Abstract
Aging is considered as one of the most important developmental processes in organisms and is closely associated with global deteriorations of epigenetic markers such as aberrant methylomic patterns. This altered epigenomic state, referred to ‘epigenetic drift’, reflects deficient maintenance of epigenetic marks and contributes to impaired cellular and molecular functions in aged cells. Epigenetic drift-induced abnormal changes during aging are scantily repaired by epigenetic modulators. This inflexibility in the aged epigenome may lead to an age-related decline in phenotypic plasticity at the cellular and molecular levels due to epigenetic drift. This perspective aims to provide novel concepts for understanding epigenetic effects on the aging process and to provide insights into epigenetic prevention and therapeutic strategies for age-related human disease.
Keywords: : aging, chromatin, DNA methylation, environment, epigenetic drift, phenotypic plasticity
Age, as the final destination in a human lifespan, is characterized by functional decline and an increased incidence of age-related degenerative diseases such as diabetes, cancers, cardiovascular diseases and dementia. In molecular aspects, aging is associated with progressive loss of physiological, organismal and cellular integrity leading to genomic instability in cells, decompensation in organisms and eventually death. Among the various hallmarks of aging, for example, irreversible telomere erosion, mitochondrial dysfunction and stem-cell exhaustion, the role of epigenetics in aging and age-related diseases has piqued interests due in part to recent discoveries of how epigenetic mechanisms alter epigenomic landscapes that contribute to functional aging [1–4]. The concepts are now generally accepted that impaired epigenetic hallmarks contribute to biological aging, chronological age and aging-associated disorders. However, the precise mechanisms by which the epigenetic machinery interacts with intrinsic and extrinsic factors in regulation of aging remain unknown. In this Perspective, we propose concepts that age-related epigenomic alterations mediate or at least contribute to the deterioration of phenotypic plasticity at the cellular and molecular level with aging. These perspectives also extend to the interaction of age-associated epigenetic drift with environmental factors and transgenerational inheritance as well as preventive or therapeutic implications of disease epigenetics.
Aging & epigenetics
The modern definition for epigenetics refers to mitotically or meiotically heritable phenotypic changes that are not derived from underlying DNA sequence change [5]. The common epigenetic mechanisms consist of DNA methylation, histone modification and noncoding RNA that are primarily involved in regulation of chromatin structure leading to gene transcription control [6]. The concept and power of epigenetics are illustrated nicely in developmental processes. During early embryonic development, a sophisticated multicellular organism composed of millions of different types of cells with different functions originally arises from a single cell, the zygote, after fertilization, whereby cellular phenotypic diversity is established during differentiation even though the DNA sequences in different cells are essentially the same (i.e., the concept of genomic equivalence). It has been well documented that developmental processes are largely regulated by epigenetic mechanisms [7]. For example, genome-wide DNA methylation reprogramming during early embryogenesis erases parental methylation markers and re-establishes a unique gene-specific methylation pattern in the progeny, which determines specific transcription through epigenetic regulation. Epigenetic mechanisms also participate in regulation of X chromosome inactivation in females and genomic imprinting [8,9]. These epigenetic patterns established during early embryonic development are then maintained throughout life in the somatic cells.
Global epigenetic patterns are fairly stable during the lifespan although fluctuant changes occur in specific loci due to environmental stimuli [10]. Recent studies focusing on the quantitative model of aging by measurements of genome-wide DNA methylation markers using Illumina HumanMethylation450K (Illumina, CA, USA) assay found specific methylomic patterns that may reflect the transcriptome during aging, which may provide biological markers to detect different rates of human aging and the role of methylation in age-related disease [11]. Inspired by these studies, we propose that the regulatory epigenetic machinery becomes gradually deregulated leading to an altered epigenetic state with age proceeding, a process called epigenetic drift, and attenuated responsiveness or adaptation to environmental stimuli, a form of loss of phenotypic plasticity in cellular and molecular levels (Figure 1). This epigenetic-mediated phenotypic plasticity decline during aging is characterized by a loss of the normal balance between the regulatory epigenetic landmarks and plasticity of response to internal and external environmental signals. Moreover, a declined- or changed-response of the aging epigenome to epigenetic-modifying agents, both preventive and therapeutic, could render an increased risk of acquiring a number of age-related diseases. We believe that the aged epigenome as well as dysregulated epigenetic regulatory machinery, such as aberrantly expressed epigenetic modulators, transcription factors and enzymatic malfunction, contributes to a progressive decline in responsiveness to such signals and thus promote the aging processes (Figure 2). Thus, epigenetic modifications can be considered as biological sensors that reflect cellular identity, health and age status. Support for this is manifested by a study in monozygous twins showing remarkable epigenetic differences in their total DNA methylation and histone acetylation between the twins at different ages [12]. Therefore, epigenetic landmarks in chromatin are altered during aging regardless of the original genetic blueprint, and this epigenetic drift results in a loss of phenotypic plasticity in response to environmental impacts leading to biological aging that affects lifespan.
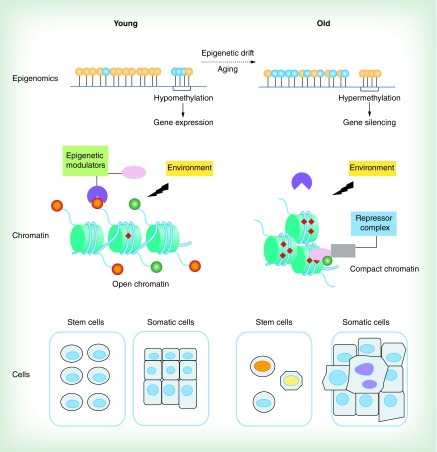
Figure 1. . A model of epigenetic drift-induced cellular phenotypic plasticity attenuation during aging.
Aging is associated with gradual deteriorations of epigenomic markers (known as epigenetic drift) such as altered methylomic patterns (top panel). Aging results in global hypomethylation in the DNA genome but induces hypermethylation in the regulatory regions of certain key genes (orange circle represents methylated CpG island and blue circle represents unmethylated CpG island, top panel). As depicted in the middle panel, ‘normal’ chromatin states are found throughout the genome in young cells. A hypothetical gene expression is activated and well controlled by the regulatory epigenetic machinery including regulations of epigenetic modulators and active epigenetic landmarks (dark orange circle, methylated histone tail; green circle, acetylated histone tail; red diamond, methylated CpG island) in response to environmental factors (middle left). With age proceeding, the regulatory epigenetic machinery becomes gradually deregulated. The aged epigenome fails to respond to environmental factors leading to an altered epigenetic state with a loss of active epigenetic markers and DNA hypermethylation, resulting in compacted chromatin, repressor complex recruitment and subsequent gene silencing (middle right). At the cellular level, young stem cells have relatively identical epigenomes (bottom left) and show robust self-renewal function. During aging, accumulated epigenetic deteriorations lead to epigenomic mosaicism in stem cells resulting in reduced regenerative capacity of stem cells and exhaustion of the stem cell pool (bottom right). In somatic cells, massive epigenetic lesions as well as accumulative genetic mutation with age lead to emergence of hyperproliferative somatic clones (represented as enlarged and irregular cells that have invaded normal tissue boundary, bottom right) in affected tissues that contributes to tumorigenesis and other age-associated diseases.
