Abstract
The group of adipokines comprises hundreds of biological active proteins and peptides released from adipose tissue. Alterations of those complex protein signatures are suggested to play a crucial role in the pathophysiology of multifactorial, metabolic diseases. We hypothesized that also the pathophysiology of type-2-diabetes is linked to the dysregulation of the adipocyte secretome. To test this, we investigated mouse models with monogenic defects in leptin signaling which are susceptible to adipositas (C57BL/6 Cg-Lepob (obob)) or adipositas with diabetes (C57BL/KS Cg-Leprdb (dbdb)) according to their genetic background. At the age of 17 weeks, visceral fat was obtained and primary murine adipocytes were isolated to harvest secretomes. Quantitative proteome analyses (LC-ESI-MS/MS) identified more than 800 potential secreted proteins. The secretome patterns revealed significant differences connected to the pathophysiology of obese mice. Pathway analyses indicated that these differences focus on exosome modelling, but failed to provide more precise specifications. To investigate the relationship of secretome data to insulin sensitivity, we examined the content of diabetogenic lipids, i.e., diacylglycerols (DAGs), identified as key players in lipid-induced insulin resistance. In contrast to obob mice, fat tissue of dbdb mice showed elevated DAG content, especially of DAG species with saturated fatty acid C16:0 and C18:0, while unsaturated fatty acid C16:1 were only changed in obob. Furthermore, DAG signatures of the models specifically correlate to secreted regulated adipokines indicating specific pathways. In conclusion, our data further support the concept that the fat tissue is an endocrine organ that releases bioactive factors corresponding to adipose tissue health status.
Keywords: primary adipocyte, mass spectrometry, healthy adipose tissue, diacylglycerol, diabetes and obesity
1. Introduction
Obesity is a worldwide health burden caused by increased energy intake and sedentary lifestyle. It increases the overall risk for life threatening comorbidities including cardiovascular risk, hypertension, pulmonary obstructive syndrome, dyslipidemia, metabolic syndrome or diabetes, and cancer [1,2,3]. Adipose tissue comprises mature adipocytes, preadipocytes and various invasive immune cells which, in sum, act as secretory organ of bioactive proteins, designated as adipokines. The secreted adipokine patterns in a certain metabolic conditions or stage of obesity are thought to reflect the state of the adipose tissue condition and “health” or its “metabolic flexibility” [4].
A recent investigation described the secretome of visceral adipose tissue from two closely related, well-characterized and metabolically healthy mouse strains, i.e., C57BL/Ks (BKS) and C57BL/6 (C57) by combining state-of-the-art protein identification and quantification tools [5]. A reference map comprising about 600 adipokines was generated (http://www.diabesityprot.org). Both commonly used experimental “wild-type” mouse strains differ in their response to metabolic stress. In contrast to animals with C57 genetic background, mice with a BKS genetic background are prone to develop diabetes under such conditions [6,7]. Therefore, mouse models with genetic defects in leptin signaling are obese (obob; loss of function mutation in leptin) or obese and diabetic (dbdb; loss of function mutation in leptin receptor), depending on genetic background.
Obesity is accompanied by disturbed lipid metabolism, elevated levels of free fatty acids (FFA) and triglycerides (TG), either due to over-nutrition or increased hepatic de novo lipid synthesis [8]. Besides adipose tissue, multiple organs, e.g., liver, skeletal muscle, pancreas, or kidneys, are affected. These organs can be the target of ectopic lipid accumulation and obesity-associated insulin resistance. The systemic overflow with increased fluxes of plasma FFA and TGs towards these tissues leads to the ectopic accumulation of lipids and obesity associated insulin resistance, ultimately altering tissue glucose metabolism and affecting blood glucose clearance. Ectopic lipid accumulation is accompanied by the accumulation of bioactive metabolites, e.g., diacylglycerol (DAG), in the various tissues [9]. DAGs are a result of several metabolic fluxes, including triglyceride hydrolysis, triglyceride synthesis, or phosphoinositide hydrolysis. In liver, it has been shown that DAG content is significantly increased in lipid-induced hepatic insulin resistance. DAGs act as second messengers activating members of novel protein kinase C (nPKC) family [10] and the role of DAGs in Golgi/ER vesicular transport is conserved. This raised our hypothesis that an excess of these bioactive metabolites alters the intracellular signaling also in adipose tissue and in consequence, the inter-organ communication in form of the adipocyte secreted protein patterns.
We intended to investigate the specific differences in the secretome of adipocytes in states of obesity and obesity with diabetes. For this, we utilized obese and obese/diabetic mouse models to compare the adipocyte-derived, not fat tissue, secretion pattern of adipokines and return the information to the adipocyte-derived DAG patterns. Our results suggest that DAG-signaling in adipose tissue acts as intermediary between healthy or diabetic state.
2. Results and Discussion
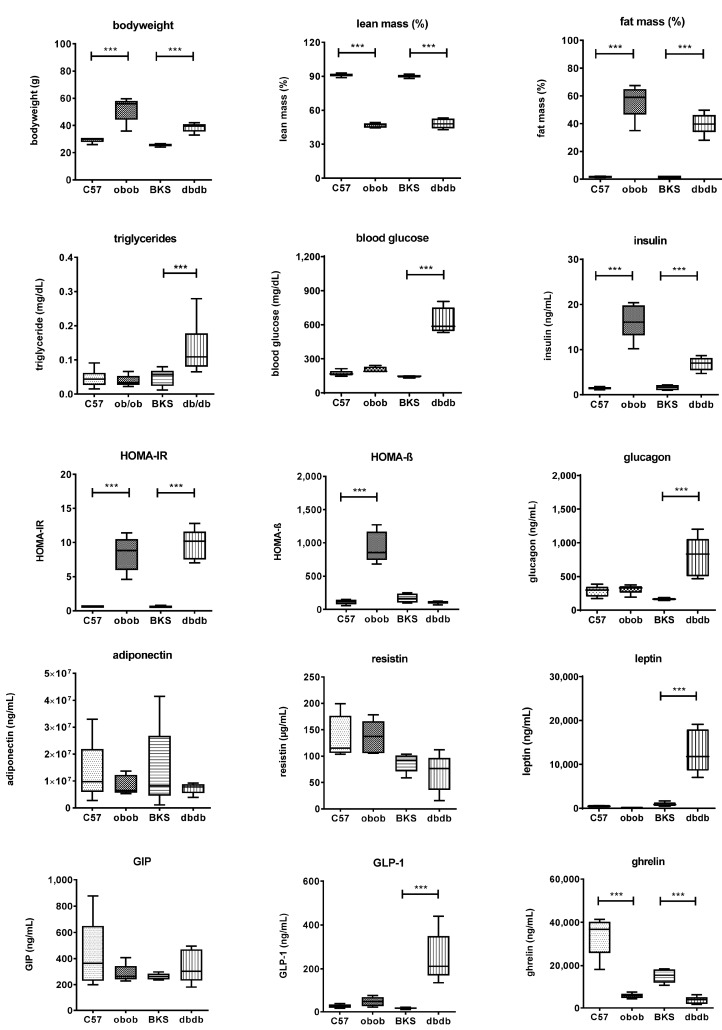
C57BL/KS.Cg-Leprdb (dbdb) mice on C57BL/KS (BKS) genetic background, a well-accepted mouse model of hyperphagia induced obesity with overt diabetes, and C57BL/6.Cg-Lepob (obob) mice on C57BL/6 (C57) background which are protected from diabetes [6,7,11,12,13,14,15] were selected for this study. The clinical characteristics of the investigated mouse models are summarized in Figure 1. The obese mouse model and the obese/diabetic mouse model showed increased body weight with more than 50% fat mass compared to the lean models. Overall, the models also showed specific differences in direct comparison to the genetic background model. Fasting glucose, triglycerides, insulin, and HOMA-IR were each significantly higher in dbdb compared to BKS mice, indicating the definitions of overt diabetes in the dbdb model compared to obob mice. In contrast, in obob mice, insulin and HOMA-IR were significantly higher compared to C57. HOMA-B were strongly elevated while glucose was normal indicating that the β cells are still capable to compensate required insulin levels. Leptin was significantly elevated only in dbdb, as expected from the genetic defect of this model. Similarly, glucagon and glucagon like peptide (GLP)-1 were significantly increased only in dbdb mice. This can be attributed to the diabetic state of these mice, which was confirmed by high HOMA-IR index and thus peripheral insulin resistance. Ghrelin was 4-fold reduced in the obese and 2-fold reduced in obese/diabetic mice, whereas glucose-dependent insulinotropic peptide (GIP), adiponectin or resistin levels did not differ significantly between models.
