Abstract
Successful pregnancy is dependent upon discrete biological events, which include embryo implantation, decidualization, and placentation. Problems associated with each of these events can cause infertility or conditions such as preeclampsia. A greater understanding of the molecular changes associated with these complex processes is necessary to aid in identifying treatments for each condition. Previous nuclear magnetic resonance spectroscopy and mass spectrometry studies have been used to identify metabolites and lipids associated with pregnancy-related complications. However, due to limitations associated with conventional implementations of both techniques, novel technology developments are needed to more fully understand the initiation and development of pregnancy related problems at the molecular level. In this perspective, we describe current analytical techniques for metabolomic and lipidomic characterization of pregnancy complications and discuss the potential for new technologies such as ion mobility spectrometry-mass spectrometry and mass spectrometry imaging to contribute to a better understanding of the molecular changes that affect the placenta and pregnancy outcomes.
Keywords: Deep placentation, Pregnancy complications, Mass spectrometry imaging, Metabolome, Lipidome, Ion mobility spectrometry-mass spectrometry, Nano-DESI
1. Introduction
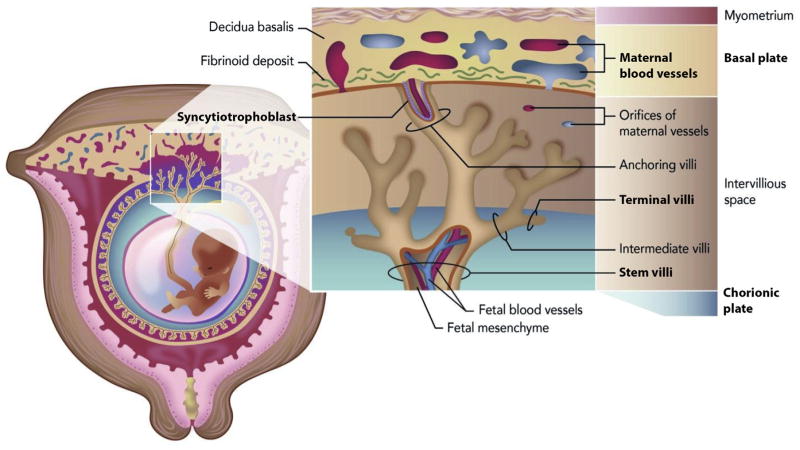
A prerequisite for successful mammalian reproduction is effective, two-way interactions between an implantation-competent blastocyst and the receptive uterus, especially since the blastocyst will only implant when molecular dialogue between these entities is established. In women, the uterus becomes receptive for only a short period, typically 7–9 days after ovulation (cycle days 21–23) during the mid-luteal phase, and after this period it becomes refractory (nonreceptive) and remains this way for the rest of the luteal phase. Unfortunately, even after implantation there are many possible problems that can occur in the uterus. Deep placentation is one of the primary pregnancy transformations that can lead to problems such as preeclampsia (PE), intrauterine growth restriction, preterm labor, and placental abruption [1]. Deep placentation normally occurs due to invasion of the placental bed by the extravillous trophoblast, involving the decidua and the inner (junctional zone) myometrium. Because the placenta is a complex organ composed of heterogeneous cell types and substructures that undergo a variety of processes for normal development (Fig. 1), multi-omic studies are required to more fully understand the molecular changes that affect the placenta and associated pregnancy.
Fig. 1. The placental regions of importance in normal pregnancies and those associated with complications.
The maternal blood vessels, basal plate, terminal villi, stem villi, chorionic plate, and syncytiotrophoblast regions are all bolded in this figure to illustrate their importance in pregnancy complications.
To better understand the molecular changes in the placenta, a previous study explored the distinct lipids found in the chorionic plate (composed primarily of fetal cells) and the basal plate (consisting of a mixture of fetal (trophoblast) and maternal cells), since different amounts of specific lipids in distinct placental regions have been indicated in pregnancy problems [2]. As expected, most lipids were common to both sides of the placenta and were present in comparable abundance. However, twelve lipids (with the majority being phosphatidylcholines (PC) and two sphingomyelins (SM)) differed significantly in their intensity between the chorionic plate and basal plate tissue with most having a higher abundance in the basal plate. This regional selectivity was thought to be from the implantation of the blastocyst in the uterine wall that is accompanied by a transformation of the endometrial lining into decidua. The decidua eventually becomes the maternal cells in the placenta, which are more common in the basal plate, resulting in higher concentrations of lipids in this area [2]. Biofluids have also been analyzed to characterize differences in the fetoplacental and maternal metabolites in pregnancies with poor outcomes compared to normal pregnancies. The goal of many of these studies was to gain insight into PE, a multi-system disorder of pregnancy that is the leading cause of maternal death [3,4]. PE is hypothesized to arise from circulating molecules derived from an unhealthy placenta, therefore many of these studies leverage metabolomics and lipidomics of plasma [5–8] and serum [9,10]. By comparing longitudinally acquired biofluid samples for both women who subsequently developed PE and those with normal pregnancies, researchers have been able to elucidate potential clues to both the etiology and pathogenesis of PE. Common findings between these studies have linked PE to dysregulation of carnitine species, amino acids, phospholipids (i.e., increased phosphatidylserine (PS) species) and sterol lipids [5,6,8,11], and other perturbed small molecules, including vitamin D metabolites and sphingolipids. Many of these molecules are also dysregulated in serum samples associated with placental abruption [12] with the pregnancies ending in poor outcomes (small gestational age infants, preterm birth, or neonatal intensive care admission) [13].
