Abstract
Single-cell, spatially resolved ‘omics analysis of tissues is poised to transform biomedical research and clinical practice. We have developed an open-source, computational multiplex image cytometry analysis toolbox (miCAT) to enable interactive, quantitative, and comprehensive exploration of individual cell phenotypes, cell-to-cell interactions, microenvironments, and morphological structures within intact tissues. We highlight the unique abilities of miCAT by analysis of highly multiplexed mass cytometry images of human breast cancer tissues.
Technological advances in multi-parametric analysis of single cells have revealed the heterogeneity of cellular phenotypes and functional states concealed in population-based studies1–3. Each cellular phenotype is defined by the interplay of its internal state and the environment in which it resides, and tissue function is the output of these coordinated cell activities. Deregulation of inter-cellular communication is central to many diseases such as cancer4. Consequently, the ability to analyze single-cell functional states with spatial resolution is key to understanding normal tissue function and disease biology and for the development of treatments of disease.
Recent techniques such as FISSEQ5, MERFISH6, cycling immunofluorescence7–9, multiplexed ion beam imaging (MIBI)10, and imaging mass cytometry (IMC)11 allow for single-cell, spatially resolved, highly multiplexed analysis of solid tissues and provide essential information on the distribution of transcripts, proteins, and their modifications within single cells, microenvironments, and entire tissues12. Despite these experimental advances no computational approach has been developed to enable comprehensive, quantitative, and interactive exploration of all levels of information within the data that results from spatially resolved, highly multiplexed measurements on tissue. Current open-source tools that provide image-linked data analysis are typically focused on the analysis of cell lines imaged with low-plex fluorescence microscopy or basic tissue histology and are not geared for analysis of highly multiplexed measurements13–15. On the other hand, tools developed to perform analyses of non-imaging, multiplexed single-cell data such as that obtained using suspension-based mass cytometry do not exploit spatial information (Supplementary Fig. 1)16,17.
In order to provide a complete picture of a tissue ecosystem, define molecular and spatial signatures necessary for analysis of tissue biology, and, in the case of disease, identify clinically relevant features, it is necessary to analyze and interrelate layers of information obtained from molecular measurements on cells, cell populations, cell-to-cell interactions, microenvironments, tissues, and experimental cohorts. Here we present a powerful, interactive computational platform called miCAT that makes quantitative analysis of highly multiplexed, single-cell-resolved tissue measurements possible (Supplementary Software 1-3). miCAT combines intuitive high-dimensional image visualization, state-of-the-art analysis methods for cell phenotype characterization, and novel algorithms for the comprehensive study of cell-to-cell interactions and the social networks of cells within complex tissues (Fig. 1). Thus, miCAT provides investigators from biology, biomedicine, and pathology a toolbox with which to investigate tissues during health, disease, and treatment.