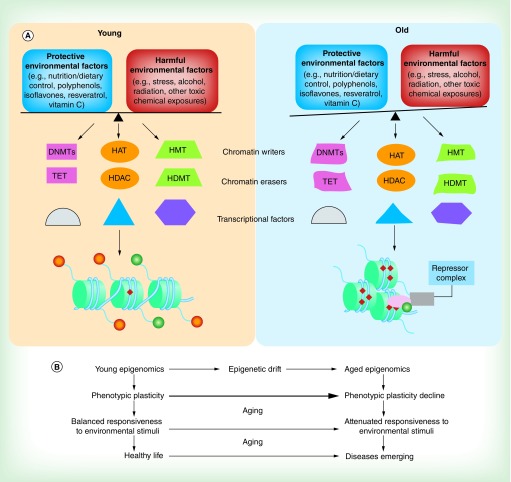
Figure 2. . The epigenomics between environment and epigenetic modulators during aging.
(A) A model of interaction of environmental factors with epigenetic modulators during aging. Environmental factors affect cell function through direct or indirect mechanisms by influencing many epigenetic modulators including chromatin writers (e.g., DNMTs, HMTs and HATs), chromatin eraser enzymes (e.g., TET, HDMTs and HDACs) and various transcriptional factors resulting in dynamic chromatin modifications, which in turn influence gene expression. Epigenetic-mediated phenotypic plasticity attenuation during aging is characterized by a loss of the normal balance between the regulatory epigenetic landmarks and plasticity of response to environmental signals. For example, in young cells, the chromatin (hypothetically) with increased active histone modifications (orange circle, methylated histone tail; green circle, acetylated histone tail) and hypomethylated DNA pattern (red diamond, methylated CpG island, left panel) leads to gene activation in response to balanced environmental stimuli. However, the gene is relatively refractory to environmentally induced activation due to age-dependent epigenetic changes in the gene regulatory region including altered epigenetic landmarks and malfunction of epigenetic modulators such as changes in gene expression, binding ability or enzymatic activity of chromatin writers, erasers and relevant transcriptional factors (represented as distorted shapes, right panel). This results in an altered chromatin structure (compacted chromatin) and regulatory machinery (repressor complex recruitment) in the loci leading to gene silencing (right panel). (B) Schematic representation of mechanistic pathway indicated in (A).
DNMT: DNA methyltransferase; HAT: Histone acetylase; HDAC: Histone deacetylase; HDMT: Histone demethylase; HMT: Histone methyltransferase; TET: Ten-eleven translocation.
DNA methylation & epigenetic drift in aging
DNA methylation is the most important epigenetic regulation that is classically linked to transcriptional repression [13]. DNA methylation involves a conserved enzymatic process including a covalent addition of a methyl group (CH3) to the 5′ site of cytosine residues in CpG dinucleotides [14]. A high density of CpGs frequently clustered into a certain region of genomic DNA is referred to CpG islands and about half of all CpG islands are located in the gene regulatory regions such as the promoter region that is closely associated with gene transcription regulation. About 40% of CpG islands are scattered in the intergenic and intragenic regions, which are not associated with annotated promoters and thus present less regulatory functions. DNA hypermethylation is normally associated with a compacted chromatin conformation and subsequent gene inactivation. In rare cases, hypermethylated transcription factor-binding sites prevent the binding of a repressor complex to the promoter, which can paradoxically induce gene activation. An excellent example of this concept is the regulation of the hTERT gene. Our studies [15–17] and many of others [18] have shown that regulation of hTERT expression is epigenetically completely counter to the normal dogma of epigenetic control.
In mammals, dynamic DNA methylation changes are observed throughout the lifespan. During early embryogenesis, most genomic DNA undergoes prevailing DNA reprogramming processes including genome-wide demethylation and de novo methylation leading to re-establishment of individually unique DNA methylation patterns in the progeny that will be maintained in the somatic cells throughout the lifespan [7]. After birth, global DNA methylation patterns are fairly stable; however, fluctuant changes occur in specific gene loci due to the accumulation of environmental stimuli [10]. Although the maintenance of methylation is in high fidelity during each cell division, aberrant DNA methylation accumulates over time as a result of DNA lesions contributing to epigenetic drift during aging.
Age-related DNA methylation drift involves a bidirectional change in the methylome including both hypermethylation and hypomethylation events. Recently, several studies indicate the existence of aging-associated differentially methylated regions (aDMRs), which contain clusters of consecutive CpG sites throughout the whole genome [19]. A recent investigation compared genome-wide DNA methylation state between newborn and nonagenarian/centenarian samples, and found a significant loss of methylated CpGs in the centenarians [20]. This age-dependent epigenetic drift in aDMRs reflects a programmed alteration in part of the methylation codes during aging, which could be associated with biological mechanisms in aging and age-related human diseases. Actually, a recent cohort study found that about 60% of aDMRs showed age-related hypomethylation in the regulatory regions of key regulatory proteins or specific chromatin modifiers, such as CTCF and Polycomb protein EZH2 [21], suggesting that these methylation changes may concomitantly contribute to development of phenotypic deterioration such as age-associated human diseases. However, one can hardly predict which portion of aDMRs is more responsible for age-related phenotypic changes due to a highly variable methylomic state between individuals. Thus, the longitudinal study focusing on individuals over time would likely acquire more information than a typical cohort study. Alternatively, aging studies focusing on certain candidate loci with potential relevance for age-related human diseases are also informative.
It has been observed that age results in global hypomethylation in the DNA genome but induces hypermethylation in a small number of individual gene loci (Figure 1), notably in the promoter regions of important cancer- and development-related genes [2–3,11,22–23]. Several studies have shown that age-associated DNA hypermethylation preferentially occurs at the promoters of those genes that are particularly associated with cancer, for example, tumor suppressors and transcription factors that are necessary for differentiation [24,25]. Aging-related methylomic changes closely approximate the impaired methylation patterns as seen during carcinogenesis, which can partially explain the high cancer incidence in aged populations [26].
