Abstract
Influenza A viral ribonucleoprotein (vRNP) complexes comprise the eight genomic negative-sense RNAs, each bound to multiple copies of viral nucleoprotein and a trimeric viral polymerase complex. The influenza virus life cycle centres on the vRNPs, which in turn, rely on host cellular processes to perform functions necessary for the successful completion of the virus life cycle. Here, we review our current knowledge about vRNP trafficking within host cells and the function of these complexes in the context of the virus life cycle, highlighting how structure contributes to function and the critical interactions with host cell pathways, as well as the information gaps that remain. An improved understanding of how vRNPs use host cell pathways is essential to identify mechanisms of virus pathogenicity, host adaptation, and ultimately, new targets for antiviral intervention.
INTRODUCTION
Influenza A virus (IAV) is an important human viral pathogen, responsible for seasonal epidemics that cause substantial human morbidity and mortality and considerable financial burden worldwide every year1–3. While vaccines are available to prevent infections in humans, the haemagglutinin (HA) proteins of circulating viruses rapidly acquire mutations (i.e., ‘antigenic drift’), which prevent their recognition by the host immune system and necessitate vaccine strain replacement every 1–3 years. Antiviral compounds are available for prophylaxis and therapeutic treatment of severe IAV infections, but antiviral resistance is a continuing problem4. IAV pandemics can occur when viruses that contain HA or neuraminidase (NA) proteins for which most humans lack pre-existing immunity are transmitted to humans from reservoir species, including birds and pigs. Because pre-existing immunity does not exist at the time of their emergence, pandemic viruses typically cause higher rates of infection and mortality among humans. The dangers posed by both seasonal and pandemic IAVs demand a complete understanding of how these viruses replicate, transmit and cause disease, so that effective countermeasures can be developed.
At the end of virus replication, viral genetic material is packaged and released from infected cells so that additional cells or hosts can be infected. The IAV genome consists of eight unique segments of negative-sense viral RNA (vRNA), each associated with multiple nucleoprotein (NP) molecules and a single, trimeric polymerase complex comprising the PB2, PB1 and PA proteins. Collectively, each subunit of vRNA-NP-polymerase is referred to as a viral ribonucleoprotein (vRNP) complex. Following IAV entry into cells, vRNPs are released from endosomes and then transported into the nucleus, where they serve as templates for genome transcription and replication; after progeny vRNPs are generated, they are transported out of the nucleus and to the plasma membrane for incorporation into newly forming virions (for a comprehensive review of the IAV life cycle, readers are referred to5–7; also, see Box 1 for an overview of vRNP trafficking and function in infected cells).
Box 1. vRNPs in the IAV Life Cycle.
(Upper Left) IAVs enter cells predominantly through endocytosis after the viral HA protein (present on the virion surface) binds to host receptor molecules at the plasma membrane. (1) Following internalization, endosomal acidification activates HA conformational changes, which leads to fusion between the virion and endosomal membranes, providing the virus genome (i.e., vRNPs) with a portal of access to the cytoplasm. Concurrently, the viral M2 ion channel promotes acidification of the virion interior, which dissociates the M1 matrix protein from the viral genome. (2) vRNPs that are released from endosomes are transported into the nucleus, and (3) primary transcription results in the production of viral mRNAs, which are exported to the cytoplasm and translated into proteins by cellular ribosomes. Newly translated viral proteins are transported to the nucleus (PB2, PB1, PA, NP, M1 and NEP) or the plasma membrane (HA, NA and M2). (4) Upon the translation and nuclear entry of PB2, PB1, PA and NP, genome replication (i.e., the production of progeny vRNPs) ensues. (5) Progeny vRNPs are then exported to the cytoplasm with the assistance of the M1 and NEP proteins. (6) Subsequently, newly exported vRNPs are trafficked to the plasma membrane on Rab11 vesicles, and (7) vRNPs are incorporated into progeny virus particles containing HA, NA, M2 and M1. Virus release from the plasma membrane is mediated by the activities of at least two virion surface proteins, M2 and NA: M2 promotes scission of budding viruses from the plasma membrane, while NA prevents virus aggregation at the cell surface. Accessory viral proteins that modulate the host response but do not contribute directly to the events illustrated in Steps 1–7 (e.g., NS1, PB1-F2, PA-X) are not included in the diagram. “NPC”, nuclear pore complex.
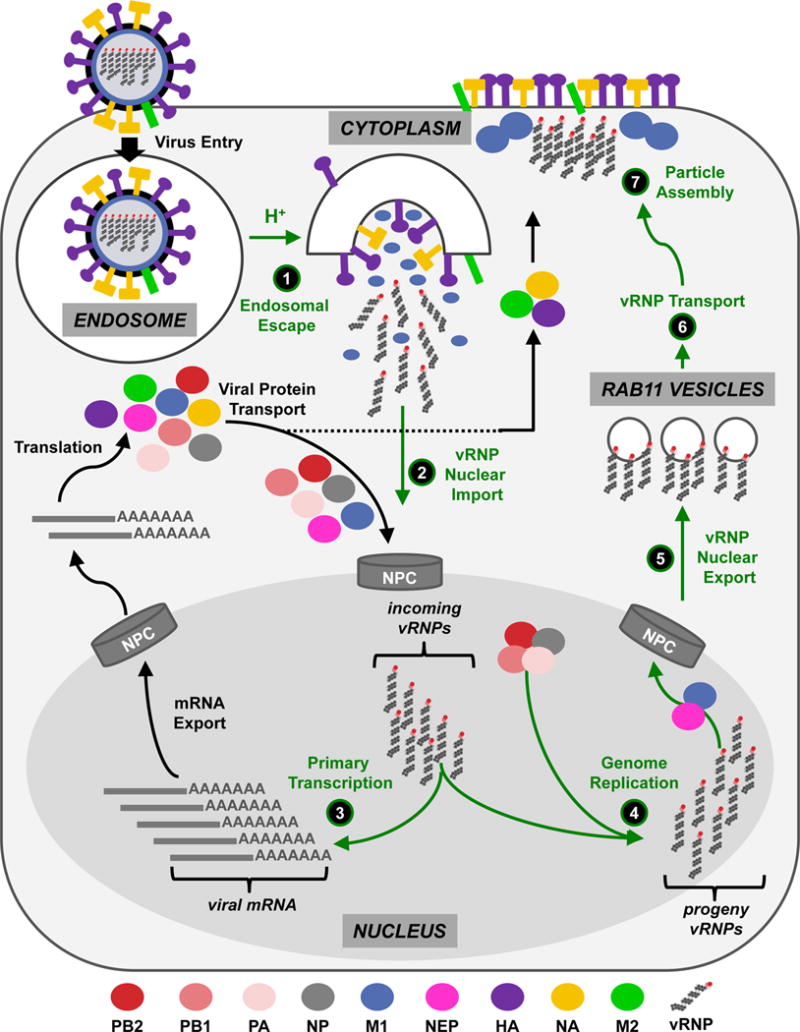
Due to IAV’s limited genetic coding capacity (8 genome segments may encode up to 14 known viral proteins), it depends on host proteins and pathways to mediate vRNP trafficking and to promote vRNP functions at all stages of the virus life cycle. Here, we review our current understanding of vRNP trafficking and functions in infected cells, emphasizing how vRNPs interface with host cell components and how vRNP structure is related to its functions. In addition, we highlight questions that remain unanswered. Because of space limitations, discussion of how the eight unique vRNPs are selectively assembled into infectious viruses, the interactions of vRNPs with the host innate immune system, and the contribution of vRNP components to host species adaptation are not discussed at length (for a review of these activities, readers are referred to6–8; also, the selective packaging of influenza vRNP segments is summarized in Box 2). Given the centrality of vRNPs to every aspect of the IAV life cycle, a clearer understanding of their fundamental roles during infection has enormous potential for revealing mechanisms of pathogenicity, providing a basis for understanding host adaptation, and for developing new methods to prevent or treat IAV disease.
Box 2. Selective Packaging of Influenza vRNP Segments.
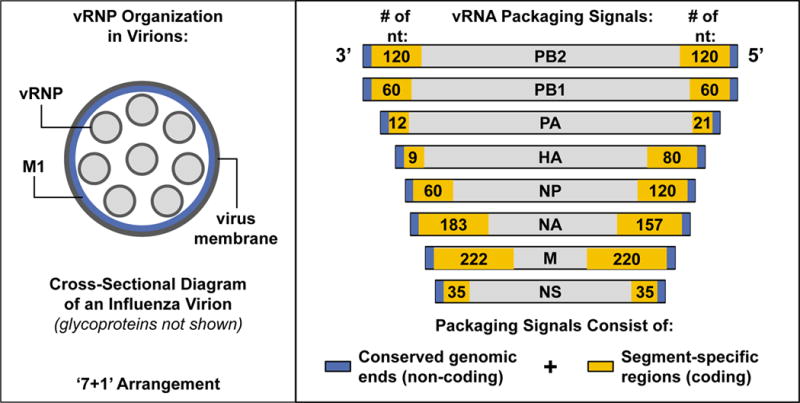
In order to preserve progeny virus infectivity, all eight unique vRNP segments – representing each influenza virus gene – must be incorporated into a budding influenza virion. Historically, two mechanisms were proposed to explain how this could occur: either packaging proceeds by a selective process, in which only one copy of each vRNP segment is selected from a mixture of all vRNPs for incorporation into a single virion; or alternatively, random packaging occurs, in which vRNP incorporation is arbitrary and any subset of any of the eight vRNP segments could be packaged together. The selective packaging model predicts that only eight vRNP segments would be present in most (or all) influenza virions released from infected cells. In contrast, the random packaging model requires the incorporation of >8 vRNP segments into each virion to maintain infectivity rates consistent with experimental observations. An elegant electron microscopy study, performed by Noda, et al.120, provided compelling evidence in support of the selective packaging model, establishing that the vast majority of influenza virions contain only eight vRNP segments, arranged in a ‘7 + 1’ formation (i.e., one central vRNP with 7 other vRNPs forming a circle around it) (a schematic representation of this arrangement is depicted in the left panel), and that the individual segments within virions differ in length in a manner consistent with differences in gene segment sizes. A more recent study by Chou et al.121, validated the observations made by Noda, et al. by using single molecule sensitivity fluorescence in situ hybridization (smFISH) with probes specific for viral genomic RNA. Although the mechanism governing how sets of eight vRNPs are selected from the cellular milieu for incorporation into virions is not well-clarified, each vRNA contains a packaging signal encompassing terminal genomic regions (which are conserved across all vRNA segments) and internal coding regions (which are unique to each segment)122–130 (packaging signals of each of the eight IAV vRNP segments, identified through deletion mapping studies in reporter constructs, are shown in the right panel; the lengths of the 3′ and 5′ coding regions [in nucleotides, nt] that are required for efficient packaging of each segment is indicated by the numbers in the gold boxes). How these packaging signals mediate selective incorporation of their cognate vRNP remains an open question. However, the recent derivation of the three-dimensional structure of vRNP sets within IAV particles suggests that intra-virion vRNP-vRNP and vRNA-vRNA interactions may occur, and that sets of vRNPs are incorporated into virus particles in a specific pattern131 (not shown in the figure panels). Other recent evidence supports the possibility of direct RNA-RNA interactions between IAV vRNAs132–136.
