Significance
Like 80–90% of land plants, legumes form endosymbioses with arbuscular mycorrhizal fungi, host endophytes, support a rhizosphere community, and are attacked by pathogens. The ability of root cells to distinguish between these soil microbes and the mixture of chitinaceous compounds they display as signal molecules is important for an appropriate plant response. We show that legumes possess very similar receptors enabling root cells to separate perception of chitin, which triggers responses to pathogens, from perception of lipochitin oligosaccharides (Nod factors), which trigger endosymbiosis with rhizobial bacteria. The chitin receptors bind chitin in biochemical assays, and inactivation of the corresponding genes impairs defense responses toward pathogens. Together this establishes a long-sought foundation for dissecting plants’ response mechanisms toward different soil microbes.
Keywords: chitin perception, plant defense, symbiosis, Lotus, Medicago
Abstract
The ability of root cells to distinguish mutualistic microbes from pathogens is crucial for plants that allow symbiotic microorganisms to infect and colonize their internal root tissues. Here we show that Lotus japonicus and Medicago truncatula possess very similar LysM pattern-recognition receptors, LjLYS6/MtLYK9 and MtLYR4, enabling root cells to separate the perception of chitin oligomeric microbe-associated molecular patterns from the perception of lipochitin oligosaccharide by the LjNFR1/MtLYK3 and LjNFR5/MtNFP receptors triggering symbiosis. Inactivation of chitin-receptor genes in Ljlys6, Mtlyk9, and Mtlyr4 mutants eliminates early reactive oxygen species responses and induction of defense-response genes in roots. Ljlys6, Mtlyk9, and Mtlyr4 mutants were also more susceptible to fungal and bacterial pathogens, while infection and colonization by rhizobia and arbuscular mycorrhizal fungi was maintained. Biochemical binding studies with purified LjLYS6 ectodomains further showed that at least six GlcNAc moieties (CO6) are required for optimal binding efficiency. The 2.3-Å crystal structure of the LjLYS6 ectodomain reveals three LysM βααβ motifs similar to other LysM proteins and a conserved chitin-binding site. These results show that distinct receptor sets in legume roots respond to chitin and lipochitin oligosaccharides found in the heterogeneous mixture of chitinaceous compounds originating from soil microbes. This establishes a foundation for genetic and biochemical dissection of the perception and the downstream responses separating defense from symbiosis in the roots of the 80–90% of land plants able to develop rhizobial and/or mycorrhizal endosymbiosis.
Legumes have an exceptionally wide range of interactions with bacteria and fungi. They form endosymbioses with rhizobia and arbuscular mycorrhizal (AM) fungi, host endophytes, support a rhizosphere community, and, like other plants, are attacked by a plethora of pathogens (1–7). Detailed studies in the two model species Lotus japonicus (hereafter, Lotus) and Medicago truncatula (hereafter, Medicago) have identified central components of the signal exchange initiating the mutual recognition processes that establish symbiosis with rhizobia and mycorrhiza. Legume roots secrete (iso)flavonoids that activate the NodD transcriptional regulator of rhizobial Nod genes (8, 9). This activation leads to synthesis of lipochitooligosaccharide signal molecules (Nod factors) that are perceived by a Nod factor receptor complex consisting of the transmembrane LysM receptor kinase proteins LjNFR1/MtLYK3 and LjNFR5/MtNFP in Lotus and Medicago, respectively (10–19). Genetic and biochemical analyses have shown that LjNFR1 and LjNFR5 bind Nod factors at physiologically relevant concentrations (20) and initiate signal transduction; one of the first consequences of this is nuclear-associated calcium oscillations (spiking) in epidermal root cells, which can be detected within minutes (21–23). In Lotus the compatibility of exopolysaccharides is subsequently assessed by LjEPR3, another LysM receptor kinase that is required for the formation of infection threads (24–26). Interestingly, exopolysaccharides also appear to play a role in endophyte invasion (6, 27).
Mycorrhiza formation is similarly guided by mutual signal exchange. Strigolactone secreted by plant roots induces spore germination and prepenetration hyphal branching in AM fungi and hyphal growth toward the root surface (28–30). AM fungi appear to signal to their host plant via short-chain chitin oligomers (CO4 and CO5), and a mixture of lipochitooligosaccharides with structural similarities to rhizobial Nod factors has also been reported to be involved (31, 32). Receptors for these different signals have not yet been identified in legumes, but results from Parasponia and tomato suggest that LysM receptor proteins may be involved, if not in the presymbiotic phase, then later during root colonization (33, 34).
Proteins carrying LysM domains were also identified as receptors of pathogen-associated molecular patterns (PAMPs) that trigger plant defense reactions to prevent or limit microbial invasion, a process called “PAMP-triggered immunity” (PTI) (35). In rice and Arabidopsis chitin is perceived by LysM proteins such as OsCEBiP and/or LysM receptor kinases OsCERK1, AtCERK1, and AtLYK5 (36–41). Similarly, two Arabidopsis LysM proteins, LYM1 and LYM3, together with AtCERK1, serve in a receptor complex perceiving peptidoglycan (42). However, this distinction between LysM receptors recognizing PAMPs and inducing PTI and LysM receptors recognizing symbiotic signals appears too simplistic. In rice OsCERK1 mediates mycorrhizal infection as well as chitin-triggered defense responses toward pathogens, indicating a functional overlap (43, 44). OsCERK1 may thus serve as a component in different receptor complexes triggering different signal pathways. Adding further complexity, in Arabidopsis AtLYK3 was reported to mediate suppression of defense reactions by lipochitin oligosaccharides (LCOs) that have no endosymbiosis with mycorrhizal fungi or rhizobia (45). Taken together, the results from Arabidopsis, rice, and legumes highlight our limited insight into the composition and mechanisms of plant receptor complexes that distinguish chitinaceous signal molecules. Since PAMP receptors and defense reactions were primarily studied in leaves, our insight into roots that must accommodate rhizobial and/or mycorrhizal symbioses is even less.
In legume roots both LCOs and COs induce calcium spiking through the common symbiosis pathway in a dose- and LCO structure-dependent manner. MtNFP was not required for CO-induced calcium spiking, and the corresponding LjNFR5 receptor from Lotus has a higher affinity for LCO than for CO, implying a difference in LCO and CO perception. This raises the question of whether CO and LCO perception is mediated by different receptor complexes and therefore are separable or whether there are functional overlaps or compensation between individual receptor components. One of the limitations for resolving this question has been a shortage of genetic tools and biochemical methods needed to distinguish the function of individual members of the large LysM gene/protein families in legumes. Compared with Arabidopsis, which has seven genes encoding predicted LysM receptor kinases, and rice, which has nine such genes, Lotus has 21 genes encoding predicted LysM receptor kinases, and Medicago has 26 (16, 46–49). Forward genetics and suppressor screens have so far assigned functions to the Nod factor and exopolysaccharide receptors LjNfr1/MtLyk3, LjNfr5/MtNfp, and LjEpr3 (12–15, 25, 26). Resources for reverse genetics have recently become available with the LORE1 and Tnt1 mutant populations and, together with new biochemical approaches, make a more comprehensive analysis of large gene families in the two model legumes now possible (50–53).
In this report, we combine biochemical and genetic approaches to identify and characterize conserved LysM receptor kinases that allow recognition of chitinaceous signals for activation of immunity signaling in both Lotus and Medicago. We show that these LysM receptors have specificity for chitin and appear not to be involved in the perception of symbiotic LCOs. Combined, these receptors play essential roles in limiting pathogens, but alone they appear to have little effect on symbiotic interactions. This work establishes the foundations for understanding how plants differentiate potential pathogens from symbionts, paving the way for detailed analyses of the downstream signal transduction pathways separating LCO responses from chitin-triggered plant responses, particularly in roots.
Results
LjLYS6 Is a Chitin-Binding Receptor Kinase.
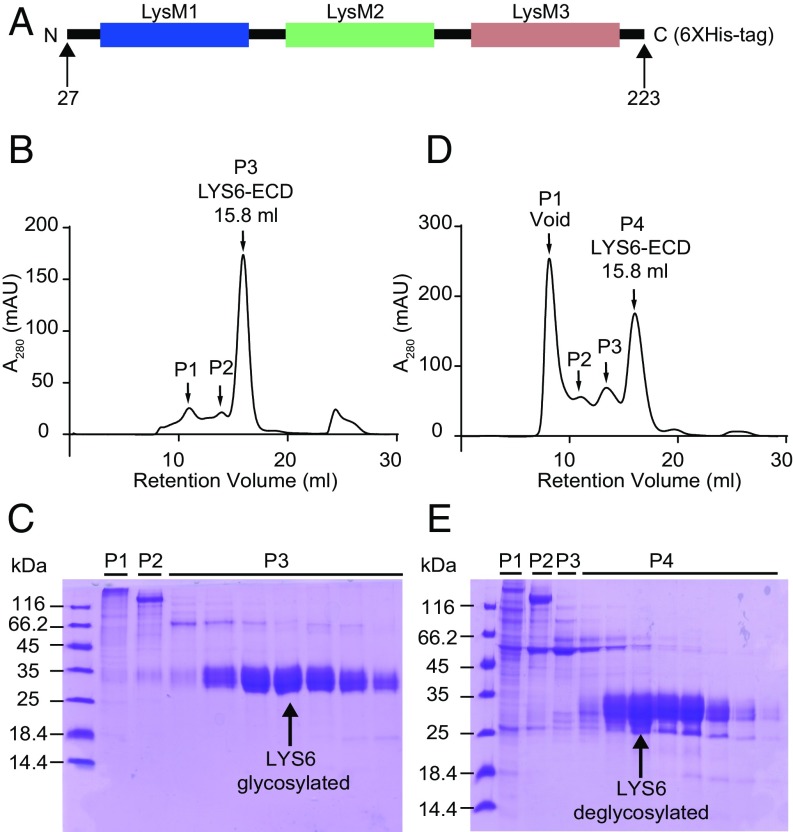
In the genome of L. japonicus four genes encode LysM receptor kinase proteins that are closely related to the LjNFR1 Nod factor receptor and belong to the same phylogenetic clade. Among these genes LjLys6 stands out as the only gene expressed at similar levels in leaves, stems, roots, and root nodules (48). This broad expression pattern suggests a role for LjLYS6 in processes that are not developmentally controlled or limited by tissue or cell specialization. We asked whether LjLYS6 as a putative pattern-recognition receptor would bind chitin, thus predicting a role in the first line of defense against pathogens. To test this possibility in vitro, affinities toward different chitin oligomers were determined biochemically. A his-tagged LjLYS6 ectodomain carrying the three LysM domains was expressed in insect cells (Fig. 1A), purified by two rounds of immobilized-metal affinity chromatography (IMAC), deglycosylated, and finally purified by size-exclusion chromatography (Fig. 1 B–E). Equilibrium dissociation constant (Kd) values for the LjLYS6 ectodomain and wheat germ agglutinin as control were determined in free solution using microscale thermophoresis as previously described (20, 54). Protein was fluorescently labeled, and binding was measured with increasing concentrations of ligands. Kd values were subsequently calculated from the binding curves (SI Appendix, Fig. S1) and are listed in Table 1. The Kd values show LjLYS6 ectodomain affinity for chitohexaose (CO6), chitoheptaose (CO7), and chitooctaose (CO8) in the lower micromolar range (∼50 µM). Binding to chitotetraose (CO4) was undetectable, whereas Kd values for chitopentaose (CO5) approached the detection limit. These direct binding measurements suggest that at least six GlcNAc residues (CO6) are required for optimal binding. Stronger binding was not apparent with CO7 and CO8. Wheat germ agglutinin binds to all chitin ligands with comparable Kd values in the low-micromolar range.