Researchers have also leveraged nuclear magnetic resonance spectroscopy and mass spectrometry-based multi-omic characterizations to identify which molecules exhibit differential expression or abundances in PE versus healthy human placental tissues [8,13,14]. Lipidomic analyses of placenta from patients with PE have shown similarities to plasma studies in that they also revealed PS as the most prevalent phospholipid species showing increased levels when compared with control placenta samples [8]. Collectively, these studies also show that lipid dysfunction could start as early as embryo implantation in pregnancies destined for PE development. Further, Dunn and colleagues reported significant changes in diglycerides, triglycerides, phospholipids, sphingolipids, fatty acids and fatty acid carnitines as the placenta develops between early and late first trimester pregnancies [13]. This same study also compared placental tissue from term-uncomplicated pregnancies with those exhibiting PE at term and found significant changes in mitochondrial metabolism (i.e. fatty acid beta-oxidation), vitamin D metabolism and oxidative stress. Oxidative stress can greatly alter the placenta metabolome [15], and some changes in placental phenotype seen in PE can be reproduced by exposing placental explant cultures to altered oxygen tension [16–18]. In this paper, we highlight analytical tools enabling researchers to create a blueprint of the lipidomic and metabolomic architecture for monitoring pregnancy and those molecular changes underlining pregnancy complications. Further, we discuss the potential of new techniques for improved future analyses.
2. Conventional metabolomics tools for understanding pregnancy complications
Analytical tools such as nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) have previously captured changes in metabolites and lipids from maternal body fluids such as plasma and serum, placental tissues and explant cultures, and syncytiotrophoblast microvesicles (Fig. 1). NMR is inherently quantitative and offers the ability to elucidate molecular structures, but suffers from low measurement sensitivity and throughput [19,20]. On the other hand, MS analyses are highly sensitive and can be used alone or in conjunction with front end separations like liquid chromatography. One of the most powerful uses of MS alone to study pregnancy complications has been with mass spectrometry imaging (MSI). MSI technologies have previously been used to characterize the high and diverse lipid content of decidua [21–24]. MSI has also been applied to normal human placentas and used to determine the specific distribution of SM(d18:1/16:0) in the stem villi and PC(16:0/20:4) in the terminal villi [25]. Alterations in the spatial distribution of these molecules have been correlated with disease. Specifically, the absence of PC(16:0/20:4) in the terminal villi and absence of SM(d18:1/16:0) in the stem villi have been linked to morphological changes in the placenta due to maternal underperfusion [26]. These observations have proven very exciting, but while MSI offers unique opportunities to study the in situ spatial distribution, it does suffer from lower sensitivity due to ionization suppression of highly concentrated species concealing low concentration molecules.
To obtain deeper coverage of many samples types, MS is commonly combined with liquid chromatography. For LC-MS, electrospray ionization (ESI) is most commonly used to introduce analyte ions to the instrument, and data are typically collected by scanning over a wide mass range (e.g. 100–1000 m/z) to detect as many molecules as possible. In most LC-MS experiments, intact molecular ions are fragmented to produce MS/MS spectra, which can be used to identify the structures of detected molecules in conjunction with libraries of reference spectra [27–29] or tentatively identify candidates using in silico fragmentation approaches [30–33]. Most LC-MS molecular characterization studies of pregnancy use either targeted or untargeted analyses. In targeted analyses only a single analyte, class of chemically similar analytes, or set of analytes whose chemistries are sufficiently similar are studied. The benefits of targeted analyses include the accurate quantification of the analyte(s) of interest, ability to quantify molecules present at low concentrations, and higher confidence in each identification. However, there are challenges to targeted approaches which include the narrow snapshot of chemistry measured, the level of effort required to fully optimize and validate the analytical pipelines, and the relatively low analysis throughput. In contrast to targeted analysis, untargeted analyses do not focus on a specific analyte but instead seek to comprehensively measure all analytes in a sample. These measurements are not biased by a priori assumptions and offer the best opportunities to discover novel markers. Caveats however include possible artifacts in the data due to the lack of method optimization, difficulty detecting very low abundance analytes, and limited confidence in identifications when reference standards are not available. Thus, the determination of additional metabolite characteristics such as molecular structure with ion mobility spectrometry (IMS) provides an opportunity to develop improved untargeted measurements.