vRNP STRUCTURE
Within the vRNP complex, NP binds stoichiometrically to vRNA (approximately one NP molecule per 24 nucleotides) with high affinity and without sequence specificity – via the phosphate backbone of RNA polymers – leaving RNA bases accessible for pairing or genome duplication9–11. Higher order double-stranded RNA structures appear to be present along the length of the vRNP, because RNA within vRNPs is partially sensitive to RNase V1 digestion12. The genomic ends of the vRNA do not associate with NP; rather, they fold back and base-pair to form a double-stranded structure that is bound by the viral polymerase complex13,14. In contrast, internal vRNA bound by NP forms a flexible, helical filament closed by a loop at the pole opposite to the genomic ends11. Two recent studies refined the structural models of the influenza vRNP complexes – derived either in native form from purified virions or from cells transfected with plasmids encoding the vRNP components – by using advanced electron microscopic analyses15,16. These studies unambiguously demonstrated that the vRNP internal region comprises a twisted, anti-parallel double helix of vRNA-NP complexes that is maintained by interactions between NP subunits. The same structures are detected within influenza virions15. A schematic representation of the vRNP structure is shown in Figure 1.
Figure 1. Influenza vRNP complex.
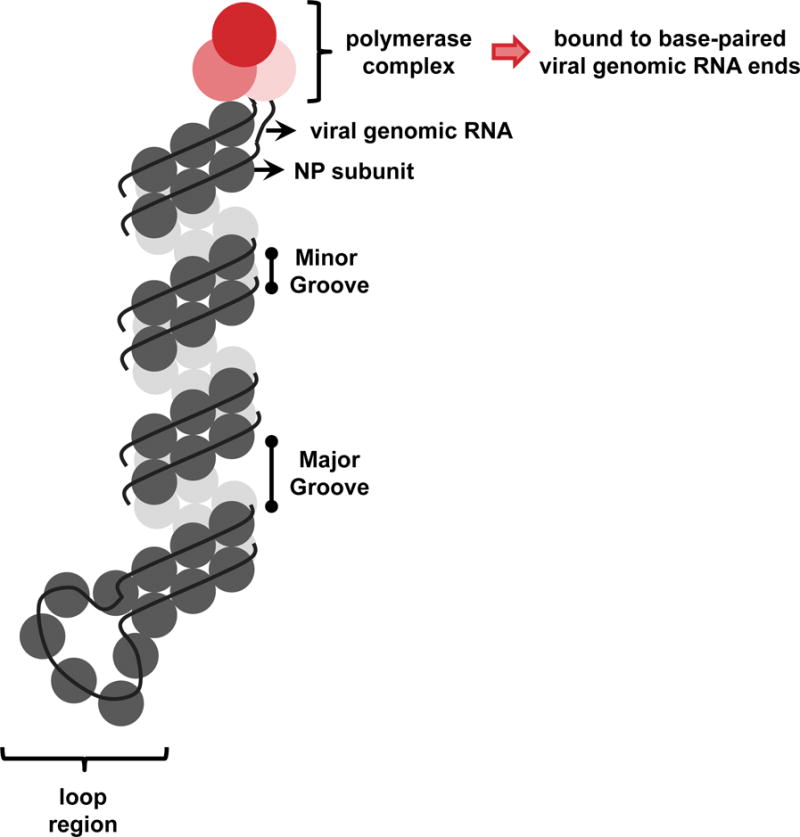
Each influenza vRNP consists of one single-stranded, negative-sense genomic RNA associated with multiple NP monomers and a single trimeric polymerase complex (composed of PB2, PB1, and PA). The 5′ and 3′ vRNA ends are complementary and base pair to form a double-stranded structure, which is bound by the polymerase complex at one end of the vRNP filament. The internal vRNA region is organized into an anti-parallel double helix, whose formation is driven by contacts between NP monomers (i.e., the ‘minor’ groove), and a loop can be observed at the end of the filament opposite to that bound by the polymerase complex.
vRNP TRAFFICKING INTO THE NUCLEUS
Influenza vRNP complexes within incoming virions are released from endosomes by the coordinated functions of the viral HA molecule and the M2 proton channel (Step 1 of Box 1). HA-mediated fusion between the viral and endosomal membranes provides an opening through which matrix-dissociated vRNPs can enter the cytoplasm. Meanwhile, M2 mediates vRNP dissociation from the virion matrix, which predominantly contains M1 protein, by acidifying the virion interior; this is necessary to make vRNPs competent for nuclear import17,18. Cytoplasmic vRNPs that have been dissociated from the M1 protein traffic into the nucleus by using the classical importin α/β1 (IMPα/β1)-dependent nuclear import pathway (Step 2 of Box 1). In this section, we describe the basic components of the IMPα/β1 pathway, the evidence that links it to IAV vRNP nuclear import, and the interactions between the vRNPs and IMPα/β1 pathway components. A model for vRNP nuclear import is provided in Figure 2.
Figure 2. Model for vRNP nuclear import.
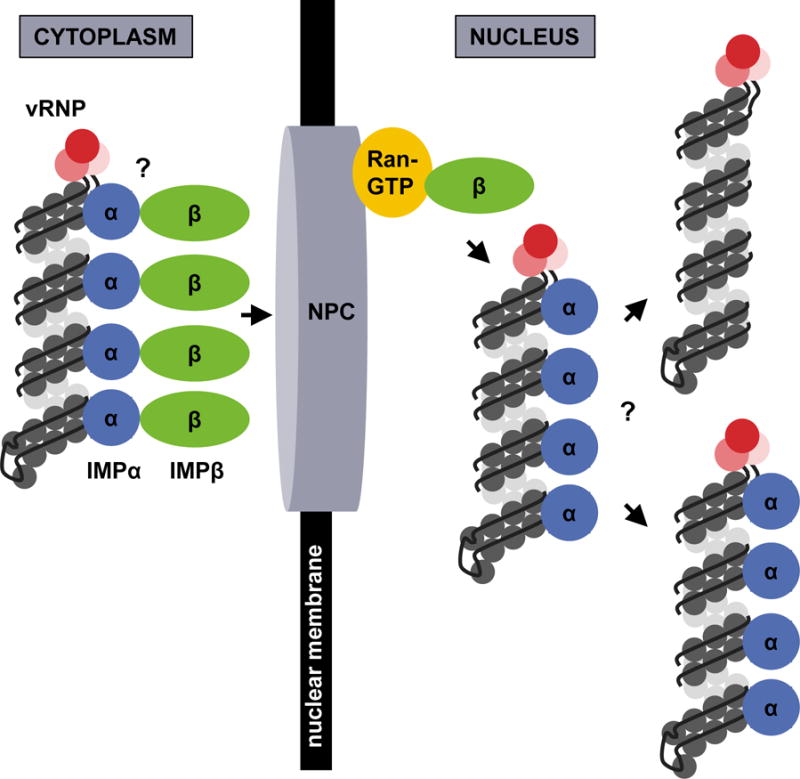
Uncoated, cytoplasmic vRNPs with exposed NP NLS motifs associate with IMPα, which in turn associates with IMPβ. The entire complex docks at the NPC and is transported into the nucleus, where Ran-GTP binds IMPβ and facilitates vRNP release into the nucleoplasm to initiate transcription and replication. Whether multiple IMPα and IMPβ molecules associate with each vRNP is unknown, as is the fate of the vRNP-associated IMPα once the vRNP cargo is released into the nucleoplasm.
IMPα/β1-mediated nuclear import
Proteins of less than 40 kDa can passively diffuse through the nuclear pore complex (NPC), whereas larger molecules and protein complexes, including vRNPs, must be transported by an active, energy-dependent process involving the recognition of nuclear localization signals (NLSs) in cargo proteins (reviewed in19). The classical pathway of cellular protein nuclear import is mediated by importin α (IMPα) adaptor proteins, which typically recognize basic (i.e., R/K-rich) NLSs, and in turn are recognized by the IMPβ transport receptor. Subsequently, the ternary IMPα-IMPβ-cargo complex traverses the NPC, and once inside the nucleus, IMPβ is dissociated by the activated form of the Ran GTPase (Ran-GTP), and IMPα-cargo complexes are released into the nucleoplasm. An additional dissociation step involving the cellular CAS protein is required to liberate IMPα from the cargo molecules20. Following cargo release, IMPα and IMPβ are individually recycled back to the cytoplasm.
vRNPs and the IMPα/β1 pathway
By using NP trafficking to the nucleus as a proxy for vRNP nuclear import at very early times post-infection, Martin and Helenius established that influenza vRNPs gain access to the nucleus via the NPC21. Studies with microinjected vRNPs further suggested that vRNPs use an active nuclear import process22. Additional studies with digitonin-permeabilized cells revealed that influenza vRNA docks at the NPC only in the presence of IMPα, IMPβ, and the viral NP, and further, that vRNA accumulation in the nucleus requires the Ran-GTPase23. Most recently, a small molecule inhibitor of the IMPβ-Ran GTPase interaction (importazole) was shown to impair vRNP nuclear transport24. These studies provide clear evidence that influenza vRNPs rely on the classical nuclear import pathway to enter the nucleus early in the virus life cycle.