Figure 1.
Metabolic characterization of C57, BKS, obob and dbdb mice used in the study. Data are expressed as mean ± SD (n = 8 of each phenotype). *** p < 0.001 by Student’s t test.
To further analyze differences between adipose tissue of these mouse models, we determined the content of secreted proteins by mass spectrometry. The overall comparison is given in Supplementary Materials Table S1. According to our experimental design, identified proteins were located outside of intact primary adipocytes. Proteins traffic through the secretory pathway according to their N-terminal signaling sequence to reach their intracellular destination, e.g., an organelle, or to ultimately be secreted. Transport throughout the endomembrane system occurs via the endoplasmic reticulum and the golgi apparatus towards the plasma membrane. Here, the release can occur passive, active channel-mediated, or driven by formation of secretory granules and exosomes. Proteins are targeted according to classical (SP(+)) or non-classical (SP(−)) signal sequences. Proteins without any known signal sequence (NP) are thought to follow e.g., pore-mediated translocation across the plasma membrane, ABC transporter-based secretion or autophagosome/endosome-based secretion [16]. This classification can also help to determine transmembrane proteins [17]. Nevertheless, we cannot completely exclude that some of the proteins might be identified due to apoptosis or autophagy.
The comparisons identified 873 non-redundant proteins. Of these, 216 were assigned to contain a SP(+) signal peptide, 290 were SP(−), thus not carrying a classical signaling peptide and 367 were NP without a signaling domain. The NP proteins contain 164 proteins, which can be assigned to mouse adipocyte exosomes. Nevertheless, we cannot completely role out that some of the identified proteins were derived from autophagy or apoptosis in vitro cultured primary adipocytes after isolation from adipose tissue. The pairwise comparison of the four animal models showed significant alteration (Supplementary Materials Table S2). The comparisons of BKS and dbdb models identified 198 upregulated and 153 downregulated proteins in dbdb (94 SP(+), 118 SP(−), 139 NP). The comparisons of C57 and obob indicated 182 upregulated and 118 downregulated proteins in obob (88 SP(+), 98 SP(−), 114 NP). The individual comparisons indicated in the lean models 108 upregulated and 136 downregulated proteins in BKS (59 SP(+), 87 SP(−), 98 NP), and in the obese and obese/diabetic models 88 upregulated and 112 downregulated proteins in dbdb (61 SP(+), 75 SP(−), 64 NP). Table 1 (see Supplementary Materials Table S2 for complete analyses) summarizes the top 10 up- and down- regulated putative secreted (SP(+), SP(−)) proteins of the comparisons. Furthermore, there were proteins specific for either genotype in the comparisons (Table 2). Additionally, there where 38 solitaire proteins in BKS vs. C57 (8 SP(+), 13 SP(−), 17 NP), 22 in C57 vs. obob (10 SP(+), 6 SP(−), 6 NP), 60 in BKS vs. dbdb (17 SP(+), 17 SP(−), 26 NP) or 10 in dbdb vs. obob (5 SP(+), 2 SP(−), 3 NP).
Table 1.
Top putatively regulated proteins (SP(+), SP(−)) in the comparisons.
| Protein Names | Protein ID | Gene Names | BKS_C57 | BKS_C57 |
|---|---|---|---|---|
| Log2 Fold Change | p-Value (Welch Test) | |||
| Complement factor D | P03953 | Cfd | −5.42 | 1.49 × 106 |
| cAMP-dependent protein kinase type II-β | P31324 | Prkar2b | −2.47 | 2.73 × 106 |
| Apolipoprotein E | P08226 | Apoe | −2.17 | 7.51 × 104 |
| Receptor expression-enhancing protein 6 | Q9JM62 | Reep6 | −2.14 | 9.00 × 104 |
| Lipoprotein lipase | P11152 | Lpl | −2.03 | 8.64 × 103 |
| Prolargin | Q9JK53 | Prelp | −1.90 | 8.59 × 104 |
| 26S protease regulatory subunit 10B | P62334 | Psmc6 | −1.89 | 6.65 × 105 |
| Coiled-coil domain-containing protein 80 | Q8R2G6 | Ccdc80 | −1.89 | 2.33 × 104 |
| Pentraxin-related protein PTX3 | P48759 | Ptx3 | −1.87 | 3.35 × 102 |
| Tenascin | Q80YX1 | Tnc | −1.86 | 6.09 × 105 |
| Inositol polyphosphate 1-phosphatase | P49442 | Inpp1 | 1.52 | 1.24 × 105 |
| Carbonic anhydrase 2 | P00920 | Ca2 | 1.57 | 1.29 × 104 |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 48 kDa subunit | O54734 | Ddost | 1.71 | 9.47 × 105 |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 2 | Q9DBG6 | Rpn2 | 1.72 | 4.48 × 104 |
| Nodal modulator 1 | Q6GQT9 | Nomo1 | 1.78 | 1.74 × 105 |
| 60S ribosomal protein L12 | P35979 | Rpl12 | 1.86 | 9.57 × 105 |
| Ketohexokinase | P97328 | Khk | 1.94 | 9.40 × 105 |
| Carbonyl reductase 3 | Q8K354 | Cbr3 | 1.98 | 1.06 × 104 |
| Carbonic anhydrase 1 | P13634 | Ca1 | 2.38 | 3.45 × 107 |
| Glutathione S-transferase θ-2 | Q61133 | Gstt2 | 3.25 | 4.80 × 109 |
| Protein Names | Protein IDs | Gene Names | obob_C57 | obob_C57 |
| Complement factor D | P03953 | Cfd | −9.46 | 5.52 × 1010 |
| Collagen α-1(XII) chain | Q60847 | Col12a1 | −6.39 | 1.12 × 109 |
| Collagen α-5(VI) chain | A6H584 | Col6a5 | −6.20 | 1.22 × 109 |
| Angiotensinogen | P11859 | Agt | −4.43 | 2.77 × 107 |
| Fructose-1,6-bisphosphatase 1 | Q9QXD6 | Fbp1 | −3.94 | 1.45 × 107 |
| Carboxypeptidase Q | Q9WVJ3 | Cpq | −3.64 | 1.15 × 106 |
| α-Amylase 1 | P00687 | Amy1 | −3.64 | 1.84 × 105 |
| Coiled-coil domain-containing protein 80 | Q8R2G6 | Ccdc80 | −3.47 | 2.67 × 107 |
| Tissue α-l-fucosidase | Q99LJ1 | Fuca1 | −3.28 | 9.84 × 107 |
| Ganglioside GM2 activator | Q60648 | Gm2a | −3.26 | 2.29 × 106 |
| Actin-related protein 2/3 complex subunit 3 | Q9JM76 | Arpc3 | 1.94 | 4.89 × 105 |