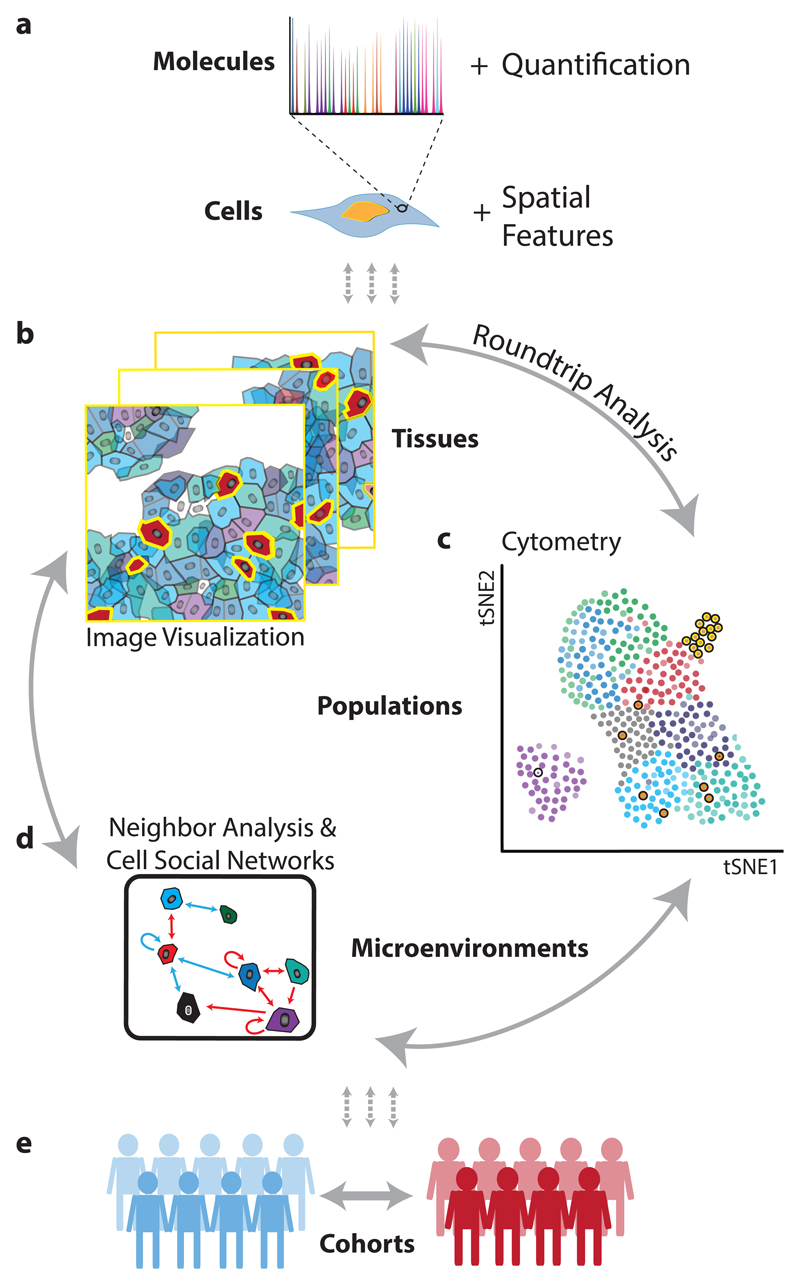
Figure 1. From molecular to clinical information: multi-scale analysis of the tissue ecosystem.
(a) Spatially resolved, high-dimension molecular measurements are aggregated using image masks to define regions corresponding to each cell. (b) Visualization of images, (c) cytometry analysis, and (d) analysis of neighbors and cellular interaction networks facilitate “round-trip” analysis through layers of information. (e) Using molecular, cellular, and spatial signatures experimental cohorts can be compared and contrasted.
In miCAT, all single-cell information, including spatial features (Fig. 1a), is linked to the corresponding multiplex image enabling visualization of images and single-cell analysis in parallel (Fig. 1b,c). miCAT uses a segmentation mask to extract single-cell data from images including abundances of all measured markers for a cell and area of interest, spatial features (like cell size and shape), and aspects of the cell’s environment such as cell neighbors and cell crowding. This information is compiled into a flow cytometry standard format (.fcs) file for further analysis inside or outside miCAT. “Round-trip” analyses, from a specific area of an image, to dataset-wide analyses of single-cell phenotypes and their interactions, and back to the visualization of unique cells on images, enables users to define and understand key cell populations and their spatial context in tissue. To enable quantitative and systematic analysis of all cell-to-cell interactions, we developed a novel algorithm to identify proximate cell-to-cell interactions that are present more frequently than expected by chance (Fig. 1d). Our algorithm enables determination of significant interactions and unique cell environments across entire datasets and within specific cohorts (Fig. 1d,e). These interactions can be represented in “social networks” of cells and is complementary to interactions inferred from the presence of ligand receptor pairs18.
To combine image-based spatial information and high-dimensional cytometry data, the miCAT GUI is divided in two parallel sections for paired image and cytometry analysis (Supplementary Note 1). In the image visualization section of miCAT, high-dimensional images (Fig. 2a) as well as cell masks, single-cell marker quantification, and cell identification labels can be visualized. In the analysis section of miCAT, image-derived marker quantification and spatial features of single-cell data are extracted for each image (Fig. 2a), combined (Fig. 2b), and visualized using multi-dimensional reduction tools such as tSNE maps16 (Fig. 2b), scatter plots (Supplementary Fig. 2), histograms, box plots, or other visualizations (Supplementary Note 1).