3. Advanced ion mobility spectrometry and nano-DESI mass spectrometry imaging capabilities
An appealing technique for enhancing current untargeted analytical methods is by incorporating IMS into the LC-MS-based metabolomics workflow for three dimensional LC-IMS-MS analyses [34–36]. In IMS, ions travel through a drift tube under the influence of a weak attractive electric field while colliding with a stationary buffer gas. In this way, ions with compact structures or small collisional cross sections (CCSs) spend less time in the drift tube compared to other equally charged ions with larger CCS values [37,38]. The CCS of each ion can be directly determined [39] and correlates with its 3-dimensional shape [40], thus providing a new metabolite characteristic to each measurement. The ability to resolve isomers that are difficult to distinguish using LC-MS alone is an inherent strength of IMS. This capability is particularly important in small molecule analyses since many metabolites and other chemicals have the same molecular formula but different chemistries, thus playing very different roles in biological systems. IMS has been used to separate some important classes of isomers [41–46], which have the same molecular formula and often co-elute in generic untargeted LC-MS methods [47]. Because IMS instruments depend only on drift cell pressure, temperature, and length, CCS values on a single instrument are extremely repeatable (<0.4% error) [48–51], allowing reliable molecular feature alignment (i.e. identity confirmation) across multiple samples. Furthermore, measurements from instruments in different laboratories have also been compared and their values normally agree within <2% error [52] and recent IMS instruments are yielding values with reproducibility precision of <1% [48,49].
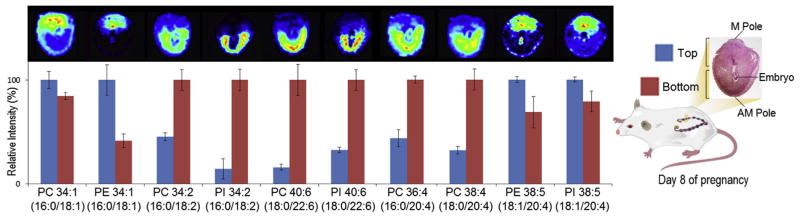
The high reproducibility and speed of IMS allows it to be easily nested between LC and MS for additional separation power and dynamic range for in depth characterization of lipid networks critical to pregnancy and information on protein and small molecule changes. Two advantages of coupling LC and IMS are that LC reduces the complexity of the samples by fractionating them prior to their introduction into the instrument, while IMS distributes the ions throughout a range of drift times so they do not all arrive simultaneously at the detector. LC-IMS-MS can also either provide greater coverage of a sample for the same LC gradient length (i.e. by increasing the dynamic range of the measurement) [53], or allow for decreasing the LC gradient length while maintaining the same measurement coverage. For example, using a LC-IMS-MS-based proteomics method allowed shortening the LC gradient from 100 to 15.5 min with no loss in coverage of 20 standard peptides spiked in a mouse plasma protein digest [54]. In a recent study, we established that LC-IMS-MS outperformed a traditional high-resolution LC-MS platform for characterizing protein expression in mouse uterine decidua due to the greater sensitivity and dynamic range of measurements [55]. We also tested the ability of LC-IMS-MS to accurately quantify the complexity of lipid distributions in early mouse pregnancy. For example, a previous MALDI-FTICR imaging publication revealed that the spatial distribution of select phospholipid species associated with mouse embryo implantation changed markedly between the distinct mesometrial (M, top) and antimesometrial (AM, bottom) poles during day 8 of pregnancy (Fig. 2) [21]. By replicating this experimental design and micro-dissecting day 8 implantation sites for the LC-IMS-MS experiments, like the previous study, our analyses found PC(16:0/18:1), PE(16:0/18:1), PE(18:1/20:4) and PI (18:1/20:4) to predominately localize to the angiogenic M pole, the presumptive site of formation of the placenta. Conversely, PC(16:0/18:2), PI(16:0/18:2), PC(18:0/22:6), PI(18:0/22:6), PC(16:0/20:4), and PC (18:0/20:4) predominately localized to decidual cells at the AM side. This agreement between the MALDI-FTICR imaging and LC-IMS-MS data demonstrates the great promise of measurements with both spatial localization and deeper coverage.
Fig. 2. Quantification of lipids using LC-IMS-MS.
Significant abundance changes of select phospholipids quantified by LC-IMS-MS data from microdissected top (M pole) and bottom (AM pole) regions of implantation sites collected on day 8 of pregnancy correlates with previously published MALDI-MSI images [21]. Figure adapted from Ref. [44].