| NADH-cytochrome b5 reductase 3 | Q9DCN2 | Cyb5r3 | 2.07 | 5.69 × 105 |
| Epoxide hydrolase 1 | Q9D379 | Ephx1 | 2.12 | 3.21 × 108 |
| Acyl-CoA dehydrogenase 9, mitochondrial | Q8JZN5 | Acad9 | 2.18 | 5.98 × 108 |
| Serum deprivation-response protein | Q63918 | Sdpr | 2.22 | 2.03 × 106 |
| Serpin H1 | P19324 | Serpinh1 | 2.25 | 3.14 × 105 |
| Galectin-3 | P16110 | Lgals3 | 2.25 | 6.50 × 107 |
| GTP:AMP phosphotransferase AK3, mitochondrial | Q9WTP7 | Ak3 | 2.28 | 2.06 × 107 |
| Apolipoprotein A-IV | P06728 | Apoa4 | 2.42 | 6.29 × 107 |
| Interleukin-6 | P08505 | Il6 | 3.19 | 6.96 × 107 |
| Protein Names | Protein IDs | Gene Names | dbdb_BKS | dbdb_BKS |
| Collagen α-5(VI) chain | A6H584 | Col6a5 | −7.87 | 3.53 × 1011 |
| Fructose-1,6-bisphosphatase 1 | Q9QXD6 | Fbp1 | −4.10 | 1.64 × 107 |
| Tissue α-l-fucosidase | Q99LJ1 | Fuca1 | −3.92 | 4.32 × 108 |
| Carboxypeptidase Q | Q9WVJ3 | Cpq | −3.34 | 1.63 × 106 |
| Complement factor D | P03953 | Cfd | −3.33 | 2.71 × 104 |
| Ganglioside GM2 activator | Q60648 | Gm2a | −3.28 | 1.18 × 106 |
| Carboxylesterase 1D | Q8VCT4 | Ces1d | −2.97 | 1.49 × 1011 |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 2 | Q9DBG6 | Rpn2 | −2.87 | 2.22 × 104 |
| Fructose-1,6-bisphosphatase isozyme 2 | P70695 | Fbp2 | −2.80 | 5.61 × 105 |
| Angiotensinogen | P11859 | Agt | −2.44 | 2.67 × 104 |
| Phospholipid transfer protein | P55065 | Pltp | 2.77 | 1.62 × 102 |
| Serum deprivation-response protein | Q63918 | Sdpr | 2.86 | 5.17 × 107 |
| Polymerase I and transcript release factor | O54724 | Ptrf | 2.93 | 1.87 × 108 |
| Platelet-activating factor acetylhydrolase | Q60963 | Pla2g7 | 2.97 | 4.03 × 103 |
| Vimentin | P20152 | Vim | 3.12 | 8.70 × 1011 |
| C-C motif chemokine 2 | P10148 | Ccl2 | 3.37 | 1.23 × 102 |
| Prolargin | Q9JK53 | Prelp | 3.39 | 1.65 × 106 |
| cAMP-dependent protein kinase type II-β | P31324 | Prkar2b | 3.70 | 4.52 × 109 |
| Growth-regulated α protein | P12850 | Cxcl1 | 4.36 | 1.13 × 106 |
| Interleukin-6 | P08505 | Il6 | 5.35 | 2.09 × 1010 |
| Protein Names | Protein IDs | Gene Names | dbdb_obob | dbdb_obob |
| Transthyretin | P07309 | Ttr | −3.06 | 9.14 × 108 |
| ATP-citrate synthase | Q91V92 | Acly | −2.98 | 4.13 × 1011 |
| Sarcosine dehydrogenase, mitochondrial | Q99LB7 | Sardh | −2.34 | 3.51 × 106 |
| GTP:AMP phosphotransferase AK3, mitochondrial | Q9WTP7 | Ak3 | −2.15 | 1.91 × 107 |
| Serpin H1 | P19324 | Serpinh1 | −2.01 | 2.23 × 104 |
| Acyl-CoA dehydrogenase 9, mitochondrial | Q8JZN5 | Acad9 | −1.99 | 2.95 × 107 |
| Vitamin D-binding protein | P21614 | Gc | −1.90 | 9.29 × 107 |
| 3-Hydroxyisobutyrate dehydrogenase, mitochondrial | Q99L13 | Hibadh | −1.86 | 3.24 × 104 |
| Pyruvate dehydrogenase E1 mitochondrial | P35486 | Pdha1 | −1.81 | 1.05 × 103 |
| Citrate synthase, mitochondrial | Q9CZU6 | Cs | −1.70 | 1.07 × 105 |
| Calmodulin | P62204 | Calm1 | 1.51 | 3.03 × 105 |
| Laminin subunit β-2 | Q61292 | Lamb2 | 1.55 | 7.46 × 107 |
| C-X-C motif chemokine 5;GCP-2(1-78);GCP-2(9-78) | P50228 | Cxcl5 | 1.77 | 4.79 × 104 |
| Prolargin | Q9JK53 | Prelp | 1.82 | 3.83 × 103 |
| Pentraxin-related protein PTX3 | P48759 | Ptx3 | 1.84 | 9.82 × 103 |
| Desmin | P31001 | Des | 2.09 | 5.46 × 107 |
| Lactotransferrin | P08071 | Ltf | 2.15 | 9.64 × 104 |
| Collagen α-1(XII) chain | Q60847 | Col12a1 | 4.45 | 1.31 × 107 |
p-Value: post hoc test (ANOVA).
Table 2.
Proteins detected in only one genotype in comparisons.
| Protein Names | Protein ID | Gene Names | SP(+) | SP(−) | NP |
|---|---|---|---|---|---|
| BKS_C57 | |||||
| Tyrosine-protein phosphatase non-receptor type 6 | P29351 | Ptpn6 | + | ||
| NADPH-cytochrome P450 reductase | P37040 | Por | + | ||
| Vacuolar protein sorting-associated protein 13C | Q8BX70 | Vps13c | + | ||
| Annexin A11 | P97384 | Anxa11 | + | ||
| Epidermal growth factor receptor substrate 15 | P42567 | Eps15 | + | ||
| Adenosine deaminase | P03958 | Ada | + | ||
| Pyruvate dehydrogenase E1 subunit α, mitochondrial | P35486 | Pdha1 | + | ||
| Carnitine O-acetyltransferase | P47934 | Crat | + | ||
| Histidine triad nucleotide-binding protein 1 | P70349 | Hint1 | + | ||
| Heat shock protein 75 kDa, mitochondrial | Q9CQN1 | Trap1 | + | ||
| COP9 signalosome complex subunit 8 | Q8VBV7 | Cops8 | + | ||
| Endothelial cell-selective adhesion molecule | Q925F2 | Esam | + | ||
| Carbonic anhydrase 4 | Q64444 | Ca4 | + | ||
| Arsenite methyltransferase | Q91WU5 | As3mt | + | ||
| Deoxyguanosine kinase, mitochondrial | Q9QX60 | Dguok | + | ||
| H-2 class I histocompatibility antigen, D-B α chain | P01899 | H2-D1 | + | ||
| Phosphoserine phosphatase | Q99LS3 | Psph | + | ||
| Mannosyl-oligosaccharide 1,2-α-mannosidase IA | P45700 | Man1a1 | + | ||
| Band 4.1-like protein 2 | O70318 | Epb41l2 | + | ||
| Glucosamine-6-phosphate isomerase 1 | O88958 | Gnpda1 | + | ||
| β-Galactosidase | P23780 | Glb1 | + | ||
| Semaphorin-7A | Q9QUR8 | Sema7a | + | ||
| Very long-chain acyl-CoA dehydrogenase, mitochondrial | P50544 | Acadvl | + | ||
| Inter-α-trypsin inhibitor heavy chain H1 | Q61702 | Itih1 | + | ||
| Glutathione S-transferase θ-1 | Q64471 | Gstt1 | + | ||
| Catenin β-1 | Q02248 | Ctnnb1 | + | ||
| BTB/POZ domain-containing protein KCTD12 | Q6WVG3 | Kctd12 | + | ||
| Retinol-binding protein 2 | Q08652 | Rbp2 | + | ||
| Small nuclear ribonucleoprotein Sm D3 | P62320 | Snrpd3 | + | ||
| Coronin-7 | Q9D2V7 | Coro7 | + | ||
| DNA topoisomerase 2-β | Q64511 | Top2b | + | ||
| Mast cell protease 2 | P15119 | Mcpt2 | + | ||
| Retinol-binding protein 1 | Q00915 | Rbp1 | + | ||
| Phosphoglycolate phosphatase | Q8CHP8 | Pgp | + | ||