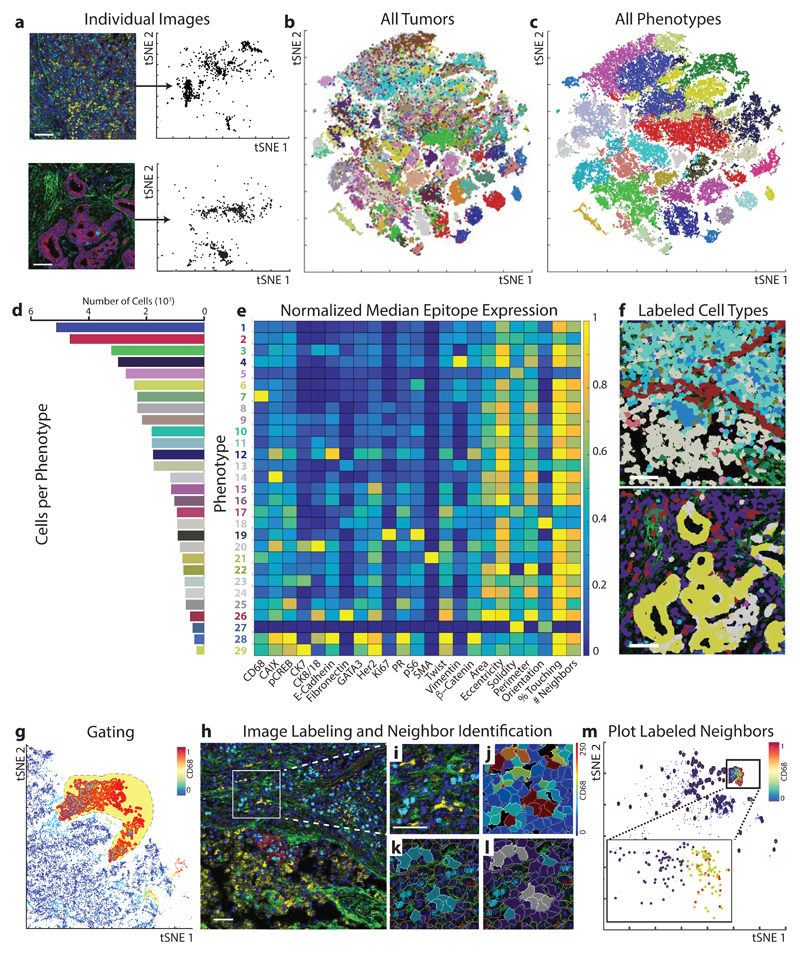
Figure 2. Round-trip analysis of unique cell types in high-dimension images of breast cancer.
(a) Left: Two representative multi-parametric images are displayed in the miCAT image window using user-defined color channels (red, E-cadherin; green, vimentin; blue, histone H3; cyan, Ki-67; magenta, cytokeratin 7; yellow, CD68). Up to six colors can be defined. Scale bar = 100 µm. Right: High-dimension single-cell data, including spatial features and all expressed markers from each segmented cell are extracted from each individual image and visualized in a t-SNE plot. (b) When the entire breast cancer dataset is visualized in one t-SNE plot, distinct colors distinguish cells of each source image. (c) Unsupervised clustering of all cells according to their marker expression throughout the dataset using PhenoGraph defines complex cell phenotypes and enables labeling of each cell phenotype cluster with a distinct color. (d) Bar plot of the total number of cells from each PhenoGraph-defined cell phenotype in the dataset. (e) Cell phenotypes can be further investigated using plotting tools such as heatmaps. (f) All single cells can be colored according to the identified phenotypes within the context of the tissue microenvironment on their original image. In this example, non-cell tissue is not labeled. (g) Quantification of an individual parameter can be heatmapped onto the t-SNE plot, and populations can be identified in a supervised manner using the gating tool. (h) All images containing cells of a subpopulation of interest can then be identified. In this example: red, E-cadherin; green, fibronectin; blue, histone H3; cyan, Ki67; magenta, cytokeratin 7; yellow, CD68. Scale bar = 100 µm. Images can be visualized using (i) pseudo-color or by (j) heatmap representing the intensity of a marker in each cell. (k) Cells of interest can be highlighted on the image (turquoise), and neighboring cells (purple or gray if representing both subpopulations) within a defined pixel range can also be identified and highlighted on (l) the image or (m) the analysis plot of the individual image (red, cell of interest; blue, neighbor; yellow, both subpopulations).
To demonstrate the potential of miCAT-powered analyses, we investigated cellular phenotypes and microenvironments of human breast cancer as visualized by IMC. By pairing classic immunohistochemistry staining, high-resolution tissue laser ablation, and mass cytometry, IMC can measure abundances of more than 40 unique metal-isotope-labeled tissue-bound antibodies simultaneously at a resolution comparable to fluorescence microscopy11. Here we analyzed images collected from 49 diverse breast cancer samples and three matched normal tissues, and an additional six normal breast tissue samples. Tissues were stained with an antibody panel tailored to identify cell lineages and to detect signaling pathway activation, proliferation, apoptosis, and clinical markers (Supplementary Fig. 3, Supplementary Tables 1-2).
