ABSTRACT
The nucleolus is a distinct compartment of the nucleus responsible for ribosome biogenesis. Mis-regulation of nucleolar functions and of the cellular translation machinery has been associated with disease, in particular with many types of cancer. Indeed, many tumor suppressors (p53, Rb, PTEN, PICT1, BRCA1) and proto-oncogenes (MYC, NPM) play a direct role in the nucleolus, and interact with the RNA polymerase I transcription machinery and the nucleolar stress response. We have identified Dicer and the RNA interference pathway as having an essential role in the nucleolus of quiescent Schizosaccharomyces pombe cells, distinct from pericentromeric silencing, by controlling RNA polymerase I release. We propose that this novel function is evolutionarily conserved and may contribute to the tumorigenic pre-disposition of DICER1 mutations in mammals.
KEYWORDS: cancer, dicer, epigenetics, nucleolus, rDNA
The nucleolus specializes in ribosomal transcription and assembly
The nucleolus is the nuclear compartment in which RNA (rRNA) is transcribed, processed, and assembled into ribosomal subunits during interphase, that are then exported to form mature ribosomes. The nucleolus is formed by the process of ribosome biogenesis1 and organizes into 3 main zones: the fibrillar(s) center(s) (FC), the dense fibrillar component (DFC), and the granular component (GC). DNA (rDNA) is present within the FC, at the periphery of which pre-rRNA is actively transcribed and elongates into the DFC. RNA processing occurs co-transcriptionally and rRNA assembles with ribosomal proteins, to form immature ribosomes in the GC2,3 (Fig. 1). A specific RNA polymerase, RNA polymerase I, specializes in transcription of the DNA (rDNA) locus. The resulting 48S pre-rRNA is processed by a series of cleavages to obtain the 18S, 5.8S and 28S mature rRNAs, while RNA polymerase III transcribes the fourth rRNA species, 5S rRNA, outside the nucleolus, where it is then imported along with small nucleolar RNAs (snoRNAs) important for rRNA processing. The major transcriptional burden of a growing cell lies in ribosome biosynthesis—rRNAs can represent 80% of cellular RNAs—and is heavily correlated with rates of protein synthesis, highlighting its central importance in cellular metabolism. Understandably, nucleolar transcription is tightly regulated according to the phase of the cell cycle, nutritional availability, environment, and growth status of the cell.4 Conversely, multiple ribosomal quality control mechanisms provide feedback for cell cycle progression.5
Figure 1.
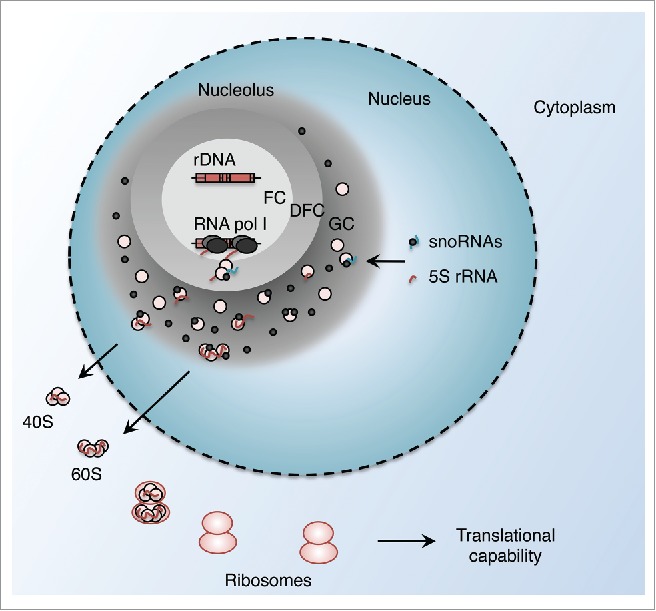
Overview of nucleolar structure and function. RNA polymerase I transcribes the rDNA repeats at the interface between the fibrillar center (FC) and the dense fibrillar component (DFC). rRNA molecules and ribosomal proteins associate in complexes in the granular component (GC), and are exported outside of the nucleolus as immature ribosomal particles, which are then assembled into ribosomes.
The rDNA promoter
The initiation of rDNA transcription requires several basal transcription factors in addition to the multi-subunit RNA polymerase I complex. Active rDNA promoters are organized in a configuration free of nucleosomes,3 bound by dimers of UBF (Upstream Binding Factor), a multi-functional HMGB-box protein which activates transcription6 and forms a structure termed the ‘enhancosome'.7 Furthermore, UBF spreads across the rDNA locus and contributes to its structural organization.8,9
The core rDNA promoter is bound by 2 basal transcription initiation factors, TIF-IA/RRN3 and TIF-IB/SL1, which are necessary for the formation of the RNA polymerase I pre-initiation complex (PIC). The TIF-IB/SL1 complex comprises the TATA-binding protein (TBP) in a specific complex with TAF proteins (TBP-associated factors: TAF1A, TAF1B, TAF1C, TAF1D),10 which confers promoter specificity. TIF-IA/RRN3 recruits initiation-competent RNA polymerase I complexes to the rDNA promoter, and interacts directly with TAF1B and TAF1C in the SL1 complex.11 Binding of SL1 to the rDNA promoter further stabilizes UBF.12 Budding yeast and fission yeast form a functionally equivalent TBP-TAF complex (RRN6, RRN7, RRN11),13 while the upstream element (UCE) of the rDNA promoter is bound by an additional complex, the Upstream Activating Factor (UAF).14 The promoter and the terminator regions of rDNA are brought in close contact via binding of TTF-I, which participates in RNA polymerase I transcription termination and re-initiation.15,16
Regulatory mechanisms of ribosomal transcription
RNA polymerase I transcription is tightly regulated to couple the translational capability of the cell to its requirements according to its growth status. The size and organization of the nucleolus is particularly dependent on the phase of the cell cycle, as well as environmental conditions.3,17 Regulation of RNA polymerase I transcription occurs at several key-points, in particular at the level of the formation of the pre-initiation complex, and it has been proposed that the rate-limiting step of RNA polymerase I transcription is promoter clearance subsequent to PIC formation,18 although other studies also pointed to transcription elongation, depending on growth conditions.19
The phosphorylation status of TIF-IA/RRN3 is regulated by nutrient availability through the TOR signaling pathway,20,21 mitogen-activated kinase (MAPK) cascades,22 and RNA polymerase I activity is modulated by several key signaling pathways such as PI3K,23 AKT,24 GSK3β,25 the RAS-family protein RasL11a.26 The inputs of these different signaling pathways converge to fine-tune RNA polymerase I transcription, integrating growth factors, nutrient availability and energetic capability.27
Another layer of regulation lies within the repetitive structure of the rDNA. The rDNA repeats form at least 3 fractions differing in their transcriptional status: active rDNA repeats undergoing RNA polymerase I transcription, silent rDNA repeats packed into heterochromatin, and poised/inactive rDNA repeats that, while not actively being transcribed, do not present heterochromatic marks and may constitute a readily-available ‘buffer' of repeats.28,29 UBF is an important player in determining the active/inactive rDNA repeat ratio.30
Two key phases of the cell cycle when RNA polymerase I activity needs to be regulated are mitosis and quiescence. During mitosis, RNA polymerase I transcription is halted until G1 via phosphorylation of UBF and the TAF1C subunit of SL1.31 Cells alternate between phases of quiescence and division, for example, to maintain stem cell populations in mammals,32 or to survive conditions of nutritional scarcity in microbial eukaryotes.33 In non-dividing quiescent cells that retain metabolic and transcriptional activity,32,34 RNA polymerase I remains essential for maintaining basal levels of rRNAs to ensure translation of transcripts necessary for quiescence maintenance, but is strongly downregulated. Quiescent fibroblasts down-regulate transcripts of genes involved in ribosomal biogenesis.35 This regulation is conserved in eukaryotes; for example, in the fission yeast Schizosaccharomyces pombe, a model organism for the cell cycle and cellular quiescence,33 rDNA occupancy of RNA polymerase I is greatly decreased in quiescent cells,36 and conversely, rDNA transcription and ribosomal proteins are upregulated upon re-entry into the cell cycle.37
Misregulation of the nucleolus is central to cancer
Misregulation of ribosome biosynthesis, the central function of the nucleolus, is a recurrent feature in cancer, as well as in a distinct set of rare genetic diseases known as ribosomopathies.38 Early on, an enlarged nucleolus has been recognized as a hallmark of cancer cells/cellular transformation,39 and higher numbers of silver-stained nucleolar organizer regions (NORs) correlate with cell proliferation rate and tumor prognosis.40 This correlation can be interpreted as a cause or as a consequence of the increased translational burden of cancer cells due to their uncontrolled proliferation. On the one hand, the inability to exit the cell cycle would leave the cell in a constant state of growth and division, requiring heavy transcription and translation and therefore large amounts of ribosomal synthesis. On the other hand, the existence of a nucleolar feedback on the cell cycle4 indicates that uncontrolled rDNA transcription and misregulation of RNA polymerase I activity could by itself be a major factor promoting cellular proliferation by providing the first step to unwarranted G0-G1 transition, a hallmark of cancer.41 Accordingly, recent evidence now clearly recognizes nucleolar misregulation as an important contributor to cancer and many key proto-oncogenes and tumor suppressors appear to play a direct role in the nucleolus and in RNA polymerase I transcription.17,42-46
Proto-oncogenes and tumor suppressors linked to nucleolar function
The proto-oncogene c-Myc is a major activator of RNA pol I transcription, is expressed in proliferating cells at a level correlating with rRNA47,48 and almost absent from quiescent cells.48 c-Myc binds directly to rDNA at the promoter region and at the transcription initiation region, as well as near a transcription termination site, and physically associates with the TBP-containing SL1 complex.48 Modulating c-Myc levels directly affects levels of SL1 and UBF binding at the rDNA.48 Another MYC family transcription factor frequently mutated in small cell lung cancer, L-Myc, also promotes pre-rRNA synthesis.49 The nucleolar function of MYC is evolutionarily conserved as its Drosophila ortholog dMyc also plays an essential role controlling rRNA synthesis during development.50 In addition to this direct role at the rDNA, MYC binds and up-regulates ribosome biogenesis genes51 and activates RNA polymerase III transcription of tRNAs and 5S rRNA genes.52,53 MYC therefore appears to be a key growth regulator that directly affects transcription by all RNA polymerases to modulate the translational capacity of the cell.