Fig. 1.
Expression and purification of the LjLYS6 ectodomain. (A) Schematic representation of the LjLYS6 construct with residue boundaries indicated. (B and D) The size-exclusion chromatography profile of glycosylated (B) and deglycosylated (D) LjLYS6 ectodomain purified on a Superdex 200 10/300 GL column. In the size-exclusion chromatography profile, both glycosylated and deglycosylated LjLYS6 ectodomains elute as a single peak (P3 and P4 respectively). (C and E) Coomassie brilliant-stained SDS–PAGE gels of the corresponding size-exclusion chromatography fractions. Glycosylated and deglycosylated LjLYS6 ectodomain from P3 (C) and P4 (E) are visible as distinct bands ∼35 or 27 kDa, respectively (indicated by a black arrow).
Table 1.
Dissociation constants of LjLYS6 ectodomain and wheat germ agglutinin toward chitin oligomers
| Ligand | Kd, µM | SD, µM | n of replicates |
| LYS6 ectodomain | |||
| Maltodextrin | N.D. | — | 2 |
| Chitotetraose | N.D. | — | 3 |
| Chitopentaose | 277 | 118 | 3 |
| Chitohexaose | 71 | 38 | 4 |
| Chitoheptaose | 26 | 16 | 3 |
| Chitooctaose | 38 | 22 | 4 |
| Wheat germ agglutinin | |||
| Maltodextrin | N.D. | — | 2 |
| Chitotetraose | 7.6 | 1.4 | 3 |
| Chitopentaose | 1.7 | 0.3 | 3 |
| Chitohexaose | 2.2 | 0.4 | 3 |
| Chitooctaose | 1.5 | 0.8 | 4 |
Dissociation constants of the LjLYS6 ectodomain and wheat germ agglutinin with various ligands as measured on a Monolith NT.115 system. Replicates for the LYS6 ectodomain are biological replicates; replicates for wheat germ agglutinin are technical replicates. Maltodextrin was used as a negative control. N.D., not detectable.
LjLYS6 Crystal Structure.
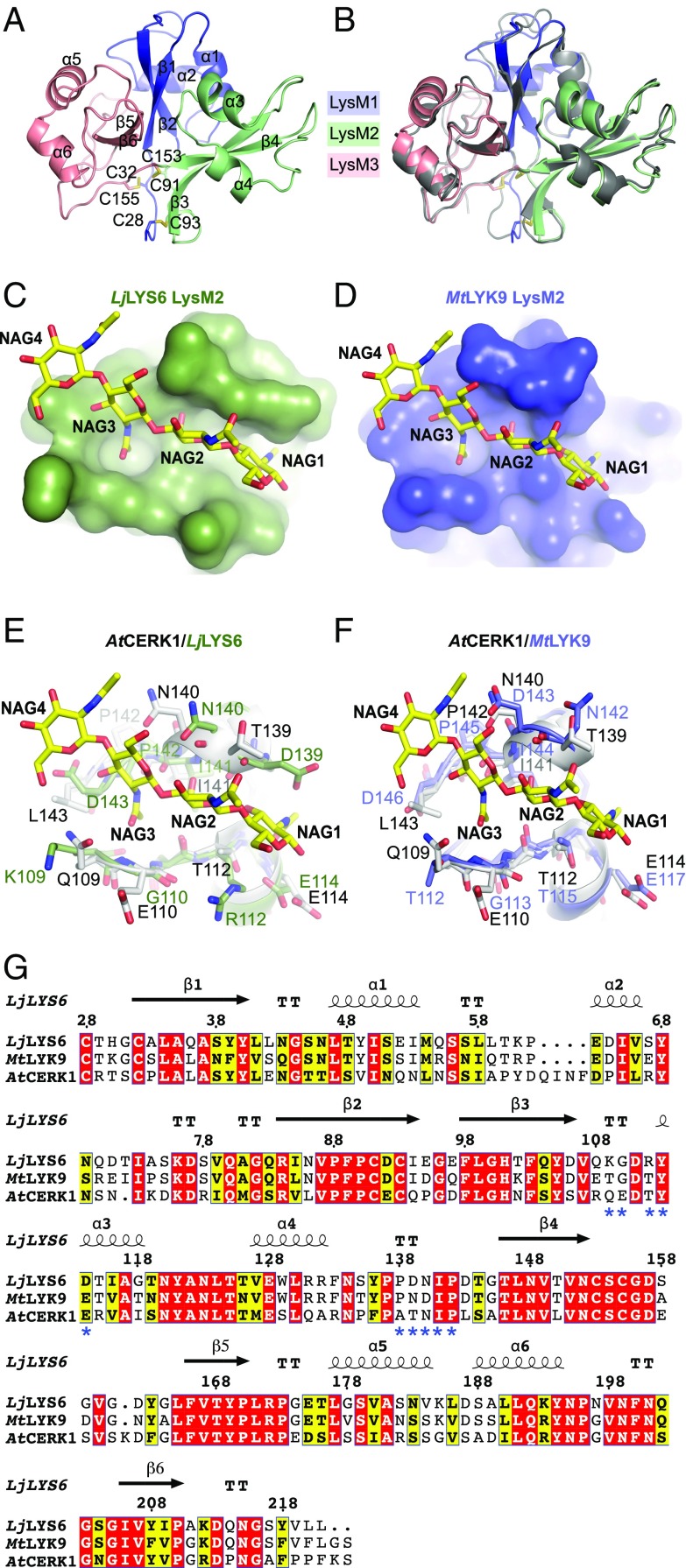
The purified LjLYS6 ectodomain from residues Lys27 to Ser223 was crystallized. The structure was solved by molecular replacement using the structure of the AtCERK1 ectodomain [Protein Data Bank (PDB) ID code: 4EBZ] as a search model and refined to 2.3-Å resolution. The crystal belongs to the P21 space group with two molecules of LjLYS6 per asymmetric unit. The data collection and refinement statistics (SI Appendix, Table S1) support the quality of the structure. Clear electron density is visible for residues of two LjLYS6 monomers in the asymmetric unit with the exception of the first and last residues and the multihistidine tag in the C terminus. Moreover, core glycosylation is clearly visible at three N-glycosylation sites on Asn46, Asn123, and Asn148 for each monomer, despite pretreatment of the protein with Peptide:N-glycosidase F (PNGase F) endoglycosidase.
The three LysM domains of each monomer are organized as an overall globular protein. Each LysM domain adopts a βααβ structure in which the two β-strands form an antiparallel β-sheet (Fig. 2A). This fold is typical for crystal structures of LysM domains from different proteins (55–57). The LysM2 and LysM3 domains are packed tightly against the LysM1 domain through two disulphide bridges established between Cys28 and Cys32 proximal to LysM1 and Cys93 and Cys155 proximal to LysM2 and LysM3, respectively. Finally, an additional disulphide bridge formed between Cys91 from LysM2 and Cys153 from LysM3 stabilizes the ectodomain structure (Fig. 2A).
Fig. 2.
Structural comparison of the ectodomains of LjLYS6, AtCERK1, and MtLYK9. (A) Cartoon representation of the crystal structure of the LjLYS6 ectodomain. The LysM1, LysM2, and LysM3 domains are colored in slate blue, pale green, and salmon pink, respectively. The disulphide bridges are represented as sticks. “C” followed by a number denotes the cysteines. The α and β signs followed by numbers indicate the numeration of α-helices and β-strands, respectively. (B) Superposition of the crystal structure of LjLYS6 onto AtCERK1 (gray; PDB ID code: 4EBZ). (C and D) Docking of a chitin molecule in the predicted LysM2-binding site of theLjLYS6 (C) and MtLYK9 (D) homology model. (E and F) Comparison of the LysM2 domain of AtCERK1, LjLYS6 crystal structures, and MtLYK9 homology model. The structures of the AtCERK1 (white), LjLYS6 (pale green), and MtLYK9 (blue) LysM2 domains are aligned, and residues involved (AtCERK1) or putatively involved (LjLYS6, MtLYK9) in chitin recognition are displayed as sticks. The chitin molecule is displayed as yellow sticks. NAG, N-acetylglucosamine. NAG1 indicates the carbohydrate at the reducing end. (G) Primary sequence alignment based on the 3D structures of LjLYS6, AtCERK1, and MtLYK9 ectodomains. The red and yellow boxes indicate strict or semi-conservation of the residues, and the blue asterisks mark residues involved in chitin recognition in AtCERK1. The secondary structures of LjLYS6 and AtCERK1 are indicated by arrows for β-strands and spirals for α-helices; the amino acids numbering corresponds to the position in the LjLYS6 primary sequence. TT, turn.
The LjLYS6 ectodomain structure is similar to that of the Arabidopsis AtCERK1 ectodomain (55). Superimposition of the main chains of LjLYS6 and AtCERK1 structures lead to an rmsd of 0.6 Å, documenting that the two are corresponding structures (Fig. 2B). Despite many attempts at cocrystallization or soaking crystals with chitin oligomers, we could not obtain a LjLYS6 structure bound to chitin. However, we could easily dock a chitin molecule in the LysM2 domain of LjLYS6 by superimposing the LysM2 domain of the AtCERK1:chitin complex crystal structure (PDB ID code: 4EBZ) onto it. To perform this docking without triggering any steric hindrance between the chitin molecule and the amino acid side chains, only two manual adjustments were made: changing the rotamers of residues Arg122 and Asp142 of LjLYS6. This docking shows that a potential chitin-binding site exists in the LysM2 domain of LjLYS6 and that the residues interacting with chitin in the AtCERK1 chitin structure are well conserved in LjLYS6 (Fig. 2C). In the AtCERK1 chitin crystal structure 10 residues are involved in the recognition of the carbohydrate via either hydrogen bonds or Van der Waals interactions. The structural and primary sequence alignments show that all these potential interactions are indeed conserved in the LjLYS6 ectodomain (Fig. 2E). The structural analysis of LjLYS6 therefore strongly supports a chitin-binding function for LjLYS6.
Ectodomain Conservation in Lotus and Medicago.