| β-Hexosaminidase subunit β | P20060 | Hexb | + | ||
| Putative hydroxypyruvate isomerase | Q8R1F5 | Hyi | + | ||
| S-adenosylmethionine synthase isoform type-2 | Q3THS6 | Mat2a | + | ||
| Bisphosphoglycerate mutase | P15327 | Bpgm | + | ||
| obob_C57 | |||||
| Semaphorin-7A | Q9QUR8 | Sema7a | + | ||
| α-Methylacyl-CoA racemase | O09174 | Amacr | + | ||
| Leukemia inhibitory factor | P09056 | Lif | + | ||
| Fructose-1,6-bisphosphatase isozyme 2 | P70695 | Fbp2 | + | ||
| Thiosulfate sulfurtransferase | P52196 | Tst | + | ||
| Eukaryotic translation initiation factor 3 subunit C | Q8R1B4 | Eif3c | + | ||
| Tyrosine-protein phosphatase non-receptor type 6 | P29351 | Ptpn6 | + | ||
| Signal transducer and activator of transcription 1 | P42225 | Stat1 | + | ||
| Fibromodulin | P50608 | Fmod | + | ||
| Growth-regulated α protein | P12850 | Cxcl1 | + | ||
| Plasminogen activator inhibitor 2, macrophage | P12388 | Serpinb2 | + | ||
| 1-Acyl-sn-glycerol-3-phosphate acyltransferase β | Q8K3K7 | Agpat2 | + | ||
| BTB/POZ domain-containing protein KCTD12 | Q6WVG3 | Kctd12 | + | ||
| C-X-C motif chemokine 3 | Q6W5C0 | Cxcl3 | + | ||
| Epidermal growth factor receptor substrate 15 | P42567 | Eps15 | + | ||
| 26S proteasome non-ATPase regulatory subunit 13 | Q9WVJ2 | Psmd13 | + | ||
| Glutathione S-transferase Mu 7 | Q80W21 | Gstm7 | + | ||
| Hereditary hemochromatosis protein homolog | P70387 | Hfe | + | ||
| Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | P50544 | Acadvl | + | ||
| Cadherin-16 | O88338 | Cdh16 | + | ||
| C-X-C motif chemokine 5 | P50228 | Cxcl5 | + | ||
| α-1-Antitrypsin 1-5 | Q00898 | Serpina1e | + | ||
| dbdb_BKS | |||||
| Tyrosine-protein phosphatase non-receptor type 6 | P29351 | Ptpn6 | + | ||
| Hereditary hemochromatosis protein homolog | P70387 | Hfe | + | ||
| Proteasome activator complex subunit 3 | P61290 | Psme3 | + | ||
| C-C motif chemokine 9 | P51670 | Ccl9 | + | ||
| Tripeptidyl-peptidase 2 | Q64514 | Tpp2 | + | ||
| Vacuolar protein sorting-associated protein 13C | Q8BX70 | Vps13c | + | ||
| Fibromodulin | P50608 | Fmod | + | ||
| Eukaryotic translation initiation factor 2 subunit 1 | Q6ZWX6 | Eif2s1 | + | ||
| Metalloproteinase inhibitor 2 | P25785 | Timp2 | + | ||
| Mannosyl-oligosaccharide 1,2-α-mannosidase IA | P45700 | Man1a1 | + | ||
| Importin-9 | Q91YE6 | Ipo9 | + | ||
| Carnitine O-acetyltransferase | P47934 | Crat | + | ||
| Small glutamine-rich tetratricopeptide repeat-containing protein α | Q8BJU0 | Sgta | + | ||
| T-complex protein 1 subunit ζ | P80317 | Cct6a | + | ||
| Epidermal growth factor receptor substrate 15 | P42567 | Eps15 | + | ||
| Arginase-1 | Q61176 | Arg1 | + | ||
| Granulocyte colony-stimulating factor | P09920 | Csf3 | + | ||
| AP-2 complex subunit mu | P84091 | Ap2m1 | + | ||
| Phosphoserine phosphatase | Q99LS3 | Psph | + | ||
| Histidine triad nucleotide-binding protein 1 | P70349 | Hint1 | + | ||
| Plasminogen activator inhibitor 2, macrophage | P12388 | Serpinb2 | + | ||
| Basigin | P18572 | Bsg | + | ||
| Dynactin subunit 2 | Q99KJ8 | Dctn2 | + | ||
| COP9 signalosome complex subunit 8 | Q8VBV7 | Cops8 | + | ||
| Coatomer subunit ζ-1 | P61924 | Copz1 | + | ||
| 6-Pyruvoyl tetrahydrobiopterin synthase | Q9R1Z7 | Pts | + | ||
| Inter-α-trypsin inhibitor heavy chain H1 | Q61702 | Itih1 | + | ||
| Annexin A11 | P97384 | Anxa11 | + | ||
| Plastin-1 | Q3V0K9 | Pls1 | + | ||
| Eosinophil cationic protein 1 | P97426 | Ear1 | + | ||
| Isopentenyl-diphosphate δ-isomerase 1 | P58044 | Idi1 | + | ||
| Cadherin-1 | P09803 | Cdh1 | + | ||
| 4-Hydroxy-2-oxoglutarate aldolase, mitochondrial | Q9DCU9 | Hoga1 | + | ||
| Nucleoside diphosphate-linked moiety X motif 19, mitochondrial | P11930 | Nudt19 | + | ||
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 48 kDa subunit | O54734 | Ddost | + | ||
| Thiosulfate sulfurtransferase | P52196 | Tst | + | ||
| Cysteine and glycine-rich protein 1 | P97315 | Csrp1 | + | ||
| N(G),N(G)-Dimethylarginine dimethylaminohydrolase 1 | Q9CWS0 | Ddah1 | + | ||
| Small nuclear ribonucleoprotein Sm D3 | P62320 | Snrpd3 | + | ||
| SUMO-conjugating enzyme UBC9 | P63280 | Ube2i | + | ||
| Band 4.1-like protein 2 | O70318 | Epb41l2 | + | ||
| Semaphorin-7A | Q9QUR8 | Sema7a | + | ||
| Adenosine deaminase | P03958 | Ada | + | ||
| Coronin-7 | Q9D2V7 | Coro7 | + | ||
| Nodal modulator 1 | Q6GQT9 | Nomo1 | + | ||
| Phosphoglycolate phosphatase | Q8CHP8 | Pgp | + | ||
| C-X-C motif chemokine 3 | Q6W5C0 | Cxcl3 | + | ||
| β-Hexosaminidase subunit β | P20060 | Hexb | + | ||
| Putative hydrolase RBBP9 | O88851 | Rbbp9 | + | ||
| Ketohexokinase | P97328 | Khk | + | ||
| Interleukin-1 receptor antagonist protein | P25085 | Il1rn | + | ||
| α-Methylacyl-CoA racemase | O09174 | Amacr | + | ||
| Protein kinase C δ-binding protein | Q91VJ2 | Prkcdbp | + | ||
| Retinol-binding protein 1 | Q00915 | Rbp1 | + | ||
| S-adenosylmethionine synthase isoform type-2 | Q3THS6 | Mat2a | + | ||
| Glutathione S-transferase θ-2 | Q61133 | Gstt2 | + | ||
| Cadherin-16 | O88338 | Cdh16 | + | ||
| Bisphosphoglycerate mutase | P15327 | Bpgm | + | ||
| C-X-C motif chemokine 5 | P50228 | Cxcl5 | + | ||
| α-1-Antitrypsin 1-5 | Q00898 | Serpina1e | + | ||
| dbdb_obob | |||||
| Deoxyguanosine kinase, mitochondrial | Q9QX60 | Dguok | + | ||
| Semaphorin-7A | Q9QUR8 | Sema7a | + | ||
| Glutathione S-transferase Mu 7 | Q80W21 | Gstm7 | + | ||
| Phosphoglucomutase-like protein 5 | Q8BZF8 | Pgm5 | + | ||
| 1-Acyl-sn-glycerol-3-phosphate acyltransferase β | Q8K3K7 | Agpat2 | + | ||
| Regulator of microtubule dynamics protein 3 | Q3UJU9 | Rmdn3 | + | ||