To gain a tissue-wide overview of cell phenotypes present in a given image set, we have incorporated two approaches into miCAT. The first approach is supervised and based on tSNE16, a data dimensionality reduction method that projects multi-marker quantification into two dimensions, grouping similar cells (as shown for image analysis of all samples in Fig. 2b,c). On the tSNE map in miCAT, expression of individual markers can be highlighted using color scales, manually gated, and annotated for cell phenotype (Supplementary Note 1). The second approach is based on the unsupervised clustering algorithm Phenograph19. In the breast cancer and normal tissue samples, PhenoGraph identified 29 phenotype clusters shared across images and clinical subgroups, which were then visualized on a tSNE map (Fig. 2c, Supplementary Fig. 3). These cell phenotypes are present at different frequencies (Fig. 2d) and were characterized by specific epitopes (e.g., vimentin, phenotype #4; and CD68, phenotype #7) and combinations of markers (e.g., proliferative Ki-67+ and phospho-S6+ phenotypes #8, #10, and #19) (Fig. 2e). Cell populations can be linked back to their source images and visualized within the context of their multi-cellular environment (Fig. 2f).
Tumor-associated macrophages (TAMs) can drive or hinder tumor progression and are therefore highly attractive biomarkers and drug targets20. To gain a deeper understanding of TAMs and their neighborhoods, we inspected PhenoGraph phenotype #7 with high CD68 signal suggestive of macrophage identity (Fig. 2e, Supplementary Fig. 2b). It is also possible to select potential macrophages by gating for CD68 expression on the tSNE plot (Fig. 2g). CD68 epitope expression was visualized in images by color (Fig. 2h,i, yellow) and heatmapped for each segmented cell (Fig. 2j). miCAT also allows CD68+ cells selected from a plot to be highlighted on source images in their original tissue context (Fig. 2k).
miCAT has two neighborhood functions that enable investigation of the microenvironment. The first function is user guided and returns a subpopulation of cells touching or proximal to a cell population of interest for visualization on images (Fig. 2l) or for downstream analysis (Fig. 2m). This analysis performed on the 52 tumor-derived images showed that distinct proliferative (Ki-67, phospho-S6) and hypoxic (carbonic anhydrase IX) epithelial tumor cells neighbor CD68+ cells (Supplementary Fig. 4). The second neighborhood function enables the unbiased and systematic study of all cell-to-cell interactions present in a tissue or all tissues of a sample cohort by using a permutation test to compare the number of interactions between all cell types in a given image to that of a matched control containing randomized cell phenotypes (Fig. 3a). This approach determines the significance of cell-to-cell interactions and reveals enrichments or depletions in cell-to-cell interactions that are indicative of cellular organization. The significance of a neighboring interaction between each pair of phenotypes is visualized as a heatmap in which rows represent the neighborhood of a cell phenotype of interest and columns the enrichment or depletion of a cell in other neighborhoods (Fig. 3b,c).