Current versions of the Lotus and Medicago genomes suggest that they encode at least 21 and 26 LysM receptor kinases, respectively (58, 59). Phylogenetic analysis shows that LjLYS6 and MtLYK9 are orthologs and their primary sequences share 79.8% amino acid identities (SI Appendix, Fig. S2A). Both the LjLYS6 and MtLYK9 proteins have three LysM modules in their ectodomains and a cytoplasmic kinase where all classical kinase domains are conserved (SI Appendix, Fig. S2C). Aligning the sequence of the LysM2 motif of LjLYS6 and the homologous MtLYK9 with that of the LysM2 motif of AtCERK1 shows an identity of 54% and 57%, respectively. Homology modeling of the MtLYK9 suggests an ectodomain structure similar to LjLYS6 and AtCERK1 and also conservation of the amino acids and structure of the LysM2 chitin-binding site (Fig. 2 D, F, and G). However, the MtLYK9 ectodomain expresses poorly, and sufficient amounts for biochemical studies were not obtained.
Ljlys6 and Mtlyk9 Mutants Are Impaired in Defense Responses.
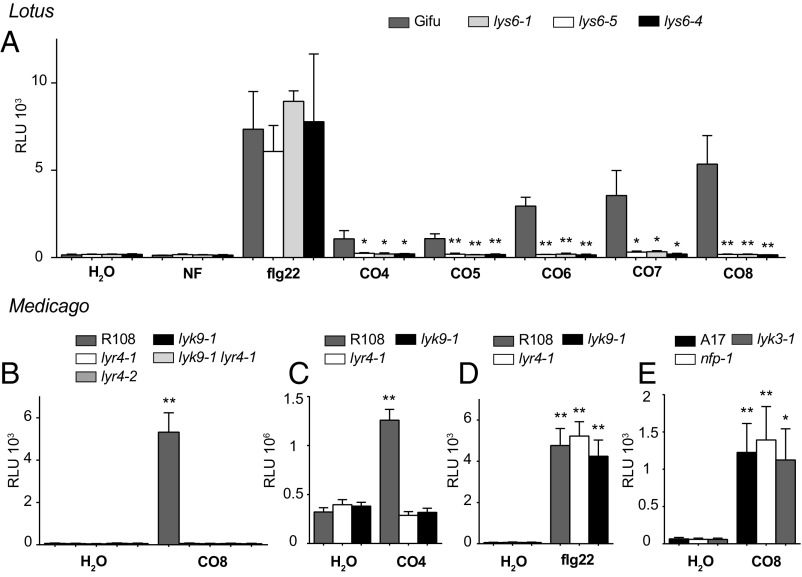
The biochemical and structural results obtained with LjLYS6 suggested that LjLYS6 and MtLYK9 function as first-line pattern recognition receptors for long chitin oligomers (n > 4). Five mutant alleles of Ljlys6 insertion mutants were therefore obtained from the Lotus LORE1 population, and three were characterized in detail. One insertion was obtained from the Medicago Tnt1 collection (SI Appendix, Fig. S2B). Wild-type controls and Ljlys6 mutants were treated with 1-µM concentrations of CO4, CO5, CO6, CO7, and CO8, which elicited a robust response, and the reactive oxygen species (ROS) responses were quantified using a fluorescence assay (Fig. 3A and SI Appendix, Fig. S3 A and B). In Lotus wild-type plants, CO8 elicited the strongest ROS response. The ROS level elicited by CO6 was intermediate; CO4 elicited a significantly lower response than CO8 (Fig. 3A), while purified Mesorhizobium loti Nod factor at 10−12 to 10−6 M did not elicit any ROS response using this assay (Fig. 3A and SI Appendix, Fig. S3 C and D). In contrast to Lotus wild-type plants, the CO8-, CO7-, CO-6, CO5-, and CO4-induced ROS responses were absent in Ljlys6 mutants. However, Ljlys6 mutants responded to flg22 similar to wild type (Fig. 3A). Similarly, the ROS response to 1 µM CO4 and CO8 in Medicago was dependent on MtLyk9 (Fig. 3 B and C), whereas the flg22 response in Mtlyk9 mutants was comparable to that in Medicago wild type (Fig. 3D). Complementation of the Mtlyk9 mutant restored the ROS response (SI Appendix, Fig. S4). In contrast to the Ljlys6 and Mtlyk9 mutants, an unchanged ROS response was observed in Lotus Nod factor receptor mutants Ljnfr1 and Ljnfr5 and in Ljnfr1 Ljnfr5 double mutants (12, 14, 17) and in the corresponding Medicago Mtnfp and Mtlyk3 mutants (Fig. 3 A and E and SI Appendix, Fig. S5) (11, 15).
Fig. 3.
Chitin-induced release of ROS is dependent on LjLys6, MtLyk9, and MtLyr4. (A) ROS production in roots of Lotus Gifu (wild type), Ljlys6-1, Ljlys6-4, and Ljlys6-5 mutants treated with 1 μM CO4, CO5, CO6, CO7, CO8, or Nod factor (NF) or 0.5 μM flg22. (B) Medicago R108 (wild type) and Mtlyk9, Mtlyr4-1, Mtlyr4-2, and Mtlyk9 Mtlyr4-1 double mutants treated with 1 μM CO8. (C) Medicago R108 and Mtlyk9, Mtlyr4-1 treated with 1 μM CO4. (D) Medicago R108 and Mtlyk9, Mtlyr4-1 treated with 1 μM flg22. (E) Medicago A17 (wild type), Mtnfp-1, and Mtlyk3-1 treated with 1 μM CO8. Peak values are plotted as relative light units (RLU), and error bars represent the SEM. *P < 0.05, **P < 0.01 indicates a significant difference (t test) between Ljlys6 mutants and wild-type Lotus or between the elicitor and the respective water-treated control of Medicago.
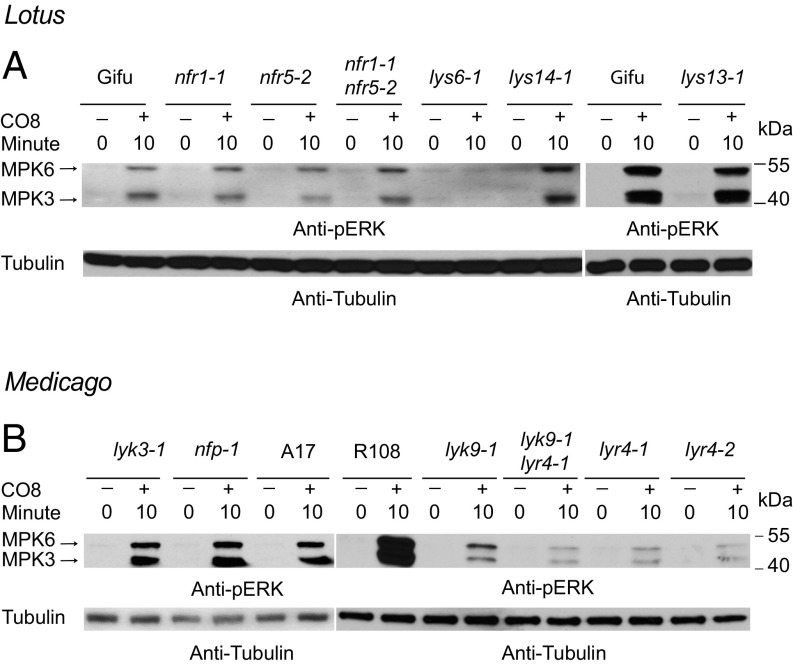
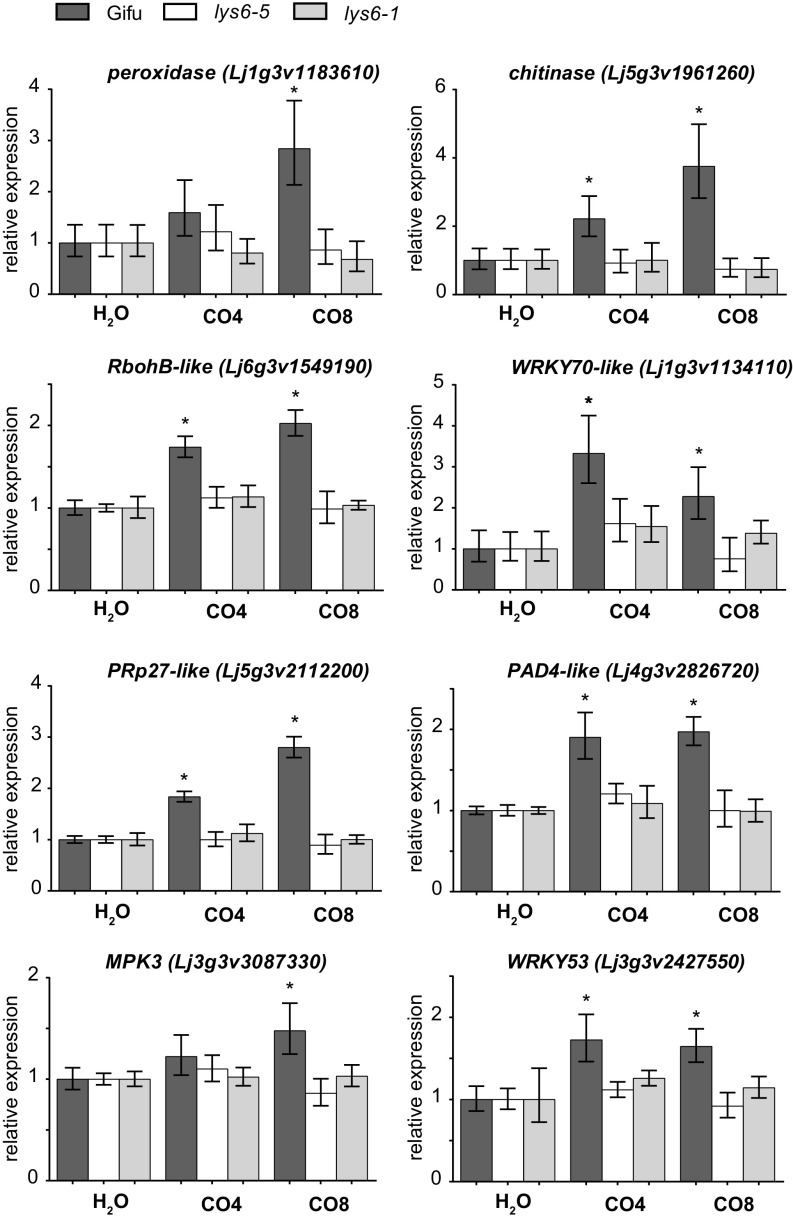
Further evidence for the role of LjLys6 and MtLyk9 in the activation of defense responses was obtained by assaying MAPK3/6 phosphorylation. A CO8-induced increase in phosphorylation of MAPK3/6 was not observed in Ljlys6, while decreased phosphorylation of MAPK3/6 was observed in the Mtlyk9 mutants (Fig. 4). In Ljlys6 there was also no detectable increase after CO4 or CO6 treatment (SI Appendix, Fig. S6). In line with the ROS results, the MAPK3/6 phosphorylation was also unchanged in Ljnfr1, Ljnfr5, Mtnfp, and Mtlyk3 mutants and thus was independent of the symbiotic pathway (Fig. 4). Compared with wild-type control plants transcriptional up-regulation of six typical defense-response genes after chitin (CO4 or CO8) treatment was impaired in Ljlys6 mutants (Fig. 5), while the flg22 response was as in wild type (SI Appendix, Fig. S7). The six genes tested are homologs of the potato respiratory burst oxidase homolog protein B (RbohB-like), Arabidopsis WRKY70 (WRKY70-like), potato pathogenesis-related protein 27 (PRp27-like), and Arabidopsis PHYTOALEXIN DEFICIENT 4 (PAD4-like), as well as the Lotus mitogen-activated protein kinase 3 (MPK3) and WRKY53 genes (60–64).