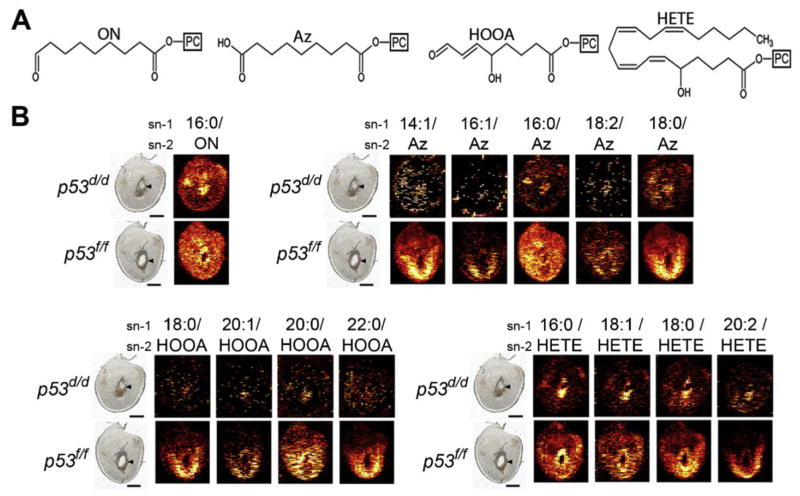
Another imaging innovation which shows promise for higher sensitivity molecular characterization is in situ nanospray desorption electrospray ionization (nano-DESI) MSI. Nano-DESI relies on localized liquid extraction of molecular targets from tissue sections, after which the sampled molecules are delivered to the MS. The solvent dissolves a tiny portion of the sample, mixing it into the liquid. The sample is then efficiently transferred to the MS and electrosprayed as highly charged sub-micron droplets that facilitate sensitive MS analysis. Three recent nano-DESI studies have illustrated the success of this technique for imaging lipids and metabolites from uterine sections under ambient conditions and without special sample pre-treatment [22–24]. In the most recent study, implantation sites from a mouse model of preterm birth (p53d/d mice) exhibited distinct spatially resolved changes demonstrating depletion of oxidized phosphatidylcholine (Ox-PC) species (Fig. 3), accumulation of DG species, and increase in species with more unsaturated acyl chains including arachidonic (20:4) and docosahexaenoic acid (22:6) by day 8 of pregnancy [24]. In this mouse model, uterine deletion of Trp53 increases the incidence of preterm birth through an increase in the cyclooxygenase 2 (COX2)/PGF synthase/PGF (2alpha) pathway [56]. Comparable signatures of decidual senescence with increased COX signaling were observed in women undergoing preterm birth, making lipid metabolism and signaling clinically relevant [57]. Characterizing abnormal molecular changes in abundance and localization of bioactive molecules such as lipids and metabolites early in disease progression is an important step toward understanding disease etiology and discovering novel targets for treatment, and the high sensitivity nano-DESI MSI measurements are essential for this capability.
Fig. 3. Intensities of selected Ox-PC species in implantation sites from a preterm birth mouse model (p53d/d) vs control littermates (p53f/f) on day 8 of pregnancy.
(A) Structures of the sn-2 acyl chain of four groups of Ox-PC species. From left to right; oxononanoyl (ON)-PC, azelayl (Az)-PC, hydroxy-oxooct-enoyl (HOOA)-PC, and hydrox-eicosa-tetra-enoyl (HETE)-PC. (B) Ion images of Ox-PC species (the acyl group in the sn-1 position of the PC/the oxidized moiety in the sn-2 position). Scale bars show 1 mm. Arrowheads denote embryos. Implantation sites are orientated with the M pole on top and the AM pole on bottom. Figure adapted from Ref. [24].
4. Future outlook
While NMR and MS-based studies have begun paving our understanding of the metabolites and lipids responsible for pregnancy related complications, there is still much to learn and the need for new technologies to gain this knowledge. Both IMS and nano-DESI MSI measurements show great promise in addressing the spatial localization and in depth coverage needed to better understand pregnancy complications. The molecular findings from these studies will enable the development of clinical tests for rapid diagnosis of pregnancy related conditions, allowing prompt treatments and better outcomes. While both technologies are expected to play a critical role in the future, we also feel developing technologies such as structures for lossless ion manipulations [58] could make an impact on the number of metabolites and lipids detected in biofluid and tissue measurements. Thus, by combining knowledge enabled by these new techniques, in the next decade we expect a much better understanding of the molecular changes that affect the placenta and pregnancy outcomes.