Nucleophosmin, a nucleolar proto-oncogene found mutated in several cancer types,54 affects the nucleolar function of MYC. In particular, c-Myc transformation is enhanced by expression of nucleophosmin,55 and nucleophosmin is essential for c-Myc nucleolar localization and rDNA transcription.56 An important partner of nucleophosmin is ARF/p14, a major tumor suppressor, which has recently been found to have a nucleolar function in addition to cell cycle inhibition via the MDM2-p53 pathway. ARF binds to the rDNA promoter,57 binds UBF in a p53-independent manner,58 and directly affects nucleolar chromatin organization by masking the nucleolar localization sequence of TTF-I.59 The other major tumor suppressors Rb, and the Rb-like protein p130, in addition to their role in G1/S cell cycle progression inhibition via sequestering E2F factors, also directly repress RNA polymerase I transcription by binding and inactivating UBF.60-62
The major tumor suppressor p53 also plays an important role coordinating ribosome biogenesis and the cell cycle/proliferation via signaling nucleolar stress. Signals such as oncogenic activation and DNA damage trigger nucleolar stress, and nucleolar disruption is a key event for p53 activation.63 Nucleolar stress triggers the nucleoplasmic export of the ribosomal proteins RPL5 and RPL11, which bind MDM2 and block MDM2-mediated p53 degradation, resulting in p53 activation, cell cycle arrest and apoptosis.64 p53 is then recruited to the FCs of the nucleolus,65 where it represses RNA polymerase I by directly binding to TBP and TAF-IC in the SL1 complex, disrupting its interaction with UBF.66,67 p53 furthermore directly represses c-Myc expression,68 along with RPL5/RPL11.69 PICT1, a nucleolar protein with mutations found in cancer, is a key DNA damage sensor at the rDNA that signals nucleolar stress through the RPL5/11-MDM2-p53 pathway70 and inhibits RNA polymerase I transcription.71
Other examples include PTEN, a tumor suppressor that inhibits cellular proliferation both by inhibiting the AKT/PI3K pathway and by affecting the rDNA occupancy of the SL1 complex, thereby repressing RNA polymerase I transcription,72 in association with GSK3β.25 It has also been reported that a PTEN isoform, PTENβ, physically associates with nucleolin and the 2 core subunits shared by RNA polymerase I and RNA polymerase III, AC40POLR1C and AC19POLR1D to repress ribosomal transcription.73 BRCA1, a key tumor suppressor, interacts directly at the rDNA repeats with UBF, the SL1 complex and RNA polymerase I, and the interaction is decreased in response to DNA damage.74 CTCF, a key insulator and multi-functional tumor suppressor involved in chromatin organization,75 also binds unmethylated DNA upstream of the rDNA promoter and interacts with UBF as well as RNA polymerase I,76 and CTCF expression correlates with pre-rRNA levels.77 In Drosophila, CTCF knock-down results in defects of rDNA silencing and nucleolar fragmentation.78 Overall, it appears that many proto-oncogenes and tumor suppressors have a nucleolar function and that generally, oncogenic proteins increase RNA polymerase I transcription, while tumor suppressors repress it.
Targeting RNA polymerase I is anti-tumorigenic
As might be expected, RNA polymerase I transcription has emerged as a key target in cancer therapy.79 For example, the RNA polymerase I inhibitors CX-5461 and CX-3543 (quarfloxin) have undergone early clinical trials after indications of anti-tumor activity.80,81 Both drugs bind and stabilize G-quadruplexes at the rDNA, which in addition to inhibiting RNA polymerase I initiation, results in the displacement of nucleolin to the nucleoplasm, where it binds to a G-quadruplex structure in the MYC promoter leading to repression of its expression.82,83 CX-5461 targets MYC in myeloma in a p53-independent manner,84 and L-Myc with strong tumor inhibition in a mouse model of small cell lung cancer.85 The G-quadruplex stabilization activity of CX-5461 and CX-3543 also leads to selective lethality in BRCA-deficient tumors, including tumors resistant to PARP inhibitors.86 Several other small molecules, BMH-9, -21, -22 and -23, which were originally identified in a screen for activators of the p53 pathway and found to do so in a DNA damage-independent manner,87 were subsequently found to inhibit RNA polymerase I via degradation of its main catalytic subunit RPA1, conferring them potent antitumoral activity across several cancer cell types.88,89
A better understanding of nucleolar pathways in cancer will improve our understanding of cross-reactions in combinatorial treatments, and devise efficient and synergistic drug combinations; for example, mTOR inhibitors also inhibit the p53-dependent nucleolar stress pathway, an effect that may not be beneficial,90 while combining RNA polymerase I inhibition with drugs targeting MYC signaling91 or ATM/ATR signaling92 has yielded promising results.
RNA interference: A new player in nucleolar function?
We have recently discovered that Dicer plays a novel essential function specifically in quiescent cells by targeting RNA polymerase I.36 The fission yeast Schizosaccharomyces pombe is an excellent model organism for quiescence,33 and mutants in RNA interference (RNAi) such as Dicer (dcr1Δ), Argonaute (ago1Δ) and RNA-dependent RNA polymerase (rdp1Δ) lose viability specifically in G0. Intriguingly, this novel function of RNAi is largely independent of heterochromatin formation at pericentromeric repeats.93 We found that in quiescence, Dicer mutants are defective in RNA polymerase I release, resulting in the accumulation of stalled RNA pol I over the rDNA repeats, DNA damage (visualized using γH2AX, which is bound by Crb253BP1), and extensive silencing of the rDNA repeats by the H3K9 methylation machinery.36 This defect has striking parallels in cycling cells, where Dicer mutants fail to release RNA polymerase II from pericentromeres, highly expressed genes and rDNA, also resulting in DNA damage.94,95 This parallel is further strengthened by the observation that mutants in corresponding subunits of RNA polymerase I (A12) and RNA polymerase II (Rpb9+TFIIS) suppress specifically Dicer's G0 and cycling cell silencing defects, respectively,36,96 prompting us to propose that the RNA interference machinery is tightly associated with, and regulates, the transcription machinery of all RNA polymerases.
Is this novel function of Dicer evolutionarily conserved? The association of RNA polymerase I transcription with cancer, and evidence that DICER1 is a tumor suppressor, suggest further investigation of this possibility. Importantly this function might be independent of Dicer's role in the biosynthesis of microRNA (miRNA), which are absent from S. pombe and most fungi. In accordance with this hypothesis, Dicer in mammalian cells promotes stem cell quiescence32 and can be found in the nucleolus binding to both active and silent rDNA repeats.97 Similarly, human AGO2 binds to numerous rRNA sites and binding is DICER-dependent.98 While AGO2 knockdown does not impact rRNA processing,98 Dicer and Drosha mutants accumulate 5.8S rRNA precursors, and Dicer but not Drosha mutants have a drastic effect on nucleolar morphology.99 Dicer also interacts with SIRT7,100 a sirtuin that associates with RNA polymerase I and UBF in mammals.101 In Drosophila, RNA interference is necessary for nucleolar organization,102 and in the model plant Arabidopsis thaliana, a specific set of RNA interference proteins comprising RDR2, DCL3, AGO4, DRD1 and the specialized RNA polymerase IV co-localize to nucleolar Cajal bodies.103,104 In Candida albicans, Dicer is necessary for 3′ETS cleavage of pre-rRNA,105 a function borne by the related RNase III enzyme Rnt1 in yeast.106 These observations strongly suggest that a nucleolar function of RNA interference—in particular of Dicer—is conserved in eukaryotic evolution.