Although DNA methylation signatures show heterogeneous changes depending on cell composition in different tissues, age-dependent DNA methylation changes are frequently detected in more homogeneous cell populations such as stem cells [27,28]. Accumulated epigenetic deteriorations throughout life may reduce regeneration capacity of stem cells as commonly seen in hematopoietic stem cells (HSCs) [27,28] and stem cells in organs such as the GI tract [29]. Recent studies link epigenetic variations to aging in stem cells. Aged HSCs have shown plasticity defects such as skewed differentiation with more myeloid and fewer lymphoid progeny [30]. It has been reported that aged HSCs have a higher incidence of site-specific DNA hypermethylation of target regions in key transcription factors such as PRC2-target genes that are important for lineage-specific gene expression [27,31]. These methylomic alterations contribute to functional decline of HSCs during aging. The depletion of self-renewal/multilineage differentiation potential can also result from hypomethylation and reactivation of germ cell-restricted expression genes, thus triggering autoimmune destruction of the stem cell pools. Therefore, it is possible that age-related methylation drift contributes to declined plasticity in aged stem cells by unmasking hidden tissue antigens through DNA hypomethylation. In the human body, the GI tract is one of the most proliferative and regenerative organs. Coincidentally, the prominent age-related changes in DNA methylation are also observed in the GI tract [22]. This could be due to a large amount of stem cells in the digestive system that are more prone to exposure, as well as sensitive to environmental stimuli. Recent studies showed an age-related DNA methylation increases from several different aged colons but heterogeneous patterns between crypts within the same colon [29,32]. Given that each colonic crypt is derived from stem cell niches, it suggests that age-related hypermethylation is a property of stem cells.
Although the overall genome-wide DNA methylation status in aged cells is relatively low, methylomic changes have shown variable, site-specific effects on gene expression. For example, aged stem cells show a significant epigenetic drift, whereas methylation signatures vary in different gene loci that may lead to considerable mosaicism in gene expression profiles in aged individuals [22,29]. Epigenetic drift profoundly affects the function of aged stem cells. Most age-related methylation drifts are targeted to Polycomb group genes and transcription factors in stem cells that are functionally involved in differentiation and development [22,33]. Aberrant key regulatory gene expression due to age-related DNA methylation abnormality limits the plasticity of stem cells, which compromises their differentiation potential and thus contributes to functional degradation in aged tissues (Figure 1). In addition, gene silencing by hypermethylation machinery in those loci essential for self-renewal leads to age-related depletion of the stem cell pool. Thus, compromised stem cell function is one of the important consequences of age-related epigenetic drift.
Epigenetic drift can also be observed in aged somatic cells and it appears as a tissue-specific epigenetic mosaic pattern [22,26]. Although many aDMRs are shared between different tissues and individuals, the overall DNA methylation level is featured as a tissue-specific and heterogeneous characteristic in young individuals and gradual deviation accumulates with aging. Methylomic deterioration also occurs in slow proliferative tissue such as the CNS tissues. A recent functional enrichment analysis revealed that the aging-related methylation module is present in multiple genes that are involved in nervous system development, neuron differentiation and neurogenesis, which may contribute to phenotypic deterioration in the CNS tissues and development of age-related CNS degenerative diseases such as Alzheimer's disease [34]. On the other hand, age-related epigenetic drift may reduce cellular defensive and repairing functions by equalizing stable gene expression variation, which reduces a powerful mechanism for natural selection and defense in response to environmental impacts. As a result, massive epigenetic lesions plus loss of natural selection as well as accumulated genetic mutations lead to emergence of hyperproliferative somatic clones in affected tissues such as cancer and other age-associated diseases (Figure 1). In summary, we believe that the major consequences of age-related epigenetic drift may include reduced stem cell function as well as reduced capacity for tissue defense and repair in somatic cells.
Histone modification in aging
Histone modifications affect the basic structure of the chromatin via post-translational modification on histone tails [35]. In most cases, histone remodeling occurs at the lysine residues (K) at the N-terminal region of histone proteins, which can be modified by diverse molecular patterns such as acetylation and methylation. Histone modifications are associated with both gene activation and gene repression. Histone modifications directly change nucleosome configuration and result in chromatin status switching either to an active or silencing status leading to gene transcription alterations. Therefore, histone modifications represent a dynamic epigenetic regulatory mechanism to provide another level of gene expression control. Histone modifications can be modulated by chromatin ‘writers’, ‘readers’ and ‘erasers’ that remodel nucleosomes along the DNA and establish specific chromatin states involved in gene transcription [36]. Chromatin ‘writers’ represent a group of epigenetic enzymes that catalyze the transfer of methyl groups, acetyl groups or other moieties to a limited set of amino acid residues on histone tails. Examples of these writers are DNA methyltransferases (DNMTs) [37], histone methyltransferases and histone acetylases (HATs) [35]. These chromatin modifications can be recognized by special proteins called ‘readers’ that can interpret the signals and facilitate chromatin structure changes [38]. Chromatin modification can be removed by ‘eraser’ enzymes such as demethylases, histone deacetylases (HDACs) and histone demethylases [35]. Thus, histone modifications involve a dynamic enzymatic process including adding and/or removing these post-translational modifications on histone tails, leading to a highly controlled histone code and gene transcription management.
Several studies have demonstrated a link between histone variants and aging development in various organisms [39,40]. It has been found that the global distribution of many histone methylation and acetylation markers changes in organismal and cellular models of aging. For example, a global loss of a repressive chromatin marker, H3K27me3, is correlated with aging and manipulation of chromatin regulators to increase H3K27me3 can result in increased longevity [41]. Evidence also shows that histone acetylation patterns change during the aging process. In yeast, decreased global levels of H3K56 acetylation (H3K56ac) and increased H4K16ac are closely associated with replicative aging [42].
Furthermore, the manipulation of histone modulators such as chromatin writers including HATs and histone methyltransferases, and erasers such as HDACs and histone demethylases can modulate longevity of model organisms. In yeast, H4K16ac is deacetylated by Sir2 known as class III HDACs and increased Sir2 dosage extends yeast lifespan [43]. In mammals, Sir2 orthologs such as SIRT6 can deacetylate H3K9ac68 and H3K56ac69, and its deficiency induces a progeroid phenotype in mice [44]. Thus, remodeling of specific histone marks by regulation of certain epigenetic modulators has shown a potential promise for aging control and longevity.