Given that the classical nuclear import pathway mediates cargo transfer through recognition of NLS motifs, it is reasonable to predict that one or more of the vRNP protein components bears the NLS (or NLSs) required for vRNP nuclear import. All four vRNP protein components are reported to contain NLS motifs, and enter the nucleus either alone (NP and PB2) or in a complex (PB1 and PA)25–31. However, while each virion-associated vRNP includes only a single trimeric polymerase complex, multiple NP subunits are present, which could provide numerous NLS motifs to promote nuclear import.
The NP protein encodes a non-classical (i.e., not R/K-rich) NLS (‘ncNLS’) within its first 13 amino acids28,29, which binds to IMPα isoforms (IMPα1 and IMPα5)29, and is essential for both vRNP nuclear import in digitonin-permeabilized cells and for influenza virus replication32. NP also binds to IMPα3 and IMPα733, although interactions with these isoforms are less well characterized. In both of the recently solved structural models of the influenza vRNP15,16, the ncNLS motif is surface exposed, suggesting that it is available for interaction with IMPα adaptor proteins in vRNP complexes. Consistent with this observation, ultrastructural analysis of the surface availability of the ncNLS on purified vRNPs revealed multiple, periodically exposed motifs along the vRNP filament34. A second, classical, bipartite NLS (bNLS) has been identified in NP30, although its contribution to vRNP nuclear import has been questioned32. The bNLS was found not to be surface exposed on one of the vRNP structural models16, and experimental assessment suggested only limited surface availability on vRNP structures34 supporting, at most, a limited role in vRNP nuclear import. Nonetheless, this substantial collection of evidence implicates NP as the vRNP component responsible for IMPα binding and vRNP nuclear import in early IAV infection, and indicates that the ncNLS is most likely the dominant NLS motif.
Open questions
Several interesting questions remain regarding vRNP trafficking into the nucleus during the early stage of IAV infection:
First, while multiple NP ncNLS motifs are exposed on the surface of incoming vRNPs, it is not clear whether multiple IMPα/β1 complexes are associated with transporting vRNPs. In addition, because Ran-GTP is required to release vRNP complexes from docking at the nuclear membrane, it is presumed that IMPβ is dissociated from vRNP transport complexes as they enter the nucleus; however, whether IMPα is also dissociated remains to be determined. Some evidence suggests that IMPα can promote genome transcription and/or replication processes in addition to its activities in vRNP nuclear import (see below). Thus, if vRNPs recruit numerous IMPα proteins in the cytoplasm and maintain their association following nuclear import, this could provide an efficient means of rapidly establishing genome transcription. The potential contribution of the PB2 protein, which binds to multiple IMPα isoforms33,35, also should be examined more closely.
Second, it is not clear whether vRNPs are transported into the nucleus as individual components, as bundles of eight vRNP complexes, or as partially disrupted bundles containing some subset of the eight vRNPs. A recent study using single molecule sensitivity fluorescence in situ hybridization (smFISH) analysis to assess the distribution of two vRNA segments within the first hour of IAV infection suggested that these two vRNAs were only separated from each other after nuclear entry24. While these data are compelling, additional studies are needed to clarify whether more than two vRNPs can undergo nuclear import together, and if so, how this impacts the vRNP nuclear import mechanism.
GENERATION AND ASSEMBLY OF PROGENY vRNPs
Once in the nucleus, the primary roles of vRNPs are to transcribe viral mRNAs for the production of viral proteins (Step 3 of Box 1) and to replicate full-length complementary genomic RNA (cRNA) for amplification of vRNA and generation of progeny vRNPs (Step 4 of Box 1). In the following sections, we describe how vRNPs achieve genome transcription and replication and how host factors contribute to these processes. A model of primary transcription and genome replication is provided in Figure 3.
Figure 3. Model for genome transcription and replication.
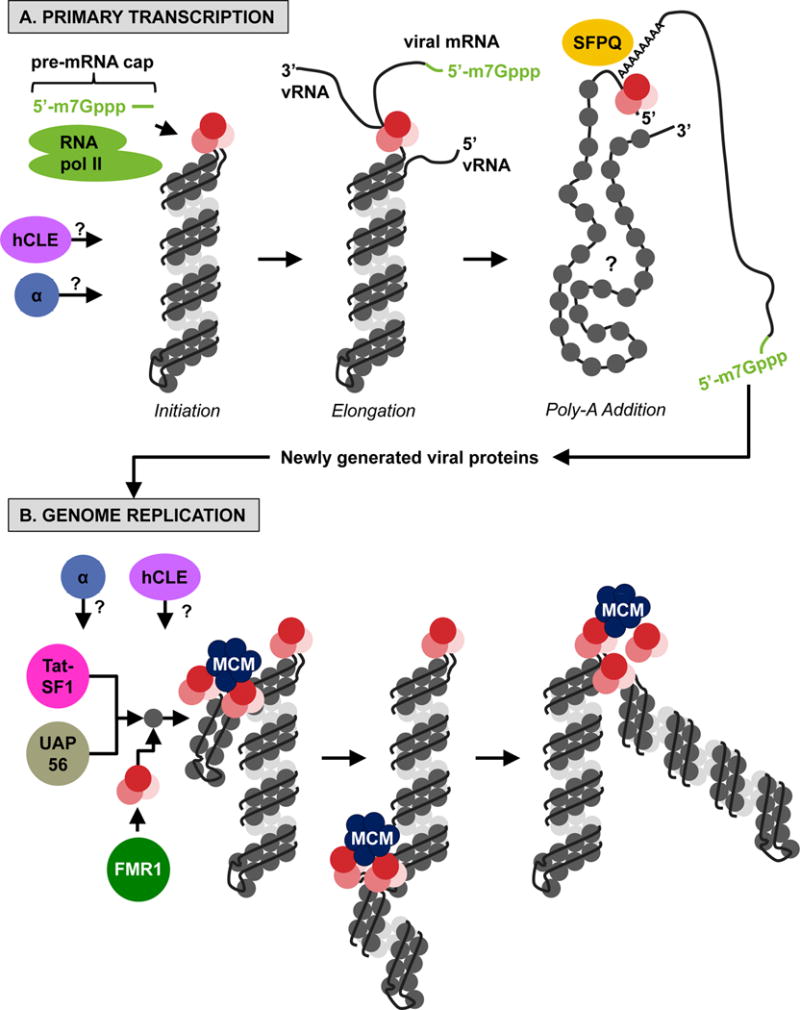
(A, Primary Transcription) Initiation: Following nuclear entry, vRNPs that were associated with incoming viruses transcribe viral mRNAs in cis using the resident polymerase complex bound to the double-stranded genomic ends and cellular pre-mRNA caps obtained by cap-snatching from cellular RNA polymerase II. Elongation: Then, the vRNA is threaded through (and copied by) the viral polymerase complex. Poly-A Addition: When the 5′ end of the vRNA is reached, it is held by the polymerase to promote the generation of the poly-A tail in conjunction with the cellular SFPQ protein. (B, Genome Replication) After new viral proteins are translated by the cellular machinery, soluble polymerases mediate genome replication in trans, promoted by the activity of the cellular MCM complex and the Tat-SF1 and UAP56 cellular proteins. The FMR1 protein stimulates the assembly of polymerase complexes and NP in the presence of vRNA. The specific contributions of hCLE and IMPα (represented as ‘α’ in both the upper and lower panels) are currently unknown.
Primary genome transcription
After nuclear import, influenza vRNPs are transcribed to produce viral mRNAs (‘primary transcription’) in a process that is independent of de novo viral protein synthesis36. Primary transcription is primer-dependent, and the primers used for this purpose, namely 7-methylguanosine-capped pre-mRNAs with 10–13 associated nucleotides, are obtained through ‘cap-snatching’ of cellular mRNAs by the viral polymerase complex37–39. Once the caps are acquired, transcripts are generated by replication from a vRNP template, by use of the native polymerase complex of the vRNP (i.e., in cis)40, and poly-A tails are produced by reiterative stuttering and copying of the poly-U sequence motif at the conserved 5′ end of the vRNA41,42. With the recent availability of the structural model of vRNPs isolated from cells, a more specific, albeit speculative, model for primary transcription has been described16. Specifically, prior to initiation of primary transcription, vRNA termini are base-paired and located adjacent to the PB1 active site. Then, after cap-snatching by the PB2 and PA proteins, the 3′ end of the vRNA is released from base-pairing and transferred into the PB1 active site. Transcription proceeds as the PA C-terminal domain feeds the vRNA strand to PB1, and after the full-length gene has been transcribed, the 5′ terminus remains associated with PB1 to facilitate polymerase stuttering and production of the poly-A tail.
Viral mRNA processing, transport, and translation
After primary transcription, viral mRNAs are exported to the cytoplasm and translated by cellular ribosomes. Subsequently, the protein components of the vRNP complex are imported into the nucleus where progeny vRNP complexes are assembled. Because the processes associated with viral mRNA post-transcriptional modifications (e.g., splicing), viral mRNA transport, and viral protein translation are not directly relevant to the function, production, or trafficking of vRNPs (aside from generating the raw components), they will not be described further here. Additional information can be found in other review articles43–45.
Genome replication and RNP assembly
The replication of full-length genomic cRNA from vRNP complexes (and similarly, replication of full-length genomic vRNA from cRNA-containing RNP complexes [cRNP]) contrasts strikingly with primary transcription. Genome replication requires newly translated NP and polymerase complex proteins to protect genomic RNA from degradation46; likely proceeds in the absence of an RNA primer due to the presence of 5′ triphosphates at the 5′ ends of both vRNA and cRNA43; and most interestingly, can be initiated by soluble polymerase complexes in trans (i.e., not by the polymerase complex that is resident to the RNP complex)40,47. Support for the trans genome replication model is provided by electron microscopy analyses of RNPs purified from cells, which showed multiple branched RNP-like structures emanating from the full-length parental RNPs16. Because the junction of these branches can be stained with antibodies that recognize the PB2 protein16, it is likely that they represent the activity of non-resident polymerase complexes in the process of generating progeny RNPs. Recently described RNP structural models further suggest that for genome copying to occur in trans, inter-strand connections between NP molecules, which form the minor groove of the RNP helical structure, may need to be locally disrupted15 (although definitive evidence for this is currently lacking); and that RNA packaging into RNP complexes is initiated by polymerase complex binding to the 5′ terminus of the nascent RNA molecule (by a polymerase complex different from the transcribing polymerase(s)), which protects the RNA and also initiates the sequential addition of NP oligomers in the 5′ → 3′ direction15,16. How polymerase complexes, NP oligomers, and the genomic RNA cooperate to achieve the final helical RNP structure remains an open question.