Fig. 4.
Chitin-induced phosphorylation of MAPK3/6 depends on LjLys6, MtLyk9, and MtLyr4. (A) CO8-induced MAPK3/6 phosphorylation in Lotus Gifu, Ljlys6-1, Ljlys13-1, Ljlys14-1, Ljnfr1-1, and Ljnfr5-2 and in Ljnfr1-1 Ljnfr5-2 double mutants. (B) CO8-induced MAPK3/6 phosphorylation in Medicago R108, A17, Mtlyk9-1, Mtlyr4-1, in Mtlyr4-2, Mtlyk9 Mtlyr4-1 double mutants, and in Mtnfp-1, Mtlyk3-1 mutants.
Fig. 5.
Up-regulation of CO4- and CO8-induced defense-response genes is impaired in Ljlys6 mutants. Transcript levels in roots treated with 1 μM CO4 or CO8 for 1 h were determined by qRT-PCR. Transcripts of a putative peroxidase, a putative chitinase, the Lotus mitogen-activated protein kinase 3 (MPK3), WRKY53, and the Lotus homologs of the potato respiratory burst oxidase homolog protein B (RbohB-like), Arabidopsis WRKY70 (WRKY70-like), potato pathogenesis-related protein 27 (PRp27-like), and Arabidopsis PHYTOALEXIN DEFICIENT 4 (PAD4-like) were measured. Lotus genome v3.0 gene identifiers (GI) are given in parentheses. Values are geometric means of three biological and three technical replicates relative to the respective water-treated control set as 1. Error bars show the 95% CI. An asterisk indicates a significant difference (P < 0.05) compared with the respective water-treated control.
Ljlys6 and Mtlyk9 Mutants Are More Susceptible to Pathogens.
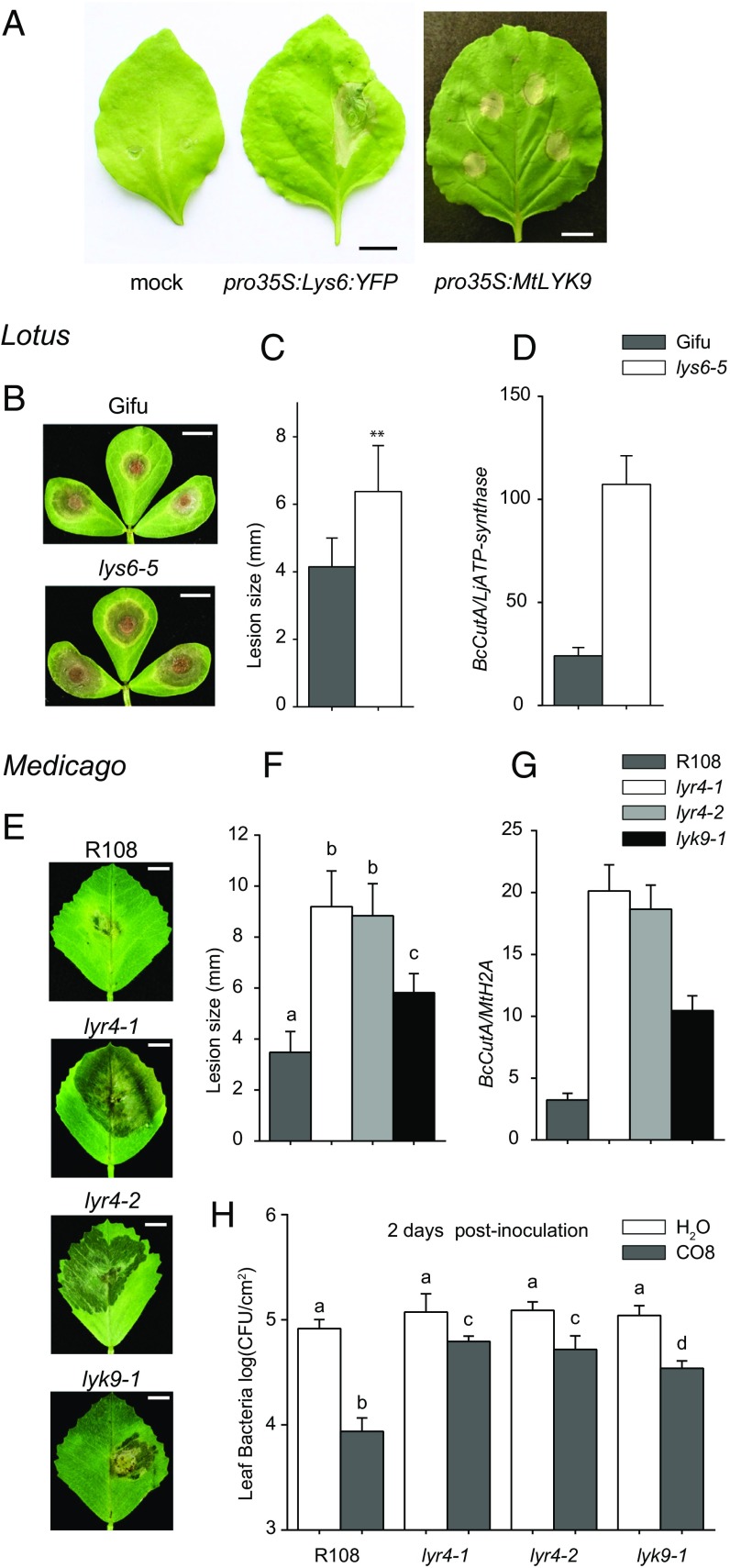
Transient expression of LjLys6 or MtLyk9 in Nicotiana benthamiana leaves induces a cell-death response, and typical tissue collapse zones, characteristic of a defense reaction against pathogens, were observed (Fig. 6A). Direct evidence of this was obtained by pathogen-infection tests of Ljlys6 and Mtlyk9 mutants. An increased lesion size on Ljlys6 and Mtlyk9 leaves infected with Botrytis cinerea and an increased colonization of mutant leaf tissue show that LjLys6 and MtLyk9 are required for resistance toward this fungal pathogen (Fig. 6 B–G). The role of MtLyk9 in plant immunity was further shown using a bacterial pathogen Pseudomonas syringae pv. tomato (Pst DC3000). Priming the plant immune system by prior CO8 treatment decreases bacterial colonization of wild-type leaf tissue, while in Mtlyk9 mutants this response is attenuated (Fig. 6H). Infection studies using a Pst hrcC− mutant show that MtLyk9 is not involved in P. syringae-triggered immunity (SI Appendix, Fig. S8).
Fig. 6.
Ljlys6, Mtlyk9, and Mtlyr4 are more susceptible to pathogens than the corresponding wild-type plants. (A) Transient expression of pro35:LjLys6:YFP and pro35S:MtLyk9 constructs in N. benthamiana leaves induces localized cell death. No cell death was observed on the mock-infiltrated leaves. (Scale bars: 1 cm.) (B, C, E, and F) Ljlys6-5 (B) and Mtlyr4-1, Mtlyr4-2, and Mtlyk9 (E) mutants are more susceptible to B. cinerea than the corresponding wild types and have significantly increased lesion sizes (C and F). (D and G) Quantification of the Botrytis genomic CutA gene relative to the Lotus ATP-synthase (D) and Medicago histone H2A (G) housekeeping genes. (H) CO8-induced priming of immunity is MtLyk9 and MtLyr4 dependent. Suppression of Pst DC3000 infection after CO8 pretreatment is impaired in Mtlyr4-1, Mtlyr4-2, and Mtlyk9 mutants. **P < 0.01 indicates significant differences (t test) in C; different letters over bars in F and H indicate significant differences (P < 0.01; t test).
Mtlyr4 Mutants Are Impaired in Defense Responses.
The Lotus LjLys13 and LjLys14 and Medicago MtLyr4 genes are all homologs of AtLYK5 (SI Appendix, Fig. S2D) known to perceive chitin in Arabidopsis (40), and chitin treatment in Lotus was shown to increase the expression of LjLys13 and LjLys14 in roots and shoots (48). This suggested that the LjLYS13 and LjLYS14 and Medicago MtLYR4 pseudokinase LysM receptors might be involved in chitin perception together with LjLYS6 and MtLYK9. Mutant alleles of Ljlys13 and Ljlys14 responded like wild type to CO4, CO8, and flg22 (SI Appendix, Fig. S9); double mutants could not be obtained since they are tightly linked neighboring genes (48). In contrast, the MtLyr4 gene is a single-copy gene. ROS responses to CO4 and CO8 in two Mtlyr4-1 and Mtlyr4-2 mutant alleles were absent, while complementation of Mtlyr4-1 restored the ROS response (Fig. 3 B and C and SI Appendix, Fig. S4). The response to flg22 was comparable to that of wild type (Fig. 3D). Further evidence for the role of MtLyr4 in the activation of defense responses was obtained by assaying MAPK3/6 phosphorylation. Decreased phosphorylation of MAPK3/6 was observed in Mtlyr4 mutants compared with wild-type plants (Fig. 4B). Supporting these results, an increased lesion size on Mtlyr4-1 and Mtlyr4-2 leaves infected with B. cinerea and an increased colonization of mutant leaf tissue show that MtLyr4 is required for resistance toward this fungal pathogen (Fig. 6 E–H). Compared with Medicago wild-type control plants, transcriptional up-regulation of two typical defense-response genes after chitin (CO4 or CO8) treatment was impaired in Mtlyr4-1 and Mtlyr4-2 mutants (SI Appendix, Fig. S10). Biochemical studies of the MtLYR4 ectodomain were not possible, as a sufficient amount of protein was not obtained from insect cell expression.
Expression Studies.