Early studies have discovered a link between DNA methylation and histone modifications mainly mediated by a group of important chromatin readers including methyl CpG-binding protein 2 and methyl CpG-binding domain proteins [45]. These proteins recognize and bind the methylated DNA promoters, then subsequently recruit a set of proteins including DNMTs, HDACs and certain transcriptional factors forming a repressor complex that silences gene expression. These studies demonstrate that DNA methylation may result in chromatin structural changes through alteration of histone modification patterns indicating an important crosstalk between DNA methylation and histone modification. The Polycomb target genes provide a good example of how histone modification and DNA methylation interact to regulate gene silencing [46]. This is mediated by an important epigenetic silencing mechanism, PRC2-based H3K27me3, catalyzed by the SET domain histone methyltransferase EZH2, as a part of the PRC2 complex. This crosstalk between DNA methylation and PRC2-H3K27me3 has been found during the processes of early embryonic development, stem cell differentiation and tumorigenesis. Dysregulation of these two epigenetic mechanisms has frequently been observed in many types of cancers and may contribute to early tumorigenesis. For example, aberrant DNA methylation patterns during tumorigenesis affect the affinity of PRC2 binding leading to redistribution of the H3K27me3 pattern and de novo methylation of relevant genes. Due to the important roles of PRC2 proteins in aging, the alteration reduces the epigenetic plasticity by eliminating the regulation of key silencing regulators that may also contribute to aging development.
Histone modification patterns fluctuate in response to a changing cellular milieu. Compared with DNA methylation, histone modifications may not represent as a good marker for age-associated epigenetic drift due to its perturbation status. We propose that loss of the control in histone modifications may also contribute to increased phenotypic heterogeneity due to epigenomic variability and lead to reduced responsiveness of aging cells to specific environmental stimuli with aging. It is likely that dysregulation of epigenetic modulators including ‘writers’, ‘readers’ and ‘erasers’ may collectively play a major role in this age-associated turbulence in histone remodeling and chromatin structures during aging (Figure 2).
Mechanisms of epigenetic drift in aging
In the effort to understand the mechanisms underlying age-related epigenetic drift, it is important to know several unique features for changes of epigenomic landmarks during aging. As for DNA methylation, age-related epigenetic drift is characterized by three important features. The first is bidirectional indicating that DNA methylation can be lost or gained throughout the genome, especially hypermethylation in regulatory regions such as the promoters and hypomethylation elsewhere [11,22]; second, epigenetic drift is highly variable between individuals of the same age suggesting discontinuity between biological age and chronological age [11–12,22]; third, age-associated epigenetic drift occurs in a cell- and tissue-specific manner in the same individual, such as a significantly higher drift in stem cells and the most proliferative tissues as compared with other somatic cells and tissues (Figure 1) [11,22,29].
This raises a question with respect to the internal motivation for epigenetic drift. Is it a passive or an active process with age? One can speculate reduced fidelity in replication-associated DNA methylation. However, a spontaneous error rate in methylation maintenance at promoter CpG islands (both gains and losses) is only at 10-4 to 10-5 in vitro [47]. Thus, a simple explanation for methylation drift as the presence of replication-dependent errors is obviously insufficient. Age-related methylation drift shows genomic-specific character. For example, Polycomb group target genes and transcription factors in stem cells that are functionally involved in differentiation and development are frequently influenced [22,33]. It is plausible that specific epigenetic changes more likely occur in particular loci, and complicated regulatory machineries involved in cis- and trans-interactions may account for these ‘site addiction’ changes. Genome-wide studies have also identified genetic variants such as SNPs that may affect the rate of DNA methylation in cis [48]. On the other hand, epigenetic drift is also dependent on trans-acting factors such as important epigenetic modulators, transcriptional factors and domain modifiers (Figure 2).
Aged tissues and cells have been shown to have certain age-associated changes in histone chromatin marks such as increased histone H4K16 acetylation, H4K20 trimethylation or H3K4 trimethylation, as well as decreased H3K9 methylation [39–42]. Recent studies provide evidence that epigenetic mechanisms allow rapid adaptations to environmental changes and respond to signal cascades in regulation of chromatin modulators leading to age-accompanied chromatin remodeling [49].
It is notable that environmental impacts play a crucial role in altering epigenomic profiles and therefore influencing phenotypic plasticity in aged organisms. Age-related epigenetic drift attenuates beneficial phenotypic plasticity, which is characterized by a loss of the normal balance between the regulatory epigenetic landmarks and plasticity of responsiveness to environmental signals (Figure 2). For example, certain genes are relatively refractory to environmentally induced regulations due to age-dependent epigenetic changes in the regulatory regions of these genes, which include altered epigenetic landmarks such as DNA methylation or histone modifications, and/or malfunction of epigenetic modulators such as changes in gene expression, binding ability and enzymatic activities of chromatin writers, erasers and relevant transcriptional factors. This altered aging epigenome leads to a deteriorated status in gene, cellular and organismal levels that is correlated with aged-related phenotypic changes such as reduced functions in stem cells as well as reduced capacity for tissue defense and repair in somatic cells (Figure 1).
An emerging hypothesis claims that epigenetic modifications can persist in each cell division and that epigenetic drift can be recognized as a ‘biological sensor’ to predict age like telomere attrition. Although some studies found it may not be accurate [11], several separate studies have reported DNA methylation-based age predictors in clinical cases or even for forensic science to determine the approximate age of a disfigured body [50,51]. Although the precise triggers for rewriting the epigenetic landmarks are still unknown, it has been reported that exposure to environmental factors could potentially change epigenomic status. We believe that environmental exposure accelerates the mitotic clock by changing the epigenetic hallmarks leading to long-lasting phonotypic effects that may be particularly relevant to aging (Figure 2).
An emerging question is whether aged somatic cells could become ‘rejuvenated’ through epigenetic reprogramming. Recent studies in stem reprogramming have been largely advanced by a technique called ‘induced pluripotent stem cells’, by which adult differentiated cells are converted back to pluripotent cells through the re-expression of pluripotency regulators [52]. Interestingly, the epigenome state in induced pluripotent stem cells has been stably reset compared with uninduced adult cells [53]. Thus, an intriguing and exciting possibility suggests that age-driven accumulation of epigenetic markers could be rewound and re-established to resemble the epigenetic patterns of embryonic stem cells, called ‘epigenetic rejuvenation’ [54]. Determining the accurate mechanisms for how epigenetic rejuvenation occurs and how to control this process will be invaluable, not only for better understanding of how lifespan trajectories evolve, but also for exploring future therapeutic strategies for age-related human disease and potential ‘rejuvenation’.
Environment, nutrition & age-related epigenetics
Numerous studies have highlighted the importance of environmental factors in the regulation of aging and longevity, including diets, lifestyles, chemicals and radiation exposures [4,6]. It has been well accepted that establishment of a good lifestyle is closely correlated with aging delay and longevity [55]. For example, the circadian clock controls many physiological and behavioral systems, and disrupting circadian rhythms negatively influences health and longevity partially through reprogramming of epigenomic profiles [56]. In addition, physical activity may influence chromatin dynamics, thus promoting overall health and potential aging delay in humans [4]. On the contrary, however, bad habits such as cigarette smoking are also involved in alteration of chromatin structures such as DNA methylation patterns, which may serve as an environmental risk factor for age-related chronic diseases [57].