Host factors in genome transcription, replication and RNP assembly
Numerous host factors have been shown to influence genome transcription, replication and RNP assembly, but in general, no unifying themes tie these host factors together. Some (including RanBP548, HSP7049, and HSP9050,51) appear to indirectly promote influenza genome replication by enhancing the nuclear import and assembly of trimeric polymerase complexes, and therefore, will not be considered further here. Others with established and direct roles in regulating RNP transcription, replication and/or vRNP assembly are discussed below.
Host factors involved in primary transcription of viral mRNAs include the RNA polymerase II (Pol II) complex and SFPQ. The viral polymerase complex interacts with actively transcribing Pol II52, presumably to acquire cellular mRNA caps through cap-snatching. However, the PB2 and PA proteins also promote Pol II degradation, an ability that is associated with increased IAV pathogenicity in mice53. SFPQ is a multi-functional splicing factor that associates with several vRNP components54,55 and promotes poly-adenylation of viral mRNA transcripts54.
Progeny vRNP generation is promoted by the MCM helicase complex, UAP56, Tat-SF1 and FMR1. Five of the six components of the MCM complex, which is involved in cellular DNA replication, associate with the PA protein in isolation and with vRNPs in infected cells56. The MCM complex promotes production of full-length viral RNA products, possibly by stabilizing interactions between nascent RNA and the viral polymerase complex56. Both UAP56, an RNA helicase, and the transcription-splicing coupling factor Tat-SF1 interact with NP alone but not NP associated with viral RNA, and both stimulate genomic RNA synthesis by promoting the formation of NP-RNA complexes57–59. The FMR1 protein is an RNA-binding protein that is required for efficient IAV replication in cells and mice60. FMR1 expression stimulates the production of all viral RNA species, transiently interacts with vRNP complexes, and stimulates interactions between NP and polymerase complex proteins in the presence of vRNA, suggesting involvement in vRNP assembly60.
Other host factors, including hCLE and IMPα isoforms, have established roles in enhancing polymerase activity, but the specific mechanism(s) are not well-clarified. hCLE, a positive modulator of Pol II activity61, interacts with several vRNP components, and its knockdown leads to impaired virus replication, decreasing both negative and positive polarity viral RNA, through an unknown mechanism62. The IMPα1 and IMPα7 cellular nuclear import receptors positively regulate viral polymerase activity in mammalian, but not avian, cells independently of nuclear import functions63,64. Although the specific mechanism is unknown, viruses expressing PB2 proteins that possess a mammalian adapting amino acid (lysine at residue 627), which significantly increases the replicative ability of the viral polymerase complex in mammalian, but not avian, cells65, exhibit impaired polymerase activity when IMPα1 or IMPα7 protein levels are reduced by use of RNA interference (RNAi), with no effect on the nuclear localization of the PB2 or NP proteins63. A similar reduction in polymerase activity was not observed for the avian PB2 variant (glutamic acid at residue 627)63, suggesting that IMPα proteins play an important role in IAV adaptation to different hosts. An additional study clearly demonstrated that IMPα7 contributes to adaptation of avian viruses in mammals33. Another mammalian adapting PB2 mutation (asparagine at residue 701, ‘701N’) indirectly enhances IAV polymerase activity in mammalian cells by increasing PB2 binding to IMPα1, thereby increasing PB2 nuclear import35. However, whether PB2 proteins containing the 701N mutation have impaired polymerase activity relative to their avian counterpart (i.e., proteins with aspartic acid at residue 701) in mammalian cells with reduced Impα1 or Impα7 expression is unknown.
Open questions
Although we have made substantial progress in understanding primary transcription and genome replication from vRNPs, one area that remains largely unexplored is the specific contribution of host factors, particularly in light of observations that vRNP components are strongly implicated in IAV adaptation to mammalian hosts8,66–69. Several recent studies have revealed an abundance of interactions between host proteins and components of the vRNP complex70–72, and it will be interesting to fully establish how these factors contribute to the role of vRNPs in the virus life cycle. Another question is whether vRNPs associate with a specific nuclear site for primary transcription and genome replication activities. Given that vRNP targeting to a specific site could increase the likelihood of encountering host factors that are important for transcription and replication, this is a worthwhile avenue for future research.
NUCLEAR EXPORT OF PROGENY vRNPs
In the late phase of infection, progeny vRNPs must be transported from the nucleus to plasma membrane sites of new virus formation. The first step in this process is nuclear export (Step 5 of Box 1). IAVs use the cellular CRM1-dependent nuclear export pathway to mediate transport of vRNPs from the nucleus to the cytoplasm. In the following sections, we describe the basic components of the CRM1 pathway, how IAV usurps this pathway to preferentially export vRNPs, the interactions between vRNPs and CRM1 pathway components, and how cellular kinase signaling may contribute to this process. The synthesis of these observations is summarized by the model of vRNP nuclear export shown in Figure 4.
Figure 4. Model for vRNP nuclear export.
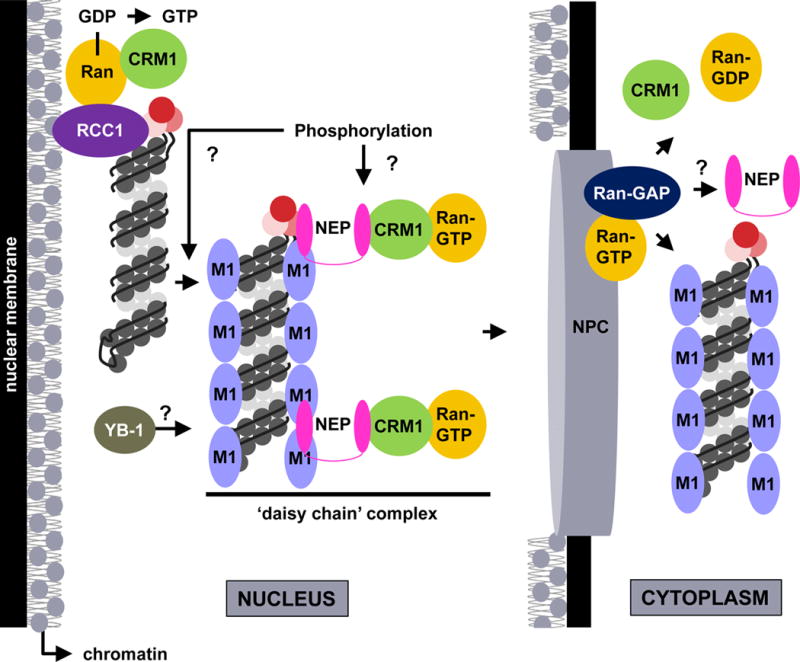
Late in infection, vRNPs localize to the chromatin at the nuclear periphery and associate with components of the CRM1 nuclear export pathway (e.g., RCC1). After a signal that may include phosphorylation of an unknown target protein, mediated by signal transduction cascades activated by HA at the plasma membrane, the vRNPs are released from the chromatin. The vRNPs are subsequently exported from the nucleus by means of a ‘daisy chain’ complex, through which the viral NEP protein acts as an adaptor between vRNP-M1 and CRM1-Ran-GTP. The cellular YB-1 protein may be co-transported with vRNPs. At the cytoplasmic side of the NPC, Ran-GTP hydrolysis releases transport factors and vRNPs into the cytoplasm, where vRNPs remain associated with M1. Whether NEP remains associated with the cytoplasmic vRNPs remains unclear.
The CRM1 nuclear export pathway
The cellular CRM1 protein is a nuclear export receptor that recognizes cargo proteins bearing leucine-rich nuclear export signals (NESs). CRM1 binds NES motifs in cargo proteins in the nucleus, and in association with the GTP-loaded Ran GTPase, transports cargo across the NPC. The Ran GTPase activating protein (RanGAP), which is tethered to the cytosolic face of the NPC, hydrolyzes Ran-GTP to Ran-GDP, and this facilitates complex dissociation and cargo release into the cytoplasm. Subsequently, Ran-GDP is transported back into the nucleus, and its GDP is exchanged for GTP by the chromatin-associated Ran guanylnucleotide exchange factor (Ran-GEF), RCC1, to enable another round of cargo protein nuclear export.
vRNPs and the CRM1 export machinery
Influenza vRNPs are retained in the nucleus of infected cells in the presence of leptomycin B (LMB; a specific CRM1 inhibitor)73–75, indicating that the CRM1 nuclear export pathway is essential for vRNP nuclear export. Interestingly, LMB treatment leads to vRNP accumulation at the nuclear periphery73, suggesting that this localization pattern represents an intermediate step in vRNP nuclear export, preceding vRNP passage through the NPC. A subset of nuclear vRNPs is tightly bound to chromatin at the nuclear periphery of infected cells76,77, and the amount of vRNPs in this region increases after LMB treatment77. These observations imply that vRNP association with chromatin could be the intermediate nuclear export step originally identified by examining vRNP localization after LMB treatment. Theoretically, chromatin association would position vRNPs in close proximity to RCC1, the Ran-GEF, which normally associates with chromatin; in support of this prediction, RCC1 and vRNPs form complexes in infected cells77. CRM1 is also recruited to chromatin during influenza virus infection, and compared to uninfected cells, complexes of CRM1-Ran-RCC1 are increased77. Given that CRM1-mediated nuclear export of cellular cargo decreases after infection despite the increased potential for CRM1 pathway activity77, it seems likely that IAV induces alterations to the host cell that result in sequestration of the CRM1 pathway machinery on chromatin at the nuclear periphery to exclude cellular cargo and provide preferential access to vRNPs.