Acknowledgments
This work was supported by the Pacific Northwest National Laboratory (PNNL) Laboratory Directed Research and Development program and is a contribution of the Microbiomes in Transition Initiative (TOM), as well as the National Institutes of Health (NIH) National Institute of Environmental Health Sciences grant R01 ES022190 to ESB and the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development grant R21 HD084788 to KEBJ. The authors would like to thank Nathan Johnson for assistance in preparing figures. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Brosens I, Pijnenborg R, Vercruysse L, Romero R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am J Obstetrics Gynecol. 2011;204(3):193–201. doi: 10.1016/j.ajog.2010.08.009. http://dx.doi.org/10.1016/j.ajog.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kedia K, Nichols CA, Thulin CD, Graves SW. Novel “omics” approach for study of low-abundance, low-molecular-weight components of a complex biological tissue: regional differences between chorionic and basal plates of the human placenta. Anal Bioanal Chem. 2015;407(28):8543–8556. doi: 10.1007/s00216-015-9009-3. http://dx.doi.org/10.1007/s00216-015-9009-3. [DOI] [PubMed] [Google Scholar]
- 3.Uzan J, Carbonnel M, Piconne O, Asmar R, Ayoubi JM. Pre-eclampsia: pathophysiology, diagnosis, and management. Vasc Health Risk Manag. 2011;7:467–474. doi: 10.2147/VHRM.S20181. http://dx.doi.org/10.2147/VHRM.S20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aabidha PM, Cherian AG, Paul E, Helan J. Maternal and fetal outcome in pre-eclampsia in a secondary care hospital in South India. J Fam Med Prim Care. 2015;4(2):257–260. doi: 10.4103/2249-4863.154669. http://dx.doi.org/10.4103/2249-4863.154669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenny LC, Broadhurst DI, Dunn W, Brown M, North RA, McCowan L, Roberts C, Cooper GJS, Kell DB, Baker PN Consortium obotSfPE. Robust early pregnancy prediction of later preeclampsia using metabolomic bio-markers. Hypertension. 2010;56(4):741–749. doi: 10.1161/HYPERTENSIONAHA.110.157297. http://dx.doi.org/10.1161/hypertensionaha.110.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K. Metabolomics and first-trimester prediction of early-onset preeclampsia. J Maternal-Fetal Neonatal Med. 2012;25(10):1840–1847. doi: 10.3109/14767058.2012.680254. http://dx.doi.org/10.3109/14767058.2012.680254. [DOI] [PubMed] [Google Scholar]
- 7.De Oliveira L, Câmara NOS, Bonetti T, Turco EGL, Bertolla RP, Moron AF, Sass N, Da Silva IDCG. Lipid fingerprinting in women with early-onset preeclampsia: a first look. Clin Biochem. 2012;45(10–11):852–855. doi: 10.1016/j.clinbiochem.2012.04.012. http://dx.doi.org/10.1016/j.clinbiochem.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Korkes HA, Sass N, Moron AF, Câmara NOS, Bonetti T, Cerdeira AS, Da Silva IDCG, De Oliveira L. Lipidomic assessment of plasma and placenta of women with early-onset preeclampsia. PLoS One. 2014;9(10):e110747. doi: 10.1371/journal.pone.0110747. http://dx.doi.org/10.1371/journal.pone.0110747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K. First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstetrics Gynecol. 2013;208(1):58.e1–58.e7. doi: 10.1016/j.ajog.2012.11.003. http://dx.doi.org/10.1016/j.ajog.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Austdal M, Skråstad RB, Gundersen AS, Austgulen R, Iversen AC, Bathen TF. Metabolomic biomarkers in serum and urine in women with preeclampsia. PLoS One. 2014;9(3):e91923. doi: 10.1371/journal.pone.0091923. http://dx.doi.org/10.1371/journal.pone.0091923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odibo AO, Goetzinger KR, Odibo L, Cahill AG, Macones GA, Nelson DM, Dietzen DJ. First-trimester prediction of Preeclampsia using metabolomic biomarkers: a discovery phase study. Prenat Diagn. 2011;31(10):990–994. doi: 10.1002/pd.2822. http://dx.doi.org/10.1002/pd.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelaye B, Sumner SJ, McRitchie S, Carlson JE, Ananth CV, Enquobahrie DA, Qiu C, Sorensen TK, Williams MA. Maternal early pregnancy serum metabolomics profile and abnormal vaginal bleeding as predictors of placental abruption: a prospective study. PLoS One. 2016;11(6):e0156755. doi: 10.1371/journal.pone.0156755. http://dx.doi.org/10.1371/journal.pone.0156755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heazell AEP, Bernatavicius G, Warrander L, Brown MC, Dunn WB. A metabolomic approach identifies differences in maternal serum in third trimester pregnancies that end in poor perinatal outcome. Reprod Sci. 2012;19(8):863–875. doi: 10.1177/1933719112438446. http://dx.doi.org/10.1177/1933719112438446. [DOI] [PubMed] [Google Scholar]