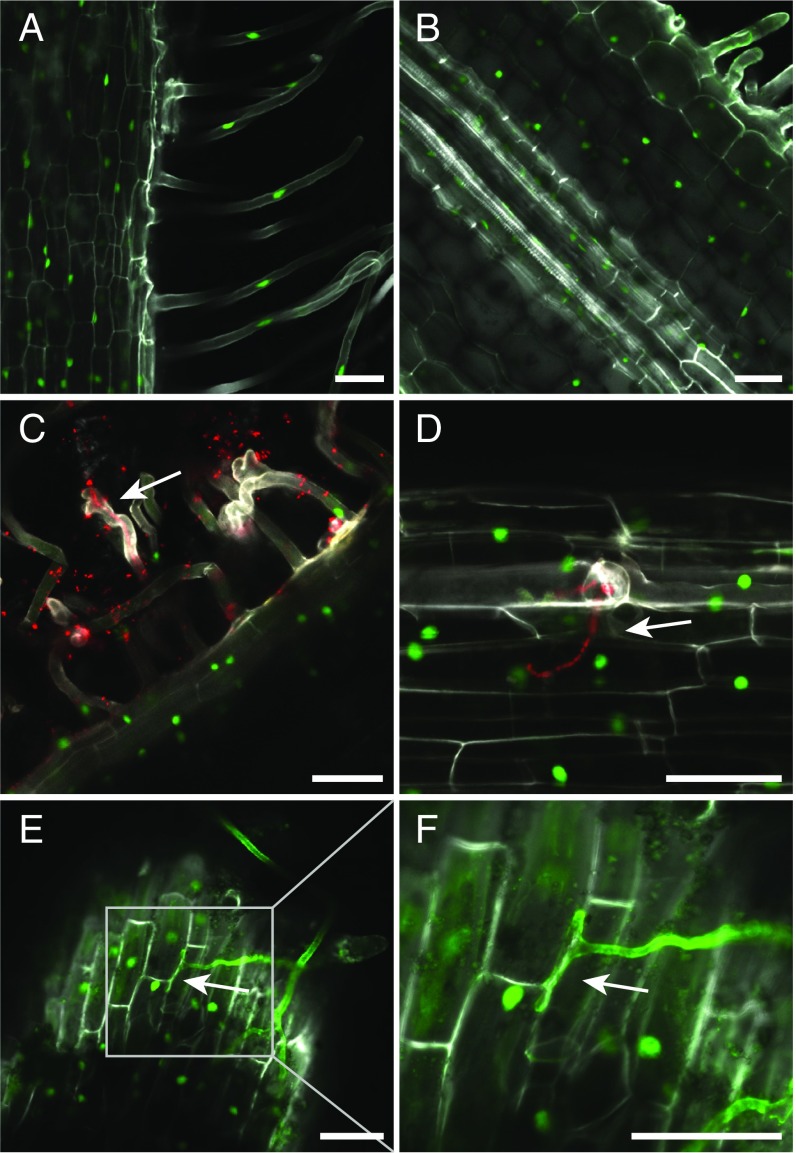
Legume roots are infected by rhizobia during the development of symbiotic nitrogen-fixing root nodules and by AM fungi forming mycorrhiza. Expression of the LjLys6 gene was therefore assayed in the root cells infected by these symbionts and/or in cells adjacent to infection sites. A proLys6:tYFP-NLS reporter construct localizing YFP in the nucleus of transgenic roots was used to resolve LjLys6 gene expression at the cellular level. The LjLys6 promoter was expressed in all root cells (Fig. 7 A and B). Inoculation with M. loti or colonization by AM fungi (Rhizophagus irregularis) did not change the expression pattern or the level of expression (Fig. 7 C–F and SI Appendix, Fig. S11). LjLys6 was expressed in all stages of fungal invasion and also in cells with arbuscules. In contrast, expression was not observed in infected root nodule cells (SI Appendix, Fig. S11), suggesting that LjLys6 is repressed in infected root nodule cells. Further qPCR analysis confirmed that, in contrast to LjNin, a typical symbiotically activated gene, LjLys6 gene expression is not regulated by Nod factor treatment and that LjNin expression is unchanged in Ljlys6 mutants. (SI Appendix, Fig. S12 A and B). Likewise, there was no detectable increase in LjLys6 gene expression following CO4 or CO8 treatment (SI Appendix, Fig. S12C).
Fig. 7.
The LjLys6 promoter is expressed in all root cells of rhizobial and mycorrhized plants. (A and B) Activity of the proLys6:NLS-tYFP reporter in uninfected root. (C and D) Activity of the proLys6:NLS-tYFP reporter in epidermal root cells inoculated with and infected by M. loti labeled with DsRed. (E and F) Activity of proLys6:NLS-tYFP reporter in epidermal root cells infected by R. irregularis. Extraradical hyphae were stained with Alexa Fluor 488 conjugate of wheat germ agglutinin. (F) Close-up of the boxed area in E. Arrows indicate an infection thread in C and D and a hypophodium in E and F. (Scale bars: 50 μM.)
Symbiotic Phenotype of Ljlys6, Mtlyk9, and Mtlyr4 Mutants.
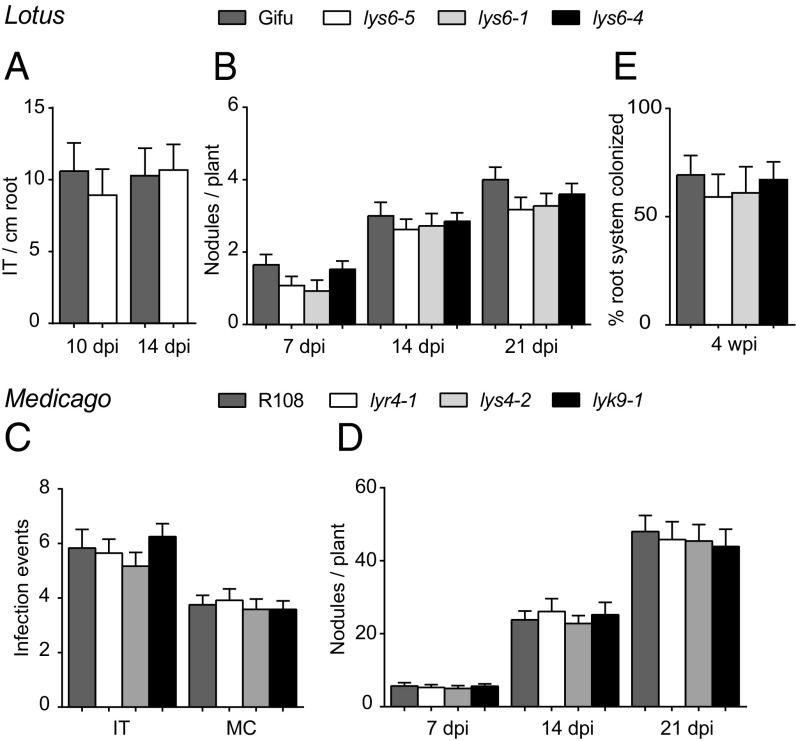
Ljlys6, Mtlyk9, and Mtlyr4 mutants were assayed for nodulation after inoculation with M. loti or Sinorhizobium meliloti, respectively. There were no changes in the number of infection threads or nodules of greenhouse-grown Ljlys6 mutants and also there was no consistent difference in nodulation on plates (Fig. 8 A and B and SI Appendix, Fig. S13). Likewise, there was no change in S. meliloti microcolony, infection thread, or nodulation numbers of Mtlyk9 and Mtlyr4 mutants (Fig. 8 C and D). Ljlys6 mutants were also assayed for mycorrhization after inoculation with AM fungal spores. No significant changes in colonization of Ljlys6 mutant roots by mycorrhizal hyphae or formation of arbuscules were observed (Fig. 8E).
Fig. 8.
Symbiotic phenotype of the Ljlys6, Mtlyk9, and Mtlyr4 mutants. (A) Infection threads (IT) and (B) nodule numbers (n = 40) of Ljlys6 mutants and Lotus wild-type plants (n = 20) inoculated with DsRed-labeled M. loti. (C and D) Microcolonies (MC) (n = 15), infection threads (n = 15) (C) and nodule numbers (n = 15) (D) of S. meliloti-inoculated Mtlyk9 and Mtlyr4 mutants and Medicago wild-type plants at the indicated times. (E) Mycorrhization of Ljlys6 and wild-type Lotus plants (n = 10). Error bars indicate 95% CI. wpi, weeks postinfection.
Discussion
Most land plants have an evolutionarily ancient endosymbiosis with AM fungi colonizing their roots to establish a long-lasting mutualistic relationship, and legumes also engage rhizobia to establish symbiotic nitrogen fixation. Transcriptome profiling in legumes has detected only a modest induction of defense reactions during initiation of endosymbiosis, and, once recognized by the host, microbial access to the root tissue is promoted (12, 14, 65–67). At the same time legumes, like other plants, defend themselves against infection from pathogens. The molecular basis of this dichotomy is one of the open questions that only recently has begun to be addressed systematically (67, 68). Here we identified and characterized chitin receptors from Lotus and Medicago with equivalent functions in plant defense. Biochemical studies show that the LjLYS6 ectodomain distinguishes short-chain CO4/CO5 from long-chain CO6/CO7/CO8 chitin oligomers and binds CO6/CO7/CO8 chitin oligomers with Kd values in the low-micromolar range. Comparable Kd values in the low-micromolar range were found for the AtCERK1 ectodomain (55), while a Kd of 82 nM was estimated for full-length AtCERK1 binding to chitin beads (69), suggesting increased avidity by interaction of full-length proteins. Expression of sufficient amounts of MtLYK9 and MtLYR4 ectodomains for similar biochemical studies has so far been unsuccessful. However, the in vitro results were corroborated using exonic insertion mutants that, like previously analyzed exonic mutants (70), are null mutants as judged by the lack of intact LjLys6, MtLyk9, and MtLyr4 transcripts (SI Appendix, Fig. S2G). Inactivation of the LjLys6, MtLyk9, and MtLyr4 genes eliminates ROS and eliminates or reduces the MAP3/6 kinase phosphorylation induced in planta. In both Lotus and Medicago wild-type plants, the amount of ROS released increases with the chain length of the chitin oligomers. CO8 induces the strongest response, whereas CO4 induces the lowest level of ROS. These in planta data support the direct biochemical binding assays, which show no detectable CO4 binding and weak CO5 binding by LjLYS6, and suggest that the full-length LjLYS6 receptor in its in planta context has a higher ligand affinity than the in vitro-expressed ectodomains used in the biochemical assays. The LjLYS6 receptor may gain avidity by functioning as a dimer or in a larger receptor complex, and this would not be reflected in vitro where the ectodomains most likely act as monomers. In our assay a ROS response to Nod factor application was not detected, and the Ljnfr1, Ljnfr5, Mtnfp, and Mtlyk3 Nod factor receptor mutants also responded to chitin oligomers like the Lotus and Medicago wild-type plants. Previous reports have detected a low, transient ROS release in root hairs of the susceptible zone following Nod factor treatment (71–75). Our assay may not have sufficient sensitivity to detect such localized effects, and we infer that only responses related to chitin-induced host defense reactions are distinguished in the conventional disease-related ROS-response assay adapted for our measurements. On this basis, we conclude that the LjLYS6, MtLYK9, and MtLYR4 receptors are pattern-recognition receptors for chitin and that they can be functionally separated from Nod factor perception. A domain-swap receptor composed of an NFR1 ectodomain and a LjLYS6 kinase was previously shown to partly rescue nodulation of Ljnfr1 mutants (76). This observation highlights the importance of the ectodomain for separating perception of chitin and Nod factor and suggests that the LjLYS6 kinase can substitute, at least in part, for the LjNFR1 kinase.