Among the vRNP complex proteins, only NP possesses a known NES. However, infected cells that lack M1 or NEP cannot export NP from the nucleus17,76, indicating that these proteins are necessary for the vRNP nuclear export process. Evidence linking M1 and NEP to vRNP nuclear export follows:
The M1 protein is directly implicated in the regulation of vRNP nuclear export, because cells microinjected with anti-M1 antibodies exhibited NP confinement to the nucleus during infection17, and because exogenous M1 expression in infected cells with impaired “late” gene expression (“late” genes include HA, M1, M2 and NA, and exhibit delayed expression relative to the NP gene; their expression can be impaired with the H7 kinase inhibitor78) rescued the ability of vRNPs to accumulate in the cytoplasm76. An NES motif has been reported for the M1 protein, and although mutation of this motif reduces the efficiency of vRNP nuclear export during infection, the same NES is not sensitive to LMB, and thus, likely does not interact with CRM179. This indicates that the M1 NES motif is not directly responsible for linking vRNPs to the CRM1 export pathway. Yet, M1 and vRNPs must interact to promote vRNP nuclear export80, so it is possible that M1 serves as an intermediary between vRNPs and an additional CRM1 association factor. Alternatively, because vRNPs are tightly associated with chromatin76,77 and M1 has also been associated with chromatin proteins77,81, another (not necessarily exclusive) possibility is that M1 binding is required to release vRNPs from chromatin to enable nuclear export progression. In addition to promoting vRNP nuclear export, the association between M1 and vRNPs prevents nuclear re-import of vRNPs82, potentially because M1 shields vRNP NLS motifs from recognition by nuclear import adaptor proteins; however, associations between M1 and cytoplasmic vRNPs are not well-established, so the precise mechanism of M1-impaired vRNP nuclear re-import requires further investigation.
The influenza NEP protein (or NS2) also plays an important role in vRNP nuclear export. Similarly to M1, when anti-NEP antibodies were microinjected into the nucleus of infected cells, NP was retained in the nucleus, suggesting that vRNP nuclear export was impaired when NEP protein functions were blocked83. Further, infectious influenza viruses lacking NEP expression could not be recovered by reverse genetics, indicating that NEP is essential to the IAV life cycle. This was attributed to a defect in vRNP nuclear export because NP was restricted to the nucleus in cells infected with the recovered, NEP-deficient virus-like particles84. NEP interacts directly with CRM1 through two N-terminal hydrophobic NES motifs, which are both sensitive to LMB and sufficient to transfer nuclear export activity to nuclear constrained proteins83–85. Mutation of either NES motif in isolation does not ablate NEP binding to CRM184,85, while mutation of both motifs in parallel results in a nearly complete loss of the NEP-CRM1 interaction85. Mutations in NES1 inhibit virus recovery by reverse genetics and strongly impede vRNP nuclear export84; whereas mutations in NES2 do not abolish virus recovery but severely impair viral growth kinetics and reduce the rate of vRNP nuclear export in infected cells85. Although these observations provide strong evidence of a role for NEP in vRNP nuclear export, no direct interaction between full-length NEP and the vRNP complex has been established83,86–89. However, since NEP can interact directly with the M1 protein86,90, which in turn interacts with vRNPs80, a ‘daisy-chain model’ for influenza vRNP nuclear export has been proposed, whereby NEP acts as an adaptor between the CRM1 nuclear export pathway and M1-vRNP complexes (Figure 4). Recent data suggests that a more refined model – in which NEP simultaneously interacts with M1, the PB1 subunit of the vRNP-associated polymerase complex, and the CRM1 nuclear export receptor89 – may be necessary to explain the mechanism of vRNP nuclear export; further work is needed to clarify this process. Additionally, whether NEP remains associated with vRNP complexes that have been transported to the cytoplasm is currently unknown.
The NP protein can undergo nuclear export in the absence of other viral proteins28,82 and encodes residues involved in nuclear export within its N-terminal 38 amino acids28. A recent study suggested that other NP NES motifs may exist91. In addition, NP binds directly to CRM1 in vitro74. These observations imply that the NP protein could mediate vRNP nuclear export through a direct interaction with CRM1; however, the contribution of specific NP NES motifs to vRNP nuclear export has never been established in the context of infected cells or with isolated vRNP complexes. Accordingly, the role of NP in vRNP nuclear export remains unclear.
Cellular kinases and vRNP nuclear export
Cellular protein kinase activity may impact vRNP nuclear export. Specifically, vRNP nuclear export is impaired in infected cells treated with a protein kinase C inhibitor76, a Raf/MEK/ERK MAPK inhibitor92, a receptor tyrosine kinase inhibitor93, and a serum- and glucocorticoid-regulated kinase 1 inhibitor94, implying that phosphorylation of viral and/or cellular factors is required for this process. Interestingly, vRNP nuclear export can be enhanced by viral HA-mediated activation of the cellular Raf/MEK/ERK MAPK signaling pathway, which occurs when HA is expressed at the plasma membrane95,96. This suggests an indirect role for HA in promoting vRNP nuclear export. In addition, because HA is abundant in virus particles, and vRNPs must ‘meet’ HA at plasma membrane sites to form progeny viruses, HA-mediated Raf/MEK/ERK induction could serve as a signal to activate vRNP nuclear export when the budding sites are ‘ready’ (i.e., when HA is present). Although specific phosphorylation events and the kinases involved in promoting vRNP nuclear export have not yet been defined, it is notable that viral proteins with direct roles in the vRNP nuclear export process (i.e., NP, M1, and NEP) all exist as phosphoproteins in infected cells97–101.
Open questions
While considerable effort has been applied towards a better understanding of the vRNP nuclear export mechanism, questions still remain:
First, the recent identification of chromatin targeting as a mechanism to facilitate vRNP access to the nuclear export machinery is interesting, and opens new avenues for research. It will be important to determine how this sequestration occurs and the extent to which it affects host nucleocytoplasmic trafficking.
Second, while the ‘daisy chain’ hypothesis can explain how influenza vRNPs are transported to the cytoplasm, tangible evidence to support complex formation in infected cells is lacking, despite being a requirement to validate this proposed mechanism. Further, it is unknown whether vRNP complexes are exported from the nucleus as individual vRNP components or as complexes of multiple vRNPs, although a few studies seem to support the former possibility24,102. Because this information could provide insights into the mechanism of incorporation of the full set of eight unique vRNPs into the assembling virions, further clarification of the nature of the transport complex is necessary.
Third, although many reports indicate that phosphorylation of some viral or cellular target(s) may be vital for vRNP nuclear export, essentially no data are available to explain this phenomenon. Additional studies are needed to identify the viral or cellular target(s) involved.
vRNP TRANSPORT THROUGH THE CYTOPLASM
Nuclear export constitutes only part of the vRNP’s trek to the IAV budding site. vRNPs must also navigate through a dense matrix of cytoplasmic structures to reach the plasma membrane (Step 6 of Box 1). Combined with the observation that IAV progeny viruses bud from discrete plasma membrane sites103, it is logical to predict that vRNP cytoplasmic transport is organized rather than random. Indeed, a recent focus on this phase of the IAV life cycle has revealed that vRNPs are transported on microtubule networks, in association with Rab11-positive recycling endosomes.
Cytoskeletal networks
Three types of cytoskeletal filaments are found in distinct networks in eukaryotic cells: actin microfilaments, microtubules, and intermediate filaments. All three contribute to the structure, stability, and integrity of the cell, but cargo-mediated transport occurs only along microfilaments and microtubules. Myosin motors transport cargo on actin filaments, which are concentrated directly under the plasma membrane. Alternatively, kinesin and dynein motors transport cargo on microtubules, which are nucleated at the microtubule organizing center (MTOC), and typically emanate from the centre of the cell to its periphery.
Microtubules and vRNPs
Shortly after nuclear export, vRNPs accumulate near the MTOC104,105, located immediately adjacent to the nucleus. Later, they are aligned with microtubule networks en route to the plasma membrane104,106. While these observations are suggestive of the use of microtubules in vRNP cytoplasmic transport, several lines of evidence provide more direct support for this concept: live cell imaging studies with fluorescently tagged vRNP components demonstrate vRNP movement along microtubule tracks106, which is characterized by intermittent, saltatory motions105–107 (this type of motion is well-established for microtubule-mediated cargo108,109); cells treated with microtubule depolymerizing agents exhibit altered vRNP distribution and reduced microtubule-like movement104,105,107; and IAV growth is reduced in cells treated with a microtubule-depolymerizing agent104. Overall, these findings strongly implicate microtubule networks in vRNP cytoplasmic transport, although no studies have yet addressed whether a specific motor complex is involved.
Rab11 and vRNPs
Membrane-bound vesicles are a common microtubule cargo in eukaryotic cells. Under normal physiological conditions, early and late endosomes and lysosomes are transported predominantly inward, towards the centre of the cell, whereas secretory vesicles derived from the Golgi complex and vesicles of the recycling endosome system are transported outward, towards the cell periphery. Both vRNA and vRNPs co-localize extensively with Rab11-positive recycling endosomes in infected cells24,105–107,110 – initially concentrated at the MTOC immediately after nuclear export, and later localized throughout the cytoplasm and near the plasma membrane – suggesting that vRNPs ‘piggy-back’ on Rab11 vesicles for transport through the cytoplasm. At the end of the IAV life cycle, vRNPs appear to transfer from Rab11-positive vesicles to the plasma membrane110, suggesting that a mechanism exists to extract vRNPs from Rab11 vesicle surfaces for incorporation into budding virions. The lack of Rab11 detection in influenza virus particles111 reinforces this premise.
In support of a specific role for Rab11 in mediating vRNP cytoplasmic transport (as opposed to ‘guilt by co-localization’), both Rab11 protein knockdown and exogenous overexpression of a dominant-negative Rab11 mutant protein impaired vRNP association with Rab11-positive vesicles, disrupted vRNP accumulation at the plasma membrane, and sharply reduced infectious progeny virus output106,110. In infected cells, Rab11 forms a complex with all vRNP protein components and with vRNA, but when complexes are treated with RNase, only the association with the polymerase complex proteins (PB2, PB1 and PA) is maintained106. Further, direct binding is observed only between Rab11 and the PB2 protein of the polymerase complex105. Because a single polymerase trimer is associated with each vRNP complex – localized with the base-paired genomic ends – these observations predict that the Rab11 GTPase recruits vRNPs to the recycling endosome membrane in a head-to-tail orientation, with polymerase-bound genomic ends attached to the membrane and the looped ends facing away from the membrane. In addition to providing a convenient mechanism for cytoplasmic transport, Rab11 vesicles may function as a platform for concentrating vRNPs (thereby reducing their diffusion) prior to virus assembly and budding, which may promote interactions necessary for creating supra-molecular complexes of vRNPs. However, this hypothesis has not been experimentally tested.