Many epigenetic changes could be due to direct or indirect epigenetic remodeling by environmental factors, thus subsequently influencing cell function and individual health status throughout the life course. However, causal mechanisms linking specific environmental factors to age-associated chromatin changes, and subsequently influences in aging and longevity, are still under intense investigation. Among those environmental factors, nutrition is believed to be one of the most important contributors to the regulation of gene expression in aging processes by impacting epigenetic pathways [58,59]. In this section, we will present perspectives on dietary impacts in regulation of age-related epigenetic profiles and how these changes promote health span and lifespan.
Diet is the key environmental factor that shapes the activity of the epigenome and determines stress-adaptive responses, energy metabolism, immune homeostasis and the physiology of the body [4,6,58–59]. In fact, both the quantity and the quality of the diet play crucial roles in the regulation of aging processes. Dietary restriction or caloric restriction is by far the best established environmental manipulation to extend lifespan and prevent the onset of age-related chronic diseases in many species [60]. Dietary restriction induces a large spectrum of protein changes including genes that regulate metabolism, stress responses, DNA repair and chromatin structure [61]. Dietary restriction also affects the chromatin landscapes [61]. For instance, in yeast, dietary restriction induces positional shifts of nucleosomes at thousands of genes targeted by the Isw2 chromatin-remodeling complex, which may result in a stress response at the transcriptional level [62]. Evidence has shown that dietary restriction is likely to recover age-induced aberrant DNA methylation patterns via reversal of both specific and global changes in the loci that may be closely correlated with metabolic pathways [60,61]. For example, our previous studies found that glucose restriction extends normal cell lifespan via epigenetic activation of hTERT, suggesting dietary restriction may interfere with aging processes through epigenetic improvement of declined telomerase activity [16]. The widespread DNA methylation changes of numerous gene loci may facilitate dietary restriction in reprogramming the DNA methylation profile, which may also explain a powerful and universal effect of this dietary intervention in influencing different aspects of human health. Recent study reveals that SIRT1, a NAD+-dependent HDAC, affects DNA methylation in particular genes that respond to dietary restriction [63]. This suggests a crosstalking process between DNA methylation and histone modifications in control of dietary restriction-associated age intervention. Studies also show that DNMT1, known to play an important role in maintaining methylation status, is significantly elevated in response to caloric restriction, as a direct result from dietary restriction, suggesting epigenetic modulator changes may also contribute to correct age-related epigenetic lesions when energy is reduced [16]. By contrast, however, high nutrient intake in humans, which will lead to increased body weight and obesity, seems to induce age-associated DNA methylation changes in the liver [64]. Thus, nutrient quantity intake has important connections to the regulation of both age and chromatin structure.
In addition to quantitative aspects of dietary intake, the effects of the dietary quality appear to be crucial to achieve the correct relationship between dietary balance and health. Several dietary compounds, such as resveratrol, tea polyphenols and flavonoids, which are commonly found in vegetables, fruits and plants, exert well-evidenced chemopreventive properties against cardiovascular, neurodegenerative disease and chronic inflammatory diseases that are correlated with aging [4,6,58–59]. Besides the classic proposed mechanisms such as antioxidant activity and antiapoptosis, some of these botanic nutrients have been identified to mediate epigenetic processes.
SIRT1 is a member of the sirtuin family of proteins, homologs of the Sir2 gene in yeast. SIRT is classified as a class III HDAC and has a strong functional link to chromatin regulation, which is required for their effect on lifespan, at least in yeast [43]. Because the enzymatic activity of SIRT largely depends on NAD/NADH ratio (nicotinamide adenine dinucleotide, NAD; NADH reduced form of NAD), SIRT is also considered as an ‘energy sensor’ that is tightly connected to the metabolic state of cells. As mentioned previously, SIRT prevents genomic instability and promotes dietary restriction-induced longevity [63]. Various natural flavonoids have been identified as activators of class III HDACs (SIRTs). For instance, resveratrol, a compound found in grapes, red wine and nuts, has been found to prevent several age-related diseases and part of the mechanism may relate to its SIRT-like property leading to increased longevity in vivo and in vitro [65].
Although many studies have focused on 5-methylcytosine, the recent discovery of different modifications of DNA methylation such as 5-hydroxymethylcytosine (5-hmC), have been found to link to biological aging [4,66]. 5-hmC represents an intermediate product during DNA demethylation processes and the conversion of 5-methylcytosine to 5-hmC is catalyzed by the ten-eleven translocation (TET) family of enzymes. Vitamin C, a potential co-factor for Fe(II) 2-oxoglutarate dioxygenase enzymes like TETs, can enhance DNA demethylation processes through increasing enzymatic activity of TETs [67]. Genome-wide epigenetic studies have found that the global level of 5-hmC is decreased in mouse-aged HSCs [68], which may contribute to silencing expression of key lineage-specific gene expression due to hypermethylation during aging. We, therefore, propose that changes in 5-hmC with aging could potentially be countered by dietary agents such as vitamin C that acts as a cofactor for the TET proteins. Further experiment will be urgently needed to verify this provocative hypothesis.
Other important health diets such as green tea, broccoli sprouts and soybean, and the bioactive compounds extracted from these diets have received extensive attention due to their profound actions on prevention of multiple age-related diseases such as cancers by reversal of the aberrant epigenetic profiles accumulated in the affected cells [4,6,58–59]. The functions of nutritional compounds have been characterized by interfering epigenetic profiles through multiple pathways. They can affect enzymatic activity of chromatin writers, readers or erasers such as DNMTs, HDACs, HATs and SIRTs, which contribute to modulate inflammatory responses, immunological senescence and carcinogenesis processes (Figure 2). For example, green tea polyphenols EGCG ([−]-epigallocatechin-3-gallate) and soybean genistein are considered as natural DNMT inhibitors: EGCG works mainly through direct binding to block the DNMT catalytic active site and indirect effects by influencing DNMT expression [69], whereas genistein affects DNMT more likely through an indirect pathway [15,58]. Natural isothiocyanates such as sulforaphane from cruciferous vegetables (e.g., broccoli) show inhibitory activities against class I HDACs and induce increases in histone acetylation in the promoters of many tumor suppressor genes leading to tumor suppression in an in vitro study of breast cancer cell lines [17]. Long-term consumption of these health diets is highly associated with a low incidence of various aging-related degenerative diseases and longevity. We believe that exposure to these bioactive diets could lead to remodeling of the epigenome in a cumulative fashion by inhibiting, delaying and reversing aberrant epigenetic machineries during aging.