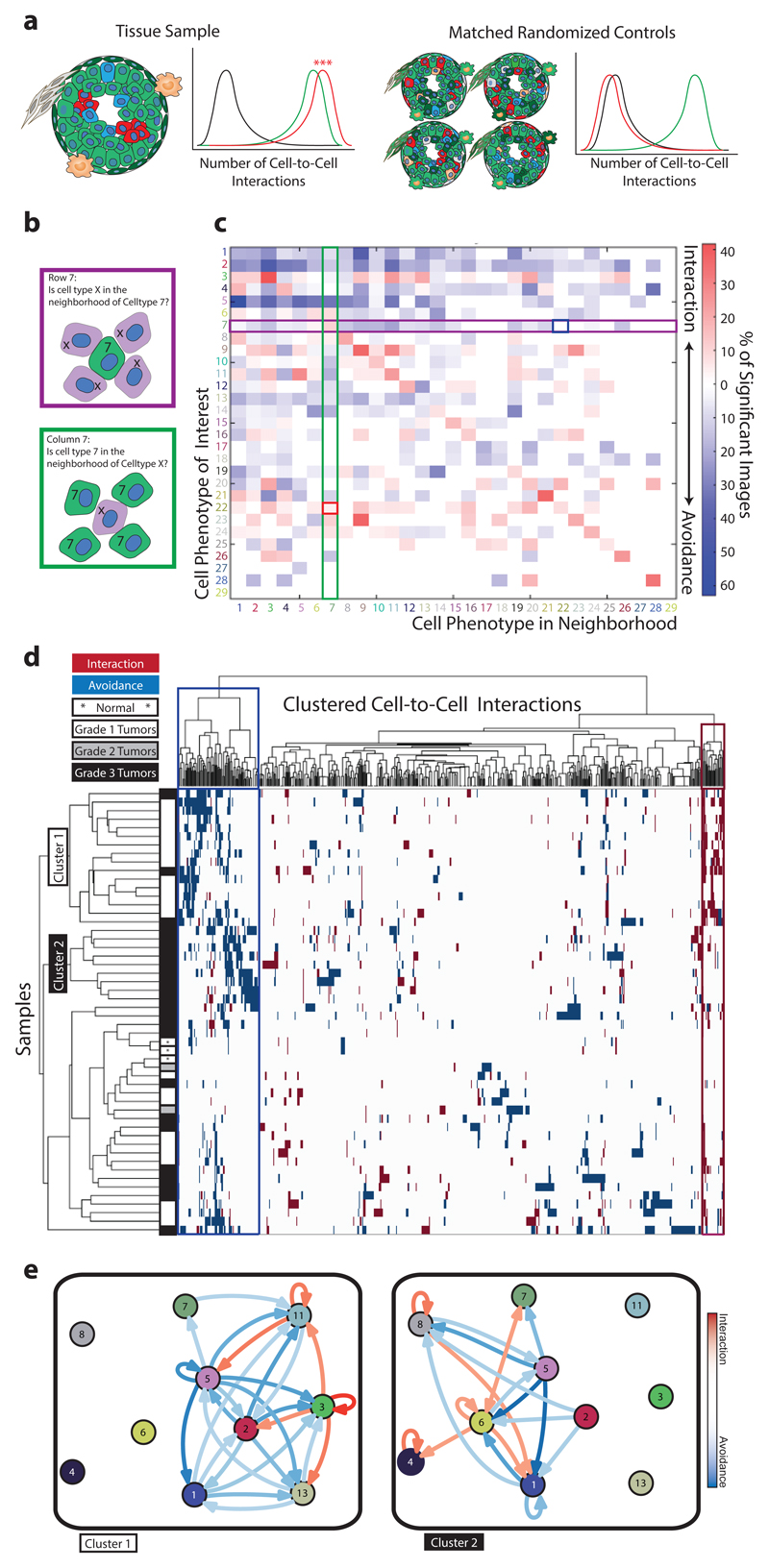
Figure 3. Neighborhood analysis of breast cancer cell phenotypes.
(a) Schematic of neighbor analysis in which the prevalence of a particular cell-to-cell interaction in an image is quantified and significance is determined by comparison to its prevalence in cell-type-randomized controls of the same image. Number of interactions between abundant green cells (green line), between rare clustered red cells (red line), and between abundant green cells and rare red cells (black line). (b) Schematic depicting directional aspects of neighbor interactions visualized in the heatmap. Rows visualize the significance of all cell types surrounding a cell type of interest. Columns visualize the significance of the cell type of interest surrounding other cell types. White represents a prevalence of less than 10%. (c) All interactions present in 49 breast tumor images and three matched normal tissue images are represented as a heatmap in which the cell type in the row is significantly neighbored (red) or avoided (blue) by the cell type in the column. Significance was determined by permutation test (p < 0.01). Highlighted squares indicate an example of a directional interaction: Stromal phenotype #7 significantly surrounds tumor cell type #22 (red square), but #22 is not surrounded by #7 (blue square). (d) Agglomerative clustering of all samples and cell-to-cell interactions according to the presence of significant (p < 0.01) phenotype interaction (red) or avoidance (blue). White represents interactions that are not present or not significant. (e) Force-directed cell interaction network graphs representing the interactions of PhenoGraph-defined cell phenotypes in Cluster 1 and Cluster 2 tumors. Circle color corresponds to PhenoGraph cluster. Red arrows indicate interaction and blue arrows avoidance, and intensities of the line color indicate significance. A connection is only visualized if the interaction or avoidance is significant in a least 30% of the grouped samples and the cell phenotypes are simultaneously present in at least 90% of the grouped samples.
We validated this strategy (Methods, Supplementary Notes 2-3) on synthetic data (Supplementary Fig. 5), investigated its robustness to variations in cell segmentation (Supplementary Fig. 6), and showed that the algorithm identified known luminal-basal cell interactions in healthy mammary ducts and alveoli (Supplementary Fig. 7). Neighborhood analyses of images obtained from 49 breast cancer samples and three matched normal tissues identified cell phenotypes that surround or are surrounded by another cell phenotype (Fig. 3b,c, Supplementary Fig. 8a,b). For TAMs (phenotype #7), unsupervised neighborhood analysis revealed that they surround multiple cell phenotypes (Fig. 2e; Fig. 3c, column 7 rows 6, 8, 22, 23, 24) identifying key TAM-interacting cells within all neighbor interactions (Supplementary Fig. 4j). Relationships and cellular crosstalk between CD68+ cells and other cells, including phospho-S6+/vimentin+ stromal cells (#6) and E-cadherin+/phospho-S6+/Twist+ tumor cells (#22), were identified, suggestive of distinct tumor microenvironments for future study.