The phenotypes of the Ljlys6, Mtlyk9, and Mtlyr4 mutants support the notion that LjLYS6, MtLYK9, and MtLYR4 receptors are pattern-recognition receptors for chitin. Short-chain CO4 and CO5 chitin oligomers have been isolated from extracts of germinating mycorrhizal spores (32). Our biochemical binding assay shows that they are poor ligands for the LjLYS6 pattern-recognition receptor, and CO4 and CO5 oligomers may therefore be presymbiotic signals, as previously suggested (32). Further support for a specialized function for the LjLYS6, MtLYK9, and MtLYR4 receptors comes from the increased susceptibility toward pathogens demonstrated in Ljlys6, Mtlyk9, and Mtlyr4 mutants. Finally, the crystal structure of LjLYS6 shows strong similarity to AtCERK1, and the putative chitin-binding site in LysM2 is well conserved, suggesting that LjLYS6 serves as a pattern-recognition receptor for chitin. Comparing the kinase domains of LjLYS6, MtLYK9, OsCERK1, and AtCERK1 shows that LjLYS6 and MtLYK9 are more similar to OsCERK1 than to AtCERK1. The AtCERK1 kinase appears to have diverged from both legume and cereal chitin receptors (76, 77). However, considering the structural and functional similarities within this group of receptors, we have renamed LjLYS6 as “LjCERK6” and the corresponding gene as “LjCerk6” and will use this nomenclature in further communications.
Understanding the signaling leading to symbiosis and defense responses in plants entering endosymbiosis is a challenge for the future. Isolation of Lotus and Medicago mutants in the chitin receptor reported here opens the way for more detailed investigation of the different signal-transduction pathways involved using combinations of mutations removing one or the other or both pathways. The involvement of OsCERK1 in mycorrhizal infection as well as in chitin-triggered defense responses highlights the importance of this approach.
Materials and Methods
LjLys6 Ectodomain Cloning, Expression, and Purification.
The LjLYS6 ectodomain was defined by secondary structure prediction performed with PSIPRED (78), whereas the signal peptide was predicted using the SignalP 4.1 server (79). The predicted ectodomain sequence, without its peptide ranging from Lys27 to Ser223, was codon-optimized for insect cell expression and synthesized with an N-terminal gp67 secretion signal peptide (GenScript). The optimized sequence was cloned into the pOET4 transfer vector (Oxford Expression Technologies) between the SacI and HindIII restriction sites. An additional hexahistidine tag was added in frame to the C terminus of the LjLYS6 ectodomain sequence.
Recombinant Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) baculoviruses were produced in Sf9 cells cultured with TNM-FH medium (Sigma Aldrich) supplemented with 10% (vol/vol) FBS (Gibco), 1% (vol/vol) chemically defined lipid concentrate (Gibco), and 1% (vol/vol) penicillin/streptomycin (Pen/Strep) (10,000 U/mL; Life Technologies). The flashBAC GOLD kit (Oxford Expression Technologies) was used according to the manufacturer’s instructions. The virus was amplified until a third-passage virus culture of 500 mL was obtained.
Protein expression was performed as follows. Sf9 cells infected with 5% (vol/vol) passage-three virus solution were cultured in suspension with serum-free MAX-XP medium (BD Bioscience) or SFX-Insect medium (HyClone) supplemented with chemically defined lipid concentrate and Pen/Strep as described above. The culture was maintained in a shaking incubator at 26 °C for 5 d. Afterward, the medium was harvested by centrifugation in a Sorvall RC5plus centrifuge (SLA-1500 rotor) at 6,000 rpm at room temperature for 25 min to remove cellular debris. The supernatant was then dialyzed against 10 volumes of buffer A [50 mM Tris⋅HCl (pH 8) and 200 mM NaCl] for 1 d at room temperature, and the buffer was exchanged at least four times. The medium was then subjected to another centrifugation step at the same conditions described above to remove insoluble particles.
The resulting supernatant was then loaded on a HisTrap excel column (GE Healthcare) equilibrated with buffer A and recirculated over 3 d at 4 °C using a peristaltic pump. The column was washed with buffer W [50 mM Tris⋅HCl (pH 8), 500 mM NaCl, and 20 mM imidazole] to remove contaminants. The proteins were then eluted with buffer B [50 mM Tris⋅HCl (pH 8), 200 mM NaCl, and 500 mM imidazole]. Imidazole was removed by dialyzing against buffer A, and the proteins were subjected to another round of IMAC purification using a HisTrap HP column (GE Healthcare). N-glycans were removed by treatment with PNGase F (home-made, 1:10, wt/wt) overnight at room temperature. The protein was then concentrated in a Vivaspin column (10-Da cutoff; Sartorius Stedim Biotech) and loaded onto a Superdex 200 10/300 GL size-exclusion column connected to an ÄKTApurifier system (both GE Healthcare) using isocratic elution with buffer A. At each purification step, yield and purity were assayed using SDS/PAGE.
Microscale Thermophoresis.
Purified LYS6 ectodomain and wheat germ agglutinin were fluorescently labeled in microscale thermophoresis (MST) labeling buffer [50 mM KH2PO4 (pH 7.5)] using the Monolith NT.115TM Protein Labeling Kit Blue NHS (NanoTemper Technologies) according to the manufacturer’s instructions. A ligand titration series (CO4–CO8 and negative control maltodextrin) was prepared in which the concentration of the labeled proteins was kept constant at 75 nM, and all experiments were performed in MST running buffer [50 mM K2PO (pH 7.5), 500 mM NaCl, 0.05% Tween-20]. Mixtures were incubated for 30 min at room temperature to facilitate complex formation. MST experiments were conducted on a Monolith NT.115 TM instrument (25 °C, blue LED at 40%, 20% MST power) using standard capillaries (both from NanoTemper Technologies). For each ligand, three or four complete datasets (biological replicates) were obtained and processed using the GraphPad Prism 6 software (GraphPad Software, Inc.) using the sigmoidal dose–response model to obtain equilibrium Kd values.
Crystallization, Data Collection, and Structure Refinement.
The deglycosylated LjLYS6 ectodomain was concentrated to 5 mg/mL after the last purification step and was crystallized in sitting drops by vapor drop diffusion mixing 0.5 μL of protein with 0.5 μL of reservoir solution containing 0.2 M ammonium sulfate, 0.1 M sodium acetate (pH 4.6), and 28% PEG-2000 monomethyl ether (MME) at 4 °C. Crystals were quickly soaked in 0.2 M ammonium sulfate, 0.1 M sodium acetate (pH 4.6), 28% PEG-2000 MME, and 10% glycerol before being cryo-cooled in liquid nitrogen. A full dataset was collected at the I911-3 beamline, MAX-Laboratory II synchrotron. The structure was solved by molecular replacement using Phaser from the Phenix package (80) and the structure of AtCERK1 (PDB ID code: 4EBZ) as a search model (55). Refinement and model quality assessment were performed with the PHENIX package (80), and model building was performed with COOT (81). Data collection and refinement statistics are reported in SI Appendix, Table S1. The structure coordinates and structure factors were deposited at the Protein Data Bank (ID code: 5LS2). The figures were prepared with the PyMOL program (www.pymol.org). Primary sequence alignments were generated with ESPript (82).
Homology Modeling.
The homology modeling of the MtLYK9 ectodomain was performed using the Phyre2 server (83) with the default parameters. The geometry of the model was further optimized manually with Coot (81) and using the phenix.geometry_minimization command from the Phenix package (80). The figure was prepared with PyMOL (www.pymol.org).
Plant Material and Growth Conditions.
The DK09-030067625 (lys6-1), DK11-030085995 (lys6-2), DK07-030053579 (lys6-3), DK13-030106162 (lys6-4), DK07-030054694 (lys6-5), DK07-030056007 (lys13-1), and DK02-030009815 (lys14-1) LORE1 insertion mutant lines were identified by searching Lotus Base (84). L. japonicus wild-type Gifu (85) and mutant seeds were sandpaper scarified and surface sterilized for 15 min with 0.5% sodium hypochlorite. Seedlings were germinated on a stack of wet filter paper (AGF 651; Frisenette ApS) in an upright position in sterile square Petri dishes at 21 °C and then were transferred to slanted agar plates solidified with 1.4% Agar Noble (Difco). Plates for nodule and infection thread counting and gene-expression studies were supplemented with nitrogen-free 1/4× Broughton and Dilworth (B&D) nutrient solution, and the surface of the slope was covered with filter paper. Hairy root transformations were done on agar slopes supplemented with 1/2× Gamborg’s B5 nutrient solution (Duchefa Biochemie) without filter paper on the agar surface. In the promoter:reporter gene experiments plants with transgenic roots were inoculated with Rhizophagus intraradices as described below or were moved to Magenta boxes (Sigma-Aldrich) filled with a 4:1 mixture of lightweight expanded clay aggregate (LECA, 2–4 mm; Saint-Gobain Weber A/S) and vermiculite (size M; Damolin A/S) supplemented with nitrogen-free 1/4× B&D nutrient solution and inoculated with Rhizobium. All plants were grown at 21 °C under 16/8-h light/dark conditions. Rhizobium inoculations were done using an OD600 = 0.02 of a M. loti R7A strain constitutively expressing the fluorescent protein DsRed (24).
The NF16753 (lyk9-1), NF10265 (lyr4-1), and NF15280 (lyr4-2) Tnt1 insertion mutant lines were acquired by reverse screening from the Samuel Roberts Noble Foundation database (52). M. truncatula were cultivated in controlled-environment rooms at 22 °C and 80% humidity (16-h photoperiod and 300 μE⋅m−2⋅s−1). Medicago seeds were scarified in 1 M H2SO4 for 10 min, then sterilized in 10% sodium hypochlorite for 2 min and rinsed in sterile distilled water after several washes. They were then placed on inverted agar plates in the dark at 4 °C for 3 d and at 22 °C for 1 d. The germinated seedlings were transferred to plates or soil according to the experiments to be performed. One-week-old Medicago seedlings were cultivated in a 1:1 mix of Terragreen/sand (Oil-Dri Corporation) and inoculated with 4 mL of Sinorhizobium meliloti 1021 phemA:LacZ at OD600 = 0.02. Fifteen plants were used to count the infection events after 1-wk inoculation, and the nodule numbers were counted at indicated times.
Mycorrhization Analysis.
Ten-day-old seedlings or plants with transgenic roots were placed on a 0.22-µm Global Soil Wetness Project (GSWP) nitrocellulose membrane (Merck Millipore), inoculated with R. intraradices spores (Symbiom), and covered with another nitrocellulose membrane. The spores were suspended in modified Long–Ashton (mLA) solution (0.75 mM MgSO4, 1 mM NaNO3, 1 mM K2SO4, 2 mM CaCl2, 3.2 µM Na2HPO4, 25 µM FeNa-EDTA, 5 µM MnSO4, 0.25 µM CuSO4, 0.5 µM ZnSO4, 25 µM H3BO3, 0.1 µM Na2MoO4). The membrane sandwich was then placed in a sterile Magenta box (Sigma-Aldrich) filled with baked sand and moistened with mLA solution. Plants were grown for 4 wk under 16/8-h light/dark conditions at 21 °C and then were harvested. The roots were cleared in 10% KOH; then the fungal structures were stained with 5% black ink (Quink black; Parker) diluted in 5% acetic acid following the method developed by Vierheilig et al. (86). Mycorrhizal colonization was determined by counting whole root systems according to the method by Trouvelot et al. (87).