| Eosinophil cationic protein 1 | P97426 | Ear1 | + | ||
| H-2 class I histocompatibility antigen, D-B α chain | P01899 | H2-D1 | + | ||
| Interleukin-1 receptor antagonist protein | P25085 | Il1rn | + | ||
| Putative hydroxypyruvate isomerase | Q8R1F5 | Hyi | + |
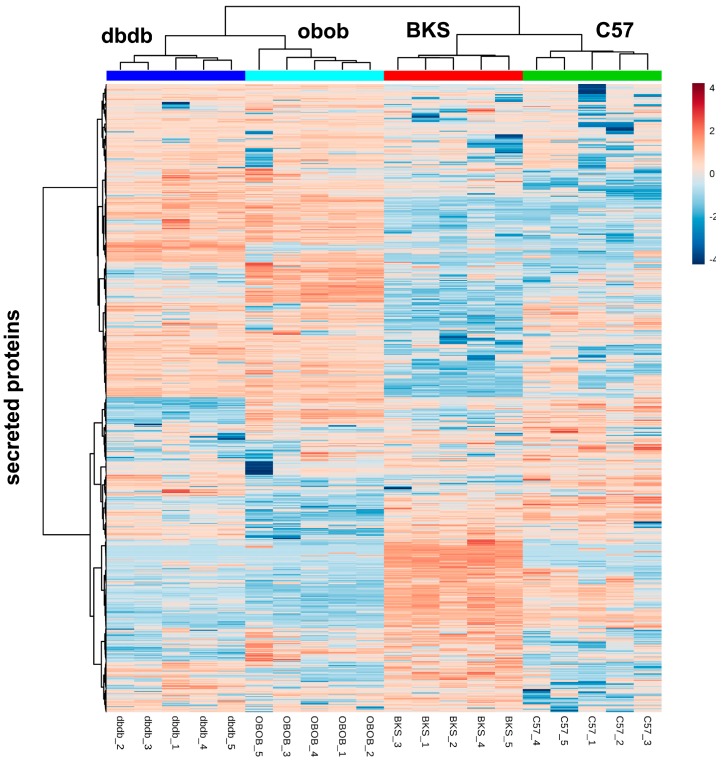
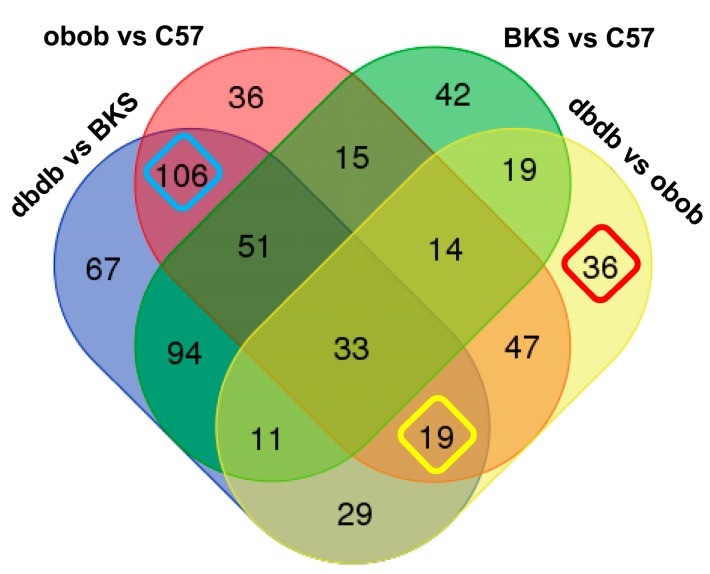
With regard to function, top regulated proteins or solitaire proteins were comprehensibly, among them proteins involved in lipid transport (e.g., ApoE, ApoA4), enzymes (e.g., Lpl, Aad9, Acadvl, Fbp1, Acyl, Ca4, Khk, Pgp), and signaling proteins (e.g., Il6, Sdpr, Gc, Esp15, Rbp-1, Cxcl-5, -3, -9) proteins. Overall, the total adipocyte secreted proteins were able to differentiate lean, obese and the obese/diabetic mouse models (Figure 2). Nevertheless, patterns do not only show overlap according to the lean or obese and obese/diabetic phenotype, but also according to genotype. So, we compared all differentially abundant proteins in the various groups (Figure 3, Supplementary Materials Table S3). With these analyses, we were able to account on any different abundance in conditions depending on genetic background. So, we identified 36 proteins that were solely differential abundant within lean and obese mice (C57 vs. obob), 67 proteins that differed in the comparison of lean to obese/diabetic mice (BKS vs. dbdb), and 42 proteins that differed in the lean background strains. These candidates might be of interest in regard to the phenotype, but still contain the genotype bias.
Figure 2.
Heatmap of all identified adipokines.
Figure 3.
Venn analyses of differential abundant proteins. Proteins with differential abundance in the comparisons C57 vs. BKS, C57 vs. obob, BKS vs. dbdb and dbdb vs. obob (>1.5 fold, one-way ANOVA posthoc p-value < 0.05) were analyzed for overlap to determine genotype specific and genotype independent alterations. Genotype independent differential abundant proteins for “obesity” (n = 106, turquoise), diabetes (n = 36, red) and diabetes despite obesity (n = 19, yellow) are highlighted. p-Value was determined by Welch test. Further information of proteins of all groups are detailed in Supplementary Materials Table S3.
Consistent with the experimental design of adipocyte secretome analyses, all comparisons in databases such as GO, KEGG or IPA annotated to keywords like “extracellular exosome” (FDR = 1.03 × 1010–7.13 × 1027), or “membrane-bounded vesicle” (FDR = 1.03 × 1010–7.08 × 1024) with the highest significance. Other keywords were rather unspecific e.g., “amide metabolism” (n = 7, FDR = 2.02 × 103), or “regulation of protein metabolic process” (n = 13, FDR = 2.02 × 103) for C57 based comparisons. BKS based comparisons also identified general metabolic pathways like “metabolic process” (n = 46, FDR = 2.86 × 105), “regulation of protein transport” (n = 10, FDR = 7.30 × 104), or “protein metabolic process” (n = 23, FDR = 8.94 × 104).
The analyses further identified proteins, that differed in both obese and obese/diabetic models compared to the lean mice (n = 106, “obesity pattern”). These proteins were related to the obese phenotype independent of genotypes investigated. Another 19 proteins differed between obese and obese/diabetic regardless of genotype, and 36 proteins were specific for diabetes despite obesity as they differ among obese and obese/diabetic (Figure 3, Supplementary Materials Table S3). In pathway analyses of these protein sets, functional annotation only indicated direct secretion or vesicle secretion, as expected from experimental design (Supplementary Materials Table S3). Functional annotation identified key words like “extracellular exosome” (n = 70, FDR = 3.26 × 1041) or “membrane-bounded vesicle” (n = 75, FDR = 3.36 × 1041) for the obesity pattern, “extracellular region” (n = 13, FDR = 3.34 × 105) for the diabetes pattern or “extracellular exosome” (n = 24, FDR = 5.89 × 1013), and “membrane-bounded vesicle” (n = 25, FDR = 3.96 × 1012) for the diabetes despite obesity pattern as best hits. Other key terms of potential interest to metabolic energy balance showed lower significance and limited numbers of assigned proteins e.g., mitochondria (BKS vs. dbdb, n = 17, FDR = 1.76 × 103; diabetes despite obesity, n = 15, FRD = 6.44 × 107), lipid metabolism (BKS genotype based differences, n = 11, FDR = 1.12 × 103), lipid catabolic process, lipid- or phospholipid binding (obesity pattern, n = 5, FDR = 3.17 × 103; n = 12, FDR = 4.68 × 104; n = 9, FDR = 4.68 × 104), or fatty acid degradation (diabetic despite obesity pattern, n = 3, FDR = 1.42 × 104) (Supplementary Materials Table S3).
In general, enrichment analyses were used to facilitate the interpretation of numerous genes or proteins which are the usual outcome of hypothesis generating experimental designs. Thus, the accumulation of candidates with known biological function or interaction, either directly experiment proven or deduced from literature, were monitored in a dataset. Knowledge based pathway annotation or gene enrichment analyses can help to classify “Omics” data, but also bares some restrictions. Next to bioinformatics, the main issue being intrinsic to the experimental setting [18]. We use secreted proteins, so enrichment of secreted proteins or related pathways with highest significance confirmed our experimental approach. The other bias, for sure is the limited number of differential proteins in the specific regulations we focus on, which hampers annotations in a general way.