Dicer is a tumor suppressor
There is now a well-described association between human DICER1 (14q32.13) mutations and cancer risk.107 Germline mutations in DICER1 (summarized in Fig. 2)108-117 result in a predisposition to develop a wide variety of otherwise rare cancers—the “DICER1 syndrome”—such as pleuropulmonary blastoma (PPB),118 cystic nephroma (CF),116,119 uterine cervix rhabdomyosarcoma,120 ovarian Sertoli-Leydig cell tumors (SL),109 thyroid cancer,121 multinodular goiter,108,122 pineoblastoma,123 ocular medulloepithelioma,110,124 and Wilms tumors (nephroblastoma).120,125 In many cases of PPB and SL, there is a frequent co-occurrence of 2 mutations;126 affected patients most frequently inherit a null allele (frameshift or stop truncation) in the germline and subsequently acquire a mutation on the second allele at hotspot residues in catalytic and metal-binding residues of the RNAse IIIb domain (Fig. 2).115,127-129 In a survey of 350 confirmed PPB cases, 66% patients carried a germline DICER1 mutation.130 Mutations in DICER1 are frequently associated with biallelic loss of TP53 in PPB131 as well as in Wilms tumors,132 suggesting that Dicer loss may trigger p53 activation resulting in increased selective pressure to mutate p53 in the tumorigenic tissue. In accordance with this, in mouse fibroblasts, Dicer deletion results in p53 activation and induction of senescence,133 and co-ablation of Dicer and p53 results in multiple skin carcinomas with increased DNA damage.134 Furthermore, Dicer has been described as a key target of the tumor suppressor p63; p63-null mice develop metastatic tumors with low Dicer and miR-130b expression, and re-expression of Dicer suppresses this phenotype.135
Figure 2.
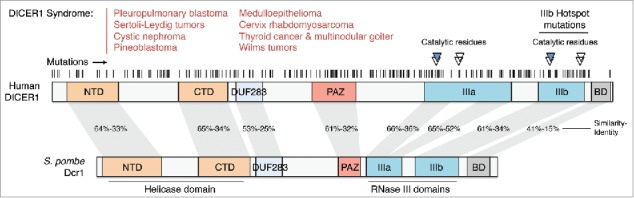
DICER1 is frequently mutated in cancer. Overview of DICER1 mutations found in several cancer types, as described in108-130 and in NCBI ClinVar. The majority of mutations are predicted to be heterozygous germline loss-of-function alleles, while the hotspot for somatic mutations is located at the RNase IIIb domain catalytic residues.
Dicer mutants are affected in development
The frequent heterozygous profile of DICER1 mutations suggests that this gene is a haploinsufficient tumor suppressor. Dicer is essential in mammals: Dicer-null mice die before gastrulation.136 In a mouse model of PPB, loss of DICER1 is sufficient for tumor development,137 and deletion of DICER1 in the female reproductive tract not only impaired its development, but also causes cystic nephromas.136 In a mouse retinoblastoma-sensitized model, monoallelic, but not biallelic, loss of Dicer increases tumorigenesis,138 and complete Dicer deletion is selected against during tumorigenesis,139-141 suggesting that low levels of Dicer protein are able to support most of its functions. In accordance with this, Dicer hypomorphic mice (10–30% expression level depending on tissue) have been reported to show few developmental defects, which were mostly limited to pancreatic morphological abnormalities.142
Conditional Dicer knockout is lethal during the development of many tissues, such as lung epithelia,143 Sertoli cells,144 male germ cells,145 haematopoietic stem cells and in erythroid lineage development,146 neuronal development,147 cerebellar development,148 and B-cell development.149 Knockdown of Dicer results in defects in oocyte maturation.150
Which molecular function underlies Dicer's tumor suppressor role?
One model to explain the tumorigeneticity of DICER1 hotspot mutations is that mutations are hypomorphic and the resulting enzyme is still able to generate miRNAs, though with a cleavage bias for one strand of the pre-miRNA. The RNase IIIa and RNase IIIb domains respectively cleave the 3′ (3p) and the 5′ (5p) end of the double-stranded RNA in the miRNA hairpin precursor, and the Dicer RNase IIIb mutants result in a strong loss of 5p miRNAs.127,151,152 Conversely, Dicer RNase IIIa mutants result in a strong loss of 3p miRNAs.151,153 Among 5p miRNAs, the let-7 family plays a tumor suppressor role by repressing several targets including MYC (reviewed in154), leading to propose that let-7 dysregulation in DICER1 mutants is at the core of its promoting cancer stem cell stemness.155 In C. elegans, let-7 regulates nucleolar size by affecting mRNA levels of ncl−1, a translational inhibitor of fibrillarin and thereby of rRNA transcription and processing.156
While the RNase IIIb domain is a clear hotspot for second-hit mutations in DICER1 tumors, screening of somatic mutations in DICER1 syndromes should not be limited to sequencing this domain as particularly interesting mutations over the rest of the gene have been detected. For example, a RNase IIIa mutation was found in one Wilms tumor,125 a homozygous G803R mutation (between the DUF283 and PAZ domains) in another Wilms tumor,117 and a compound mutation with both alleles truncating the protein before the RNase IIIa domain in pineoblastoma.123 These mutations suggest that the oncogenicity of DICER1 hypomorphic mutants may be more complex than reduction of 5p miRNAs.
DICER1 is able to cleave a variety of double-stranded RNA (dsRNA) substrates including endogenous siRNAs,157 as well as secondary structure-forming RNAs such as RNAs containing triplet-repeats,158 tRNAs,159 snoRNAs,160,161 the 7SL RNA component of the signal recognition particle (SRP) complex,162,163 and was shown recently to also play a role in DNA repair via DNA damage-induced small RNAs (ddRNAs).164,165 Importantly, the biologic function of many of these substrates is not well understood. Phenotypically, that DICER1 can have miRNA-independent roles is also shown by its requirement for murine oocyte development, while DROSHA is not required.166 Canonical miRNA production is abolished in DROSHA mutant cells, while in the absence of Dicer many miRNAs are still produced by loading pre-miRNAs directly into Argonaute, where they are trimmed.167
Dicer in the nucleolus: A potential link underlying its tumor suppressor role
Is there a link between the conserved role of Dicer in the nucleolus and the pro-tumorigenic consequences of nucleolar misregulation? The selection for hypomorphic RNase IIIb mutants in DICER1 cancers is presumably explained by the fact that Dicer is essential for cell survival, precluding biallelic loss-of-function i.e. it acts as a haploinsufficient tumor suppressor.139 But in the presence of reduced Dicer function in polymerase release, RNA polymerase I may undergo stalling and DNA damage at the rDNA, triggering activation of the nucleolar stress pathway and p53. Selective pressure to inactivate p53 could result in the frequent appearance of p53 null-mutants in Dicer hypomorphs, as has been observed.131,132 Similarly, several ribosomopathies display both sustained activation of p53 and cancer predisposition.4
To assess this possibility, a particularly interesting line of study would be to assay the phenotypic consequences of conserved RNase IIIa and RNase IIIb mutations found in cancer on nucleolar functions. In particular, as fission yeast has no miRNA, mutants could be assayed in fission yeast for RNA polymerase I stalling and formation of DNA damage at the rDNA. In that regard, while the dual catalytic-mutant (IIIa + IIIb, dcr1–5 allele in S. pombe) has an identical phenotype to the full deletion dcr1Δ,36 the contribution of each RNase III domain independently has not yet been assayed. However, it is also possible that specific miRNAs underlie the nucleolar function of Dicer, especially given that miRNA analogs are predicted along the primary rRNA transcript.168 A set of miRNAs is found within the nucleolus, although they appear to be Dicer- and RNA polymerase I-independent.169 Besides, many miRNA precursors form a structure similar to the C/D box of snoRNAs, and are able to bind fibrillarin, including let-7 g.170
Targeting Dicer's nucleolar function in cancer
One novel strategy for specific targeting of cancer cells is by taking advantage of negative epistatic interactions (synthetic lethals).171 In other words, a pathway that is not essential in wild-type can become essential in a cancerous cell; targeting this pathway would specifically kill the cancer cell (negative epistasis is often referred to as “oncogene addiction”). This strategy has been successful in the identification of PARP inhibitors for targeting tumors with BRCA1/2 mutations, leading to FDA approval of the drug Olaparib for ovarian cancer.172 Several new negative epistasis candidates have been proposed, through large screens using yeast and mammalian cells in a combinatorial approach.171