To identify cellular landscapes, we clustered images based on all significant cell-to-cell interactions (Fig. 3d). Distinct subgroups enriched in Grade 1 and Grade 3 tumors became apparent as well as a mixed grade subgroup. To facilitate the visualization and comparison of cell-to-cell interactions over large datasets, we used our permutation-based neighborhood algorithm to identify social networks of cells that are specific to tumor grade (Fig. 3e). Separation of the Grade 1 tumor subgroup was driven by tumor cell phenotypes (phenotypes #3, #9, #11) that interact with surrounding stromal cells (phenotypes #1, #5, #13), a cellular organization that reflects tissue tubularity, a major pathology grading criteria (Fig. 3d, e, Supplementary Fig. 8a,b, Supplementary Fig. 9). The more advanced lesions of Grade 3 samples contained hypoxic cells (phenotype #14), interacting proliferative cells (phenotypes #8, #10, #19), and interactions with “active” stroma (phospho-S6+ and vimentin+, phenotype #6) including macrophages (phenotype #7) (Fig. 3d,e, Supplementary Fig. 8c,d, Supplementary Fig. 9). Clustering of images based on significant cell interactions defined groups of tissues (in this case, the cancer samples) that have similar organization and revealed pathology grade-associated cellular ecosystems that may distinguish unique disease states.
By combining cytometry, image analysis, and novel algorithms for cell-to-cell interaction network analysis, miCAT is able to define complex cell types using multiplexed measurements and spatial features as parameters and elucidate patterns of cellular interactions within heterogeneous tissues. The use of round-trip analyses between single-cell data and source images using machine learning and community-finding algorithms within an intuitive user interface will enhance our understanding of tissue structure at the cellular level. Combined with focused, hypothesis-driven datasets, future investigations of multiplexed imaging cytometry data using miCAT could reveal cell types and cell interactions that drive disease. miCAT is open source: we invite the community to further develop this toolbox for the analysis of next-generation imaging and pathology data.
Online Methods
Preparation and staining of breast cancer tissue specimens
Formalin-fixed paraffin-embedded tissue samples from patients treated at the University Hospital Zurich between 1991 and 2005 were retrieved from the archives of the Institute of Surgical Pathology. This project was approved by the local Commission of Ethics (ref. no. StV 12-2005). H&E stained sections of all tumors were re-evaluated by a pathologist for their suitability for tissue microarray construction prior to array construction as previously described21.
Tissues were stained as previously described11. Briefly, tissue sections were de-waxed overnight in xylene and rehydrated in a graded series of alcohol (ethanol:deionized water 100:0, 90:10, 80:20, 70:30, 50:50, 0:100; 5 min each). Heat-induced epitope retrieval was conducted in a water bath at 95 °C in Tris-EDTA buffer at pH 9 for 20 min. After immediate cooling, the microarrays were blocked with 3% BSA in TBS for 1 hour. Samples were incubated overnight at 4 °C in primary antibody at 7.5 g/L diluted in TBS/0.1% Triton X-100/1% BSA (clones in Supplementary Table 1, Supplementary Note 4). Panel design and antibody database management was done in AirLab22. Samples were then washed twice with TBS/0.1% Triton X-100 and twice with TBS and dried before imaging mass cytometry measurement.
Imaging mass cytometry
Antibody staining of tissue sections was quantified through the combination of laser ablation using a modified ArF excimer GeoLas C laser system (Coherent) to ablate tissues in a rastered pattern at 20 Hz and to direct aerosol transportation of the sample to a CyTOF mass cytometer (Fluidigm) as described previously23. All raw data processing was performed using in-house Matlab routines as described and provided previously11.
Segmentation
Segmentation was performed using Ilastik 1.1.924 and CellProfiler 2.1.115. Ilastik was used to classify pixels into three classes (nuclei, membrane, and background) and to generate probability maps. CellProfiler was used to segment probability maps to generate segmentation masks. A combination of channels was used to classify the background and membrane25. These masks were combined with the individual tiff files to extract single-cell information from each individual image.
To quantitatively assess the segmentation quality, we used four intuitive segmentation constraints for a segmentation score25: (1) mask should overlap with membrane signal; (2) mask should not overlap with nuclei signal; (3) segmented cells should contain maximally one nucleus; (4) mask should approximate the expected number of cells based on cell radius. We also analyzed the effect of segmentation by using segmentation masks generated by five independent users on three test images (Supplementary Fig. 6). The complexities and difficulties of image segmentation are discussed in our Supplementary Note 3.