Epigenetic transgenerational regulation in aging
Several phenotypes that are associated with stress and metabolic states can influence chromatin states in germ cells, which can be inherited in a transgenerational manner allowing the transmission of a previous environmental state through generations [4]. Certain exposures during crucial time windows of early development can shift the normal differentiation process and lead to a persistent effect on the epigenome that may be transmitted to several generations [70]. This altered epigenomic state induced by nutritional or environmental factors leads to altered epigenetic hallmarks in parental somatic and germline cells that may be transmitted through mitosis and meiosis during gametogenesis and the following embryogenesis. Studies have found that various chromatin marks such as H3K9me3, H3K27me3, PRC2 and DNA methylation status can be transmitted as an epigenetic memory to the next generation [71,72]. Thus, the phenotypic alterations will manifest in the progeny long after the exposure disappears, which could represent a beneficial mechanism for natural evolution by establishing adaptive response to environmental perturbations such as food restriction in the generations.
The good examples are the studies of human populations following famine such as the Dutch Hunger Winter cohort indicating individuals whose mothers were exposed to famine in WWII during their first trimester showed a higher incidence of cardiovascular diseases and a poorer glucose tolerance than those born the year before the time of nutritional insult [73]. Thus, the maternal nutrition status greatly impacts several pathologic variations in the offspring. Imbalanced maternal nutrition is known to be highly associated with increased risk of Type 2 diabetes, obesity and other metabolic syndromes, which may be due to incorrect epigenetic programming during their early development [74]. Hereby, specific environmental intervention during early life may induce site-specific changes in chromatin signatures at critical regulatory sites that may influence gene expression and subsequently affect the risk of age-associated pathologies.
Recently, a concept known as ‘transgenerational prevention’ has emerged that emphasizes the importance of timing for intervention [70]. It is likely that the earlier the interference, the more effective the results may be. For instance, the timing for early-life intervention can range from preconception, prenatal (pregnancy), lactation and neonatal life. This idea has been verified by a recent longitudinal twin study comparing DNA methylation profiles from birth to 18 months of age, which showed that age-associated epigenetic drift initiates immediately after birth and the changes are particularly prominent in early life [75]. This result indicates that the epigenome might be particularly sensitive to environmental stressors during early-life exposures and certain interventions during this particular period could exert more impacts than any other life period on influencing epigenetic drift.
Various dietary exposures during these critical developmental stages may be vitally important in regulating epigenetic reprogramming and determining health beneficial effects such as prevention, delay or diminishing the effect of many diseases that are associated with age. Early consumption of these environmental compounds including nutritional factors such as methyl donors (e.g., folate) and dietary compounds with properties in regulating epigenetic processes (e.g., green tea polyphenols, soybean isoflavone and broccoli sprouts sulforaphane) has been linked to a decreased incidence in many age-associated diseases such as diabetes and cancers [70]. In addition, a recent study reporting that dietary vitamin C promotes TET activity leading to a rapid and global increase of 5-hmC in mouse embryonic stem cells [76] stimulates another important hypothesis that early exposure to vitamin C may play a crucial role in transgenerational effects by inducing TET-dependent DNA demethylation process during early embryonic development.
Although the exact mechanism that explains how environmental factors regulate epigenetic reprogramming during early development are not fully understood, it is noteworthy that better understanding of this process will further help to develop novel early-life intervention approaches against various diseases in adult life by recovering age-associated epigenetic landmarks in an effective time frame.
Age-associated human diseases & epigenetic therapies
It is clear that the epigenome is altered by an age-associated epigenetic drift, whereby normal chromatin patterns including DNA methylation and histone modifications become deregulated with age leading to genomic instability and diseases. An aged epigenome is closely linked to reduced plasticity of stem cells, limited stem cell pools and deteriorated function of somatic cells in which alterations could result in significant pathologic changes. Thus, most age-related diseases have an epigenetic component.
Numerous studies have demonstrated a widespread epigenetic variation in the age-related human diseases. The most common age-related human diseases are noncommunicable diseases. Noncommunicable diseases refer to a group of chronic human diseases with noninfectious and nontransmissible medical conditions such as cancer, diabetes and cardiovascular diseases with defined genetic predisposition. For example, age-related epigenetic variation could induce long-term alterations in metabolic regulation that contribute to Type 2 diabetes [77]. Several genome-wide studies also demonstrated a highly statistically significant overlap between age-associated epigenetic drift and epigenetic alterations associated with cancer [25,26]. Thus, cancer is characterized by coincident genetic and epigenetic damage in aging, and age-related epigenetic drift may be the key factor in explaining the rise in incidence of cancer as humans age.
The intriguing link between age-associated epigenetic drift and the epigenetic changes seen in cancer suggests a causal role for epigenetic mechanisms in disease initiation and may also extend to other age-associated diseases. Although environmental stimuli such as carcinogen-induced genetic changes (e.g., mutation) are well recognized as a key contributor to carcinogenesis, epigenetics provides an added layer of variation that might mediate the relationship between genotype and environmental factors [4,6].
As epigenetic mechanisms have been identified in aging development, it triggers an exciting therapeutic avenue known as epigenetic therapy, which is focusing on repairing damaged epigenetic markers in various human diseases. For example, two classes of epigenome-modifying agents such as DNMT inhibitors, decitabine and HDAC inhibitors such as suberoylanilide hydroxamic acid are currently used for cancer therapy in clinical trials [78]. Recent clinical trials indicate antitumor activity of HDAC inhibitors against certain hematological malignancies such as cutaneous T-cell lymphoma; however, their therapeutic potential in solid tumors remains unclear [79]. Numerous studies have also identified HDAC inhibitors as candidate drugs for the treatment of age-related neurodegenerative disorders such as Parkinson's disease and Alzheimer's disease due to their effects on ameliorating deficits in synaptic plasticity and promoting learning and memory in brain [80]. These studies highlight epigenetic therapy as a promising candidate for antiaging strategy. However, due to their effects on global epigenetic profiles and lack of specific targets, the overall efficacy for these epigenetic drugs is still under consideration before routine use in the treatment plans for cancers and other age-related diseases. As proposed previously, a more healthy and effective way to delay aging and prevent age-related diseases is to consume a healthy diet consisting of active dietary components, such as resveratrol, tea polyphenols and flavonoids that may help to change the aged epigenome to a much younger epigenetic profile. In addition to that, determining an appropriate exposure time may also help to maximize the preventive effects of these bioactive dietary compounds.