ROS Measurements.
Lotus and Medicago seedlings were grown, respectively, on a stack of wet filter paper (AGF 651; Frisenette ApS) for 7 d or on 1.5% water agar for 5 d after germination. Roots were cut into 0.5-cm pieces, collected into white 96-well flat-bottomed polystyrene plates (Greiner Bio-One), and kept overnight in sterile water. Six Lotus roots or one Medicago root were used per well. For the ROS assay, the water on the root pieces was replaced by a reaction mixture consisting of 20 µM luminol (Sigma) or 0.5 mM L-012 (Wako Chemicals), 5 (for Lotus) or 10 (for Medicago) µg/mL horseradish peroxidase (Sigma), and the respective elicitor. The chitin elicitors tetra-N-acetyl-chitotetraose (CO4), penta-N-acetyl-chitopentaose (CO5), hexa-N-acetyl-chitohexaose (CO6), hepta-N-acetyl-chitoheptaose (CO7), and octa-N-acetyl-chitooctaose (CO8) were used at 1-µM concentrations. flg22, the 30–51 22-amino acid peptide of Pseudomonas aeruginosa flagellin, was used at 0.5 µM (for Lotus) or 1 µM (for Medicago). For Lotus, CO4, CO5, and CO6 were obtained from Megazyme, CO7 and CO8 from IsoSep, and flg22 from Alpha Diagnostic. For Medicago, CO4 and CO8 were synthesized by Sébastien Fort, Université Grenoble Alpes, Grenoble, France, and flg22 was obtained from EZBiolab. In the negative-control wells the elicitor was replaced by water. Luminescence in the 15-min assays was recorded with a Varioskan Flash Multimode Reader (Thermo Scientific). In one repetition, at least three wells were measured for every treatment for every genotype. At least three repetitions were conducted with similar results.
MAPK3/6 Phosphorylation Analyses.
For Lotus, scarified and surface-sterilized seeds were imbibed overnight in sterile water with gentle shaking at 4 °C. Twenty imbibed seeds were placed in a sterile polystyrene plant container (In Vitro A/S) on a specially fabricated metal grid allowing the roots to grow up to 4 cm. Sterile water on the bottom of the container and a stack of wet filter paper on top of the seeds maintained humidity. The roots were grown for 7 d in complete darkness at 21 °C before the treatment. Treatments were done by placing the metal grid holding the roots in elicitor solution followed immediately by freezing in liquid nitrogen. For Medicago, plants were grown on modified Fahraeus plant medium (FP, pH 7.5) for 1 wk. Then 50- to 100-mg plant roots were collected, cut into 0.5-cm strips, and incubated in sterile water overnight. The next day the root pieces were treated with the respective elicitor and then frozen in liquid nitrogen; 1 µM CO4, CO6, or CO8 or 0.5 µM flg22 was used as elicitor, and sterile water was the negative control. Treatment time was 10 min with COs and water or 15 min with flg22, which peaks later. Total root protein was isolated in a buffer containing 50 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 15 mM EGTA (pH 7.5), 10 mM MgCl2, 0.1% Triton X-100, 1 mM NaF, 30 mM β-glycerophosphate, 5 mM DTT, 0.5 mM PMSF, 1% protease inhibitor mixture (P9599; Sigma), 1% phosphatase inhibitor mixture 2 (P5726; Sigma), and 1% phosphatase inhibitor mixture 3 (P0044; Sigma) for Lotus or 50 mM Hepes-KOH (pH 7.5), 150 mM KCl, 1 mM EDTA, 0.5% Triton-X 100, 1 mM DTT, complete protease inhibitors (Roche), and phosphatase inhibitors (Roche) for Medicago. Equal amounts of total protein were electrophoresed by 10% SDS–PAGE. Twin gels were run to visualize the phospho-MAPK3/6 and α/β-tubulin bands using anti–phospho-p44/42 MAPK (no. 4370; Cell Signaling) and anti–α/β-tubulin (no. 2148; Cell Signaling) for Lotus or anti–α/β-tubulin (Sigma) for Medicago.
Gene-Expression Analysis.
Germinated Lotus seedlings were grown on 1/4× B&D medium slopes covered with filter paper. The roots were kept in darkness by placing a metal spacer below the hypocotyl of the plants inside the Petri dish and aluminum foil around the outside of the lower part of the Petri dish, while the shoots were kept under 16-h/8-h light/dark conditions. Ten-day-old plants were treated with 10 nM M. loti R7A Nod factors for 24 h or with 1 µM CO4 or CO8 for 1 h in parallel with water controls. Root messenger RNAs were isolated using the Dynabeads mRNA DIRECT kit (Ambion) followed by cDNA synthesis with RevertAid M-MuLV Reverse Transcriptase (Fermentas) using oligo(dT) primer. Genomic DNA contamination was tested with primers specific for the Nin gene promoter region. The real-time qPCR reactions were performed in a Lightcycler480 instrument (Roche) using the LightCycler 480 SYBR Green I master reaction mix (Roche). ATP-synthase (ATP), ubiquitin-conjugating enzyme (UBC), and protein phosphatase 2A (PP2A) were used as internal control genes. Primer specificity was tested by melting curve analysis and sequencing the PCR products. Relative quantification was done with LightCycler 480 Software (release 1.5.0; Roche) to determine the efficiency-corrected relative transcript levels normalized to a calibrator sample and then the geometric mean, and the corresponding 95% CI of three biological and three technical replicates was calculated.
M. truncatula R108, lyr4-1, and lyr4-2 plants were grown on BNM plates for 5 d and then were carefully transferred to liquid buffered nodulation medium (BNM) with DMSO (control) or elicitors (100 nM flg22, 100 nM CO4, or 100 nM CO8) for 6 h. Roots were harvested and snap-frozen in liquid nitrogen. Total RNA was extracted using the Qiagen RNeasy Plant mini kit. One microgram of total RNA was used for cDNA synthesis with the iScript cDNA Synthesis Kit (Bio-Rad). Gene expression was measured by an ABI 7500 instrument using SYBR Green PCR master mix (Bio-Rad). The M. truncatula histone H2A gene was used as endogenous control, and fold induction was calculated for the different treatments relative to the DMSO-treated wild-type R108.
The complete list of primers used for RT-PCR is shown in SI Appendix, Table S2.
Promoter Activity Analysis.
A putative promoter region of 2,496 bp upstream of the translation start of Lys6 was used to drive a triple YFP reporter carrying a nuclear localization signal on the C terminus. The construct was assembled in a pIV10 vector backbone using the Golden Gate method (88). Agrobacterium rhizogenes-mediated hairy root transformation was used to introduce the pIV10:proLys6:tYFP-NLS construct into L. japonicus Gifu plants using the strain AR1193 (89, 90). Plants with transgenic roots were inoculated with either M. loti R7A DsRed or R. intraradices as described above. For better visualization, extraradical hyphae on the mycorrhized roots were stained with wheat germ agglutinin Alexa Fluor 488 conjugate (Invitrogen). Root and nodule sections (60- to 100-µm thick) were prepared in 4% water agar with a VT1000 S vibrating-blade microtome (Leica). Imaging was done with a Zeiss LSM 780 laser-scanning confocal microscope. For image processing Zen (black edition) 2011 software was used (Zeiss).
Mutant Complementation.
For the complementation experiments synthesized coding sequences of Medicago Lyr4 or Lyk9 under the control of their own promoter (1,245 bp and 2,340 bp, respectively) and a 35S terminator were assembled in a Golden Gate vector carrying a DsRed selection marker. The constructs were transformed in M. truncatula roots by hairy root transformation. After 3 wk 10 individually transformed roots were cut into 0.5-cm fragments and tested in a ROS assay.
Transient Gene Expression in N. benthamiana Leaves.
The cauliflower mosaic virus (CaMV) 35S promoter was used to express the Lotus LYS6 and Medicago LYK9 receptors in N. benthamiana leaves. A. tumefaciens AGL1 was used for the infiltration, and AGL1 without an expression vector was included for the mock control. Liquid cultures of A. tumefaciens AGL1 were grown to high densities. Cells were harvested by centrifugation and suspended in infiltration buffer consisting of 10 mM Mes, 10 mM MgCl2, and 450 µM acetosyringone. Final concentrations of OD600 = 0.2 for the clones carrying the respective expression construct (or the mock control) were used together with OD600 = 0.02 concentration of an A. tumefaciens strain expressing the silencing suppressor p19. Leaves of 3- to 4-wk-old N. benthamiana plants were infiltrated with a syringe and analyzed 3 d after infiltration.
Phylogenetic Analysis.
Phylogenetic analysis was done using the MEGA6 package (91). Full-length amino acid sequences were aligned with ClustalW. Maximum likelihood phylogeny estimation was done using the Jones–Taylor–Thornton (JTT) matrix-based model with discrete Gamma distribution (+G, five categories). One thousand bootstrap replications were performed.
Pathogen Inoculation.
Plants were cultivated in nutrient-rich soil for 6 wk. Pst DC3000 and its hrcC− derivative strain were grown in King’s B (KB) medium (proteose peptone 29 g/L, K2HPO4 1.5 g/L, glycerol 10 mL/L, MgSO4 0.74 g/L) for 2 d. A similar-sized terminal leaflet of each plant was syringe infiltrated with the indicated bacteria strain at OD600 = 0.008; 20 leaves were used as replicates. Leaflets were then transferred to a growth chamber with high humidity. For the CO8 treatment, the plant leaves were infiltrated with 1 µM CO8 for 24 h before inoculation with Pst DC3000. The leaf punches were collected at the indicated time points and ground in sterile water. The suspension liquid then was diluted to different concentrations with water and spread on KB plates. Colonies were counted after 2 d incubation at 30 °C.
B. cinerea strain B05.10 was cultivated on Difco potato dextrose agar (Thermo Fisher) 39 g/L medium as described (92). Spores were suspended in 1/4× potato dextrose broth with 0.02% Tween 80. Six-week-old Medicago or Lotus leaves were detached and put on 0.6% water agar with 5-µL droplet suspensions of spores (2.5 × 105 spores/mL) on the center of the leaf discs. The lesion size was documented and measured at 3 d postinfection (dpi). To quantify fungal growth, total DNA was isolated from leaf discs, and copy numbers of the Botrytis Cutinase A gene were determined relative to Lotus ATP-synthase or Medicago histone H2A housekeeping genes, respectively (93).
Supplementary Material
Acknowledgments
We thank Finn Pedersen and Karina Kristensen for taking care of plants in the greenhouse. This work was supported by Danish National Research Foundation Grant DNRF79, by the Bill and Melinda Gates Foundation as part of Engineering the Nitrogen Symbiosis for Africa, and by Biotechnology and Biological Sciences Research Council Grant BB/J004553/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5LS2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706795114/-/DCSupplemental.