Single-cell feature extraction
miCAT uses Matlab’s regionprops function to extract shape and pixel value measurements. Additionally, by step-wise pixel expansion, miCAT creates a network of neighbors surrounding each cell at a range of defined distances. In most cases, expansion in the range of 1 to 6 pixels was chosen. Cells were expanded using a rectangular membrane shape. All cells within the defined range were considered neighbors. The distances between centroids were used to define cell-to-cell distances. The number of neighbors and the percent of membrane in contact with a neighboring cell were both determined with modified CellProfiler 1.0 modules15.
Data transformation
Raw measurements were used for the presented data. miCAT also offers arcsinh transformation with a variable cofactor input.
Normalization
All images were segmented and single-cell measurements were extracted from all available channels using the mean pixel values for each segmented cell. The presented data were not normalized, but miCAT features Z-score normalization across all samples or across a subgroup as a module. We used 99th percentile normalized data for t-SNE and PhenoGraph as suggested16,19. Heat maps were visualized using scaled values from 0-1 for individual columns in analysis plots as well as on images for individual masks.
miCAT
miCAT can be downloaded either as a Matlab 2014b app or as a stand-alone application for Mac OS12 (Supplementary Software 1), Windows 7 (Supplementary Software 2), and Windows 10 (Supplementary Software 3) from https://github.com/BodenmillerGroup/miCAT. Documentation, user manual, and development versions of miCAT can also be found on the project page. All modules, if not differently stated, were written in Matlab 2014b, and the GUI was designed in Matlab 2014b using Matlab's GUI development environment (GUIDE). miCAT is built modularly to enable addition of new features without need for changes to the existing structure. In general, features in miCAT must include only two basic scripts: callback from the GUI and the script executing the function. The main functions are not linked to the GUI and can be run independently.
All data necessary to perform any function for the current session can be retrieved from the GUI handles or included manually without the GUI. Throughout a session, the data are kept in the fcs-format structure. There is one main matrix containing a column for each channel and a row for each individual cell of each image. This matrix is continuously updated during the session and will therefore also contain the custom gates and channels. The corresponding channel names for each image are saved in a cell array. All individual tiff files and corresponding masks are stored in a multidimensional matrix structure.
bh-t-SNE
We used the Barnes-Hut t-SNE implementation in miCAT16. Data were 99th-percentile normalized before the analysis, and we used the default t-SNE parameters (initial dimensions: 110; perplexity: 30; and theta: 0.5). The random seeds for the individual runs can be recorded.
PhenoGraph
PhenoGraph version 0.2 was used19. Data were 99th-percentile normalized before the analysis, and default parameters with nearest neighbors of 75 were used. This parameter was chosen based on prior knowledge of the underlying cell types. Lower values for nearest neighbors result in an over clustering and higher values an under clustering. The random seeds for the individual runs can be recorded.
Neighborhood analysis – permutation test
The neighborhood analysis uses basic statistical methods to find significantly enriched interactions between or within cell phenotypes. First, cells are manually or automatically classified. Manual classification can be done by manual gating on biaxial/t-SNE plots. Automatic classification uses the PhenoGraph19 algorithm, which consistently performs well for datasets with multiple cell populations26.
Once classified, pairwise interactions at a user-defined distance (6 pixels) between and within cell phenotypes are calculated for each single cell with its neighbors. A neighbor is defined as a cell within the pixel distance selected during the loading process. Pairwise interactions between and within cell phenotypes are compared to a random distribution using a permutation test. This test provides us with a p-value for each one-tailed test which represents interaction or avoidance. These p-values represent the likelihood of a neighborhood interaction being enriched or delimited in comparison to a randomized version of the same tissue. The comparison to a matched randomized tissue for every individual image controls for both the distinct connectivity and the specific cell types in that tissue. Equation 1 describes our approach using a permutation test with Monte Carlo sampling. We run this test twice to calculate the p-value for each tail.
| (Equation 1) |
Agglomerative hierarchical clustering using inner squared distance (minimum variance algorithm employing Ward’s method) as linkage criteria and Euclidean distance as the distance metric are applied to display similar interaction signatures in a dendrogram of all samples.
