T, Quesnel B, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: A study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28:3717–3723. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- 29.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, Späth D, Kayser S, Zucknick M, Götze K, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 30.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan SM, Majeti R. Role of DNMT3A, TET2 and IDH1/2 mutations in pre-leukemic stem cells in acute myeloid leukemia. Int J Hematol. 2013;98:648–657. doi: 10.1007/s12185-013-1407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.V97.5.1172. [DOI] [PubMed] [Google Scholar]
- 35.Abdel-Wahab O, Pardanani A, Rampal R, Lasho TL, Levine RL, Tefferi A. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia. 2011;25:1219–1220. doi: 10.1038/leu.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan Y, Wu CJ. SF3B1 mutations in chronic lymphocytic leukemia. Blood. 2013;121:4627–4634. doi: 10.1182/blood-2013-02-427641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasho TL, Finke CM, Hanson CA, Jimma T, Knudson RA, Ketterling RP, Pardanani A, Tefferi A. SF3B1 mutations in primary myelofibrosis: Clinical, histopathology and genetic correlates among 155 patients. Leukemia. 2012;26:1135–1137. doi: 10.1038/leu.2011.320. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwaederlé M, Ghia E, Rassenti LZ, Obara M, Dell'Aquila ML, Fecteau JF, Kipps TJ. Subclonal evolution involving SF3B1 mutations in chronic lymphocytic leukemia. Leukemia. 2013;27:1214–1217. doi: 10.1038/leu.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, Fangazio M, Vaisitti T, Monti S, Chiaretti S, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: Association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasseur F, Sendid B, Broly F, Gower-Rousseau C, Sarazin A, Standaert-Vitse A, Colombel JF, Poulain D, Jouault T. The CARD8 p.C10X mutation associates with a low anti-glycans antibody response in patients with Crohn's disease. BMC Med Genet. 2013;14:35. doi: 10.1186/1471-2350-14-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenu EW, Ashton KJ, Batovska J, Staines DR, Marshall-Gradisnik SM. High-throughput sequencing of plasma microRNA in chronic fatigue syndrome/myalgic encephalomyelitis. PLoS One. 2014;9:e102783. doi: 10.1371/journal.pone.0102783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collares CV, Evangelista AF, Xavier DJ, Rassi DM, Arns T, Foss-Freitas MC, Foss MC, Puthier D, Sakamoto-Hojo ET, Passos GA, Donadi EA. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res Notes. 2013;6:491. doi: 10.1186/1756-0500-6-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuokkanen S, Chen B, Ojalvo L, Benard L, Santoro N, Pollard JW. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol Reprod. 2010;82:791–801. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andréasson B, Swolin B, Kutti J. Patients with idiopathic myelofibrosis show increased CD34+ cell concentrations in peripheral blood compared to patients with polycythaemia vera and essential thrombocythaemia. Eur J Haematol. 2002;68:189–193. doi: 10.1034/j.1600-0609.2002.01610.x. [DOI] [PubMed] [Google Scholar]