- 14.Kedia K, Smith SF, Wright AH, Barnes JM, Tolley HD, Esplin MS, Graves SW. Global “omics” evaluation of human placental responses to preeclamptic conditions. Am J Obstetrics Gynecol. 2016;215(2):238.e1–238.e20. doi: 10.1016/j.ajog.2016.03.004. http://dx.doi.org/10.1016/j.ajog.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Tissot van Patot MC, Murray AJ, Beckey V, Cindrova-Davies T, Johns J, Zwerdlinger L, Jauniaux E, Burton GJ, Serkova NJ. Human placental metabolic adaptation to chronic hypoxia, high altitude: hypoxic preconditioning. American Journal of Physiology - Regulatory. Integr Comp Physiol. 2010;298(1):R166–R172. doi: 10.1152/ajpregu.00383.2009. http://dx.doi.org/10.1152/ajpregu.00383.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heazell AEP, Brown M, Dunn WB, Worton SA, Crocker IP, Baker PN, Kell DB. Analysis of the metabolic footprint and tissue metabolome of placental villous explants cultured at different oxygen tensions reveals novel redox biomarkers. Placenta. 2008;29(8):691–698. doi: 10.1016/j.placenta.2008.05.002. http://dx.doi.org/10.1016/j.placenta.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Dunn WB, Brown M, Worton SA, Crocker IP, Broadhurst D, Horgan R, Kenny LC, Baker PN, Kell DB, Heazell AEP. Changes in the metabolic footprint of placental explant-conditioned culture medium identifies metabolic disturbances related to hypoxia and pre-eclampsia. Placenta. 2009;30(11):974–980. doi: 10.1016/j.placenta.2009.08.008. http://dx.doi.org/10.1016/j.placenta.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Horgan RP, Broadhurst DI, Dunn WB, Brown M, Heazell AEP, Kell DB, Baker PN, Kenny LC. Changes in the metabolic footprint of placental explant-conditioned medium cultured in different oxygen tensions from placentas of small for gestational age and normal pregnancies. Placenta. 2010;31(10):893–901. doi: 10.1016/j.placenta.2010.07.002. http://dx.doi.org/10.1016/j.placenta.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Nagana Gowda GA, Gowda YN, Raftery D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal Chem. 2014 doi: 10.1021/ac503651e. http://dx.doi.org/10.1021/ac503651e. [DOI] [PMC free article] [PubMed]
- 20.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78(13):4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 21.Burnum KE, Cornett DS, Puolitaival SM, Milne SB, Myers DS, Tranguch S, Brown HA, Dey SK, Caprioli RM. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J lipid Res. 2009;50(11):2290–2298. doi: 10.1194/jlr.M900100-JLR200. http://dx.doi.org/10.1194/jlr.M900100-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanekoff I, Burnum-Johnson K, Thomas M, Short J, Carson JP, Cha J, Dey SK, Yang P, Prieto Conaway MC, Laskin J. High-speed tandem mass spectrometric in situ imaging by nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2013;85(20):9596–9603. doi: 10.1021/ac401760s. http://dx.doi.org/10.1021/ac401760s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanekoff I, Burnum-Johnson K, Thomas M, Cha J, Dey SK, Yang P, Prieto Conaway MC, Laskin J. Three-dimensional imaging of lipids and metabolites in tissues by nanospray desorption electrospray ionization mass spectrometry. Anal Bioanal Chem. 2015;407(8):2063–2071. doi: 10.1007/s00216-014-8174-0. http://dx.doi.org/10.1007/s00216-014-8174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanekoff I, Cha J, Kyle JE, Dey SK, Laskin J, Burnum-Johnson KE. Trp53 deficient mice predisposed to preterm birth display region-specific lipid alterations at the embryo implantation site. Sci Rep. 2016;6:33023. doi: 10.1038/srep33023. http://dx.doi.org/10.1038/srep33023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi Y, Hayasaka T, Setou M, Itoh H, Kanayama N. Comparison of phospholipid molecular species between terminal and stem villi of human term placenta by imaging mass spectrometry. Placenta. 2010;31(3):245–248. doi: 10.1016/j.placenta.2009.12.026. http://dx.doi.org/10.1016/j.placenta.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki K, Masaki N, Kohmura-Kobayashi Y, Yaguchi C, Hayasaka T, Itoh H, Setou M, Kanayama N. Decrease in sphingomyelin (d18:1/16:0) in stem villi and phosphatidylcholine (16:0/20:4) in terminal villi of human term placentas with pathohistological maternal malperfusion. PLoS One. 2015;10(11):e0142609. doi: 10.1371/journal.pone.0142609. http://dx.doi.org/10.1371/journal.pone.0142609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. MassBank: a public repository for sharing mass spectral data for life sciences. J mass Spectrom JMS. 2010;45(7):703–714. doi: 10.1002/jms.1777. http://dx.doi.org/10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 28.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 29.Vinaixa M, Schymanski EL, Neumann S, Navarro M, Salek RM, Yanes O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: state of the field and future prospects. Trac-Trend Anal Chem. 2016;78:23–35. http://dx.doi.org/10.1016/j.trac.2015.09.005. [Google Scholar]