References
- 1.Oldroyd GED, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 2.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 3.Bordenave CD, et al. Defense responses in two ecotypes of Lotus japonicus against non-pathogenic Pseudomonas syringae. PLoS One. 2013;8:e83199. doi: 10.1371/journal.pone.0083199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben C, et al. Natural diversity in the model legume Medicago truncatula allows identifying distinct genetic mechanisms conferring partial resistance to Verticillium wilt. J Exp Bot. 2013;64:317–332. doi: 10.1093/jxb/ers337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben C, et al. MtQRRS1, an R-locus required for Medicago truncatula quantitative resistance to Ralstonia solanacearum. New Phytol. 2013;199:758–772. doi: 10.1111/nph.12299. [DOI] [PubMed] [Google Scholar]
- 6.Zgadzaj R, et al. A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genet. 2015;11:e1005280. doi: 10.1371/journal.pgen.1005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zgadzaj R, et al. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc Natl Acad Sci USA. 2016;113:E7996–E8005. doi: 10.1073/pnas.1616564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peck MC, Fisher RF, Long SR. Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J Bacteriol. 2006;188:5417–5427. doi: 10.1128/JB.00376-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper JE. Early interactions between legumes and rhizobia: Disclosing complexity in a molecular dialogue. J Appl Microbiol. 2007;103:1355–1365. doi: 10.1111/j.1365-2672.2007.03366.x. [DOI] [PubMed] [Google Scholar]
- 10.Dénarié J, Debellé F, Promé J-C. Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 11.Catoira R, et al. The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development. 2001;128:1507–1518. doi: 10.1242/dev.128.9.1507. [DOI] [PubMed] [Google Scholar]
- 12.Madsen EB, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 13.Limpens E, et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 14.Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 15.Amor BB, et al. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003;34:495–506. doi: 10.1046/j.1365-313x.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- 16.Arrighi J-F, et al. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madsen LH, et al. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haney CH, et al. Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell. 2011;23:2774–2787. doi: 10.1105/tpc.111.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madsen EB, et al. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 2011;65:404–417. doi: 10.1111/j.1365-313X.2010.04431.x. [DOI] [PubMed] [Google Scholar]
- 20.Broghammer A, et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 22.Oldroyd GED, Downie JA. Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol. 2006;9:351–357. doi: 10.1016/j.pbi.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Miwa H, Sun J, Oldroyd GED, Downie JA. Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant Microbe Interact. 2006;19:914–923. doi: 10.1094/MPMI-19-0914. [DOI] [PubMed] [Google Scholar]
- 24.Kelly SJ, et al. Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol Plant Microbe Interact. 2013;26:319–329. doi: 10.1094/MPMI-09-12-0227-R. [DOI] [PubMed] [Google Scholar]
- 25.Kawaharada Y, et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature. 2015;523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 26.Kawaharada Y, et al. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun. 2017;8:14534. doi: 10.1038/ncomms14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meneses CHSG, Rouws LFM, Simoes-Araujo JL, Vidal MS, Baldani JI. Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol Plant Microbe Interact. 2011;24:1448–1458. doi: 10.1094/MPMI-05-11-0127. [DOI] [PubMed] [Google Scholar]
- 28.Buee M, Rossignol M, Jauneau A, Ranjeva R, Bécard G. The pre-symbiotic growth of arbuscular mycorrhizal fungi is induced by a branching factor partially purified from plant root exudates. Mol Plant Microbe Interact. 2000;13:693–698. doi: 10.1094/MPMI.2000.13.6.693. [DOI] [PubMed] [Google Scholar]
- 29.Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell. 2005;17:3489–3499. doi: 10.1105/tpc.105.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 31.Maillet F, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 32.Genre A, et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 33.Op den Camp R, et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331:909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- 34.Buendia L, Wang T, Girardin A, Lefebvre B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 2016;210:184–195. doi: 10.1111/nph.13753. [DOI] [PubMed] [Google Scholar]
- 35.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 36.Kaku H, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan J, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu T, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y, et al. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife. 2014;3:e03766. doi: 10.7554/eLife.03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carotenuto G, et al. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017;214:1440–1446. doi: 10.1111/nph.14539. [DOI] [PubMed] [Google Scholar]
- 42.Willmann R, et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA. 2011;108:19824–19829. doi: 10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyata K, et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014;55:1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, et al. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015;81:258–267. doi: 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- 45.Liang Y, et al. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science. 2013;341:1384–1387. doi: 10.1126/science.1242736. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X-C, et al. Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007;144:623–636. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiu S-H, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lohmann GV, et al. Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol Plant Microbe Interact. 2010;23:510–521. doi: 10.1094/MPMI-23-4-0510. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X-C, Cannon SB, Stacey G. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 2009;9:183. doi: 10.1186/1471-2148-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbański DF, Małolepszy A, Stougaard J, Andersen SU. Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J. 2012;69:731–741. doi: 10.1111/j.1365-313X.2011.04827.x. [DOI] [PubMed] [Google Scholar]
- 51.Fukai E, et al. Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J. 2012;69:720–730. doi: 10.1111/j.1365-313X.2011.04826.x. [DOI] [PubMed] [Google Scholar]
- 52.Tadege M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 53.Małolepszy A, et al. The LORE1 insertion mutant resource. Plant J. 2016;88:306–317. doi: 10.1111/tpj.13243. [DOI] [PubMed] [Google Scholar]
- 54.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 55.Liu T, et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336:1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 56.Wong JEMM, et al. An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase. Acta Crystallogr D Biol Crystallogr. 2015;71:592–605. doi: 10.1107/S139900471402793X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sánchez-Vallet A, et al. Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. Elife. 2013;2:e00790. doi: 10.7554/eLife.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato S, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang H, et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics. 2014;15:312. doi: 10.1186/1471-2164-15-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshioka H, et al. Induction of plant gp91 phox homolog by fungal cell wall, arachidonic acid, and salicylic acid in potato. Mol Plant Microbe Interact. 2001;14:725–736. doi: 10.1094/MPMI.2001.14.6.725. [DOI] [PubMed] [Google Scholar]
- 61.Li J, Brader G, Palva ET. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi X, Tian Z, Liu J, van der Vossen EAG, Xie C. A potato pathogenesis-related protein gene, StPRp27, contributes to race-nonspecific resistance against Phytophthora infestans. Mol Biol Rep. 2012;39:1909–1916. doi: 10.1007/s11033-011-0937-5. [DOI] [PubMed] [Google Scholar]
- 63.Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wan J, Zhang S, Stacey G. Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol. 2004;5:125–135. doi: 10.1111/j.1364-3703.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 65.Limpens E, et al. Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS One. 2013;8:e64377. doi: 10.1371/journal.pone.0064377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cabeza RA, et al. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N2 fixation before emerging P deficiency reaches the nodules. J Exp Bot. 2014;65:6035–6048. doi: 10.1093/jxb/eru341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berrabah F, Ratet P, Gourion B. Multiple steps control immunity during the intracellular accommodation of rhizobia. J Exp Bot. 2015;66:1977–1985. doi: 10.1093/jxb/eru545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giovannetti M, Mari A, Novero M, Bonfante P. Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Front Plant Sci. 2015;6:480. doi: 10.3389/fpls.2015.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iizasa E, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pedersen CT, et al. N-glycan maturation mutants in Lotus japonicus for basic and applied glycoprotein research. Plant J. 2017;91:394–407. doi: 10.1111/tpj.13570. [DOI] [PubMed] [Google Scholar]
- 71.Bueno P, et al. Time-course of lipoxygenase, antioxidant enzyme activities and H2O2 accumulation during the early stages of Rhizobium–legume symbiosis. New Phytol. 2001;152:91–96. doi: 10.1046/j.0028-646x.2001.00246.x. [DOI] [PubMed] [Google Scholar]
- 72.Santos R, Hérouart D, Sigaud S, Touati D, Puppo A. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol Plant Microbe Interact. 2001;14:86–89. doi: 10.1094/MPMI.2001.14.1.86. [DOI] [PubMed] [Google Scholar]
- 73.Ramu SK, Peng H-M, Cook DR. Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol Plant Microbe Interact. 2002;15:522–528. doi: 10.1094/MPMI.2002.15.6.522. [DOI] [PubMed] [Google Scholar]
- 74.Peleg-Grossman S, Volpin H, Levine A. Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J Exp Bot. 2007;58:1637–1649. doi: 10.1093/jxb/erm013. [DOI] [PubMed] [Google Scholar]
- 75.Cárdenas L, Martínez A, Sánchez F, Quinto C. Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs) Plant J. 2008;56:802–813. doi: 10.1111/j.1365-313X.2008.03644.x. [DOI] [PubMed] [Google Scholar]
- 76.Nakagawa T, et al. From defense to symbiosis: Limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J. 2011;65:169–180. doi: 10.1111/j.1365-313X.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- 77.De Mita S, Streng A, Bisseling T, Geurts R. Evolution of a symbiotic receptor through gene duplications in the legume-Rhizobium mutualism. New Phytol. 2014;201:961–972. doi: 10.1111/nph.12549. [DOI] [PubMed] [Google Scholar]
- 78.Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 80.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mun T, Bachmann A, Gupta V, Stougaard J, Andersen SU. Lotus base: An integrated information portal for the model legume Lotus japonicus. Sci Rep. 2016;6:39447. doi: 10.1038/srep39447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2:487–496. [Google Scholar]
- 86.Vierheilig H, Coughlan AP, Wyss U, Piché Y. Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol. 1998;64:5004–5007. doi: 10.1128/aem.64.12.5004-5007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trouvelot A, Kough JL, Gianinazzi-Pearson V. Mesure du taux de mycorhization VA d’un systeme radiculaire. Recherche de methods d’estimation ayant une signification fonctionnelle. In: Gianinazzi-Pearson V, Gianinazzi S, editors. Physiological and Genetical Aspects of Mycorrhizae. INRA; Paris: 1986. pp. 217–221. [Google Scholar]
- 88.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stougaard J, Abildsten D, Marcker KA. The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol Gen Genet. 1987;207:251–255. [Google Scholar]
- 90.Hansen J, Jørgensen JE, Stougaard J, Marcker KA. Hairy roots–A short cut to transgenic root nodules. Plant Cell Rep. 1989;8:12–15. doi: 10.1007/BF00735768. [DOI] [PubMed] [Google Scholar]
- 91.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lloyd SR, Schoonbeek H-J, Trick M, Zipfel C, Ridout CJ. Methods to study PAMP-triggered immunity in Brassica species. Mol Plant Microbe Interact. 2014;27:286–295. doi: 10.1094/MPMI-05-13-0154-FI. [DOI] [PubMed] [Google Scholar]
- 93.Gachon C, Saindrenan P. Real-time PCR monitoring of fungal development in Arabidopsis thaliana infected by Alternaria brassicicola and Botrytis cinerea. Plant Physiol Biochem. 2004;42:367–371. doi: 10.1016/j.plaphy.2004.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.