Identifying genes that are negatively epistatic with DICER1 mutations would be an interesting approach toward therapeutic prevention of DICER1 syndrome. Examples from fission yeast suggest that DNA repair, particularly homologous recombination repair, represents a significant negative epistatic target in Dicer mutant cycling cells.94,173 A similar approach could be taken during cellular quiescence, which could underlie Dicer's tumor suppressor function in regulation of RNA polymerase I. Candidates could then be tested in mammalian cancer models.171 For example, we have found that the G0 defects of Dicer mutants can be rescued by alleviating RNA polymerase I stalling. One way to do so is via lowering RNA pol I recruitment to the rDNA, for example in the TBP mutant tbp1-D156Y.36 dcr1Δtbp1-D156Y double-mutants show a strong reduction in DNA damage at rDNA repeats and suppress the synthetic sickness/lethality between dcr1Δ and rad51Δ in both dividing and non-dividing cells.36,94 Suppression was also conferred by deleting the non-essential subunit Spp27 of the rDNA UAF complex (Upstream Activation Factor), a SWI/SNF domain-containing protein that is involved in RNA polymerase I transcription initiation (unpublished observations). Another way to alleviate stalling is to destabilize RNA polymerase I itself, using a deletion mutant of its non-essential subunit A12, which suppresses dcr1Δ G0 defects.36 Stalling of RNA polymerase I results in the unchecked silencing of rDNA repeats, resulting in accumulation of H3K9 methylation. Targeting this latter step using mutants in the H3K9 methylation pathway, H3K9R histone substitution mutants, or HP1 mutants also suppresses G0 lethality, although it occurs downstream and cells therefore still undergo RNA polymerase I stalling and DNA damage. Conversely, overexpression of the H3K9 methyltransferase Clr4 (ortholog of SUV39H1) enhances the phenotype, and is lethal in quiescent cells.36
Interestingly, no epistasis was observed between dcr1Δ and the deletion mutant of the yeast ortholog of UBF, hmo1Δ (unpublished observations), suggesting suppression might be limited to the TBP and UAF complexes. In an adenocarcinomal cell line, knock-down of Dicer results in loss of cisplatin resistance,174 potentially indicating that Dicer mutants may act independently of UBF for RNA polymerase I transcription regulation. If this hypothesis is true, then Dicer mutant cells may be more sensitive to targeting the SL1 complex, or RNA polymerase I itself. Reducing the accumulation of stalled polymerases may alleviate the tumorigenicity of Dicer-mutated cancer cells. Therefore, drugs that inhibit RNA polymerase I transcription by targeting the SL1 complex, such as 9-hydroxyellipticin,175 or by triggering degradation of RPA1, such as BMH-9, -21, -22 and -23,88,89 would therefore be of particular interest to test on Dicer mutant cells. As 9-hydroxyellipticin is p53-independent175 while BMH compounds trigger p53 activation,88,89 their action could be synergistic, and bypass the frequent p53 loss found in Dicer tumors.131,132
Dicer has distinct roles in both quiescence and proliferation
Epistatic interactions can be different in cycling and G0 cells. For example, we found a very strong inhibition of growth in double-mutants of Dicer and TTF-I, but this did not affect the viability of quiescent cells.36 The mechanism underlying this epistatic interaction is likely linked to DNA replication; TTF-I bound to the rDNA terminator sites provides an unidirectional block of the DNA replication fork, insuring that rDNA replication and rRNA transcription occur in the same direction during S-phase to avoid replication-transcription collisions.176,177 Dicer is involved in collision resolution in fission yeast, and could become essential for timely completion of replication in the absence of TTF-1.94,95 It is therefore important to assay epistatic interactions during both growth and quiescence. Ideally, targets can be combined to achieve growth inhibition in both actively-dividing and non-dividing cells. In the context of a tumor, if the treatment of the majority of actively-dividing cells does not result in complete tumor elimination, it is important to specifically target non-dividing and slow-dividing cancer cells responsible for relapse and metastasis.178 In this regard fission yeast would be an ideal model system for conducting such a screen and identifying negative epistasis targets of nucleolar functions of RNA interference and DICER1, in both cycling and quiescent cells, which would provide potential targets of therapeutic significance.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the S. pombe community for discussion, and sharing strains, the Pombase database for providing a very useful resource, Jie Ren for discussion, Jean-Sébastien Parent, Andrea Schorn, Michael Gutbrod and Sarah Herridge for discussion and suggestions on the manuscript. We apologize to researchers whose work was not cited due to size constraints.
Funding
This work was funded by NIH grant R01 GM076396-08, the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation Plant Biology Investigator Program (to R.M.), and the Agence Nationale de la Recherche grant ANR-13-BSV8-0018 (to B.A.). The authors acknowledge support from the Chaire Blaise Pascal (Fondation de I'École Normale Supérieure, France).
References
- [1].McClintock B. The relation of a particular chromosomal element to the development of nucleoli in Zea mays. Zeitschrift für Zellforschung und Mikroskopische Anatomie 1934;2:294-326. doi: 10.1007/BF00374060 [DOI] [Google Scholar]
- [2].Hernandez-Verdun D. Structural organization of the nucleolus as a consequence of the dynamics of ribosome biogenesis In: Olson MOJ, editor. The nucleolus. Protein reviews 15 New York: Springer-Verlag; 2011. p. 3-28. [Google Scholar]
- [3].Derenzini M, Pasquinelli G, O'Donohue MF, Ploton D, Thiry M. Structural and functional organization of ribosomal genes within the mammalian cell nucleolus. J Histochem Cytochem 2006;54:131-45. doi: 10.1369/jhc.5R6780.2005. PMID:16204224 [DOI] [PubMed] [Google Scholar]
- [4].Voit R, Grummt I. The RNA polymerase I transcription machinery In: Olson MOJ, editor. The nucleolus. Protein reviews 15 New York: Springer-Verlag; 2011. p. 107-34. [Google Scholar]
- [5].Gérus M, Caizergues-Ferrer M, Henry Y, Henras A. Crosstalk between ribosome synthesis and cell cycle progression and its potential implications in human diseases In: Olson MOJ, editor. The nucleolus. Protein reviews 15 New York: Springer-Verlag; 2011. p. 157-84. [Google Scholar]
- [6].Kuhn A, Grummt I. Dual role of the nucleolar transcription factor UBF: Trans-activator and antirepressor. Proc Natl Acad Sci USA 1992;89:7340-4. PMID:1502143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleid Acids Res 2001;29:3241-7. PMID:11470882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 2002;22:657-68. doi: 10.1128/MCB.22.2.657-668.2002. PMID:11756560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wright JE, Mais C, Prieto JL, McStay B. A role for upstream binding factor in organizing ribosomal gene chromatin. Biochem Soc Symp 2006;73:77-84. doi: 10.1042/bss0730077. PMID:16626289 [DOI] [PubMed] [Google Scholar]
- [10].Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JC. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J 2007;26:1560-8. doi: 10.1038/sj.emboj.7601601. PMID:17318177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JC. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J 2001;20:1373-82. doi: 10.1093/emboj/20.6.1373. PMID:11250903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Friedrich JK, Panov KI, Cabart P, Russell J, Zomerdijk JC. TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J Biol Chem 2005;280:29551-8. doi: 10.1074/jbc.M501595200. PMID:15970593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Knutson BA, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae RNA polymerase I core factor complex. Nat Struct Mol Biol 2014;21:810-6. doi: 10.1038/nsmb.2873. PMID:25132180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hontz RD, French SL, Oakes ML, Tongaonkar P, Nomura M, Beyer AL, Smith JS. Transcription of multiple yeast ribosomal DNA genes requires targeting of UAF to the promoter by Uaf30. Mol Cell Biol 2008;28:6709-19. doi: 10.1128/MCB.00703-08. PMID:18765638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Längst G, Becker PB, Grummt I. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J 1998;17:3135-45. doi: 10.1093/emboj/17.11.3135. PMID:9606195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Diermeier SD, Németh A, Rehli M, Grummt I, Längst G. Chromatin-specific regulation of mammalian rDNA transcription by clustered TTF-I-binding sites. PLoS Genet 2013;9:e1003786. doi: 10.1371/journal.pgen.1003786. PMID:24068958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Diesch J, Hannan RD, Sanij E. Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease. Cell Biosci 2014;4:43. doi: 10.1186/2045-3701-4-43. PMID:25949792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Panov KI, Friedrich JK, Zomerdijk JC. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol Cell Biol 2001;21:2641-9. doi: 10.1128/MCB.21.8.2641-2649.2001. PMID:11283244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hung SS, Lesmana A, Peck A, Lee R, Tchoubrieva E, Hannan KM, Lin J, Sheppard KE, Jastrzebski K, Quinn LM, et al.. Cell cycle and growth stimuli regulate different steps of RNA polymerase I transcription. Gene 2017;612:36-48. doi: 10.1016/j.gene.2016.12.015. PMID:27989772 [DOI] [PubMed] [Google Scholar]