So, we decided to focus on our initial working hypothesis, i.e., to identify alterations in the adipocyte “communication” with regard to specific physiological states. Adipose tissue controls systemic energy storage and needs to expand in regard to metabolic needs. In healthy conditions, this can be due to hyperplasia, but in metabolically affected adipose tissue as in obesity or diabetes, increased ad libitum storage of fatty acids occurs, even in non-adipose tissues. Increased lipid load in these cells favors accumulation of fatty acids (FA)-derived metabolites such as fatty acyl-CoA or DAG which initiated cellular processes via PKC signaling [19]. As chronical process, the cells get insulin resistance with dysfunctional mitochondria resulting in the development of obesity and diabetes. With regard to adipocyte function, the combination of both should alter the DAG patterns in adipocytes, like observed in other insulin-sensitive tissues as liver, skeletal muscle or even pancreas [9,20,21,22].
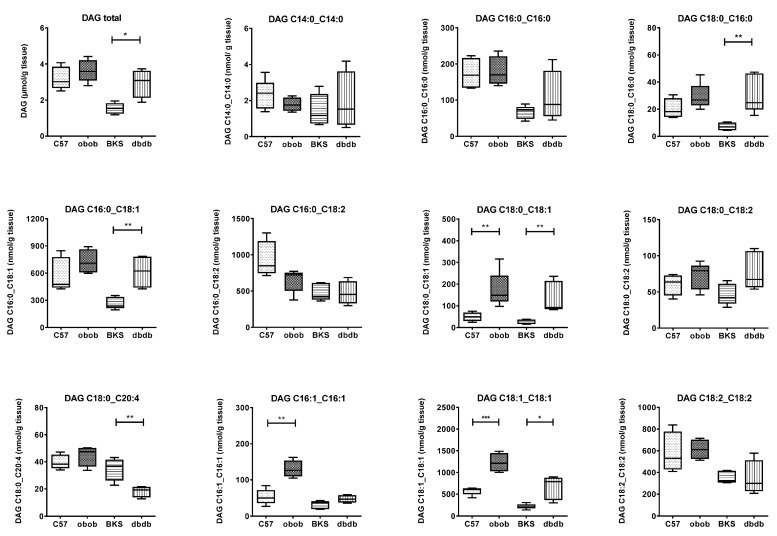
Adipose tissue DAGs were determined by mass spectrometry (Figure 4). In contrary to the obese obob, there was an increase in total DAG content in obese/diabetic dbdb mice compared to their backgrounds. Nevertheless, the comparison of the distinct DAGs revealed that in both obese models the DAG species with the fatty acid C18:1 were equally regulated (C18:0_18:1; C18:1_18:1). DAG species with saturated fatty acid C16:0 and C18:0 were only changed in dbdb (C16:0_C18:0; C16:0_C18:1, C18:0_C20:4), whereas the DAG species with unsaturated fatty acid C16:1 were only changed in obob (C16:1_C16:1).
Figure 4.
Diacylglycerol pattern in adipose tissue of C57, BKS, obob and dbdb mice. Data are given in (μmol or nmol/g tissue) and expressed as mean ± SD (n = 5 of each phenotype). * p < 0.05, ** p < 0.01, *** p < 0.001 by Student’s t test.
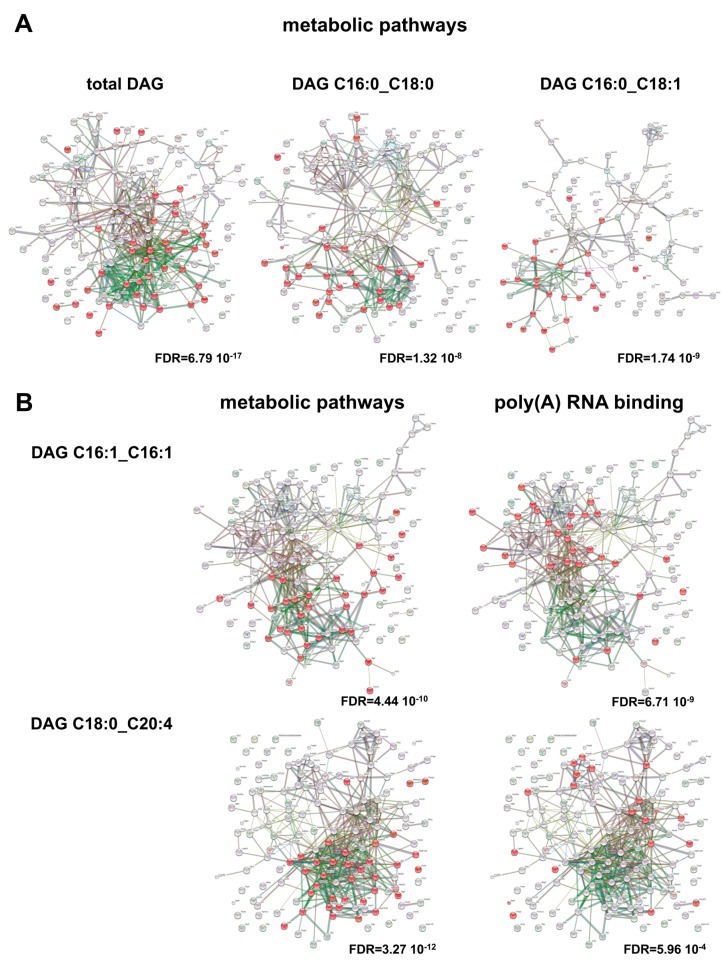
According to our hypothesis, intensities of a vast amount of adipose secreted proteins correlated to the total adipose derived DAGs (n = 152; 23 SP(+), 53 SP(−), 76 NP). Of these 105 proteins (20 SP(+), 36 SP(−), 49 NP) also showed differential abundance in either comparison of mouse phenotypes. In addition, specific DAG species correlate to adipocyte-secreted proteins (Figure 5, Supplementary Materials Table S4). Here, all secreted correlated proteins can be assigned to metabolic active proteins with the highest prevalence. Of note, DAG species specific for obob shows poly(A) RNA binding proteins as highest annotation (poly(A)RNA binding, n = 30, FDR = 6.71 × 109, RNA binding, n = 31, FDR = 4.10 × 107). This is also observed, if not in highest position with DAG species specific for dbdb (n = 20, FDR = 5.96 × 104) (Figure 5). This is of interest as it focused the differences in obesity and obesity/diabetes to the concept of moonlighting enzymes in metabolic control. Moonlighting proteins or gene sharing defines various functions of a certain gene and are independent to alternative splicing, posttranslational modification or multifunctionality. Especially ancestral and conserved proteins in central metabolic processes show moonlighting functions, e.g., glycolysis or tricarboxylic acid cycle enzymes [23]. This process of metabolic regulation can account for expression levels, differential localization, protein interactions and is mediated by binding of RNA species to a distinct, but not necessary active domain of an enzyme. Best known examples of metabolic enzymes regulated by binding of RNA species are GOT2, FASN, or GAPDH [24]. We identified Adk, Aldh6A1, Aldoa, Eno, Lta4h and Hsd17B10 to be secreted from adipocytes and to correlate to DAG species C16:1_C16:1 or DAG species C18:1_C20:4. All of these proteins were previously identified in RNA interaction studies and implicated to have moonlighting functions [24]. For example, the metabolic enzyme fructose-1,6-bisphosphate aldolase (Aldoa) which catalyzes the reversible cleavage of fructose-1,6-bisphosphate to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate in glycolysis and gluconeogenesis pathways, has been shown to regulate insulin-dependent glucose transporter GLUT4 in mouse adipocyte cell lines 3T3-L1 [25]. Furthermore, enolase (Eno) catalyzes the dehydrolyzation of 2-phospho-d-glycerate to phosphoenolpyruvate in glycolysis, but has also been shown to bind plasminogen and to mediate its cell surface peptidase activity [26,27]. So, one could speculate that alterations in such regulatory processes might interfere with the subcellular localization and trafficking of proteins, depending on which functions is favored, and are also an essential target in the overall picture of metabolic regulation.
Figure 5.