Conclusion
Epigenetic marks undergo a significant drift during aging, and this perturbation of chromatin landmarks leads to age-associated diseases, which can directly affect lifespan. This drift results in epigenetic-mediated phenotypic plasticity loss in aging, which is characterized by a loss of normal balance between the regulatory machinery and phenotypic plasticity of response to internal and external environmental signals. This epigenetic rigidity reduces adaptive response to environmental stimuli that substantially contributes to aging pathologies. Because environmental exposure throughout life is an important driver to induce epigenomic instability in aging, it is noteworthy that improved understanding of age-associated epigenetic drift will allow us to manipulate the epigenome and reverse age-related epigenetic aberrations. Considerable interests are currently focused on studying the effects of nutrition intervention in modulation of abnormal epigenetic alterations during aging. To prevent or to reverse adverse epigenetic alterations associated with age-associated diseases, numerous dietary components have been identified as epigenetic modulators and used as an effective approach to prevent age-related human diseases and improve general health benefits. To maximize the preventive/therapeutic effect, we suggest that exploration of appropriate combinatorial approaches by introducing different nutrients and determining an effective exposure window will be an important future direction in the field of aging research.
Future perspective
Age-related epigenetic drift leads to loss of phenotypic plasticity and gene expression variation that could be a key contributor to age-associated pathologic changes. Better understanding of this process and manipulating the epigenome hold enormous promise for preventing and treating common human illness in aging. Epigenetic information integrates environmental input with genetic background during aging. However, the changes of epigenomic landmarks during aging are still unknown. To better understand these integrative changes during aging, epigenome maps throughout the lifespan would be particularly helpful. Such an aging epigenomic roadmap should help to develop better strategies for delaying age or reversing age-related diseases. The plasticity of chromatin states and individual phenotype is a result of gene–environment interactions. Various dietary nutrients that target chromatin modifiers have been identified to slow aging and prevent age-related diseases that collectively contribute to longevity. Future research will be necessary to evaluate the optimal dose and exposure window for specific dietary compounds to elicit maximal epigenetic benefits against age and its associated diseases, and improve the overall quality of life of the human population.
Executive summary.
Aging & epigenetics
Aging processes are largely regulated by epigenetic mechanisms.
The regulatory epigenetic machinery becomes gradually deregulated leading to an altered epigenetic state during aging, a process we call epigenetic drift.
The epigenetic drift results in a loss of phenotypic plasticity in response to environmental impacts, which contributes to biological aging.
DNA methylation & epigenetic drift in aging
Aging results in global hypomethylation in the DNA genome but induces hypermethylation in the promoter regions of certain important cancer- and development-related genes.
Age-related epigenetic drift in DNA methylation contributes to compromised stem cell function and hyperproliferative somatic clones resulting in cancer and other age-associated diseases.
Histone modification in aging
Histone modifications contribute to increased phenotypic heterogeneity in aging due to epigenomic variability.
It is likely that dysregulation of epigenetic modulators including ‘writers’, ‘readers’ and ‘erasers’ may play a major role in this age-associated turbulence in histone remodeling and chromatin structures during aging.
Mechanisms of epigenetic drift in aging
Age-related specific epigenetic changes more likely occur in certain particular loci and complicated regulatory machineries involved in cis- and trans-interactions may account for these ‘site addiction’ changes.
Environmental exposure accelerates the mitotic clock by changing the epigenetic hallmarks leading to long-lasting phonotypic effects that may be particularly relevant to aging.
Determining the accurate mechanisms for how epigenetic rejuvenation occurs and how to control this process will be invaluable for exploring future strategies for ‘age prevention’ and ‘rejuvenation’ in humans.
Environment, nutrition & age-related epigenetics
Diet is one of the most important environmental factors in regulation of age-related epigenetic profiles that may influence health span and lifespan.
Both the quantity and the quality of the diet play crucial roles in the regulation of aging process.
Several dietary compounds, such as resveratrol, tea polyphenols and flavonoids, which are commonly found in vegetables, fruits and plants, exert well-evidenced chemopreventive properties against many age-related human diseases through, at least in part, regulation of epigenetic mechanisms.
We propose that changes in 5-hydroxymethylcytosine with aging could potentially be countered by dietary agents such as vitamin C that acts as a co-factor for the ten-eleven translocation proteins.
Epigenetic transgenerational regulation in aging
Several phenotypes can influence chromatin states in germ cells, which can be inherited in a transgenerational manner allowing the transmission of a previous environmental state through generations.
The epigenome might be particularly sensitive to environmental stressors during early-life exposures, and transgenerational prevention shows great promise in late-life disease prevention through early intervention.
Early consumption of certain dietary factors with properties in regulating epigenetic processes has been linked to a decreased incidence in many age-associated diseases such as diabetes and cancers.
We hypothesize that early exposure to vitamin C may play a crucial role in transgenerational effects by interfering with DNA demethylation process during early embryonic development.
Age-associated human diseases & epigenetic therapies
A significant overlap between age-associated epigenetic drift and epigenetic alterations associated with cancer suggests epigenetic drift could play a causal role in disease initiation during aging.
Epigenetic therapy focusing on repairing damaged epigenetic markers could serve as an exciting therapeutic avenue for age-related human diseases.
Future perspective
We believe that age-related epigenetic drift leads to loss of phenotypic plasticity and gene expression variation that could be a key contributor to age-associated pathologic changes.
We suggest future research will be necessary to evaluate the optimal dose and exposure window for specific environmental intervention such as bioactive dietary compounds to elicit maximal epigenetic benefits against age-associated diseases.
Acknowledgements
The authors appreciate R Kala for proving insights for this manuscript.
Footnotes
Financial & competing interests disclosure
This work was supported by grants from the National Cancer Institute (R01 CA178441; R03 CA176766) and the American Institute for Cancer Research (316184). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Creative Commons Attribution-NonCommercial 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Hum. Mol. Genet. 2013;22(R1):R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Issa JP. Aging and epigenetic drift: a vicious cycle. J. Clin. Invest. 2014;124(1):24–29. doi: 10.1172/JCI69735. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• One of the most important papers in review of aging and epigenetic drift.
- 4.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015;16(10):593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann. NY Acad. Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- 6.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 7.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 2013;20(3):282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 8.Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2001;2(1):59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 9.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13(8):323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 10.Kanherkar R, Bhatia-Dey N, Csoka A. Epigenetics across the human lifespan. Front. Cell Dev. Biol. 2014;2:49–68. doi: 10.3389/fcell.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• One of the most comprehensive studies on investigation of quantitative methylomic markers in age-related human diseases.
- 12.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An important human study demonstrating environmental impacts on epigenetic diversity.