- [20].Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell 2004;15:946-56. doi: 10.1091/mbc.E03-08-0594. PMID:14595104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev 2004;18:423-34. doi: 10.1101/gad.285504. PMID:15004009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhao J, Yuan X, Frödin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell 2003;11:405-13. doi: 10.1016/S1097-2765(03)00036-4. PMID:12620228 [DOI] [PubMed] [Google Scholar]
- [23].James MJ, Zomerdijk JC. Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J Biol Chem 2004;279:8911-8. https//doi.org/ 10.1074/jbc.M307735200. PMID:14688273 [DOI] [PubMed] [Google Scholar]
- [24].Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M, et al.. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal 2011;4:ra56. doi: 10.1126/scisignal.2001754. PMID:21878679 [DOI] [PubMed] [Google Scholar]
- [25].Vincent T, Kukalev A, Andäng M, Pettersson R, Percipalle P. The glycogen synthase kinase (GSK) 3β represses RNA polymerase I transcription. Oncogene 2008;27:5254-9. doi: 10.1038/onc.2008.152. PMID:18490923 [DOI] [PubMed] [Google Scholar]
- [26].Pistoni M, Verrecchia A, Doni M, Guccione E, Amati B. Chromatin association and regulation of rDNA transcription by the Ras-family protein RasL11a. EMBO J 2010;29:1215-24. doi: 10.1038/emboj.2010.16. PMID:20168301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kusnadi EP, Hannan KM, Hicks RJ, Hannan RD, Pearson RB, Kang J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 2015;556:27-34. doi: 10.1016/j.gene.2014.11.010. PMID:25447905 [DOI] [PubMed] [Google Scholar]
- [28].Lyapunova NA, Veiko NN, Porokhovnik LN. Human rDNA genes: Identification of four fractions, their functions and nucleolar location In: O'Day DH, Catalano A, editors. Proteins of the Nucleolus New York: Springer-Verlag; 2013. p. 95-118. [Google Scholar]
- [29].Santoro R. The epigenetics of the nucleolus: Structure and function of active and silent ribosomal RNA genes In: Olson MOJ, editor. The nucleolus. Protein reviews 15 New York: Springer-Verlag; 2011. p. 57-82. [Google Scholar]
- [30].Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, et al.. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol 2008;183:1259-74. doi: 10.1083/jcb.200805146. PMID:19103806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Klein J, Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci USA 1999;96:6096-101. doi: 10.1073/pnas.96.11.6096. PMID:10339547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 2013;14:329-40. doi: 10.1038/nrm3591. PMID:23698583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yanagida M. Cellular quiescence: Are controlling genes conserved? Trends Cell Biol 2009;19:705-15. doi: 10.1016/j.tcb.2009.09.006. PMID:19833516 [DOI] [PubMed] [Google Scholar]
- [34].Roche B, Arcangioli B, Martienssen RA. Transcriptional reprogramming in cellular quiescence. RNA Biol 2017;12:1-11. doi: 10.1080/15476286.2017.1327510. PMID:28497998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Johnson EL, Robinson DG, Coller HA. Widespread changes in mRNA stability contribute to quiescence-specific gene expression patterns in a fibroblast model of quiescence. BMC Genomics 2017;18:123. doi: 10.1186/s12864-017-3521-0. PMID:28143407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Roche B, Arcangioli B, Martienssen RA. RNA interference is essential for cellular quiescence. Science 2016;354:aah5651. doi: 10.1126/science.aah5651. PMID:27738016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shimanuki M, Chung SY, Chikashige Y, Kawasaki Y, Uehara L, Tsutsumi C, Hatanaka M, Hiraoka Y, Nagao K, Yanagida M. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells 2007;12:677-92. doi: 10.1111/j.1365-2443.2007.01079.x. PMID:17535257 [DOI] [PubMed] [Google Scholar]
- [38].Narla A, Ebert BL. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010;115:3196-205. doi: 10.1182/blood-2009-10-178129. PMID:20194897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gani R. The nucleoli of cultured human lymphocytes. I. Nucleolar morphology in relation to transformation and the DNA cycle. Exp Cell Res 1976;97:249-58. doi: 10.1016/0014-4827(76)90614-5. PMID:1248517 [DOI] [PubMed] [Google Scholar]
- [40].Derenzini M, Trerè D. Importance of interphase nucleolar organizer regions in tumor pathology. Virchows Arch B Cell Pathol Incl Mol Pathol 1991;61:1-8. doi: 10.1007/BF02890399. PMID:1683059 [DOI] [PubMed] [Google Scholar]
- [41].O'Farrell PH. Quiescence: Early evolutionary origins and universality do not imply uniformity. Philos Trans R Soc Lond B Biol Sci 2011;366:3498-507. doi: 10.1098/rstb.2011.0079. PMID:22084377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol 2008;173:301-10. doi: 10.2353/ajpath.2008.070752. PMID:18583314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Montanaro L, Treré D, Derenzini M. The emerging role of RNA polymerase I transcription machinery in human malignancy: A clinical perspective. Onco Targets Ther 2013;6:909-16. doi: 10.2147/OTT.S36627. PMID:23888116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB. Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta 2013;1829:342-60. doi: 10.1016/j.bbagrm.2012.10.014. PMID:23153826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin Cancer Biol 2016;37–38:36-50. doi: 10.1016/j.semcancer.2015.12.004. PMID:26721423 [DOI] [PubMed] [Google Scholar]
- [46].Derenzini M, Montanaro L, Treré D. Ribosome biogenesis and cancer. Acta Histochem 2017;119:190-7. doi: 10.1016/j.acthis.2017.01.009. PMID:28168996 [DOI] [PubMed] [Google Scholar]
- [47].Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, Fahlén S, Hydbring P, Söderberg O, Grummt I, et al.. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 2005;7:303-10. doi: 10.1038/ncb1225. PMID:15723053 [DOI] [PubMed] [Google Scholar]
- [48].Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 2005;7:311-8. doi: 10.1038/ncb1224. PMID:15723054 [DOI] [PubMed] [Google Scholar]
- [49].Kim DW, Wu N, Kim YC, Cheng PF, Basom R, Kim D, Dunn CT, Lee AY, Kim K, Lee CS, et al.. Genetic requirement for Mycl and efficacy of RNA pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30:1289-99. doi: 10.1101/gad.279307.116. PMID:27298335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 2005;7:295-302. doi: 10.1038/ncb1223. PMID:15723055 [DOI] [PubMed] [Google Scholar]
- [51].Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voûte PA, Schwab M, et al.. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 2001;20:1383-93. doi: 10.1093/emboj/20.6.1383. PMID:11250904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle 2003;2:181-4. doi: 10.4161/cc.2.3.375. PMID:12734418 [DOI] [PubMed] [Google Scholar]
- [53].Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature 2003;421:290-4. doi: 10.1038/nature01327. PMID:12529648 [DOI] [PubMed] [Google Scholar]
- [54].Scott DD, Oeffinger M. Nucleolin and nucleophosmin: Nucleolar proteins with multiple functions in DNA repair. Biochem Cell Biol 2016;94:419-32. doi: 10.1139/bcb-2016-0068. PMID:27673355 [DOI] [PubMed] [Google Scholar]
- [55].Li Z, Boone D, Hann SR. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci USA 2008;105:18794-9. doi: 10.1073/pnas.0806879105. PMID:19033198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li Z, Hann SR. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene 2013;32:1988-94. doi: 10.1038/onc.2012.227. PMID:22665062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ayrault O, Andrigue L, Larsen CJ, Seite P. Human Arf tumor suppressor specifically interacts with chromatin containing the promoter of rRNA genes. Oncogene 2004;23:8097-104. doi: 10.1038/sj.onc.1207968. PMID:15361825 [DOI] [PubMed] [Google Scholar]
- [58].Ayrault O, Andrigue L, Fauvin C, Eymin B, Gazzeri S, Séité P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 2006;25:7577-86. doi: 10.1038/sj.onc.1209743. PMID:16924243 [DOI] [PubMed] [Google Scholar]
- [59].Lessard F, Morin F, Ivanchuk S, Langlois F, Stefanovsky V, Rutka J, Moss T. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol Cell 2010;38:539-50. doi: 10.1016/j.molcel.2010.03.015. PMID:20513429 [DOI] [PubMed] [Google Scholar]
- [60].Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 1995;374:177-80. doi: 10.1038/374177a0. PMID:7877691 [DOI] [PubMed] [Google Scholar]
- [61].Hannan KM, Hannan RD, Smith SD, Jefferson LS, Lun M, Rothblum LI. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene 2000;19:4988-99. doi: 10.1038/sj.onc.1203875. PMID:11042686 [DOI] [PubMed] [Google Scholar]
- [62].Ciarmatori S, Scott PH, Sutcliffe JE, McLees A, Alzuherri HM, Dannenberg JH, te Riele H, Grummt I, Voit R, White RJ. Overlapping functions of the pRb family in the regulation of rRNA synthesis. Mol Cell Biol 2001;21:5806-14. doi: 10.1128/MCB.21.17.5806-5814.2001. PMID:11486020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 2003;22:6068-77. doi: 10.1093/emboj/cdg579. PMID:14609953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, Itoh B, Wang J, Komatsu Y, Yang YR, et al.. Regulation of the MDM2-p53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med 2011;17:944-51. doi: 10.1038/nm.2392. PMID:21804542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Karni-Schmidt O, Zupnick A, Castillo M, Ahmed A, Matos T, Bouvet P, Cordon-Cardo C, Prives C. p53 is localized to a sub-nucleolar compartment after proteasomal inhibition in an energy-dependent manner. J Cell Sci 2008;121:4098-105. doi: 10.1242/jcs.030098. PMID:19033390 [DOI] [PubMed] [Google Scholar]