Other host factors and vRNP cytoplasmic transport
Several additional host proteins have been ascribed potential functions in the cytoplasmic transport of vRNP complexes. Two components of host ribonucleoprotein complexes that regulate translation of cellular mRNAs – YB-1 and STAU1 – are associated with influenza vRNPs during late-stage infection112,113. The YB-1 protein translocates to the nucleus in infected cells, where it localizes with vRNA and later can be found in complexes with α-tubulin, Rab11, and vRNP components in the cytoplasm112, suggesting that it undergoes nuclear export in complex with vRNPs. YB-1 overexpression also stimulates the production of infectious progeny viruses in a Rab11-dependent manner112. On the basis of these observations, it has been suggested that YB-1 facilitates the delivery of vRNPs to Rab11 endosomes at the MTOC following nuclear export, but direct evidence in support of this hypothesis is lacking. The role of STAU1 is less clear, although this factor is thought to promote a late aspect of the influenza virus life cycle because STAU1 knockdown via RNAi does not impair viral protein expression or vRNP nuclear export113. The HIV rev binding protein (HRB), which is a host endosomal protein114,115 and an NEP interaction partner83, is essential for efficient influenza virus replication, and can be visualized in association with influenza vRNPs at the MTOC after vRNP nuclear export116. RNAi-mediated HRB knockdown results in the retention of vRNPs in the perinuclear region, suggesting that HRB plays a role in vRNP transport from the MTOC to the plasma membrane.
Viral proteins and vRNP recruitment to the budding site
As noted above, the Rab11 protein does not appear to be incorporated into budding virus particles, implying that another mechanism exists for vRNP recruitment at the budding site (Step 7 of Box 1). Several reports have suggested that the M2 protein is required for this process117–119, and that it occurs through a direct interaction between M1 and the M2 cytoplasmic tail119. As already noted, the M1 protein is required for vRNP nuclear export17,76 and prevents vRNP nuclear re-import in late-stage infection, suggesting that M1 remains associated with vRNPs that have been transported to the cytoplasm. However, M1 association with vRNPs on Rab11 vesicles has not been demonstrated. Nonetheless, these observations suggest that M1-coated vRNPs are recruited by the M2-M1 interaction once Rab11 has delivered vRNPs to the plasma membrane.
Open questions
Recent data have unambiguously implicated microtubules and Rab11 in vRNP transport through the cytoplasm. Yet, how the vRNPs are recruited to the Rab11 vesicles, how these vesicles are attached to microtubules, and the signals that promote Rab11 vesicle trafficking to the cellular periphery remain open questions.
PERSPECTIVE AND FUTURE DIRECTIONS
The multi-dimensional trafficking and functions of vRNP complexes are essential to the IAV life cycle and, although complex, merit the considerable effort that has been applied towards their improved understanding. However, much remains to be learned, and additional work is required to elucidate the full spectrum of vRNP activities in infected cells. Key open questions have already been described throughout this review. Below, we elaborate on the most important of these questions, emphasizing crucial experimental approaches that are required for generating the most comprehensive model of vRNP function during IAV infection.
A significant and unsolved problem in IAV biology is the mechanism by which a complete set of eight vRNP segments is selected and assembled for incorporation into progeny viruses. This process is likely to involve aspects of vRNP structure; the accessibility of vRNAs within vRNP structures, which could allow for interactions with other vRNAs, proteins or cellular RNA species; inter-segment interactions between unique vRNPs (driven either by vRNA-vRNA or protein-based associations) (see Box 2 for additional details); and intracellular trafficking mechanisms that promote interactions between vRNPs. A more advanced understanding of these processes will require the use of cutting-edge technologies in structural analysis (e.g., crystallization of native RNP structures), high-resolution electron microscopy, innovative light microscopy approaches (e.g., super-resolution methods in combination with live cell imaging and/or smFISH), and the application of these technologies to realistic models of IAV infection (e.g., primary cells or tissue explants). Increased knowledge about the mechanism of vRNP assembly will provide insights into how IAVs undergo reassortment (and cause pandemics), and further, may open novel channels for antiviral development (e.g., molecules that interfere with vRNP-vRNP interactions). Thus, resolving the mechanism of vRNP assembly should be pursued in earnest.
Another major gap in knowledge is the lack of a comprehensive understanding of the sizeable network of host proteins that interact with the IAV polymerase complex and/or vRNPs, and how this network contributes to vRNP activities. An even greater paucity of information exists for how avian and mammalian vRNP components differentially interact with this host protein network, and whether putative differences could impact inter-species virus transmission. The continued use of RNAi and other mechanisms to interfere with expression or activity of specific host factors within this network will be essential for defining a particular factor’s role in regulating vRNP functions, and when possible, studies should be extended into animal models of IAV disease, including knockout mice. In addition, computational strategies could be more effectively harnessed in combination with available and new high throughput datasets (e.g., protein-protein interaction data, genome-wide knockdown studies and ‘OMICs’ analyses of infected cells or tissues) to identify host factor candidates with the strongest regulatory influence over the virus life cycle and/or inter-species transmission.
As we have already described, there is a critical need for development of novel antiviral strategies for treatment of severe influenza virus disease in humans. Currently, two classes of antiviral drugs are available: adamantanes, which impair viral M2 ion channel activity, and neuraminidase inhibitors, such as oseltamivir and zanamivir. However, because of the remarkable propensity of influenza viruses to adapt under selective pressure, use of these compounds has resulted in the widespread emergence of virus strains that are resistant to one or both inhibitor classes4. An alternative antiviral development strategy, targeting an essential host factor (rather than a viral protein), may reduce the emergence of resistant variants because it may be difficult for the virus to bypass the use of such a factor. Since vRNPs are central to the IAV life cycle and depend on host factors for their functions, knowledge gained from a deeper understanding of vRNP-host machinery interactions could yield unique strategies for host-targeted antiviral development, and therefore, this knowledge must be further expanded.
Altogether, the combination of extensive previous work and future efforts aimed at clarifying mechanisms that regulate vRNP functions in IAV-infected cells will not only improve our basic scientific knowledge, but may also reveal novel mechanisms of IAV pathogenicity and adaptation to mammalian hosts, and further, could lead to identification of novel druggable targets (viral or cellular) that might be exploited to prevent or treat IAV disease.
ONLINE SUMMARY.
This paper describes the trafficking and functions of influenza A virus (IAV) viral ribonucleoproteins (vRNPs), the genetic material of IAV, within the host cell. We emphasize how vRNPs interact with and depend on host factors and pathways, how vRNP structure contributes to its function and the key open questions that still need to be solved.
-
▪
The structure of vRNPs in their native form is described.
-
▪
The mechanism of vRNP nuclear import is explained, including a discussion of how vRNP components interact with the cellular importin α/β1 nuclear import pathway.
-
▪
Primary genome transcription and genome replication is described, focusing on how host factors contribute to these processes.
-
▪
vRNP nuclear export, which is mediated by the cellular CRM1 nuclear export pathway, is illustrated.
-
▪
Recent work implicating the cellular Rab11 vesicle transport system and microtubules in vRNP cytoplasmic transport in late stage infection is described.
-
▪
We highlight important open questions and suggest methods that could be used to address these gaps in knowledge.
Acknowledgments
The authors are grateful to Susan Watson for scientific editing of the manuscript. This work was supported by grants-in-aid from the Ministry of Health, Labour, and Welfare, Japan, by ERATO (Japan Science and Technology Agency), and by National Institute of Allergy and Infectious Diseases Public Health Service research grants.
Biographies
Amie J. Eisfeld is a staff scientist at the Influenza Research Institute at the University of Wisconsin-Madison. She performed her doctoral dissertation work on varicella-zoster virus and obtained her Ph.D. from the University of Pittsburgh, School of Medicine. In her post-doctoral studies with Yoshihiro Kawaoka, she identified novel host-pathogen interactions that facilitate influenza vRNP trafficking in host cells. Her current work focuses on host-pathogen interplay and systems biology approaches for dissecting the mechanisms of viral pathogenesis.
Gabriele Neumann is a Research Professor at the Influenza Research Institute at the University of Wisconsin-Madison. She obtained her Ph.D. from Justus-Liebig University, Giessen, Germany. Her post-doctoral work with Yoshihiro Kawaoka led to the establishment of reverse genetics systems for influenza and Ebola viruses. Her research interests include the evolution and pathogenesis of influenza and Ebola viruses.
Yoshihiro Kawaoka is a Professor of Pathobiological Sciences at the Influenza Research Institute at the University of Wisconsin-Madison; and a Professor of Virology at the University of Tokyo, where he directs the International Research Center for Infectious Diseases. He performed graduate work on Yersinia enterocolitica and post-doctoral work with Robert Webster at St. Jude Children’s Research Hospital in Memphis, Tennessee. His laboratories conduct research on evolution, pathogenesis, transmission and systems biology of negative sense RNA viruses, including influenza A virus and Ebola virus.
Footnotes
AUTHOR CONTRIBUTIONS STATEMENT
AJE, GN, and YK wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.WHO. Influenza (Seasonal) Fact sheet N°211. 2009 < http://www.who.int/mediacentre/factsheets/fs211/en/index.html>.