Analyses of synthetic data (Supplementary Fig. 5), variations in single cell masks (Supplementary Fig. 6), and organized normal breast tissue (Supplementary Fig. 7) validated and highlighted the robustness of our neighborhood analysis. In the case of rare cell populations (i.e., low cell number) our algorithm is able to detect significantly enriched interactions (Supplementary Note 2-3). In contrast, avoidance in rare cell populations will rarely be detected due to the minimal interactions among the rare cell population in the random shuffled control. The absolute cell number or the size of the image does not have a direct effect on the neighborhood analysis, as described above; only the frequencies of cell types present and the relative quantities are important. The expected average interactions can be described by equation 2, which includes the amount of cell type A multiplied by the fraction of cell type B and by the average connectivity of the image. If the average connectivity in an image is high, the expected interactions are high and vice versa.
| (Equation 2) |
Supplementary Material
Acknowledgements
We would like to thank the Bodenmiller lab for support and fruitful discussions. This work was supported by the Swiss National Science Foundation (SNSF) R’Equip grant 316030-139220, a SNSF Assistant Professorship grant PP00P3-144874, a Swiss Cancer League grant, the PhosphonetPPM and MetastasiX SystemsX grants, and funding from the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement n. 336921. D Schapiro was supported by the Forschungskredit of the University of Zurich, grant FK-74419-01-01. HWJ and D Schulz are supported by European Molecular Biology Organization (EMBO) Long Term Fellowships co-funded by the European Commission (LTFCOFUND2013 and 2014), grants ALTF-711 2015 and ALTF-970 2014, respectively.
Footnotes
Contributions
D Schapiro, HWJ, and BB conceived of the project and software. HWJ, CG, and RCF collected samples and validated antibodies. ZV assembled, classified, and provided tumor samples. HWJ completed the staining and image acquisition. D Schapiro, SR, and JF wrote the code. D Schapiro, HWJ, and D Schulz tested software on multiple data sources. D Schapiro, HWJ, and VZ analyzed the images and single cell data. D Schapiro, HWJ, and BB prepared the figures and wrote the manuscript. BB directed the project.
Competing Financial Interests
The authors declare no competing financial interests.
Data availability
All software and code that produced the findings of the study including Mac OS12 (Supplementary Software 1), Windows 7 (Supplementary Software 2), and Windows 10 (Supplementary Software 3) versions of miCAT are available at https://github.com/BodenmillerGroup/miCAT and http://www.bodenmillerlab.org/research-2/micat/. All raw data, miCAT sessions, and interactive graphs that support the findings of the study are available at http://www.bodenmillerlab.org/research-2/micat/.
References
- 1.Tirosh I, et al. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendall SC, et al. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodenmiller B, et al. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabassum DP, Polyak K. Nat Rev Cancer. 2015;15:473–483. doi: 10.1038/nrc3971. [DOI] [PubMed] [Google Scholar]
- 5.Lee JH, et al. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J-R, Fallahi-Sichani M, Sorger PK. Nat Commun. 2015;6:8390. doi: 10.1038/ncomms9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubert W, et al. Nat Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 9.Gerdes MJ, et al. Proc Natl Acad Sci U S A. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angelo M, et al. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giesen C, et al. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 12.Bodenmiller B. Cell Systems. 2016;2:225–238. doi: 10.1016/j.cels.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Ding H, Wang C, Huang K, Machiraju R. BMC Bioinformatics. 2015;16(Suppl 11):S10. doi: 10.1186/1471-2105-16-S11-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck AH, et al. Sci Transl Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 15.Jones TR, et al. BMC Bioinformatics. 2008;9:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir ED, et al. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shekhar K, Brodin P, Davis MM, Chakraborty AK. Proc Natl Acad Sci U S A. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rieckmann JC, et al. Nat Immunol. 2017;18:583–593. doi: 10.1038/ni.3693. [DOI] [PubMed] [Google Scholar]
- 19.Levine JH, et al. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostuni R, Kratochvill F, Murray PJ, Natoli G. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Kononen J, et al. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 22.Catena R, Özcan A, Jacobs A, Chevrier S, Bodenmiller B. Genome Biol. 2016;17:142. doi: 10.1186/s13059-016-1006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang HAO, et al. Anal Chem. 2013;85:10107–10116. doi: 10.1021/ac400996x. [DOI] [PubMed] [Google Scholar]
- 24.Sommer C, Straehle C, Köthe U, Hamprecht FA. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on (ed. IEEE); 2011. pp. 230–233. [Google Scholar]
- 25.Schüffler PJ, et al. Cytometry A. 2015;87:936–942. doi: 10.1002/cyto.a.22702. [DOI] [PubMed] [Google Scholar]
- 26.Weber LM, Robinson MD. Cytometry A. 2016;89:1084–1096. doi: 10.1002/cyto.a.23030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.