- [66].Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene 1999;18:1119-24. doi: 10.1038/sj.onc.1202402. PMID:10023689 [DOI] [PubMed] [Google Scholar]
- [67].Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol 2000;20:5930-8. doi: 10.1128/MCB.20.16.5930-5938.2000. PMID:10913176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ho JS, Ma W, Mao DY, Benchimol S. p53-dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol 2005;25:7423-31. doi: 10.1128/MCB.25.17.7423-7431.2005. PMID:16107691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Liao JM, Zhou X, Gatignol A, Lu H. Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene 2014;33:4916-23. doi: 10.1038/onc.2013.430. PMID:24141778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chen H, Han L, Tsai H, Wang Z, Wu Y, Duo Y, Cao W, Chen L, Tan Z, Xu N, et al.. PICT-1 is a key nucleolar sensor in DNA damage response signaling that regulates apoptosis through the RPL11-MDM2-p53 pathway. Oncotarget 2016;7:83241-57. doi: 10.18632/oncotarget.13082. PMID:27829214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen H, Duo Y, Hu B, Wang Z, Zhang F, Tsai H, Zhang J, Zhou L, Wang L, Wang X, et al.. PICT-1 triggers a pro-death autophagy through inhibiting rRNA transcription and AKT/mTOR/p70S6K signaling pathway. Oncotarget 2016;7:78747-63. doi: 10.18632/oncotarget.12288. PMID:27729611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhang C, Comai L, Johnson DL. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol Cell Biol 2005;25:6899-911. doi: 10.1128/MCB.25.16.6899-6911.2005. PMID:16055704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Liang H, Chen X, Yin Q, Ruan D, Zhao X, Zhang C, McNutt MA, Yin Y. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat Commun 2017;8:14771. doi: 10.1038/ncomms14771. PMID:28332494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Johnston R, D'Costa Z, Ray S, Gorski J, Harkin DP, Mullan P, Panov KI. The identification of a novel role for BRCA1 in regulating RNA polymerase I transcription. Oncotarget 2016;7:68097-110. doi: 10.18632/oncotarget.11770. PMID:27589844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Fiorentino FP, Giordano A. The tumor suppressor role of CTCF. J Cell Physiol 2012;227:479-92. doi: 10.1002/jcp.22780. PMID:21465478 [DOI] [PubMed] [Google Scholar]
- [76].van de Nobelen S, Rosa-Garrido M, Leers J, Heath H, Soochit W, Joosen L, Jonkers I, Demmers J, van der Reijden M, Torrano V, et al.. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin 2010;3:19. doi: 10.1186/1756-8935-3-19. PMID:21059229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Huang K, Jia J, Wu C, Yao M, Li M, Jin J, Jiang C, Cai Y, Pei D, Pan G, et al.. Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J Biol Chem 2013;288:26067-77. doi: 10.1074/jbc.M113.486175. PMID:23884423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Guerrero PA, Maggert KA. The CCCTC-binding factor (CTCF) of Drosophila contributes to the regulation of the ribosomal DNA and nucleolar stability. PLoS One 2011;6:e16401. doi: 10.1371/journal.pone.0016401. PMID:21283722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 2010;50:131-56. doi: 10.1146/annurev.pharmtox.010909.105844. PMID:20055700 [DOI] [PubMed] [Google Scholar]
- [80].Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, et al.. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res 2009;69:7653-61. doi: 10.1158/0008-5472.CAN-09-1304. PMID:19738048 [DOI] [PubMed] [Google Scholar]
- [81].Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, et al.. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res 2011;71:1418-30. doi: 10.1158/0008-5472.CAN-10-1728. PMID:21159662 [DOI] [PubMed] [Google Scholar]
- [82].González V, Hurley LH. The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry 2010;49:9706-14. doi: 10.1021/bi100509s. PMID:20932061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Brooks TA, Hurley LH. Targeting MYC expression through G-quadruplexes. Genes Cancer 2010;1:641-9. doi: 10.1177/1947601910377493. PMID:21113409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lee HC, Wang H, Baladandayuthapani V, Lin H, He J, Jones RJ, Gu D, Wang Z, Ma W, Lim J, et al.. RNA polymerase I inhibition with CX-5461 as a novel therapeutic strategy to target MYC in multiple myeloma. Br J Haematol 2017;177:80-94. doi: 10.1111/bjh.14525. PMID:28369725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kim DW, Wu N, Kim YC, Cheng PF, Basom R, Kim D, Dunn CT, Lee AY, Kim K, Lee CS, et al.. Genetic requirement for Mycl and efficacy of RNA pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30:1289-99. doi: 10.1101/gad.279307.116. PMID:27298335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O'Neil NJ, Santos ND, Silvester J, Wei V, Garcia J, et al.. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun 2017;8:14432. doi: 10.1038/ncomms14432. PMID:28211448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Peltonen K, Colis L, Liu H, Jäämaa S, Moore HM, Enbäck J, Laakkonen P, Vaahtokari A, Jones RJ, Af Hällström TM, et al.. Identification of novel p53 pathway activating small-molecule compounds reveals unexpected similarities with known therapeutic agents. PLoS One 2010;5:e12996. doi: 10.1371/journal.pone.0012996. PMID:20885994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM, Bai B, Rudek MA, Bieberich CJ, Laiho M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 2014;25:77-90. doi: 10.1016/j.ccr.2013.12.009. PMID:24434211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Peltonen K, Colis L, Liu H, Jäämaa S, Zhang Z, Af Hällström T, Moore HM, Sirajuddin P, Laiho M. Small molecule BMH-compounds that inhibit RNA polymerase I and cause nucleolar stress. Mol Cancer Ther 2014;13:2537-46. doi: 10.1158/1535-7163.MCT-14-0256. PMID:25277384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Goudarzi KM, Nistér M, Lindström MS. mTOR inhibitors blunt the p53 response to nucleolar stress by regulating RPL11 and MDM2 levels. Cancer Biol Ther 2014;15:1499-514. doi: 10.4161/15384047.2014.955743. PMID:25482947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Rebello RJ, Kusdani E, Cameron DP, Pearson HB, Lesmana A, Devlin JR, Drygin D, Clark AK, Porter L, Pedersen J, et al.. The dual inhibition of RNA pol I transcription and PIM kinase as a new therapeutic approach to treat advanced prostate cancer. Clin Cancer Res 2016;22:5539-52. doi: 10.1158/1078-0432.CCR-16-0124. PMID:27486174 [DOI] [PubMed] [Google Scholar]
- [92].Quin J, Chan KT, Devlin JR, Cameron DP, Diesch J, Cullinane C, Ahern J, Khot A, Hein N, George AJ, et al.. Inhibition of RNA polymerase I transcription initiation by CX-5461 activates non-canonical ATM/ATR signaling. Oncotarget 2016;7:49800-18. doi: 10.18632/oncotarget.10452. PMID:27391441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Castel SE, Martienssen RA. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 2013;14:100-12. doi: 10.1038/nrg3355. PMID:23329111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marín L, Chang AY, Goto D, et al.. RNAi promotes heterochromatic silencing through replication-coupled release of RNA pol II. Nature 2011;479:135-8. doi: 10.1038/nature10501. PMID:22002604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Castel SE, Ren J, Bhattacharjee S, Chang AY, Sánchez M, Valbuena A, Antequera F, Martienssen RA. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 2014;159:572-83. doi: 10.1016/j.cell.2014.09.031. PMID:25417108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 2011;18:1132-8. doi: 10.1038/nsmb.2122. PMID:21892171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS One 2010;5:e12175. doi: 10.1371/journal.pone.0012175. PMID:20730047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Atwood BL, Woolnough JL, Lefevre GM, Saint Just Ribeiro M, Felsenfeld G, Giles KE. Human Argonaute 2 is tethered to ribosomal RNA through microRNA interactions. J Biol Chem 2016;291:17919-28. doi: 10.1074/jbc.M116.725051. PMID:27288410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Liang XH, Crooke ST. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleid Acids Res 2011;39:4875-89. doi: 10.1093/nar/gkr076. PMID:21321021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhang PY, Li G, Deng ZJ, Liu LY, Chen L, Tang JZ, Wang YQ, Cao ST, Fang YX, Wen F, et al.. Dicer interacts with SIRT7 and regulates H3K18 deacetylation in response to DNA damaging agents. Nucleic Acids Res 2016;44:3629-42. doi: 10.1093/nar/gkv1504. PMID:26704979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics 2012;11:60-76. doi: 10.1074/mcp.A111.015156. PMID:22586326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 2007;9:25-35. doi: 10.1038/ncb1514. PMID:17159999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 2006;126:93-106. doi: 10.1016/j.cell.2006.05.032. PMID:16839879 [DOI] [PubMed] [Google Scholar]
- [104].Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 2006;126:79-92. doi: 10.1016/j.cell.2006.05.031. PMID:16839878 [DOI] [PubMed] [Google Scholar]