- 30.Hufsky F, Scheubert K, Bocker S. Computational mass spectrometry for small-molecule fragmentation. Trac-Trend Anal Chem. 2014;53:41–48. http://dx.doi.org/10.1016/j.trac.2013.09.008. [Google Scholar]
- 31.Vaniya A, Fiehn O. Using fragmentation trees and mass spectral trees for identifying unknown compounds in metabolomics. Trac-Trend Anal Chem. 2015;69:52–61. doi: 10.1016/j.trac.2015.04.002. http://dx.doi.org/10.1016/j.trac.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duhrkop K, Shen H, Meusel M, Rousu J, Bocker S. Searching molecular structure databases with tandem mass spectra using CSI: Finger ID. Proc Natl Acad Sci U S A. 2015;112(41):12580–12585. doi: 10.1073/pnas.1509788112. http://dx.doi.org/10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J Cheminform. 2016;8:3. doi: 10.1186/s13321-016-0115-9. http://dx.doi.org/10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burnum-Johnson KE, Nie S, Casey CP, Monroe ME, Orton DJ, Ibrahim YM, Gritsenko MA, Clauss TR, Shukla AK, Moore RJ, Purvine SO, Shi T, Qian W, Liu T, Baker ES, Smith RD. Simultaneous proteomic discovery and targeted monitoring using liquid chromatography, ion mobility spectrometry, and mass spectrometry. Mol Cell Proteomics. 2016;15(12):3694–3705. doi: 10.1074/mcp.M116.061143. http://dx.doi.org/10.1074/mcp.M116.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prost SA, Crowell KL, Baker ES, Ibrahim YM, Clowers BH, Monroe ME, Anderson GA, Smith RD, Payne SH. Detecting and removing data artifacts in Hadamard transform ion mobility-mass spectrometry measurements. J Am Soc Mass Spectrom. 2014;25(12):2020–2027. doi: 10.1007/s13361-014-0895-y. http://dx.doi.org/10.1007/s13361-014-0895-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentine SJ, Liu X, Plasencia MD, Hilderbrand AE, Kurulugama RT, Koeniger SL, Clemmer DE. Developing liquid chromatography ion mobility mass spectometry techniques. Expert Rev Proteomics. 2005;2(4):553–565. doi: 10.1586/14789450.2.4.553. http://dx.doi.org/10.1586/14789450.2.4.553. [DOI] [PubMed] [Google Scholar]
- 37.Suhr H. Plasma chromatography. In: Carr TW, editor. Berichte der Bunsenge-sellschaft für physikalische Chemie. Plenum Press; New York, London: 1984. p. 924. [Google Scholar]
- 38.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. John Wiley and Sons; New York: 1988. p. 701. [Google Scholar]
- 39.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. J mass spectrom: JMS. 2008;43(1):1–22. doi: 10.1002/jms.1383. http://dx.doi.org/10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 40.Revercomb HE, Mason EA. Theory of plasma chromatography gaseous electrophoresis - review. Anal Chem. 1975;47(7):970–983. http://dx.doi.org/10.1021/ac60357a043. [Google Scholar]
- 41.May JC, McLean JA. Ion mobility-mass spectrometry: time-dispersive instrumentation. Anal Chem. 2015;87(3):1422–1436. doi: 10.1021/ac504720m. http://dx.doi.org/10.1021/ac504720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams JP, Bugarcic T, Habtemariam A, Giles K, Campuzano I, Rodger PM, Sadler PJ. Isomer separation and gas-phase configurations of organoruthenium anticancer complexes: ion mobility mass spectrometry and modeling. J Am Soc Mass Spectrom. 2009;20(6):1119–1122. doi: 10.1016/j.jasms.2009.02.016. http://dx.doi.org/10.1016/j.jasms.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 43.Wu C, Siems WF, Klasmeier J, Hill HH. Separation of isomeric peptides using electrospray ionization/high-resolution ion mobility spectrometry. Anal Chem. 2000;72(2):391–395. doi: 10.1021/ac990601c. http://dx.doi.org/10.1021/ac990601c. [DOI] [PubMed] [Google Scholar]
- 44.Kyle JE, Zhang X, Weitz KK, Monroe ME, Ibrahim YM, Moore RJ, Cha J, Sun X, Lovelace ES, Wagoner J, Polyak SJ, Metz TO, Dey SK, Smith RD, Burnum-Johnson KE, Baker ES. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst. 2016;141(5):1649–1659. doi: 10.1039/c5an02062j. http://dx.doi.org/10.1039/c5an02062j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng X, Zhang X, Schocker NS, Renslow RS, Orton DJ, Khamsi J, Ashmus RA, Almeida IC, Tang K, Costello CE, Smith RD, Michael K, Baker ES. Enhancing glycan isomer separations with metal ions and positive and negative polarity ion mobility spectrometry-mass spectrometry analyses. Anal Bioanal Chem. 2016 doi: 10.1007/s00216-016-9866-4. http://dx.doi.org/10.1007/s00216-016-9866-4. [DOI] [PMC free article] [PubMed]