Functional network of adipokines correlated to DAG species. Adipokines with significant correlation to total DAG content or indicated DAG species were used for over representation analyses. (A) Adipokines correlated to total DAGs, DAG C16:0_C18:0 DAG C16:0_C18:1 species are enriched in metabolic pathways (highlighted in red); (B) Adipokines correlating to DAG C16:1_C16:1 and DAG C18:0_C20:4 are enriched in metabolic pathways or poly(A) RNA binding (highlighted in red). Enrichment FDR is given. Graphs show results of interaction analyses (https://string-db.org).
3. Materials and Methods
3.1. Mouse Models
C57BL/6 (C57), C57BL/KS (BKS), C57BL/KS.Cg-Leprdb (dbdb) and C57BL/KS.Cg-Lepob (obob) mice were bred and maintained in a regular 12 h light/dark cycle under constant temperature, humidity (22 ± 1 °C, 50 ± 5% humidity), with free access to water and standard laboratory food (Ssniff, Soest, Germany). Mice were sacrificed by CO2 asphyxiation at 17 weeks of age. Mice (n = 5 each genotype, males) were sacrificed at 7 a.m. after 6 h food restriction and visceral adipose tissue was removed [5,28]. Adipose tissue was either processed directly to isolate adipocytes or was snap frozen in liquid nitrogen and stored at −80 °C for lipid analyses. Serum was collected by left ventricular punctuation. The Animal Care Committee of the University Duesseldorf approved all animal care and procedures (Approval#50.05-240-35/06, August 2006).
3.2. Metabolic Characterization of the Mouse Models
Blood parameters were measured at 17 weeks of age (n = 8). Blood glucose was measured with Freestyle™ and leptin, insulin as well as glucagon levels were determines using quantitative Bio-Plex Pro Mouse Diabetes 8-Plex Assay (Bio-Rad, Munich, Germany) according to the manufacturer’s instructions. Data were collected and analyzed using a BioPlex 200 instrument equipped with BioManager analysis software (Bio-Rad). To determine insulin resistance and pancreatic β cell function the surrogate parameters HOMA-IR (homeostatic model assessment of insulin resistance) and HOMA-β (homeostatic model assessment of β cell function) were used. Body composition was measured using nuclear magnetic resonance (n = 9–23/per genotype, Whole Body Composition Analyzer; Echo MRI, Houston, TX, USA).
3.3. Secretome Profiling by Liquid Chromatography (LC)-Electrospray Ionization (ESI)-MS/MS and Data Analyses
Murine mature adipocytes from visceral fat isolated by collagenase digestion were cultured for 24 h (DMEM/F12 without FCS supplementation (Thermofisher Scientific, Darmstadt, Germany)), and secretomes were harvested as described [29]. Data of all mouse models were acquired in parallel as described in detail [5]. In brief, secretome samples were tryptically digested and analyzed using LC-ESI mass spectrometry using an Ultimate 3000 Rapid Separation liquid chromatography system (Dionex/Thermo Scientific, Idstein, Germany). Afterwards, mass spectrometry was carried out (Orbitrap Elite high resolution instrument, Thermo Scientific, Bremen, Germany). For the comparison of mouse strains, log2 PSM values were used. MaxQuant (version 1.4.1.2, Max Planck Institute for Biochemistry, Munich, Germany) was used for protein and peptide identification and quantification with default parameters if not otherwise stated. Searches were carried out using 16.671 mouse sequences from the Swiss-Prot part of UniProtKB (release 9.7.2014) applying the following parameters: mass tolerance precursor (Orbitrap): mass tolerance precursor: 20 ppm firt search and 4.5 ppm after recalibration (Orbitrap), mass tolerance fragment spectra: 0.4 Da (linear ion trap), trypsin specific cleavage (maximum of one missed cleavage site), fixed modification: carbamidomethyl, variable modifications: methionine oxidation and N-terminal acetylation. For peptide and protein acceptance, the false discovery rate (FDR) was set to 1%, only proteins with at least two identified peptides were used for protein assembly. Quantification was carried out using the label-free quantification algorithm implemented in MaxQuant using a minimal ratio count of 2 and the “match between runs” option enabled.
3.4. Lipid Analysis of Adipose Tissue
Extraction, purification and analysis of DAGs from frozen adipose tissue samples was conducted using an LC-MS/MS approach [21]. In brief, 20 mg of adipose tissue was homogenized in 20 mM Tris/HCL, 1 mM EDTA 0.25 mM EGTA, pH 7.4, using a tight-fitting glass douncer (Wheaton Lab Supplies, Birmingham, UK). Internal standard (d517:0-DAG; Avanti Polar Lipids, Alabaster, AL, USA) was added and lipids were extracted according to Folch et al., [30]. Diacylglycerols were separated from triglycerides using solid phase extraction (Sep Pak Diol Cartridegs; Waters, Milford, MA, USA). The resulting lipid phase was dried under a gentle flow of nitrogen and re-suspended in methanol. Diacylglycerols were separated using a Phenomenex Luna Omega column (1.6 µm 100 A; Phenomenex, Torrance, CA, USA) on an Infinity 1290 HPLC system (Agilent Technologies, Waldbronn, Germany) and analyzed by multiple reaction monitoring on a triplequadrupole mass spectrometer (Agilent 6495; Agilent Technologies), operated in positive ion mode.
3.5. Prediction and Annotation of Secretory Proteins
Secretory protein prediction and functional annotation was done using different independent methods. First, protein information of all identified proteins was extracted from the Swiss-Prot database (http://www.uniprot.org/). To assess secretory properties, protein sequences were analysed by SignalP 4.1 [17]; (http://www.cbs.dtu.dk/services/SignalP/), SecretomeP 2.0. [31]; (http://www.cbs.dtu.dk/services/SecretomeP/) and Exocarta (http://www.exocarta.org/) [32]. Literature screening was performed with NCBI/Pubmed (http://www.ncbi.nlm.nih.gov/pubmed) and protein-protein interaction analyses with https://string-db.org.
3.6. Web-Based Functional Annotation
The identification types were uniprot swissprot accession or gene ID, respectively. Information driven analyses including functional annotation was performed with String v10.5 (https://string-db.org) [33], David Bioinformatics Resources 6.8 (https://david.ncifcrf.gov) [34,35], and IPA (IngenuityTM, Qiagen, Hilden, Germany). For differential protein sets expression analyses, expression fold change (1.5×) and expression differences (p-value < 0.05) were analyzed following the core analyses modules. Differential abundant proteins (1.5× fold difference, p-value < 0.05 (one-way ANOVA, post hoc) were analyzed separately for C57 vs. BKS, C57 vs. obob, BKS vs. dbdb and dbdb vs. obob.
3.7. Statistical Methods
Statistical analyses were performed in GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA) and SPSS 22 (IBM, Armonk, NY, USA). Data are given as mean ± standard deviation (SD) and data were directly compared with an unpaired Student’s t test. Figure legends indicate the statistical tests applied for each experiment in detail.
4. Conclusions
We showed that genetic mouse models, which are susceptible to obesity or obesity/diabetes according to their genetic background genotype show phenotype-specific differences in primary adipocyte adipokinome in quantitative proteome analyses. Knowledge based annotation of identified differentially regulated adipokinome did not add much further information. According to the predictive value of DAG-species for lipid metabolism and insulin resistance in liver and skeletal muscle [9], we determined DAG levels also as classifying parameter for lipid metabolism and insulin resistance in adipose tissue. Adipose tissue DAG patterns differ in obesity and obesity/diabetes especially of DAG species with saturated fatty acid C16:0 and C18:0 in diabetes and unsaturated fatty acid C16:1 in obesity or unsaturated fatty acid C20:4 in obesity/diabetes. Our study provides evidence that the analyses of one “Omics”-like secretome might not be sufficient to get insight in a complex phenotypical problem.
Here, the combination of specific DAG species and the holistic pattern of primary adipocyte-secreted proteins helped to get hints to an interacting mechanism and to unravel RNA-binding proteins involved in metabolic control differing in obesity and obesity/diabetes.
Acknowledgments
The work was supported by the German Diabetes Center (DDZ), which is funded by the German Federal Ministry of Health and the Ministry of Innovation, Science, Research and Technology of the state North Rhine-Westphalia. This study was supported in part by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e. V.).
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/9/1928/s1.