- [105].Bernstein DA, Vyas VK, Weinberg DE, Drinnenberg IA, Bartel DP, Fink GR. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proc Natl Acad Sci USA 2012;109:523-8. doi: 10.1073/pnas.1118859109. PMID:22173636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ETS but not the 5′ETS. RNA 1999;5:909-17. doi: 10.1017/S135583829999026X. PMID:10411134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Cai S, Zhao W, Nie X, Abbas A, Fu L, Bihi S, Feng G, Liu T, Lv Y, Ma X, et al.. Multimorbidity and genetic characteristics of DICER1 syndrome based on systematic review. J Pediatr Hematol Oncol 2017;39:355-61. doi: 10.1097/MPH.0000000000000715. PMID:27906793 [DOI] [PubMed] [Google Scholar]
- [108].Fernández-Martinez L, Villegas JA, Santamaría I, Pitiot AS, Alvarado MG, Fernández S, Torres H, Á Paredes, Blay P, Balbín M. Identification of somatic and germ-line DICER1 mutations in pleuropulmonary blastoma, cystic nephroma and rhabdomyosarcoma tumors within a DICER1 syndrome pedigree. BMC Cancer 2017;17:146. doi: 10.1186/s12885-017-3136-5. PMID:28222777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Rossing M, Gerdes AM, Juul A, Rechnitzer C, Rudnicki M, Nielsen FC, Vo Hansen T. A novel DICER1 mutation identified in a female with ovarian Sertoli-Leydig cell tumor and multinodular goiter: A case report. J Med Case Rep 2014;8:112. doi: 10.1186/1752-1947-8-112. PMID:24708902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Durieux E, Descotes F, Nguyen AM, Grange JD, Devouassoux-Shisheboran M. Somatic DICER1 gene mutation in sporadic intraocular medulloepithelioma without pleuropulmonary blastoma syndrome. Hum Pathol 2015;46:783-7. doi: 10.1016/j.humpath.2015.01.020. PMID:25791583 [DOI] [PubMed] [Google Scholar]
- [111].Durieux E, Descotes F, Mauduit C, Decaussin M, Guyetant S, Devouassoux-Shisheboran M. The co-occurrence of an ovarian Sertoli-Leydig cell tumor with a thyroid carcinoma is highly suggestive of a DICER1 syndrome. Virchows Arch 2016;468:631-6. doi: 10.1007/s00428-016-1922-0. PMID:26983701 [DOI] [PubMed] [Google Scholar]
- [112].Rutter MM, Jha P, Schultz KA, Sheil A, Harris AK, Bauer AJ, Field AL, Geller J, Hill DA. DICER1 mutations and differentiated thyroid carcinoma: Evidence of a direct association. J Clin Endocrinol Metab 2016;101:1-5. doi: 10.1210/jc.2015-2169. PMID:26555935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Fremerey J, Balzer S, Brozou T, Schaper J, Borkhardt A, Kuhlen M. Embryonal rhabdomyosarcoma in a patient with a heterozygous frameshift variant in the DICER1 gene and additional manifestations of the DICER1 syndrome. Fam Cancer 2017;16:401-5. doi: 10.1007/s10689-016-9958-5. PMID:27896549 [DOI] [PubMed] [Google Scholar]
- [114].Conlon N, Schultheis AM, Piscuoglio S, Silva A, Guerra E, Tornos C, Reuter VE, Soslow RA, Young RH, Oliva E, et al.. A survey of DICER1 hotspot mutations in ovarian and testicular sex cord-stromal tumors. Mod Pathol 2015;28:1603-12. doi: 10.1038/modpathol.2015.115. PMID:26428316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, Fejes AP, Chow C, Tone A, Kalloger SE, et al.. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 2012;366:234-42. doi: 10.1056/NEJMoa1102903. PMID:22187960 [DOI] [PubMed] [Google Scholar]
- [116].Bahubeshi A, Bal N, Rio Frio T, Hamel N, Pouchet C, Yilmaz A, Bouron-Dal Soglio D, Williams GM, Tischkowitz M, Priest JR, et al.. Germline DICER1 mutations and familial cystic nephroma. J Med Genet 2010;47:863-6. doi: 10.1136/jmg.2010.081216. PMID:21036787 [DOI] [PubMed] [Google Scholar]
- [117].Palculict TB, Ruteshouser EC, Fan Y, Wang W, Strong L, Huff V. Identification of germline DICER1 mutations and loss of heterozygosity in familial Wilms tumour. J Med Genet 2016;53:385-8. doi: 10.1136/jmedgenet-2015-103311. PMID:26566882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, et al.. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009;325:965. doi: 10.1126/science.1174334. PMID:19556464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Doros LA, Rossi CT, Yang J, Field A, Williams GM, Messinger Y, Cajaiba MM, Perlman EJ, A Schultz K, Cathro HP, et al.. DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod Pathol 2014;27:1267-80. doi: 10.1038/modpathol.2013.242. PMID:24481001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Foulkes WD, Bahubeshi A, Hamel N, Pasini B, Asioli S, Baynam G, Choong CS, Charles A, Frieder RP, Dishop MK, et al.. Extending the phenotypes associated with DICER1 mutations. Hum Mutat 2011;32:1381-4. doi: 10.1002/humu.21600. PMID:21882293 [DOI] [PubMed] [Google Scholar]
- [121].Khan NE, Bauer AJ, Schultz KAP, Doros L, Decastro RM, Ling A, Lodish MB, Harney LA, Kase RG, Carr AG, et al.. Quantification of thyroid cancer and multinodular goiter risk in the DICER1 syndrome: A family-based cohort study. J Clin Endocrinol Metab 2017;102:1614-22. doi: 10.1210/jc.2016-2954. PMID:28323992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Rio Frio T, Bahubeshi A, Kanellopoulou C, Hamel N, Niedziela M, Sabbaghian N, Pouchet C, Gilbert L, O'Brien PK, Serfas K, et al.. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA 2011;305:68-77. doi: 10.1001/jama.2010.1910. PMID:21205968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].de Kock L, Sabbaghian N, Druker H, Weber E, Hamel N, Miller S, Choong CS, Gottardo NG, Kees UR, Rednam SP, et al.. Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol 2014;128:583-95. doi: 10.1007/s00401-014-1318-7. PMID:25022261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sahm F, Jakobiec FA, Meyer J, Schrimpf D, Eberhart CG, Hovestadt V, Capper D, Lambo S, Ryzhova M, Schüller U, et al.. Somatic mutations of DICER1 and KMT2D are frequent in intraocular medulloepitheliomas. Genes Chromosomes Cancer 2016;55:418-27. doi: 10.1002/gcc.22344. PMID:26841698 [DOI] [PubMed] [Google Scholar]
- [125].Wu MK, Sabbaghian N, Xu B, Addidou-Kalucki S, Bernard C, Zou D, Reeve AE, Eccles MR, Cole C, Choong CS, et al.. Biallelic DICER1 mutations occur in Wilms tumours. J Pathol 2013;230:154-64. doi: 10.1002/path.4196. PMID:23620094 [DOI] [PubMed] [Google Scholar]
- [126].Seki M, Yoshida K, Shiraishi Y, Shimamura T, Sato Y, Nishimura R, Okuno Y, Chiba K, Tanaka H, Kato K, et al.. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res 2014;74:2742-9. doi: 10.1158/0008-5472.CAN-13-2470. PMID:24675358 [DOI] [PubMed] [Google Scholar]
- [127].Wang Y, Chen J, Yang W, Mo F, Senz J, Yap D, Anglesio MS, Gilks B, Morin GB, Huntsman DG. The oncogenic roles of DICER1 RNase IIIb domain mutations in ovarian Sertoli-Leydig cell tumors. Neoplasia 2015;17:650-60. doi: 10.1016/j.neo.2015.08.003. PMID:26408257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Brenneman M, Field A, Yang J, Williams G, Doros L, Rossi C, Schultz KA, Rosenberg A, Ivanovich J, Turner J, et al.. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in DICER1 syndrome: A unique variant of the two-hit tumor suppression model. F1000Res 2015;4:214. doi: 10.12688/f1000research.6746.1. PMID:26925222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Witkowski L, Mattina J, Schönberger S, Murray MJ, Choong CS, Huntsman DG, Reis-Filho JS, McCluggage WG, Nicholson JC, Coleman N, et al.. DICER1 hotspot mutations in non-epithelial gonadal tumours. Br J Cancer 2013;109:2744-50. doi: 10.1038/bjc.2013.637. PMID:24136150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Messinger YH, Stewart DR, Priest JR, Williams GM, Harris AK, Schultz KA, Yang J, Doros L, Rosenberg PS, Hill DA, et al.. Pleuropulmonary blastoma: A report on 350 central pathology-confirmed pleuropulmonary blastoma cases by the International Pleuropulmonary Blastoma Registry. Cancer 2015;121:276-85. doi: 10.1002/cncr.29032. PMID:25209242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Pugh TJ, Yu W, Yang J, Field AL, Ambrogio L, Carter SL, Cibulskis K, Giannikopoulos P, Kiezun A, Kim J, et al.. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene 2014;33:5295-302. doi: 10.1038/onc.2014.150. PMID:24909177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang TC, Khokhar S, Wickiser JE, Karandikar NJ, Malter JS, et al.. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun 2014;2:4802. doi: 10.1038/ncomms5802. PMID:25190313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol 2008;181:1055-63. doi: 10.1083/jcb.200802105. PMID:18591425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Lyle S, Hoover K, Colpan C, Zhu Z, Matijasevic Z, Jones SN. Dicer cooperates with p53 to suppress DNA damage and skin carcinogenesis in mice. PLoS One 2014;9:e100920. doi: 10.1371/journal.pone.0100920. PMID:24979267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, et al.. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010;467:986-90. doi: 10.1038/nature09459. PMID:20962848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Pastorelli LM, Wells S, Fray M, Smith A, Hough T, Harfe BD, McManus MT, Smith L, Woolf AS, Cheeseman M, et al.. Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome 2009;20:140-51. doi: 10.1007/s00335-008-9169-y. PMID:19169742 [DOI] [PubMed] [Google Scholar]