- 2.Molinari NA, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 3.Simonsen L. The global impact of influenza on morbidity and mortality. Vaccine. 1999;17(Suppl 1):S3–10. doi: 10.1016/s0264-410x(99)00099-7. [DOI] [PubMed] [Google Scholar]
- 4.van der Vries E, Schutten M, Fraaij P, Boucher C, Osterhaus A. Influenza virus resistance to antiviral therapy. Adv Pharmacol. 2013;67:217–246. doi: 10.1016/B978-0-12-405880-4.00006-8. [DOI] [PubMed] [Google Scholar]
- 5.Matsuoka Y, et al. A comprehensive map of the influenza A virus replication cycle. BMC systems biology. 2013;7:97. doi: 10.1186/1752-0509-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw ML, Palese P. In: Fields Virology Vol. Section II: Specific Virus Families. Knipe DM, Howley P, editors. Lippincott Williams and Wilkins; 2013. [Google Scholar]
- 7.Wright PF, Neumann G, Kawaoka Y. In: Fields Virology Vol. Section II: Specific Virus Families. Knipe DM, Howley P, editors. Lippincott Williams and Wilkins; 2013. [Google Scholar]
- 8.Gabriel G, Fodor E. Molecular Determinants of Pathogenicity in the Polymerase Complex. Current topics in microbiology and immunology. 2014 doi: 10.1007/82_2014_386. [DOI] [PubMed] [Google Scholar]
- 9.Scholtissek C, Becht H. Binding of ribonucleic acids to the RNP-antigen protein of influenza viruses. The Journal of general virology. 1971;10:11–16. doi: 10.1099/0022-1317-10-1-11. [DOI] [PubMed] [Google Scholar]
- 10.Baudin F, Bach C, Cusack S, Ruigrok RW. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. The EMBO journal. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Compans RW, Content J, Duesberg PH. Structure of the ribonucleoprotein of influenza virus. Journal of virology. 1972;10:795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamanaka K, Ishihama A, Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. The Journal of biological chemistry. 1990;265:11151–11155. [PubMed] [Google Scholar]
- 13.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor E, Seong BL, Brownlee GG. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. The Journal of general virology. 1993;74(Pt 7):1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 15.Arranz R, et al. The structure of native influenza virion ribonucleoproteins. Science. 2012;338:1634–1637. doi: 10.1126/science.1228172. This paper describes the three-dimensional structure of native RNPs derived from influenza virions. [DOI] [PubMed] [Google Scholar]
- 16.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. A companion to the paper cited in reference 15, this paper describes the three-dimensional structure of native RNPs derived from cells expressing influenza RNP complex components. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. This paper was the first to establish that the influenza M1 protein regulates both nuclear import and nuclear export of influenza vRNPs. [DOI] [PubMed] [Google Scholar]
- 18.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. Journal of virology. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nature reviews Molecular cell biology. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 20.Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 21.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. Journal of virology. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. This paper was the first to show that influenza vRNPs enter the nucleus through the nuclear pore complex by using an active mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemler I, Whittaker G, Helenius A. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology. 1994;202:1028–1033. doi: 10.1006/viro.1994.1432. [DOI] [PubMed] [Google Scholar]
- 23.O’Neill RE, Jaskunas R, Blobel G, Palese P, Moroianu J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. The Journal of biological chemistry. 1995;270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 24.Chou YY, et al. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS pathogens. 2013;9:e1003358. doi: 10.1371/journal.ppat.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarendeau F, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature structural & molecular biology. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 26.Nath ST, Nayak DP. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1 N1) Molecular and cellular biology. 1990;10:4139–4145. doi: 10.1128/mcb.10.8.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. Journal of virology. 2004;78:9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann G, Castrucci MR, Kawaoka Y. Nuclear import and export of influenza virus nucleoprotein. Journal of virology. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Palese P, O’Neill RE. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. Journal of virology. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber F, Kochs G, Gruber S, Haller O. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology. 1998;250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 31.Nieto A, de la Luna S, Barcena J, Portela A, Ortin J. Complex structure of the nuclear translocation signal of influenza virus polymerase PA subunit. The Journal of general virology. 1994;75(Pt 1):29–36. doi: 10.1099/0022-1317-75-1-29. [DOI] [PubMed] [Google Scholar]
- 32.Cros JF, Garcia-Sastre A, Palese P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic. 2005;6:205–213. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- 33.Gabriel G, et al. Differential use of importin-alpha isoforms governs cell tropism and host adaptation of influenza virus. Nature communications. 2011;2:156. doi: 10.1038/ncomms1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu WW, Weaver LL, Pante N. Ultrastructural analysis of the nuclear localization sequences on influenza A ribonucleoprotein complexes. Journal of molecular biology. 2007;374:910–916. doi: 10.1016/j.jmb.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS pathogens. 2008;4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mark GE, Taylor JM, Broni B, Krug RM. Nuclear accumulation of influenza viral RNA transcripts and the effects of cycloheximide, actinomycin D, and alpha-amanitin. Journal of virology. 1979;29:744–752. doi: 10.1128/jvi.29.2.744-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouloy M, Plotch SJ, Krug RM. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4886–4890. doi: 10.1073/pnas.75.10.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plotch SJ, Bouloy M, Krug RM. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 40.Jorba N, Coloma R, Ortin J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS pathogens. 2009;5:e1000462. doi: 10.1371/journal.ppat.1000462. This paper describes a new model for influenza virus RNA transcription and replication. Specifically, the data suggest that primary transcription is carried out by the resident polymerase complex bound to the double-stranded end of the vRNP, while genome replication is carried out by soluble polymerase complexes, and yet another polymerase complex directs the encapsidation of progeny vRNPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson JS, Schubert M, Lazzarini RA. Polyadenylation sites for influenza virus mRNA. Journal of virology. 1981;38:157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poon LL, Pritlove DC, Fodor E, Brownlee GG. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. Journal of virology. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fodor E. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta virologica. 2013;57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 44.Yanguez E, Nieto A. So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus research. 2011;156:1–12. doi: 10.1016/j.virusres.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 45.Schneider J, Wolff T. Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export? Vaccine. 2009;27:6312–6316. doi: 10.1016/j.vaccine.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Vreede FT, Jung TE, Brownlee GG. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. Journal of virology. 2004;78:9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.York A, Hengrung N, Vreede FT, Huiskonen JT, Fodor E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4238–4245. doi: 10.1073/pnas.1315068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng T, et al. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. Journal of virology. 2006;80:11911–11919. doi: 10.1128/JVI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manzoor R, et al. Heat Shock Protein 70 Modulates Influenza A Virus Polymerase Activity. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M113.507798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Momose F, et al. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. The Journal of biological chemistry. 2002;277:45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- 51.Naito T, Momose F, Kawaguchi A, Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. Journal of virology. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engelhardt OG, Smith M, Fodor E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. Journal of virology. 2005;79:5812–5818. doi: 10.1128/JVI.79.9.5812-5818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Llompart CM, Nieto A, Rodriguez-Frandsen A. Specific residues of PB2 and PA influenza virus polymerase subunits confer the ability for RNA polymerase II degradation and virus pathogenicity in mice. Journal of virology. 2014 doi: 10.1128/JVI.02263-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landeras-Bueno S, Jorba N, Perez-Cidoncha M, Ortin J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS pathogens. 2011;7:e1002397. doi: 10.1371/journal.ppat.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorba N, et al. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics. 2008;8:2077–2088. doi: 10.1002/pmic.200700508. [DOI] [PubMed] [Google Scholar]
- 56.Kawaguchi A, Nagata K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. The EMBO journal. 2007;26:4566–4575. doi: 10.1038/sj.emboj.7601881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naito T, et al. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18235–18240. doi: 10.1073/pnas.0705856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Momose F, et al. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. Journal of virology. 2001;75:1899–1908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawaguchi A, Momose F, Nagata K. Replication-coupled and host factor-mediated encapsidation of the influenza virus genome by viral nucleoprotein. Journal of virology. 2011;85:6197–6204. doi: 10.1128/JVI.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Z, et al. Fragile X mental retardation protein stimulates ribonucleoprotein assembly of influenza A virus. Nature communications. 2014;5:3259. doi: 10.1038/ncomms4259. [DOI] [PubMed] [Google Scholar]
- 61.Perez-Gonzalez A, Rodriguez A, Huarte M, Salanueva IJ, Nieto A. hCLE/CGI-99, a human protein that interacts with the influenza virus polymerase, is a mRNA transcription modulator. Journal of molecular biology. 2006;362:887–900. doi: 10.1016/j.jmb.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez A, Perez-Gonzalez A, Nieto A. Cellular human CLE/C14orf166 protein interacts with influenza virus polymerase and is required for viral replication. Journal of virology. 2011;85:12062–12066. doi: 10.1128/JVI.00684-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hudjetz B, Gabriel G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS pathogens. 2012;8:e1002488. doi: 10.1371/journal.ppat.1002488. This paper demonstrated that importin α1 and α7 are positive regulators of human-like, but not avian-like, influenza polymerase activity, independent of importin nuclear import activities, suggesting that importins regulate IAV transmission between reservoir species and humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Resa-Infante P, et al. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PloS one. 2008;3:e3904. doi: 10.1371/journal.pone.0003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shinya K, et al. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology. 2004;320:258–266. doi: 10.1016/j.virol.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 66.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 67.Li Z, et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. Journal of virology. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. Journal of virology. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manz B, Schwemmle M, Brunotte L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. Journal of virology. 2013;87:7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shapira SD, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tafforeau L, et al. Generation and comprehensive analysis of an influenza virus polymerase cellular interaction network. Journal of virology. 2011;85:13010–13018. doi: 10.1128/JVI.02651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bradel-Tretheway BG, et al. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. Journal of virology. 2011;85:8569–8581. doi: 10.1128/JVI.00496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma K, Roy AM, Whittaker GR. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology. 2001;282:215–220. doi: 10.1006/viro.2001.0833. [DOI] [PubMed] [Google Scholar]
- 74.Elton D, et al. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. Journal of virology. 2001;75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe K, et al. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus research. 2001;77:31–42. doi: 10.1016/s0168-1702(01)00263-5. [DOI] [PubMed] [Google Scholar]