- 13.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 14.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Liu L, Andrews LG, Tollefsbol TO. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int. J. Cancer. 2009;125(2):286–296. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Liu L, Tollefsbol TO. Glucose restriction can extend normal cell lifespan and impair precancerous cell growth through epigenetic control of hTERT and p16 expression. FASEB J. 2010;24(5):1442–1453. doi: 10.1096/fj.09-149328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS ONE. 2010;5(7):e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int. J. Cancer. 2002;101(4):335–341. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- 19.Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Bürkle A, Caiafa P. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl Acad. Sci. USA. 2012;109(26):10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An important human study in comparable methylomic states in young and aged populations.
- 21.McClay JL, Aberg KA, Clark SL, et al. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum. Mol. Genet. 2014;23(5):1175–1185. doi: 10.1093/hmg/ddt511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maegawa S, Hinkal G, Kim HS, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An important study comparing age-related methylation states in various tissues.
- 23.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakyan VK, Down TA, Maslau S, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JPJ. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;158(23):5489–5494. [PubMed] [Google Scholar]
- 26.Waki T, Tamura G, Sato M, Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene. 2003;22(26):4128–4133. doi: 10.1038/sj.onc.1206651. [DOI] [PubMed] [Google Scholar]
- 27.Beerman I, Bock C, Garrison BS, et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12(4):413–425. doi: 10.1016/j.stem.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Sun D, Luo M, Jeong M, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata D. Inferring human stem cell behavior from epigenetic drift. J. Pathol. 2009;217(2):199–205. doi: 10.1002/path.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5(8):e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh J, Lee YD, Wagers AJ. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat. Med. 2014;20(8):870–880. doi: 10.1038/nm.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yatabe Y, Tavaré S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl Acad. Sci. USA. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvageau M, Sauvageau G. Polycomb group proteins: multi faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13(10):R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO. Rep. 2011;12(7):647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585(13):2041–2048. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Sullivan RJ, Karlseder J. The great unravelling: chromatin structure romatin as a modulator of the aging process. Trends Biochem. Sci. 2012;37(11):466–476. doi: 10.1016/j.tibs.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang W, Steffen KK, Perry R, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 44.Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6 . Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 45.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 46.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 47.Rose JA, Yates PA, Simpson J, Tischfield JA, Stambrook PJ, Turker MS. Biallelic methylation and silencing of mouse Aprt in normal kidney cells. Cancer Res. 2000;60(13):3404–3408. [PubMed] [Google Scholar]
- 48.Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E. Genomic surveys by methylationsensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40(7):904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 49.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem. Sci. 2014;39(7):310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Koch CM, Wagner W. Epigenetic-aging-signature to determine age in different tissues. Aging. 2011;3:1018–1027. doi: 10.18632/aging.100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bocklandt S, Lin W, Sehl ME, et al. Epigenetic predictor of age. PLoS ONE. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 53.Wahlestedt M, Norddahl GL, Sten G, et al. An epigenetic component of hematopoietic stem cell aging amenable to reprogramming into a young state. Blood. 2013;121(21):4257–4264. doi: 10.1182/blood-2012-11-469080. [DOI] [PubMed] [Google Scholar]
- 54.Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fontana L, Partridge L, Longo VD. Extending healthy life span – from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguilar-Arnal L, Sassone-Corsi P. The circadian epigenome: how metabolism talks to chromatin remodeling. Curr. Opin. Cell Biol. 2013;25(2):170–176. doi: 10.1016/j.ceb.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front. Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr. Med. Chem. 2010;17(20):2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szarc vel Szic K, Declerck K, Vidaković M, Vanden Berghe W. From inflammaging to healthy aging by dietary lifestyle choices: is epigenetics the key to personalized nutrition? Clin. Epigenetics. 2015;7(1):33. doi: 10.1186/s13148-015-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Daniel M, Tollefsbol TO. Epigenetic regulation of caloric restriction in aging. BMC Med. 2011;9:98. doi: 10.1186/1741-7015-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23:1849–1869. doi: 10.1101/gad.1807009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dang W, Sutphin GL, Dorsey JA, et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 2014;19(6):952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guarente L, Picard F. Calorie restriction – the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 64.Horvath S, Erhart W, Brosch M, et al. Obesity accelerates epigenetic aging of human liver. Proc. Natl Acad. Sci. USA. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agarwal B, Baur JA. Resveratrol and life extension. Ann. NY Acad. Sci. 2011;1215:138–143. doi: 10.1111/j.1749-6632.2010.05850.x. [DOI] [PubMed] [Google Scholar]
- 66.van den Hove DL, Chouliaras L, Rutten BP. The role of 5-hydroxymethylcytosine in aging and Alzheimer's disease: current status and prospects for future studies. Curr. Alzheimer Res. 2012;9(5):545–549. doi: 10.2174/156720512800618008. [DOI] [PubMed] [Google Scholar]
- 67.Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (TET) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013;288(19):13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng Y, Xie N, Jin P, Wang T. DNA methylation and hydroxymethylation in stem cells. Cell Biochem. Funct. 2015;33(4):161–173. doi: 10.1002/cbf.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63(22):7563–7570. [PubMed] [Google Scholar]
- 70.Li Y, Saldanha SN, Tollefsbol TO. Impact of epigenetic dietary compounds on transgenerational prevention of human diseases. AAPS J. 2014;16(1):27–36. doi: 10.1208/s12248-013-9538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348(6230):1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaydos LJ, Wang W, Strome S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345(6203):1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lumey LH, Stein AD. Transgenerational effects of prenatal exposure to the Dutch famine. BJOG. 2009;116(6):868. doi: 10.1111/j.1471-0528.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 74.Attig L, Gabory A, Junien C. Nutritional developmental epigenomics: immediate and long-lasting effects. Proc. Nutr. Soc. 2010;69(2):221–231. doi: 10.1017/S002966511000008X. [DOI] [PubMed] [Google Scholar]
- 75.Martino D, Loke YJ, Gordon L, et al. Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biol. 2013;14:R42. doi: 10.1186/gb-2013-14-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blaschke K, Ebata KT, Karimi MM, et al. Vitamin C induces TET-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mack GS. Epigenetic cancer therapy makes headway. J. Natl Cancer Inst. 2006;98:1443–1444. doi: 10.1093/jnci/djj447. [DOI] [PubMed] [Google Scholar]
- 79.Rangwala S, Zhang C, Duvic M. HDAC inhibitors for the treatment of cutaneous T-cell lymphomas. Future Med. Chem. 2012;4(4):471–486. doi: 10.4155/fmc.12.6. [DOI] [PubMed] [Google Scholar]
- 80.Vaiserman AM, Pasyukova EG. Epigenetic drugs: a novel anti-aging strategy? Front. Genet. 2012;3:224. doi: 10.3389/fgene.2012.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]