- [137].Wagh PK, Gardner MA, Ma X, Callahan M, Shannon JM, Wert SE, Messinger YH, Dehner LP, Hill DA, Wikenheiser-Brokamp KA. Cell- and developmental stage-specific Dicer1 ablation in the lung epithelium models cystic pleuropulmonary blastoma. J Pathol 2015;236:41-52. doi: 10.1002/path.4500. PMID:25500911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, Dyer MA, Marine JC. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ 2010;17:633-41. doi: 10.1038/cdd.2009.202. PMID:20019750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 2009;23:2700-4. doi: 10.1101/gad.1848209. PMID:19903759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Arrate MP, Vincent T, Odvody J, Kar R, Jones SN, Eischen CM. MicroRNA biogenesis is required for Myc-induced B-cell lymphoma development and survival. Cancer Res 2010;70:6083-92. doi: 10.1158/0008-5472.CAN-09-4736. PMID:20587524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yoshikawa T, Otsuka M, Kishikawa T, Takata A, Ohno M, Shibata C, Kang YJ, Yoshida H, Koike K. Unique haploinsufficient role of the microRNA-processing molecule Dicer1 in a murine colitis-associated tumorigenesis model. PLoS One 2013;8:e71969. doi: 10.1371/journal.pone.0071969. PMID:24023722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Morita S, Hara A, Kojima I, Horii T, Kimura M, Kitamura M, Ochiya T, Nakanishi K, Matoba R, Matsubara K, et al.. Dicer is required for maintaining adult pancreas. PLoS One 2009;4:e4212. doi: 10.1371/journal.pone.0004212. PMID:19148298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA 2006;103:2208-13. doi: 10.1073/pnas.0510839103. PMID:16452165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Kim GJ, Georg I, Scherthan H, Merkenschlager M, Guillou F, Scherer G, Barrionuevo F. Dicer is required for Sertoli cell function and survival. Int J Dev Biol 2010;54:867-75. doi: 10.1387/ijdb.092874gk. PMID:19876815 [DOI] [PubMed] [Google Scholar]
- [145].Korhonen HM, Meikar O, Yadav RP, Papaioannou MD, Romero Y, Da Ros M, Herrera PL, Toppari J, Nef S, Kotaja N. Dicer is required for haploid male germ cell differentiation in mice. PLoS One 2011;6:e24821. doi: 10.1371/journal.pone.0024821. PMID:21949761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Buza-Vidas N, Cismasiu VB, Moore S, Mead AJ, Woll PS, Lutteropp M, Melchiori L, Luc S, Bouriez-Jones T, Atkinson D, et al.. Dicer is selectively important for the earliest stages of erythroid development. Blood 2012;120:2412-6. doi: 10.1182/blood-2011-10-383653. PMID:22869792 [DOI] [PubMed] [Google Scholar]
- [147].Li Z, He X, Feng J. Dicer is essential for neuronal polarity. Int J Dev Neurosci 2012;30:607-11. doi: 10.1016/j.ijdevneu.2012.08.002. PMID:22982054 [DOI] [PubMed] [Google Scholar]
- [148].Swahari V, Nakamura A, Baran-Gale J, Garcia I, Crowther AJ, Sons R, Gershon TR, Hammond S, Sethupathy P, Deshmukh M. Essential function of Dicer in resolving DNA damage in the rapidly dividing cells of the developing and malignant cerebellum. Cell Rep 2016;14:216-24. doi: 10.1016/j.celrep.2015.12.037. PMID:26748703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood 2012;119:767-76. doi: 10.1182/blood-2011-05-355412. PMID:22117047 [DOI] [PubMed] [Google Scholar]
- [150].Liu HC, Tang Y, He Z, Rosenwaks Z. Dicer is a key player in oocyte maturation. J Assist Reprod Genet 2010;27:571-80. doi: 10.1007/s10815-010-9456-x. PMID:20827505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Gurtan AM, Lu V, Bhutkar A, Sharp PA. In vivo structure-function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA 2012;18:1116-22. doi: 10.1261/rna.032680.112. PMID:22546613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Anglesio MS, Wang Y, Yang W, Senz J, Wan A, Heravi-Moussavi A, Salamanca C, Maines-Bandiera S, Huntsman DG, Morin GB. Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol 2013;229:400-9. doi: 10.1002/path.4135. PMID:23132766 [DOI] [PubMed] [Google Scholar]
- [153].Ohishi K, Nakano T. A forward genetic screen to study mammalian RNA interference: Essential role of RNase IIIa domain of Dicer1 in 3′ strand cleavage of dsRNA in vivo. FEBS J 2012;279:832-43. doi: 10.1111/j.1742-4658.2012.08474.x. PMID:22221880 [DOI] [PubMed] [Google Scholar]
- [154].Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 pathway in cancer. Front Genet 2017;8:31. doi: 10.3389/fgene.2017.00031. PMID:28400788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Wang XJ, Jiang FZ, Tong H, Ke JQ, Li YR, Zhang HL, Yan XF, Wang FY, Wan XP. Dicer1 dysfunction promotes stemness and agression in endometrial carcinoma. Tumour Biol 2017;39:1010428317695967. doi: 10.1177/1010428317695967. PMID:28381177 [DOI] [PubMed] [Google Scholar]
- [156].Yi YH, Ma TH, Lee LW, Chiou PT, Chen PH, Lee CM, Chu YD, Yu H, Hsiung KC, Tsai YT, et al.. A genetic cascade of let-7-ncl-1-fib-1 modulates nucleolar size and rRNA pool in Caenorhabditis elegans. PLoS Genet 2015;11:e1005580. doi: 10.1371/journal.pgen.1005580. PMID:26492166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008;22:2773-85. doi: 10.1101/gad.1705308. PMID:18923076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Johanson TM, Lew AM, Chong MM. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol 2013;3:130144. doi: 10.1098/rsob.130144. PMID:24153005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009;15:2147-60. doi: 10.1261/rna.1738409. PMID:19850906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with miRNA-like functions. Mol Cell 2008;32:519-28. doi: 10.1016/j.molcel.2008.10.017. PMID:19026782 [DOI] [PubMed] [Google Scholar]
- [161].Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA 2009;15:1233-40. doi: 10.1261/rna.1528909. PMID:19474147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Ren YF, Li G, Wu J, Xue YF, Song YJ, Lv L, Zhang XJ, Tang KF. Dicer-dependent biogenesis of small RNAs derived from 7SL RNA. PLoS One 2012;7:e40705. doi: 10.1371/journal.pone.0040705. PMID:22808238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Ren YF, Li G, Xue YF, Zhang XJ, Song YJ, Lv L, Wu J, Fang YX, Wang YQ, Shi KQ, et al.. Decreased dicer expression enhances SRP-mediated protein targeting. PLoS One 2013;8:e56950. doi: 10.1371/journal.pone.0056950. PMID:23468895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012;488:231-5. doi: 10.1038/nature11179. PMID:22722852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Francia S, Cabrini M, Matti V, Oldani A, d'Adda di Fagagna F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J Cell Sci 2016;129:1468-76. doi: 10.1242/jcs.182188. PMID:26906421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Yuan S, Ortogero N, Wu Q, Zheng H, Yan W. Murine follicular development requires oocyte DICER, but not DROSHA. Biol Reprod 2014;91:39. doi: 10.1095/biolreprod.114.119370. PMID:24990804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci USA 2016;113:E1881-9. doi: 10.1073/pnas.1602532113. PMID:26976605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Yoshikawa M, Fujii YR. Human ribosomal RNA-derived resident microRNAs as the transmitter of information upon the cytoplasmic cancer stress. Biomed Res Int 2016;2016:7562085. doi: 10.1155/2016/7562085. PMID:27517048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Bai B, Liu H, Laiho M. Small RNA expression and deep sequencing analyses of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Biol 2014;4:441-9. doi: 10.1016/j.fob.2014.04.010. PMID:24918059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleid Acids Res 2011;39:3879-91. doi: 10.1093/nar/gkq1355. PMID:21247878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Srivas R, Shen JP, Yang CC, Sun SM, Li J, Gross AM, Jensen J, Licon K, Bojorquez-Gomez A, Klepper K, et al.. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol Cell 2016;63:514-25. doi: 10.1016/j.molcel.2016.06.022. PMID:27453043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Kim G, Ison G, McKee AE, Zhang H, Tang S, Gwise T, Sridhara R, Lee E, Tzou A, Philip R, et al.. FDA approval summary: Olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res 2015;21:4257-61. doi: 10.1158/1078-0432.CCR-15-0887. PMID:26187614 [DOI] [PubMed] [Google Scholar]
- [173].Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al.. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 2008;322:405-10. doi: 10.1126/science.1162609. PMID:18818364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Pouliot LM, Shen DW, Suzuki T, Hall MD, Gottesman MM. Contributions of microRNA dysregulation to cisplatin resistance in adenocarcinoma cells. Exp Cell Res 2013;319:566-74. doi: 10.1016/j.yexcr.2012.10.012. PMID:23137650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Andrews WJ, Panova T, Normand C, Gadal O, Tikhonova IG, Panov KI. Old drug, new target: Ellipticines selectively inhibit RNA polymerase I transcription. J Biol Chem 2013;288:4567-82. doi: 10.1074/jbc.M112.411611. PMID:23293027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Gerber JK, Gögel E, Berger C, Wallisch M, Müller F, Grummt I, Grummt F. Termination of mammalian rDNA replication: Polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 1997;90:559-67. doi: 10.1016/S0092-8674(00)80515-2. PMID:9267035 [DOI] [PubMed] [Google Scholar]
- [177].Sánchez-Gorostiaga A, López-Estraño C, Krimer DB, Schvartzman JB, Hernández P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol 2004;24:398-406. doi: 10.1128/MCB.24.1.398-406.2004. PMID:14673172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007;7:834-46. doi: 10.1038/nrc2256. PMID:17957189 [DOI] [PMC free article] [PubMed] [Google Scholar]