- 76.Bui M, Wills EG, Helenius A, Whittaker GR. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. Journal of virology. 2000;74:1781–1786. doi: 10.1128/jvi.74.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chase GP, et al. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS pathogens. 2011;7:e1002187. doi: 10.1371/journal.ppat.1002187. This paper describes a strong association between influenza vRNPs and cellular chromatin, which precedes vRNP nuclear export in late-stage infection. The authors hypothesize that influenza virus uses this mechanism to gain preferential access to nuclear export machinery to enhance vRNP nuclear export and, ultimately, replicative ability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurokawa M, Ochiai H, Nakajima K, Niwayama S. Inhibitory effect of protein kinase C inhibitor on the replication of influenza type A virus. The Journal of general virology. 1990;71(Pt 9):2149–2155. doi: 10.1099/0022-1317-71-9-2149. [DOI] [PubMed] [Google Scholar]
- 79.Cao S, et al. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication. Journal of virology. 2012;86:4883–4891. doi: 10.1128/JVI.06586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakaguchi A, Hirayama E, Hiraki A, Ishida Y, Kim J. Nuclear export of influenza viral ribonucleoprotein is temperature-dependently inhibited by dissociation of viral matrix protein. Virology. 2003;306:244–253. doi: 10.1016/s0042-6822(02)00013-2. [DOI] [PubMed] [Google Scholar]
- 81.Zhirnov OP, Klenk HD. Histones as a target for influenza virus matrix protein M1. Virology. 1997;235:302–310. doi: 10.1006/viro.1997.8700. [DOI] [PubMed] [Google Scholar]
- 82.Whittaker G, Bui M, Helenius A. Nuclear trafficking of influenza virus ribonuleoproteins in heterokaryons. Journal of virology. 1996;70:2743–2756. doi: 10.1128/jvi.70.5.2743-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. The EMBO journal. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. This paper was the first to establish that the influenza NEP protein regulates vRNP nuclear export, and to identify a region of the NEP protein that acts like a nuclear export signal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neumann G, Hughes MT, Kawaoka Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. The EMBO journal. 2000;19:6751–6758. doi: 10.1093/emboj/19.24.6751. This paper established the importance of the NEP protein in vRNP nuclear export in the context of viral infection and identified the most critical residues of the NEP nuclear export signal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang S, et al. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. Journal of virology. 2013;87:767–778. doi: 10.1128/JVI.06519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Akarsu H, et al. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2) The EMBO journal. 2003;22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Imai M, Watanabe S, Odagiri T. Influenza B virus NS2, a nuclear export protein, directly associates with the viral ribonucleoprotein complex. Archives of virology. 2003;148:1873–1884. doi: 10.1007/s00705-003-0166-x. [DOI] [PubMed] [Google Scholar]
- 88.Robb NC, Smith M, Vreede FT, Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. The Journal of general virology. 2009;90:1398–1407. doi: 10.1099/vir.0.009639-0. [DOI] [PubMed] [Google Scholar]
- 89.Brunotte L, et al. The Nuclear Export Protein of H5N1 Influenza A Viruses Recruits Matrix 1 (M1) Protein to the Viral Ribonucleoprotein to Mediate Nuclear Export. The Journal of biological chemistry. 2014;289:20067–20077. doi: 10.1074/jbc.M114.569178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shimizu T, Takizawa N, Watanabe K, Nagata K, Kobayashi N. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS letters. 2011;585:41–46. doi: 10.1016/j.febslet.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 91.Yu M, et al. Identification and characterization of three novel nuclear export signals in the influenza A virus nucleoprotein. Journal of virology. 2012;86:4970–4980. doi: 10.1128/JVI.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pleschka S, et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nature cell biology. 2001;3:301–305. doi: 10.1038/35060098. This paper established the importance of the Raf/MEK/ERK pathway to efficient influenza virus replication and identified the nuclear export of influenza vRNPs as the life cycle step that is affected by Raf/MEK/ERK inhibition, indicating that cellular kinase pathways are required for vRNP nuclear export. [DOI] [PubMed] [Google Scholar]
- 93.Kumar N, Liang Y, Parslow TG. Receptor tyrosine kinase inhibitors block multiple steps of influenza a virus replication. Journal of virology. 2011;85:2818–2827. doi: 10.1128/JVI.01969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alamares-Sapuay JG, et al. Serum- and glucocorticoid-regulated kinase 1 is required for nuclear export of the ribonucleoprotein of influenza A virus. Journal of virology. 2013;87:6020–6026. doi: 10.1128/JVI.01258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marjuki H, et al. Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. The Journal of biological chemistry. 2006;281:16707–16715. doi: 10.1074/jbc.M510233200. [DOI] [PubMed] [Google Scholar]
- 96.Marjuki H, et al. Higher polymerase activity of a human influenza virus enhances activation of the hemagglutinin-induced Raf/MEK/ERK signal cascade. Virology journal. 2007;4:134. doi: 10.1186/1743-422X-4-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reinhardt J, Wolff T. The influenza A virus M1 protein interacts with the cellular receptor of activated C kinase (RACK) 1 and can be phosphorylated by protein kinase C. Veterinary microbiology. 2000;74:87–100. doi: 10.1016/s0378-1135(00)00169-3. [DOI] [PubMed] [Google Scholar]
- 98.Petri T, Dimmock NJ. Phosphorylation of influenza virus nucleoprotein in vivo. The Journal of general virology. 1981;57:185–190. doi: 10.1099/0022-1317-57-1-185. [DOI] [PubMed] [Google Scholar]
- 99.Kistner O, Muller K, Scholtissek C. Differential phosphorylation of the nucleoprotein of influenza A viruses. The Journal of general virology. 1989;70(Pt 9):2421–2431. doi: 10.1099/0022-1317-70-9-2421. [DOI] [PubMed] [Google Scholar]
- 100.Richardson JC, Akkina RK. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Archives of virology. 1991;116:69–80. doi: 10.1007/BF01319232. [DOI] [PubMed] [Google Scholar]
- 101.Hutchinson EC, et al. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS pathogens. 2012;8:e1002993. doi: 10.1371/journal.ppat.1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Takizawa N, Kumakura M, Takeuchi K, Kobayashi N, Nagata K. Sorting of influenza A virus RNA genome segments after nuclear export. Virology. 2010;401:248–256. doi: 10.1016/j.virol.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 103.Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 104.Momose F, Kikuchi Y, Komase K, Morikawa Y. Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes and infection / Institut Pasteur. 2007;9:1422–1433. doi: 10.1016/j.micinf.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 105.Amorim MJ, et al. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. Journal of virology. 2011;85:4143–4156. doi: 10.1128/JVI.02606-10. This paper was the first to identify overlap between the Rab11 recycling endosome compartment and the localization of cytoplasmic vRNPs in late-stage influenza virus infection. In addition, the PB2 protein of the polymerase complex was shown to have a direct interaction with the Rab11 protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Momose F, et al. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PloS one. 2011;6:e21123. doi: 10.1371/journal.pone.0021123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Avilov SV, Moisy D, Naffakh N, Cusack S. Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein. Vaccine. 2012;30:7411–7417. doi: 10.1016/j.vaccine.2012.09.077. [DOI] [PubMed] [Google Scholar]
- 108.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rietdorf J, et al. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nature cell biology. 2001;3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 110.Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. Journal of virology. 2011;85:6117–6126. doi: 10.1128/JVI.00378-11. This paper established a clear and tight relationship between Rab11 vesicle trafficking and the trafficking of nuclear exported influenza vRNPs between the MTOC and the plasma membrane in late-stage infection. Additionally, Rab11 expression and functional GTPase activity were shown to be crucial for influenza vRNP cytoplasmic transport. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS pathogens. 2008;4:e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kawaguchi A, Matsumoto K, Nagata K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. Journal of virology. 2012;86:11086–11095. doi: 10.1128/JVI.00453-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Lucas S, Peredo J, Marion RM, Sanchez C, Ortin J. Human Staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication. Journal of virology. 2010;84:7603–7612. doi: 10.1128/JVI.00504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chaineau M, Danglot L, Proux-Gillardeaux V, Galli T. Role of HRB in clathrin-dependent endocytosis. The Journal of biological chemistry. 2008;283:34365–34373. doi: 10.1074/jbc.M804587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pryor PR, et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 2008;134:817–827. doi: 10.1016/j.cell.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eisfeld AJ, Neumann G, Kawaoka Y. Human immunodeficiency virus rev-binding protein is essential for influenza a virus replication and promotes genome trafficking in late-stage infection. Journal of virology. 2011;85:9588–9598. doi: 10.1128/JVI.05064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. Journal of virology. 2005;79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iwatsuki-Horimoto K, et al. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. Journal of virology. 2006;80:5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. Journal of virology. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Noda T, et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. This paper was the first to describe the organization pattern of influenza vRNPs in virus particles and provided strong support for the selective vRNP packaging model. [DOI] [PubMed] [Google Scholar]
- 121.Chou YY, et al. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9101–9106. doi: 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. This paper was the first to identify segment-specific packaging signals for an influenza vRNP, which supported the selective incorporation model of vRNP packaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Watanabe T, Watanabe S, Noda T, Fujii Y, Kawaoka Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. Journal of virology. 2003;77:10575–10583. doi: 10.1128/JVI.77.19.10575-10583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ozawa M, et al. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. Journal of virology. 2007;81:30–41. doi: 10.1128/JVI.01434-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liang Y, Hong Y, Parslow TG. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. Journal of virology. 2005;79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liang Y, Huang T, Ly H, Parslow TG. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. Journal of virology. 2008;82:229–236. doi: 10.1128/JVI.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. Journal of virology. 2008;82:11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gog JR, et al. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic acids research. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Muramoto Y, et al. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. Journal of virology. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. Journal of virology. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Noda T, et al. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nature communications. 2012;3:639. doi: 10.1038/ncomms1647. This paper was the first to describe the three dimensional structure of influenza vRNPs within virus particles. The data suggest that inter-segment vRNP-vRNP interactions occur within virions, and that vRNPs are incorporated into progeny virus particles in a specific pattern. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fournier E, et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic acids research. 2012;40:2197–2209. doi: 10.1093/nar/gkr985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fournier E, et al. Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine. 2012;30:7359–7367. doi: 10.1016/j.vaccine.2012.09.079. [DOI] [PubMed] [Google Scholar]
- 134.Gavazzi C, et al. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: comparison with a human H3N2 virus. Nucleic acids research. 2013;41:1241–1254. doi: 10.1093/nar/gks1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Essere B, et al. Critical role of segment-specific packaging signals in genetic reassortment of influenza A viruses. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3840–3848. doi: 10.1073/pnas.1308649110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gavazzi C, et al. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16604–16609. doi: 10.1073/pnas.1314419110. This paper provides an intriguing set of experiments that may suggest the existence of a bona fide inter-segment vRNA-vRNA interaction that occurs in IAV-infected cells. [DOI] [PMC free article] [PubMed] [Google Scholar]