Author Contributions
Simon Goeddeke, Gereon Poschmann, Daniel F. Markgraf, Christina Preuss, Waltraud Passlack, Ulrike Nitzgen, Sylvia Jacob, Sonja Hartwig, researched the data. Birgit Knebel and Jorg Kotzka designed experiments, analyzed data and wrote the manuscript. Hans-Dieter Dicken curates the database (http://www.diabesityprot.org). Kai Stühler and Stefan Lehr contributed to design of the secretome analyses and supervised the adipokine identification. All authors have confirmed the final version of the manuscript. Jorg Kotzka is the guarantor of the work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Meldrum D.R., Morris M.A., Gambone J.C. Obesity pandemic: Causes, consequences, and solutions-but do we have the will? Fertil. Steril. 2017;107:833–839. doi: 10.1016/j.fertnstert.2017.02.104. [DOI] [PubMed] [Google Scholar]
- 2.Hall J.E., do Carmo J.M., da Silva A.A., Wang Z., Hall M.E. Obesity-induced hypertension: Interaction of neurohumoral and renal mechanisms. Circ. Res. 2015;116:991–1006. doi: 10.1161/CIRCRESAHA.116.305697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohmann A.E., Goodwin P.J., Chlebowski R.T., Pan K., Stambolic V., Dowling R.J. Association of obesity-related metabolic disruptions with cancer risk and Outcome. J. Clin. Oncol. 2016;34:4249–4255. doi: 10.1200/JCO.2016.69.6187. [DOI] [PubMed] [Google Scholar]
- 4.Ouchi N., Parker J.L., Lugus J.J., Walsh K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011;11:85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwig S., Goeddeke S., Poschmann G., Dicken H.D., Jacob S., Nitzgen U., Passlack W., Stühler K., Ouwens D.M., Al-Hasani H., et al. Identification of novel adipokines differential regulated in C57BL/Ks and C57BL/6. Arch. Physiol. Biochem. 2014;120:208–215. doi: 10.3109/13813455.2014.970197. [DOI] [PubMed] [Google Scholar]
- 6.Coleman D.L., Hummel K.P. Symposium IV: Diabetic syndrome in animals. Influence of genetic background on the expression of mutations at the diabetes locus in the mouse. II. Studies on background modifiers. Isr. J. Med. Sci. 1975;11:708–713. [PubMed] [Google Scholar]
- 7.Hummel K.P., Coleman D.L., Lane P.W. The influence of genetic background on expression of mutations at the diabetes locus in the mouse. I. C57BL-KsJ and C57BL-6J strains. Biochem. Genet. 1972;7:1–13. doi: 10.1007/BF00487005. [DOI] [PubMed] [Google Scholar]
- 8.Unger R.H., Clark G.O., Scherer P.E., Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim. Biophys. Acta. 2010;1801:209–214. doi: 10.1016/j.bbalip.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Petersen M.C., Shulmann G.I. Roles of Diacylglycerols and Ceramides in Hepatic Insulin Resistance. Trends Pharmacol. Sci. 2017;38:649–665. doi: 10.1016/j.tips.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson S.R., Cooper D.R. Specific protein kinase C isoforms as transducers and modulators of insulin signaling. Mol. Genet. Metab. 2006;89:32–47. doi: 10.1016/j.ymgme.2006.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leiter E.H., Chapman H.D., Coleman D.L. The influence of genetic background on the expression of mutations at the diabetes locus in the mouse. V. Interaction between the db gene and hepatic sex steroid sulfotransferases correlates with gender-dependent susceptibility to hyperglycemia. Endocrinology. 1989;124:912–922. doi: 10.1210/endo-124-2-912. [DOI] [PubMed] [Google Scholar]
- 12.Leiter E.H., Reifsnyder P.C., Flurkey K., Partke H.-J., Junger E., Herberg L. NIDDM genes in mice. Deleterious synergism by both parental genomes contributes to diabetic thresholds. Diabetes. 1998;47:1287–1295. doi: 10.2337/diab.47.8.1287. [DOI] [PubMed] [Google Scholar]
- 13.Schattenberg P., Gallem R. Steatohepatitis: Of Mice and Man Animal Models of Non-Alcoholic Liver and Metabolic Syndrome. Dig. Dis. 2010;28:247–254. doi: 10.1159/000282097. [DOI] [PubMed] [Google Scholar]
- 14.Friedman J.M., Leibel R.L., Siegel D.S., Walsh J., Bahary N. Molecular mapping of the mouse ob mutation. Genomics. 1991;11:1054–1062. doi: 10.1016/0888-7543(91)90032-A. [DOI] [PubMed] [Google Scholar]
- 15.Wang B., Chandrasekera P.C., Pippin J.J. Leptin- and leptin receptor-deficient rodent models: Relevance for human type 2 diabetes. Curr. Diabetes Rev. 2014;10:131–145. doi: 10.2174/1573399810666140508121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabouille C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017;27:230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 18.Sung J., Wang Y., Chandrasekaran S., Witten D.M., Price N.D. Molecular signatures from omics data: From chaos to consensus. Biotechnol. J. 2012;8:946–957. doi: 10.1002/biot.201100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jornayvaz F.R., Shulman G.I. Diacylglycerol activation of protein kinase Cε and hepatic insulin resistance. Cell Metab. 2012;15:574–584. doi: 10.1016/j.cmet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itani S.I., Ruderman N.B., Schmieder F., Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IκB-α. Diabetes. 2002;51:2005–2011. doi: 10.2337/diabetes.51.7.2005. [DOI] [PubMed] [Google Scholar]
- 21.Kumashiro N., Erion D.M., Zhang D., Kahn M., Beddow S.A., Chu X., Still C.D., Gerhard G.S., Han X., Dziura J., et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA. 2011;108:16381–16385. doi: 10.1073/pnas.1113359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magkos F., Su X., Bradley D., Fabbrini E., Conte C., Eagon J.C., Varela J.E., Brunt E.M., Patterson B.W., Klein S. Intrahepatic diacylglycerol content is associated with hepatic insulin resistance in obese subjects. Gastroenterology. 2012;142:1444–1446. doi: 10.1053/j.gastro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sriram G., Martinez J.A., McCabe E.R., Liao J.C., Dipple K.M. Single-gene disorders: What role could moonlighting enzymes play? Am. J. Hum. Genet. 2005;76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castello A., Hentze M.W., Preiss T. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol. Metab. 2015;12:746–757. doi: 10.1016/j.tem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao A.W., Noda Y., Johnson J.H., Pessin J.E., Saltiel A.R. Aldolase mediates the association of F-actin with the insulin-responsive glucose transporter GLUT4. J. Biol. Chem. 1999;274:17742–17747. doi: 10.1074/jbc.274.25.17742. [DOI] [PubMed] [Google Scholar]
- 26.Miles L.A., Dahlberg C.M., Plescia J., Felez J., Kato K., Plow E.F. Role of cell-surface lysines in plasmin(ogen)-binding to cells: Identification of α-enolase as a candidate plasminogen receptor. Biochemistry. 1991;30:1682–1691. doi: 10.1021/bi00220a034. [DOI] [PubMed] [Google Scholar]
- 27.Redlitz A., Fowler B.J., Plow E.F., Miles L.A. The role of an enolase-related molecule in plasminogen binding to cells. Eur. J. Biochem. 1995;227:407–415. doi: 10.1111/j.1432-1033.1995.tb20403.x. [DOI] [PubMed] [Google Scholar]
- 28.Knebel B., Hartwig S., Haas J., Lehr S., Goeddeke S., Susanto F., Bohne L., Jacob S., Koellmer C., Nitzgen U., et al. Peroxisomes compensate hepatic lipid overflow in mice with fatty liver. Biochim. Biophys. Acta. 2015;1851:965–976. doi: 10.1016/j.bbalip.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Göddeke S., Kotzka J., Lehr S. Investigating the adipose tissue secretome: A protocol to generate high-quality samples appropriate for comprehensive proteomic profiling. Methods Mol. Biol. 2015;1295:43–53. doi: 10.1007/978-1-4939-2550-6_4. [DOI] [PubMed] [Google Scholar]
- 30.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 31.Bendtsen J.D., Jensen L.J., Blom N., von Heijne G., Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 32.Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N., et al. ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.