- 46.Baker ES, Hong JW, Gidden J, Bartholomew GP, Bazan GC, Bowers MT. Diastereomer assignment of an olefin-linked bisparacyclophane by ion mobility mass spectrometry. J Am Chem Soc. 2004;126(20):6255–6257. doi: 10.1021/ja039486k. http://dx.doi.org/10.1021/ja039486k. [DOI] [PubMed] [Google Scholar]
- 47.Schymanski EL, Singer HP, Longree P, Loos M, Ruff M, Stravs MA, Ripolles Vidal C, Hollender J. Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry. Environ Sci Technol. 2014;48(3):1811–1818. doi: 10.1021/es4044374. http://dx.doi.org/10.1021/es4044374. [DOI] [PubMed] [Google Scholar]
- 48.Fjeldsted J, Kurulugama RT, Mordehai A, Rennie EE, Darland E, Stafford GC, May JC, Stow SM, McLean JA, Causon TJ, Mairinger T, Hann S, Baker ES. Highly Accurate Collision Cross Section Measurements for Comprehensive High Throughput Applications. 64th Annual Meeting of the American Mass Spectrometry Society; 2016; San Antonio, TX, USA. [Google Scholar]
- 49.Causon TJ, Mairinger T, Hung LS, Stow SM, May JC, McLean JA, Baker ES, Zheng X, Smith RD, Kurulugama RT, Rennie EE, Mordehai A, Darland E, Stafford GC, Fjeldsted JC, Hann S. Addition of drift-tube ion mobility to liquid chromatography-mass spectrometry workflows: examining the potential for cellular metabolomics. 12th Annual Conference of the Metabolomics Society; 2016 June 30, 2016; Dublin, Ireland. [Google Scholar]
- 50.Stephan S, Hippler J, Kohler T, Deeb AA, Schmidt TC, Schmitz OJ. Contaminant screening of wastewater with HPLC-IM-qTOF-MS and LC+LC-IM-qTOF-MS using a CCS database. Anal Bioanal Chem. 2016;408(24):6545–6555. doi: 10.1007/s00216-016-9820-5. http://dx.doi.org/10.1007/s00216-016-9820-5. [DOI] [PubMed] [Google Scholar]
- 51.Wyttenbach T, Bowers MT. Gas-phase conformations: the ion mobility/ion chromatography method. Top Curr Chem. 2003;225:207–232. http://dx.doi.org/10.1007/b10470. [Google Scholar]
- 52.Baker ES, Clowers BH, Li F, Tang K, Tolmachev AV, Prior DC, Belov ME, Smith RD. Ion mobility spectrometry-mass spectrometry performance using electrodynamic ion funnels and elevated drift gas pressures. J Am Soc Mass Spectrom. 2007;18(7):1176–1187. doi: 10.1016/j.jasms.2007.03.031. http://dx.doi.org/10.1016/j.jasms.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Valentine SJ, Plasencia MD, Trimpin S, Naylor S, Clemmer DE. Mapping the human plasma proteome by SCX-LC-IMS-MS. J Am Soc Mass Spectrom. 2007;18(7):1249–1264. doi: 10.1016/j.jasms.2007.04.012. http://dx.doi.org/10.1016/j.jasms.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker ES, Livesay EA, Orton DJ, Moore RJ, Danielson WF, 3rd, Prior DC, Ibrahim YM, LaMarche BL, Mayampurath AM, Schepmoes AA, Hopkins DF, Tang K, Smith RD, Belov ME. An LC-IMS-MS platform providing increased dynamic range for high-throughput proteomic studies. J proteome Res. 2010;9(2):997–1006. doi: 10.1021/pr900888b. http://dx.doi.org/10.1021/pr900888b. Epub 2009/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burnum KE, Hirota Y, Baker ES, Yoshie M, Ibrahim YM, Monroe ME, Anderson GA, Smith RD, Daikoku T, Dey SK. Uterine deletion of Trp53 compromises antioxidant responses in the mouse decidua. Endocrinology. 2012;153(9):4568–4579. doi: 10.1210/en.2012-1335. http://dx.doi.org/10.1210/en.2012-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest. 2010;120(3):803–815. doi: 10.1172/JCI40051. http://dx.doi.org/10.1172/JCI40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cha J, Bartos A, Egashira M, Haraguchi H, Saito-Fujita T, Leishman E, Bradshaw H, Dey SK, Hirota Y. Combinatory approaches prevent preterm birth profoundly exacerbated by gene-environment interactions. J Clin Invest. 2013;123(9):4063–4075. doi: 10.1172/JCI70098. http://dx.doi.org/10.1172/JCI70098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng L, Ibrahim YM, Baker ES, Aly NA, Hamid AM, Zhang X, Zheng X, Garimella SVB, Webb IK, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Smith RD. Ion mobility separations of isomers based upon long path length structures for lossless ion manipulations combined with mass spectrometry. ChemistrySelect. 2016;1(10):2396–2399. doi: 10.1002/slct.201600460. http://dx.doi.org/10.1002/slct.201600460. [DOI] [PMC free article] [PubMed] [Google Scholar]